Abstract
Previous observational studies have suggested an association between tryptophan (TRP)–kynurenine (KYN) pathway and inflammatory bowel disease (IBD). However, whether there is a causal relationship among them remains unclear. Therefore, a two-sample Mendelian randomization (MR) study was conducted to explore the potential causal effects of crucial metabolites in TRP–KYN pathway on IBD and its subtypes. Using summary data from genome-wide association studies, a two-sample MR was employed to evaluate the genetic associations between TRP and KYN as exposures and IBD as an outcome. The inverse variance weighted method was used as the primary MR analysis, with MR-Egger, weighted mode, simple mode, and weighted median methods as complementary analyses. The odds ratios (OR) and 95% confidence intervals (CI) were determined for TRP–IBD (OR 0.739, 95% CI [0.697; 0.783]), TRP–UC (OR 0.875, 95% CI [0.814; 0.942]), TRP–CD (OR 0.685, 95% CI [0.613; 0.765]), KYN–IBD (OR 4.406, 95% CI [2.247; 8.641]), KYN–UC (OR 2.578, 95% CI [1.368; 4.858], and KYN–CD (OR 13.516, 95% CI [4.919; 37.134]). Collectively, the MR analysis demonstrated a significant protective association between TRP and IBD, whereas KYN was identified as a risk factor for IBD.
Subject terms: Immunological disorders, Immunology, Microbiology, Diseases, Gastroenterology
Introduction
Inflammatory bowel disease (IBD) is a group of immune-mediated disorders that predominantly affect the gastrointestinal tract, including ulcerative colitis (UC) and Crohn’s disease (CD). These disorders are commonly associated with clinical manifestations such as abdominal pain, diarrhea, mucopurulent bloody stool, and other symptoms. In severe cases, IBD can result in malnutrition and intestinal perforation1. This chronic condition is currently incurable and necessitates lifelong medication. The global prevalence rate of IBD exceeds 0.3%2. The impact of IBD on patients’ quality of life is significant, and its high prevalence represents a substantial economic and medical burden to society. Therefore, investigating the risk factors and pathogenesis is of paramount importance for IBD.
Tryptophan (TRP) is an important component of the human diet and plays a crucial role in inflammatory responses and gastrointestinal health3. TRP deficiency can lead to dysbiosis of the gut microbiota, resulting in gastrointestinal and even systemic inflammation3,4. In the human body, the major catabolic pathway of TRP is the kynurenine (KYN) pathway (KP), which accounts for 95% of total TRP degradation5. TRP can regulate intestinal inflammation6–8 and thus affect the occurrence of inflammatory bowel disease (IBD). Clinical studies have found that the serum TRP levels in IBD patients are significantly lower than those in normal control individuals, and there is a significant increase in the KYN/TRP ratio9. Furthermore, these levels are closely related to the degree of endoscopic inflammation and disease activity10,11. TRP deficiency and metabolic abnormalities may promote the development of IBD9. Previous studies have also indicated the crucial role of TRP–KYN metabolism dysregulation in the occurrence and progression of IBD12. In gene expression research, current studies have revealed the involvement of several gene loci associated with the TRP–KYN metabolic pathway in the susceptibility and progression of inflammatory bowel disease (IBD). Through genome-wide association studies, polymorphisms in key gene loci, including IL23R13, NOD2/CARD1514, and ATG16L113, are significantly associated with the risk of developing IBD. These gene loci are functionally linked to the TRP–KYN metabolic pathway, and their encoded proteins play critical roles in immune response regulation, inflammation signaling pathways, and autophagy, among other processes15–18. The association between TRP–KYN metabolites and IBD, however, could be biased due to confounding variables19,20, and the genetic and causal relationships are still unclear. Additionally, most of the above studies are observational and may have potential detection errors and confounding factors, which may reverse causality.
Therefore, we employed the Mendelian randomization (MR), a method of causal inference using genetic variation, to exclude genetic and causal relationships between crucial metabolites in TRP–KYN pathway and IBD. By utilizing the invariance of individual genotypes and Mendelian laws of inheritance (random allocation of alleles during gamete formation), it can avoid interference from common confounding factors, such as the postnatal environment, socioeconomic status, and behavioral habits21.
Materials and methods
Study design
This study investigated the causal relationship between TRP–KYN metabolites as exposures and the outcome of IBD by adhering to the fundamental principles and core assumptions of MR. Three assumptions required for MR analysis were met: (1) the genetic instrumental variables (IVs) used in the study were strongly associated with TRP and KYN; (2) the genetic IVs were not associated with any confounding factors related to IBD; and (3) the genetic IVs influenced IBD only through their effect on TRP and KYN. This study utilized publicly available datasets that had already received ethical approval and informed consent; therefore, additional ethical approval or informed consent was unnecessary.
Data sources
We utilized the most up-to-date and comprehensive Genome-Wide Association Studies (GWAS) datasets currently available for investigating metabolites in TRP–KYN pathway and IBD. The GWAS summary statistics for IBD (including CD and UC) were obtained from The IEU GWAS database (https://gwas.mrcieu.ac.uk/datasets/), while the data for metabolites in TRP–KYN pathway (only TRP and KYN were available) were obtained from the GWAS catalog (https://www.ebi.ac.uk/gwas/). All of these datasets can be downloaded. To prevent bias, only individuals of European origin were included in this Mendelian randomization study. Tables S1 summarizes the study population, including the number of genetic variants (i.e., SNPs available, Ncase) used in the analysis (Table 1).
Table 1.
Characteristics of the study population.
| Phenotype | GWAS ID | SNPs available | Ncase | Ncontrol | Year published | Population |
|---|---|---|---|---|---|---|
| IBD | finn-b-K11_IBD_STRICT | 16,380,455 | 3753 | 210,300 | 2021 | European |
| UC | finn-b-ULCERNAS | 16,380,457 | 2207 | 210,300 | 2021 | European |
| CD | finn-b-K11_KELACROHN | 16,380,466 | 940 | 217,852 | 2021 | European |
| TRP22 | – | 15,430,214 | 8299 | – | 2022 | European |
| KYN23 | – | 2,545,666 | 7824 | – | 2014 | European |
Selection of genetic IVs
We implemented strict quality control criteria to filter the SNPs from the GWAS summary data. SNPs significantly associated with the risk factor at the genome-wide level (p value < 5 × 108) were identified based on prior GWAS standards24. Subsequently, SNPs demonstrating independent inheritance and minimal linkage disequilibrium (LD) (r2 < 0.001, kb = 10,000) were selected. Ambiguous or palindromic SNPs were further excluded. To avoid the risk of weak instrumental bias, the F statistic (F = beta2/se2) was performed to evaluate the strength of the IV25,26. When F > 10, the association between the IV and exposures was deemed to be sufficiently robust, thereby safeguarding the results of the MR analysis against potential weak instrumental bias.
To measure the strength of the IVs, we calculated the F-statistic for each SNP, with those SNPs having an F-statistic < 10 being excluded as weak instruments. The MR-PRESSO test was further performed to detect and exclude any SNPs with potential pleiotropy27 and PhenoScanner (http://www.phenoscanner.medschl.cam.ac.uk/) was introduced to identify and remove SNPs with potential associations with confounding factors that might violate the independence assumption.
After several rounds of rigorous filtering, a set of eligible instrumental variables for the subsequent MR analysis were obtained.
Statistical analysis
To examine the relationship between exposures and outcome, multiple MR approaches were employed. The inverse variance weighted (IVW) method was the primary approach used, given its ability to produce unbiased estimates and avoid confounding factors in the absence of horizontal pleiotropy28. Moreover, the MR-Egger, weighted mode, simple mode, and weighted median methods were also used for supplementary and substitution analysis29. To ensure the quality and robustness of our research results, we conducted various analyses, including pleiotropy, heterogeneity, and sensitivity analyses. MR-Egger regression was used to assess the presence of horizontal pleiotropy. Cochran’s Q-test statistic was used to examine heterogeneity among all SNPs in each database30. Finally, a leave-one-out sensitivity analysis was conducted to verify the stability of the results. The analyses were conducted using RStudio software version 4.2.2 and the packages ‘TwoSampleMR’ and ‘MRPRESSO’.
Results
To investigate the causative influence of TRP on IBD risk, significant at the genome-wide level (p < 5 × 10−8) and independently inherited (r2 < 0.001, kb = 10,000) from the 15,430,214 SNPs, 173 SNPs were tentatively selected as IVs for IBD. Based on the findings from the PhenoScanner database, we excluded 28 SNPs associated with schizophrenia due to their potential impact on IBD occurrence31, such as rs12211045. Finally, a total of 145 SNPs were approved for MR analysis to assess the causal impact of TRP on IBD risk. Using the same methodology, we identified a total of 43 SNPs strongly associated with kynurenine (KYN) from a pool of 2,545,666 SNPs. Additionally, we excluded 6 SNPs associated with risk factors for IBD, including smoking (rs10774625, rs11065987, rs17630235, rs11066188), celiac disease (rs3184504, rs653178), and IBD (rs3184504, rs653178)32–34. Taking into account the differential effects of smoking on UC and CD33,35, we conducted MR analysis ultimately using 41 SNPs to assess the causal influence of KYN on UC risk, and 37 SNPs to investigate the causal effects of KYN on IBD and CD risk. All of the IVs had F-statistics > 10 (ranging from 29.76 to 39.70 for TRP and ranging from 29.58 to 111.13 for KYN). Supplementary Tables S1 contain comprehensive information regarding all the IVs.
Causal effects of TRP on IBD, UC, and CD
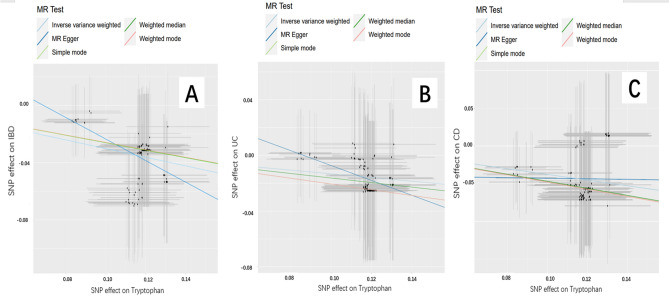
The Mendelian randomization analysis demonstrated a significant association between TRP and IBD outcomes (Fig. 1), indicating that TRP has a protective effect against IBD (odds ratio [OR]IVW = 0.739, 95% confidence interval (CI) [0.697–0.783]; P < 0.05), including CD ([OR]IVW = 0.685, 95% CI [0.613–0.765]; P < 0.05) and UC ([OR]IVW = 0.875, 95% CI [0.814–0.942]; P < 0.05). These results were consistent across IVW, MR-Egger, weighted median, simple mode, and weighted mode methods. Table 2 shows the details of the MR analysis investigating the causal effects of genetically predicted TRP on IBD, UC, and CD.
Figure 1.
Scatter plots of the genetic associations with TRP against IBD risk using different MR methods. (A) TRP (tryptophan) against IBD (inflammatory bowel disease) risk, (B) TRP (tryptophan) against UC (ulcerative colitis) risk, and (C) TRP (tryptophan) against CD (Crohn’s disease) risk. The slopes of each line represent the causal association for each method.
Table 2.
Mendelian randomization estimates for causal effects of TRP on IBD.
| Exposure | Outcome | Method | OR | 95% CI | P value |
|---|---|---|---|---|---|
| TRP | IBD | IVW | 0.739 | 0.697–0.783 | 1.19e−24 |
| MR-Egger | 0.471 | 0.295–0.753 | 2.09e−03 | ||
| Weighted median | 0.770 | 0.713–0.831 | 2.10e−11 | ||
| Simple mode | 0.771 | 0.625–0.952 | 1.72e−02 | ||
| Weighted mode | 0.771 | 0.635–0.938 | 1.04e−02 | ||
| UC | IVW | 0.875 | 0.814–0.942 | 3.95e−04 | |
| MR-Egger | 0.586 | 0.323–1.063 | 8.13e−02 | ||
| Weighted median | 0.849 | 0.774–0.932 | 6.17e−04 | ||
| Simple mode | 0.814 | 0.644–1.028 | 8.68e−02 | ||
| Weighted mode | 0.814 | 0.636–1.041 | 1.04e−01 | ||
| CD | IVW | 0.685 | 0.613–0.765 | 2.56e−11 | |
| MR-Egger | 0.962 | 0.390–2.373 | 9.34e−01 | ||
| Weighted median | 0.625 | 0.539–0.725 | 5.87e−10 | ||
| Simple mode | 0.619 | 0.421–0.912 | 1.67e−02 | ||
| Weighted mode | 0.619 | 0.420–0.913 | 1.71e−02 |
IBD inflammatory bowel disease, UC ulcerative colitis, CD Crohn’s disease, TRY tryptophan.
Causal effects of KYN on IBD, UC, and CD
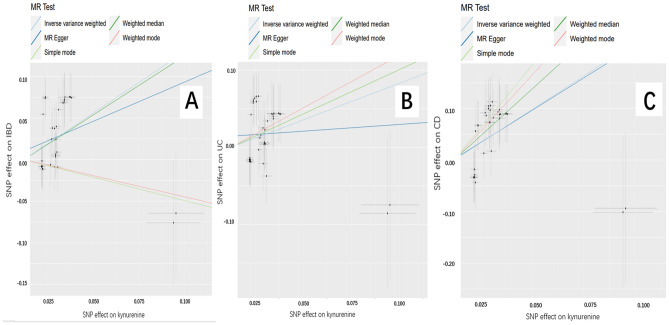
Figure 2 shows the results of estimating the causal effect of KYN on IBD, UC and CD. The MR results from the IVW showed a significant correlation between KYN and an increased risk of IBD ([OR]IVW = 4.406, 95% CI [2.247–8.641]; P < 0.05).There was a relationship between KYN and UC, with OR > 1 and P < 0.05 in the IVW, indicating that KYN is a risk factor for UC ([OR]IVW = 2.578, 95% CI [1.368–4.858]; P < 0.05).All MR methods showed a significant increase in the risk of CD with KYN ([OR]IVW = 13.516, 95% CI [4.919–37.134]; P < 0.05). Details of the MR analysis investigating the causal effects of genetically predicted KYN on IBD (including UC and CD) are provided in Table 3.
Figure 2.
Scatter plots of the genetic associations with KYN(Kynurenine) against IBD (inflammatory bowel disease) risk using different MR methods. (A) KYN (Kynurenine) against IBD risk, (B) KYN (Kynurenine) against UC (ulcerative colitis) risk, and (C) KYN (Kynurenine) against CD (Crohn’s disease) risk. The slopes of each line represent the causal association for each method.
Table 3.
Mendelian randomization estimates for causal effects of KYN on IBD.
| Exposure | Outcome | Method | OR | 95% CI | P value |
|---|---|---|---|---|---|
| KYN | IBD | IVW | 4.406 | 2.247–8.641 | 1.58 e−05 |
| MR-Egger | 2.586 | 0.558–11.986 | 2.32 e−01 | ||
| Weighted median | 4.163 | 1.767–9.808 | 1.10 e−03 | ||
| Simple mode | 0.568 | 0.048–6.682 | 6.56 e−01 | ||
| Weighted mode | 0.599 | 0.037–9.559 | 7.19 e−01 | ||
| UC | IVW | 2.578 | 1.368–4.858 | 3.38 e−03 | |
| MR-Egger | 1.190 | 0.270–5.237 | 8.18 e−01 | ||
| Weighted median | 3.156 | 1.249–7.976 | 1.50 e−02 | ||
| Simple mode | 3.156 | 0.427–23.315 | 2.66 e−01 | ||
| Weighted mode | 3.895 | 0.687–22.074 | 0.13 e−01 | ||
| CD | IVW | 13.516 | 4.919–37.134 | 4.42 e−07 | |
| MR-Egger | 12.443 | 1.221–126.765 | 4.03 e−02 | ||
| Weighted median | 38.857 | 9.166–164.720 | 6.81 e−07 | ||
| Simple mode | 131.837 | 7.579–2293.046 | 1.90 e−03 | ||
| Weighted mode | 79.732 | 4.190–1517.157 | 6.11 e−03 |
IBD inflammatory bowel disease, UC ulcerative colitis, CD Crohn’s disease, KYN kynurenine.
Sensitivity analyses
To ensure the validity of the aforementioned results, we conducted additional analyses to assess pleiotropy, heterogeneity, and sensitivity. Assessment using the MR-Egger intercept and the MR–PRESSO global test showed no evidence of horizontal pleiotropy (all P > 0.05). As statistical heterogeneity was detected for KYN in IBD (p < 0.05), we employed the IVW approach in a random-effects model. Heterogeneity tests using MR-Egger and IVW methods for the remaining results did not reveal significant heterogeneity. Finally, our leave-one-out sensitivity analysis confirmed the robustness of our causal estimates of the effect of genetically predicted TRP on IBD. The results of the sensitivity analysis are presented in Table 4 and Supplementary Figs. S1–S6.
Table 4.
Heterogeneity and pleiotropy analysis of TRP and KYN with IBD, UC and CD.
| Exposure | Outcome | MR-Egger | IVW | ||||
|---|---|---|---|---|---|---|---|
| Intercept | Pleiotropy | Cochran’s Q | Heterogeneity | Cochran’s Q | Heterogeneity | ||
| p value | Statistic | p value | Statistic | p value | |||
| TRP | IBD | 0.051 | 0.060 | 16.817 | 1 | 20.406 | 1 |
| UC | 0.046 | 0.185 | 3.665 | 1 | 5.437 | 1 | |
| CD | − 0.039 | 0.457 | 16.951 | 1 | 17.506 | 1 | |
| KYN | IBD | 0.012 | 0.452 | 59.876 | 5.50e−3 | 60.861 | 5.93e−3 |
| UC | 0.014 | 0.264 | 36.081 | 0.603 | 37.361 | 0.589 | |
| CD | 0.001 | 0.938 | 36.929 | 0.379 | 36.935 | 0.452 | |
IBD inflammatory bowel disease, UC ulcerative colitis, CD Crohn’s disease, TRY tryptophan, KYN kynurenine.
Discussion
This study provides a comprehensive analysis of the causal relationships between TRP–KYN pathway and IBD using summary GWAS data. Due to a lack of GWAS data for all metabolites in this pathway in public databases, we opted to focus our research on GWAS data specifically for the key metabolites in this pathway, namely TRP and KYN.
By performing MR analysis, we can circumvent cumbersome and confounder-prone steps to estimate causal relationships between exposure and outcome, providing genetic support for the physiological and pathological processes between TRP and IBD. Our MR analysis revealed that TRP is a protective factor for both UC and CD, while KYN is a risk factor for IBD. These findings were consistent across different Mendelian tools and statistical models, and no evidence of horizontal pleiotropy was detected in any of the analyses. Although there was some heterogeneity observed in one result, we utilized statistical methods to optimize our analysis, ensuring that our conclusions are reliable.
TRP is an essential amino acid in mammals and is a biosynthetic precursor of numerous microbial and host metabolites. It is mainly metabolized through the KYN, serotonin, and indole pathways36, with 95% of TRP being metabolized via the KYN pathway under the action of rate-limiting enzymes, such as indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO)5. IDO is a key enzyme in the metabolism of TRP, and IDO directs TRP toward KYN degradation. Additionally, increased IDO activity reduces the availability of TRP for other pathways37. IDO activity can also be stimulated by inflammatory cytokines, leading to increased consumption of TRP, especially in inflammatory states38.
In patients with IBD, there are significant alterations in the levels of TRP and KYN. Numerous clinical studies have shown that the expression of IDO, the concentration of KYN, and the KYN/TRP ratio are higher in the colonic and ileal lesions of IBD patients than in those of normal control individuals39. These indices are positively correlated with disease severity40. However, in nonlesion tissue of IBD patients, these indices are lower and similar to those of healthy control individuals41. In addition, the concentration of KYN in serum samples of IBD patients is also significantly increased42,43. Steroids, salicylates, and antitumor necrosis factor (TNF) biologics, such as infliximab, significantly reduce IDO expression and reverse the changes in KYN concentration and the KYN/TRP ratio in the treatment of IBD12,19,40,41.
Previous studies have found that TRP and its metabolites regulate intestinal inflammation9 and may exert anti-inflammatory effects through mechanisms such as regulating cellular immune function6, regulating the homeostasis of the intestinal microbiota8, and maintaining the balance and stability of the intestinal mucosa7,44. For example, KYN and indole can activate the aryl hydrocarbon receptor (AhR) by binding to it in a series of processes known as the TRP-AhR pathway. The activated TRP-AhR pathway can induce the expression of downstream cytokines such as interleukin-22 (IL-22) and interleukin-17 (IL-17)45, regulate T-cell proliferation, and thereby regulate intestinal immunity46. The TRP-AhR pathway plays an important role in regulating intestinal inflammation12, and Card9, as a susceptibility gene for inflammatory bowel disease (IBD), is closely related to this pathway. Studies have found that the intestinal microbiota of Card9 gene knockout mice cannot metabolize TRP into AhR ligands, resulting in reduced production of IL-22 and increased susceptibility to colitis47. When Card9 gene knockout mice were inoculated with three strains of lactobacilli that can metabolize TRP or treated with an AhR agonist, intestinal inflammation in mice was reduced8. Researchers have fed wild-type and AhR gene knockout mice diets containing either no TRP or 0.5% TRP. They found that the TRP diet improved colitis symptoms and severity in wild-type mice but not in AhR gene knockout mice. The TRP diet reduced the expression of inflammatory cytokines in the wild-type group and increased the expression of IL-22 and Stat3 mRNA in the colon, which protected the integrity of the epithelium48. In a mouse colitis model induced by dextran sulfate sodium (DSS), mice that lacked a TRP diet showed significantly increased susceptibility to inflammation and decreased levels of antimicrobial peptides in the gut49, which could be improved by adding TRP. In a mouse model of DSS-induced colitis, mice in the TRP group had less weight loss, reduced frequency of bloody stools, and improved histological changes in colonic tissue compared to mice in the no-TRP diet group50. In a piglet model of DSS-induced colitis, adding extra TRP to their regular diet improved clinical symptoms of colitis, increased piglet weight, and reduced intestinal permeability, which was possibly related to the reduced expression of proinflammatory cytokines (tumor necrosis factor-α, interleukin (IL)-6, interferon (IFN)-γ) and apoptosis initiators (caspase-8, Bax)51. A reduction in AhR ligand production has also been observed in the microbiota of IBD individuals, and a TRP-rich diet can improve treatment by increasing AhR ligands52, which ensures normal gut metabolism46,53. These results suggest that supplementation with TRP can improve intestinal inflammation and regulate epithelial homeostasis. Thus, TRP may be an effective immunomodulator for the treatment of IBD.
Current research indicates that KYN has a protective effect on intestinal inflammation through the AhR pathway53,54, which contradicts our research findings that suggest that KYN may be a potential risk factor for IBD. Some researchers have suggested that the activation of the KYN pathway during an inflammatory state is part of a physiological negative feedback compensation mechanism aimed at counteracting disease symptoms19,55. In summary, the potential mechanisms underlying the relationships among TRP, KYN, and IBD are complex and require further investigation. Based on current research, changes in TRP and KYN levels may make the KYN/TRP ratio a potential surrogate marker for disease activity in IBD patients, which could aid in predicting treatment response or relapse and better monitoring of the disease54.
Targeted therapy and modulation of the TRP–KYN pathway have primarily been the focus of cancer research56. Regulating key enzymes of TRP metabolism, such as IDO and TDO, can have therapeutic effects on diseases. In colorectal cancer cells, overexpression of IDO inhibits the infiltration of immune cells (CD3+ T, CD8+ T, CD3+ CD8+ T, and CD57+ NK cells), leading to immune escape, distant metastasis, consumption of local TRP, and the production of pro-apoptotic factors, significantly promoting disease progression and shortening overall survival of patients57–59. The IDO inhibitor 1-L-methyltryptophan (1-L-MT) reduces the transcription and proliferation of human colorectal cancer cells, induces mitochondrial damage, and causes cancer cell apoptosis60. When combined with chemotherapy and targeted drugs, 1-L-MT improves the overall survival rate of cancer patients61. Our research has revealed a clear causal relationship between TRP and KYN and IBD. We believe that further investigation of the mechanism between TRP–KYN metabolism and IBD may have great potential for targeted therapy of IBD.
Given the above, our study has several strengths. First, we used a large-scale GWAS dataset for MR analysis, allowing us to conduct robust MR analyses with a population of individuals of European ancestry, thus minimizing the impact of population stratification bias. Second, we used independent and strong genetic variants as instrumental variables (IVs) to mitigate the effects of linkage disequilibrium (LD) and weak instrument bias. Third, we used multiple MR methods, providing robust support for exploring the causal effects of TRP and KYN on IBD.
However, our study also has several limitations. First, although we used a GWAS dataset, we were unable to analyze different stages of IBD (e.g., active disease vs. remission) due to a lack of relative studies. Further MR analyses are needed to estimate the causal relationship between TRP and KYN and IBD at different stages.
Second, although our results suggest that TRP plays a protective role for IBD outcomes and KYN is a risk factor for IBD, the results of MR analyses are only based on genetic evidence. Additionally, definitive causal relationships require further mechanistic studies and randomized controlled trials. Third, although the populations we studied were all European, they were not from the same country, which may introduce some bias. Additionally, due to genetic, environmental, and dietary differences between Eastern and Western populations, these results need to be validated in other ethnic groups. Recent epidemiological surveys have shown that non-European populations are becoming new victims of IBD1,62. In Asia, the incidence of IBD has increased rapidly in the past 20 years63. We performed MR analysis using IBD13 and tryptophan22 data from East and South Asian populations. However, due to the lack of large sample sizes and high-quality GWAS data in Asian populations, we did not obtain any significant conclusions. Thus, the support of future large-scale GWAS data is needed to conduct relevant MR studies involving other ethnic groups. Fourth, although we matched all selected SNPs with the PhenoScanner database to avoid potential confounding factors and related horizontal pleiotropy, this measure cannot completely eliminate the influence of horizontal pleiotropy because the exact biological functions of many genetic variants are still unknown.
Conclusions
This MR study provides new genetic evidence for the causal relationship between key metabolites in TRP–KYN pathway (including TRP and KYN) and the risk of IBD (including UC and CD). We recommend giving due attention to TRP supplementation in IBD patients and propose that disease evaluation and assessment in these patients can be conducted by monitoring the KYN/TRP ratio. Further experiments or population-based observational studies are needed to elucidate the potential mechanisms underlying the relationships among TRP, KYN, and IBD and to explore the possibility of targeted therapy for IBD based on the TRP–KYN metabolic pathway.
Supplementary Information
Acknowledgements
We thank all of the investigators of the FinnGen, GWAS Catalog and IEU OpenGWAS project for sharing summary-level data on GWAS for IBD, TRP and KYN.
Author contributions
F.Q.Y., T.D.Y. and C.L. conceived and designed the study and reviewed and edited the manuscript. F.Q.Y. and W.M.L. acquired the data and analyzed the results. Z.H.Y., S.L. and H.Y.Z. proofread the results. F.Q.Y., D.L. and W.B.F. provided the methodology. F.Q.Y. and D.L. wrote the paper, and D.L., S.M.L., X.G.L. reviewed and edited the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (82074522), the R&D Program of Guangdong Province Drug Administration (2021YDZ06), the Research Fund for Zhaoyang Talents of Guangdong Provincial Hospital of Chinese Medicine (No. ZY2022KY08), the Scientific Research Project of Traditional Chinese Medicine Bureau of Guangdong Province (20221159), Guangdong Provincial Natural Science Fund (2022A1515010430), Guangxi Natural Science Foundation under Grant (No. 2020GXNSFBA159046) and Guangdong Provincial Special Project for the High-Quality Development, Inheritance, Innovation, and Enhancement of Traditional Chinese Medicine ([2022]102).
Data availability
We used publicly available databases, and all data mentioned in the manuscript can be found on the website provided in the article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiang-Guang Li, Email: xgli@gdut.edu.cn.
Ding Luo, Email: doctorluoding@163.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-50990-9.
References
- 1.Kaplan GG. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015;12(12):720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 2.Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet. 2017;390(10114):2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 3.Yusufu I, Ding K, Smith K, et al. A tryptophan-deficient diet induces gut microbiota dysbiosis and increases systemic inflammation in aged mice. Int. J. Mol. Sci. 2021 doi: 10.3390/ijms22095005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He F, Wu C, Li P, et al. Functions and signaling pathways of amino acids in intestinal inflammation. Biomed. Res. Int. 2018;2018:9171905. doi: 10.1155/2018/9171905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badawy AA. Kynurenine pathway of tryptophan metabolism: Regulatory and functional aspects. Int. J. Tryptophan Res. 2017;10:517487138. doi: 10.1177/1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michaudel C, Danne C, Agus A, et al. Rewiring the altered tryptophan metabolism as a novel therapeutic strategy in inflammatory bowel diseases. Gut. 2022 doi: 10.1136/gutjnl-2022-327337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zelante T, Iannitti RG, Cunha C, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Lamas B, Richard ML, Leducq V, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22(6):598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolaus S, Schulte B, Al-Massad N, et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology. 2017;153(6):1504–1516. doi: 10.1053/j.gastro.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 10.Sofia MA, Ciorba MA, Meckel K, et al. Tryptophan metabolism through the kynurenine pathway is associated with endoscopic inflammation in ulcerative colitis. Inflamm. Bowel Dis. 2018;24(7):1471–1480. doi: 10.1093/ibd/izy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta NK, Thaker AI, Kanuri N, et al. Serum analysis of tryptophan catabolism pathway: Correlation with Crohn’s disease activity. Inflamm. Bowel Dis. 2012;18(7):1214–1220. doi: 10.1002/ibd.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai W, Huang Z, Li S, et al. Kynurenine pathway metabolites modulated the comorbidity of IBD and depressive symptoms through the immune response. Int. Immunopharmacol. 2023;117:109840. doi: 10.1016/j.intimp.2023.109840. [DOI] [PubMed] [Google Scholar]
- 13.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47(9):979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buning C, Molnar T, Nagy F, et al. NOD2/CARD15 gene polymorphism in patients with inflammatory bowel disease: Is Hungary different? World J. Gastroenterol. 2005;11(3):407–411. doi: 10.3748/wjg.v11.i3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Steensel MA, Badeloe S, Winnepenninckx V, et al. Granulomatous rosacea and Crohn’s disease in a patient homozygous for the Crohn-associated NOD2/CARD15 polymorphism R702W. Exp. Dermatol. 2008;17(12):1057–1058. doi: 10.1111/j.1600-0625.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- 16.Georgy J, Arlt Y, Moll JM, et al. Tryptophan (W) at position 37 of murine IL-12/IL-23 p40 is mandatory for binding to IL-12Rbeta1 and subsequent signal transduction. J. Biol. Chem. 2021;297(5):101295. doi: 10.1016/j.jbc.2021.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linares R, Fernandez MF, Gutierrez A, et al. Endocrine disruption in Crohn’s disease: Bisphenol A enhances systemic inflammatory response in patients with gut barrier translocation of dysbiotic microbiota products. FASEB J. 2021;35(7):e21697. doi: 10.1096/fj.202100481R. [DOI] [PubMed] [Google Scholar]
- 18.Dudzinska E, Szymona K, Kloc R, et al. Increased expression of kynurenine aminotransferases mRNA in lymphocytes of patients with inflammatory bowel disease. Ther. Adv. Gastroenterol. 2019;12:321897128. doi: 10.1177/1756284819881304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wnorowski A, Wnorowska S, Kurzepa J, et al. Alterations in kynurenine and NAD(+) salvage pathways during the successful treatment of inflammatory bowel disease suggest HCAR3 and NNMT as potential drug targets. Int. J. Mol. Sci. 2021 doi: 10.3390/ijms222413497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin-Subero M, Anderson G, Kanchanatawan B, et al. Comorbidity between depression and inflammatory bowel disease explained by immune-inflammatory, oxidative, and nitrosative stress; tryptophan catabolite; and gut-brain pathways. CNS Spectr. 2016;21(2):184–198. doi: 10.1017/S1092852915000449. [DOI] [PubMed] [Google Scholar]
- 21.Smith GD, Ebrahim S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Lu T, Pettersson-Kymmer U, et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat. Genet. 2023;55(1):44–53. doi: 10.1038/s41588-022-01270-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: An expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35(22):4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011;40(3):755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 26.Feng R, Lu M, Xu J, et al. Pulmonary embolism and 529 human blood metabolites: Genetic correlation and two-sample Mendelian randomization study. BMC Genom. Data. 2022;23(1):69. doi: 10.1186/s12863-022-01082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verbanck M, Chen CY, Neale B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowden J, Spiller W, Del GMF, et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the radial plot and radial regression. Int. J. Epidemiol. 2018;47(6):2100. doi: 10.1093/ije/dyy265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowden J, Del GMF, Minelli C, et al. Improving the accuracy of two-sample summary-data Mendelian randomization: Moving beyond the NOME assumption. Int. J. Epidemiol. 2019;48(3):728–742. doi: 10.1093/ije/dyy258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian L, He X, Gao F, et al. Estimation of the bidirectional relationship between schizophrenia and inflammatory bowel disease using the Mendelian randomization approach. Schizophrenia (Heidelb) 2022;8(1):31. doi: 10.1038/s41537-022-00244-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozich JJ, Holmer A, Singh S. Effect of lifestyle factors on outcomes in patients with inflammatory bowel diseases. Am. J. Gastroenterol. 2020;115(6):832–840. doi: 10.14309/ajg.0000000000000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015;12(4):205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 34.Yuan S, Kim JH, Xu P, et al. Causal association between celiac disease and inflammatory bowel disease: A two-sample bidirectional Mendelian randomization study. Front. Immunol. 2022;13:1057253. doi: 10.3389/fimmu.2022.1057253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carreras-Torres R, Ibanez-Sanz G, Obon-Santacana M, et al. Identifying environmental risk factors for inflammatory bowel diseases: A Mendelian randomization study. Sci. Rep. 2020;10(1):19273. doi: 10.1038/s41598-020-76361-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23(6):716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Zhai L, Ladomersky E, Lenzen A, et al. IDO1 in cancer: A Gemini of immune checkpoints. Cell Mol. Immunol. 2018;15(5):447–457. doi: 10.1038/cmi.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strasser B, Becker K, Fuchs D, et al. Kynurenine pathway metabolism and immune activation: Peripheral measurements in psychiatric and co-morbid conditions. Neuropharmacology. 2017;112(Pt B):286–296. doi: 10.1016/j.neuropharm.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 39.Ferdinande L, Demetter P, Perez-Novo C, et al. Inflamed intestinal mucosa features a specific epithelial expression pattern of indoleamine 2,3-dioxygenase. Int. J. Immunopathol. Pharmacol. 2008;21(2):289–295. doi: 10.1177/039463200802100205. [DOI] [PubMed] [Google Scholar]
- 40.Zhou L, Chen H, Wen Q, et al. Indoleamine 2,3-dioxygenase expression in human inflammatory bowel disease. Eur. J. Gastroenterol. Hepatol. 2012;24(6):695–701. doi: 10.1097/MEG.0b013e328351c1c2. [DOI] [PubMed] [Google Scholar]
- 41.Wolf AM, Wolf D, Rumpold H, et al. Overexpression of indoleamine 2,3-dioxygenase in human inflammatory bowel disease. Clin. Immunol. 2004;113(1):47–55. doi: 10.1016/j.clim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 42.Forrest CM, Youd P, Kennedy A, et al. Purine, kynurenine, neopterin and lipid peroxidation levels in inflammatory bowel disease. J. Biomed. Sci. 2002;9(5):436–442. doi: 10.1007/BF02256538. [DOI] [PubMed] [Google Scholar]
- 43.Forrest CM, Gould SR, Darlington LG, et al. Levels of purine, kynurenine and lipid peroxidation products in patients with inflammatory bowel disease. Adv. Exp. Med. Biol. 2003;527:395–400. doi: 10.1007/978-1-4615-0135-0_46. [DOI] [PubMed] [Google Scholar]
- 44.Lai W, Xian C, Chen M, et al. Single-cell and bulk transcriptomics reveals M2d macrophages as a potential therapeutic strategy for mucosal healing in ulcerative colitis. Int. Immunopharmacol. 2023;121:110509. doi: 10.1016/j.intimp.2023.110509. [DOI] [PubMed] [Google Scholar]
- 45.Ma N, Guo P, Zhang J, et al. Nutrients mediate intestinal bacteria-mucosal immune crosstalk. Front. Immunol. 2018;9:5. doi: 10.3389/fimmu.2018.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun M, Ma N, He T, et al. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR) Crit. Rev. Food Sci. Nutr. 2020;60(10):1760–1768. doi: 10.1080/10408398.2019.1598334. [DOI] [PubMed] [Google Scholar]
- 47.Lamas B, Richard ML, Sokol H. Caspase recruitment domain 9, microbiota, and tryptophan metabolism: Dangerous liaisons in inflammatory bowel diseases. Curr. Opin. Clin. Nutr. Metab. Care. 2017;20(4):243–247. doi: 10.1097/MCO.0000000000000382. [DOI] [PubMed] [Google Scholar]
- 48.Islam J, Sato S, Watanabe K, et al. Dietary tryptophan alleviates dextran sodium sulfate-induced colitis through aryl hydrocarbon receptor in mice. J. Nutr. Biochem. 2017;42:43–50. doi: 10.1016/j.jnutbio.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 49.Hashimoto T, Perlot T, Rehman A, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487(7408):477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shizuma T, Mori H, Fukuyama N. Protective effect of tryptophan against dextran sulfate sodium- induced experimental colitis. Turk. J. Gastroenterol. 2013;24(1):30–35. doi: 10.4318/tjg.2013.0558. [DOI] [PubMed] [Google Scholar]
- 51.Kim CJ, Kovacs-Nolan JA, Yang C, et al. l-Tryptophan exhibits therapeutic function in a porcine model of dextran sodium sulfate (DSS)-induced colitis. J. Nutr. Biochem. 2010;21(6):468–475. doi: 10.1016/j.jnutbio.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 52.He P, Yu L, Tian F, et al. Dietary patterns and gut microbiota: The crucial actors in inflammatory bowel disease. Adv. Nutr. 2022;13(5):1628–1651. doi: 10.1093/advances/nmac029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Zhang ZH, Zabed HM, et al. An insight into the roles of dietary tryptophan and its metabolites in intestinal inflammation and inflammatory bowel disease. Mol. Nutr. Food Res. 2021;65(5):e2000461. doi: 10.1002/mnfr.202000461. [DOI] [PubMed] [Google Scholar]
- 54.Ala M. Tryptophan metabolites modulate inflammatory bowel disease and colorectal cancer by affecting immune system. Int. Rev. Immunol. 2022;41(3):326–345. doi: 10.1080/08830185.2021.1954638. [DOI] [PubMed] [Google Scholar]
- 55.Joisten N, Ruas JL, Braidy N, et al. The kynurenine pathway in chronic diseases: A compensatory mechanism or a driving force? Trends Mol. Med. 2021;27(10):946–954. doi: 10.1016/j.molmed.2021.07.006. [DOI] [PubMed] [Google Scholar]
- 56.Xue C, Li G, Zheng Q, et al. Tryptophan metabolism in health and disease. Cell Metab. 2023 doi: 10.1016/j.cmet.2023.06.004. [DOI] [PubMed] [Google Scholar]
- 57.Brandacher G, Perathoner A, Ladurner R, et al. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: Effect on tumor-infiltrating T cells. Clin. Cancer Res. 2006;12(4):1144–1151. doi: 10.1158/1078-0432.CCR-05-1966. [DOI] [PubMed] [Google Scholar]
- 58.Zamanakou M, Germenis AE, Karanikas V. Tumor immune escape mediated by indoleamine 2,3-dioxygenase. Immunol. Lett. 2007;111(2):69–75. doi: 10.1016/j.imlet.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 59.Ferdinande L, Decaestecker C, Verset L, et al. Clinicopathological significance of indoleamine 2,3-dioxygenase 1 expression in colorectal cancer. Br. J. Cancer. 2012;106(1):141–147. doi: 10.1038/bjc.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X, Zhou W, Zhang X, et al. 1-L-MT, an IDO inhibitor, prevented colitis-associated cancer by inducing CDC20 inhibition-mediated mitotic death of colon cancer cells. Int. J. Cancer. 2018;143(6):1516–1529. doi: 10.1002/ijc.31417. [DOI] [PubMed] [Google Scholar]
- 61.Wainwright DA, Chang AL, Dey M, et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin. Cancer Res. 2014;20(20):5290–5301. doi: 10.1158/1078-0432.CCR-14-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gearry RB. IBD and environment: Are there differences between east and west. Dig. Dis. 2016;34(1–2):84–89. doi: 10.1159/000442933. [DOI] [PubMed] [Google Scholar]
- 63.Mak WY, Zhao M, Ng SC, et al. The epidemiology of inflammatory bowel disease: East meets west. J. Gastroenterol. Hepatol. 2020;35(3):380–389. doi: 10.1111/jgh.14872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We used publicly available databases, and all data mentioned in the manuscript can be found on the website provided in the article.