Abstract
The FliM protein of Escherichia coli is required for the assembly and function of flagella. Genetic analyses and binding studies have shown that FliM interacts with several other flagellar proteins, including FliN, FliG, phosphorylated CheY, other copies of FliM, and possibly MotA and FliF. Here, we examine the effects of a set of linker insertions and partial deletions in FliM on its binding to FliN, FliG, CheY, and phospho-CheY and on its functions in flagellar assembly and rotation. The results suggest that FliM is organized into multiple domains. A C-terminal domain of about 90 residues binds to FliN in coprecipitation experiments, is most stable when coexpressed with FliN, and has some sequence similarity to FliN. This C-terminal domain is joined to the rest of FliM by a segment (residues 237 to 247) that is poorly conserved, tolerates linker insertion, and may be an interdomain linker. Binding to FliG occurs through multiple segments of FliM, some in the C-terminal domain and others in an N-terminal domain of 144 residues. Binding of FliM to CheY and phospho-CheY was complex. In coprecipitation experiments using purified FliM, the protein bound weakly to unphosphorylated CheY and more strongly to phospho-CheY, in agreement with previous reports. By contrast, in experiments using FliM in fresh cell lysates, the protein bound to unphosphorylated CheY about as well as to phospho-CheY. Determinants for binding CheY occur both near the N terminus of FliM, which appears most important for binding to the phosphorylated protein, and in the C-terminal domain, which binds more strongly to unphosphorylated CheY. Several different deletions and linker insertions in FliM enhanced its binding to phospho-CheY in coprecipitation experiments with protein from cell lysates. This suggests that determinants for binding phospho-CheY may be partly masked in the FliM protein as it exists in the cytoplasm. A model is proposed for the arrangement and function of FliM domains in the flagellar motor.
FliG, FliM, and FliN are proteins of the bacterial flagellum that have multiple functions (25, 34, 35; reviewed in references 14, 15, and 24). All three proteins are essential for flagellar assembly, and all are involved in controlling motor switching between clockwise (CW) and counterclockwise (CCW) rotation. FliG also functions directly in torque generation by the motor (7, 12, 13). Genetic suppression studies by Yamaguchi et al. (34, 35) first provided evidence that FliG, FliM, and FliN function together in a complex, which has been termed the “switch complex.” All three proteins were subsequently localized to the flagellar basal body by immunoelectron microscopy (4, 5, 8, 9, 36–38). Binding of FliM to FliN, FliG, and other copies of FliM was detected in experiments using the yeast two-hybrid system (16, 17). These and additional binding interactions were also observed in coprecipitation experiments using glutathione S-transferase (GST) fusion proteins (29). A FliM-FliN fusion protein can support assembly and also some motor function, consistent with the proposal that FliM and FliN function within the same complex (10). Most recently, Toker and Macnab (31) used affinity blotting to demonstrate binding of FliM to FliG, FliN, and phospho-CheY and to examine the effects of deletions in FliM upon each of these interactions. FliM may also interact weakly with MotA, a stator component that functions in transmembrane proton conduction (29), and with FliF, the protein that forms the membrane-embedded MS ring of the flagellar basal body (19).
The complex containing FliG, FliM, and FliN is thought to reside on the rotating part (the rotor) of the flagellar motor (4, 5, 16, 17, 27–29, 36–38). Among the three proteins, FliM has an especially large role in controlling the direction of motor rotation. Many point mutations in FliM affect CW-CCW switching (25), and binding studies using purified proteins showed that FliM can bind to phospho-CheY (2, 3, 32), the chemotactic signaling molecule that triggers switching to the CW direction (20, 33). FliM appears to have little if any direct role in torque generation per se. Although certain mutations of FliM can give a nonmotile, flagellated phenotype (25, 30), some motility is restored when either the mutant FliM protein or one of the other switch complex proteins is overexpressed (12, 30).
Here, we report the effects of several deletions and linker insertion mutations in FliM on the functions of the protein in flagellar assembly, motility, and switching and on its binding to FliN, FliG, CheY, and phospho-CheY. The results provide insight into the domain organization of FliM and identify segments of the protein involved in interactions with some of its partners. A model is proposed for the function and arrangement of FliM domains in the flagellar motor.
MATERIALS AND METHODS
Strains, media, and plasmids.
The strains and plasmids used are listed in Table 1. Transformations, plasmid isolation, and DNA manipulations used standard procedures (23). Most linker insertions were made in plasmid pHT32, whose parent is pTM30 (18), a high-copy-number plasmid that expresses cloned genes from the tac promoter. Plasmid pHT32 was made by inserting a BamHI fragment encoding fliM, obtained by PCR amplification of the cloned fliM gene (27), into the unique BamHI site in pTM30. It encodes a translational fusion with the residues MLNDPH fused to the N terminus of FliM and complements the fliM null strain DFB190 (27) to wild-type motility on swarm plates when induced with 60 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Plasmids expressing GST fusions to FliN and FliG have been described elsewhere (29). A plasmid expressing a GST-CheY fusion (pHT121) was made by replacing a BamHI segment of the GST-FliM expression vector pHT86 (29) with a BamHI segment encoding cheY, which was obtained from plasmid pHT111, a pTM30 derivative that encodes cheY. Plasmid pHT121 complemented a smooth-swimming, nonchemotactic cheY deletion strain (RP5232) to give frequent tumbles and good chemotaxis. The GST-only plasmid used for negative controls (pHT100) has been described elsewhere (29).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or property | Source or reference |
|---|---|---|
| RP437 | Wild type for motility and chemotaxis | J. S. Parkinson |
| DFB190 | fliM null strain | 27 |
| RP3098 | De(flhC-flhA) (expresses no chromosomal flagellar genes) | J. S. Parkinson |
| RP5232 | De(cheY) | J. S. Parkinson |
| BL21(DE3) | Host for T7 expression vectors | 26 |
| pTM30 | Protein expression vector, parent of pHT32, pDFB72 and pMn linker-insertion plasmids | 18 |
| pDFB72 | FliM expression vector; Apr | 27 |
| pHT32 | FliM expression vector; Apr | This work |
| pHT41 | fliM in pAlter-1, vector for site-directed mutagenesis | 27 |
| pHT96 | GST-FliN expression vector; Kmr | 29 |
| pHT97 | GST-FliG expression vector; Kmr | 29 |
| pHT100 | GST-only expression vector; Kmr | 29 |
| pHT111 | CheY expression vector; Apr | This work |
| pHT121 | GST-CheY expression vector; Kmr | This work |
| pM16 | Insertion of Pro linker and linker L43 at fliM codon 16 in pDFB72; Apr | This work |
| pM38 | Linker (L42) insertion at fliM codon 38 (BsiWI site) in pHT32; Apr | This work |
| pM60 | Linker (L43) insertion at fliM codon 60 (Eco47III site) in pHT32; Apr | This work |
| pM81 | Pro linker insertion at fliM codon 81 in pHT32; Apr | This work |
| pM111 | Insertion of Pro linker and linker L43 at fliM codon 111 in pHT32; Apr | This work |
| pM132 | Pro linker insertion at fliM codon 132 in pHT32; Apr | This work |
| pM144 | Linker (L40) insertion at fliM codon 144 (NruI site) in pHT32; Apr | This work |
| pM163 | Pro linker insertion at fliM codon 163 in pHT32; Apr | This work |
| pM212 | Insertion of Pro linker and linker L43 at fliM codon 212 in pHT32; Apr | This work |
| pM227 | Pro linker insertion at fliM codon 227 in pHT32; Apr | This work |
| pM241 | Linker (L43) insertion at fliM codon 241 in pHT32; codon 241 also changed from Asn to Asp; Apr | This work |
| pM258 | Linker (L40) insertion at fliM codon 258 (PvuII site) in pHT32; Apr | This work |
| pM267 | Linker (L40) insertion at fliM codon 267 (EcoRV site) in pDFB32; Apr | This work |
| pM282 | Linker (L41) insertion at fliM codon 282 (AatII site) in pHT32; Apr | This work |
| pM310 | Insertion of Pro linker and linker L43 at fliM codon 310 in pHT32; Apr | This work |
| pDFB81 | FliMΔ1–38 expression vector; Apr | This work |
| pHT17 | FliMΔ1–60 expression vector; Apr | This work |
| pHT67 | FliMΔ61–144 expression vector; Apr | This work |
| pHT134 | FliMΔ145–241 expression vector; Apr | This work |
| pHT126 | FliMΔ241–334 expression vector; Apr | This work |
| pHT127 | FliMΔ1–241 expression vector; Apr | This work |
| pMAM1 | FliMΔ61–334 expression vector; Apr | This work |
| pMAM2 | FliMΔ81–334 expression vector; Apr | This work |
| pMAM3 | FliMΔ112–334 expression vector; Apr | This work |
| pMAM4 | FliMΔ145–334 expression vector; Apr | This work |
| pMAM5 | FliMΔ61–240 expression vector; Apr | This work |
| pPB3 | FliMΔ1–247 expression vector; Apr | This work |
Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl) was the medium used for routine culture growth and plasmid transformations. For assays of swarming and swimming motility, cells were grown in tryptone broth (TB) (1% tryptone, 0.5% NaCl). Where appropriate, ampicillin and kanamycin were used at 100 and 50 μg/ml, respectively. IPTG was prepared as a 0.1 M stock in water and used at 100 μM unless otherwise indicated in the figures.
FliM linker insertions.
Four 12-residue oligonucleotides, which are wholly or partially self-complementary, were used to make a series of linker insertion mutations in fliM. They are as follows: L40, 5′-GCTCCCGGGAGC-3′ (SmaI linker); L41, 5′-GCCCGGGCACGT-3′ (AatII to SmaI adapter); L42, 5′-GTACCCCCGGGG-3′ (BsiWI to SmaI adapter); and L43, 5′-CCCCTCGAGGGG-3′ (XhoI linker). In some cases, single proline codons rather than 12-residue linkers were inserted into fliM, as detailed below. Plasmids expressing the linker insertion mutant FliM proteins were named pMn, with n specifying the fliM codon after which the linker was inserted. Fifteen linker insertions, which fell into three groups according to the method of construction, were made. The first group includes pM38, pM60, pM144, pM258, pM267, and pM282, which were made by inserting one of the linkers into existing restriction sites in the fliM gene, resulting in the introduction of the nonnative residues specified in Table 2. The second group includes pM81, pM132, pM163, and pM227, which were made by inserting a single proline codon at the sites indicated. This modification was made either by inserting the triplet CCG or CCC into C/GG or /GGG sequences in the native fliM sequence (slashes indicate sites of insertion) or, in the case of pM163, by replacing nucleotide A at position 489 with the nucleotides CCCG. These mutations were made by using the Altered Sites procedure (Promega) on the fliM gene cloned in plasmid pHT41. These mutations generated SmaI sites (CCCGGG), which were confirmed by restriction digests. The proline insertion mutations were then transferred into pHT32 by exchange of a restriction fragment bordered by a BsiWI site in the fliM coding region and a HindIII site in the downstream polylinker. Each of these single-proline insertions disrupted FliM function, as judged by swarming assays, and so no further insertions were made. The third group includes pM16, pM111, pM212, pM241, and pM310. At these positions, except for pM241, single proline insertions were first made and subcloned into pHT32, as described above. In making the proline insertion at codon 16, a silent mutation was introduced in codon 16 (AAT→AAC). In the case of pM241, a SmaI site was introduced by changing codons 239 through 241 from TCG CGT AAT to TCC CGG GAT, which resulted in the substitution Asn241→Asp. Swarming was not significantly affected by these proline insertions or by the Asn→Asp mutation at residue 241, and so a 12-bp oligonucleotide (L43) was then inserted into each site, by using the SmaI site generated in the first step.
TABLE 2.
Effects of linker insertions in FliM on its function and binding to other flagellar proteins
| Linker position (codon no.) | Introduced sequence | Swarminga rate (mm/h) | Swimminga speed (relative) | Flagellationa (no./cell) | Binding tob:
|
|||
|---|---|---|---|---|---|---|---|---|
| FliN | FliG | CheY | CheY-P | |||||
| 16 | PPSRG | 5.3 | 0.95 | 3.1 | ND | ND | + | ++ |
| 38 | PRGY | 0.4 | 0.8 | 3.0 | + | + | + | + |
| 60 | R60→PPRGG | 0.0 | 0.0 | 0.0 | + | + | + | ++ |
| 81 | P | 0.8 | 0.7 | 2.9 | + | + | + | ++ |
| 111 | PPSRG | 4.3 | 0.9 | 3.1 | ND | ND | ND | ND |
| 132 | P | 0.0 | 0.0 | 0.0 | + | ± | + | ++ |
| 144 | LPGA | 0.4 | 0.55 | 2.6 | + | + | + | + |
| 163 | P | 0.0 | (0.0)c | 1.0 | + | + | ND | ND |
| 212 | PPSRG | 0.4 | 0.7 | 2.9 | + | + | + | ++ |
| 227 | P | 1.0 | 0.5 | 3.2 | + | + | + | ++ |
| 241 | R240N241→PPRGGD | 6.0 | 0.9 | 3.0 | ND | ND | ND | ND |
| 258 | APGS | 0.6 | (0.0) | 3.0 | + | + | ND | ND |
| 267 | APGS | Trailsd | (0.0) | (0.0) | − | + | ND | ND |
| 282 | VPGH | Trails | (0.0) | (0.0) | − | + | ND | ND |
| 310 | PPSRG | 4.0 | 0.65 | 2.8 | ND | ND | ND | ND |
| Wild type | None | 7.0 | 1.0 | 3.1 | + | + | + | + |
a The pMn plasmids carrying the mutant fliM genes expressed from the tac promoter were transformed into the fliM null strain DFB190, and swarming rates, motility in liquid culture, and numbers of flagella were assayed as described in Materials and Methods. In two cases (positions 60 and 241), the linkers replaced some residues, as indicated. Three different wild-type controls were used (the wild-type strain RP437, DFB190[pDFB72] induced with 25 μM IPTG, and DFB190[pHT32] induced with 60 μM IPTG). All gave similar results, and average values are reported. Swimming speeds are expressed relative to wild-type controls and are average values for 48 cells.
b Binding experiments were carried out as described in Materials and Methods. The symbols ++, +, and ± indicate three different levels of binding that differed in the amount of coprecipitated FliM by factors of about 2; − indicates no significant binding. ND, not determined.
c Parentheses indicate that most cells were nonmotile or nonflagellate but that a small number (one among hundreds) of cells were motile or had a single flagellum.
d Trails, satellite microcolonies were observed on swarm plates.
fliM deletion constructs.
fliM deletions were constructed in either pHT32 or one of the pHT32-derived pMn plasmids. Plasmid pHT17 (encoding FliMΔ1–60) was made by deleting a segment extending from the PstI site in the upstream linker to an Eco47III site at codon 60. The PstI site was blunted with mung bean nuclease before ligation. Plasmid pHT67 (FliMΔ60–144) was made by deleting a 252-bp segment extending from the Eco47III site at codon 60 to an NruI site at codon 144. Plasmid pHT134 (FliMΔ145–241) was made from the Asn241→Asp mutant plasmid, which contains an introduced SmaI site at codon 240, by deleting a 287-bp segment extending from the NruI site at nucleotide 431 to the SmaI site at nucleotide 718 and inserting in its place an 8-bp SalI linker (GGTCGACC) to restore the reading frame. Plasmid pHT126 (FliMΔ241–334) was also made from the Asn241→Asp mutant plasmid, by deleting the segment between the SmaI site and an EcoRI site in the downstream polylinker. The EcoRI end was blunted with mung bean nuclease before ligation. Plasmid pHT127 (FliMΔ1–241) was made from the Asn241→Asp mutant plasmid by deleting a segment extending from BamHI in the upstream polylinker to the SmaI site at nucleotide 718. The BamHI end was made blunt before ligation, by using T4 DNA polymerase and deoxynucleoside triphosphates. Plasmid pDFB81 (FliMΔ1–38) was made by first changing the BsiWI site at codon 38 in fliM to a BamHI site and then moving codons 38 to 334 of fliM into pTM30, by using the introduced BamHI site and a SmaI site in a downstream polylinker. Plasmid pMAM4 (FliMΔ145–334) was made by deleting a segment of pHT32 between the NruI site at nucleotide 431 and an EcoRV site in the downstream polylinker. Plasmid pPB3 (FliMΔ1–247) was constructed by inserting a segment of fliM encoding residues 248 through 334, obtained by PCR amplification of the cloned fliM gene (27) with primers that introduced an NdeI site at the 5′ end and a BamHI site at the 3′ end, into the T7 expression vector pAED4 (27).
Flagellation, motility, and swarming.
To determine the phenotypes of the fliM linker insertion mutants, the pMn plasmids were transformed into the fliM null strain DFB190. Staining and counting of flagella were carried out as described elsewhere (27). For swimming-speed measurements, overnight cultures were grown in TB and the appropriate antibiotic, diluted 100-fold into fresh TB containing various concentrations of IPTG, and then cultured for 4 h at 32°C. Cells swimming close to the coverslip were observed though a phase-contrast microscope and recorded on videotape. The paths of individual cells were measured by marking their positions on transparencies at intervals of 1/10 s (three video frames), by using a manual frame-advance feature of the recorder. Each cell was measured for about 1/2 s. Reported swimming speeds are averages for 48 cells.
Measurements of swarming rate in soft agar (TB and 0.28% Bacto Agar) were carried out as described elsewhere (27). Plots of swarm diameter versus time were fitted to a line, and the slopes are reported in millimeters per hour. Swarm assays and flagellar staining were done in medium containing the concentration of IPTG that gave maximal swarming rate when wild-type fliM was expressed from the plasmid (60 μM IPTG for derivatives of pHT32 and 25 μM IPTG for derivatives of pDFB72).
GST fusion coprecipitation procedure.
Coprecipitation experiments were carried out essentially as described elsewhere (29). These experiments employed an flhDC strain (RP3098) that expresses no flagellar proteins except those encoded on plasmids. In experiments to probe interactions of FliM with FliN or FliG, this strain was transformed with two plasmids, one that expresses a GST fusion protein and another that expresses FliM or its mutant variants. The transformants were cultured overnight in TB containing the appropriate antibiotics and 100 μM IPTG. Cells were harvested and lysed by sonication as described elsewhere (29). The following modifications were made to the earlier procedure in order to minimize proteolytic degradation of mutant FliM proteins. First, the 30-min incubation on ice prior to sonication was omitted, and the cells were instead lysed immediately after being resuspended. Second, the incubation of cell lysates with glutathione-Sepharose 4B beads was shortened from 30 to 15 min, and the elution step was shortened from 10 to 1 min. Finally, all steps were done either on ice or in a cold room.
The coprecipitation assay was modified further for studies of FliM binding to CheY or phospho-CheY. The levels of some of the mutant FliM proteins were significantly decreased when GST-CheY was coexpressed in the cells, and so these experiments used two strains, one expressing GST-CheY and the other expressing FliM or its mutant variants. Cells were cultured overnight at 37°C in LB broth plus antibiotics, aliquots (0.5 ml) were added to 120 ml of LB broth containing antibiotics and 100 μM IPTG, and growth was continued for 10 h at 37°C. Absorbance at 600 nm (A600) was measured, and cells were pelleted by centrifugation (3,000 × g, 5 min) and resuspended in phosphate-buffered saline (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) containing 5 mM EDTA, 0.2 mM APMSF (4-amidinophenylmethanesulfonyl fluoride), and 0.1% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate), by using 2 ml of buffer per A600 U to adjust to the same final cell density. The cells were then frozen in 0.5-ml aliquots and stored at −70°C.
For the assay, an aliquot of cells that expressed GST-CheY (or GST alone as a negative control) and another of cells that expressed FliM or its mutant variants were thawed; mixed; combined with 100 μl of a lysozyme solution (5 mg/ml in 50% glycerol), 10 μl of APMSF (10 mM stock in methanol), 60 μl of 1 M MgCl2, and 100 μl of either water (nonphosphorylating conditions) or 0.5 M acetyl phosphate (final concentration, 40 mM); and then lysed by sonication (Branson Model 450 sonifier, Power 3, duty cycle 50%, three times for 50 s each). Debris was pelleted at 4°C (16,000 × g, 15 min). Fifty microliters of the supernatant was saved for use in estimating the amount of FliM present before addition of affinity beads. The rest (ca. 1 ml) was transferred to a clean tube, mixed with 100 μl of a 50% slurry of glutathione-Sepharose 4B (Pharmacia) prepared according to the manufacturer’s directions, and incubated at 4°C for 30 min to allow binding. The Sepharose beads were then pelleted by a 5-s microcentrifuge spin, washed with 1 ml of phosphate-buffered saline, and pelleted again by a brief spin. This wash step was repeated twice more. The beads were then incubated with 50 μl of elution buffer (50 mM reduced glutathione in 50 mM Tris-HCl [pH 8]) at room temperature for 1 min, with occasional mixing to release the protein. Beads were then pelleted, and the supernatant was collected for analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting, as described elsewhere (29). Coprecipitated material was quantitated by immunoblots of serially diluted samples, calibrated by a standard curve constructed with known amounts of purified FliM. Densitometry employed a video frame-capture system and the analysis program NIH Image, version 1.52.
Secondary structure prediction.
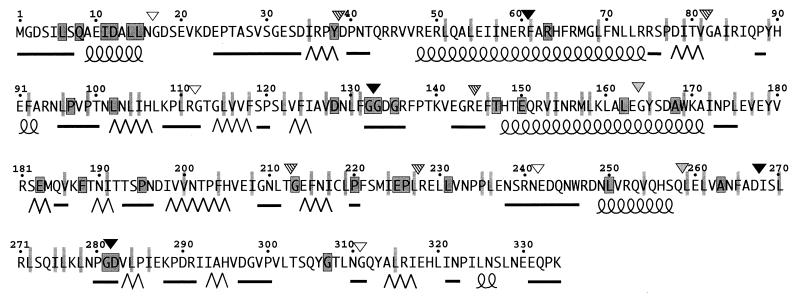
Secondary-structure prediction of FliM used the neural network algorithm of Rost and Sander (21, 22), accessed via electronic mail to the web site maintained by the European Molecular Biology Laboratory in Heidelberg, Germany (http://www.embl-heidelberg.de/predictprotein/). Structure prediction was carried out by using all of the sequence data in an alignment of FliM proteins from Escherichia coli, Bacillus subtilis, Borrelia burgdorferi, and Caulobacter crescentus and also by using just the sequence from E. coli. Figure 5 displays the elements of secondary structure clearly predicted in both cases.
FIG. 5.
Patterns of sequence conservation, predicted secondary structures, and effects of linker insertions in FliM. The alignment used FliM sequences from E. coli, B. subtilis, B. burdorferi, and C. crescentus. The sequence from Agrobacterium tumefaciens is also known, but it is quite different from all of the others and was not included. Outlined and shaded boxes indicate residues that are identical in the four sequences, and nonoutlined, lightly shaded bars indicate positions where residues with hydrophobic character are found in all four sequences. Secondary structures were predicted by the neural net algorithm of Rost and Sander (21, 22), by using the information from all of the sequences. The α-helices are represented by coiled lines, the β-strands are represented by zigzag lines, and segments of nonregular secondary structure are represented by straight lines. Segments where predictions were ambiguous are left blank. Inverted triangles indicate positions and phenotypes of linker insertion mutations: solid, nonflagellate; stippled, flagellate but most cells nonmotile; striped, motile but nonswarming because of a strong CCW bias; and open, close to wild-type swarming. (See Table 2 for exact phenotypes and sequences of the insertions.)
RESULTS
FliM linker-insertion mutants.
The fliM gene was mutagenized by inserting short oligonucleotide linkers in 15 approximately regularly spaced locations. Each linker encoded from one to five nonnative residues and included at least one proline. The positions of the insertions and the residues introduced by the linkers are listed in Table 2. The linkers will presumably affect function significantly when they are inserted into interior segments of the protein that are important for folding or in exposed segments that contact other proteins. In segments that are not important for folding or function, insertions should have relatively minor effects. The effects of the linker insertions on FliM function were determined by expressing the mutant proteins from plasmids in the fliM null strain DFB190 and measuring numbers of flagella, rates of swarming in soft agar, and swimming speeds in liquid culture. The results are summarized in Table 2.
Four linker insertions, at codons 16, 111, 241, and 310, did not disrupt FliM function significantly. When expressed in the fliM null strain, these four proteins supported normal flagellation and nearly normal swarming in soft agar. These four positions are thus not critical to the folding of the protein, its incorporation into flagella, or its functions in motor rotation and switching.
Five linker insertions affected flagellar assembly to various extents. The insertions at residues 60 and 132 prevented flagellar assembly completely, and those at residues 267 and 282 prevented assembly almost completely, so that only one cell among hundreds had a single flagellum. Cells expressing the residue 267 and 282 insertion proteins also produced satellite microcolonies on swarm plates (data not shown), indicating that the flagella that occasionally were assembled were functional. The linker insertion at codon 163 reduced the number of flagella per cell to about one-third of normal, and it also caused serious defects in swimming and swarming (Table 2).
The other six linker insertion mutants had normal numbers of flagella but swarmed poorly. One of these (at codon 258) nearly eliminated motility. The other five (at codons 38, 81, 144, 212, and 227) allowed good swimming but affected the CW-CCW rotational bias of the motor, so that the cells swam smoothly and did not tumble.
The linker insertions did not prevent synthesis or folding of the FliM protein. Each of the mutant proteins accumulated in cells to approximately normal levels, as judged by immunoblots of proteins in fresh cell lysates. When the cell lysates were left at room temperature for 2 h, however, most of the mutant proteins were significantly degraded, whereas the wild-type protein was not (data not shown).
FliM deletion mutants.
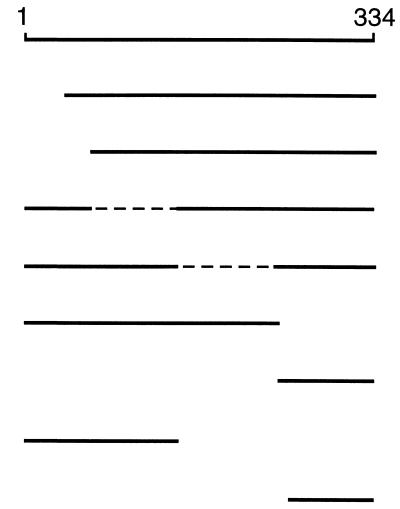
To obtain additional insight into the domain organization of FliM, we also constructed several deletion mutants. The choice of segments for deletion was based on the phenotypes of linker insertion mutants, available restriction sites, and sequence comparisons and predictions of secondary structure (see Fig. 5). Effects of the FliM deletions on function were assessed by measuring numbers of flagella, rates of swarming in soft agar, and motility of cells in liquid culture. The results are summarized in Table 3.All of the deletions tested gave a nonflagellate phenotype, with the exception of a 38-residue N-terminal deletion that gave a motile but nonchemotactic phenotype. Cells of the FliMΔ1–38 mutant swam smoothly, with few or no tumbles, indicating that their motors rotated with a strong CCW bias. The nonflagellate phenotype of most deletions was not caused by destabilization of the FliM protein, since most of the FliM fragments were stable enough to accumulate in cells. Stable FliM variants included a large C-terminal deletion (Δ145–334) and a medium-sized N-terminal deletion (Δ1–60). Some larger N-terminal deletions, which are not included in Table 3 but are described below, destabilized the protein so that it did not accumulate to a detectable level. A large N-terminal deletion of 241 residues left a fairly small (93-residue) C-terminal fragment that was stable under certain circumstances, as described below. Because this C-terminal fragment was marginally stable and its binding to other flagellar proteins proved especially interesting, we also constructed a plasmid that directs higher-level expression of a slightly smaller C-terminal fragment (residues 248 to 334). When overexpressed, this 87-residue C-terminal fragment accumulated in cells and was readily detectable on immunoblots. This fragment was used in some of the binding experiments described below.
TABLE 3.
Effects of deletions in FliM on its function and binding to other proteinsa
a Summary of FliM deletions studied, their phenotypes, and the binding of FliM fragments to FliN, FliG, CheY, and phospho-CheY. Phenotypes of the deletions were determined by expressing the proteins in the fliM null strain DFB190. Binding was examined in coprecipitation experiments using the one-cell protocol and induction by 200 μM IPTG (interactions with FliN or FliG) or the two-cell protocol and induction by 100 μM IPTG (interactions with CheY or phospho-CheY). The symbols ++, +, +/−, and +/−− indicate four levels of binding, differing from each other by a factor of about 2 in the amount of coprecipitated FliM; ND, not determined; WT, wild type. Broken lines indicate internal deletions in FliM.
Interactions with FliN or FliG.
Insertions in FliM that disrupt function might disrupt interactions with other proteins. To test this possibility, we examined the binding of the mutant FliM proteins to other proteins with which FliM is known to interact. These experiments used GST fusions and coisolation assays developed previously to study binding interactions among the switch complex proteins (29). Because high-level overexpression of FliM might lead to nonspecific binding, we first used Coomassie blue-stained gels to determine whether FliM was highly overexpressed under the conditions used for binding experiments. Under the conditions typically used in binding experiments (induction of pHT32 or its variants with 100 μM IPTG), no band assignable to FliM was observed on gels of whole-cell proteins, indicating that FliM was not among the more abundant proteins present. FliM was readily detected on immunoblots of proteins from the same cultures (data not shown).
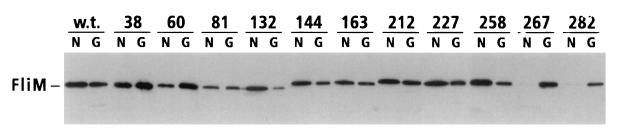
Binding of the insertion-mutant FliM proteins to FliN and FliG was then examined in coprecipitation experiments with GST-FliN and GST-FliG. These experiments tested only the 11 insertions in FliM that disrupt function. Representative results are shown in Fig. 1, and binding data are summarized in Table 2. In agreement with the previous binding study (29), wild-type FliM was coprecipitated with both GST-FliN and GST-FliG. FliM was not coprecipitated with GST alone (example shown in Fig. 4) (see reference 29). Binding to FliN was eliminated by two adjacent linker insertions at residues 267 and 282 in FliM and was weakened somewhat by insertions at residues 60 and 81. The other seven insertions did not measurably affect the FliM-FliN binding. Binding of FliM to FliG was not affected strongly by any of the linker insertions. The insertion at residue 132 weakened FliG binding somewhat, and the insertion at residue 81 may have had a small effect. The other nine insertions in FliM had no significant effect on binding to FliG.
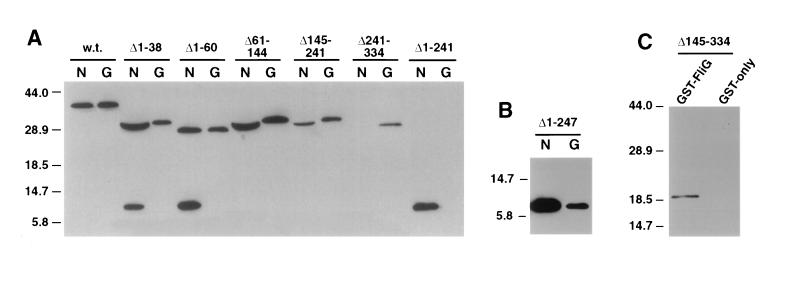
FIG. 1.
Coprecipitation of linker insertion mutant FliM proteins with GST-FliN (lanes labeled N) or GST-FliG (lanes labeled G). Positions of the linker insertions in FliM are indicated at the top. The experiment used the one-cell protocol (Materials and Methods). Coprecipitated material was analyzed on immunoblots probed with anti-FliM antiserum. Immunoblots of samples not exposed to the glutathione beads showed that all of the insertion mutant FliM proteins were present in cell lysates at levels comparable to that of the wild-type (w.t.) protein (data not shown).
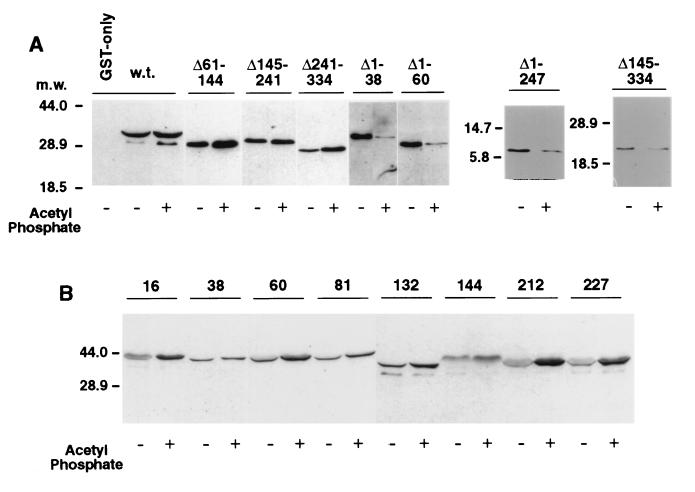
FIG. 4.
Effects of deletions or linker insertions in FliM on binding to CheY in the presence or absence of the phosphorylating agent acetyl phosphate (all experiments contained Mg2+). (A) Coprecipitation of FliM deletion-mutant proteins with GST-CheY. The FliM deletions are indicated at the top. For the experiment using wild-type FliM protein, the GST-only negative control is also shown (first lane in panel A); all other lanes used the GST-CheY fusion protein. Negative controls for each of the FliM fragments showed that none was coprecipitated with GST alone (data not shown). Blots were typically exposed to film for 5 min after addition of the chemiluminescence reagents, but with the Δ1–38 and Δ1–60 mutants, the binding was somewhat weaker in the lanes without acetyl phosphate and significantly weaker in the lanes with acetyl phosphate, so overnight exposure was used. (The densitometry results in Table 3 used uniform exposures and a range of protein concentrations.) (B) Coprecipitation of FliM linker insertion proteins with GST-CheY. Positions of the linker insertions are indicated at the top. w.t., wild type. Numbers to the left of each panel show molecular mass (m.w.) in kilodaltons.
Next, we measured binding of the FliM deletion constructs to FliN and to FliG, again by coprecipitation experiments with GST-FliN and GST-FliG. Sample gels are shown in Fig. 2, and the results are summarized in Table 3. All except one of the FliM fragments studied were stable enough to accumulate in the cells, as determined by immunoblots of cell lysates prior to addition of the glutathione beads (data not shown). The exception was a C-terminal fragment consisting of residues 242 to 334, which accumulated to detectable levels when coexpressed with FliN or with the GST-FliN fusion protein but not when expressed alone or with GST-FliG.
FIG. 2.
Coprecipitation of FliM fragments with GST-FliN (lanes labeled N) and GST-FliG (lanes labeled G). The parts of FliM deleted are indicated at the top of each lane. (A) An initial set of FliM deletions, which together span the protein. The C-terminal fragment FliM242–334 (labeled Δ1-241 over the two rightmost lanes) was not stable except in the presence of FliN or GST-FliN, and so its failure to coprecipitate with GST-FliG is inconclusive. (B) Coprecipitation of an 87-residue C-terminal fragment of FliM with both GST-FliN and GST-FliG. This fragment accumulated in cells to detectable levels even in the absence of FliN (see text). (C) Coprecipitation of a 144-residue N-terminal fragment of FliM with GST-FliG. In negative-control experiments, neither FliM nor any of the FliM fragments was coprecipitated with GST alone (example control for a FliM fragment is shown here, and that for full-length FliM is shown in Fig. 4). w.t., wild type. Numbers to the left of each panel show molecular mass in kilodaltons.
Full-length FliM was coprecipitated with GST-FliN (Fig. 2) (see reference 29), whereas in negative controls with GST alone neither FliM nor any of the FliM deletion constructs was coprecipitated (example control for the fragment FliM1–144 is shown). The C-terminal fragment FliM242–334 bound well to FliN, as did several other FliM constructs that contained this C-terminal domain. A slightly smaller C-terminal fragment (FliM248–334) was also tested, and it was also coprecipitated with GST-FliN (Fig. 2B). Because the FliM248–334 fragment was expressed from the T7 promoter, this experiment required the use of a different strain and the two-cell protocol. A large N-terminal fragment (FliM1–240) was stable enough to accumulate in cells but was not coprecipitated by GST-FliN. These results show that a ca. 90-residue C-terminal fragment of FliM is necessary and sufficient for binding FliN.
Full-length FliM was coprecipitated with GST-FliG in good yield (Fig. 2) (see reference 29). The binding of FliM to FliG was not prevented by any of the FliM deletions studied. Together, these deletions cover the entire FliM protein, and their endpoints coincide with the positions of linker insertions that also did not abolish binding to FliG. These results suggest that multiple, noncontiguous segments of FliM bind to FliG.
To test this proposal and to localize further the parts of FliM that bind to FliG, we made additional FliM deletion constructs. FliM fragments consisting of residues 1 to 60, 1 to 80, or 1 to 111 are evidently unstable, as they did not accumulate to detectable levels. A FliM fragment consisting of residues 1 to 144 did accumulate, although its level was lower than that of the more stable FliM fragments. The FliM1–144 fragment was coprecipitated with GST-FliG (Fig. 2C). As noted, the C-terminal 93-residue fragment of FliM accumulated in cells only when coexpressed with FliN or GST-FliN, and so its binding to FliG could not be tested. The smaller C-terminal fragment FliM248–334 did accumulate, to a level detectable on immunoblots but not on Coomassie blue-stained gels, and it was coprecipitated with GST-FliG (Fig. 2B).
The FliM proteins with deletions of residues 1 to 38 or 1 to 60 gave rise to an additional band at about 10 kDa, which appears to be a FliM breakdown product. The breakdown product was not observed in experiments using wild-type FliM or FliM fragments with normal N termini. Its size on gels was indistinguishable from the C-terminal FliM fragment consisting of residues 241 to 334, and it was coprecipitated with GST-FliN but not GST-FliG. These observations suggest that the 10-kDa fragment is the C-terminal domain of FliM and that the site of proteolysis is near residue 240. N-terminal deletions of FliM thus appear to affect the protease susceptibility of the protein in the vicinity of residue 240.
Binding of FliM to CheY and phospho-CheY.
Phospho-CheY is the signaling molecule that triggers switching of the motor to CW rotation. Binding of purified FliM to phospho-CheY has been demonstrated previously in coisolation assays in which CheY was covalently linked to Sepharose beads (32) and by chemical cross-linking (2, 3). These studies showed that binding was significantly stronger in the presence of agents that phosphorylate CheY (acetyl phosphate and Mg2+) than in the absence of these agents.
Binding of FliM to CheY was examined in coprecipitation assays, by using a GST-CheY fusion and the two-cell procedure. FliM was coprecipitated with GST-CheY in significant amounts even in the absence of acetyl phosphate and Mg2+, and the amount of coprecipitated FliM was not significantly increased by the addition of these agents (Fig. 3). This contrasts with previous reports, in which binding was significantly stronger when CheY was phosphorylated than when it was not (2, 32).
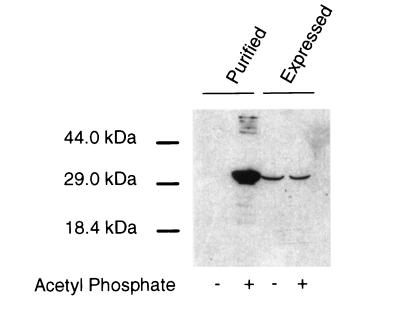
FIG. 3.
Coprecipitation of FliM with GST-CheY in the presence or absence of agents that phosphorylate CheY, with FliM from two sources. (Left lanes) Experiment using FliM that was purified as described elsewhere (19, 32). Purified FliM was added to a suspension of RP3098 cells (which express no flagellar proteins) to give a final FliM level similar to that in other binding experiments. These FliM-supplemented cells were mixed with cells expressing GST-CheY, the cells were lysed, and a binding experiment was performed by the two-cell protocol. (Right lanes) A binding experiment done in the same way, except that the FliM was expressed within the RP3098 cells, and the samples were not supplemented with purified FliM.
The previous studies used purified FliM and CheY proteins. We therefore purified FliM according to published procedures, which included denaturation and refolding steps (2, 3, 19, 32), and used this purified FliM in coprecipitation experiments with GST-CheY. When purified FliM was used, little of the protein was coprecipitated with GST-CheY under nonphosphorylating conditions, but much more was coprecipitated under phosphorylating conditions (Fig. 3). The GST-CheY coprecipitation assay thus reproduces the principal result of previous studies when purified FliM protein is used.
Binding of GST-CheY to each of the FliM deletion constructs was then tested. These experiments, and the others described below, used fresh cell lysates rather than purified FliM. Sample gels are shown in Fig. 4, and the results are summarized in Table 3. FliM molecules lacking residues 61 to 144, 145 to 241, or 241 to 334 bound to CheY about as well as did full-length FliM under nonphosphorylating conditions. In contrast to full-length FliM, however, the binding of these deletion proteins was somewhat enhanced by the addition of phosphorylating agents. Short N-terminal deletions of 38 or 60 residues had different effects. These weakened the binding to CheY significantly under nonphosphorylating conditions and reversed the phosphorylation effect so that binding was significantly weaker in the presence of the phosphorylating agents (Fig. 4).
These results suggest that some determinants for binding phospho-CheY are located near the N terminus of FliM but that other parts of FliM are also involved. Binding of multiple segments of FliM to CheY was confirmed in coprecipitation experiments with the N-terminal fragment FliM1–144 and the C-terminal fragment FliM248–334. The fragment FliM1–144 was coprecipitated with GST-CheY, in both the presence and the absence of phosphorylating agents. The fragment FliM248–334 was also coprecipitated with GST-CheY, with the yield being significantly less in the presence of phosphorylating agents (Fig. 4).
We next examined the binding of CheY and phospho-CheY to some of the FliM proteins with linker insertions (at residues 16, 38, 60, 81, 132, 144, 212, and 227). None of the linker insertions eliminated binding to CheY; all of the mutant proteins were coprecipitated with GST-CheY under both nonphosphorylating and phosphorylating conditions. Some of the insertion mutants significantly enhanced the phosphorylation effect. Unlike wild-type FliM, proteins with insertions at positions 16, 60, 81, 212, and 227 bound significantly more CheY when phosphorylating agents were present (Fig. 4 and Table 2).
Sequence conservation and predicted secondary structure of FliM.
The fliM genes from several species have been cloned and sequenced. Figure 5 displays patterns of sequence conservation as determined by an alignment of FliM sequences from four species and shows the principal elements of secondary structure predicted by a neural net algorithm (21, 22). These analyses give clues to the overall organization of the protein and will provide a useful framework for discussing the results presented here and in other studies of sequence-function relationships in FliM (2, 25, 30, 31). Features of interest are as follows. A short segment near the N terminus (residues 8 to 15) is well conserved and is predicted to be mainly α-helical. The bulk of the protein is predicted to be organized into domains with mixed α and β secondary structure, joined by sizable segments that are poorly conserved and predicted to have nonregular secondary structure and which might be interdomain linkers (residues 17 to 34, 136 to 145, and 237 to 247). Adjacent to one of these putative linkers is a short segment (residues 132 to 135) that contains two invariant Gly residues and a third Gly that is present in all species but at slightly different positions. The most C-terminal part of the protein, from about residue 287 to the end, is relatively poorly conserved.
DISCUSSION
The phenotypes of fliM linker insertion mutations.
Four linker insertions in FliM, at residues 16, 111, 240, and 310, allowed nearly normal function. This result can be rationalized in terms of the predicted secondary structures and patterns of sequence conservation (open inverted triangles in Fig. 5). Residue 16 is at the N-terminal boundary of a segment that is poorly conserved and predicted to have nonregular secondary structure. This region might be a linker between the well-conserved N-terminal segment (residues 8 to 15) and the rest of the protein. Residue 111 is in a segment predicted to be a loop between two β-strands. This loop is evidently not on a functionally important surface of the protein. Residue 240 is near the middle of a segment (residues 237 to 247) with several properties suggestive of an interdomain linker—poor conservation, polar character, and nonregular secondary structure. Residue 310 is in a segment that is relatively poorly conserved and is predicted to have nonregular secondary structure.
Only two linker insertions completely prevented flagellar assembly, one in the conserved Gly-rich segment near residue 132 and the other in a strongly predicted α-helix at residues 48 to 75. Two other insertions (at residues 267 and 282) made flagella very rare, probably by disrupting binding to FliN (see below). The scarcity of insertions that prevent flagellar assembly contrasts with the 10-residue deletions studied by Toker et al. (30), most of which (21 of 34) gave a nonflagellate phenotype (31). This finding suggests that small deletions disrupt the protein structure more than short linker insertions, at least in most cases. The 10-residue deletions that did not prevent flagellar assembly (30) follow a pattern that is consistent with the secondary structure proposed in Fig. 5: most occur near the N terminus, the C terminus, or the putative linker around residue 240, in segments that are poorly conserved and predicted to have nonregular secondary structure.
Five of the linker insertions affected motor switching, causing a strong CCW bias. Just two linker insertions gave a nearly Mot− phenotype, in which flagella were assembled but most cells were nonmotile. Previous studies of spontaneous fliM mutants also suggested that mutations giving an aberrant-switching phenotype are more common than those giving a paralyzed phenotype (25). The insertions that gave strong CCW bias did not weaken binding of FliM to CheY or phospho-CheY. They nevertheless prevented normal CCW→CW switching, possibly by impeding conformational changes normally triggered by binding of phospho-CheY. Insertion mutations giving the various phenotypes did not cluster according to any obvious pattern, except that the adjacent insertions at residues 267 and 282 both gave a nearly nonflagellate phenotype and led to the formation of satellite microcolonies on swarm plates.
A C-terminal FliN-binding domain.
A number of observations indicate that a ca. 90-residue C-terminal domain of FliM is necessary and sufficient for binding FliN. C-terminal FliM fragments accumulated in cells when they were coexpressed with FliN (or GST-FliN) or when they were highly overexpressed, but not otherwise. These FliM fragments bound FliN in coprecipitation experiments, whereas a large N-terminal fragment (residues 1 to 240) did not. Binding to FliN was abolished by two adjacent linker insertions in the C-terminal domain of FliM, at residues 267 and 282. As noted, the C-terminal domain of FliM is joined to the rest of the protein by a segment (residues 237 to 247) that might function as an interdomain linker.
Our results concerning the FliM-FliN interaction are consistent with those of Marykwas et al. (17), who used the two-hybrid system to show that FliN binds to full-length FliM, but not to FliM with 52 residues deleted from the C terminus. Our results also agree for the most part with the affinity blot study of Toker and Macnab (31) but differ in certain details. Their study suggested that the main determinants of FliN binding extend from ca. residue 270 to residue 320 and that the segment from residue 230 to residue 270 might also be important. The present results suggest that the main determinants of FliN binding are more localized, probably to between residues 260 and 300. Binding to FliN was not affected by a linker insertion at residue 258, and flagellar assembly and function were not seriously affected by linker insertions at residue 241 or 310. (Binding was not tested for these insertions.) Since the linker insertions were spaced at some distance and since each might affect structure only locally, our analysis may have missed some determinants of FliN binding. Alternatively, some of the 10-residue deletions used in the affinity blot study may have altered the protein conformation enough to disrupt FliN binding indirectly, leading to an overestimate of the extent of the FliN-binding site.
Bischoff and Ordal isolated a gene from B. subtilis, dubbed fliY, whose product shows similarity to both FliN and FliM (1). FliY appears to function mainly in the role of FliN, because a plasmid-borne fliY gene restores motility to a Salmonella fliN mutant and a ca. 100-residue segment at the C terminus of FliY shows strong homology to FliN. FliY also shows strong homology to FliM, in a short segment near the N terminus (FliY residues 6 to 15 are 90% identical to FliM residues 6 to 15). FliY shows weak homology to FliM elsewhere, including the C-terminal domain that is strongly homologous to FliN. The C-terminal domain of FliY thus resembles both FliN (strongly) and FliM (weakly). This suggests a possible evolutionary relationship between FliN and parts of FliM. To determine whether similarity between FliN and C-terminal parts of FliM is also seen in other species, we carried out pairwise sequence alignments of FliN and C-terminal domains of FliM for species for which both sequences are known. Some homology was observed in all species, weak in some (including E. coli) but significant in others (Fig. 6). The similarity to FliN is greatest in FliM residues 259 to 286 (in E. coli numbering), which is also the segment implicated in binding to FliN. Although homology is weak in some species, in all species there is a conserved pattern in the positions of hydrophobic residues, suggesting that these parts of FliN and FliM might have a similar fold. FliM and FliN are both components in the C ring of the flagellum (5, 37, 38). FliN and the C-terminal domain of FliM might occupy quasiequivalent positions within this ring (see Fig. 7 and the discussion below), which might require that they share some structural features.
FIG. 6.
Sequence alignments of segments of FliN with segments in the C-terminal domain of FliM, for species where both sequences are known (B. subtilis, Treponema pallidum, B. burgdorferi, C. crescentus, and E. coli). (The sequences are also known for Salmonella but are not significantly different from those of E. coli.) In B. subtilis and T. pallidum, the FliN homolog is called FliY and is a much larger protein that shows close homology to FliM in a short segment near the N terminus (1) (see the text). Residue numbers are not given for FliM from T. pallidum because the entire sequence is not known. Darkly shaded boxes indicate residues identical in FliM and FliN (or its homolog FliY) from the same species. Lightly shaded bars indicate positions where a sizable hydrophobic residue is found in both proteins from all species. Arrows indicate positions of the linker insertions in FliM that disrupted its binding to FliN.
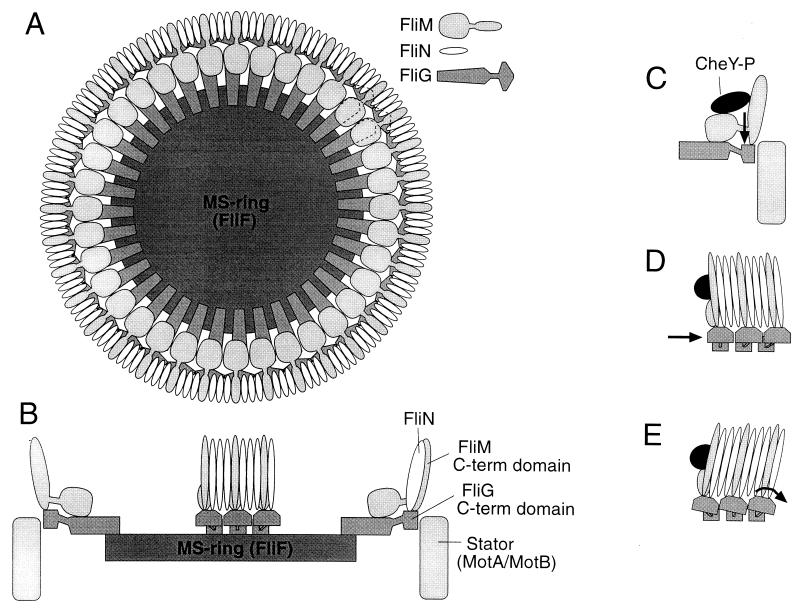
FIG. 7.
Model of the arrangement of the FliG, FliM, and FliN proteins in the flagellar motor. (A) The FliG-FliM-FliN assembly mounted on the MS ring, as viewed from the cytoplasm. Subunit stoichiometries are approximate and are based on immunoblots of proteins in isolated flagellar structures (37, 38); FliN-FliM stoichiometries of 2:1 or 4:1 are also possible. The location of FliN in the C ring is based on immunoelectron microscopy (5, 38). Results of the present study suggest that FliN and the C-terminal domain of FliM occupy similar positions in the structure. The C-terminal domain of FliG is placed at the rotor-stator interface on the basis of mutational studies of FliG and of the stator proteins MotA and MotB (6, 7, 12, 13, 39, 40). (B) Side view of the FliG-FliM-FliN assembly. For clarity, only a subset of the proteins is shown. The cytoplasm is toward the top, and the periplasm is toward the bottom. The MS ring and MotA-MotB complexes are located in the cytoplasmic membrane, which is not pictured. (C to E) Hypotheses for the movements that might be triggered by binding of phospho-CheY to FliM, to cause switching to the CW direction of motor rotation. In each case, switching is suggested to involve a change in the position or orientation of the FliG C-terminal domain, relative to the stator and/or other parts of the rotor. (C) Binding of phospho-CheY might cause a domain of FliM, and the attached domain of FliG, to move up or down in a direction parallel to the rotation axis of the motor. (D) Phospho-CheY might induce subunits of the C ring to tilt, causing attached domains of FliG to move tangentially relative to other components of the rotor. Here and in panel E, the view is rotated 90° relative to that in panel C. (E) Phospho-CheY might induce tilting of both the C-ring subunits and the attached FliG domains, changing the angular orientation of the FliG domains.
The FliM-FliN interaction appears important for flagellar assembly, because linker insertions that disrupt this interaction gave a nearly nonflagellate phenotype. Both FliM and FliN are essential for flagellar assembly (27, 28), and a recent report suggests that both must be present in order for either to be incorporated into the flagellum (11). The FliM-FliN interaction is probably not directly important for CW-CCW motor switching, because point mutations that affect switching are rare in FliN (7), and although they occur at high frequency in FliM they are not found in the C-terminal domain (25), nor are linker insertions that affect switching found there. The FliM-FliN interaction is also not likely to be directly important for torque generation, because the mutations in FliM and FliN that give a Mot− phenotype (7, 25) appear to affect the installation of proteins in the flagellar motor rather than torque generation per se (12).
Site(s) of FliG binding.
Both deletions and insertions in FliM had surprisingly small effects on its binding to FliG (Tables 2 and 3), suggesting that this interaction might involve multiple, noncontiguous segments of the protein. This prediction was confirmed in experiments with the N-terminal fragment FliM1–144 and the C-terminal fragment FliM248–334, both of which were coprecipitated with GST-FliG (Fig. 2). Our conclusions concerning the FliG-binding site thus differ from those of Toker and Macnab (31), whose affinity blot study suggested that determinants for FliG binding are located in the middle of the protein, in the segment between residues 140 and 220. We cannot rule out binding of the middle part of FliM to FliG, because fragments lacking large segments from the N terminus were unstable (data not shown), but our results suggest that the middle of FliM is not the only part involved in binding FliG.
The only insertion mutation that measurably reduced binding to FliG was a proline introduced after residue 132. This mutation also abolished flagellation. The FliM-FliG interaction remained strong when residues 60 through 144 were deleted, however, which implies that the segment containing residue 132 does not form a sole binding site for FliG. This segment may contribute to one among multiple sites for FliG binding, or it may influence FliG binding indirectly, by altering the conformation of FliM. As noted, three Gly residues are conserved in the segment from residues 132 to 135 in FliM, suggesting a special structural role for this part of the protein. This segment also contains about half of the mutations that gave a paralyzed phenotype in extensive mutational studies of FliM from Salmonella strains (25). These Mot− mutations of fliM can be partially suppressed by overexpressing FliN, which suggests that they affect the installation of FliN (or FliM-FliN complexes) into the motor (12). Toker and Macnab (31) found that a FliM protein lacking residues 131 to 140 was stable but did not bind FliN, a result that is surprising given the location of the FliN-binding site much nearer the C terminus. Collectively, these observations suggest that the segment near residue 132 is an important determinant of FliM conformation.
The binding site for CheY.
Small N-terminal deletions in FliM weakened its binding to CheY and reversed the effect of CheY phosphorylation so that binding was weaker in the presence of phosphorylating agents. FliM with a 38-residue N-terminal deletion conferred a smooth-swimming phenotype, indicating that motor switching is impaired. These results suggest that residues near the N terminus of FliM are important for binding to CheY, and particularly for binding to phospho-CheY. This conclusion agrees with the recent study of Bren and Eisenbach (2), who used peptides in competition experiments to show that the first 16 residues of FliM contain determinants for binding phospho-CheY. Evidence of CheY binding to the N-terminal part of FliM was also obtained in the deletion study of Toker and Macnab (31). A linker inserted after residue 16 did not interfere with flagellar assembly, motor rotation, chemotaxis, or CheY binding. This finding implies that residues immediately C-terminal to residue 16 are not important for binding to CheY or for any conformational changes that accompany motor switching.
Although the extreme N-terminal part of FliM appears important for binding CheY, it does not form the sole CheY-binding site, because proteins lacking 38 or 60 N-terminal residues were still coprecipitated with GST-CheY. At least one additional site for binding CheY is located in the C-terminal domain of FliM (FliM248–334), which bound to GST-CheY in coprecipitation experiments (Fig. 4). This binding was relatively weak, however, and its importance remains to be established, given the absence of any mutations that affect switching in this domain. Also, our results do not rule out binding sites for CheY in the middle of FliM. Whatever the exact FliM segments involved, the binding of CheY to multiple parts of FliM suggests that motor switching might involve a relative movement of FliM domains. In this context, it may be relevant that N-terminal deletions of 38 or 60 residues seem to affect the protease susceptibility of FliM in the vicinity of residue 240 (Fig. 2).
Our results point to an important difference between FliM in cell lysates and FliM purified by published procedures. FliM in cell lysates bound weakly to both CheY and phospho-CheY. In contrast, purified FliM bound weakly to CheY but much more strongly to phospho-CheY, as was observed in previous studies using the purified protein (32). Most FliM in cells is found in the cytoplasm, not in the flagella (27). Our results suggest that cytoplasmic FliM exists in a state that is different from that of purified FliM. The cytoplasmic FliM is probably also different from FliM in the flagellar motors, which should presumably bind phospho-CheY strongly so that the motors can respond sensitively to changes in phospho-CheY level. Weak binding of cytoplasmic FliM to phospho-CheY might be necessary for sensitive chemotaxis: if phospho-CheY bound strongly to FliM in the cytoplasm, the phospho-CheY might be prevented from reaching its binding sites in the flagellar motors. In the free cytoplasmic FliM, binding sites for phospho-CheY might be present but be masked by other parts of the protein. This could account for the observation that several different linker insertions and deletions in FliM significantly enhance its binding to phospho-CheY.
Assembly of FliM into the flagellum.
A model for the arrangement of FliM and the other switch complex proteins in the flagellum is presented in Fig. 7. Key features of the hypothesis are as follows. FliN and the C-terminal domain of FliM bind to each other and together form the bulk of the C ring (5, 38). FliN and FliM alternate in a regular pattern, which is suggested to be three FliN molecules per FliM molecule on the basis of present estimates of subunit stoichiometry (ca. 110 FliN and 35 FliM molecules per flagellum [38]). Ratios of 4:1 or 2:1 are also possible and would not alter the essentials of the model. The C ring contacts another ring formed from FliG. Because FliG is present in 25 to 50 copies per flagellum (37), whereas FliM and FliN together total more than 100 copies, the FliG subunits most likely contact only some of the subunits in the C ring. Both N-terminal and C-terminal domains of FliM are pictured binding to FliG. The C ring does not contribute directly to the site of torque generation, but it is important for positioning the C-terminal domain of FliG there and for ensuring that directional switching occurs synchronously in all parts of the rotor (Fig. 7C to E). Binding to phospho-CheY is also suggested to involve multiple domains of FliM. This binding might induce a relative movement of FliM domains, and of the FliG domain(s) to which they are attached, causing changes at the rotor-stator interface that lead to CW rotation (Fig. 7C to E).
ACKNOWLEDGMENTS
We thank Stephenie Billings, Xun Wang, and Patrick Thronson for assistance with flagellar staining, DNA preparation, and swimming-speed measurements; Perry Brown for construction of plasmid pPB3 and discussions of flagellar protein stoichiometry; and Melinda Hill and Sergei Bibikov for assistance with the neural network-based secondary structure prediction algorithm.
This work was supported by grant MCB-9513486 from the National Science Foundation. M.A.A.M. received support from training grant 5T32-GM08537 from the National Institute of General Medical Sciences. The Protein-DNA Core Facility at the University of Utah receives support from the National Cancer Institute (5P30 CA42014).
REFERENCES
- 1.Bischoff D S, Ordal G W. Identification and characterization of FliY, a novel component of the Bacillus subtilis flagellar switch complex. Mol Microbiol. 1992;6:2715–2723. doi: 10.1111/j.1365-2958.1992.tb01448.x. [DOI] [PubMed] [Google Scholar]
- 2.Bren A, Eisenbach M. The N-terminus of the flagellar switch protein, FliM, is the binding domain of the chemotactic response regulator, CheY. J Mol Biol. 1998;278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 3.Bren A, Welch M, Blat Y, Eisenbach M. Signal termination in bacterial chemotaxis: CheZ mediates dephosphorylation of free rather than switch-bound CheY. Proc Natl Acad Sci USA. 1996;93:10090–10093. doi: 10.1073/pnas.93.19.10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francis N R, Irikura V M, Yamaguchi S, DeRosier D J, Macnab R M. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci USA. 1992;89:6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francis N R, Sosinsky G E, Thomas D, DeRosier D J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 6.Garza A G, Biran R, Wohlschlegel J, Manson M D. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J Mol Biol. 1996;258:270–285. doi: 10.1006/jmbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- 7.Irikura V M, Kihara M, Yamaguchi S, Sockett H, Macnab R M. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol. 1993;175:802–810. doi: 10.1128/jb.175.3.802-810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan I H, Reese T S, Khan S. The cytoplasmic component of the bacterial flagellar motor. Proc Natl Acad Sci USA. 1992;89:5956–5960. doi: 10.1073/pnas.89.13.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan S, Khan I H, Reese T S. New structural features of the flagellar base in Salmonella typhimurium revealed by rapid-freeze electron microscopy. J Bacteriol. 1991;173:2888–2896. doi: 10.1128/jb.173.9.2888-2896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kihara M, Francis N R, DeRosier D J, Macnab R M. Analysis of a FliM-FliN flagellar switch fusion mutant of Salmonella typhimurium. J Bacteriol. 1996;178:4582–4589. doi: 10.1128/jb.178.15.4582-4589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubori T, Yamaguchi S, Aizawa S-I. Assembly of the switch complex onto the MS-ring complex of Salmonella typhimurium does not require any other flagellar proteins. J Bacteriol. 1997;179:813–817. doi: 10.1128/jb.179.3.813-817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd S A, Tang H, Wang X, Billings S, Blair D F. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol. 1996;178:223–231. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd S A, Blair D F. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol. 1997;266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 14.Macnab R. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:129–156. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 15.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 16.Marykwas D L, Berg H C. A mutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J Bacteriol. 1996;178:1289–1294. doi: 10.1128/jb.178.5.1289-1294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marykwas D L, Schmidt S A, Berg H C. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J Mol Biol. 1996;256:564–576. doi: 10.1006/jmbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- 18.Morrison T B, Parkinson J S. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5485–5489. doi: 10.1073/pnas.91.12.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oosawa K, Ueno T, Aizawa S-I. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J Bacteriol. 1994;176:3683–3691. doi: 10.1128/jb.176.12.3683-3691.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson J S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1993;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Schuster S C, Khan S. The bacterial flagellar motor. Annu Rev Biophys Biomol Struct. 1994;23:509–539. doi: 10.1146/annurev.bb.23.060194.002453. [DOI] [PubMed] [Google Scholar]
- 25.Sockett H, Yamaguchi S, Kihara M, Irikura V M, Macnab R M. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol. 1992;174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 27.Tang H, Blair D F. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang H, Billings S, Wang X, Sharp L, Blair D F. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J Bacteriol. 1995;177:3496–3503. doi: 10.1128/jb.177.12.3496-3503.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang H, Braun T F, Blair D F. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 30.Toker A S, Kihara M, Macnab R M. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J Bacteriol. 1996;178:7069–7079. doi: 10.1128/jb.178.24.7069-7079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toker A S, Macnab R M. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J Mol Biol. 1997;273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 32.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfe A J, Conley M P, Kramer T J, Berg H C. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi S, Fujita H, Ishihara A, Aizawa S-I, Macnab R M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986;166:187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi S, Aizawa S-I, Kihara M, Isomura M, Jones C J, Macnab R M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao R, Schuster S C, Khan S. Structural effects of mutations in S. typhimurium flagellar switch complex. J Mol Biol. 1995;251:400–412. doi: 10.1006/jmbi.1995.0443. [DOI] [PubMed] [Google Scholar]
- 37.Zhao R, Amsler C D, Matsumura P, Khan S. FliG and FliM distribution in the Salmonella typhimurium cell and flagellar basal bodies. J Bacteriol. 1996;178:258–265. doi: 10.1128/jb.178.1.258-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao R, Pathak N, Jaffe H, Reese T S, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Blair D F. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol. 1997;273:428–439. doi: 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Lloyd S A, Blair D F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]