Abstract
Rationale and objectives
Plexiform neurofibromas (PNs) are peripheral nerve tumors that occur in 25–50 % of patients with neurofibromatosis type 1. PNs may have complex, diffused, and irregular shapes. The objective of this work was to develop a volumetric quantification method for PNs as clinical assessment is currently based on unidimensional measurement.
Materials and methods
A semi-automatic segmentation technique based on mean magnetic resonance imaging (MRI) intensity thresholding (SSTMean) was developed and compared to a similar and previously published technique based on minimum image intensity thresholding (SSTMini). The performance (volume and computation time) of the two techniques was compared to manual tracings of 15 tumors of different locations, shapes, and sizes. Performance was also assessed using different MRI sequences. Reproducibility was assessed by inter-observer analysis.
Results
When compared to manual tracing, quantification performed with SSTMean was not significantly different (mean difference: 1.2 %), while volumes computed by SSTMini were significantly different (p < .0001, mean difference: 13.4 %). Volumes quantified by SSTMean were also significantly different than the ones assessed by SSTMini (p < .0001). Using SSTMean, volumes quantified with short TI inversion recovery, T1-, and T2-weighted imaging were not significantly different. Computation times used by SSTMean and SSTMini were significantly lower than for manual segmentation (p < .0001). The highest difference measured by two users was 8 cm3.
Conclusion
Our method showed accuracy compared to a current gold standard (manual tracing) and reproducibility between users. The refined segmentation threshold and the possibility to define multiple regions-of-interest to initiate segmentation may have contributed to its performance. The versatility and speed of our method may prove useful to better monitor volumetric changes in lesions of patients enrolled in clinical trials to assessing response to therapy.
Keywords: Plexiform neurofibroma (PN), Magnetic resonance imaging (MRI), Image segmentation, Volumetric tumor quantification
1. Introduction
Neurofibromatosis type 1 (NF1) is an autosomal-dominant disease that affects 1 in 3000–4000 individuals worldwide [1,2]. NF1 is associated with a wide variety of physical, psychological, and cognitive manifestations. Among physical manifestations of NF1, plexiform neurofibromas (PNs) are peripheral nerve tumors that occur in 25–50 % of patients [3,4]. Plexiform neurofibromas are particularly pathognomonic for NF1 with diffuse involvement along a nerve segment and its branches. PNs affect several areas of the body (e.g., head, neck, and trunk) and can cause deformity, limit function, and may lead to significant compression of vital organs depending on the size and location of the tumor [[5], [6], [7], [8]].
Due to the various comorbidities associated with these slowly progressing lesions, several clinical trials have been initiated over the last decade. Recent studies focused on evaluating the response to treatment with targeted therapy, including MEK inhibitors [[9], [10], [11]]. Response assessment of PNs is typically evaluated with magnetic resonance imaging (MRI) [12]. Tumor measurements follow uniform guidelines that allow the assessment of changes in the tumor at specific time points. Currently, the RECIST criteria are used to assess solid tumor response in most clinical trials [13,14]. The RECIST assessment is based on the characterization of anatomical changes in tumors in one dimension. This is an important limitation for PNs as these tumors often have complex and irregular shapes. Previous studies using RECIST criteria showed poor reproducibility in assessing therapeutical response for similar tumors [15,16].
Volumetric methods to assess changes in PNs using MRI data have been implemented previously [8,15,[17], [18], [19], [20], [21]]. When compared to unidimensional assessment, volumetric approaches allow the detection of early changes in tumors and the possibility of evaluating asymmetric growth [16]. These approaches were based on volumetric tumor segmentation methods including intensity-based histograms [17] and level set methods [22]. Semi-automatic segmentation techniques typically required user interaction to initially localize the tumor in the body. Also, these approaches necessitated high computation times associated to the segmentation process of tumors with complex shapes [8,19]. Some of these methods also needed manual corrections when the segmentation failed [8].
In this study, our objective was to develop a semi-automatic segmentation technique based on mean image intensity thresholding (defined as SSTMean) that requires minimal user interaction to initially define the tumor region and that further automatically segments PN lesions with no manual correction. SSTMean was also conceived to allow the user to initiate the tumor segmentation by defining multiple regions-of-interest. SSTMean was compared with a semi-automatic segmentation technique based on minimum image intensity thresholding (defined as SSTMini) that is similar to a previous approach [8,17]. The performance and accuracy of the two techniques were compared with manual tracings of 15 tumors of different locations, shapes, and sizes. Inter-observer analysis of the performance of SSTMean was used to assess its reproducibility.
2. Material and methods
2.1. Patients
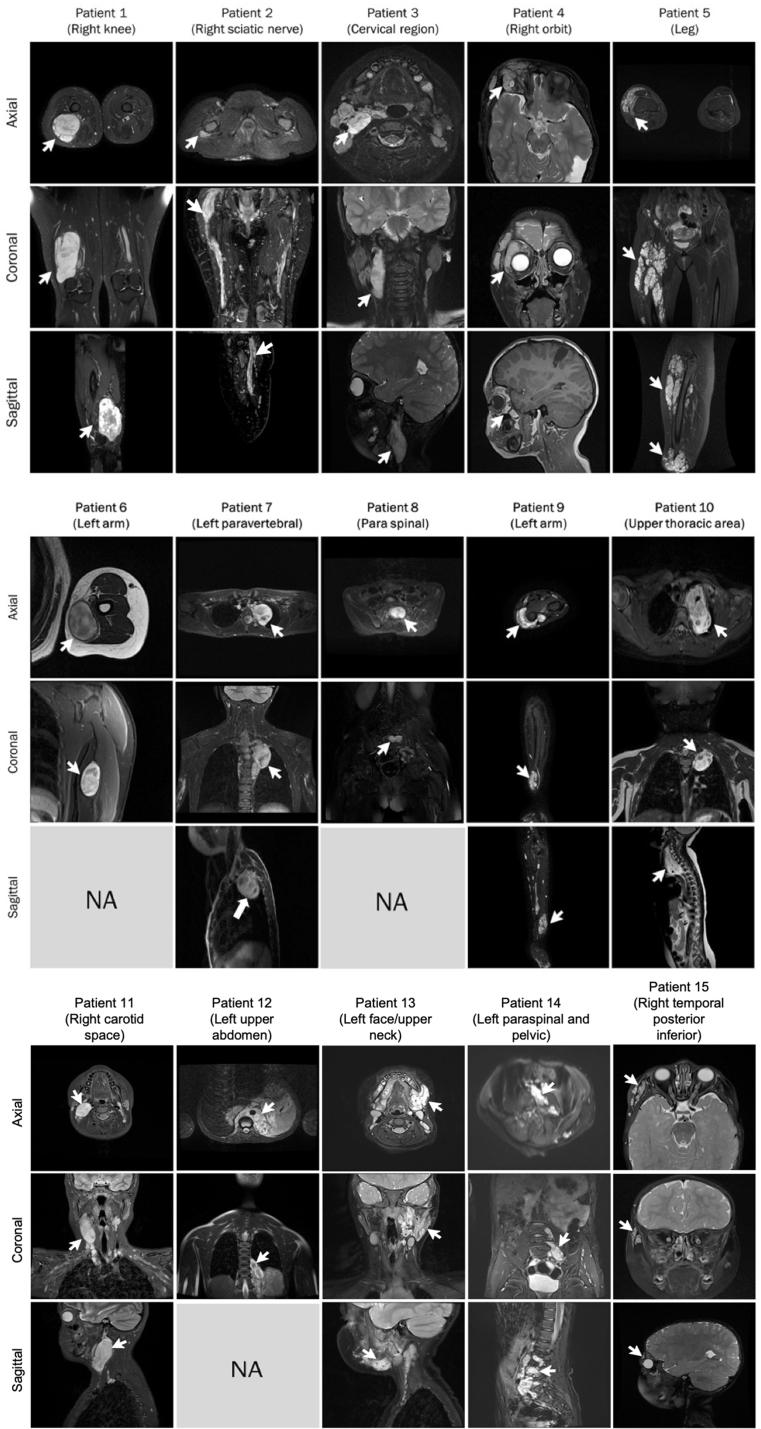
Fifteen patients diagnosed with PNs were selected to assess the performance of the SSTMean and SSTMini methods. These patients were enrolled as part of an ongoing clinical trial with trametinib (NIH clinical trial #NCT03363217) [23]. The study (#MP-21-2018-1742) was approved by the institutional review board of our institution. Parental written informed consent was obtained for each patient. For this study, patients were selected based on the shape, location, and size of their tumors to represent a heterogeneous set of lesions in different organs, representative of a variety of clinical scenarios. Supplementary Fig. 1 shows each tumor in the three orientations using either T1-weighted, T2-weighted, or short TI inversion recovery (STIR) images (except for Patients 6, 8 and 12 where MRI images were not prescribed in the sagittal plane). MRI imaging may have been performed with gadolinium.
2.2. Data acquisition
Based on the clinical MRI protocol, PNs were imaged with STIR, T1-and T2-weighted imaging sequences. Imaging parameters and sequences depended on the location, shape and size of the tumor and were not identical for all patients. Imaging parameters for STIR were: repetition time (TR, ms) = [2681–5716], echo time (TE, ms) = [23–65], inversion time (TI, ms) = [120−170], and slice number = [18–78]. For T1-weighted imaging, TR = [5–3000] ms, TE = [1–107] ms, and slice number = [20–75]. For T2-weighted imaging, TR = [1328–5880] ms, TE = [60−221] ms, and slice number = [34–83].
2.3. Segmentation technique
The SSTMean method consisted of 4 main steps that were implemented in a Matlab script (Version 9.9.0, Natick, MA).
-
(1)
Selection of the region-of-interest (ROI): This process was initiated by loading MRI image files (in PNG format) and selecting the sequence of interest (STIR, T1-or T2-weighted) and the orientation (axial, coronal, or sagittal). For each slice, a region was drawn by the operator using a drawing tool, which further generated a circular ROI around the lesion. This tracing allowed the algorithm to exclude healthy tissue surrounding the tumor as in previous works [8,17] To optimize the segmentation performance, the ROI was defined to maximize the lesion areas and minimize the healthy tissue content. For images with no tumoral tissue, no ROI was necessary. The tool also allowed the operator to trace multiple overlapping or separate ROIs when tumors appeared in multiple disconnected locations on the same slice (see an example in Supplementary Fig. 2), which was not specified in previous tools [8,17].
-
(2)
Smoothing of the lesion boundaries: In the ROI, a filtering method was applied to detect the edges (boundaries) of the lesion. As in previous studies [8,17], a Gaussian smoothing filter with standard deviation σ = 0.5 was used and applied in both the horizontal and vertical directions to reduce the noise level of the image.
-
(3)
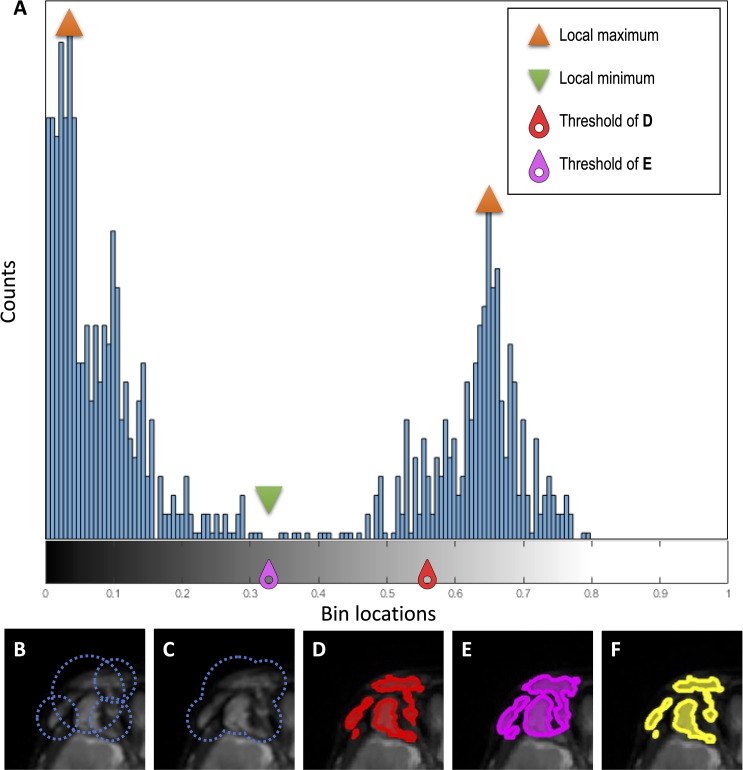
Automatic calculation of the intensity threshold: As in previous algorithms [8,17], a histogram of the ROI signal intensity was performed to delineate pixels corresponding to the tumor and pixels associated with healthy tissue (see an example in Supplementary Fig. 2). On STIR and T2-weighted images, pixels corresponding to the tumor were typically hyperintense while for T1-weighted, they were hypointense [24]. The algorithm performed the following steps to determine the image intensity threshold to differentiate healthy and tumoral tissues: 1) plot the ROI signal intensity histogram using 256 intensity bins, 2) calculate the mean of the ROI signal intensity histogram and divide it by two, 3) ensure that this value is located between the two local maxima of the image (associated with healthy and tumoral tissues, respectively), and 4) assign this value as the image intensity threshold for this slice and ROI (e.g., see the red threshold in Supplementary Fig. 2A and its corresponding segmentation in Supplementary Fig. 2D). Except for the segmentation threshold based on the mean image intensity (step 2), these steps were similar to previous tools [8,17].
-
(4)
Generation of the lesion mask: A binary mask was generated with pixels corresponding to values above the threshold [8,17]. This step also allowed the generation of a mask that included multiple tumors. For granular tumors, the mask may include healthy tissues distributed and diffused between the tumoral aggregate. For multiple ROIs, individual masks were subsequently merged to yield a global binary mask, which was not described in previous works [8,17].
To segment the tumors with the SSTMini method, the image intensity threshold defined in step 3 was modified by the local minimum located between the two maxima of the image (see magenta threshold in Supplementary Fig. 2A and corresponding segmentation in Supplementary Fig. 2E). This thresholding method was similar to a previous approach [8,17].
2.4. Volumetric quantification
To determine the tumor volume, the boundaries of two consecutive slices were linearly interpolated and the tumor was reconstructed as a 3D volume. This reconstruction was based on the slice spacing, slice thickness, and spatial resolution in the slice plane. Total volume corresponded to the sum of volumes from all merged slices.
2.5. Manual tumor tracing
There is no gold standard for the volumetric quantification of PNs. Tumor volumes generated by SSTMean and SSTMini methods were then compared with manual tracing of the tumors (e.g., Supplementary Fig. 2F). For each MRI slice, the tumor was identified by drawing its contour(s) using the pencil tool from FSLeyes [25]. The voxels included into the contour regions and corresponding to the tumor were identified and a mask was generated using the FSLeyes mask saving tool. After completing the entire MRI dataset, the masks were opened in the Matlab script, where only the volumetric quantification was performed.
2.6. Statistical analysis
Bland-Altman plots with 90 % two-sided confidence intervals were used to visualize the differences between volumes generated by the semi-automatic segmentation methods (SSTMean and SSTMini) and manual tracing. Wilcoxon signed-rank tests were also used to statistically compare the volumetric quantification of the semi-automatic segmentation methods with manual tracing. Computations times were compared between the three techniques using Wilcoxon signed-rank tests. For a subset of four patients, an inter-observer analysis was performed using the SSTMean method and differences between volumes generated by the two observers were reported in a Bland-Altman plot with 90 % two-sided confidence intervals.
3. RESULTS
Fig. 1 shows 4 examples (Patient 4, 5, 6, and 10) of the segmented lesion performed by SSTMean (left column, red) and SSTMini (middle column, magenta) in comparison with manual tracing (right column, yellow). MRI STIR images were used for these examples.
FIG [1].
Examples of lesion segmentation in Patient 4 (A, B and C), Patient 5 (D, E and F), Patient 6 (G, H and I), and Patient 10 (J, K and L) were performed by SSTMean (in red), SSTMini (in magenta), and manual tracing (in yellow) for a single 2D short TI inversion recovery image.
Table 1 reports tumor volumes quantified by SSTMean, SSTMini, and manual tracing for all patients, as well as computation times. Volume percentage differences between SSTMean and SSTMini with manual tracing are also provided. For each patient, volume quantification was performed using the three sequences (STIR, T1-and T2-weighted imaging). When compared to manual tracing, volume quantification performed with SSTMean was not significantly different (p = .32), while volumes computed by SSTMini were significantly different (p < .0001). Volumes quantified by SSTMean were also significantly different than the ones assessed by SSTMini (p < .0001). Using SSTMean, volumes quantified with the STIR, T1-and T2-weighted imaging sequences were not significantly different. Differences between MRI orientations were not assessed since orientations were prescribed based on the location of the tumor. Computation times used by SSTMean and SSTMini were significantly different than times used by manual tracing (p < .0001).
Table 1.
Volumetric quantification of tumors performed by SSTMean and SSTMini with differences (%) with manual tracing and corresponding computation times.
| Patients | SSTMean |
SSTMini |
Manual tracing |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Volume (cm3) | Comp. time (Min) | Difference* (%) | Volume (cm3) | Comp. time (Min) | Difference* (%) | Volume (cm3) | Comp. time (Min) | ||
| Patient 1 | Sag STIR | 377.1 | 7 | 0.6 | 411.3 | 7 | 9.3 | 374.8 | 53 |
| Ax T1 | 375.6 | 6 | −0.5 | 416.2 | 6 | 9.8 | 377.4 | 50 | |
| Cor T2 | 376.4 | 7 | 0.4 | 412.5 | 7 | 9.6 | 374.8 | 55 | |
| Patient 2 | Ax STIR | 16.6 | 7 | −2.4 | 19.9 | 7 | 15.7 | 17 | 59 |
| Cor T1 | 16.2 | 8 | −3.0 | 19.6 | 8 | 16 | 16.7 | 56 | |
| Sag T2 | 16 | 5 | −3.1 | 19.2 | 5 | 15.1 | 16.5 | 53 | |
| Patient 3 | Ax STIR | 158.2 | 6 | 0.6 | 160.1 | 6 | 1.8 | 157.3 | 43 |
| Sag T1 | 159.5 | 7 | 0.7 | 161.7 | 7 | 2.1 | 158.4 | 50 | |
| Cor T2 | 158 | 6 | 0.6 | 159.1 | 6 | 1.3 | 157.1 | 48 | |
| Patient 4 | Ax STIR | 46.7 | 5 | 1.7 | 48.1 | 5 | 4.8 | 45.9 | 46 |
| Sag T1 | 45.8 | 7 | 2.0 | 46.3 | 7 | 3.1 | 44.9 | 50 | |
| Cor T2 | 47.2 | 6 | 1.9 | 49.2 | 6 | 6.3 | 46.3 | 45 | |
| Patient 5 | Ax STIR | 442 | 8 | 1.1 | 461.8 | 8 | 5.7 | 437 | 56 |
| Sag T1 | 444 | 8 | 1.4 | 463.2 | 8 | 5.8 | 438 | 58 | |
| Cor T2 | 437 | 7 | 1.2 | 458 | 7 | 6.0 | 432 | 55 | |
| Patient 6 | Cor STIR | 31.5 | 5 | −1.3 | 42.1 | 5 | 27.6 | 31.9 | 31 |
| Cor T1 | 31.1 | 7 | −1.3 | 42 | 7 | 28.6 | 31.5 | 38 | |
| Ax T2 | 31.6 | 6 | −0.3 | 42.7 | 6 | 29.6 | 31.7 | 36 | |
| Patient 7 | Cor STIR | 58.3 | 4 | −0.3 | 59.5 | 4 | 1.7 | 58.5 | 28 |
| Sag T1 | 58.1 | 6 | −0.2 | 58.5 | 6 | 0.5 | 58.2 | 31 | |
| Ax T2 | 58.5 | 5 | −0.3 | 59.1 | 5 | 0.7 | 58.7 | 30 | |
| Patient 8 | Ax STIR | 37.7 | 4 | 2.7 | 39.4 | 4 | 7.1 | 36.7 | 25 |
| Ax T1 | 37.4 | 6 | −1.9 | 39.2 | 6 | 2.8 | 38.1 | 32 | |
| Cor T2 | 37.3 | 5 | −2.1 | 39.2 | 5 | 2.8 | 38.1 | 30 | |
| Patient 9 | Ax STIR | 26.2 | 4 | −0.4 | 26.4 | 4 | 0.4 | 26.3 | 23 |
| Cor T1 | 25.8 | 6 | −0.4 | 26.9 | 6 | 3.8 | 25.9 | 28 | |
| Sag T2 | 26 | 5 | −0.8 | 26.7 | 5 | 1.9 | 26.2 | 25 | |
| Patient 10 | Ax STIR | 99.4 | 9 | −0.8 | 103.2 | 9 | 3 | 100.2 | 57 |
| Sag T1 | 99 | 10 | −0.4 | 100.6 | 10 | 1.2 | 99.4 | 59 | |
| Cor T2 | 99.3 | 10 | −0.5 | 101.7 | 10 | 1.9 | 99.8 | 58 | |
| Patient 11 | Ax STIR | 67.5 | 8 | −1.3 | 89.1 | 8 | 26.3 | 68.4 | 53 |
| Cor T2 | 64.9 | 5 | −1.1 | 89.2 | 5 | 30.5 | 65.6 | 50 | |
| Sag T2 | 65.4 | 7 | −0.9 | 87.6 | 7 | 28.1 | 66 | 55 | |
| Patient 12 | Ax STIR | 158.6 | 6 | 1.1 | 179.6 | 6 | 13.6 | 156.8 | 54 |
| Cor T2 | 159.5 | 7 | −1.2 | 180.1 | 7 | 11 | 161.4 | 54 | |
| Patient 13 | Ax T1 | 215.8 | 6 | 1 | 247.1 | 6 | 11.6 | 211.1 | 65 |
| Sag T2 | 213.9 | 6 | 1.5 | 247.6 | 6 | 16.1 | 210.8 | 60 | |
| Cor T2 | 216.2 | 7 | 2.2 | 240.3 | 7 | 15.7 | 214 | 58 | |
| Patient 14 | Ax T1 | 43.3 | 6 | −1.6 | 61 | 6 | 32.4 | 44 | 58 |
| Sag STIR | 42.8 | 7 | −1.2 | 60.9 | 7 | 33.8 | 43.3 | 54 | |
| Cor T2 | 43.9 | 6 | 2.8 | 62.2 | 6 | 37.2 | 42.7 | 52 | |
| Patient 15 | Ax STIR | 46.7 | 6 | −0.6 | 69.3 | 6 | 38.3 | 47 | 63 |
| Sag T1 | 46.1 | 6 | −2.6 | 68.5 | 6 | 36.6 | 47.3 | 65 | |
| Cor T2 | 47.9 | 8 | −1.2 | 69.9 | 8 | 36.1 | 48.5 | 65 | |
| Mean (SD)‡ | NA | NA | 6.4 (1.4) | 1.2 (0.8) | NA | 6.4 (1.4) | 13.4 (12.6) | NA | 48 (12.5) |
Notes: *Volumetric percentage differences between the semi-automatic segmentation methods and manual tracing. ‡Mean and standard deviation of absolute percentage differences and computation times. Abbreviations: Comp, computation; STIR, short TI inversion recovery; T1, T1-weighted imaging; T2, T2-weighted imaging; Ax, axial; Cor, coronal; Sag, sagittal; NA, not applicable.
Bland-Altman plots from Fig. 2 show differences between tumoral volumes segmented by SSTMean and manual tracing(Fig. 2A) as well as between SSTMini and manual tracing (Fig. 2B). The maximum difference between volumes segmented by SSTMean and manual tracing was 4.11 cm3 compared to 38.23 cm3 for the comparison between SSTMini and manual tracing.
FIG [2].
Bland-Altman plots with 90 % confidence intervals (CI) of differences between volumes generated by the semi-automatic segmentation methods (A, SSTMean and B, SSTMini) and manual tracing.
Fig. 3 shows differences between tumoral volumes generated by the two observers using the SSTMean method. The maximum difference is 8 cm3.
FIG [3].
Bland-Altman plot with 90 % confidence intervals (CI) of differences between volumes generated by the two observers.
4. Discussion
The objective of this work was to develop a fast, reproducible, and accurate method to quantify the volume of PNs in the setting of a pediatric clinical trial. The proposed segmentation method required only one minimal user interaction for each MRI slice which resulted in fast computation time (4–10 min) compared to manual tracing (23–65 min). The possibility to define multiple ROIs that merge in a single one allowed to better enclose granular and disconnected tumors. Lesion volumes quantified by the SSTMean method were not statistically different to that of manual segmentation, which was the gold standard. The performance of SSTMean was accurate for a wide range of lesion sizes, from small (∼16 cm3) to large (∼444 cm3) lesions. The performance of SSTMean was independent of the structural MRI input sequence and provided similar volumetric assessments for T1-weighted, T2-weighted and STIR imaging data. The inter-observer analysis also showed small volumetric differences between users.
The technique developed in this study was compared to another algorithm (SSTMini) which was based on a similar published approach [17]. Compared to SSTMini, the algorithm provided more comparable volumes to manual quantification (mean difference of 1.2 % compared to 13.4 %). This difference may be due to the thresholding specificity of SSTMean and the possibility to define multiple ROIs. However, it was not possible to replicate exactly the published method as some methodological details were not fully described in the original article [17]. In particular, the identification of the PN lesion peak was based on a predefined percentage of normal tissue that was not available. The published method also performed a connected component analysis to remove lesions that were considered too small. There was no detail on this predetermined size. For granular and disconnected tumors, our approach considered all lesions of all sizes which were enclosed in multiple ROIs. This strategy, coupled with a finer thresholding technique may explain differences in accuracy when compared to that of SSTMini. The assessment of all lesions of any size may help global surveillance of a patient during treatment.
Other segmentation approaches were previously developed for solid tumors such as PNs [8,15,[17], [18], [19], [20], [21]]. Some of these algorithms were implemented in commercial visualization and analysis software including MEDx [14,16,17,20], Vitrea2 workstation [20], and 3DQI software [21], or separately programmed as PNist [8,19]. These techniques showed great performance for simple and regular tumors. However, some of these methods requested an additional step known as manual adjustment to produce accurate volumetric measurements. Manual correction was not used in our method to minimize user interaction and computation time. The finer thresholding technique with the possibility to segment complex shapes with disconnected elements may have helped the algorithm to provide acceptable volumes without the need of manual adjustments. However, manual correction could potentially improve the segmentation of the most complex PNs, while user input for corrections may also introduce additional volumetric variability.
Algorithms mentioned above and ours were semi-automatic and required user involvement to initially locate the tumor in the image. However, their segmentation procedure differed. In the method described by Cai et al. [15] the thresholding approach was dynamic and based on a 3D level set method to automatically segment tumors >5 cm3. The method described by Weizman et al. [19] was interactive and based on histogram tumor models from a training set. In their process, the user was required to draw a scribble over the tumor region that provided a seed for segmentation. This interactive approach was simpler and more reliable, particularly for complex, disconnected tumors, even if, for some tumors, manual correction was inevitable. Their approach was accurate (6.8 %) and fast (7 min) when compared to manual tracing [19]. Our method was slightly more accurate and consumed similar computational resources.
Our algorithm can be used with different MRI sequences and for all three orientations compared to previous methods, which reported results with one or two sequences in one or two orientations [8,15,[17], [18], [19], [20], [21]]. The computation time was also shorter than previous software (MEDx and 3DQI). These characteristics may prove useful in the clinical setting where MRI sequences and prescribed orientations may be limited due to imaging time constrains which can be critical especially in children. In addition, the use of several orientations and sequences may help to validate volume assessment for complex tumors with irregular shapes.
Our algorithm has several limitations. The selection of patients was biased. It would have been possible to select the first 15 consecutive patients from our clinical trial (NCT03363217) or select them randomly. However, this approach would have not ensured a heterogeneous set of lesions affecting different organs, and therefore limiting the generalization of our technique. As in previous approaches, the need for user interaction to initially define the ROI in each single slice slowed volume quantification. This limitation is amplified for tumors with multiple disconnected lesions as multiple ROIs were defined. Novel approaches such as using large datasets to train data and mathematical models to predict volume changes are promising and may allow avoiding any user interaction [8,19]. In SSTMean, the ROI has to be defined such as it minimizes the presence of healthy tissue. When arbitrary large ROI is selected, the algorithm performs as similar methods [8,17]. The accuracy of SSTMean was also limited by artifacts and low MRI image quality, which can affect the ROI selection and subsequent thresholding calculation based on mean image intensity. Because there is no current gold standard for tumor volume quantification, the use of manual segmentation to compare our approach was intrinsically a limitation. To the best of our knowledge, manual tracing remains the most accurate standard for comparison. For the comparison with the published approach [8,17], the use of the same PNs images would have also allowed a direct assessment. Novel techniques based on machine learning approaches could also potentially improve the precision, effectiveness, and reproducibility of tumor detection [26,27]. However, the use of these methods requires large amounts of high-quality training data and specialised expertise to select features and classifiers.
To implement a volumetric assessment tool such as the one presented here in the clinical environment, several considerations must be analyzed including security and service availability. One approach could be the implementation of a computer-aided diagnosis tool or a clinical decision support system to assist the activities of clinicians. Service continuity could be secured by implementing self-organised systems based on a non-fixed structure allowing organisational changes and flexibility to adapt to internal and external environment changes. Concerning security, service-oriented networks ensuring that communication nodes are appropriately and flawlessly connected could be used to defend against foreign invaders, attacks, and dysfunctions [28].These approaches would allow a secure and robust system able to provide uninterrupted service availability, which is mandatory in a clinical environment.
The algorithm could also be integrated into an imaging platform such as picture archiving and communication systems (PACS). This integration would avoid the need to extract, transfer, and anonymize MRI data before volumetric assessment. This algorithm has the potential to be used for various peripheral solid tumors as well as for central nervous systems tumors. Due to fast computation, volumetric analysis of response to therapy or tumor progression could be analyzed and reported at the time of the patient follow-up visits.
5. Conclusion
Volumetric analysis and MRI image segmentation are essential for the characterization of PNs and to following their progression. To quantify tumor volume, we proposed a semi-automatic segmentation technique based on mean image intensity thresholding and compared it with manual tracing. Our method showed accuracy, reproducibility and versatility. Our method was used to quantify lesions from 16 to 444 cm3. Our technique was fast and necessitated only one manual step associated with the ROI selection, which is promising for further clinical integration and use in clinical trials assessing the response to therapy. This integration would have to consider security and service availability.
Funding/support
The authors would like to acknowledge funding from the Natural Sciences and Engineering Council of Canada (NSERC) Discovery grant RGPIN-2015-04672, the Fonds de Recherche du Québec-Santé (FRQS) grant 32600 [to M.D.], the Canadian Institutes of Health Research (CIHR) grant PJT165976 [to S.P.], and the collaboration between the “Association de la Neurofibromatose du Québec” and “Fondation du grand défi Pierre Lavoie” [to S.P. and M.D.].
Declarations
This study was reviewed and approved by the Institutional Review Board of the Comité d’Éthique de la Recherche at CHU Sainte-Justine, named by the Quebec Government (#FWA00021692), in accordance with Quebec and Canada Laws, and the Code of Federal Regulations in the United States, with the approval number: MP-21-2018-1742. All patients (or their proxies/legal guardians) provided informed consent to participate in the study.
The authors declare no potential conflicts of interest relevant to this article to disclose.
Data availability statement
Data associated with this study are not currently available into a publicly available repository because the clinical trial recruiting patients for whom MRI data were collected is not yet completed. We will share our MRI data (completed or not) upon reasonable request.
CRediT authorship contribution statement
Dorsa Sadat Kiaei: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology, Investigation, Formal analysis, Data curation. Ramy El Jalbout: Visualization, Validation, Software, Methodology, Data curation. Jean-Claude Décarie: Visualization, Validation, Methodology, Data curation. Sébastien Perreault: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Mathieu Dehaes: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We would like to thank the patients and families who consented for the use of their imaging exams as well as the staff, nurses and physicians at participating Centres including colleagues in the Departments of Neurology and Radiology. Special thanks to research coordinators from each of the participating Centres.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e23445.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
MRI data of patients prescribed with either T1-, T2-, and/or short TI inversion recovery images in the three orientations (except for Patient 6,8 and 12). Arrows indicate the location of the lesions. Abbreviation: NA, not available.
figs2.
Histogram analysis (A) of MRI voxels included in multiple regions-of-interest (B) and its merged version (C) used to segment the PN located in the right eye orbit (Patient 4). Segmentation of the lesion area using SSTMean (D), SSTMini (E), and manual tracing (F).
References
- 1.Jett K., Friedman J.M. Clinical and genetic aspects of neurofibromatosis 1. Genet. Med. Jan. 2010;12(1):1–11. doi: 10.1097/GIM.0b013e3181bf15e3. [DOI] [PubMed] [Google Scholar]
- 2.Williams V.C., Lucas J., Babcock M.A., Gutmann D.H., Bruce B., Maria B.L. Neurofibromatosis type 1 revisited. J. Pediatr. Jan. 2009;123(1):124–133. doi: 10.1542/peds.2007-3204. [DOI] [PubMed] [Google Scholar]
- 3.Gross A.M., et al. Association of plexiform neurofibroma volume changes and development of clinical morbidities in neurofibromatosis 1. J. Neuro Oncol. Apr. 2018;20(12):1643–1651. doi: 10.1093/neuonc/noy067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Struemph K.L., et al. Adolescents and young adults with neurofibromatosis type 1: a descriptive study of adaptive functioning. Am. J. Med. Genet. Nov. 2021;188(2):488–497. doi: 10.1002/ajmg.a.62544. [DOI] [PubMed] [Google Scholar]
- 5.Akshintala S., et al. Longitudinal evaluation of peripheral nerve sheath tumors in neurofibromatosis type 1: growth analysis of plexiform neurofibromas and distinct nodular lesions. J. Neuro Oncol. 2020;22(9):1368–1378. doi: 10.1093/neuonc/noaa053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulanger J.M., Larbrisseau A. Neurofibromatosis type 1 in a pediatric population: ste-Justine’s experience. Can. J. Neurol. Sci. May 2005;32(2):225–231. doi: 10.1017/S0317167100004017. [DOI] [PubMed] [Google Scholar]
- 7.Gosein M., Ameeral A., Banfield R., Mosodeen M. Plexiform neurofibroma of the wrist: imaging features and when to suspect malignancy. Radiol. Case Rep. 2013;2013 doi: 10.1155/2013/493752. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weizman L., et al. Interactive segmentation of plexiform neurofibroma tissue: method and preliminary performance evaluation. Med. Biol. Eng. Comput. 2012;50(8):877–884. doi: 10.1007/s11517-012-0929-1. [DOI] [PubMed] [Google Scholar]
- 9.Gross A.M., et al. Selumetinib in children with inoperable plexiform neurofibromas. N. Engl. J. Med. Apr. 2020;382(15):1430–1442. doi: 10.1056/nejmoa1912735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller A., Staquet M., Winkler A., Junqueira A.C.C. Reporting results of cancer treatment. Cancer. Jan. 1981;47(1):207–214. doi: 10.1002/1097-0142(19810101)47:1<207::aid-cncr2820470134>3.0.co;2-6. 10.1002/1097-0142(19810101)47:1<207::AID-CNCR2820470134>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Therasse P., et al. New guidelines to evaluate the response to treatment in solid tumors. Bull. Cancer. Feb. 2000;92(3):205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 12.Ahlawat S., et al. Imaging evaluation of plexiform neurofibromas in neurofibromatosis type 1: a survey-based assessment. J. Neurol. Aug. 2021;97(7 Suppl 1):111–119. doi: 10.1212/WNL.0000000000012437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rustin G.J.S., et al. Definitions for response and progression in ovarian cancer clinical trials incorporating recist 1.1 and CA 125 agreed by the gynecological cancer intergroup (GCIG) Int. J. Gynecol. Cancer. Feb. 2011;21(2):419–423. doi: 10.1097/IGC.0b013e3182070f17. [DOI] [PubMed] [Google Scholar]
- 14.Semiglazov V. RECIST for response (clinical and imaging) in neoadjuvant clinical trials in operable breast cancer. J. Natl. Cancer Inst. Monogr. 2015;51:21–23. doi: 10.1093/jncimonographs/lgv021. 2015. [DOI] [PubMed] [Google Scholar]
- 15.Cai W., et al. Tumor burden in patients with neurofibromatosis types 1 and 2 and schwannomatosis: determination on whole-body MR images. Radiology. Mar. 2009;250(3):665–673. doi: 10.1148/radiol.2503080700. [DOI] [PubMed] [Google Scholar]
- 16.Jaffe C.C. Response assessment in clinical trials: implications for sarcoma clinical trial design. Oncol. 2008;13(suppl 2):14–18. doi: 10.1634/theoncologist.13-s2-14. [DOI] [PubMed] [Google Scholar]
- 17.Solomon J., Warren K., Dombi E., Patronas N., Widemann B. Automated detection and volume measurement of plexiform neurofibromas in neurofibromatosis 1 using magnetic resonance imaging. Comput Med Imaging Graph. Jul. 2004;28(5):257–265. doi: 10.1016/j.compmedimag.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Dombi E., et al. NF1 plexiform neurofibroma growth rate by volumetric MRI Relationship to age and body weight. Neurology. Feb. 2007;68(9):643–647. doi: 10.1212/01.wnl.0000250332.89420.e6. [DOI] [PubMed] [Google Scholar]
- 19.Weizman L., et al. PNist: interactive volumetric measurements of plexiform neurofibromas in MRI scans. Int. J. Comput. Assist. Radiol. Surg. Jul. 2014;9(4):683–693. doi: 10.1007/s11548-013-0961-0. [DOI] [PubMed] [Google Scholar]
- 20.Dombi E., et al. Recommendations for imaging tumor response in neurofibromatosis clinical trials. Neurology. Nov. 2013;81(21 Suppl 1):33–40. doi: 10.1212/01.wnl.0000435744.57038.af. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai W., et al. Volumetric MRI analysis of plexiform neurofibromas in neurofibromatosis Type 1: comparison of two methods. Acad. Radiol. Feb. 2018;25(2):144–152. doi: 10.1016/j.acra.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khosravanian A., Rahmanimanesh M., Keshavarzi P., Mozaffari S. Fast level set method for glioma brain tumor segmentation based on Superpixel fuzzy clustering and lattice Boltzmann method. Comput. Methods Programs Biomed. Jan. 2021;198(17) doi: 10.1016/j.cmpb.2020.105809. [DOI] [PubMed] [Google Scholar]
- 23.Perreault S., et al. A phase 2 study of trametinib for patients with pediatric glioma or plexiform neurofibroma with refractory tumor and activation of the MAPK/ERK pathway: TRAM-01. BMC Cancer. Dec. 2019;19(1):1250. doi: 10.1186/s12885-019-6442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laffan E.E., Ngan B.-Y., Navarro O.M. Pediatric soft-tissue tumors and pseudotumors: MR imaging features with pathologic correlation. Radiographics. 2009;29(3):887–906. doi: 10.1148/rg.e36. [DOI] [PubMed] [Google Scholar]
- 25.https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSLeyes
- 26.Seo H., et al. Machine learning techniques for biomedical image segmentation: an overview of technical aspects and introduction to state-of-art applications. Med. Phys. Jun. 2020;47(5):148–167. doi: 10.1002/mp.13649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranjbarzadeh R., Bagherian Kasgari A., Jafarzadeh Ghoushchi S., Anari S., Naseri M., Bendechache M. Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images. Sci. Rep. May 2021;11(1) doi: 10.1038/s41598-021-90428-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conti V., Militello C., Rundo L., Vitabile S. A novel bio-inspired approach for high-performance management in service-oriented networks. IEEE Trans Emerg Top Comput. 2021;9(4):1709–1722. doi: 10.1109/TETC.2020.3018312. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study are not currently available into a publicly available repository because the clinical trial recruiting patients for whom MRI data were collected is not yet completed. We will share our MRI data (completed or not) upon reasonable request.



![FIG [1]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8510/10761559/2bf38d713400/gr1.jpg)
![FIG [2]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8510/10761559/0fce8c78989e/gr2.jpg)
![FIG [3]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8510/10761559/4d6bc2daac58/gr3.jpg)