Abstract
The Asian ladybeetle, Coccinella septempunctata, is an important insect of predatory natural enemy, which has a strong control effect and application prospects for aphids, whiteflies, mealybugs, and other small-sized pests of agriculture and forestry crops. Gut microbiota composition posed impacts on development of insects. In order to clarify the effect of artificial feed feeding on the intestinal microbial species and structure of C. septempunctata, we compared the intestinal microbial flora of C. septempunctata reared on bean aphids and artificial food for 15 days. Results show that Proteobacteria was the dominant component in all groups at phylum level, Rhodobacter, Methylovigula, Burkholderia, and Bradyrhizobium were the dominant bacteria among all groups at genus level. As to the differences in bacterial community structure and diversity, there is no significant difference between Shannon index and Simpson index, the principal components analysis of the bacterial communities, and the samples were roughly distributed in different regions. After 15 days of feeding, artificial diet did not significantly reduce the microbial diversity of the gut of C. septempunctata compared to the aphid group, and there was no significant effect on the abundance of dominant flora in the gut of C. septempunctata, C. septempunctata gut has a similar core microbiota. This study clarifies the effects in intestinal microbial diversity and composition structure of the C. septempunctata with artificial diet, and provides a theoretical basis for understanding the intestinal microorganisms and optimizating the artificial diet of C. septempunctata.
Subject terms: Entomology, Microbial ecology
Introduction
The Asian ladybeetle, Coccinella septempunctata (Linnaeus) (Coleoptera: Coccinellidae), is widely distributed in all provinces and regions of China, which is an important insect of predatory natural enemy in agriculture1. C. septempunctataa is a generalist predator that prey on aphids, whiteflies, thrips, and scale insects. Aphids (Hemiptera: Aphididae) are among the most destructive and abundant pests of agricultural crops worldwide2. Pea aphid Aphis craccivora (Hemiptera: Aphididae) is one of the key pests of pulse crops worldwide, which has a broad host range, infesting crops such as faba bean, lupine, alfalfa, lentil, chickpea, grass pea and pea3, is a typical prey of C. septempunctata4, which constantly secrete a large amount of nectar by feeding on plant juices to meet their own needs. In this process, pea aphid can also use plant viruses as vectors to spread their own viruses in an effective manner, making them as one of the most destructive pests5 .
C. septempunctata is a polyphagous insect, which is a natural enemy for comprehensive pest control due to its wide variety of food6. At present, C. septempunctata is fed on aphids that are reared at large scales. This conventional feeding method is expensive and labor-intensive7. Studies on artificial diet for ladybeetles started in the 1950s and continue to date8. In 1958, the first formulated artificial diet was successfully used to raise a variety of predatory lady beetles with dry prey powder as the main component9. Subsequently, research on the artificial diet of lady beetles shifted from prey components to non-prey components, mestic and foreign biocontrol scholars have studied many kinds of artificial feeds for predatory ladybirds10–12. With artificial feeds being used to multiply predator numbers, biological control is becoming more efficient. However, the amount and the quality of food have a direct impact on the growth and reproduction of insects13, therefore, determining the effects of different diets on the development and population maintenance of C. septempunctata is a crucial part of mass production and biocontrol.
In recent years, attention has been focused on the intestinal microbiota. During the long history of evolution, a reciprocal relationship exists between insects and their gut microbes, and this interaction undoubtedly drives insect evolution14. Firstly, insect provides the foundation for gut microbes' growth, providing the necessary habitats and nutrition, in the other hand, the gut intestinal microbiota also has ramifications for the host. Gut microbiota not only promotes digestion and absorption of food by the host15, and it also plays a significant role in host development16, immunity17, aging18, and defense against pathogen invasion19.
Numerous studies have suggested that diet affects the composition and structure of gut microbiota. Food plays a crucial role in regulating the composition and structure of gut microbiota, both short- and long-term effects are evident20. In hosts with flexible diets, such as humans and mice, diet has the potential to alter gut microbiomes over a short period of time21. In insects, herbivorous, arboreal species of ants harbour a greater abundance of bacteria than omnivorous, ground-dwelling species of ants22. Insects' gut microbiota was shaped by their diet.
Insects are the world’s most diverse and abundant animals in terms of species diversity and body mass in all ecological habitats23. Many studies have shown that intestinal microbes play an important role in insect, intestinal microbes play an important role in insect development24. However, only a few studies have examined the gut microbiota of C. septempunctata exposed to different diets and most of these studies focused on the efficiency of biological control methods25–29.
In this study, we are comparing the composition and structure of the gut microbiota between C. septempunctata that were fed on live pea aphids and C. septempunctata that were fed on artificial diet. High-throughput sequencing technology was used to sequence the gut microbiota of C. septempunctata, and predict the function of the gut microbiome, in order to provide a theoretical basis for further in‐depth research on the gut microbes of ladybeetles, and optimize artificial feed components.
Results
Summary of the sequencing data
Deep sequencing of eight gut samples from C. septempunctata yielded 209,107 sequences with a total length of 304,078,811 bp and an average length of 1449.35 bp. In total, 21 phyla, 37 classes, 60 orders, 97 families, 137 genera, 145 species, and 6370 OTUs were detected from gut samples at a 97% similarity level (Table 1). Good’s coverage index showed that the estimated values of all samples were over 97%, indicating that all samples reached an appropriate sequencing depth and values could represent the overall structure and composition of the ladybeetles' gut microbiota (Table 2).
Table 1.
OTU clustering and classification level statistics in the gut of adult Coccinella septempunctata.
| Sample | Reads num | OUT num | Classification | |||||
|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | Species | |||
| ArM | 16,943 | 1404 | 8 | 12 | 25 | 39 | 59 | 46 |
| ArF | 13,330 | 1946 | 8 | 12 | 25 | 33 | 48 | 32 |
| ApF | 16,239 | 1421 | 9 | 14 | 26 | 46 | 62 | 48 |
| ApM | 14,299 | 2175 | 12 | 14 | 28 | 41 | 54 | 40 |
| All | 60,811 | 6370 | 21 | 37 | 60 | 97 | 137 | 145 |
OTU (Operational Taxonomic Units) is a set of identical markers artificially assigned to a taxonomic unit (strain, genus, species, grouping, etc.) for the purpose of analysis in phylogenetic or population genetics research. In order to obtain the species classification information corresponding to each OTU, the uclust algorithm was used to perform taxonomic analysis on the representative sequences of OTUs, and the community composition of each sample was statistically analyzed at each classification level.
ArM male adults fed with artificial diet for 15 days, ArF female adults fed with artificial diet for 15 days, ApM male adults fed with aphids for 15 days, ApF female adults fed with aphids for 15 days.
Table 2.
Alpha diversity index statistics in the gut of adult Coccinella septempunctata.
| Sample | Coverage | Alpha diversity | |||
|---|---|---|---|---|---|
| Ace | Chao | Shannon | Simpson | ||
| ArM | 0.9738 | 2019.88 ± 818.77 | 1870.28 ± 812.11 | 2.54 ± 0.06 | 0.46 ± 0.02 |
| ArF | 0.9498 | 2760.49 ± 604.09 | 2352.20 ± 544.69 | 3.67 ± 0.28 | 0.29 ± 0.03 |
| ApF | 0.9707 | 2077.92 ± 714.66 | 1803.31 ± 624.35 | 2.97 ± 0.28 | 0.37 ± 0.02 |
| ApM | 0.9472 | 3060.83 ± 490.91 | 2686.05 ± 429.84 | 3.14 ± 0.19 | 0.43 ± 0.06 |
The diversity of bacterial communities is reflected through the Shannon and Simpson indices, and the richness of the community is reflected by the Chao1 and ACE indices.
ArM male adults fed with artificial diet for 15 days, ArF female adults fed with artificial diet for 15 days, ApM male adults fed with aphids for 15 days, ApF female adults fed with aphids for 15 days.
Composition in gut microbiota diversity in ladybeetles reared by different diets
The diversity of bacterial communities is reflected through the Shannon and Simpson indices, and the richness of the community is reflected by the Chao1 and ACE indices. Ace and Chao showed that the ACE index and Chao l index of the artificially fed groups were generally higher than those of the aphid group, but the difference was not statistically significant (p > 0.05). A Shannon diversity method and a Simpson diversity method were used to evaluate species diversity. There is no significant difference between the Shannon index and Simpson index (p > 0.05). In addition, in artificial diet group, there was significant difference of Shannon and Simpson indices between female and male ladybirds (p < 0.05), female adults with higher diversity of bacterial communities than males. In aphid group, there was no significant difference in Shannon and Simpson indices between female and male ladybirds (p > 0.05) (Table 2).
Composition of the gut microbiota in ladybeetles through different feeding methods
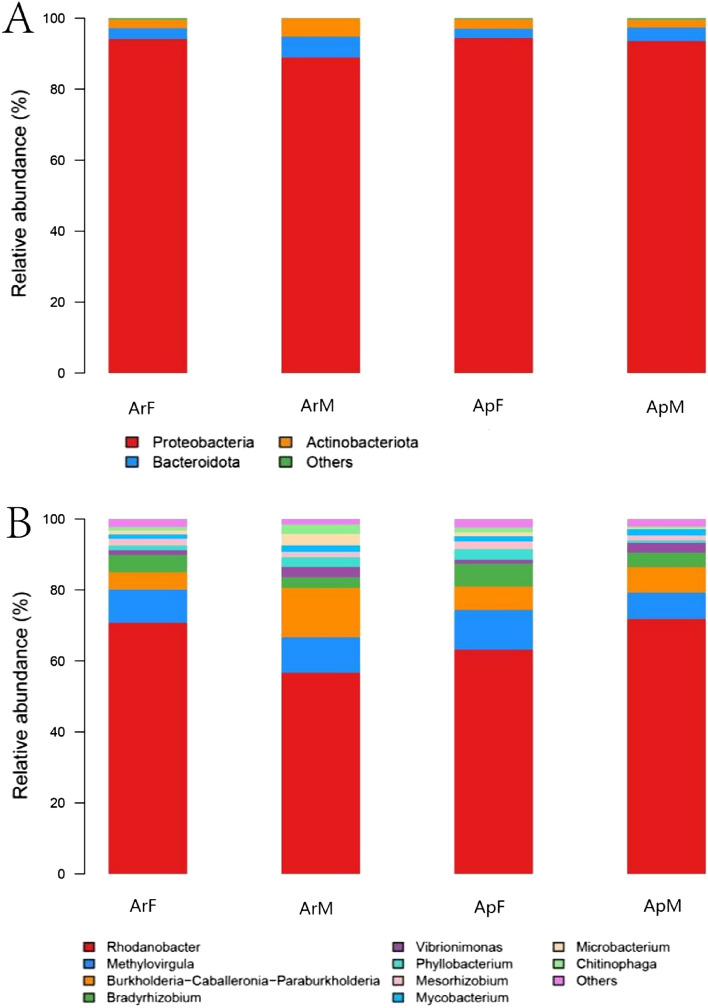
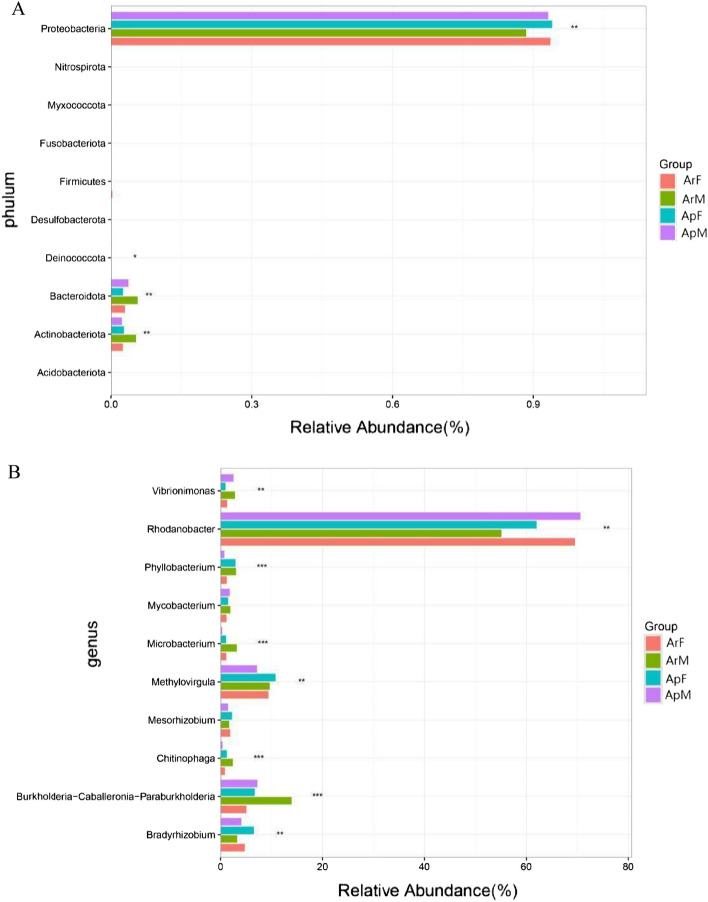
At phylum level of classification, the gut microbiota of ladybeetles consisted mainly of the following bacteria: Proteobacteria, Bacteroidota, Actinobacteriota, Firmicutes, Deinoccota, Myxococcota, Deinoccota, Desulfobacterota, Acidobacteriota, Planctomycetota and Chloroflexi (Fig. 1A). In this study, the top 10 phyla in the guts of ladybeetles from different habitats showed no significant differences in relative abundances. Proteobacteria had the highest relative abundance in ApF (94.12%) and the lowest abundance in ArF (88.63%). Bacteroidetes are influential and consistent component of the ladybeetle's gut, and their relative abundance was highest in ArF (6.02%) and lowest in ApF (2.67%). The relative abundance of actinobacteria was highest in ArF (5.21%) and lowest in ApM (2.39%). In addition, Firmicutes had the highest relative abundance in ArM (0.31%) and the lowest abundance in ArF (0.05%). Furthermore, the relative abundances of the following bacteria were relatively insignificant, not exceeding one percent: Deinococcota, Myxococcota, Deinococca, Acidobacteriota, Planctomycetota and Chloroflexi (Fig. 2B and Table 3).
Figure 1.
Relative abundance of the dominant phyla (A) and genera (B) of bacteria in gut of adult Coccinella septempunctata in this study. ArM male adults fed with artificial diet for 15 days, ArF female adults fed with artificial diet for 15 days, ApM male adults fed with aphids for 15 days, ApF female adults fed with aphids for 15 days. Different colors represent different types of dominant bacterial..
Figure 2.
Relative abundance of the top ten phyla (A) and genera (B) in gut of adult Coccinella septempunctata. ArM male adults fed with artificial diet for 15 days, ArF female adults fed with artificial diet for 15 days, ApM male adults fed with aphids for 15 days, ApF female adults fed with aphids for 15 days. The vertical axis represents the genus level of classification and the corresponding column length represents the average relative abundance of the gut microbiota species from the four treatment groups. The different colors represent different Groups. On the far right is the value of P, *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001.
Table 3.
Top 10 abundant phyla of bacteria in gut of adult Coccinella septempunctata.
| Phylum | Different habitats (%) | |||
|---|---|---|---|---|
| ArM | ArF | ApF | ApM | |
| Proteobacteria | 93.80 | 88.63 | 94.12 | 93.19 |
| Bacteroidota | 3.12 | 6.02 | 2.67 | 3.95 |
| Actinobacteriota | 2.63 | 5.21 | 2.84 | 2.39 |
| Firmicutes | 0.31 | 0.05 | 0.19 | 0.13 |
| Deinococcota | 0.09 | 0.05 | 0.13 | 0.30 |
| Myxococcota | 0.01 | 0 | 0.01 | 0.01 |
| Desulfobacterota | 0.01 | 0.01 | 0 | 0.01 |
| Acidobacteriota | 0 | 0 | 0 | 0 |
| Planctomycetota | 0 | 0 | 0 | 0.01 |
| Chloroflexi | 0 | 0 | 0 | 0 |
| Others | 0.03 | 0.03 | 0.04 | 0.01 |
ArM male adults fed with artificial diet for 15 days, ArF female adults fed with artificial diet for 15 days, ApM male adults fed with aphids for 15 days, ApF female adults fed with aphids for 15 days.
Results of this study indicated that there were 35 genera with a relative abundance higher than 1% in the gut of ladybeetles (Fig. 1B). The gut microbiota of ladybeetles consisted mainly of the following bacteria: Rhodanobacter, Methylovirgula, Burkholderia-Caballeronia-Paraburkholderia, Bradyrhizobium, Phyllobacterium, Vibrionimonas, Mesorhizobium, Mycobacterium, Microbacterium and Chitinophaga. Within the top 10 gut microbiota of ladybeetles, there were significant differences in the relative abundance of gut microbiota at the genus level of ladybeetles reared on different diets (Table 3). Among them, the genera with the highest relative abundances was Rhodanobacter, which was highest (70.73%) in ApM group, and lowest (55.22%) in ArF group. The relative abundance of Methylovirgula was highest (10.92%) in ApF group and lowest (7.25%) in ApM group. Burkholderia-Caballeronia-Paraburkholderia were most abundant (14.02%) in ArF group and least abundant (5.17%) in ArM group and the relative abundance of Bradyrhizobium was highest (6.68%) in ApF group and lowest (3.38%) in ArF group. The relative abundance of Phyllobacterium was highest (3.12%) in ArF group and lowest (0.85%) in ApM group. Vibrionimonas relative abundance was highest (5.08%) in ArM group and lowest (1.07%) in ApF group, Mesorhizobium was most prevalent (2.35%) in ApF group, whilst, the ArM group had the lowest relative abundance (0.09%). The relative abundance of Mycobacterium was highest (2.06%) in ArM group and lowest (1.59%) in ApF group, Microbacterium relative abundance was highest (3.31%) in ArF group and lowest (0.02%) in ArM group, and Chitinophaga relative abundance was highest (2.50%) in ArF group and lowest (0.49%) in ApM group (Fig. 2A and Table 4).
Table 4.
Top 10 abundant genera of bacteria in gut of adult Coccinella septempunctata.
| Genera | Different habitats (%) | |||
|---|---|---|---|---|
| ArM | ArF | ApF | ApM | |
| Rhodanobacter | 69.65 | 55.22 | 62.12 | 70.73 |
| Methylovirgula | 9.51 | 9.75 | 10.92 | 7.25 |
| Burkholderia-Caballeronia-Paraburkholderia | 5.17 | 14.02 | 6.84 | 7.33 |
| Bradyrhizobium | 4.86 | 3.38 | 6.68 | 4.20 |
| Phyllobacterium | 1.39 | 3.12 | 3.01 | 0.85 |
| Vibrionimonas | 5.08 | 2.94 | 1.07 | 2.66 |
| Mesorhizobium | 0.09 | 1.77 | 2.35 | 1.58 |
| Mycobacterium | 1.93 | 2.06 | 1.59 | 1.91 |
| Microbacterium | 0.02 | 3.31 | 1.17 | 0.39 |
| Chitinophaga | 1.35 | 2.50 | 1.35 | 0.49 |
| Others | 6.08 | 10.07 | 8.09 | 4.21 |
ArM male adults fed with artificial diet for 15 days, ArF female adults fed with artificial diet for 15 days, ApM male adults fed with aphids for 15 days, ApF female adults fed with aphids for 15 days.
Beta diversity of gut bacteria
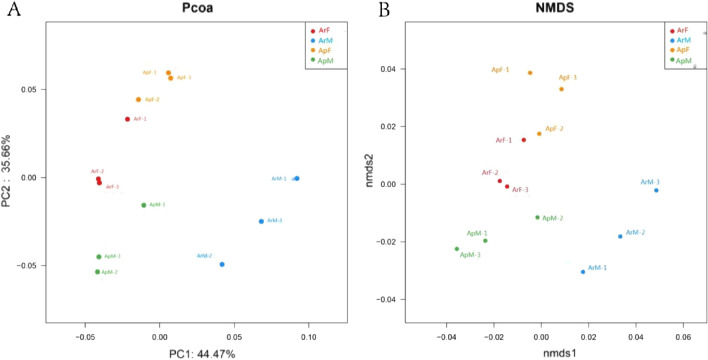
The gut microbiota was analyzed based on the weighted UniFrac distance of a principal coordinates analysis (PCoA) and BrayCurtis distance of the non-metric multidimensional scaling (NMDS) method for different sample. PCoA and NMDS analyses revealed that the compositions of the gut microbiota of C. septempunctata at different diets were varied from one another (Fig. 3). In PCoA, PC1 accounted for 63.5% of the total variance, and PC2 accounted for 29.18%. Through the PCoA calculated based on the weighted unifrac distance, we can find that each group of C. septempuncta samples is relatively discrete. This indicates that the similarity of intestinal microbial diversity among samples in each group is low, and there are differences among individuals (Fig. 3).
Figure 3.
PCA analyses (A) and NMDS diagram (B) analysis in the intestine of adult Coccinella septempunctata. ArM male adults fed with artificial diet for 15 days, ArF female adults fed with artificial diet for 15 days, ApM male adults fed with aphids for 15 days, ApF female adults fed with aphids for 15 days. The gut microbiota was analyzed based on the weighted UniFrac distance of a principal coordinates analysis (PCoA) and Bray Curtis distance of the non-metric multidimensional scaling (NMDS) method for different Sample.
Venn diagram analysis was used to obtain the overlap of OTUs under different treatments. Results showed that the number of OTUs in artificially fed group was much less than that in aphid group. There were 471 OTUs in artificially fed group, 637 OTUs in aphid group, and 74 OTUs were the same in all groups. This indicated that the bacterial community composition of the intestines of the artificially fed group and the aphid group differed greatly, and the aphid group was richer in flora. In addition, under the same feeding conditions, there were greatly differences in the composition of gut microbiota between females and males (Fig. 4).
Figure 4.
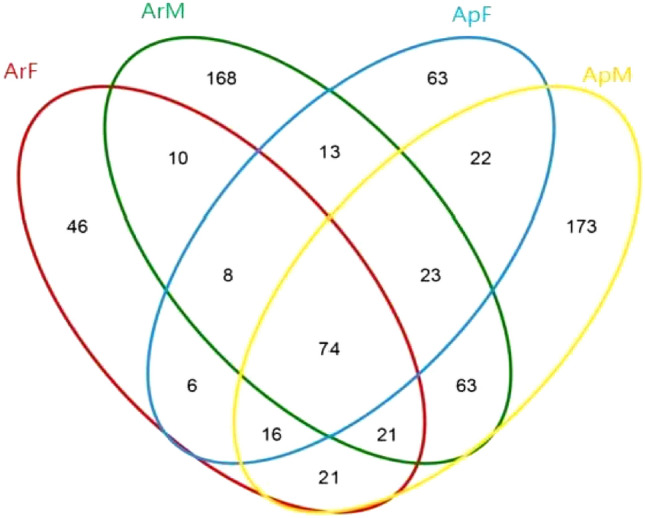
Venn diagram of OTUs of bacteria in the gut of adult Coccinella septempunctata. ArM male adults fed with an artificial diet for 15 days, ArF female adults were fed with artificial diet for 15 days, ApM male adults fed with aphids for 15 days, ApF female adults were fed with aphids for 15 days. Different colors represent different samples, areas where two circles of different colors overlap represent the number of shared OTUs, and areas where no circles overlap represent the number of unique OTUs.
Functional analysis of gut microbiota
In order to compare the potentially functional capacity of different feeding gut microbial communities of ladybeetles, the abundance of different sample flora at the level of the KEGG primary and secondary pathways was predicted using PICRUSt (Table 2). All annotated to KEGG first-level pathway: metabolism (69.82%), genetic information processing (12.51%), environmental information processing (10.35%), cellular processes (0.45%), genetic information processing (14.06%), human diseases (0.16%) and organismal systems (0.12%), respectively. The abundance of metabolic functional pathways accounted for more than half of the abundance of all primary pathways, therefore this shows that the gut bacteria of C. septempunctata mainly perform the function of metabolizing various substances (Table 5).
Table 5.
KEGG level 1 pathway abundance of bacteria in the gut of adult Coccinella septempunctata.
| Pathway name | ArM | ArF | ApF | ApM |
|---|---|---|---|---|
| Metabolism | 21,313,853 | 18,199,839 | 21,270,829 | 18,023,851 |
| Genetic information processing | 3,806,576 | 3,020,611 | 3,649,855 | 3,202,181 |
| Environmental information Processing | 3,203,499 | 2,907,971 | 3,320,681 | 2,711,319 |
| Cellular processes | 1,406,282 | 1,132,220 | 1,379,373 | 1,196,700 |
| Organismal systems | 389,121 | 324,413 | 380,687 | 331,329 |
| Human diseases | 510,615 | 455,972 | 532,221 | 426,250 |
The abundance of different sample flora at the level of the KEGG primary pathways was predicted using PICRUSt. All annotated to KEGG first-level pathway.
ArM male adults fed with artificial diet for 15 days, ArF female adults fed with artificial diet for 15 days, ApM male adults fed with aphids for 15 days, ApF female adults fed with aphids for 15 days.
A total of 11 secondary KEGG pathways were identified by mapping and annotating the secondary pathways in the metabolic pathways of the adult intestinal flora (Table 3). The corresponding functional names of these secondary pathways and their percentage of the total sample were: carbohydrate metabolism (22.55%), amino acid metabolism (21.32%), energy metabolism (13.42%), metabolism of cofactors and vitamins (9.07%), nucleotide metabolism (7.91%), biodegradation and metabolism of xenobiotics (6.50%), lipid metabolism ( 6.60%), metabolism of other amino acids (5.34%), metabolism of terpenoids and polyketides (3.21%), and Biosynthesis and metabolism of sugars (2.97%), and biosynthesis of other secondary metabolites (2.05%). This indicates that the adult gut flora of C. septempunctata are mainly involved in the metabolism of carbohydrates and amino acids, while a small proportion is involved in the metabolism of sugars, terpenoids, and polyketides. The abundance of these functions showed significant differences, consistent with the level 1 KEGG pathways (Table 6).
Table 6.
KEGG pathway (metabolism pathway) abundance of bacteria in the gut of adult Coccinella septempunctata.
| Pathway name | ArM | ArF | ApF | ApM |
|---|---|---|---|---|
| Carbohydrate metabolism | 4,533,895 | 3,923,237 | 4,549,245 | 3,823,709 |
| Biosynthesis of other secondary metabolites | 437,249 | 363,536 | 425,913 | 372,277 |
| Xenobiotics biodegradation and metabolism | 1,467,149 | 1,404,482 | 1,580,870 | 1,256,733 |
| Energy metabolism | 2,863,891 | 2,342,822 | 2,817,641 | 2,394,541 |
| Lipid metabolism | 1,407,662 | 1,247,730 | 1,429,264 | 1,197,205 |
| Nucleotide metabolism | 1,687,921 | 1,362,463 | 1,635,365 | 1,421,351 |
| Amino acid metabolism | 4,523,006 | 3,881,668 | 4,501,857 | 3,842,628 |
| Metabolism of other amino acids | 1,139,079 | 963,649 | 1,130,662 | 968,481 |
| Glycan biosynthesis and metabolism | 633,354 | 507,886 | 600,090 | 539,422 |
| Metabolism of cofactors and vitamins | 1,934,564 | 1,609,146 | 1,910,542 | 1,623,616 |
| Metabolism of terpenoids and polyketides | 686,083 | 593,221 | 689,380 | 583,888 |
The abundance of different sample flora at the level of the KEGG secondary pathways was predicted using PICRUSt. All annotated to KEGG level 2 pathway (metabolism pathway).
ArM male adults fed with artificial diet for 15 days, ArF female adults fed with artificial diet for 15 days, ApM male adults fed with aphids for 15 days, ApF female adults fed with aphids for 15 days.
Discussion
C. septenuctata is an important predatory insect that preys on aphids, whiteflies, mites and eggs/young larvae of lepidopterans, and its gut microbiota plays an instrumental role in its growth and reproduction1,30,31. Studies have shown that when there is a decline in the amount of beneficial flora, this increases the chance of viral infections and ultimately decreases the survival rate of the host32. Gut microbiota is of critical importance for the rearing of C. septempunctata. Microbes were significantly correlated with host development, aging, and social behavior33,34. Sex, feeding, different developmental stages, and adaptation to the environment of insects are all factors that affect the diversity and composition of gut microorganisms in insects35–37. This study found that artificial feed affected the composition and structure of the gut microbiota of C. septempunctata, but there was no significant difference in its diversity. In addition, many studies have shown that diet is one of the most significant variables influencing the gut microbial community38–40. Insects gut microbiota composition is affected by different diets and the type and number of gut microbes can change, as well as the dominant flora41. Despite this, not much research has been conducted on C. septempunctata's gut microbes using various diets. Despite this, there are not many studies on the intestinal microbial diversity of the ladybug under different feeding conditions, which is not conducive to the protection and cultivation of the ladybug. Therefore, this study used high-throughput sequencing technology to explore the gut microbes of C. septempunctata on different diets.
In this study, based on the high-throughput sequencing technology of 16S rDNA, the gut bacterial community diversity and divergence of two adult populations of aphid-feeding C. septempunctata and artificially fed C. septempunctata were analysised. In total there were 26 phyla, 64 classes, 154 orders, 247 families and 433 genera. Alpha diversity analysis revealed that both the diversity and richness of gut bacterial communities were higher in aphid-feeding C. septempunctata compared to artificially fed C. septempunctata. Microbiota can influence nutrient extraction from dietary sources, and diet can also impact gut microbiota composition and function at the same time42. Therefore, it is speculated that the reason for this may be that aphids are natural prey of insects and therefore the nutrients obtained from this source are more suitable for growth and development in ladybeetles. Research show that aphids, such as A. pisum, can be handled and consumed more easily by predators, thus making them as a more suitable prey in terms of net energy gain43. This was further supported by results obtained in PCA analysis of two populations, which exhibited significant differences in the composition and structure of the microbial community.
The diversity of gut microorganisms varies between insects of different sexes44. The alpha diversity analysis in this experiment showed that the diversity of intestinal bacteria in female C. septempunctata was higher than that in male C. septempunctata in the artificial diet group, indicating that the diversity of intestinal bacteria of C. septempunctata is also affected by gender differences. The same phenomenon has also been found, in intestines of male and female silkworms, the abundance of 23 bacterial genera in male is more than 1.5 times that of female, while the abundance of 7 bacterial genera in female is more than 1.5 times that of male45. There are also significant differences in the diversity of gut microorganisms between male and female adults of Chrysoperla sinica46. Gender differences may also lead to different risks of autoimmune (type I) diabetes in female and male mice47. However, in aphid group, alpha diversity analysis showed no significant difference in gut microbial diversity between male and female ladybirds. Therefore, we speculate that it may be due to aphids fed have more nutrients suitable for the growth and development of ladybirds, and the food plays a major role in the changes of the gut microbial community in the ladybirds.
This experiment found that Proteobacteria was the most prevalent bacteria in both the aphid diet-fed and artificial diet-fed feeding groups, which was found throughout nature, including in animals and plants48. Proteobacteria is associated with diverse metabolisms such as decomposition, fermentation of complex sugars, and the production of vitamins and degraded aromatic compounds, boosting their hosts' absorption of nutrients49. Previous study show that Proteobacteria dominate the gut microbiome of Gypsy moths50, Bombyx51and cotton bollworms52, similar findings also have been reported in many other species of insects in the family Cerambycidae53,54. The aforementioned studies suggested that Proteobacteria are widely found in gut microbiota of insects and the dominant role of Proteobacteria in the insect gut microbiota may be a distinctive feature of insect gut microbiota composition. In addition, in comparison with previous studies on the intestinal flora of C. septempunctata, we found that the intestinal core flora of C. septempunctata is mainly composed of Proteobacteria, Firmicutes, and Bacteroides53. Another study reported the species of Cerambycidae adult gut bacteria of Korea and found the dominant phyla were Proteobacteria, Firmicutes and Bacteroidetes55. It has been found that Firmicutes have the ability to encode enzymes for energy metabolism, biosynthesize vitamin B, and can produce enzymes for digestion and absorption of various substances, thereforehelping their hosts to digest and absorb nutrients56. Microbes of the Bacteroidetes are primarily responsible for decomposing carbohydrates through carbohydrate-active enzymes57, Microbes of the Bacteroidetes are primarily responsible for decomposing carbohydrates through carbohydrate-active enzymes57.
At the genus level, we discovered that Rhodanobacter was the most dominant bacterial genera in ArM, ArF, ApF and ApM groups. Among them, the ApM group had the highest abundance. Studies have revealed that Rhodanobacter genera can utilize various carbon sources, including cellobiose58. In larvae of longhorned beetles that feed on plants rich in carbohydrates (cellulose and hemicellulose) and lignin, Rhodanobacter can help the larvae digest more carbon nutrients through carbon sequestration54. We speculate that the highest abundance of Rhodanobacter bacteria in the ApM group may come from the aphid fed diet, which contains many sources of carbon. Thus, the function of Rhodanobacter genera may be associated with nutrient metabolism in the insect gut. Furthermore, Methylovirgula, Burkholderia- Caballeronia-Paraburkholderia, Mesorhizobium and Bradyrhizobium are also the dominant genera of bacteria in each group. As far as we know, Methylovirgula has not been identified in C. septempunctata and its biological function is unknown. Methylovirgula is ubiquitous in soil and has been found in many soil samples as a major species producing carbon activity, scholars have found that the microorganism has the highest content in mixed peat swamp forest systems and has the effect of harnessing and reducing methane59. Burkholderia, Mesorhizobium and Bradyrhizobium are commonly found in plant roots and they all have nitrogen fixation abilities. Burkholderia belongs to the class of β-proteobacteria and is a bona fide genus of archaebacteria that separates itself from group II of the Pseudomonas rRNA gene60. The genus was originally proposed by Yabuuchi and over 80 species of Burkholderia have been reported so far. A Burkholderia bacterium is known for its ability to fix nitrogen, promote plant growth, and degrade recalcitrant carbohydrates and aromatic compounds like L-phenylalanine61. Strains of Burkholderia have been reported in citrus root samples, and evidence suggests that these strains may be able to trigger the expression of genes associated with disease resistance62,63. However, the biological effects of Burkholderia on the C. septempunctata is unclear, and it is speculated that it may participate in the metabolism of carbohydrates. Rhizobia, a group of soil bacteria with which legume plants interact symbiotically, forms nitrogen-fixing root nodules on the roots of legume plants as a result of their symbiotic interactions4. It is important to note that Mesorhizobium and Bradyrhizobium are also two other types of nodules, these nodules fix the diazine in the atmosphere into ammonia, which is used as a nitrogen source by the host plant64. As far as this genus of bacteria is concerned, most of what we know about them revolves around their capacity to fix nitrogen in plant roots, and their role on insects remains unclear. We speculate that the presence of Mesorhizobium and Bradyrhizobium in ladybird beetles may come from their aphid food source, in particular when the aphids feed on broad beans.
Insects digest and absorb food in their guts, and intestinal flora plays an important role in their metabolism and nutrition65. In this study, the potential functions of ladybeetle gut bacteria under different food sources were predicted by PICRUSt. In addition, the intestinal flora of C. septempunctata mainly metabolizes carbohydrates, amino acids, and other substances to provide adequate nutrition for it. This study also predicted the metabolic functions of gut bacteria for sugars, exogenous chemicals, terpenoids, and polyketides, suggesting that these gut bacteria are involved in the metabolism of sugars, terpenoids, and cellulose, sugar promotes the growth, development, and reproduction of ladybugs66. In our research, Serratia was one of the main genera, it has been shown that Serratia are dominant bacteria in the guts of many insects and can help host insects efficiently degrade substances such as cellulose, monoterpenoids or diterpenes67,68.
In this study, we completed a comparative analysis of the composition and diversity of adult gut microbes through different feeding in C. septempunctata using high-throughput sequencing technology to predict the potential functions of gut microbes. The results showed that after 15 days of artificial feeding, there was no significant change in the gut microbial diversity and dominant bacterial species of the C. septempunctata. C. septempunctata gut has a similar core microbiota, the dominant phyla at the level of the intestinal flora are Proteobacteria, Firmicutes and Bacteroidetes, and the dominant genera are Rhodanobacter, Bradyrhizobium, Ralstonia, Asinibacterium and Burkholderia. By PICRUSt prediction analysis, the gut microorganisms of C. septempunctata in this study mainly functioned to metabolize various substances, mainly carbohydrates and amino acids, and to a lesser extent sugars, terpenoids and polyketides. However, more studies were needed to further analyze the interaction between host and microbiota in different rations, as well as the structure and function of microorganisms. Follow-up studies will be conducted to screen and validate the intestinal tract with these bacteria. This will be done to further clarify the functions of the relevant intestinal microbes and their mechanisms, in order to provide theoretical basis and ideas for the subsequent in-depth study on the gut microorganisms of heptastar and optimization of artificial feed components.
Methods
Insect sampling
Both aphids and C. septempunctata were obtained from the insect laboratory at Institute of Plant Protection, Sichuan Academy of Agricultural Sciences, China. The main components of the artificial diet were dried bee pupae powder, sucrose, dried pig liver powder, rape pollen, pea aphid powder, β-carotene (2% active ingredient) and dried banana powder. In this study, two diet regimes were used and a total of four groups were divided by distinguished between male and female and feeding time. (1) Females were fed with an artificial diet for 15 days (ArF), (2) Males were fed with an artificial diet for 15 days (ArM), (3) Males were fed with aphids for 15 days (ApM), (4) Females were fed with aphides for 15 days (ApF).
Collect the eggs of C. septempunctata reared with aphids, place them in a cage containing live aphids, and until them hatch. The larvae feed on aphids until they enter the adult stage. Then collect the adult worms into a 1000 ml plastic culture box (30 heads per box), a total of 8 groups, for different feeding methods. After feeding for 15 days, immerse the worm body in 70% ethanol for 1 min (pipette 1 ml into a 1.5 ml sterile centrifuge tube), repeatedly invade it 3 times, and then rinse it with sterile water 5 times to remove the surface DNA of the worm. Then write the name of the sample on the sterile tube or centrifuge tube, record all sample information, and place the collection tube in liquid nitrogen for temporary storage; Finally, it is stored in a dry ice box and sent to the sequencing company. Groups ApM and ApF were fed ~ 150–170 aphid heads per day to prevent C. septempunctata from feeding on each other. The artificial fed groups, ArF and ArM, were fed with both dry powder and 20% sucrose liquid (sucrose: purified water = 2:8) supplied separately. Place five grams of dry powder in a plastic sheet, place the sucrose solution on the cotton wolf, roll it into a ball, place it in the bottle cap, and then put it together in a feeding box. The food was chaged daily using steritle equipment. Visually observe and record every 24 h. All insects were kept indoors under the following environmental conditions: temperature (25 ± 1) °C, relative humidity 75% ± 7%, photoperiod 10 L:14 D, light intensity 18,000 LX.
DNA extraction and PCR amplification
Microbial DNA was extracted from C. septempunctata samples using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to manufacturer’s protocols. The V1-V9 region of the bacteria 16S ribosomal RNA gene were amplified by PCR (95 °C for 2 min, followed by 27 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s and a final extension at 72 °C for 5 min. The primers primers 27F 5′-AGRGTTYGATYMTGGCTCAG-3′ and 1492R 5′-RGYTACCTTGTTACGACTT-3′, were used which included an eight-base barcode sequence unique to each sample. PCR reactions were performed in triplicate using a 20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA. Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.) according to the manufacturer’s instructions.
Library construction and sequencing
SMRTbell libraries were prepared from the amplified DNA by blunt-ligation according to the manufacturer’s instructions (Pacific Biosciences). Purified SMRTbell libraries from Zymo and HMP mock communities were sequenced on dedicated PacBio Sequel II 8 M cells using the Sequencing Kit 2.0 chemistry. Purified SMRTbell libraries from the pooled and barcoded samples were sequenced on a single PacBio Sequel II cell. All amplicon sequencing was performed by Shanghai Biozeron Biotechnology Co. Ltd (Shanghai, China).
Processing of sequencing data
PacBio raw reads were processed using the SMRT Link Analysis software version 9.0 to obtain demultiplexed circular consensus sequence (CCS) reads with the following settings: minimum number of passes = 3, minimum predicted accuracy = 0.99. Raw reads were processed through SMRT Portal to filter sequences for length (< 800 or > 2500 bp) and quality.Sequences were further filtered by removing barcodes, primer sequences, chirmas and sequences if they contained 10 consecutive identical bases.
OTUs were clustered with 98.65% similarity cutoff using UPARSE (version 7.1 http://drive5.com/uparse/) and chimeric sequences were identified and removed using UCHIME. The phylogenetic affiliation of each 16S rRNA gene sequence was analyzed by RDP Classifier (http://rdp.cme.msu.edu/) against the silva (SSU132) 16S rRNA database using confidence threshold of 70%69.
Alpha- and beta-diversity analyses
The rarefaction analysis based on Mothur v.1.21.1 was conducted to reveal the diversity indices, including the Chao, ACE, and Shannon diversity indices. The beta diversity analysis was performed using UniFrac60 to compare the results of the principal component analysis (PCA) using the community ecology package, R-forge (Vegan 2.0 package was used to generate a PCA figure). Mantel tests were carried out to examine the Spearman’s rank correlation between the factor and the bacterial community similarity using Bray–Curtis distance matrices with 999 permutations, using the vegan package in R. Multivariate analysis of variance (MANOVA) was conducted to further confirm the observed differences. The Spearman’s correlation coefficients were assessed to determine the relationships between microbiota and chemical factors such as signaling molecules. Correlation was considered significant when the absolute value of Spearman’s rank correlation coefficient (Spearman’s r) was > 0.6 and statistically significant (P < 0.05). All statistic analysis were performed by R stats package. The R (pheatmap package) and Cytoscape (http://www.cytoscape.org) were applied to visualize the relationships through correlation heatmap and network diagrams, respectively. Redundancy analysis (RDA) was employed to explore the relationship between environmental factors and bacterial communities. One-way analysis of variance (ANOVA) tests were performed to assess the statistically significant difference of diversity indices between samples. Differences were considered significant at p < 0.05. Venn diagrams were drawn using online tool “Draw Venn Diagram” (http://bioinformatics.psb.ugent.be/webtools/Venn) to analyze overlapped and unique OTUs during the treatment processes. One-way permutational analysis of variance (PERMANOVA) was performed using R vegan package to assess the statistically significant effects of treatment processes on bacterial communities.
Functional prediction of the microbial genes
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (http://picrust.github.io/picrust/tutorials/genome_prediction.html) program based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to predict the functional alteration of microbiota in different samples. The OTU data obtained were used to generate BIOM files formatted as input for PICRUSt v1.1.09 with the make.biom script usable in the Mothur. OTU abundances were mapped to Greengenes OTU IDs as input to speculate about the functional alteration of microbiota (Supplementary Tables S1–S3).
Supplementary Information
Acknowledgements
This research was funded by Sichuan Academy of Agricultural Sciences (2019QYXK032), Tobacco Company of Sichuan (SCYC202212), and Tea Innovation Team of National Modern Agricultural Industry Technology System (sccxtd-2020-10) to D.Q.P.
Author contributions
L.Q.C., S.J.W., H.H.G. and D.Q.P. conceived and designed the experiments; J.M.Y., X.L.W., M.L., and H.L.L. collected the specimens; L.Q.C., S.J.W., P.C. and D.Q.P. performed the experiments and analyzed the data; L.Q.C. and D.Q.P. wrote the paper and discussed the results.
Data availability
The datasets generated and/or analysed during the current study are available in PRJNA963975 repository, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA963975.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Heng-Guo He, Email: hengguohe@163.com.
De-Qiang Pu, Email: pdqpudeqiang@163.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-49885-6.
References
- 1.Deligeorgidis PN, et al. Predatory effect of Coccinella Septempunctata on thrips tabaci and trialeurodes vaporariorum. J. Appl. Entomol. 2005;129:246–249. doi: 10.1111/j.1439-0418.2005.00959.x. [DOI] [Google Scholar]
- 2.Goggin FL. Plant aphid interactions: molecular and ecological perspectives. Curr. Opin. Plant. Biol. 2007;10:399–408. doi: 10.1016/j.pbi.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Castro C, et al. Effect of different diets on biology, reproductive variables and life and fertility tables of Harmonia Axyridis (Pallas) (Coleoptera, Coccinellidae) Rev. Bras. Entomol. 2016;60:260–266. doi: 10.1016/j.rbe.2016.03.003. [DOI] [Google Scholar]
- 4.Ruben GO, et al. Modular traits of the rhizobiales root microbiota and their evolutionary rela tionship with symbiotic rhizobia. Cell. Host. Microbe. 2018;24:155–167. doi: 10.1016/j.chom.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabbinge R, et al. Damage effects of cereal aphids in wheat. European J. Plant. Path. 1981;87:217–232. [Google Scholar]
- 6.Guedes CF, Lúcia MA. The potential of different fruit species as food for Harmonia Axyridis (Pallas, 1773) (Coleoptera: Coccinellidae) Rev. Bras. Frutic. 2013;35:1025–1031. doi: 10.1590/S0100-29452013000400013. [DOI] [Google Scholar]
- 7.Bonte M, Samih MA, Clercq PD. Development and reproduction of adalia bipunctata on factitious and artificial foods. Biocontrol. 2010;55:485–491. doi: 10.1007/s10526-010-9266-1. [DOI] [Google Scholar]
- 8.Singh P. Artificial diets for insects, mites, and spiders. Bull. ESA. 1978;24:123–124. [Google Scholar]
- 9.Smirnoff WA. An artificial diet for rearing coccinellid beetles. Can. Entomol. 1958;90:563–565. doi: 10.4039/Ent90563-9. [DOI] [Google Scholar]
- 10.Luca S, et al. Biological and biochemical characteristics for quality control of Harmonia Axyridis (Pallas) (Coleoptera, Coccinellidae) reared on a liver-based diet. Arch. Insect. Biochem. 2008;68:26–39. doi: 10.1002/arch.20233. [DOI] [PubMed] [Google Scholar]
- 11.Intazar A, et al. Artificial diet development and its effect on the reproductive performances of Propylea Japonica and Harmonia Axyridis. J. Asia-pac. Entomol. 2016;19:289–293. doi: 10.1016/j.aspen.2016.03.005. [DOI] [Google Scholar]
- 12.Ashraf M, et al. A study on laboratory rearing of lady bird beetle (Coccinella Septempunctata) to observe its fecundity and longevity on natural and artificial diets I. Irobp. Medline. 2010;2:1. [Google Scholar]
- 13.Evans EW, Dixon A. Insect predator-prey dynamics: ladybird beetles and biological control. Rev. Biol. 2000;38:38–2149. [Google Scholar]
- 14.Armitage SAO, et al. Tripartite interactions: How immunity, microbiota and pathogens interact and affect pathogen virulence evolution. Curr. Opin. Insect. 2022;50:100871. doi: 10.1016/j.cois.2021.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Cummings. & John, H. Microbial Digestion of Complex Carbohydrates in Man. P. Nutr. Soc. 43, 35–44 (1984). [DOI] [PubMed]
- 16.Sommer F, Bckhed F. The gut microbiota-masters of host development and physiology. Nat. Rev. Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 17.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Toole PW, Ian BJ. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 19.Pamer, E. G. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science352, 535–538 (2016). [DOI] [PMC free article] [PubMed]
- 20.Philipp E, Moran NA. The gut microbiota of insects diversity in structure and function. FEMS. Micro. Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 21.Faith JJ, et al. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosa TF, et al. The effects of high-severity fires on the arboreal ant community of a neotropical savanna. Oecologia. 2021;196:1–11. doi: 10.1007/s00442-021-04922-x. [DOI] [PubMed] [Google Scholar]
- 23.Nagarajan VM, et al. Status of important coastal habitats of North Tamil Nadu: Diversity, current threats and approaches for conservation. Reg. Stud. Mar. Sci. 2022;49:102–106. [Google Scholar]
- 24.Sharon G, Segal D, Ringo JM. Commensal bacteria play a role in mating preference of Drosophila melanogaster. PNAS. 2010;107:20051–20056. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeed, R., Razaq, M. & Hardy, I. The importance of alternative host plants as reservoirs of the cotton leaf hopper, Amrasca devastans, and its natural enemies. J. P. Sci.88, 517–531 (2015).
- 26.Bozsik A. Susceptibility of adult Coccinella septempunctata (Coleoptera: Coccinellidae) to insecticides with different modes of action. Pest. Manag. Sci. 2006;62:651–654. doi: 10.1002/ps.1221. [DOI] [PubMed] [Google Scholar]
- 27.Hodek I, Michaud JP. Why is Coccinella septempunctata so successful? (A point-of-view) Eur. J. Entomol. 2008;105:1–12. doi: 10.14411/eje.2008.001. [DOI] [Google Scholar]
- 28.Stark, J. D., Vargas, R. & Banks, J. E. Incorporating ecologically relevant measures of pesticide effect for estimating the compatibility of pesticides and biocontrol agents. J. Econ. Entomol.100, 1027–1032 (2007). [DOI] [PubMed]
- 29.Thaynah F, et al. The effects of high-severity fires on the arboreal ant community of a Neotropical savanna. Oecologia. 2021;196:53–18. doi: 10.1007/s00442-021-04922-x. [DOI] [PubMed] [Google Scholar]
- 30.Borer E, et al. Predators, parasitoids, and pathogens: A cross-cutting examination of intraguild predation theory. Ecology. 2007;88:2681–2688. doi: 10.1890/06-1707.1. [DOI] [PubMed] [Google Scholar]
- 31.Raymann K, Nancy AM. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect. Sci. 2018;26:97–104. doi: 10.1016/j.cois.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng YC, et al. Antibiotics induced changes in intestinal bacteria result in the sensitivity of honey bee to virus. Environ. Pollut. 2022;314:1. doi: 10.1016/j.envpol.2022.120278. [DOI] [PubMed] [Google Scholar]
- 33.Dong ZX, Li HY, Chen YF, Wang F. Colonization of the gut microbiota of honey bee (Apis Mellifera) workers at different developmental stages. Microbiol. Res. 2020;231:126–370. doi: 10.1016/j.micres.2019.126370. [DOI] [PubMed] [Google Scholar]
- 34.Jia Y, et al. Gut microbiome modulates Drosophila aggression through octop amine signaling. Nat. Commun. 2021;12:2698–2698. doi: 10.1038/s41467-021-23041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell JA, et al. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. PANS. 2009;106:50. doi: 10.1073/pnas.0907926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolouli K, et al. How the mighty have adapted: Genetic and microbiome changes during laboratory adaptation in the key pest Drosophila suzukii. Entomol. Gen. 2022;42:723–732. doi: 10.1127/entomologia/2022/1517. [DOI] [Google Scholar]
- 37.Bai S, et al. Gut fungal community and its probiotic effect on Bactroera dorsalis. Insect Sci. 2021;29:286–301. doi: 10.1111/1744-7917.12868. [DOI] [PubMed] [Google Scholar]
- 38.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2020;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higginbottom SK, Smits SA, Sonnenburg ED. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang, Q. et al. Metagenomic insights into the microbe-mediated B and K2 vitamin biosynthesis in the gastrointestinal: Microbiome of ruminants. Microbiol. 1, 109–109 (2022). [DOI] [PMC free article] [PubMed]
- 41.Dillon R, Charnley K. Mutualism between the desert locust Schistocerca Gregaria and its gut microbiota. Res. Microbiol. 2002;153:503–509. doi: 10.1016/S0923-2508(02)01361-X. [DOI] [PubMed] [Google Scholar]
- 42.Semova I, et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell. Host. Microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang ZD, et al. Predation and functional response of the multi-coloured Asian ladybeetle Harmonia axyridis on the adult Asian citruspsyllid Diaphorina citri. Biocontrol. Sci. Techn. 2019;29:293–307. doi: 10.1080/09583157.2018.1553028. [DOI] [Google Scholar]
- 44.Wei XY, et al. Research progress on influencing factors of microbial diversity in insects. J. Biol. 2019;28:170–176. [Google Scholar]
- 45.Xu G, et al. Analysis of diversity of silkworm gut bacteria based on 16S rRNA gene sequence. Sericul. Sci. 2015;41:641–649. [Google Scholar]
- 46.Zhao H, et al. Analysis of the composition of symbiotic bacteria in the Chinese cimex litoralis by 16S rDNA cloning library method. Chin. J. Bio. Control. 2017;33:849–856. [Google Scholar]
- 47.Markle JG, et al. Sex differences in thegut microbiome drive hormone-dependent regulation of aut oimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 48.Haldar S, Sanghamitra S. Impact of plant development on the rhizobacterial population Ofarachi Hypogaea: A multifactorial analysis. J. Basic. Microb. 2015;55:922–928. doi: 10.1002/jobm.201400683. [DOI] [PubMed] [Google Scholar]
- 49.Nicola M, Reid SL, Addison LJ, Macdonald GL. Biodiversity of active and inactive bacteria in the gutflora of wood feeding huhu beetle larvae (Prionoplus Reticularis) Appl. Environ. Microb. 2011;77:7000–7006. doi: 10.1128/AEM.05609-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng JY, et al. Ambient temperature ediated enzymic activities and intestinal microflora in lymantria dispar larvae. Arch. Insect. Biochem. 2019;102:2. doi: 10.1002/arch.21597. [DOI] [PubMed] [Google Scholar]
- 51.Kumar D, et al. Bombyx mori bidensovirus infection alters the intestinalmicroflora of fifth instar silkworm (Bombyx Mori) Larvae. J. Invertebr. Pathol. 2019;163:48–63. doi: 10.1016/j.jip.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Ranjith MT, Harish ER, Girija D, Nazeem PA. Bacterial communities associated with the gut of tomato fruit borer, Helicoverpa Armigera (Hübner) (Lepidoptera: Noctuidae) based on illumina next-generation sequencing. J. Asia-pac. Entomol. 2016;19:333–340. doi: 10.1016/j.aspen.2016.03.007. [DOI] [Google Scholar]
- 53.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. AEM. 2009;75:37–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grünwald S, Pilhofer M. Microbial associations in gut systems of wood- and barkinhabiting longhorned beetles (Coleoptera: Cerambycidae) Syst. Appl. Microbiol. 2010;33:25–34. doi: 10.1016/j.syapm.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Kim, J. H., Lee, J. W, Lee, Y. S. & Byun M W. Effects of combined treatment of gamma irradiation and addition of rosemary extract powder on ready-to-eat hamburger steaks: I. microbiological quality and shelf-life. JKSFSN. 33, 694–699 (2004).
- 56.Flint, H. J. et al. Forano E. Microbial degradation of complex carbohydrates in the gut. Gut. Microbes. 3, 289–306 (2012). [DOI] [PMC free article] [PubMed]
- 57.Abdessamad E, et al. The abundance and variety of carbohydrate active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 58.Renaud, N. et al. Rhodanobacter Lindaniclasticus Gen. Nov., Sp. Nov., a Lindane-Degrading Bacterium. IJSB. 49, 19–23. (1999). [DOI] [PubMed]
- 59.Dom SP, et al. Linking prokaryotic community composition to carbon biogeochemical cycling across a tropical peat dome in sarawak, malaysia. Sci. Rep. 2021;11:6416–6416. doi: 10.1038/s41598-021-81865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yabuuchi, E. et al. Proposal of Burkholderia gen. nov.and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. Nov. Microbiol Immunol. 36, 1251–1275 (1992). [DOI] [PubMed]
- 61.Dias GM, et al. Comparative genomics of Paraburkholderia kururiensis and its potential in bioremediation, biofertilization, and biocontrol of plant pathogens. Microbiologyopen. 2019;8:8. doi: 10.1002/mbo3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trivedi P, Duan Y, Wang N. Huanglongbing, a systemic disease, restructures the bacterial community associated with citrus roots. Appl. Environ. Microbiol. 2010;76:3427–3436. doi: 10.1128/AEM.02901-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, et al. Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbio. 2017;5:97. doi: 10.1186/s40168-017-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Safirah TN, et al. Rhizobia use a pathogenic-like effector to hijack leguminous nodulation sign alling. Sci. Rep. 2021;11:1. doi: 10.1038/s41598-021-81598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Philipp E, Moran NA. The gut microbiota of insects -diversity in structure and function. Fems. Microbiol. Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 66.Yan L, et al. The effect of different dietary sugars on the development and fecundity of Harmonia axyridis. Front. physiol. 2020;11:574–851. doi: 10.3389/fphys.2020.574851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berasategui A, et al. Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Mol. Ecol. 2017;26:4099–4110. doi: 10.1111/mec.14186. [DOI] [PubMed] [Google Scholar]
- 68.Amato KR, et al. Habitat degradation impacts black howler monkey (Alouatta Pigra) gastroin testinal microbio mes. ISME. J. 2013;7:1344–1353. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kathy L, Iverson MC, Bromel AW, Anderson P. Bacterial Symbionts in the Sugar Beet Root Maggot: Tetanops Myopaeformis (Von Rder) AEM. 1994;47:22–27. doi: 10.1128/aem.47.1.22-27.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in PRJNA963975 repository, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA963975.