Abstract
Epigenetic mechanisms, primarily mediated through histone and DNA modifications, play a pivotal role in orchestrating the functional identity of a cell and its response to environmental cues. Similarly, the spatial arrangement of chromatin within the three- dimensional (3D) nucleus has been recognized as a significant factor influencing genomic function. Investigating the relationship between epigenetic regulation and 3D chromatin structure has revealed correlation and causality between these processes, from the global alignment of average chromatin structure with chromatin marks to the nuanced correlations at smaller scales. This review aims to dissect the biological significance and the interplay between the epigenome and 3D chromatin structure, while also exploring the underlying molecular mechanisms. By synthesizing insights from both experimental and modeling perspectives, we seek to provide a comprehensive understanding of cellular functions.
Keywords: 3D chromatin modeling, 3D chromatin structure, Chromatin modifications, Chromatin perturbation, Epigenome
INTRODUCTION
While our understanding of genomic research has expanded considerably, the fundamental question remains: how does the nuanced role of epigenetic regulation through chromatin modifications intersect with 3D chromatin organization in managing cellular functionality and identity? Histones, far more than mere DNA-packaging proteins, undergo a spectrum of post- translational modifications, such as methylation and acetylation. Additionally, DNA itself is subject to a crucial modification known as DNA methylation, wherein methyl groups are added to the DNA itself. Each of these modifications casts a subtle influence on chromatin structure and gene expression. This interplay both dictates the cellular processes through varying scales of chromatin structures like Topologically Associating Domains (TADs) and compartments and becomes vitally pertinent when considering diseases like cancer and neurological disorders. This review navigates the interplay between epigenome and genomic organization, exploring their interactions and potential casualties while dissecting molecular mechanisms through experimental and modeling perspectives.
EPIGENETIC REGULATION
Epigenetics is the study of how gene expression can be changed, without changing the DNA sequence itself (1). A clear example of epigenetic gene regulation is observed in the specific expression of genes in various tissues. Although every cell in an organism possesses the same DNA sequence, they exhibit diverse functions and characteristics, due to variations in their gene activity (2). Furthermore, as these cells replicate and divide, they faithfully preserve the unique gene expression patterns specific to their cell type. Epigenetic mechanisms play a crucial role in maintaining this tissue-specific gene expression during the numerous rounds of DNA replication that occur during early development, and that persist throughout an individual’s life in many tissues.
Post-translational modifications (PTMs) of histones and DNA methylation constitute a fundamental mechanism of epigenetic regulation. In eukaryotic cells, chromatin consists of 147 bp of DNA wrapped around histone proteins, forming nucleosomes that resemble beads on a string (3). These histone proteins assemble into octamers, each consisting of two copies of H2A, H2B, H3, and H4 (4). Each of these histone proteins is decorated by various post-translational modifications that include methylation, acetylation, phosphorylation, ubiquitination, SUMOylation, citrullination, and ADP-ribosylation (5). These PTMs are referred to as chromatin marks that can regulate different DNA- templated processes, such as DNA replication, repair, and gene transcription. In the context of transcription, the outcome— whether gene activation or repression—depends on the specific residue and its associated modifications. Such changes in gene expression can be a direct consequence of the biochemical properties that are intrinsic to a specific mark. For example, histone acetylation and phosphorylation serve to decrease the positive charge of histones, potentially disrupting electrostatic interactions between histones and DNA (6). This decrease leads to a more relaxed chromatin structure, which in turn enhances DNA accessibility for protein complexes involved in transcription, ultimately resulting in increased gene expression (7). In mammalian genomes, DNA methylation is an epigenetic process where a methyl group is added to cytosine at the C5 position, forming 5-methylcytosine (8). This primarily occurs in CpG islands (CGI), regions with high G/C content, especially near gene promoter regions. DNA methylation intricately regulates gene expression by modulating the interactions between DNA and various chromatin proteins, as well as specific transcription factors (8). These interactions can lead to either the activation or repression of gene expression, depending on the chromatin context.
Modifications of DNA and histone proteins are introduced, maintained, and reset through the actions of specialized proteins, referred to as “writers”, “readers”, or “erasers”. Writers are enzymes that are responsible for adding modifications to histones or DNA and are categorized into different classes based on the specific types of modifications they influence (9). Similarly, erasers are enzymes that remove specific modifications from histone substrates or DNA and are categorized into classes corresponding to the modifications they target (9). Lastly, readers are dedicated protein factors that can recognize either post-translational marks on histones, covalent modifications of DNA, or a combination of marks and histone variants (9). These readers play a role in directing specific chromatin mark-dependent transcriptional outcomes. For example, the basal transcription factor TFIID “reads” and directly binds to H3K4me3 modification, typically located in active promoter regions, thereby promoting transcription (10). Furthermore, the specific depletion of H3K4me3 results in diminished transcription from a particular set of promoters, and reduces the binding of TFIID (10). Another reader protein, the BET protein Brd4, recognizes acetylated chromatin and controls gene expression by recruiting transcriptional regulatory complexes (11).
Beyond their role in regulating gene expression through nucleosome-level condensation via electrostatic effects, or the recruitment of effector proteins, chromatin modifications also exert control by shaping the global chromatin landscapes. As previously discussed, DNA is wrapped around histone proteins to form nucleosomes, which are further organized into higher- order structures that are known as chromatin. This chromatin can be categorized into euchromatin and heterochromatin, each characterized by a unique histone PTM signature (12). Euchromatin regions, rich in active genes and marked by active chromatin modifications, tend to exhibit lower levels of condensation. In contrast, heterochromatin regions, marked by repressive modifications, such as H3K9 and H3K27 methylation, are predominantly compact, and serve as barriers to transcriptional machinery (13). Thus, chromatin modifications play a role in controlling gene expression by delineating the genome into domains of “loosened and accessible” euchromatin, or “compact and inaccessible” heterochromatin.
Moreover, chromatin is non-randomly arranged within the nucleus. Although we discuss genome organization in detail, it is worth mentioning the potential influence of chromatin modifications in positioning the genome. Typically, heterochromatin localizes to the nuclear periphery, while euchromatin occupies the interior part of the nucleus. In mammals, HP1 (heterochromatin protein 1) proteins, known as readers with an affinity for H3K9me3, are involved in anchoring heterochromatin to the nuclear lamina (14). In C. elegans embryos, chromodomain protein CEC-4 anchors heterochromatin to perinuclear through the recognition of methylated H3K9, and CEC-4 mutants fail to commit fully to cell-type-specific differentiation (15). This tethering of heterochromatin to the periphery is hypothesized to be a crucial mechanism in regulating gene expression, and ongoing research continues to explore this intriguing phenomenon.
3D CHROMATIN ORGANIZATION
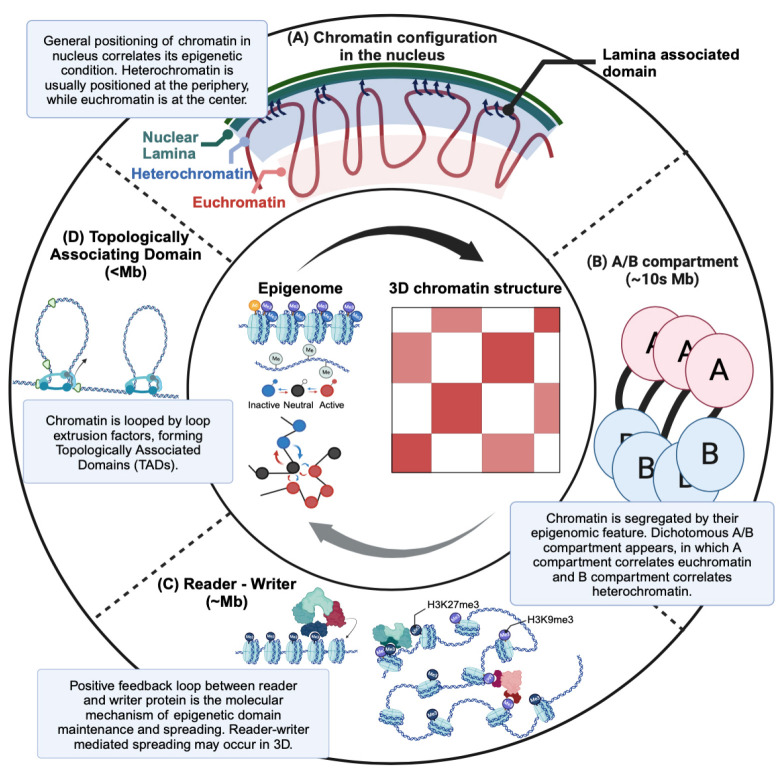
In eukaryotes, chromatin exhibits hierarchical organization. At the highest level, a dichotomous interaction pattern emerges, recognized as the A and B compartments, associated with euchromatin and heterochromatin, respectively (16). Studies have revealed that genomic segments in the A compartment predominantly interact with other A compartment regions, mirroring the interaction pattern within B compartments (17). These interactions establish distinct territories within the nucleus. The precise folding and spatial arrangement of chromatin appear to hold biological significance, posing implications in transcription and other forms of regulation (18). Notably, a gene’s relative position within the nucleus aligns with its regulatory pattern: transcriptionally active genes tend to reside nearer the nuclear center (19), whereas inactive genes are located at the periphery (20). For example, transcriptionally inactive genes colocalize with lamin at the nuclear periphery, forming the Lamina Associated Domain (LAD) (21). Although the precise mechanisms governing these compartmental patterns remain elusive, interactions involving proteins and RNA are potential contributing factors.
At the sub-megabase scale, another structural motif, TADs, emerges (22, 23). TADs share certain characteristics with compartments, including frequent intra-domain interactions, and limited inter-domain interactions. However, TADs are primarily defined by their boundaries, rather than the proteins and chromatin marks within them (24, 25). The underlying principle of TAD formation has been attributed to the chromatin loop extrusion mechanism, where the cohesin ring binds to DNA and extrudes it to form a loop until it encounters boundaries established by the CTCF proteins (26). Successive loop formations dictate the chromatin structure over extended genomic regions, often spanning hundreds of kilobases (27). TADs also play important roles in regulating gene expression by frequently housing the specific interactions between enhancers and their target promoters within their boundaries (28).
Importantly, loop extrusion and compartmentalization operate through distinct mechanisms, and perturbing one may influence, though not necessarily abolish, the other (29-33). For example, the depletion of cohesin eliminates loop domains and TADs, while it rather strengthens compartments, leading to the formation of compartments at a finer scale (32). Nevertheless, when examining finer subdivisions of compartments, the relationship between compartmentalization and loop extrusion becomes particularly nuanced (34).
CORRELATION BETWEEN CHROMATIN STRUCTURE AND EPIGENOME
Global correlation between chromatin structure and chromatin marks
Recent literature increasingly suggests a correlation between 3D chromatin structure and chromatin modifications, particularly regarding the alignment of euchromatin/heterochromatin with A/B compartments. This alignment suggests a link between epigenetic domains and chromatin structures (35-39). The A and B compartments are subdivided into several smaller compartments, and each of the subdomains tends to be enriched with specific chromatin marks, such as H3K9ac, H4K4me1, and H3K27me3 (40). Also, imaging techniques show that different epigenetic domains, such as transcriptionally active chromatin, transcriptionally inactive chromatin, and Polycomb-repressed domains, display distinct physical properties and interaction patterns (41).
However, the degree of this correlation varies, depending on the length scale under consideration. Notably, while there is general concordance in the alignment between the A/B compartment and euchromatin/heterochromatin domains, some inconsistencies arise at the finer subdivision of the A/B compartment (40). For example, some subdivisions of B compartments are enriched in active histone marks such as H3K4me1, H3K4me3, H3K9ac, and H3K27ac, while, at the same time, show a lack of repressive marks like H3K9me3 and H3K27me3. The limited correlation at smaller scales may be attributed to the challenge of delineating distinct boundaries between different chromatin states in certain regions, which can be complicated by the finer-scale switching of chromatin marks between active and repressive states. Thus, achieving a clear correlation between 3D chromatin structure and chromatin modifications requires combinatorial considerations of chromatin modifications, which remains a challenge (42, 43).
Correlation in TAD level
TAD formation is thought to occur through the loop extrusion of chromatin fibers facilitated by cohesin and CTCF, whereas compartmentalization results from the homotypic interactions among distinctively modified sets of nucleosomes. This difference in underlying mechanisms may contribute to the variations observed in the structural and epigenomic landscapes of TAD regions. While TADs and sub-compartments both exhibit higher contact frequencies within their domain and reduced contacts with neighboring domains, TADs are less cell type- specific (44) and can exist independently of compartments (45). This suggests that TADs may have a lower correlation with cell type-specific epigenomic patterns. Additionally, it’s worth noting that finer compartments that form after cohesin deletion align more closely with chromatin marks compared to the coarser compartments in the wild-type (32). This suggests that the loop extrusion mechanism for the establishment of TADs might interfere with the compartmentalization controlled by epigenetic factors.
CAUSALITY BETWEEN CHROMATIN STRUCTURE AND EPIGENOME
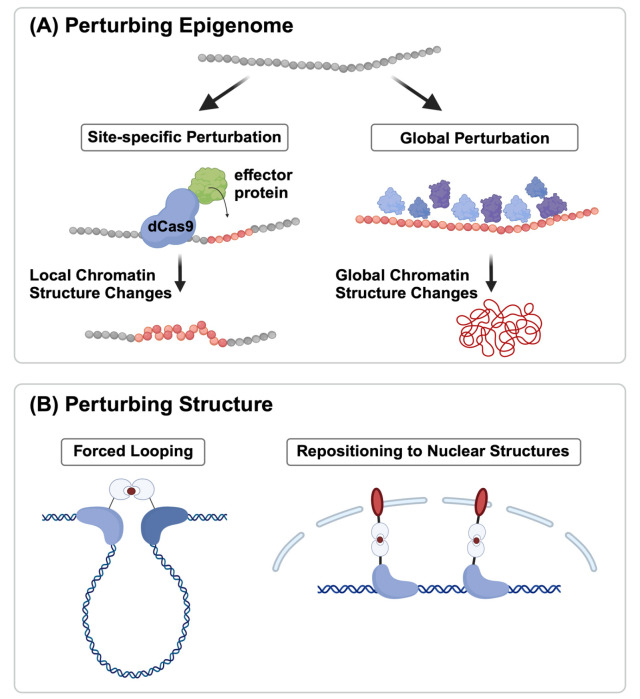
Advancement in technique has allowed us to probe genome- wide interaction frequencies within the 3D chromatin structure (16), and even at the single-cell level (46). Furthermore, imaging techniques have been used to reconstruct high-resolution 3D chromatin structures (47-49). However, due to the lack of a direct method to perturb and measure chromatin marks in tandem with the 3D chromatin structure, our grasp of the connection between the epigenome and chromatin structure remains largely correlational. Despite being in its infancy, the study of causality between chromatin structure and epigenome has seen some initial attempts. Two distinct approaches have emerged in the field: one branch explores how changes in chromatin modifications affect the 3D organization of chromatin, while the other examines how alterations of the 3D chromatin structure influence the epigenomic landscape (Fig. 1).
Fig. 1.
Tools for perturbing epigenome and structure. (A) Epigenomic patterns can be altered either site-specifically (by using DNA binding proteins like ZF or CRISPR-Cas9) or globally by depleting/ introducing various chromatin modifications. (B) 3D chromatin structure can be changed through forced looping or repositioning certain genomic regions to the nuclear structures.
Perturbing epigenome
Experimental studies have been conducted to investigate the impact of altering epigenomes on changes in the 3D organization of the genome (Fig. 1A). Histone H3 lysine 9 (H3K9) methylation, catalyzed by distinct SET domain enzymes, such as Suv39h1/Suv39h2, Eset1/Eset2, and G9a/Glp, plays a pivotal role in defining heterochromatin across various organisms (50). Depletion of all six SET domain lysine methyltransferase (KMT) genes in mouse embryonic fibroblasts (MEFs) results in the loss of H3K9 methylation, leading to disrupted heterochromatin organization, derepression of repeat elements, and compromised genomic stability (50). These effects underscore the critical function of H3K9 methylation in safeguarding heterochromatin structure and genome integrity, highlighting its importance in mammalian chromatin. Additionally, the inhibition of methyltransferases G9a and GLP has been shown to reduce H3K9me2 predominantly in A (active) genomic compartments, while residual H3K9me2 modifications are closely associated with B (inactive) genomic regions (51). Chemical inhibition of G9a/GLP in mouse hepatocytes leads to the effects of decreased chromatin-lamina interactions at G9a/GLP- sensitive regions, heightened genomic compartmentalization, and upregulation of genes linked to 3D chromatin alterations (51).
Besides the disruption of H3K9me3, the de novo establishment of this mark confirmed its significance in 3D genome organization. Feng et al. developed the EpiGo-KRAB system that was capable of introducing H3K9me3 modifications at multiple loci spanning megabases on human chromosome 19 (52). Through Hi-C analysis, they observed that the introduction of H3K9me3 can induce the remodeling of existing compartments, primarily at the boundaries between compartments (52). This study provides evidence for the ability of H3K9me3 to reshape the 3D organization of the genome at specific genomic regions, in particular at compartment boundaries.
Beyond H3K9me3, investigations into the intrinsic impact of DNA methylation on chromatin structure and function have been explored using budding yeast. Budding yeast lacks the machinery for DNA methylation and thus provides an ideal system to study the direct impact of DNA methylation (53). To examine this, murine DNA methyltransferases were expressed in Saccharomyces cerevisiae, and the correlations between DNA methylation, nucleosome positioning, gene expression, and 3D genome organization were analyzed (53). The study revealed that DNA methylation induces increased chromatin condensation in peri-centromeric regions, reduces overall DNA flexibility, and promotes the formation of a heterochromatin state. Furthermore, McLaughlin et al. showed that the overall DNA methylation status guides the polycomb-dependent 3D arrangement of mouse embryonic stem cells (mESCs), and likely, early blastocysts (54). Their results underscore the significant involvement of DNA methylation and its impact on polycomb-mediated mechanisms, influencing key aspects of 3D genome organization within stem cells.
Perturbing structure
While most current technologies for structural perturbation primarily focus on assessing their impact on gene transcription, there is a valuable opportunity to employ these methods to investigate how changes in 3D genome structure influence chromatin states (Fig. 1B). This approach is particularly relevant as histone and DNA modifications have a direct influence on transcriptional regulation (55).
Several systems have been engineered to manipulate chromatin looping and perturb 3D genome organization (56-59). An early study using the zinc-finger (ZF) protein aimed to control DNA looping at the natural β-globin location. ZF proteins were combined with the looping factor LDB1 (LIM Domain Binding 1) or LDB1’s self-association domain (SA) to create DNA loops between promoters and locus-control regions (LCRs) (58). This method effectively promoted long-range connections between enhancers and promoters, triggering gene activation for β-globin and fetal γ-globin in erythroid cells. In some studies, dCas9 heterodimers were utilized to facilitate chromatin looping between two distinct genomic regions. In one approach, two orthogonal dCas9 proteins derived from the Streptococcus pyogenes (Sp) and Streptococcus thermophilus (St) CRISPR systems were employed. This allowed the DNA-binding specificity of each dCas9 component in the heterodimer to be individually programmed using specific cognate sgRNAs. These orthogonal dCas9 proteins, when fused to leucine zippers (dCas9_Zip), formed heterodimers, which in turn induced chromatin looping (57). Additionally, another system known as CLOuD9 also harnessed two orthogonal dCas9 proteins to independently target distinct genomic regions (56). However, in this system, chromatin looping was established reversibly through the use of a chemically inducible mechanism. Forced looping via CLOuD9 increased the expression of genes such as the globin gene and Oct4, accompanied by elevated H3K4me3 levels, an active mark usually found in gene promoters. This approach demonstrates the potential to modulate gene expression and induce changes in chromatin states. In addition to a chemically inducible system, Kim et al. engineered a light-activated dynamic looping (LADL) system, where in response to blue light, spatial colocalization of two CRISPR targeted regions is induced (59).
As an alternative to forced looping, genome repositioning offers a promising technique for perturbing 3D genome structure. This approach involves using engineered proteins to tether specific genomic loci to different nuclear compartments. This tethering can be achieved using a variety of methods, such as the CRISPR-genome organization (CRISPR-GO) system, which uses dCas9 proteins to target genomic loci and the abscisic acid-inducible heterodimerization system to induce their translocation to the nuclear periphery (60). Another tool to note is a CRISPR/dCas9-system that enables gene positioning in the nucleus called CRISPR-PIN, which uses a pair of proteins that are always interacting to tether genomic regions together (61). This tool does not require any external signals to work, making it a non-inducible system. Genome repositioning employing such tools has been used to study the relationship between 3D genome organization and gene transcription in a variety of cell types and organisms.
MOLECULAR MECHANISMS UNDERLYING THE INTERPLAY BETWEEN CHROMATIN MARKS AND CHROMATIN ARCHITECTURE
Moving forward in our discussion of the interplay between the epigenome and chromatin structure, we transition from the fundamental observation of structural compartments aligning with chromatin states, to a deeper examination of the molecular mechanisms at play. The proteins responsible for establishing and maintaining specific chromatin states can potentially influence the local 3D configuration of chromatin, leading to its further compartmentalization. While experiments have yet to conclusively verify this hypothesis, it has been widely explored using computational methods. In this section, we delve into both experimental and computational approaches that illuminate the molecular mechanisms underlying the formation and maintenance of chromatin states, offering potential explanations for their association with the structural organization of chromatin.
Molecular mechanism: experimental approaches
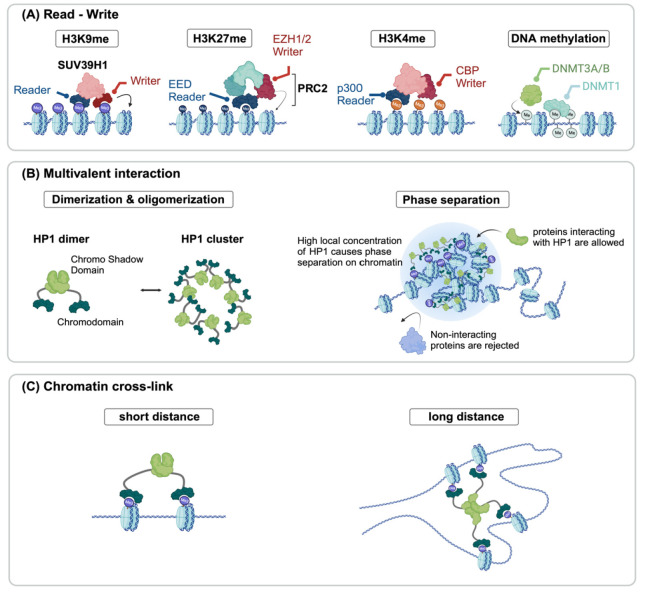
While the intricate mechanisms facilitating the establishment and propagation of chromatin modifications remain an active area of research, a shared molecular mechanism has been observed across different types of marks, notably H3K9me3 and H3K27me3.
The initial step in the assembly of chromatin states involves a nucleation stage, during which chromatin regulators are targeted to specific sequences through the action of DNA- binding proteins or RNAs (62). One extensively studied example of sequence-dependent recruitment is the nucleation of heterochromatin in fission yeast. In this process, the H3K9 methyltransferase Clr4 is recruited to specific genomic sites through the involvement of the RNA interference machinery and ATF/ CREB family transcription factors (63). Similarly, the nucleation of the PRC2 complex, responsible for H3K27 methylation in metazoans, involves a range of factors that include accessory proteins such as JARID2 and/or MTF2 (64, 65), RNAs (66), and the presence of hypomethylated CGIs (67), which direct the assembly of the PRC2 complex at the target locations (68). When PRC2 is in association with accessory proteins like JARID2, MTF2 (also called non-core subunits of PRC2 complex), or other cell-type-dependent factors, it identifies specific nucleation sites (64). At these sites, initial deposition of H3K27me2 occurs, which subsequently transforms into H3K27me3 once a “critical” concentration of PRC2 is achieved (65). This process involves the stable binding of PRC2, highlighting the coordinated interplay between PRC2 and its accessory proteins in the dynamic regulation of H3K27 methylation (65). As with accessory proteins, RNA was found to play a crucial role in the chromatin localization of the PRC2 complex in human pluripotent stem cells (66).
Following nucleation, the established mark is propagated further along the nucleosome through a read-write mechanism. Here, existing modifications are recognized by “reader”, and “writer” enzymes are subsequently recruited to modify new nucleosomes (69-75). This mechanism is seen in several heritable histone modifications, which include histone deacetylation, H3K9 methylation, and H3K27 methylation.
For example, Clr4 in fission yeast, a protein that can both read and write H3K9 methylation (76), can bind to pre-existing H3K9 methylation, a histone mark associated with heterochromatin, while it can also place a new H3K9 methylation on neighboring nucleosomes. This dual function of reading and writing is essential for the spread of heterochromatin. Similarly, in H3K27 methylation, the EED subunit of the PRC2 complex binds to H3K27me3, ensuring the recruitment of PRC2 to similarly modified nucleosomes (73). Notably, the carboxy- terminal domain of EED exhibits a specific affinity for histone tails bearing trimethyl-lysine residues associated with repressive chromatin marks (77). This interaction results in the allosteric activation of the methyltransferase activity of EZH1/2 within the PRC2 complex (78,79). This read-write mechanism ensures the faithful replication and inheritance of H3K27 methylation (Fig. 2A).
Fig. 2.
Potential molecular mechanisms explaining epigenome-structure relationship. (A) Reader-writer models consist of protein complexes containing both reading and writing chromatin mark abilities, initiating and maintaining the epigenetic domain. (B) Multivalent interaction also plays a role in structural changes. HP1 dimerization and oligomerization driven by H3K9me3 may induce phase separation in the heterochromatin, excluding non-interacting proteins from the region. (C) HP1 may alter the 3D chromatin structure by making crosslinks.
Although less extensively studied, it appears that active marks also undergo propagation and inheritance through read-write mechanisms (71). One study investigated the changes in H3K4me3 distribution on leading and lagging DNA strands over time, from the early S phase to the late S phase, to the G2 phase. The study proposes that a read-write mechanism might be one of the molecular processes responsible for restoring H3K4me3 levels following DNA replication. This mechanism may aid in preserving the levels of H3K4me3 and the transcriptional states. Furthermore, recent cryogenic electron microscopy studies have illuminated key aspects of the histone acetylation read-write mechanism (70). These experiments revealed that p300/CREB-binding protein (CBP) recognizes and extends acetylation on the histone H4 N-terminal tail (NT) within a nucleosome. This recognition involves the bromodomain pocket, responsible for reading H4NTac, as well as interactions with DNA minor grooves adjacent to the pocket, guiding the catalytic activity of p300/CBP to non-H4 histone NTs (70). Notably, this read-write process primarily targets H2BNT, and its acetylation promotes H2A-H2B dissociation from the nucleosome, facilitating context-dependent gene transcription through localized nucleosome destabilization.
Furthermore, a synthetic design approach has further substantiated the significance of this read-write mechanism. Park et al. established an orthogonal epigenetic regulatory system in mammalian cells, utilizing N6-methyladenine (6mA), a DNA modification rarely found in metazoan epigenomes (74). This system employed proteins responsible for writing and reading 6mA and demonstrated the spatial propagation of 6mA closely resembling the patterns observed in the spreading of H3K9me3 within HP1-mediated heterochromatin. Therefore, this orthogonal system demonstrates that ‘read-write’ is a fundamental and sufficient molecular mechanism that allows the spreading of the chromatin mark.
Notably, HP1 serves multifunctional roles, extending beyond its canonical involvement in facilitating the reading and writing of H3K9me3 (Fig. 2B). The chromodomain of HP1α serves as a reader for H3K9me3, while its chromo shadow domain acts as a writer for the same mark, by recruiting additional proteins with catalytic activity for H3K9me3. This coordinated action facilitates the propagation of heterochromatin domains (75). Interestingly, the chromo shadow domain of HP1α undergoes dimerization, enabling multivalent interactions (80). The multivalency of HP1α further increases by engaging in non-stoichiometric interactions and liquid phase separation (81-84). These multivalent interactions result in the formation of highly concentrated HP1 domains and play a crucial role in enabling fast and efficient competition for binding sites in a crowded nuclear environment. In these distinct regions, an environment that is conducive to the accumulation of proteins and RNA essential for the formation of heterochromatin is established, thereby promoting biological processes with enhanced efficiency. The ability of the HP1 protein to engage in multivalent interactions both compacts DNA in neighboring regions into mechanically stable domains and promotes interactions between regions bearing the same mark, even when they are at a distance, implying their pivotal role in bridging the epigenetic landscape with the structural organization of chromatin.
Gao et al. recently tested this multifaceted role of HP1 with a synthetic approach, the CRISPR-engineered chromatin organization (CRISPR-EChO) platform (85). They positioned human HP1α across large genomic regions, observing its impact on chromatin state, gene expression, and dynamic behavior in live cells. Their findings demonstrate that HP1α establishes novel contacts with natural heterochromatin, promotes long-range chromatin interactions, and can reversibly shift chromatin from a diffuse to a compact state (Fig. 2C).
Several studies have also explored the long-range bridging of H3K27me3 domains and chromatin compaction mediated by the PRC2 complex (72, 86). One such study utilized a combination of H3K27me3 HiChIP and optical reconstruction of chromatin architecture (ORCA) sequential DNA FISH imaging technique, along with genetic interventions targeting H3K27me3- associated loop anchors and EZH2 binding (72). This study aimed to uncover the establishment of long-range genomic connections associated with the PRC2 complex, and their role in spreading H3K27me3 to distant locations. The findings revealed that PRC2 facilitates long-range Polycomb-associated DNA contacts, spanning extensive genomic regions that encompass multiple TADs. Additionally, disruption of Polycomb- associated loops was observed when PRC2 binding was lost, while deletion of loop anchors led to changes in H3K27me3 distribution and gene expression.
Furthermore, another investigation involved the knockout and subsequent rescue of EED in mESCs (65). Upon EED re- expression, PRC2 was initially attracted to specific regions known as “nucleation hubs”, from which the H3K27me2/3 mark spreads to neighboring regions, both in proximity and at a distance. These long-range interactions occurred within sub- nuclear foci of Polycomb activity. Another study demonstrated that regions of DNA occupied by Polycomb group (PcG) proteins and enriched with the histone modification H3K27me3 can form loops, and physically interact with each other around a single gene in mammalian cells (87).
These experimental approaches provide valuable insights that bring us closer to unraveling the intricate molecular mechanisms, including read-write processes and associated factors, that underlie the interplay between the epigenome and chromatin structure.
Molecular mechanism: modeling approaches
In studying the interplay between the epigenome and chromatin structure, computer modeling has emerged as a cornerstone technique, primarily due to the limited direct experimental methodologies that can simultaneously measure chromatin marks and chromatin structure in vivo. For example, computational models have provided valuable insights into the epigenome-driven phase-separation of chromatin, and its subsequent role in determining nuclear configuration (88); while another study addressed the underlying principles governing the role of loop extrusion in the formation of sub-megabase TAD structures (25). However, it is worth noting that these modeling approaches exhibit notable differences in their underlying assumptions and the level of detail. We categorize these modeling strategies into two primary approaches: the bottom-up approach, and the data-driven approach (Fig. 3).
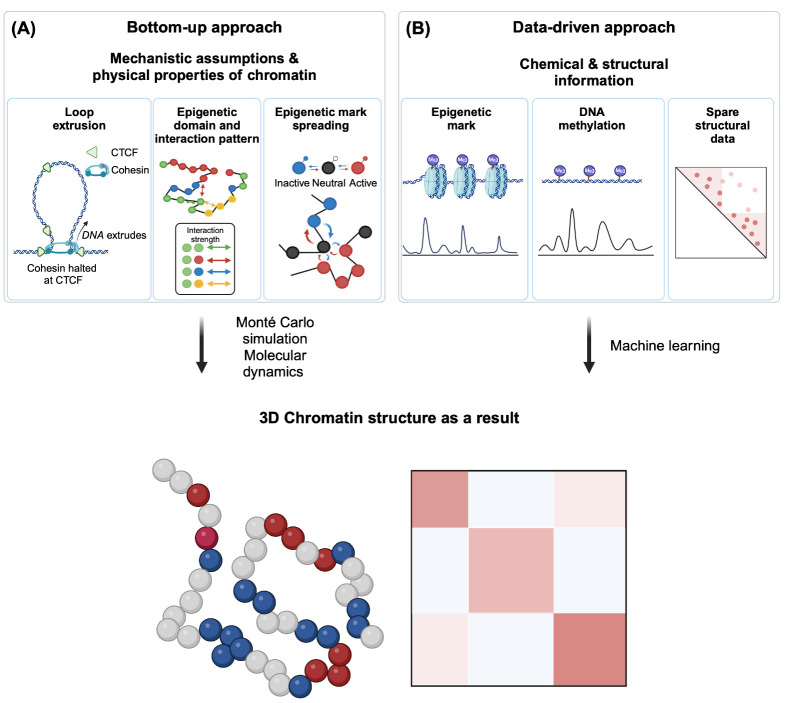
Fig. 3.
Two approaches of computational modeling. (A) A bottom- up approach is based on some assumptions and physical properties of chromatin: loop extrusion mechanism, epigenetic domain, and their interaction strength, and spreading dynamics of chromatin marks. Monté Carlo simulation or molecular dynamics can be used to simulate chromatin fiber in silico, generating 3D chromatin structure based on the assumptions made. (B) The data- driven approach does not assume anything, but uses experimentally measured chemical & structural information such as 1D chromatin mark distribution (ChIP-Seq), DNA methylation pattern, and sparse structural data. Machine learning algorithms can utilize this information to generate a 3D chromatin structure.
Bottom-up approaches
Bottom-up approaches in chromatin modeling begin by grounding themselves in foundational assumptions about molecular mechanisms (Fig. 3A). These mechanistic models encompass diverse elements to reflect various epigenetic behaviors, including the reader-writer mechanism for chromatin mark propagation, and homotypic interaction among chromatin states.
Current models often depict chromatin using the beads-on-a- string analogy. Here, each bead represents multiple nucleosomes, conceptualized as a genomic bin, while the string corresponds to the linker chromatin (89). Due to its considerable length, chromatin can be approximated as an ideal chain. In this representation, consecutive beads are positioned independently, meaning that the position of one bead is not influenced by the position of the previous one.
Chromatin can be modeled as either a “homopolymer” or “heteropolymer”. In homopolymer models, the chromatin structure consists solely of identical monomers, while heteropolymer models incorporate a diverse array of monomer types. More precisely, these models consider chromatin as a chain with N monomers, where each monomer represents a set of nucleosomes that span tens of kilobases. While homopolymer models facilitate the simulation of global chromatin structure features, they may not accurately capture nuanced epigenome- structure correlations (90). Recent models have therefore shifted towards addressing heteropolymeric chromatin, which accounts for varying monomer types, each defined by specific chromatin marks and associated protein interactions. In these models, interactions among proteins responsible for specific chromatin states are depicted as either attractive or repulsive forces between monomers, following a mechanistic model based on homotypic interactions. The precise strength of these epigenome-driven attractions or repulsions can be fine-tuned using experimental data, such as Hi-C and FISH. By adjusting the interaction strengths between monomer pairs, these models can simulate the emergence of large structural domains, including TADs and compartments, illuminating the role of interactions among distinct epigenomic states in shaping chromatin structure (90, 91).
Although the models mentioned earlier effectively explain the overall chromatin structural patterns influenced by the epigenome in steady-state chromatin (92), the dynamic nature inherent in the transition of epigenetic states, which governs the establishment and maintenance of epigenetic memory, necessitates the incorporation of chromatin state transitions. Chromatin states, initially categorized as active, inactive, or neutral, are thought to progress from one state to another, based on the states of neighboring monomers located in proximity. These transitions in chromatin states subsequently reshape interaction patterns in the 3D chromatin configuration, which in turn further influence chromatin states. This interplay— between the spread of chromatin marks (or chromatin state transitions) and the associated changes in 3D chromatin structure—is effectively modeled through iterative calculations of both read-write-mediated spreading and structural changes (93-95). Building on this set-up, computational models have evolved to better reproduce the intricate, real-time interactions between chromatin marks, and the 3D structure of chromatin. Early heteropolymer models predominantly relied on interactions derived from predetermined chromatin state distributions (90, 92). Subsequent iterations introduced state-to-state transitions, tracing the full temporal progression of chromatin marks from their inception, based on the aforementioned proximity influence.
Experimental approaches and bottom-up modeling are closely intertwined. Insights gained from experimental methods inform the mechanistic assumptions used in modeling, while experiments also play a crucial role in validating these models. The initiation and sustained maintenance of the epigenetic state are theorized to depend on the recruitment of histone- modifying enzymes (HMEs) to specific genomic states (96-98). Such recruitment is facilitated either through collaborations with DNA-binding proteins or via RNA interference (RNAi) pathways (99). Some models integrate sequence-specific nucleation and its interaction with the reader-writer process within a 3D context (100). Empirical validations of these models have yielded encouraging results, exemplified by phenomena like the formation of γH2AX domains near double-stranded breaks, the evolution of heterochromatin domain around the insertion sites of Transposable Elements (TE) in mammals, and the memory retention of heterochromatin in fission yeast (101, 102). Furthermore, mathematical modeling of the Polycomb system’s repressive mark domain, which includes site-specific HME (PRC2) recruitment and mark propagation, has demonstrated its capability to accurately predict the experimental condition of chromatin mark domains in both wild-type and mutant cells (103).
Data-driven approaches
In contrast to bottom-up approaches, which are based on established paradigms or presupposed hypotheses that explain the behavior of smaller components to derive insights to comprehend the larger system, as exemplified by the reader-writer mechanism-based modeling to explain the behavior of chromatin mark spreading (99, 104), data-driven models often evolve from an extensive analysis of datasets, and do not necessarily rely on pre-existing hypotheses or theoretical frameworks (Fig. 3B). Instead, data-driven models leverage a holistic understanding of the dataset to uncover patterns and correlations that can guide the formulation of new hypotheses.
A primary data source for extracting insights into the principle of chromatin structure and organization is Hi-C. Hi-C experiments characteristically produce data in the form of a contact probability matrix, where the frequency of pairwise interactions corresponds directly to the spatial proximity probability between two loci. Such encounter probabilities can be harnessed either as an energy function or as constraints that restrict the degrees of freedom of specific chromatin polymers. By optimizing chromatin coordinates within these constraints, it becomes possible to reconstruct a 3D structure, grounding it firmly on the provided experimental contacts (105, 106). Through iterative simulations, a range of 3D chromatin structures can be generated, all of which are in agreement with the properties that are observed experimentally (107). Parameters that delineate the precise constraints can be fine-tuned to minimize discrepancies with the experimental data. The principal aim of these modeling efforts is twofold: to elucidate biological insights from the refined parameters, and to delve into the more realistic 3D behavior of chromatin polymers (108).
Data-driven strategies also incorporate machine learning processes to provide enriched insights. These machine-learning models are instrumental in predicting structural outcomes influenced by epigenetic factors, utilizing datasets that extend beyond conventional structural information (109-114). Specifically, models like MEGABASE (112) and CoRNN (114) rely exclusively on biochemical information, e.g., ChIP-Seq, to make structural predictions. Given the extensive availability of ChIP- Seq tracks spanning varied samples, including diverse cell lines and tissues (115), these models unfold extensive application and validation potentials across numerous experiments. Beyond ChIP-Seq, other resourceful inputs include whole-genome bisulfite sequencing (113), RNA-Seq (109), and moderate coverage inter-chromosomal Hi-C (110).
Data-driven models offer a pivotal insight: the 1D epigenome can encapsulate sufficient information to elucidate the broader 3D structure of chromosomes (112, 116, 117). In essence, this suggests that chemical modification information alone may contain enough detail to instruct overall chromatin structure, thereby promoting future experiments that aim to decipher the epigenetic underpinnings of chromatin’s 3D structural patterns. Perturbation experiments applied to the epigenome can serve to validate the predictions of these models. Furthermore, insights derived from such models are instrumental in distinguishing structural changes driven by chromatin modifications from random noise, thereby enhancing our understanding of the interplay between the structural and epigenomic landscapes.
CONCLUSION
This review has explored the intricate interplay between chromatin modifications, 3D chromatin structures, and their impact on cellular function and identity (Fig. 4). While experimental methods have been essential to uncovering key insights, computational approaches have complemented our understanding, particularly in deciphering the dynamic nature of these interactions. The integration of innovative experimental techniques that establish direct causal links between the epigenome and chromatin structure holds great promise for future discoveries. Moving forward, the convergence of experimental and computational approaches will continue to advance our knowledge in this field, potentially unveiling the novel molecular mechanisms that govern this interplay.
Fig. 4.
Overview of the interplay between the epigenome and 3D chromatin structure. (A) The correlation between the epigenome and 3D chromatin structure manifests at various scales. At the whole- genome level, heterochromatin typically resides at the periphery of the nucleus and interacts with the nuclear lamina, while euchromatin is preferably centrally located. (B) On a tens-of-megabases scale, a dichotomous pattern known as A/B compartments emerges, with A compartments associated with euchromatin and B compartments with heterochromatin. Chromatin regions within the same compartment tend to be spatially clustered. (C) On the few megabases scale, epigenetic domains align with structural domains, with the epigenetic domain formed and maintained through the spreading of marks facilitated by a reader-writer positive feedback mechanism potentially in the context of 3D chromatin structure. (D) Lastly, on the sub-megabase scale, topologically associating domains (TADs) arise through the action of loop extrusion factors.
Funding Statement
ACKNOWLEDGEMENTS This work was supported by National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. NRF2022R1A5A102641311), NRF-2022R1C1C1004100, NRF-2019R1A6A1A10073887, KAIST UP Program, POSCO Science Fellowship of POSCO TJ Park Foundation.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Waterland RA. Epigenetic mechanisms and gastrointestinal development. J Pediatr. 2006;149:S137–S142. doi: 10.1016/j.jpeds.2006.06.064. [DOI] [PubMed] [Google Scholar]
- 2.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity. 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/S0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 4.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quina AS, Buschbeck M, Di Croce L. Chromatin structure and epigenetics. Biochem Pharmacol. 2006;72:1563–1569. doi: 10.1016/j.bcp.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillette TG, Hill JA. Readers, writers, and erasers: chromatin as the whiteboard of heart disease. Circ Res. 2015;116:1245–1253. doi: 10.1161/CIRCRESAHA.116.303630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermeulen M, Mulder KW, Denissov S, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 11.Shi J, Vakoc Christopher R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millán-Zambrano G, Burton A, Bannister AJ, Schneider R. Histone post-translational modifications - cause and consequence of genome function. Nat Rev Genet. 2022;23:563–580. doi: 10.1038/s41576-022-00468-7. [DOI] [PubMed] [Google Scholar]
- 13.Tamaru H. Confining euchromatin/heterochromatin territory: jumonji crosses the line. Genes Dev. 2010;24:1465–1478. doi: 10.1101/gad.1941010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padeken J, Methot SP, Gasser SM. Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance. Nat Rev Mol Cell Biol. 2022;23:623–640. doi: 10.1038/s41580-022-00483-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Sandoval A, Towbin BD, Kalck V, et al. Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell. 2015;163:1333–1347. doi: 10.1016/j.cell.2015.10.066. [DOI] [PubMed] [Google Scholar]
- 16.Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17:661–678. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- 18.Leidescher S, Ribisel J, Ullrich S, et al. Spatial organization of transcribed eukaryotic genes. Nat Cell Biol. 2022;24:327–339. doi: 10.1038/s41556-022-00847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osborne CS, Chakalova L, Brown KE, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 20.Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet. 2013;14:67–84. doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Su JH, Beliveau BJ, et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016;353:598–602. doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nora EP, Lajoie BR, Schulz EG, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szabo Q, Bantignies F, Cavalli G. Principles of genome folding into topologically associating domains. Sci Adv. 2019;5:eaaw1668. doi: 10.1126/sciadv.aaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085.b477553eadc94d389ad5dd178e7b4d42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naumova N, Imakaev M, Fudenberg G, et al. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoenfelder S, Fraser P. Long-range enhancer- promoter contacts in gene expression control. Nat Rev Genet. 2019;20:437–455. doi: 10.1038/s41576-019-0128-0. [DOI] [PubMed] [Google Scholar]
- 29.Nora EP, Goloborodko A, Valton AL, et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169:930–944. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci U S A. 2018;115:E6697–E6706. doi: 10.1073/pnas.1717730115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao SSP, Huang SC, Glenn St Hilaire B, et al. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–320. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarzer W, Abdennur N, Goloborodko A, et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wutz G, Várnai C, Nagasaka K, et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017;36:3573–3599. doi: 10.15252/embj.201798004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spracklin G, Abdennur N, Imakaev M, et al. Diverse silent chromatin states modulate genome compartmentalization and loop extrusion barriers. Nat Struct & Mol Biol. 2023;30:38–51. doi: 10.1038/s41594-022-00892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichols MH, Corces VG. Principles of 3D compartmentalization of the human genome. Cell Rep. 2021;35:109330. doi: 10.1016/j.celrep.2021.109330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fortin JP, Hansen KD. Reconstructing A/B compartments as revealed by Hi-C using long-range correlations in epigenetic data. Genome Biol. 2015;16:180. doi: 10.1186/s13059-015-0741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mourad R, Cuvier O. Predicting the spatial organization of chromosomes using epigenetic data. Genome Biol. 2015;16:182. doi: 10.1186/s13059-015-0752-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pancaldi V, Carrillo-de-Santa-Pau E, Javierre BM, et al. Integrating epigenomic data and 3D genomic structure with a new measure of chromatin assortativity. Genome Biol. 2016;17:152. doi: 10.1186/s13059-016-1003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tolsma TO, Hansen JC. Post-translational modifications and chromatin dynamics. Essays Biochem. 2019;63:89–96. doi: 10.1042/EBC20180067. [DOI] [PubMed] [Google Scholar]
- 40.Tan ZW, Guarnera E, Berezovsky IN. Exploring chromatin hierarchical organization via Markov State Modelling. PLoS Comput Biol. 2019;14:e1006686. doi: 10.1371/journal.pcbi.1006686.bc70ba955f9e48409c54544a03897131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boettiger AN, Bintu B, Moffitt JR, et al. Super- resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimura A, Horikoshi M. Partition of distinct chromosomal regions: negotiable border and fixed border. Genes Cells. 2004;9:499–508. doi: 10.1111/j.1356-9597.2004.00740.x. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Lawry ST, Cohen AL, Jia S. Chromosome boundary elements and regulation of heterochromatin spreading. Cell Mol Life Sci. 2014;71:4841–4852. doi: 10.1007/s00018-014-1725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao SS, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gassler J, Brandão HB, Imakaev M, et al. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 2017;36:3600–3618. doi: 10.15252/embj.201798083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagano T, Lubling Y, Stevens TJ, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bintu B, Mateo LJ, Su JH, et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 2018;362:1783–1791. doi: 10.1126/science.aau1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mateo LJ, Murphy SE, Hafner A, Cinquini IS, Walker CA, Boettiger AN. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature. 2019;568:49–54. doi: 10.1038/s41586-019-1035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu M, Lu Y, Yang B, et al. Multiplexed imaging of nucleome architectures in single cells of mammalian tissue. Nat Commun. 2020;11:2907. doi: 10.1038/s41467-020-16732-5.23a002745d5b4796868be007bae1e134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montavon T, Shukeir N, Erikson G, et al. Complete loss of H3K9 methylation dissolves mouse heterochromatin organization. Nat Commun. 2021;12:4359. doi: 10.1038/s41467-021-24532-8.ffb762bd6b22494b91032351da989456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Z, Ji L, Huo X, et al. G9a/GLP-sensitivity of H3K9me2 demarcates two types of genomic compartments. Genomics Proteomics Bioinformatics. 2020;18:359–370. doi: 10.1016/j.gpb.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng Y, Wang Y, Wang X, et al. Simultaneous epigenetic perturbation and genome imaging reveal distinct roles of H3K9me3 in chromatin architecture and transcription. Genome Biol. 2020;21:296. doi: 10.1186/s13059-020-02201-1.0f0e8451da42416fa2e9117869a58941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buitrago D, Labrador M, Arcon JP, et al. Impact of DNA methylation on 3D genome structure. Nat Commun. 2021;12:3243. doi: 10.1038/s41467-021-23142-8.d5ee7c9961b041b8a4708bc652b9e485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLaughlin K, Flyamer IM, Thomson JP, et al. DNA Methylation directs polycomb-dependent 3D genome re-organization in naive pluripotency. Cell Rep. 2019;29:1974–1985. doi: 10.1016/j.celrep.2019.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, Han M, Qi LS. Engineering 3D genome organization. Nat Rev Genet. 2021;22:343–360. doi: 10.1038/s41576-020-00325-5. [DOI] [PubMed] [Google Scholar]
- 56.Morgan SL, Mariano NC, Bermudez A, et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun. 2017;8:15993. doi: 10.1038/ncomms15993.5309d42ed5b04e42a18e02ba03f78364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hao N, Shearwin KE, Dodd IB. Programmable DNA looping using engineered bivalent dCas9 complexes. Nat Commun. 2017;8:1628. doi: 10.1038/s41467-017-01873-x.74f9626a97ce451fafaa5aa6b162cad8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng W, Lee J, Wang H, et al. Controlling long- range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JH, Rege M, Valeri J, et al. LADL: light- activated dynamic looping for endogenous gene expression control. Nat Methods. 2019;16:633–639. doi: 10.1038/s41592-019-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H, Xu X, Nguyen CM, et al. CRISPR-mediated programmable 3D genome positioning and nuclear organization. Cell. 2018;175:1405–1417. doi: 10.1016/j.cell.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin JL, Ekas H, Deaner M, Alper HS. CRISPR- PIN: modifying gene position in the nucleus via dCas9- mediated tethering. Syn and Sys Biotech. 2019;4:73–78. doi: 10.1016/j.synbio.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 63.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Højfeldt JW, Hedehus L, Laugesen A, et al. Non- core subunits of the PRC2 complex are collectively required for its target-site specificity. Mol Cell. 2019;76:423–436. doi: 10.1016/j.molcel.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oksuz O, Narendra V, Lee CH, et al. Capturing the onset of PRC2-mediated repressive domain formation. Mol Cell. 2018;70:1149–1162. doi: 10.1016/j.molcel.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Long Y, Hwang T, Gooding AR, et al. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat Genet. 2020;52:931–938. doi: 10.1038/s41588-020-0662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu JR, Lee CH, Oksuz O, Stafford JM, Reinberg D. PRC2 is high maintenance. Genes Dev. 2019;33:903–935. doi: 10.1101/gad.325050.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blackledge NP, Rose NR, Klose RJ. Targeting polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16:643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brickner JH. Inheritance of epigenetic transcriptional memory through read-write replication of a histone modification. Ann N Y Acad Sci. 2023;1526:50–58. doi: 10.1111/nyas.15033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kikuchi M, Morita S, Wakamori M, et al. Epigenetic mechanisms to propagate histone acetylation by p300/CBP. Nat Commun. 2023;14:4103. doi: 10.1038/s41467-023-39735-4.acaf383597024ad590d456fce9a48fc8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Serra-Cardona A, Duan S, Yu C, Zhang Z. H3K4me3 recognition by the COMPASS complex facilitates the restoration of this histone mark following DNA replication. Science Adv. 2022;8:eabm6246. doi: 10.1126/sciadv.abm6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kraft K, Yost KE, Murphy SE, et al. Polycomb- mediated genome architecture enables long-range spreading of H3K27 methylation. Proc Natl Acad Sci U S A. 2022;119:e2201883119. doi: 10.1073/pnas.2201883119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lövkvist C, Mikulski P, Reeck S, Hartley M, Dean C, Howard M. Hybrid protein assembly-histone modification mechanism for PRC2-based epigenetic switching and memory. Elife. 2021;10:e66454. doi: 10.7554/eLife.66454.sa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park M, Patel N, Keung AJ, Khalil AS. Engineering epigenetic regulation using synthetic read-write modules. Cell. 2019;176:227–238. doi: 10.1016/j.cell.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang K, Mosch K, Fischle W, Grewal SIS. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 77.Margueron R, Justin N, Ohno K, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee SH, Li Y, Kim H, et al. The role of EZH1 and EZH2 in development and cancer. BMB Rep. 2022;55:595–601. doi: 10.5483/BMBRep.2022.55.12.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee CH, Yu JR, Kumar S, et al. Allosteric activation dictates PRC2 activity independent of its recruitment to chromatin. Mol Cell. 2018;70:422–434. doi: 10.1016/j.molcel.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kilic S, Bachmann AL, Bryan LC, Fierz B. Multivalency governs HP1α association dynamics with the silent chromatin state. Nat Commun. 2015;6:7313. doi: 10.1038/ncomms8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Larson AG, Elnatan D, Keenen MM, et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grewal SIS. The molecular basis of heterochromatin assembly and epigenetic inheritance. Mol Cell. 2023;83:1767–1785. doi: 10.1016/j.molcel.2023.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang L, Gao Y, Zheng X, et al. Histone Modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol Cell. 2019;76:646–659. doi: 10.1016/j.molcel.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 85.Gao Y, Han M, Shang S, Wang H, Qi LS. Interrogation of the dynamic properties of higher-order heterochromatin using CRISPR-dCas9. Mol Cell. 2021;81:4287–4299. doi: 10.1016/j.molcel.2021.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ngan CY, Wong CH, Tjong H, et al. Chromatin interaction analyses elucidate the roles of PRC2-bound silencers in mouse development. Nat Genet. 2020;52:264–272. doi: 10.1038/s41588-020-0581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tiwari VK, McGarvey KM, Licchesi JDF, et al. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biology. 2008;6:e306. doi: 10.1371/journal.pbio.0060306.eaec9ec67a2e48f1bfe73d4f56fde3e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Falk M, Feodorova Y, Naumova N, et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature. 2019;570:395–399. doi: 10.1038/s41586-019-1275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mirny LA. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011;19:37–51. doi: 10.1007/s10577-010-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haddad N, Jost D, Vaillant C. Perspectives: using polymer modeling to understand the formation and function of nuclear compartments. Chromosome Res. 2017;25:35–50. doi: 10.1007/s10577-016-9548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.MacPherson Q, Beltran B, Spakowitz AJ. Bottom-up modeling of chromatin segregation due to epigenetic modifications. Proc Natl Acad Sci U S A. 2018;115:12739–12744. doi: 10.1073/pnas.1812268115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 93.Erdel F, Greene EC. Generalized nucleation and looping model for epigenetic memory of histone modifications. Proc Natl Acad Sci U S A. 2016;113:E4180–E4189. doi: 10.1073/pnas.1605862113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Michieletto D, Orlandini E, Marenduzzo D. Polymer model with epigenetic recoloring reveals a pathway for the de novo establishment and 3D organization of chromatin domains. Physical Review X. 2016;6:041047. doi: 10.1103/PhysRevX.6.041047.eff39e6da81643309cb1f507c0516c4b [DOI] [Google Scholar]
- 95.Jeremy AO, Dino O, Leonid AM. Design principles of 3D epigenetic memory systems. bioRxiv. 2022:2022.2009.2024.509332. doi: 10.1101/2022.09.24.509332. [DOI] [Google Scholar]
- 96.Cheng TH, Gartenberg MR. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 2000;14:452–463. doi: 10.1101/gad.14.4.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ragunathan K, Jih G, Moazed D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015;348:1258699. doi: 10.1126/science.1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Laprell F, Finkl K, Müller J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science. 2017;356:85–88. doi: 10.1126/science.aai8266. [DOI] [PubMed] [Google Scholar]
- 99.Abdulla AZ, Salari H, Tortora MMC, Vaillant C, Jost D. 4D epigenomics: deciphering the coupling between genome folding and epigenomic regulation with biophysical modeling. Curr Opin Genet Dev. 2023;79:102033. doi: 10.1016/j.gde.2023.102033. [DOI] [PubMed] [Google Scholar]
- 100.Abdulla AZ, Vaillant C, Jost D. Painters in chromatin: a unified quantitative framework to systematically characterize epigenome regulation and memory. Nucleic Acids Res. 2022;50:9083–9104. doi: 10.1093/nar/gkac702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wells JN, Feschotte C. A field guide to eukaryotic transposable elements. Annu Rev Genet. 2020;54:539–561. doi: 10.1146/annurev-genet-040620-022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rebollo R, Karimi MM, Bilenky M, et al. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 2011;7:e1002301. doi: 10.1371/journal.pgen.1002301.83d45773e2f74a8090ed73f9132f3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Newar K, Abdulla AZ, Salari H, Fanchon E, Jost D. Dynamical modeling of the H3K27 epigenetic landscape in mouse embryonic stem cells. PLoS Comput Biol. 2022;18:e1010450. doi: 10.1371/journal.pcbi.1010450.c836880648214550a3d683c03ccc4dfd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Imakaev MV, Fudenberg G, Mirny LA. Modeling chromosomes: beyond pretty pictures. FEBS Lett. 2015;589:3031–3036. doi: 10.1016/j.febslet.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baù D, Marti-Renom MA. Genome structure determination via 3C-based data integration by the Integrative Modeling Platform. Methods (San Diego, Calif.) 2012;58:300–306. doi: 10.1016/j.ymeth.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 106.Marti-Renom MA, Mirny LA. Bridging the resolution gap in structural modeling of 3D genome organization. PLoS Comput Biol. 2011;7:e1002125. doi: 10.1371/journal.pcbi.1002125.af265b7146114274bffefb28f04c710e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tjong H, Li W, Kalhor R, et al. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proc Natl Acad Sci U S A. 2016;113:E1663–E1672. doi: 10.1073/pnas.1512577113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shukron O, Holcman D. Transient chromatin properties revealed by polymer models and stochastic simulations constructed from chromosomal capture data. PLoS Comput Biol. 2017;13:e1005469. doi: 10.1371/journal.pcbi.1005469.ed27e443586a4d8191183b3a4736956d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dodero-Rojas E, Mello MF, Brahmachari S, Oliveira Junior AB, Contessoto VG, Onuchic JN. PyMEGABASE: predicting cell-type-specific structural annotations of chromosomes using the epigenome. J Mol Biol. 2023;435:168180. doi: 10.1016/j.jmb.2023.168180. [DOI] [PubMed] [Google Scholar]
- 110.Xiong K, Ma J. Revealing Hi-C subcompartments by imputing inter-chromosomal chromatin interactions. Nat Commun. 2019;10:5069. doi: 10.1038/s41467-019-12954-4.fe7973997a0e4faa966b3de9f6798712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qi Y, Zhang B. Predicting three-dimensional genome organization with chromatin states. PLoS Comput Biol. 2019;15:e1007024. doi: 10.1371/journal.pcbi.1007024.fd2a42834ec54bb6bcb55fb9fd0eebe0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Di Pierro M, Cheng RR, Lieberman Aiden E, Wolynes PG, Onuchic JN. De novo prediction of human chromosome structures: epigenetic marking patterns encode genome architecture. Proc Natl Acad Sci U S A. 2017;114:12126–12131. doi: 10.1073/pnas.1714980114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ashoor H, Chen X, Rosikiewicz W, et al. Graph embedding and unsupervised learning predict genomic sub-compartments from HiC chromatin interaction data. Nat Commun. 2020;11:1173. doi: 10.1038/s41467-020-14974-x.c528a5276a3e4bc18703af1d3cef5bed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zheng S, Thakkar N, Harris HL, et al. Predicting A/B compartments from histone modifications using deep learning. bioRxiv. 2022:2022.2004.2019.488754. doi: 10.1101/2022.04.19.488754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luo Y, Hitz BC, Gabdank I, et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020;48:D882–d889. doi: 10.1093/nar/gkz1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Di Pierro M, Zhang B, Aiden EL, Wolynes PG, Onuchic JN. Transferable model for chromosome architecture. Proc Natl Acad Sci U S A. 2016;113:12168–12173. doi: 10.1073/pnas.1613607113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Contessoto VG, Cheng RR, Onuchic JN. Uncovering the statistical physics of 3D chromosomal organization using data-driven modeling. Curr Opin Struct Biol. 2022;75:102418. doi: 10.1016/j.sbi.2022.102418. [DOI] [PubMed] [Google Scholar]