Abstract
In the nature, Candida species are normal inhabitants and can be observed in a wide variety of vertebrates. In humans, especially for cancer patients who fall prey to opportunistic pathogens, this group of susceptible multi-drug resistant and biofilm-forming yeasts, are among the commonest ones. In this study, Candida species in 76 oral lesion samples from Vietnamese nasopharyngeal-cancer patients were isolated, morphologically identified using CHROMagar™, germ tube formation, and chlamydospore formation tests, and molecularly confirmed by PCR-RFLP. The drug susceptibility of these isolates was then tested, and the gene ERG11 was DNA sequenced to investigate the mechanism of resistance.
The results showed that Candida albicans remained the most prevalent species (63.16% of the cases), followed by Candida glabrata, Candida tropicalis, and Candida krusei. The rates of resistance of non-albicans Candida for tested drugs were 85.71%, 53.57%, and 57.14% to fluconazole, clotrimazole, and miconazole, respectively. Although the drug-resistance rate of Candida albicans was lower than that of non-albicans Candida, it was higher than expected, suggesting an emerging drug-resistance phenomenon. Furthermore, ERG11 DNA sequencing revealed different mutations (especially K128T), implying the presence of multiple resistance mechanisms. Altogether, the results indicate an alarming drug-resistance situation in Candida species in Vietnamese cancer patients and emphasize the importance of species identification and their drug susceptibility prior to treatment.
Keywords: Candida species, Drug-resistance susceptibility, Antifungal agents, ERG11
1. Introduction
Oral fungal infection is a common opportunistic disease in the human oral cavity caused by the overgrowth of different kinds of fungi. Most of them belong to the family of Candida species with the commonest being Candida albicans, which is responsible for over 50% infection cases [1,2]. Except Candida albicans, other Candida species can be also detected and termed non-albicans Candida, such as Candida glabrata (Nakaseomyces glabrataa), Candida tropicalis or Candida krusei (Pichia kudriavzevii) [3]. These non-albicans Candida are more frequent in old patients than in other groups and are widely recognized as harder to treat than Candida albicans (> 80 years old) [4,5]. In addition, these two types of pathogenic yeast appear in the list of fungal priority pathogens according to the World Health Organization [6].
Pathologically, oral candidiasis is considered the commonest infection caused by fungi and normally presented as white, soft, slightly elevated plaques on the tongue and buccal mucosa [7]. In terms of available treatments for oral candidiasis, there are four main groups of antifungal agents that are usually used, including polyene, azole, alkylamines thiocarbamates, and new drugs such as caspofungin [8]. Among these antifungal groups, the azole group is the most widely used in oral candidiasis. Its mechanism of action is to inhibit the synthesis of ergosterol, thereby disrupting fungal cell membrane. Within this group, fluconazole is preferably used due to its higher concentration in saliva compared to other members [9,10]. Although there are available effective treatment strategies for oral candidiasis, the emergence of drug resistance, especially for that of the azole group, is lowering treatment effectiveness and has drawn the attention of the scientific community [11]. Multiple mechanisms of azole resistance in C. albicans, especially in the case of fluconazole, are being studied. Firstly, the point mutations in the enconding gen ERG11 reduce the affinity between the target enzyme 14α-demethylase and azoles, increasing their minimum inhibitory concentration (MIC) [12]. Secondly, Candida drug resistance genes (CDR1 and CDR2) and multiple drug resistance ones (MDR1) were also deemed culprits of such resistance in the litterature [13,14].
In particular, the rates of drug resistance are particularly pronounced in immunodeficient patients, such as people living with HIV/AIDS or cancer, due to long-term use of preventative azole [15,16]. For instance, an Indian study has pointed out that the rate of fluconazole and itraconazole resistance in cancer patients undergoing chemo/radiotherapy can be up to 14% [17]. This fungal infection decreases dramatically the quality of life of cancer patients due to the pain caused by lesions in the oral cavity, leading to malnutrition, systematic infections and significantly increasing the mortality rate [[17], [18], [19]]. Besides, candidiasis has also been proven significantly associated with a higher risk of COVID-19 [20]. Furthermore, the azole-resistance phenomenon has also been recorded in the nature, especially in birds population. For instance, a study in France in 2016 pointed out that the seagulls may be a natural reservoir for C. glabrata, which can distribute both directly and indirrectly this pathogen to humans via animal hosts or environmental contamination [21]. In another study of Castelo-Branco DSCM et al., authors reconfirmed the connection between animals and humans in the development of azole resistance via efflux pumps and emphasized the necessity of one health solutions to tackle this problem [22].
Considering the alarming situation of drug resistance in candidiasis, this study is carried out to investigate the prevalence of different Candida species, the resistance rate of azole antifungal drugs in cancer patients undergoing radio/chemotherapy in Vietnam and the molecular characteristics of the ERG11 gene of some high-level fluconazole-resistant C. albicans in order to identify mutations of fluconazole resistance that may play the main role in Vietnamese patients.
2. Materials and methods
2.1. Materials
CHROMagar™ Candida, Muller-Hinton Agar (MHA), Sabouraud dextrose agar (SDA), Corn Meal Agar (CMA), RPMI 1640 were purchased from HiMedia (Kelton, PA, USA). Tween 80, sodium hydrocarbonate, sodium hydroxide were purchased from Merck (New Jersey, USA).
Fluconazole, clotrimazole, miconazole and nystatin, fluconazole powder were provided by an EU-GMP pharmaceutical factory in Vietnam with a purity superior to 99.99% (Boston Pharma., Ho Chi Minh, Vietnam).
2.2. Sampling method
Seventy-six samples of oral cancerous lesion in cancer patients (cohort characteristics detailed in Table S1.) undergoing radiotherapy and/or chemotherapy and without any fungal infection's symptoms at the Ho Chi Minh City Oncological Hospital were sampled using the swab method. The samples were stored in a physicological buffer at 20 °C prior to subsequent tests. The storage time is not over 24 h.
2.3. Isolation and morphological identification of Candida species
Candida species were isolated and identified using CHROMagar™ and conventional methods (germ tube test and chlamydospore formation).
CHROMagar™ method: Samples were directly streaked on CHROMagar™ Candida agar plates and incubated at 37 °C for 48 h. The results were observed, and the type of Candida was identified according to the manufacturer's instructions. In each test, the reference strain Candida albicans ATCC 10231, Candida tropicalis ATCC 13803, Candida glabrata ATCC 2001, Candida parapsilosis ATCC 22019, Candida krusei ATCC 14014 were used for quality control. Colonies were inoculated on Sabouraud Dextrose Agar plates (Peptone Casein 10 g/L, Dextrose 20 g/L, Agar 15 g/L) at 37 °C for 48 h before storing in 50% glycerol at −80 °C for further experiments.
Germ Tube Formation test: One colony of Candida spp. was dispersed in an Eppendorf containing 500 μL human serum, incubated at 37 °C for 2–4 h. After incubation time, a drop of yeast was transferred on a slide and stained with Fuchsin reactive for 10 min. The sample was finally checked with an optical microscope to determine whether a germ tube was formed (Positive) or not (Negative).
Chlamydospore formation test: one colony of Candida spp. was taken by an inoculating needle and then a straight line was drawn through the CMA petri dish (corn meal infusion 2 g/L, tween 80 7 mL/L, agar 15 g/L). The coverslip at the top of the line was flamed to reduce oxygen tension, this condition stimulates chlamydospore production. After being sealed with a paraffin tape, the plates were incubated aerobically at ambient temperature for 48–72 h. The chlamydospores formation was checked with an optical microscope.
2.4. Fungal culture conditions
The yeasts were grown by streaking frozen stocks, stored at −80 °C on SDA plates, followed by 36–48 h of incubation at 37 °C. One or two colonies were subsequently selected and used to inoculate a test tube containing 5 mL of SBD, which was then incubated overnight at 37 °C until the stationary phase was reached. The cell density was measured by a spectrophotometer at 530 nm. The OD of the tested samples was in a range of 1–3.
2.5. Fungal classification by PCR-RFLP
DNA from each obtained isolate was extracted using a modified method which was previously described by Retno Wahyuningsih et al. [23]. Briefly, one loop of yeast was harvested from SDA plate and diluted into 100 μL de-ionized water; then 455 μL lysis buffer (240 mM NaOH, 2.7 mM EDTA, 74% EtOH) was added into the mixture. The mixture was heated to 80 °C for 10 min and centrifuged at 13200 rpm for 10 min. The supernatant was discarded, and the pellet was collected and dispersed in 100 μL of Tris solution (0.1 mM EDTA, 50 mM Tris-HCl pH 8.0, 1% Triton-X100, 0.5% Tween 20). The final suspension was homogenously mixed and centrifuged at 12000 rpm for 15 min. The aqueous layer in the epperndorf tube was transferred into a fresh tube. DNA concentration of the sample was determined using UV–Vis spectrophotometer (Genequant 1300, Biochrom, USA). DNA of each Candida isolate was diluted to obtain a working concentration of 200 ng/μL.
The internal transcribed spacer (ITS) 1,4 was amplified by PCR reaction solution, consisting 17.3 μL NF-Q1, 2.5 μL iTag buffer, 1 mM dNTPs, 250 μM of each primer, 100 ng of target genomic DNA, and 1.0 U Taq polymerase. PCR reactions were performed with ITS1 fungal primers (5′- TTC GTA GGT GAA CCT GCG G - 3′) and ITS4 primers (5′ – TCC TCC GCT TAT TGA TAT GC - 3′) for forward and reverse primers, respectively. The PCR reactions were perfomed in SimpliAmp Thermal Cycler (Thermo Fisher Scientific, MA, USA). The system was programmed for 30 cycles. The PCR products were digested by restriction enzyme HpaII at 37 °C for 15 min. Afterwards, all obtained samples were analyzed with the 100 bp Opti-DNA (Applied Biological Materials Inc., Canada) by gel electrophoresis through a 10% polyacrygamide gel and revealed by a ultraviolet camera (TFX-20 LC, Vilber Lourmat, France). The isolates are classified according to the size of ITS band and products after digestion with HpaII (shown in Table S2 and Figure S1).
2.6. Determination of antifungal drug susceptibility of isolated Candida species
The drug susceptibility of nystatin and three azoles, including clotrimazole, fluconazole, and miconazole was determined for these isolated strains using the dish diffusion test (according to the M27 protocol), according to the reference method of the National Committee for Clinical Laboratory Standards (CLSI) [24,25]. After the incubation time of 48 h at 37 °C, the antifungal zones were analyzed and determined by the Fiji software [26].
2.7. Determine the fluconazole susceptibility of isolated Candida species
The rate of fluconazole resistance of the obtained strains was determined through their minimum inhibitory concentrations (MIC). To do so, the MIC of each fluconazole-resistant Candida albicans strain, which was identified by the dish diffusion test (Section 2.6), was determined using the microdilution method (according to the M27 protocol) in accordance with the reference method of the National Committee for Clinical Laboratory Standards (CLSI) to determine the susceptibility. The culture medium was RPMI 1640 with MOPS and 0.2% glucose and the results were performed after an incubation period of 48 h at 37 °C. The MIC of fluconazole is determined as the lowest concentration of fluconazole where there is no detected growth of Candida species [24,25].
2.8. Polymerase chain reaction amplification and gene sequences of the ERG11 gene
The ERG11 genes of all high fluconazole resistant (MIC >128 μL/mL) were amplified using PCR method. The PCR reaction solution consisted 17.3 μL NF-Q1, 2.5 μL iTag buffer, 1 mM dNTPs, 250 μM of each primer, 100 ng of target genomic DNA, and 0.5 U Taq polymerase. PCR reactions were performed using ERG11 primers: 5′- TTA GTG TTT TAT TGG ATT CCT TGG TT- 3′ and 5′ -TTA AAA CAT ACA AGT TTC TCT TTT TTC CCA AAT G- 3′, as forward and reverse ones, respectively. The PCR results were then analyzed with the 1000 bp DNA ladder (iNtRON Biotechnology Inc., Korea) by the gel electrophoresis method using 1% agarose gel, and results were revealed by a ultraviolet camera (TFX-20 LC, Vilber Lourmat, France). ERG11 DNA sequences were analyzed by SANGER sequencing on ABI3500 system (Thermalfisher, USA). The data of DNA sequences were treated and verified by Lasergene v7.0 software. The data of obtained ERG11 DNA sequences were verified by MEGA software v11.0 to detect eventual mutations.
2.9. Ethicals aspects
All sampling procedures constructed in the current study were reviewed and accepted by the ethics committee of the Ho Chi Minh City Oncological Hospital.
3. Results
3.1. Identification of clinical Candida species in Vietnamese nasopharyngeal-cancer patients
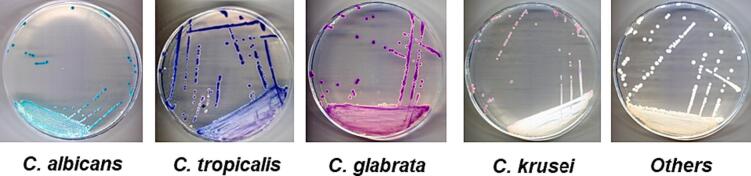
A total of 76 samples were successfully collected from oral lesions in Vietnamese cancer patients undergoing radiotherapy and/or chemotherapy at the Ho Chi Minh City Oncological Hospital. CHROMagar™ Candida, a selective chromogenic culture medium for the direct qualitative detection and identification of Candida spp. based on the color of colonies, was used in this study [27]. As shown in Fig. 1, each fungal colony was presented with a specific color, revealing its type. Indeed, Candida albicans was detected by green colonies and those of Candida tropicalis, Candida glabrata and Candida krusei were in metallic blue, brown, and pink, respectively. In total, five types of Candida spp. were identified in our samples. In addition, other kinds of Candida spp., which were usually present in a negligible proportion of Candida spp. in public, were also detected in our samples and presented as white colonies.
Fig. 1.
Candida spp. colonies color on CHROMagar™ Candida plates.
In the second step, the proportion of each type of Candida spp. was determined and presented in.
Table 1. The results revealed that C. albicans remained the most prominent species with 63.16%, followed by C. krusei (11.84%), C. glabrata (9.21%), C. tropicalis (6.58%) and other species (9.21%). The total proportion of non-albicans Candida was determined to be 36.84%.
Table 1.
Color of colonies for each kind of Candida spp. on CHROMagar™ Candida and its proportion.
| Type of Candida spp. | Number of samples |
|---|---|
| Candida albicans | 48 (63.16%) |
| Candida krusei | 9 (11.84%) |
| Candida glabrata | 7 (9.21%) |
| Candida tropicalis | 5 (6.58%) |
| Other species | 7 (9.21%) |
To further confirm the results obtained with the CHROMagar™ method, germ tube formation and chlamydospores tests, the two most conventional methods, were deployed, mainly to differentiate Candida albicans from non-albicans Candida.
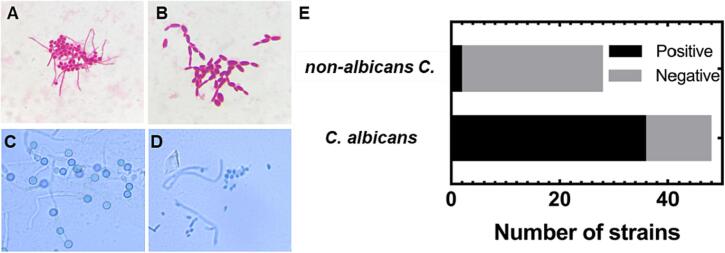
In the germ tube formation test, the results were interpreted as positive when the filamentous outgrowth extending from yeast cells was detected (Fig. 2A). In that case, the main type of yeast present in the tested sample was Candida albicans or Candida dubliniensis. Otherwise, if there was no filamentous extension or only short filamentous extensions limited at the central point were detected, the result was interpreted as negative (as shown in Fig. 2B), and the corresponding type of yeast was C. tropicalis, C. glabrata or other non-albicans Candida spp. [28]. For a total of 76 tested samples, the rate of positive-negative for this test was 1:1 (38 positive results and 38 negative results). In comparison of these results to that of the CHROMagar™ method, a difference in the rate of Candida albicans was detected. Specifically, among the strains that had green colonies on CHROMagar™, there were 12 out of 36 strains (33.33%) that had a negative result in the germ tube formation test.
Fig. 2.
Morphological identification of Candida species using the germ tube formation method. (A) Image of a positive germ tube of Candida albicans. (B) Image of a negative germ tube of non-albicans Candida. (C) Image of chlamydospores of Candida albicans. (D) Image of yeast and hyphae, lacking chlamydospores. (E) Numbers of isolated stains showing positive and negative germ tubes when compared with results on CHROMagar™ Candida.
As a third method of identification, the capacity of producing chlamydospores of the obtained clinical strains was studied. Indeed, the chlamydospores (as shown in Fig. 2C and D) are thick-wall cells that arise on elongated suspensor cells, situated on pseudohyphae or hyphae by pathogen Candida albicans [29]. To efficiently induce chlamydospores, cells were distributed at low cell concentrations on nutrient-poor media supplemented with detergents, such as Tween 80, and incubated at a low temperature (25 to 30 °C) for 48 h. Chlamydospore formation was especially abundant on solid media under glass coverslips, providing microaerophilic conditions (Dalmau inoculation technique) [29,30]. Herein, Corn Meal Agar with Tween 80 to support Candida albicans producing chlamydospores was used as culture medium. As shown in Fig. 2E, the results indicated that all Candida albicans samples (producing green colonies on CHROMagar™) produced chlamydopores after 48 h at ambiant temperature and all non-albicans Candida samples detected by the CHROMagar™ method did not produce chlamydopores.
Finally, to confirm the identification of 76 strains of Candida spp., the PCR-RFLP method using the ITS gene and the HpaII enzyme was used. The results were shown in Table 2. Indeed, all C. albicans samples detected with CHROMagar™ were reconfirmed in the PCR-RFLP method. Whereas only 38 were identified by the germ tube formation test. Furthermore, a good specificity of CHROMagar™ for non-albicans Candida was also demonstrated, especially in the case of C. glabrata (seven strains).
Table 2.
Number of Candida strains identified by CHROMagar™ Candida, germ tube formation and PCR-RFLP methods.
| Name | CHROMagar™ Candida | Germ tube formation test | PCR-RFLP |
|---|---|---|---|
| C. albicans | 48 | 38 | 48 |
| C. tropicalis | 5 | 38 | 8 |
| C. krusei | 9 | 4 | |
| C. glabrata | 7 | 8 | |
| C. parasilosis | 0 | 8 | |
| Others | 7 | 0 |
3.2. Evaluation of antifungal drug-resistance rate of clinical Candida species in Vietnamese cancer patients
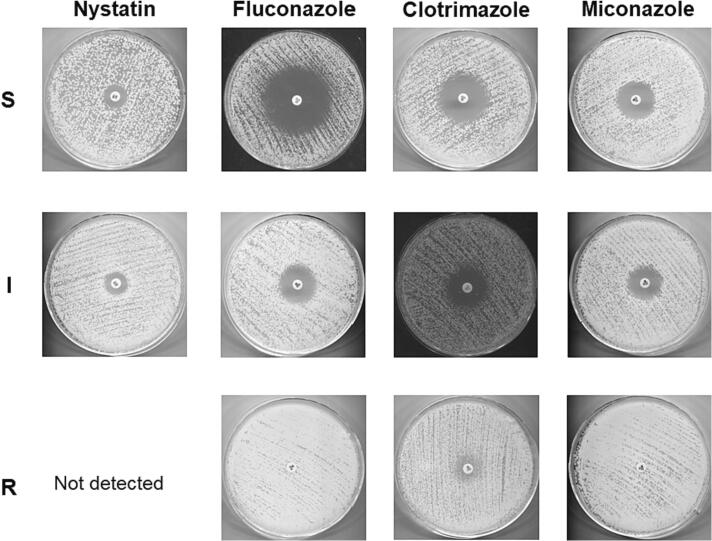
The M44 method, an antifungal disc diffusion test for yeasts, was used in this study to evaluate the drug susceptibility of the obtained samples with four main antifungal drugs which are widely used in oral candidiasis treatment, including nystatin, fluconazole, clotrimazole, and miconazole [24,25]. The susceptibility level of each drug in our samples was identified in accordance with the reference method of the National Committee for Clinical Laboratory Standards (CLSI) and divided into three levels, including sensitive, intermediate and resistant depending on the antifungal zone diameter (Fig. 3) [24,25].
Fig. 3.
Determination of drug susceptibility of the tested strains with nystain, fluconazole, clotrimazole, and miconazole using the M27 method.
As shown in Table 3, in both kinds of yeasts (C. albicans or non-albicans Candida), the resistance rate was the most pronounced for fluconazole (52.08% and 85.71%), followed by clotrimazole (6.25% and 53.57%) and miconazole (2.08% and 57.14%), respectively. Nystatin remained the only antifungal drug not resisted by both kinds of yeasts. However, the intermediate level was relatively high (33.33% and 42.86% for C. albicans and non-albicans Candida, respectively), revealing a shift from the sensitive state to the resistant state. The results also clearly indicated a difference in drug susceptibility between C. albicans and non-albicans Candida. Indeed, a higher resistance rate for non-albicans Candida than that of C. albicans was recorded for all drugs in the azole group, including fluconazole (85.71% vs 52.08%), clotrimazole (53.57% vs 6.25%), and miconazole (57.14% vs 2.08%), respectively.
Table 3.
Drug susceptibility of the tested clinical strains with nystatin, fluconazole, clotrimazole and miconazole using the M27 method (S: Sensitive, I: Intermediate, R: Resistant).
| Drug | Test Conc./ disk |
Percentatage of susceptibility C. albicans (%) |
Percentatage of susceptibility non-albicans Candida (%) |
||||
|---|---|---|---|---|---|---|---|
| S | I | R | S | I | R | ||
| Nystatin | 100 IU | 66.67% (32/48) | 33.33% (16/48) | 0 | 57.14% (16/28) |
42.86% (12/28) |
0 |
| Fluconazole | 25 μg | 43.75% (21/48) |
4.17% (2/48) |
52.08% (25/48) |
7.14% (2/28) |
7.14% (2/28) |
85.71% (24/28) |
| Clotrimazole | 10 μg | 45.83% (22/48) |
47.92% (23/48) |
6.25% (3/48) |
28.57% (8/28) |
17.86% (5/28) |
53.57% (15/28) |
| Miconazole | 10 μg | 18.75% (9/48) |
79.17% (38/48) |
2.08% (1/48) |
0 | 42.86% (12/28) |
57.14% (16/28) |
3.3. Molecular characteristic of the ERG11 gene of clinical Candida albicans
Among the tested antifungal agents, fluconazole showed the highest resistant rate on both Candida albicans and non-albicans Candida (52.08% and 85.71%, respectively). Since fluconazole is the most widely used drug in clinical settings, the following section focuses more on the resistance mechanism of this drug in the tested samples. We used the microdilution method that is in accordance with the M27 guidance to determine the minimum inhibitory concentration (MIC) of fluconazole on 76 strains and to determine their drug susceptibility (Table 4). Indeed, the numbers of sensive, intermediate and resistant strains were 30, 6, and 40, respectively. For sensitive strains, most of them were C. albicans (28 versus 2 of non-albicans Candida). For resistant strains, 11 were C. albicans, whereas 24 samples were non-albicans Candida, revealing a more alarming resistant situation for non-albicans Candida. Especially, all non-albicans Candida resistant samples had an MIC superior to 128 μg/mL.
Table 4.
The drug susceptibility pattern of isolated Candida spp. according to their MIC with fluconazole (S: Sensitive, I: Intermediate, R: Resistant).
| MIC of fluconazole (μg/mL) | Number of samples |
Total | ||
|---|---|---|---|---|
| C. albicans | non-albicans Candida | |||
| S | ≤ 0.25 | 4 | 0 | 30 |
| 0.5–1 | 16 | 0 | ||
| 2 | 8 | 2 | ||
| I | 4 | 4 | 2 | 6 |
| R | 8 | 4 | 0 | 40 |
| 16 | 0 | 0 | ||
| 32 | 1 | 0 | ||
| 64 | 0 | 0 | ||
| ≥ 128 | 11 | 24 | ||
According to the level of resistance, 11 strains of Candida albicans which had an MIC superior to 128 μL/mL were analyzed for their ERG11 sequencing in an attempt to understand their mechanism of resistance (Fig. 4). In comparison with the two sequences of fluconazole-sensitive Candida albicans strains, one from ATCC 10231 of the NCBI genbank and the other from our clinical strains, two point mutations were detected. The first mutation is K128T (K128 changed to T128), corresponding to a codon change (at 382–384) from AAA to ACA. The second one is E448G (E448 changed to G448), corresponding to a codon change (at 1342–1344) from GAG to GGG. However, the mutaion E448G happened not only on resistant strains but also on sensitive strains, and is, therefore, not likely to be involved in the fluconazole-resistant mechanism. Only the mutation K128T seemed to be linked to azole resistance, as also reported in past studies. Among 11 resistant strains, there were only three (15XL, 109, and 128) where the K128T mutation was observed.
Fig. 4.
ERG11 DNA sequences of fluconazole-sensitive and fluconazole-resistant Candida albicans strains. KM881482.1: standard Candida albicans ATCC 10231 from the NCBI genbank. Species 27: clinical fluconazole-sensitive C. albicans strain. Species 94, 53, 26, 55, 92–2, 109, 32, 128, 15XL, 99, 38: clinical fluconazole-resistant C. albicans strains with MIC >128 μg/mL.
4. Discussion
4.1. The population distribution of Candida spp. in the oral lesion of Vietnamese cancer patients undergoing chemotherapy/radiotherapy
Candida spp. are the leading opportunistic agents of fungal infections. Currently, it is widely recognized that the most prevalent Candida species in both healthy and infected human mouths is C. albicans, which is present in >80% of oral fungal isolates [31]. C. albicans is followed in descending order by C. glabrata, C. krusei, C. tropicalis, C. guilliermondii, C. kefyr and C. parapsilosis [31]. Although the symptoms of infections by non-albicans Candida are smiliar to those of C. albicans, non-albicans Candida species show high virulence factors and a higher rate of drug resistance. Therefore, it is important to determine the population of clinical Candida strains in order to decide the most appropriate treatment strategy. Recently, there is a shift in the methods used for classifying Candida clinical strains: from classic methods to DNA amplification-based ones. Nevertheless, conventional diagnostic methods still remain the most widely used thanks to their ease of use and low costs [32]. In contrary, these conventional methods are labor-intensive and time-consuming. In addition, they cannot identify precisely all species of Candida and do not have a high accuracy level like molecular methods [33]. In traditional clinical laboratories, the presumptive identification of isolates, the germ tube test is more frequent than the CHROMagar™ due to its quite good sensitivity, rapidity, and low cost [34]. However, the sensitivity and specificity of CHROMagar™ was shown to be better [34]. The only weakness of the CHROMagar™ method was that at earlier times, C. tropicalis isolates can give false-positive results [34]. Other methods for the identification of pathogenic yeasts such as MALDI-ToF MS analysis can also be used [35]. However, due to its non-availability in Vietnam, this method was not applied in our research. In this study, only CHROMagar™ and the germ tube test were applied and their difference was detected, suggesting that an appropriate combination of different methods or more sensitive ones is needed to further confirm the results obtained with these methods. As a result, two conventional methods (CHROMagar and chladyospore reproduction) and a rapid method (germ tube test) were firstly used to identify the population proportion of clinical Candida strains. Afterwards, PCR-RFLP, a molecular method, was used to reconfirm the results obtained with the conventional methods.
As shown in the results of CHROMagar™ (Table 1) and further confirmed by the germ tube formation method (Fig. 2), although a similar order of the population distribution of Candida spp. has been found, there was a drastic decrease in the proportion of C. albicans (from >80% to 63.16% or 46.05%) and a significant increase of non-albicans Candida proportion (from <20% to 36.84% or 53.95%) in our samples. On the one hand, this result revealed that there was a change in the population distribution of Candida spp. in cancer patients, and the population of non-albicans Candida tends to increase in such immune-compromised patients. In the literature, a similar phenomenon was observed around the world. For instance, in a study in 2007 in children and adolescents with cancer in Venezuela, C. albicans was also present in only 42.55% of oral candidiasis cases [36]. In another similar study carried out in India and published in 2016, whose objects were cancer patients undergoing chemo/radiotherapy, a proportion of 58% of C. albicans was recorded [17]. In other studies, this proportion was also found to be around 50% [37]. All these results suggest that there is an increase in the proportion of non-albicans Candida in cancer patients. This situation may be explained by the daily use of antifungal agents by these patients as a prevention method to candidiasis [38]. However, this trend is highly alarming as the drug-resistance rate of non-albicans Candida is higher than that of C. albicans [39]. Moreover, as non-albicans Candida is intrinsically resistant to azole agents, the only choice of treatment is to use another group of drugs, which is more expensive and more toxic. In particular, C. krusei and C. glabrata, which are resistant to fluconazole (a global resistance rate of 78.3%), were found to be present in >20% of our clinical samples [39]. Furthermore, upon comparing our results with those of a Tanzanian study published in 2006 on HIV-infected patients with oral candidiasis, we observed a significant increase in the population of non-albicans Candida. To be specific, in that study, C. albicans was present in 84.5% of patients but was only around 50% in our study or in the aforementioned studies for cancer patients [40]. This remark may indicate that the population distribution of Candida spp. is different between HIV/AIDS patients and cancer patients, and may be useful for physicians to decide the prevention method for oral candidiasis. In another study in 2018 of Narges Aslani et al., a lower proportion of C. albicans (50.6%) and a higher proportion of non-albicans Candida (47.3%) were observed for Iranian cancer patients [41]. This difference may be explained by the divergence in geological and environmental parameters. To the best of our knowledge, there is no current publication on the population distribution of Candida spp. in oral candidiasis in Vietnamese cancer patients. Therefore, our study may be useful for Vietnamese physicians to better manage oral candidiasis in cancer patients in Vietnam.
4.2. Drug susceptibility of C. albicans and non-albicans Candida in clinical Vietnamese cancer patients
Nowadays, the emergence of antifungal resistance is highly alarming and considered one of the biggest concerns in the fight against fungal infections in humans [42]. Among available antifungal drugs for the treatment of Candidiasis, the azole group is the most used but also the most resisted. Therefore, a thorough understanding of the drug resistance rate of this group of drugs is indispensible. In the current study, no isolated strain was resistant to nystatin, which is a good sign. The rate of drug resistance is ascending in the following order: nystatin < miconazole < clotrimazole < fluconazole. As a result, the resistance rate of fluconazole, which is also the most used drug for oral candidiasis, is the highest (51% for C. albicans and 86% for non-albicans Candida). In a recent study in India and in Iran, this rate was only around 20% [37,43]. This result demonstrates that fluconazole resistance in Vietnamese patients was relatively high compared to other countries. Our study also suggests that it is important to identify the Candida species before determining the treatment. For example, if the main source of oral infections is Candida non-albicans, it would be better to use nystatin than azoles. Besides, a higher resistance rate to azole drugs of non-albicans Candida than that of C. albicans was also shown in the current study. These results suggest the necessity of Candida species determination before deciding the treatment strategy.
4.3. Mechanism of fluconazole resistance of Candida albicans in Vietnamese cancer patients
In order to better understand the mechanism of resistance of C. albicans in Vietnamese cancer patients, the ERG11 gene of our clinical fluconazole-resistant C. albicans strains was sequenced and compared to that of clinical fluconazole-sensitive C. albicans strains and the standard from the NCBI genbank. In total, two point mutations, including K128T (at codon 382–384) and E448G (at codon 1342–1344), were detected in our clinical samples in comparision with the standard strain. Only the K128T mutation was shown to be connected with the resistance phenomenon and it was observed in three out of 11 tested samples. On the one hand, this finding was quite typical for the Vietnamese population, as K128T was not listed as a mutation related directly to fluconazole resistance in a study of Urbanek et al. [44]. In the litterature, several mutations have been proven to be direclty linked to fluconazole resistance in C. albicans, such as K143R, G464S, G465S, V488I, S412T, R469K, G487T, T916C, Y18D, E243D, L370S, P375H [40,[44], [45], [46], [47]]. This finding suggests that the K128T mutation may play also a role in the development of fluconazole resistance, especially for the Vietnamese cancer patients. On the other hand, the presence of this mutation in only three out of a total of 11 resistant strains suggests that other unknown mechanisms of resistance might play the main role in fluconazole resistance in Vietnamese cancer patients and need further clarification.
5. Conclusion
As both Candida albicans and non-albicans Candida species are currently listed in the WHO fungal priority pathogens list, the current study is the first study in Vietnam that has focused on the population distribution of Candida spp. in Vietnamese cancer patients as well as their drug susceptibility and its potential mechanism. Several results have been pointed out in this study, as follows.
Firstly, the study has reconfirmed the significant increase of non-albicans Candida in oral candidiasis in Vietnamese cancer patients, which threatens the treatment effectiveness of the azole group, currently the first choice of treatment for oral candidiasis in Vietnam. Besides, the effectiveness of CHROMagar™ Candida, a conventional method for Candida identification, was reconfirmed by PCR-RFLP. However, a combination of these methods remains necessary in order to enhance the outcomes of the treatment.
Secondly, the drug resistance rate of these clinical samples highlighted the importance of identifying Candida strains before the treatment strategy is decided, especially when azole drugs are prescribed.
Eventually, using the molecular method, our study revealed for the first time in Vietnam that the K128T mutation was connected with the fluconazole resistance in C. albicans in Vietnamese cancer patients but further confirmation is still needed.
Declaration of Competing Interest
The authors declare no conflicts of interest. All the experiments undertaken in this study comply with the current laws of the country where they were performed.
Acknowledgments
This research is funded by Vietnam National University Ho Chi Minh City (VNU-HCM) under grant number No. C2023-44-16. We thank Dr. Nguyen Thi Thu Hoai, Research Center for Infectious Diseases, International University, Vietnam National University Ho Chi Minh City, for providing her expertise in experiments with yeasts.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2023.100659.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- 1.Akpan A., Morgan R. Oral candidiasis. Postgrad. Med. J. 2002;78:455–459. doi: 10.1136/pmj.78.922.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh A., Verma R., Murari A., Agrawal A. Oral candidiasis: an overview. J. Oral Maxillofac. Pathol. JOMFP. 2014;18:S81–S85. doi: 10.4103/0973-029X.141325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borman A.M., Johnson E.M. Name Changes for Fungi of Medical Importance, 2018 to 2019. J. Clin. Microbiol. 2021;59:e01811–e01820. doi: 10.1128/JCM.01811-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Černáková L., Líšková A., Lengyelová L., Rodrigues C.F. Prevalence and antifungal susceptibility profile of Oral Candida spp. isolates from a Hospital in Slovakia. Med. Kaunas Lith. 2022;58:576. doi: 10.3390/medicina58050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benito-Cruz B., Aranda-Romo S., López-Esqueda F.J., de la Rosa-García E., Rosas-Hernández R., Sánchez-Vargas L.O. Oral Candida isolates and fluconazole susceptibility patterns in older Mexican women. Arch. Gerontol. Geriatr. 2016;65:204–210. doi: 10.1016/j.archger.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 6.WHO TEAM WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action. 2022. https://www.who.int/publications/i/item/9789240060241
- 7.A.N. R, N.B. Rafiq . StatPearls. StatPearls Publishing; Treasure Island (FL): 2022. Candidiasis.http://www.ncbi.nlm.nih.gov/books/NBK560624/ (accessed July 22, 2022) [Google Scholar]
- 8.Quindós G., Gil-Alonso S., Marcos-Arias C., Sevillano E., Mateo E., Jauregizar N., Eraso E. Therapeutic tools for oral candidiasis: current and new antifungal drugs. Med. Oral Patol. Oral Cir. Bucal. 2019;24:e172–e180. doi: 10.4317/medoral.22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pappas P.G., Kauffman C.A., Andes D., Benjamin D.K., Calandra T.F., Edwards J.E., Filler S.G., Fisher J.F., Kullberg B.-J., Ostrosky-Zeichner L., Reboli A.C., Rex J.H., Walsh T.J., Sobel J.D., Infectious Diseases Society of America Clinical practice guidelines for the management of candidiasis: 2009 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2009;48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rai A., Misra S.R., Panda S., Sokolowski G., Mishra L., Das R., Lapinska B. Nystatin effectiveness in Oral candidiasis treatment: a Systematic Review & Meta-Analysis of clinical trials. Life. 2022;12:1677. doi: 10.3390/life12111677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whaley S.G., Berkow E.L., Rybak J.M., Nishimoto A.T., Barker K.S., Rogers P.D. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front. Microbiol. 2016;7:2173. doi: 10.3389/fmicb.2016.02173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang M.-J., Liu J.-Y., Ni P.-H., Wang S., Shi C., Wei B., Ni Y.-X., Ge H.-L. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 2013;13:386–393. doi: 10.1111/1567-1364.12042. [DOI] [PubMed] [Google Scholar]
- 13.Tsao S., Rahkhoodaee F., Raymond M. Relative contributions of the Candida albicans ABC transporters Cdr1p and Cdr2p to clinical azole resistance. Antimicrob. Agents Chemother. 2009;53:1344–1352. doi: 10.1128/AAC.00926-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White T.C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 1997;41:1482–1487. doi: 10.1128/AAC.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaitán-Cepeda L.A., Sánchez-Vargas O., Castillo N. Prevalence of oral candidiasis in HIV/AIDS children in highly active antiretroviral therapy era. A literature analysis. Int. J. STD AIDS. 2015;26:625–632. doi: 10.1177/0956462414548906. [DOI] [PubMed] [Google Scholar]
- 16.Chitapanarux I., Wongsrita S., Sripan P., Kongsupapsiri P., Phakoetsuk P., Chachvarat S., Kittidachanan K. An underestimated pitfall of oral candidiasis in head and neck cancer patients undergoing radiotherapy: an observation study. BMC Oral Health. 2021;21:353. doi: 10.1186/s12903-021-01721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayachandran A.L., Katragadda R., Thyagarajan R., Vajravelu L., Manikesi S., Kaliappan S., Jayachandran B. Oral candidiasis among Cancer patients attending a tertiary Care Hospital in Chennai, South India: an evaluation of Clinicomycological association and antifungal susceptibility pattern. Can. J. Infect. Dis. Med. Microbiol. J. Can. Mal. Infect. Microbiol. Medicale. 2016;2016:8758461. doi: 10.1155/2016/8758461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeGregorio M.W., Lee W.M., Ries C.A. Candida infections in patients with acute leukemia: ineffectiveness of nystatin prophylaxis and relationship between oropharyngeal and systemic candidiasis. Cancer. 1982;50:2780–2784. doi: 10.1002/1097-0142(19821215)50:12<2780::aid-cncr2820501215>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 19.Jain M., Shah R., Chandolia B., Mathur A., Chauhan Y., Chawda J., Mosby S., Bhagalia S. The Oral carriage of Candida in Oral Cancer patients of Indian origin undergoing radiotherapy and/or chemotherapy. J. Clin. Diagn. Res. JCDR. 2016;10:ZC17–20. doi: 10.7860/JCDR/2016/15702.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz J. Prevalence of candidiasis and oral candidiasis in COVID-19 patients: a cross-sectional pilot study from the patients’ registry in a large health center. Quintessence Int. Berl. Ger. 2021;1985(52):714–718. doi: 10.3290/j.qi.b1491959. [DOI] [PubMed] [Google Scholar]
- 21.Al-Yasiri M.H., Normand A.-C., L’Ollivier C., Lachaud L., Bourgeois N., Rebaudet S., Piarroux R., Mauffrey J.-F., Ranque S. Opportunistic fungal pathogen Candida glabrata circulates between humans and yellow-legged gulls. Sci. Rep. 2016;6:36157. doi: 10.1038/srep36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castelo-Branco D.D.S.C.M., Paiva M.D.A.N., Teixeira C.E.C., Caetano É.P., Guedes G.M.D.M., Cordeiro R.D.A., Brilhante R.S.N., Rocha M.F.G., Sidrim J.J.C. Azole resistance in Candida from animals calls for the one health approach to tackle the emergence of antimicrobial resistance. Med. Mycol. 2020;58:896–905. doi: 10.1093/mmy/myz135. [DOI] [PubMed] [Google Scholar]
- 23.Wahyuningsih R., Freisleben H.-J., Sonntag H.-G., Schnitzler P. Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis. J. Clin. Microbiol. 2000;38:3016–3021. doi: 10.1128/JCM.38.8.3016-3021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fothergill A.W. In: Interact. Yeasts Moulds Antifung. Hall G.S., editor. Agents, Humana Press; Totowa, NJ: 2012. Antifungal susceptibility testing: Clinical laboratory and standards institute (CLSI) methods; pp. 65–74. [DOI] [Google Scholar]
- 25.Nielsen Bookdata, Place of Publication Not Identified. 3rd edition. 2018. M44: method for antifungal disk diffusion susceptibility testing of yeasts. [Google Scholar]
- 26.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borman A.M., Fraser M., Johnson E.M. CHROMagarTM Candida plus: a novel chromogenic agar that permits the rapid identification of Candida auris. Med. Mycol. 2021;59:253–258. doi: 10.1093/mmy/myaa049. [DOI] [PubMed] [Google Scholar]
- 28.Matare T., Nziramasanga P., Gwanzura L., Robertson V. Experimental germ tube induction in Candida albicans : an evaluation of the effect of sodium bicarbonate on morphogenesis and comparison with pooled human serum. Biomed. Res. Int. 2017;2017:1–5. doi: 10.1155/2017/1976273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonneborn A., Bockmühl D.P., Ernst J.F. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 1999;67:5514–5517. doi: 10.1128/IAI.67.10.5514-5517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingle S., Kodgire S., Shiradhone A., Patil R., Zore G. Chlamydospore specific proteins of Candida albicans. Data. 2017;2:26. doi: 10.3390/data2030026. [DOI] [Google Scholar]
- 31.Taylor M., Brizuela M., Raja A. StatPearls. StatPearls Publishing; Treasure Island (FL): 2023. Oral candidiasis.http://www.ncbi.nlm.nih.gov/books/NBK545282/ (accessed May 6, 2023) [Google Scholar]
- 32.Pincus D.H., Orenga S., Chatellier S. Yeast identification – past, present, and future methods. Med. Mycol. 2007;45:97–121. doi: 10.1080/13693780601059936. [DOI] [PubMed] [Google Scholar]
- 33.Burton M.J., Swiatlo E., Shah P. Misidentification of Candida parapsilosis as C famata in a clinical case of vertebral osteomyelitis. Am J Med Sci. 2011;341:71–73. doi: 10.1097/MAJ.0b013e3181f54dab. [DOI] [PubMed] [Google Scholar]
- 34.Merlino J., Tambosis E., Veal D. Chromogenic tube test for presumptive identification or confirmation of isolates as Candida albicans. J. Clin. Microbiol. 1998;36:1157–1159. doi: 10.1128/JCM.36.4.1157-1159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser M., Brown Z., Houldsworth M., Borman A.M., Johnson E.M. Rapid identification of 6328 isolates of pathogenic yeasts using MALDI-ToF MS and a simplified, rapid extraction procedure that is compatible with the Bruker Biotyper platform and database. Med. Mycol. 2015:myv085. doi: 10.1093/mmy/myv085. [DOI] [PubMed] [Google Scholar]
- 36.González Gravina H., González de Morán E., Zambrano O., Lozano Chourio M., Rodríguez de Valero S., Robertis S., Mesa L. Oral candidiasis in children and adolescents with cancer. Identification of Candida spp. Med. Oral Patol. Oral Cir. Bucal. 2007;12:E419–E423. [PubMed] [Google Scholar]
- 37.Talukdar A., Barman R., Sarma A., Krishnatreya M., Sharma J., Kataki A. Fungal profile and antifungal susceptibility pattern of candidiasis in esophageal cancer patients. J. Cancer Res. Ther. 2020;16:209. doi: 10.4103/jcrt.JCRT_581_18. [DOI] [PubMed] [Google Scholar]
- 38.Worthington H., Eden O., Clarkson J. The Cochrane Collaboration (Ed.), Cochrane Database Syst. Rev. John Wiley & Sons, Ltd; Chichester, UK: 2004. Interventions for preventing oral candidiasis for patients with cancer receiving treatment. CD003807.pub2. [DOI] [PubMed] [Google Scholar]
- 39.Sanguinetti M., Posteraro B., Lass-Flörl C. Antifungal drug resistance among Candida species: mechanisms and clinical impact. Mycoses. 2015;58:2–13. doi: 10.1111/myc.12330. [DOI] [PubMed] [Google Scholar]
- 40.Hamza O.J., Matee M.I., Moshi M.J., Simon E.N., Mugusi F., Mikx F.H., Helderman W., Rijs A.J., van der Ven A.J., Verweij P.E. Species distribution and in vitro antifungal susceptibility of oral yeast isolates from Tanzanian HIV-infected patients with primary and recurrent oropharyngeal candidiasis. BMC Microbiol. 2008;8:135. doi: 10.1186/1471-2180-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aslani N., Janbabaei G., Abastabar M., Meis J.F., Babaeian M., Khodavaisy S., Boekhout T., Badali H. Identification of uncommon oral yeasts from cancer patients by MALDI-TOF mass spectrometry. BMC Infect. Dis. 2018;18:24. doi: 10.1186/s12879-017-2916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carvalhinho S., Costa A.M., Coelho A.C., Martins E., Sampaio A. Susceptibilities of Candida albicans mouth isolates to antifungal agents, essentials oils and mouth rinses. Mycopathologia. 2012;174:69–76. doi: 10.1007/s11046-012-9520-4. [DOI] [PubMed] [Google Scholar]
- 43.Mahdavi Omran S., Rezaei Dastjerdi M., Zuashkiani M., Moqarabzadeh V., Taghizadeh-Armaki M. In vitro antifungal susceptibility of Candida species isolated from Iranian patients with denture stomatitis. Biomed. Res. Int. 2018;2018:1–6. doi: 10.1155/2018/3086586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urbanek A.K., Łapińska Z., Derkacz D., Krasowska A. The role of ERG11 point mutations in the resistance of Candida albicans to fluconazole in the presence of lactate. Pathog. Basel Switz. 2022;11:1289. doi: 10.3390/pathogens11111289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul S., Kannan I., Mohanram K. Extensive ERG11 mutations associated with fluconazole-resistant Candida albicans isolated from HIV-infected patients. Curr. Med. Mycol. 2019;5:1–6. doi: 10.18502/cmm.5.3.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y., Sheng F., Zhao J., Chen L., Li C. ERG11 mutations and expression of resistance genes in fluconazole-resistant Candida albicans isolates. Arch. Microbiol. 2015;197:1087–1093. doi: 10.1007/s00203-015-1146-8. [DOI] [PubMed] [Google Scholar]
- 47.Odiba A.S., Durojaye O.A., Ezeonu I.M., Mgbeahuruike A.C., Nwanguma B.C. A new variant of mutational and polymorphic signatures in the ERG11 gene of fluconazole-resistant Candida albicans. Infect. Drug Resist. 2022;15:3111–3133. doi: 10.2147/IDR.S360973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.