Abstract
Ataxia Telangiectasia (A-T) is an inherited autosomal recessive disorder characterized by cerebellar neurodegeneration, radiosensitivity, immunodeficiency and a high incidence of lymphomas. A-T is caused by mutations in the ATM gene. While loss of ATM function in DNA repair explains some aspects of A-T pathophysiology such as radiosensitivity and cancer predisposition, other A-T features such as neurodegeneration imply additional roles for ATM outside the nucleus. Emerging evidence suggests that ATM participates in cellular response to oxidative stress, failure of which contributes to the neurodegeneration associated with A-T. Here, we use fibroblasts derived from A-T patients to investigate whether DEAD Box 1 (DDX1), an RNA binding/unwinding protein that functions downstream of ATM in DNA double strand break repair, also plays a role in ATM-dependent cellular response to oxidative stress. Focusing on DDX1 target RNAs that are associated with neurological disorders and oxidative stress response, we show that ATM is required for increased binding of DDX1 to its target RNAs in the presence of arsenite-induced oxidative stress. Our results indicate that DDX1 functions downstream of ATM by protecting specific mRNAs in the cytoplasm of arsenite-treated cells. In keeping with a role for ATM and DDX1 in oxidative stress, levels of reactive oxygen species (ROS) are increased in ATM-deficient as well as DDX1-depleted cells. We propose that reduced levels of cytoplasmic DDX1 RNA targets sensitizes ATM-deficient cells to oxidative stress resulting in increased cell death. This sensitization would be especially detrimental to long-lived highly metabolically active cells such as neurons providing a possible explanation for the neurodegenerative defects associated with A-T.
Keywords: Ataxia Telangiectasia, Oxidative stress, Neurodegeneration, DEAD box protein 1, ATM, RNA protection
Graphical abstract
Highlights
-
•
DDX1 target RNAs encode proteins required for stress recovery and neuronal health.
-
•
Arsenite-treated A-T fibroblasts show reduced binding of DDX1 to its target RNAs.
-
•
ATM deficiency reduces the levels of DDX1 target RNAs in cells under stress.
-
•
ATM and DDX1 function in the same RNA protection pathway.
-
•
DDX1 or ATM loss increases ROS levels and decreases survival in cells under stress.
Abbreviations
- (ALS)
Amyotrophic Lateral Sclerosis
- (ALS2)
Alsin Rho guanine nucleotide exchange factor
- (AMPK)
AMP-activated protein kinase
- (As)
Arsenite
- (A-T)
Ataxia Telangiectasia
- (ATM)
A-T Mutated
- (ATXN2)
Ataxin 1
- (DDX)
DEAD Box
- (DCF-DA)
Dichlorodihydrofluorescein diacetate
- (EWSR1)
Ewing sarcoma RNA binding protein 1
- (FMR1)
Fragile X messenger ribonucleoprotein
- (FXR1)
FMR1 autosomal homolog 1
- (FXTAS)
Fragile X-associated Tremor Ataxia Syndrome
- (m6A)
N6-methyladenosine
- (NQO1)
NAD(P)H quinone dehydrogenase 1
- (OGDH)
Oxoglutarate dehydrogenase
- (OXSR1)
Oxidative stress responsive kinase 1
- (PSEN1)
Presenilin 1
- (RIPseq)
RNA immunoprecipitation and sequencing
- (SG)
Stress granules
- (STAU2)
Staufen double-stranded RNA binding protein 2
- (SOD1)
Superoxide dismutase 1
- (STK11)
Serine/threonine kinase 11
- (TAF15)
TATA-box binding protein associated factor 15
- (TDP43)
TAR DNA binding protein
- (TXN)
Thioredoxin
1. Introduction
Ataxia Telangiectasia (A-T) is a rare autosomal recessive disorder that affects multiple systems in the body and is typically diagnosed in childhood. A-T is characterized by progressive cerebellar ataxia affecting movement coordination and balance, oculocutaneous telangiectasia, extreme radiosensitivity, immunodeficiency, premature aging, predisposition to certain malignancies, especially lymphoma and leukemia, and respiratory diseases [1,2]. While patients often die from cancer or respiratory infections, progressive neurodegeneration is the root cause of this disease. Studies have shown that oxidative stress plays a significant role in the pathogenesis of A-T, contributing to the progressive cerebellar ataxia and neurodegeneration seen in affected individuals [3,4].
Oxidative stress is a physiological imbalance between the production of reactive oxygen species (ROS) and free radicals, and the ability of the cells to detoxify them. Cellular ROS are mainly generated from oxidative phosphorylation in mitochondria, with exogenous factors such as xenobiotic compounds, pollutants, alcohols, many drugs and radiation also producing ROS [5,6]. Excessive ROS production can overwhelm the cellular antioxidant defense system leading to cellular dysfunction and damage. Due to their high oxygen demands and production of highly peroxidizable substrates, neurons are especially vulnerable to ROS. Elevated ROS in neurons may result in neuroinflammation, impaired mitochondrial function, and neuronal cell death [7]. Increased ROS levels and/or impaired antioxidant systems have been implicated in the pathogenesis of cancer and neurodegenerative diseases, including Alzheimer's disease, Parkinson's disease, and Ataxia Telangiectasia [4,[8], [9], [10]].
A-T is caused by mutation in the ATM gene (Ataxia Telangiectasia Mutated), which encodes a key protein in DNA damage response and repair [11]. Upon introduction of DNA double-strand breaks (DSBs), the MRN complex is recruited to sites of DNA damage [12], which in turn activates ATM by autophosphorylation resulting in its recruitment to DSBs. ATM then phosphorylates and recruits repair proteins to the DSB site [13,14]. The RNA helicase DEAD Box 1 (DDX1) is also phosphorylated by ATM and recruited to a subset of DSBs where it facilitates DSB repair by clearing RNA from these sites [[15], [16], [17]]. DDX1 is a member of the DEAD box protein family of RNA helicases implicated in the unwinding, splicing, stability and transport of RNAs [[18], [19], [20], [21], [22], [23], [24]]. DDX1 is co-amplified with MYCN in pediatric cancers and high levels of DDX1 have been associated with a poor prognosis in breast cancer [25,26]. DDX1 is also essential for embryonic development in mice where it plays a role in the control of calcium-dependent mitochondrial activity [27,28]. DDX1 has been associated with stress-induced regulation of splicing in Drosophila melanogaster [29].
ATM has previously been shown to be activated by autophosphorylation upon oxidative stress resulting in up-regulation of signaling pathways involved in cellular homeostasis [[30], [31], [32], [33]]. These studies have revealed elevated ROS levels and neuronal cell death associated with absence of ATM activity. A role for cytoplasmic DDX1 in mitochondria, stress granules and oxidative stress response has also been reported by our lab and others [28,34,35]. We have recently used DDX1-RNA immunoprecipitation to identify DDX1 mRNA targets in the osteosarcoma cell line U2OS and DDX1-amplified neuroblastoma cell line BE(2)-C. We found that DDX1 stabilizes its target RNAs in cells exposed to oxidative stress and facilitates the resolution of stress granules [35].
Here, we investigate the relationship between ATM and DDX1 in the cellular response to oxidative stress in A-T patient fibroblasts. We find that depletion of either DDX1 or ATM in arsenite-treated fibroblasts inhibits their recovery from oxidative stress and increases ROS levels. We also characterize several DDX1 RNA targets that encode proteins involved in oxidative stress response that are associated with neurological disorders and are involved in oxidative stress response, including TDP43, TAF15, FXR1, ALS2 and ATXN2. Binding of DDX1 to these RNAs is increased when cells are under oxidative stress, thereby protecting these RNAs from degradation. We show that ATM deficiency or depletion results in reduced cytoplasmic levels of DDX1 RNA targets. Furthermore, cell survival after oxidative stress is reduced upon depletion of either DDX1 or ATM. Our results demonstrate a role for ATM in maintaining cellular homeostasis via DDX1 in fibroblasts under oxidative stress.
2. Materials and methods
2.1. Cells and treatments
A-T fibroblasts: AT2BE (from a 7-year old female) and AT3BI (from a 4-year old male) were derived from skin biopsies of A-T patients and are ATM negative [36]. AT2BE was purchased from the American Type Culture Collection, Rockville, MD, and AT3BI was kindly provided by Dr. A. Lehmann, University of Sussex, Brighton, UK. GM38 are normal human fibroblasts derived from a 9-year old female [36]. GM10 is a normal fibroblast culture of human fetal origin [37]. GM38, GM10 and A-T fibroblasts were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 °C with 5% CO2. For acute oxidative stress, cells were treated with 0.5 mM sodium arsenite (Sigma-Aldrich) for 45 min (unless specified otherwise). This concentration of arsenite is commonly used to induce oxidative stress in cells, including human fibroblasts [34,35,38]. For chronic oxidative stress, cells were treated with 2 μM sodium arsenite for 16 h.
2.2. DDX1 and/or ATM knockdowns
DDX1 and ATM were knocked down in fibroblasts by siRNA transfection using lipofectamine RNAiMax (Thermo Fisher Scientific) and 10 nM siRNAs. For negative controls, scrambled siRNAs (Low GC and Medium GC) were used. For DDX1 knockdowns, two different siRNAs were used, with single siRNAs used for each transfection: si1DDX1 (CAGGCUGAAUCUAUCCCAUUGAUCU) and si2DDX1 (UACACCAUGUUGUU GUCCCAGUAAA) in two rounds of transfection as described [35]. For ATM knockdown, si1ATM (CAAGGCUAUUCAGUGUGCGAGACAA) and si2 ATM (AGUCUAGUACUUAAUGAUCUGCUUA) were used. Cells were analyzed 72 h after transfection.
2.3. Measurement of reactive oxygen species
Reactive Oxygen Species (ROS) were analyzed using the following methods: (i) the cell-permeable, non-fluorescent probe 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA, Molecular probes) which is converted into fluorescent 2,7-dichlorofluorescein in the presence of oxidizing species and (ii) the superoxide indicator MitoSOX-Red (Thermo Fisher Scientific) which specifically measures mitochondrial ROS. Untreated and arsenite-treated cells were trypsinized, washed and stained with 10 μM H2DCF-DA for 30 min at 37 °C or 5 μM MitoSOX-Red in HBSS for 30 min at 37 °C. Fluorescence was measured using a flow cytometer (FACS Canto II, BD Biosciences) with the excitation wavelength set at 488 nm and the emission wavelength set at 535 nm for DCF-DA or 590 nm for MitoSOX-Red. The data were analyzed using BD FACS Diva software. Unstained cells served as a negative control for autofluorescence. Mean fluorescence was used as representative of ROS levels in the cells.
2.4. Fluorescence microscopy
Cells cultured on coverslips were fixed and processed as previously described [15,35]. Cells were incubated with a rabbit anti-DDX1 antibody (batch 2923; 1:1000 dilution) [35,39] followed by Alexa 488-conjugated donkey anti-rabbit secondary antibody (Life Technologies). Coverslips were mounted onto slides in polyvinyl alcohol (Calbiochem)-based mounting medium containing 1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI). Images were acquired using a Zeiss LM710 confocal microscope, exported as TIFF files using ZEN and assembled using Photoshop software.
2.5. RNA immunoprecipitation (RIP) and quantitative RT-PCR
Fibroblasts were either treated with 0.5 mM sodium arsenite for 45 min or left untreated. Cells were washed with PBS and RNAs were crosslinked to proteins using UV crosslinker at 200 mJ/cm2. Whole cell lysates were prepared by resuspending the cells in lysis buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 0.5% NP-40, 0.5% sodium deoxycholate, 5 mM EDTA, 2 mM DTT, 0.5 U/μl RNase inhibitor and 1X Complete protease inhibitor), followed by centrifugation at 12,000 rpm for 10 min and collecting the supernatant. Five hundred μg cell lysate was used to pull-down DDX1-bound RNAs with anti-DDX1 antibody (batch 2910) [16,35] for 1.5 h at 4 °C followed by incubation with Protein A Sepharose beads (GE Healthcare) for another 1.5 h at 4 °C with end-to-end rotation. Rabbit IgG was used as the negative control. Unbound fractions were removed, and beads were washed with wash buffer (50 mM Tris-Cl pH7.5, 1 M NaCl, 1% NP-40, and 1% sodium deoxycholate). Any contaminating DNA was removed by incubating the beads with 12 units of Turbo DNase (Thermo Fisher Scientific) at 37 °C for 30 min. Bound RNAs were separated from beads by digesting the proteins with Proteinase K (Roche) at 37 °C for 30 min. RNAs were extracted with phenol:chloroform (1:1), ethanol precipitated and dissolved in 20 μl RNase-free water.
To prepare cDNA, 5 μl RNA eluted from the RNA-immunoprecipitations was reverse transcribed using Superscript IV reverse transcriptase (Thermo Fisher Scientific) and random hexamer primers. Quantitative RT-PCR was performed with SYBR Green qPCR mix (Applied Biological Materials Inc.) and the primers listed in Table S1. For normalization purposes, RNA was extracted from cells grown under the same conditions as those used for RNA immunoprecipitations. Total RNA was reverse transcribed and used to determine cellular levels of each RNA targets under each condition. RNA enrichment in RNA immunoprecipitations was normalized against individual RNA levels under the same conditions. GAPDH served as the internal control.
2.6. Cell fractionation and measurement of cytoplasmic RNA and protein levels
Cytoplasmic and nuclear fractions of fibroblasts were prepared as described before [35]. Briefly, untreated and arsenite-treated fibroblasts were washed, trypsinized and lysed in cytoplasmic extraction buffer (25 mM Tris-HCl pH 7.5, 10 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, 2 mM EDTA, 2 mM DTT, 1X cOmplete protease inhibitor cocktail and 0.2 U/μl RNase inhibitor) for 5 min at 4 °C with end-to-end rotation, vortexed, kept on ice for 1 min and centrifuged at 1500 rpm for 5 min. The supernatant was collected as cytoplasmic fraction. For protein analysis, 30 μg of each cytoplasmic fraction was loaded on an 8% SDS-PAGE gel, transferred to nitrocellulose membrane, and immunoblotted with anti-DDX1 antibody (1:5000, batch 2910) or anti-GAPDH antibody (1:2000, Thermo Fisher Scientific) as cytoplasmic marker. For ATM, we used low-bis acrylamide SDS-PAGE gels. The anti-ATM (1:1000) antibody used for western blotting has been previously described [16]. For RNA analysis, total RNA from each cytoplasmic fraction was extracted with Trizol LS (Thermo Fisher Scientific). Purified RNA was dissolved in 20 μl of RNase-free water and 2 μg of RNA was used to prepare cDNA by reverse transcription using Superscript II reverse transcriptase and oligo-dT primers. RT-qPCR was performed using SYBR Green qPCR mix to determine the levels of cytoplasmic RNAs under different conditions. GAPDH served as the internal control.
2.7. In situ cell death (TUNEL) assay
To check the viability of cells under control and stress conditions, apoptosis was examined using the Cell Death Detection kit, Fluorescein (Roche) using the manufacturer's protocol. Fibroblasts untreated or treated with 0.5 mM arsenite for 45 min were trypsinized, washed and pelleted. The cells were fixed in 2% paraformaldehyde for 1 h at room temperature with constant rotation, washed with PBS and permeabilized in 0.1% Triton X-100 for 2 min on ice. The cells were then washed and stained with a mixture of TdT-terminal transferase enzyme and TUNEL assay buffer including dUTP-fluorescein isothiocyanate (FITC) for 1 h at 37 °C. Negative controls were prepared by incubating cells in assay buffer only. Positive controls were prepared by treating the DNA with DNase I for 10 min at 37 °C to cause DNA breaks prior to staining with TdT-terminal transferase and TUNEL assay buffer mixture. The cells were washed with PBS and resuspended in 500 μl PBS and acquired on FACS Canto II (BD Biosciences) at 488 nm excitation and 520 nm emission. Analysis was done using BD FACS Diva software.
2.8. Cell survival assay
Cell survival was analyzed using the clonogenic assay [40]. Cells were counted using a hemocytometer and 100 cells/well were seeded in 6-well plates in DMEM supplemented with 10% fetal calf serum. After 24 h, cells were treated with increasing concentrations of arsenite (0, 0.1, 0.2, 0.3, 0.4 and 0.5 mM) in duplicate wells for 45 min. Arsenite-containing medium was removed, and cells were washed twice with PBS. The cells were cultured in fresh medium at 37 °C with 5% CO2 and colonies allowed to form over 2–3 weeks. For staining, the medium was removed, and cells were washed with PBS. Cells were stained with 1% (w/v) crystal violet prepared in 70% ethanol for at least 30 min. The staining solution was removed, and the plates were washed with water. The plates were dried, and the number of colony-forming units was estimated by counting the number of colonies per well. Percent plating efficiency (PE) or colony forming units for each culture was calculated as follows:
2.9. Statistical analysis
Each experiment was done at least three times (N = 3). The p-value was calculated using two-sided Student's t-test. p<0.05 was used as a cut-off for significance.
3. Results
3.1. ATM and DDX1 deficiency increases ROS levels in fibroblasts
Neurodegenerative diseases and cancers have been linked to both genotoxic and environmental stress. While the studies carried out to date in A-T have mostly centered around DNA repair, recent studies have implicated elevated oxidative stress in the pathogenesis of A-T. We used fibroblasts derived from two different A-T patients (AT2BE and AT3BI) and from a child with no obvious physical or mental health problems (GM38; defined as normal) to explore their response to oxidative pressure by analyzing ROS accumulation under normal and stress conditions. The fibroblasts were either left untreated or were treated with 0.5 mM arsenite for 45 min to induce oxidative stress. Cells were stained with DCF-DA to measure total cellular ROS levels and analyzed by flow cytometry. Arsenite-treated GM38 fibroblasts had 1.5-fold higher levels of ROS compared to untreated cells. Consistent with previous reports [4,9], untreated AT2BE and AT3BI showed increased ROS levels (1.2-fold) compared to normal fibroblasts. Furthermore, A-T fibroblasts treated with arsenite underwent increased ROS accumulation compared to arsenite-treated GM38 fibroblasts (1.9-fold compared to 1.5-fold) (Fig. 1A). To verify that increased susceptibility to oxidative stress in A-T fibroblasts is due to the absence of ATM, we depleted ATM in normal GM38 fibroblasts using siRNA transfection (Fig. 1B) followed by arsenite exposure. The increase in ROS levels in ATM-depleted cells was similar to that observed in A-T fibroblasts upon arsenite treatment (Fig. 1A). These results indicate that depletion of ATM renders fibroblasts more vulnerable to oxidative stress.
Fig. 1.
ATM and DDX1 control accumulation of ROS in fibroblasts. Normal and ATM-deficient fibroblasts were treated with 0.5 mM arsenite for 45 min and stained with DCF-DA to measure ROS accumulation (mean fluorescence) by flow cytometry. (A) Bar diagrams showing mean DCF-DA fluorescence in untreated versus arsenite-treated GM38, AT2BE, AT3BI and ATM-depleted GM38 fibroblasts using two different ATM siRNAs. Increased ROS accumulation was associated with ATM deficiency. (B) Western blot analysis showing depletion of ATM in AT2BE, AT3BI and ATM siRNA-depleted GM38 fibroblasts. No changes in DDX1 levels were observed in ATM-depleted cells. GAPDH served as the loading control. (C) Bar diagrams showing mean DCF-DA fluorescence in untreated versus treated GM38 and A-T control and DDX1 knockdown cells using two different DDX1 siRNAs. Increased ROS accumulation was observed in arsenite-treated GM38 upon DDX1 depletion, but not in arsenite-treated DDX1-depleted A-T cells. Error bars depict standard deviation. Individual experiments are indicated by black circles.
*p<0.05, **p<0.01, ns: not significant (p≥0.05) (N = 3).
As DDX1 is a substrate of ATM and previously implicated in stress responses [16,35], we depleted DDX1 in GM38 using two different siRNAs, followed by arsenite treatment. Similar to what was observed in ATM-depleted cells, DDX1 knockdown in untreated cells resulted in a 1.2-fold increase in ROS levels under control conditions, and a 1.8-fold increase in ROS levels in arsenite-treated cells (Fig. 1C). We also investigated whether knockdown of DDX1 in ATM-deficient A-T fibroblasts might exacerbate oxidative stress. No significant changes in ROS accumulation were observed in arsenite-treated DDX1-depleted A-T fibroblasts compared to A-T fibroblasts. These results indicate that both ATM and DDX1 are involved in the regulation of cellular ROS levels and suggest that ATM and DDX1 may function in the same pathway, with ATM residing upstream of DDX1. ATM deficiency did not affect DDX1 levels in either A-T fibroblasts or ATM siRNA-depleted GM38 fibroblasts (Fig. 1B).
Next, we measured ROS levels in mitochondria using MitoSOX-Red. We observed a 1.3-fold increase in mitochondrial ROS in untreated A-T fibroblasts and ATM-depleted GM38 fibroblasts compared to untreated GM38 fibroblasts (Supplementary Fig. 1A). Upon arsenite treatment, mitochondrial ROS accumulation in fibroblasts increased in a dose-dependent manner. There was a 1.2-fold increase in ROS accumulation in both ATM-proficient GM38 and ATM-deficient fibroblasts in cells treated with 0.1 mM arsenite (Supplementary Fig. 1A). When cells were treated with 0.4 mM arsenite, mitochondrial ROS levels were increased 1.6-fold for ATM-proficient GM38 and 1.9-fold in ATM-deficient fibroblasts. Arsenite treatment at 0.5 mM concentration resulted in a 2.2 to 2.4-fold increase in mitochondrial ROS levels in ATM-depleted cells (GM38 and A-T fibroblasts) compared to 1.6-fold in arsenite-treated GM38 cells. All subsequent experiments were carried out with 0.5 mM arsenite, as commonly used to induce acute oxidative stress [34,35,38].
Similar to ATM depletion, mitochondrial ROS levels increased 1.3-fold upon depletion of DDX1 in GM38 fibroblasts, but not in DDX1-depleted A-T fibroblasts (Supplementary Fig. 1B). These results are in agreement with the results obtained with DCF-DA, and suggest roles for both ATM and DDX1 in ROS homeostasis, with DDX1 residing downstream of ATM.
We also examined mitochondrial ROS accumulation induced by chronic stress, with cells treated with 2 μM arsenite for 16 h. In GM38 cells, chronic stress led to a 1.2-fold increase in mitochondrial ROS levels (Supplementary Fig. 2). In contrast, DDX1-depleted GM38 cells showed a 1.4- to 1.5-fold increase in mitochondrial ROS levels. Similar increases (1.4-1.6-fold) in mitochondrial ROS levels were observed in A-T fibroblasts treated with 2 μM arsenite. DDX1 depletion in A-T fibroblasts did not cause further increases in mitochondrial ROS levels (Supplementary Fig. 2). Our combined results indicate that DDX1 and ATM are involved in the regulation of mitochondrial ROS accumulation in cells exposed to either acute or chronic stress.
We next asked whether the subcellular distribution of DDX1 was affected by ATM deficiency in control and arsenite-treated cells. DDX1 subcellular distribution was indistinguishable in untreated GM38, AT2BE and AT3BI fibroblasts, with a predominantly nuclear localization and localization to nuclear bodies (Supplementary Fig. 3A). As previously reported, arsenite treatment resulted in DDX1 accumulation in cytoplasmic stress granules [34,35]. No differences were observed in the subcellular localization of DDX1 in ATM-proficient GM38 fibroblasts compared to ATM-deficient AT2BE and AT3BI fibroblasts upon treatment with 0.5 mM arsenite (Supplementary Fig. 3B).
3.2. Identification of DDX1 target RNAs and effect of arsenite on DDX1-RNA binding
RNA binding proteins have been implicated in the pathogenesis of many neurodegenerative diseases, including Alzheimer's disease, Parkinson's disease, and Ataxia Telangiectasia [[41], [42], [43]]. Previous work from our lab identified mRNAs bound to and protected by the RNA helicase, DDX1, under oxidative stress conditions [35]. DDX1 bound RNAs were immunoprecipitated from U2OS cells using IgG (as negative control) or anti-DDX1 antibody, and DDX1-bound RNAs were sequenced and analyzed for differential binding [35]. Many of the identified RNAs were found to encode proteins with role(s) in the maintenance of cellular homeostasis under stress conditions. DDX1 depletion resulted in increased stress-induced reductions in the levels of DDX1 RNA targets, suggesting a role for DDX1 in protecting cells from oxidative stress and death.
Here, we extend the above study by asking whether there is a link between ATM and DDX1-RNA binding activity in Ataxia Telangiectasia. We selected 13 putative DDX1-RNA targets (labeled ‘targets’) with a demonstrated role in oxidative stress response in neurodegenerative diseases (ATXN2, STAU2, ALS2, PSEN1, OXSR1, FMR1, FXR1, TAF15, TDP43, DDX3X, ATM, EWSR1, and TXN) [41,44,45]. Targets were selected based on >10-fold sequence enrichment in DDX1-RNA immunoprecipitates compared to IgG control. Five genes were selected as negative controls based on similar read counts in DDX1-RNA immunoprecipitates compared to IgG control (SOD1, NQO1, STK11, AMPK, and OGDH; labeled ‘controls’). We first examined DDX1 binding to these 18 mRNAs in GM38 fibroblasts using either anti-DDX1 or IgG for RNA immunoprecipitations, followed by RT-qPCR analysis. RNA enrichment in the DDX1 immunoprecipitates compared to IgG control (set at 1) varied from ∼6X (for TXN) to ∼27X (for TAF15) for the 13 targets, with eleven of 13 targets showing >10-fold enrichment compared to the IgG control (Fig. 2A). Consistent with the sequencing data, all five controls showed enrichment of <5-fold compared to IgG control (Fig. 2A).
Fig. 2.
Increased binding of DDX1 to target RNAs upon arsenite treatment in GM38 fibroblasts. (A) RNA-DDX1 immunoprecipitation was used to measure levels of 13 target RNAs and 5 control RNAs bound to DDX1 in GM38 fibroblasts. Immunoprecipitated RNAs were quantified by RT-qPCR. Bar diagrams show fold RNA enrichment in DDX1-RIP compared to IgG-RIP (set at 1). (B) Bar diagrams showing fold enrichment in DDX1-RNA binding (relative to IgG control) in GM38 fibroblasts untreated (taken from A) and GM38 fibroblasts treated with arsenite for 45 min. Fold enrichment for arsenite-treated cells was normalized by taking account the change in RNA levels observed upon RT-qPCR analysis of total RNA extracted from treated cells relative to untreated cells. Inset shows western blot analysis of DDX1 in DDX1-RIP and IgG-RIP. 10% of input, IgG or DDX1 immunoprecipitates were loaded in the indicated lanes. Individual experiments are indicated by black circles. Error bars represent standard deviation. *p<0.05, **p<0.01, ***p<0.001 (N = 3).
We next tested the effect of oxidative stress on DDX1-RNA binding by exposing GM38 fibroblasts to arsenite, followed by DDX1 or control IgG RNA immunoprecipitation. To offset any effects that arsenite might have on levels of target or control RNAs, we normalized RNA enrichment in DDX1 RNA-immunoprecipitates to the levels of each RNA present in total RNA extracted from arsenite-treated cells. Nine of the 13 target RNAs showed a ≥2-fold increase in DDX1 binding upon arsenite treatment compared to RNA binding in control cells, with all values shown in relation to the IgG control (Fig. 2B). For example, binding of ATXN2 RNA to DDX1 was ∼17X higher than binding to IgG (Fig. 2B; dark blue bar). Upon arsenite treatment, DDX1-bound ATXN2 RNA levels increased to ∼38X (light blue bar), a 2.2X increase over control cells. The most dramatic change was the 7.6X increase in TDP43 RNA binding observed upon arsenite treatment. Two of our 13 selected targets, EWSR1 and TXN, showed no increase in DDX1 binding upon arsenite exposure. There was no significant increase in the binding of DDX1 to the five control RNAs upon exposure to arsenite. Of note, control RNAs were selected because of their documented roles in oxidative stress response, indicating that only a subset of stress related RNAs bind to DDX1 and are induced upon stress.
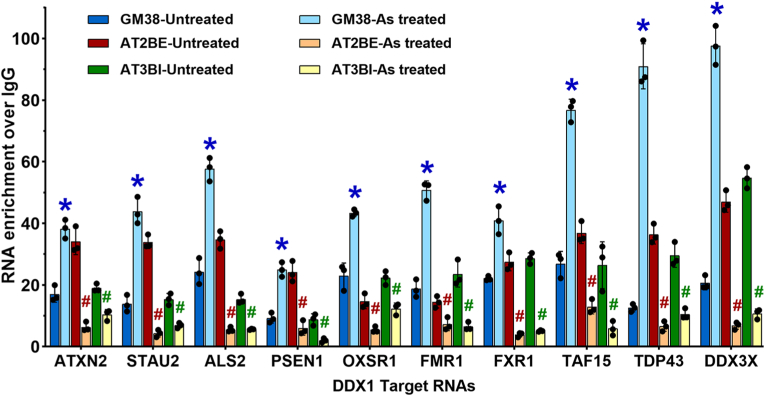
3.3. DDX1-RNA binding is defective in cells derived from A-T patients
The underlying cause of A-T is loss of activity of the well-characterized DNA repair protein, ATM. ATM has also been implicated in cellular response to environmental stressors such as oxidative stress, pointing to a possible mechanism for the neurological disorders associated with A-T [10,46]. As DDX1 is a substrate of ATM and has previously been implicated in oxidative stress response, we examined DDX1's RNA binding activity in A-T fibroblasts from two different patients, AT2BE and AT3BI. RNA immunoprecipitations were carried out with DDX1 and IgG (control), followed by RT-qPCR analysis of 10 selected targets. Three RNA targets were left out of this analysis: ATM as this gene is deficient in A-T cells, and EWSR1 and TXN, as binding to these two RNAs was not significantly altered upon arsenite-treated in GM38 fibroblasts. Similar to that observed in GM38, DDX1 effectively bound to its target RNAs in both A-T fibroblast cultures [Fig. 3, blue (GM38), red (AT2BE) and green (AT3BI) bars]. In contrast to GM38, exposure of A-T fibroblasts to arsenite resulted in a marked reduction in RNA binding, by 50% or greater (≥2-fold) for all RNAs [Fig. 3, light blue (GM38), orange (AT2BE) and yellow (AT3BI) bars] with the exception of ATXN2 RNA in AT3BI which was reduced by 45% (1.8-fold reduction) [Fig. 3, green (untreated AT3BI) versus yellow (arsenite treated AT3BI) bars]. In AT2BE, STAU2, ALS2, FXR1 and DDX3X binding to DDX1 was reduced by >85% [Fig. 3, red (untreated AT2BE) versus orange (arsenite treated AT2BE) bars]. In AT3BI, PSEN1, FXR1, TAF15 and DDX3X binding was reduced by >75% [Fig. 3, green (untreated AT3BI) versus yellow (arsenite treated AT3BI) bars]. These results suggest a link between the absence of ATM in A-T fibroblasts and reduced binding of DDX1 to its target RNAs under oxidative stress.
Fig. 3.
Defective DDX1-RNA binding in A-T patient-derived fibroblasts. GM38 and A-T fibroblasts (AT2BE and AT3BI) were treated with 0.5 mM arsenite or left untreated and used for RIP using IgG or anti-DDX1 as described before. Bar diagrams show fold RNA enrichment in DDX1-RIP compared to IgG-RIP (set at 1). GM38-untreated and GM38-arsenite treated values are taken from Fig. 2. Normalizations for AT2BE and AT3BI samples were as described in Fig. 2 using total RNA extracts. Blue asterisks represent significant increases in RNA binding in GM38 upon arsenite treatment compared to untreated GM38 cells, whereas red and green hashtags represent significant decreases in RNA binding in AT2BE and AT3BI, respectively, upon arsenite treatment compared to untreated fibroblasts. Individual experiments are indicated by black circles. Error bars represent standard deviation. * or #p<0.01 (N = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. ATM deficiency reduces DDX1-RNA binding under stress
We next addressed whether the dramatic decrease in DDX1 binding to its target RNAs observed in A-T fibroblasts might be a consequence of ATM deficiency in these cells. We have already shown interdependency between ATM and DDX1 through ATM phosphorylation of DDX1 and recruitment of DDX1 to DNA DSBs in cells exposed to radiation [15,16]. We therefore knocked down ATM in GM38 fibroblasts using two different siRNAs to test the effect of ATM deficiency on binding of DDX1 to its target RNAs upon oxidative stress in normal fibroblasts. As previously reported in Fig. 2, RT-qPCR analysis showed efficient binding of DDX1 to its RNA targets in control GM38 fibroblasts (siControl), with increased binding observed upon arsenite treatment (Fig. 4, dark blue versus light blue bars). Importantly, arsenite treatment resulted in a significant loss in DDX1-RNA binding in ATM-depleted GM38 fibroblasts (Fig. 4, brown versus orange and purple versus pink bars), in keeping with the results observed in A-T fibroblasts. To ensure that the observed effects were not specific to GM38 fibroblasts, we repeated these experiments with a second normal human fibroblast culture, GM10, and obtained similar results upon ATM depletion (Supplementary Fig. 4). The loss in DDX1-RNA binding observed in A-T cells under oxidative stress suggests a direct link to ATM deficiency.
Fig. 4.
ATM is required for optimal DDX1-RNA binding in arsenite-treated normal fibroblasts.ATM was knocked down in GM38 using two siRNAs (si1ATM and si2ATM). Control and ATM-depleted cells were left untreated or treated with arsenite, and RIP carried out as described above. Bar diagrams show fold RNA enrichment in DDX1-RIP compared to IgG-RIP (set at 1). Normalizations were as described in Fig. 2 using total RNA extracts from the relevant cultures. Blue asterisks represent a significant increase in RNA binding in GM38 upon arsenite treatment compared to untreated cells. Brown and purple hashtags represent significant decreases in RNA binding in ATM knockdown cells upon arsenite treatment compared to their untreated controls. Individual experiments are indicated by black circles. Error bars represent standard deviation. * or #p<0.01 (N = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Although there was no morphological indication of cell death in our arsenite-treated A-T cultures, we considered the possibility that loss of DDX1 RNA-binding activity in A-T fibroblasts upon arsenite treatment might be due to cell death. To address this possibility, we carried out the TUNEL assay, an apoptosis assay based on DNA fragmentation. The TUNEL assay was performed in A-T fibroblasts and GM38 cells transfected with either vehicle control or DDX1 or ATM siRNAs, followed by exposure to 0.5 mM arsenite for 45 min. Similar fluorescence intensities were observed in all cell populations, with or without arsenite treatment (Supplementary Fig. 5). We used DNase I-treated GM38 cells as a positive control for the TUNEL assay. The absence of any visible signs of cell death based on microscopy combined with the TUNEL assay indicate that loss of DDX1-RNA binding activity in arsenite-treated cells is not due to cell death.
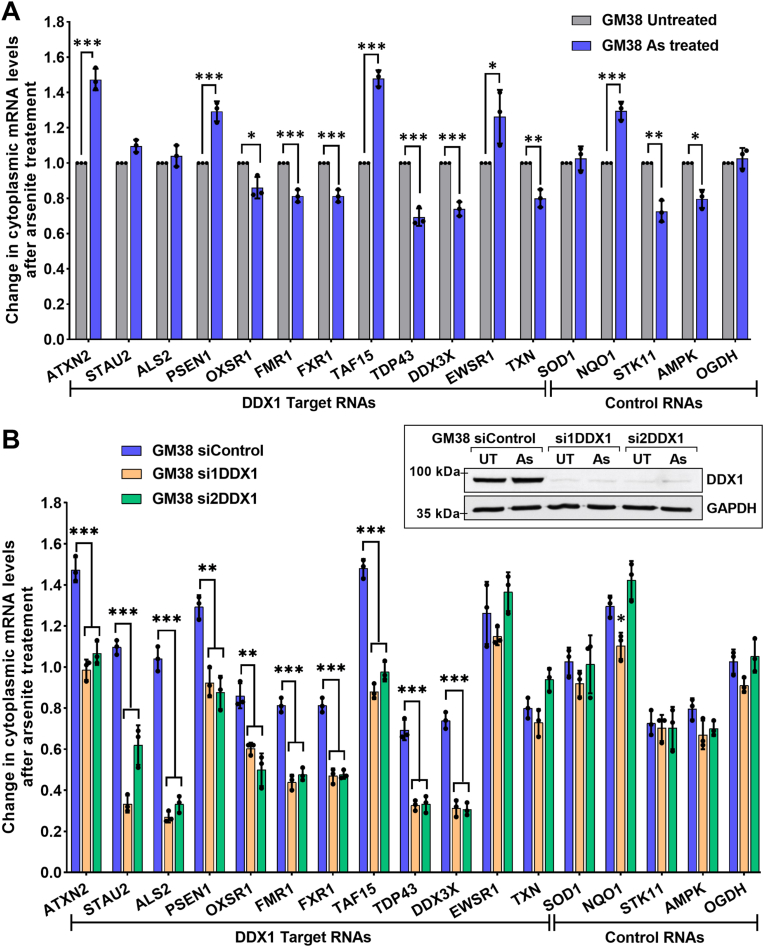
3.5. DDX1 selectively protects its target RNAs in normal fibroblasts under stress
We previously showed that binding of DDX1 to its target RNAs protects cytoplasmic RNAs from degradation in cancer cells that are under oxidative stress [35]. Here, we carry out a similar experiment in normal GM38 fibroblasts treated with vehicle control or arsenite. Cells were fractionated into cytoplasmic and nuclear fractions, and the cytoplasmic fraction was used for RNA extraction. RT-qPCR analysis was carried out using 12 target RNAs (ATM not included) and 5 control RNAs, with untreated GM38 fibroblasts serving as the normalization control (set at 1). In contrast to cancer cells, where we observed reduced cytoplasmic levels of all DDX1 RNA targets tested upon arsenite treatment, variable effects were observed in GM38 fibroblasts, with some targets up-regulated, some targets down-regulated and no change observed for other RNA targets (Fig. 5A).
Fig. 5.
DDX1 protects its target RNAs in the cytoplasm of arsenite-treated normal fibroblasts. Transfection control (siControl) and DDX1-knockdown GM38 (si1DDX1 and si2DDX1) were treated with arsenite or left untreated. Cytoplasmic RNA was extracted, reverse transcribed and used for RNA quantification by RT-qPCR. GAPDH was used for normalization. (A) Relative levels of RNA in the cytoplasm of untreated (set at 1) versus arsenite-treated GM38 cells. (B) RNA levels in the cytoplasm of arsenite-treated control and DDX1-depleted GM38 cells relative to their untreated counterparts (set at 1). Individual experiments are indicated by black circles. Inset shows western blot analysis of cytoplasmic DDX1 upon DDX1-knockdown in GM38 fibroblasts. GAPDH was used as the loading control. UT: Untreated; As: arsenite treated. Error bars depict standard deviation. *p<0.05, **p<0.01, ***p<0.001 (N = 3).
We next examined cytoplasmic RNA levels in DDX1 siRNA-transfected cells. To offset any changes that DDX1 knockdown might have on cytoplasmic RNA levels, we defined the cytoplasmic levels of each DDX1 target RNA in non-stressed siControl-, si1DDX1-and si2DDX1-transfected cells as 1, then compared cytoplasmic DDX1 target RNA levels in arsenite-treated siControl-, si1DDX1-and si2DDX1-transfected cells to their respective untreated counterparts. Decreases of 30-75% in the cytoplasmic levels of 10 out of 12 RNA targets were observed upon depletion of DDX1 in arsenite-treated GM38 fibroblasts (Fig. 5B), with ALS2 showing a 3-4-fold reduction, and STAU2, TDP43 and DDX3X showing 2-3-fold reduction in RNA levels. In keeping with the RNA binding results reported in Fig. 2B, cytoplasmic levels of EWSR1 and TXN RNAs, as well as control RNA levels, were not affected by DDX1 depletion, indicating that these RNAs are not protected by DDX1 under oxidative stress. Our combined results suggest a correlation between elevated DDX1-RNA binding and cytoplasmic RNA protection under oxidative stress conditions.
3.6. DDX1 fails to protect its target RNAs in A-T fibroblasts
Binding of DDX1 to its RNA targets is dramatically reduced in ATM-deficient A-T fibroblasts under oxidative stress. To investigate how DDX1-mediated RNA protection might be affected by ATM deficiency, we examined levels of cytoplasmic RNAs in A-T fibroblasts cultured under normal or arsenite conditions, with or without DDX1 depletion. We included control GM38 fibroblasts in Fig. 6 for comparison, with values shown for control GM38 fibroblasts (arsenite-treated compared to untreated) taken directly from Fig. 5B. Cytoplasmic levels of all ten DDX1 RNA targets (EWSR1 and TXN were excluded from these experiments) were lower in arsenite-treated A-T fibroblasts (by 20-70%) compared to arsenite-treated GM38 cells (ATM + ve, DDX1+ve) (Fig. 6A-J, Supplementary Fig. 6), with all 10 RNA targets in AT2BE and 8/10 RNA targets in AT3BI showing depletion of ≥30%. DDX1 depletion in arsenite-treated A-T fibroblasts showed no further decrease in target RNA levels except for ALS2 in AT2BE and PSEN1 and TAF15 in AT3BI (Fig. 6C,D,H). There was no significant difference in the levels of control RNAs in A-T cells transfected with either siControl or DDX1-depleted arsenite-treated A-T fibroblasts (Supplementary Fig. 6).
Fig. 6.
Cytoplasmic levels of DDX1 RNA targets are reduced in DDX1-depleted arsenite-treated A-T fibroblasts. DDX1 was knocked down in AT2BE and AT3BI (ATM-ve, DDX1+ve) using two siRNAs (si1DDX1 and si2DDX1), and cells were treated with 0.5 mM arsenite for 45 min or left untreated (mock treated). Cytoplasmic RNA was extracted from untreated and arsenite-treated cells. (A-J) Relative changes in the levels of 10 DDX1 target RNAs in arsenite-treated versus untreated cells (set as 1). DDX1 depletion in A-T fibroblasts had the strongest effect on levels of PSEN1 and TAF15 RNA in arsenite-treated cells, with significant reductions in RNA target levels also observed for STAU2, ALS2, FMR1. Minor effects were observed in ATXN2, OXSR1, TDP43 and DDX3X RNA levels. (K) Western blot analysis of cytoplasmic DDX1 protein levels in untreated and arsenite-treated GM38, AT2BE (control and DDX1 depleted) and AT3BI (control and DDX1 depleted). Individual experiments are indicated by black circles. Error bars depict standard deviation. *p<0.05, **p<0.01, ***p<0.001 (N = 3). GAPDH served as the loading control.
3.7. ATM is required for DDX1-mediated RNA protection
The above data from A-T fibroblasts suggest a role for ATM in DDX1-related RNA binding and protection in fibroblasts under oxidative stress. To further investigate a possible relationship between ATM and DDX1-RNA binding activity, we compared DDX1 RNA target levels in DDX1-depleted versus ATM-depleted arsenite-treated GM38 fibroblasts. Similar to DDX1 depletion, ATM depletion significantly reduced levels of DDX1 RNA targets in arsenite-treated GM38 (by 20-55%), with >30% reductions observed for 8 out of the 10 target RNAs (Supplementary Figs. 7 and 8). The levels of 5 of the 10 DDX1 RNA targets were similarly reduced in ATM- and DDX1-depleted GM38 cells, with the remaining five RNA targets showing greater reduction in DDX1-depleted compared to ATM-depleted fibroblasts (Supplementary Figs. 7B,C,D,G,I).
If ATM and DDX1 function in the same pathway, one would expect co-depletion of DDX1 and ATM in GM38 cells to generate similar reductions in DDX1 RNA target levels as depletions observed in either DDX1 or ATM in arsenite-treated GM38 fibroblasts. Levels of most DDX1 RNA targets were similar in DDX1-depleted compared to DDX1/ATM-co-depleted GM38 fibroblasts (Fig. 7; grey zones demarcate the upper and lower ranges of RNA levels in DDX1-knockdown cells). These results are in keeping with cytoplasmic levels of most DDX1 RNA targets being governed through an ATM/DDX1 pathway, although we cannot discount the possibility that additional factors could also influence the levels of a subset of DDX1 RNA targets.
Fig. 7.
ATM depletion results in loss of DDX1-RNA protection in arsenite-treated GM38 fibroblasts. GM38 (ATM + ve, DDX1+ve) were used to knockdown DDX1 (si1DDX1, si2DDX1), ATM (si1ATM, si2ATM), or both ATM and DDX1 siRNAs. Cells were treated with 0.5 mM arsenite for 45 min or left untreated (mock treated), and cytoplasmic RNA was extracted, reverse transcribed and used to determine the effect of arsenite treatment on the levels of 10 DDX1 target genes. For comparison, we used GM38 (ATM + ve, DDX1+ve) transfection control treated with arsenite or left untreated. (A-J) Relative changes in the levels of 10 DDX1 target RNAs in arsenite-treated versus untreated cells (set at 1). Similar reductions in cytoplasmic levels of DDX1 target RNAs were observed for ATXN2, OXSR1, FMR1, TAF15, TDP43, and DDX3X upon ATM depletion, DDX1 depletion, or co-ATM/DDX1 depletion. Small but significant differences in DDX1 RNA target levels were observed upon depletion of ATM compared to DDX1 depletion in the case of STAU2, ALS2, PSEN1 and FXR1. Individual experiments are indicated by black circles. Error bars depict standard deviation. The shaded areas in A-J are used to demarcate the upper and lower ranges of RNA levels observed upon DDX1 knockdown in GM38 fibroblasts. *p<0.05, **p<0.01, ***p<0.001 (N = 3). (K) Western blot analysis of cytoplasmic DDX1 protein levels in untreated and arsenite-treated GM38, DDX1-depleted, ATM-depleted, or co-DDX1/ATM-depleted. GAPDH served as the loading control. Please note that no ATM signal was detected in cytoplasmic extracts.
3.8. Depletion of ATM and DDX1 decreases colony forming ability in arsenite-treated cells
Failure to protect RNAs that encode proteins involved in oxidative stress response may impair the long-term survival of DDX1-and ATM-depleted fibroblasts. To address the effect of ATM and DDX1 depletion on survival after oxidative stress, we used the clonogenic assay [40]. Percentage colony formation was calculated based on the number of colony forming units (CFUs) per 100 cells. The plating efficiency of control GM38, AT2BE and AT3BI was 11.3 ± 0.6%, 6.8 ± 0.8% and 7.7 ± 0.6%, respectively (Fig. 8A). The reduced plating efficiency of A-T fibroblasts compared to normal fibroblasts has been previously reported [47,48]. GM38 and A-T fibroblasts treated with increasing concentrations of arsenite for 45 min showed greatly reduced colony forming ability, with none of the fibroblast cultures forming colonies at the highest concentration of arsenite tested (0.5 mM). A-T fibroblasts were more susceptible to arsenite than GM38, with cell survival dropping to ≤10% at 0.3 mM arsenite compared to 25% cell survival for GM38 (Fig. 8A).
Fig. 8.
ATM and DDX1 promote the survival of arsenite-treated fibroblasts. Cell survival after arsenite treatment was measured using the clonogenic assay. One hundred cells were seeded in duplicate in 6-well plates followed by treatment with increasing concentrations of arsenite (0, 0.1, 0.2, 0.3, 0.4 and 0.5 mM). After 45 min, cells were washed and cultured in fresh medium for 2–3 weeks. Colonies (>50 cells) were stained with crystal violet and the number of colonies in each well counted manually. Individual experiments are indicated by circles (N = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Depletion of either ATM or DDX1 in GM38 also resulted in reduced plating efficiency compared to control knockdown cells (Fig. 8B). Furthermore, upon arsenite treatment, ATM- and DDX1-depleted cells had a greatly compromised ability to recover from oxidative stress, with few, if any, colonies observed in cells treated with 0.3 mM arsenite. At 0.2 mM arsenite, the survival of ATM-depleted and DDX1-depleted cells dropped to ∼20% and ∼25%, respectively, compared to 50% in control GM38 cells (Fig. 8B). Overall, these data indicate a role for ATM and DDX1 in cell survival upon arsenite treatment which could be explained by the decrease in the levels of cytoplasmic mRNAs encoding proteins involved in oxidative stress response.
4. Discussion
The sensitivity of A-T cells to ionizing radiation is well-documented; however, the underlying cause for the debilitating neuronal degeneration associated with A-T is not well-understood. There is evidence suggesting that excessive production of reactive oxygen species (ROS), a measure of oxidative stress, is a key factor in the pathogenesis of A-T [4,9]. Accumulation of ROS in A-T cells causes DNA damage, mitochondrial dysfunction, and activation of apoptotic pathways [10]. ATM has well-characterized roles in the repair of DNA DSBs with emerging roles in maintenance of ROS homeostasis [10,49]. Thus, it is predicted that loss of ATM in A-T patients will on one hand sensitize cells to DNA damaging agents, with concomitant increases in carcinogenesis, and on the other hand, deregulate ROS levels with consequences to cell survival, particularly in highly metabolically active neuronal cells already prone to producing high levels of ROS [50]. In line with this, correction of ATM mutations in A-T patient cells restores both normal DNA DSB repair and ROS production [51].
ATM has hundreds of phosphorylation substrates and interacting partners, many of which have been shown to have functions related to DNA DSB repair [52]. However, there is very little information on how ATM might maintain cellular homeostasis in cells exposed to oxidative stress. Here, we investigate whether ATM's function in arsenite-treated fibroblasts is mediated through DDX1, an ATM-interacting partner implicated in both DNA DSB repair [15,16] and oxidative stress response [35]. Using an RNA-IP sequencing (RIP-seq) approach, we previously identified >1000 putative DDX1 RNA targets, many of which encode proteins involved in oxidative stress response [35]. Using a subset of putative DDX1 RNA targets implicated in both oxidative stress and neurodegeneration, we show that ATM and DDX1 play interdependent roles in oxidative stress response, with ATM residing upstream of DDX1. Upon DDX1 depletion, we observe an increase in ROS levels in ATM-proficient fibroblasts (GM38), but not in ATM-deficient fibroblasts (AT2BE and AT3BI). Arsenite treatment leads to increased binding of DDX1 to its target RNAs in ATM-proficient but not ATM-deficient fibroblasts. We also show that DDX1-mediated protection of its target RNAs requires functional ATM. The similar reductions in levels of DDX1 target RNAs observed upon arsenite treatment of DDX1 knockdown, ATM knockdown or DDX1/ATM double knockdown fibroblasts suggests that ATM and DDX1 mostly function in the same pathway, although additional pathways may also be involved in a more limited capacity.
It is generally accepted that RNAs encoding proteins involved in stress response need to be protected for the cell to recover from stress [53]. The RNA targets examined in our study all encode proteins previously implicated in neurodevelopmental/neurodegeneration disorders and oxidative stress response [44,46,54,55]. For example, mutations in TDP43, ATXN2, ALS2, TAF15, DDX3X, all of which encode RNA binding proteins, are observed in Amyotrophic Lateral Sclerosis (ALS) and Frontotemporal Dementia (FTD) [45,[56], [57], [58]]. Mutations in PSEN1 have been correlated with loss of mitochondrial function and increased ROS production in Alzheimer's disease [59]. FMR1 mutations orchestrate the ataxia, neuropathy, and cognitive disability in Fragile X-associated Tremor Ataxia Syndrome (FXTAS) [60], with elevated ROS levels observed in the astrocytes of FMR1 knockdown mice and fibroblasts derived from FMR1 premutation carrier patients [61,62]. Moreover, FMR1, FXR1, ATXN2, STAU2, TAF15, TDP43, DDX3X along with DDX1, all localize to stress granules which are formed when cells are under oxidative stress and play key roles in RNA processing [34,55,57,63,64]. Thus, alterations in the expression of DDX1 RNA targets resulting from loss of ATM activity may prevent the maintenance of homeostasis after oxidative stress in A-T patients.
A-T is primarily characterized by depletion of Purkinje and granule neurons in the cerebellum [2,65,66], suggesting that these cells are particularly sensitive to loss of ATM. Interestingly, at least five of the DDX1 targets included in our analyses have been implicated in the function and survival of Purkinje cells. For example, mutation of ATXN2 in Spinocerebellar ataxia type 2 (SCA2) results in decreased firing rates of Purkinje neurons and impaired Purkinje-granule cell synapses leading to Purkinje cell death and reduced motor coordination [67,68]. In turn, mutations or knockdown of ALS2, FMR1 and STAU2 have been associated with altered synaptic structures and defective functions of Purkinje cells and motor neurons [[69], [70], [71], [72]]. Alzheimer's patients and mice with PSEN1 mutations have reduced numbers of cerebellar Purkinje cells, and demonstrate neurodegeneration and impaired motor coordination [[73], [74], [75]]. Purkinje cells are particularly large neurons able to integrate considerable amounts of information, with granule cells connecting to Purkinje cells. Purkinje cells are some of the most metabolically active cells in the brain, resulting in high energy demands and high ROS production [76]. It therefore seems reasonable that the impaired redox system in A-T patients would preferentially target Purkinje cells [77]. We propose that elevated oxidative stress and downregulation of DDX1-RNA targets in A-T cells may adversely affect the synaptic activity and survival of Purkinje cells and their connecting granule cells.
It is interesting to note that DDX1 has previously been shown to localize to cytoplasmic RNA granules located in the dendrites and axons of neurons [19]. RNA granules contain mixtures of RNAs and proteins required for localized translation, allowing protein production in the areas where they are needed [19,78]. In light of our findings, we propose that DDX1 may also be playing an RNA protective role in neuronal RNA granules, possibly related to protection against ROS-induced damage. Similar to DNA damage, RNA damage can be highly detrimental to cells. In fact, studies have shown that oxidative stress causes more damage to single-stranded cytoplasmic RNA than to double-stranded DNA [79]. RNA damage caused by oxidative stress has been associated with neurodegenerative disorders such as Alzheimer's disease [[80], [81], [82]], Parkinson's disease [83,84] and ALS [85]. RNAs bound by DDX1 may therefore be protected against ROS-induced RNA damage, resulting in increased cytoplasmic levels of DDX1-bound RNAs in cells under oxidative stress.
Loss of ATM has been linked to increased ROS production and mitochondrial dysfunction, with ATM detected in mitochondria by electron microscopy and mitochondria purification [3,86,87]. As well, DDX1 has been shown to localize to mitochondria, with loss of DDX1 increasing ROS production and mitochondrial membrane potential in early-stage mouse embryos [28]. The increased mitochondrial ROS production observed in ATM-deficient and DDX1-depleted fibroblasts under both chronic and acute arsenite treatment may well result in mitochondrial dysfunction, with implications for possible ATM-DDX1 interaction in mitochondria. Although we were not able to address the latter because of the extremely low levels of ATM in the cytoplasm of fibroblast cells, one might expect the effect of mitochondrial dysfunction to be exacerbated in long-lasting and highly metabolically active neuronal cells.
While we do not address the mechanism driving ATM-dependent increased DDX1-RNA interaction in this study, our data show that ATM depletion does not affect overall levels of cytoplasmic DDX1. Past studies have shown that DDX1 is phosphorylated by ATM in cells undergoing genotoxic stress [16,88]. As DDX1 has multiple putative ATM phosphorylation sites, one possibility is that different DDX1 sites are phosphorylated by ATM depending on the type of stress that the cell is exposed to. Thus, absence of phosphorylation of DDX1 by ATM in ATM-deficient cells could explain the reduction in DDX1-RNA target binding observed upon arsenite treatment. Furthermore, enhanced binding of ATM-phosphorylated DDX1 to its RNA targets under oxidative stress may be mediated through modification of RNAs. It's well-recognized that some mRNAs undergo methylation; e.g. 2′-O-methylation or N6-methyladenosine (m6A)-modification, both of which are induced by oxidative stress [[89], [90], [91], [92]]. DDX1 is a known reader of 2′-O-methylated RNAs [93], with DDX1 also shown to interact with readers and writers of m6A modification [92,94,95]. Future investigations will involve analysis of ATM-dependent DDX1 phosphorylation and modification of DDX1 RNA targets in cells undergoing oxidative stress.
In conclusion, using fibroblasts derived from A-T patients, we show that deficiency in either ATM or DDX1 results in accumulation of ROS and decreased cell survival in fibroblasts. We provide evidence that ATM resides upstream of DDX1 in the protection of mRNA targets in cells undergoing oxidative stress. The RNAs protected by DDX1 have been linked to neurological disorders and oxidative stress, suggesting that maintenance of their levels may be important for the recovery of cells exposed to oxidative stress. We propose that the ATM/DDX1 pathway may be especially important for neuronal cell survival given their high oxygen consumption and inability to regenerate, thereby providing a possible explanation for the neurodegenerative pathophysiology associated with A-T.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Razmik Mirzayans for the AT2BE, AT3BI, GM10 and GM38 fibroblasts and Joan Turner for the anti-ATM antibody. This work was supported by a grant from the Canadian Institutes of Health Research grant number 162157 (to R.G.) and a postdoctoral fellowship (to M.G.) from the Women's and Children's Health Research Institute.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102988.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Boder E., Sedgwick R.P. Ataxia-telangiectasia; a familial syndrome of progressive cerebellar ataxia, oculocutaneous telangiectasia and frequent pulmonary infection. Pediatrics. 1958;21:526–554. [PubMed] [Google Scholar]
- 2.Rothblum-Oviatt C., Wright J., Lefton-Greif M.A., McGrath-Morrow S.A., Crawford T.O., Lederman H.M. Ataxia telangiectasia: a review. Orphanet J. Rare Dis. 2016;11:159. doi: 10.1186/s13023-016-0543-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maciejczyk M., Mikoluc B., Pietrucha B., Heropolitanska-Pliszka E., Pac M., Motkowski R., Car H. Oxidative stress, mitochondrial abnormalities and antioxidant defense in Ataxia-telangiectasia, Bloom syndrome and Nijmegen breakage syndrome. Redox Biol. 2017;11:375–383. doi: 10.1016/j.redox.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watters D.J. Oxidative stress in ataxia telangiectasia. Redox Rep. 2003;8:23–29. doi: 10.1179/135100003125001206. [DOI] [PubMed] [Google Scholar]
- 5.Ray P.D., Huang B.W., Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valko M., Rhodes C.J., Moncol J., Izakovic M., Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Singh A., Kukreti R., Saso L., Kukreti S. Oxidative stress: a key modulator in neurodegenerative diseases. Molecules. 2019;24 doi: 10.3390/molecules24081583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim G.H., Kim J.E., Rhie S.J., Yoon S. The role of oxidative stress in neurodegenerative diseases. Exp Neurobiol. 2015;24:325–340. doi: 10.5607/en.2015.24.4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichenbach J., Schubert R., Schindler D., Muller K., Bohles H., Zielen S. Elevated oxidative stress in patients with ataxia telangiectasia. Antioxidants Redox Signal. 2002;4:465–469. doi: 10.1089/15230860260196254. [DOI] [PubMed] [Google Scholar]
- 10.Stagni V., Cirotti C., Barila D. Ataxia-telangiectasia mutated kinase in the control of oxidative stress, mitochondria, and autophagy in cancer: a maestro with a large orchestra. Front. Oncol. 2018;8:73. doi: 10.3389/fonc.2018.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D.A., Smith S., Uziel T., Sfez S., Ashkenazi M., Pecker I., Frydman M., Harnik R., Patanjali S.R., Simmons A., Clines G.A., Sartiel A., Gatti R.A., Chessa L., Sanal O., Lavin M.F., Jaspers N.G., Taylor A.M., Arlett C.F., Miki T., Weissman S.M., Lovett M., Collins F.S., Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 12.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 13.Falck J., Coates J., Jackson S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 14.Oh J., Symington L.S. Role of the Mre11 complex in preserving genome integrity. Genes. 2018;9 doi: 10.3390/genes9120589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L., Germain D.R., Poon H.Y., Hildebrandt M.R., Monckton E.A., McDonald D., Hendzel M.J., Godbout R. DEAD box 1 facilitates removal of RNA and homologous recombination at DNA double-strand breaks. Mol. Cell Biol. 2016;36:2794–2810. doi: 10.1128/MCB.00415-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L., Monckton E.A., Godbout R. A role for DEAD box 1 at DNA double-strand breaks. Mol. Cell Biol. 2008;28:6413–6425. doi: 10.1128/MCB.01053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L., Poon H.Y., Hildebrandt M.R., Monckton E.A., Germain D.R., Fahlman R.P., Godbout R. Role for RIF1-interacting partner DDX1 in BLM recruitment to DNA double-strand breaks. DNA Repair. 2017;55:47–63. doi: 10.1016/j.dnarep.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Chen H.C., Lin W.C., Tsay Y.G., Lee S.C., Chang C.J. An RNA helicase, DDX1, interacting with poly(A) RNA and heterogeneous nuclear ribonucleoprotein K. J. Biol. Chem. 2002;277:40403–40409. doi: 10.1074/jbc.M206981200. [DOI] [PubMed] [Google Scholar]
- 19.Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 20.Lin M.H., Sivakumaran H., Jones A., Li D., Harper C., Wei T., Jin H., Rustanti L., Meunier F.A., Spann K., Harrich D. A HIV-1 Tat mutant protein disrupts HIV-1 Rev function by targeting the DEAD-box RNA helicase DDX1. Retrovirology. 2014;11:121. doi: 10.1186/s12977-014-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Gonzalez A., Pazo A., Navajas R., Ciordia S., Rodriguez-Frandsen A., Nieto A. hCLE/C14orf166 associates with DDX1-HSPC117-FAM98B in a novel transcription-dependent shuttling RNA-transporting complex. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popow J., Jurkin J., Schleiffer A., Martinez J. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature. 2014;511:104–107. doi: 10.1038/nature13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribeiro de Almeida C., Dhir S., Dhir A., Moghaddam A.E., Sattentau Q., Meinhart A., Proudfoot N.J. RNA helicase DDX1 converts RNA G-quadruplex structures into R-loops to promote IgH class switch recombination. Mol. Cell. 2018;70:650–662 e658. doi: 10.1016/j.molcel.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson-Anderson R.M., Wang J., Edgcomb S.P., Carmel A.B., Williamson J.R., Millar D.P. Single-molecule studies reveal that DEAD box protein DDX1 promotes oligomerization of HIV-1 Rev on the Rev response element. J. Mol. Biol. 2011;410:959–971. doi: 10.1016/j.jmb.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Germain D.R., Graham K., Glubrecht D.D., Hugh J.C., Mackey J.R., Godbout R. DEAD box 1: a novel and independent prognostic marker for early recurrence in breast cancer. Breast Cancer Res. Treat. 2011;127:53–63. doi: 10.1007/s10549-010-0943-7. [DOI] [PubMed] [Google Scholar]
- 26.Godbout R., Li L., Liu R.Z., Roy K. Role of DEAD box 1 in retinoblastoma and neuroblastoma. Future Oncol. 2007;3:575–587. doi: 10.2217/14796694.3.5.575. [DOI] [PubMed] [Google Scholar]
- 27.Hildebrandt M.R., Germain D.R., Monckton E.A., Brun M., Godbout R. Ddx1 knockout results in transgenerational wild-type lethality in mice. Sci. Rep. 2015;5:9829. doi: 10.1038/srep09829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Yasmin L., Li L., Gao P., Xu X., Sun X., Godbout R. DDX1 vesicles control calcium-dependent mitochondrial activity in mouse embryos. Nat. Commun. 2022;13:3794. doi: 10.1038/s41467-022-31497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Germain D.R., Li L., Hildebrandt M.R., Simmonds A.J., Hughes S.C., Godbout R. Loss of the Drosophila melanogaster DEAD box protein Ddx1 leads to reduced size and aberrant gametogenesis. Dev. Biol. 2015;407:232–245. doi: 10.1016/j.ydbio.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 31.Kozlov S.V., Waardenberg A.J., Engholm-Keller K., Arthur J.W., Graham M.E., Lavin M. Reactive oxygen species (ROS)-Activated ATM-dependent phosphorylation of cytoplasmic substrates identified by large-scale phosphoproteomics screen. Mol. Cell. Proteomics. 2016;15:1032–1047. doi: 10.1074/mcp.M115.055723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rotman G., Shiloh Y. Ataxia-telangiectasia: is ATM a sensor of oxidative damage and stress? Bioessays. 1997;19:911–917. doi: 10.1002/bies.950191011. [DOI] [PubMed] [Google Scholar]
- 33.Shackelford R.E., Innes C.L., Sieber S.O., Heinloth A.N., Leadon S.A., Paules R.S. The Ataxia telangiectasia gene product is required for oxidative stress-induced G1 and G2 checkpoint function in human fibroblasts. J. Biol. Chem. 2001;276:21951–21959. doi: 10.1074/jbc.M011303200. [DOI] [PubMed] [Google Scholar]
- 34.Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L., Garg M., Wang Y., Wang W., Godbout R. DEAD box 1 (DDX1) protein binds to and protects cytoplasmic stress response mRNAs in cells exposed to oxidative stress. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.102180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirzayans R., Sabour M., Paterson M.C. Enhanced bioreduction of 4-nitroquinoline 1-oxide by cultured ataxia telangiectasia cells. Carcinogenesis. 1988;9:1711–1715. doi: 10.1093/carcin/9.9.1711. [DOI] [PubMed] [Google Scholar]
- 37.Mirzayans R., Middlestadt M.V., Paterson M.C. Cytotoxic and mutagenic effects of methylnitrosourea in two human fetal fibroblast strains differing in O6-methylguanine-DNA methyltransferase activity. Carcinogenesis. 1992;13:1185–1190. doi: 10.1093/carcin/13.7.1185. [DOI] [PubMed] [Google Scholar]
- 38.Orru S., Coni P., Floris A., Littera R., Carcassi C., Sogos V., Brancia C. Reduced stress granule formation and cell death in fibroblasts with the A382T mutation of TARDBP gene: evidence for loss of TDP-43 nuclear function. Hum. Mol. Genet. 2016;25:4473–4483. doi: 10.1093/hmg/ddw276. [DOI] [PubMed] [Google Scholar]
- 39.Bleoo S., Sun X., Hendzel M.J., Rowe J.M., Packer M., Godbout R. Association of human DEAD box protein DDX1 with a cleavage stimulation factor involved in 3'-end processing of pre-MRNA. Mol. Biol. Cell. 2001;12:3046–3059. doi: 10.1091/mbc.12.10.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franken N.A., Rodermond H.M., Stap J., Haveman J., van Bree C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 41.Cookson M.R. RNA-binding proteins implicated in neurodegenerative diseases. Wiley Interdiscip Rev RNA. 2017;8 doi: 10.1002/wrna.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinoshita C., Kubota N., Aoyama K. Interplay of RNA-binding proteins and microRNAs in neurodegenerative diseases. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22105292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolozin B., Ivanov P. Stress granules and neurodegeneration. Nat. Rev. Neurosci. 2019;20:649–666. doi: 10.1038/s41583-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi F., Meng Q., Hayashi I., Kobayashi J. FXR1 is a novel MRE11-binding partner and participates in oxidative stress responses. J. Radiat. Res. 2020;61:368–375. doi: 10.1093/jrr/rraa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue Y.C., Ng C.S., Xiang P., Liu H., Zhang K., Mohamud Y., Luo H. Dysregulation of RNA-binding proteins in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 2020;13:78. doi: 10.3389/fnmol.2020.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choy K.R., Watters D.J. Neurodegeneration in ataxia-telangiectasia: multiple roles of ATM kinase in cellular homeostasis. Dev. Dynam. 2018;247:33–46. doi: 10.1002/dvdy.24522. [DOI] [PubMed] [Google Scholar]
- 47.Kasten-Pisula U., Tastan H., Dikomey E. Huge differences in cellular radiosensitivity due to only very small variations in double-strand break repair capacity. Int. J. Radiat. Biol. 2005;81:409–419. doi: 10.1080/09553000500140498. [DOI] [PubMed] [Google Scholar]
- 48.Shiloh Y., Tabor E., Becker Y. Colony-forming ability of ataxia-telangiectasia skin fibroblasts is an indicator of their early senescence and increased demand for growth factors. Exp. Cell Res. 1982;140:191–199. doi: 10.1016/0014-4827(82)90169-0. [DOI] [PubMed] [Google Scholar]
- 49.Xie X., Zhang Y., Wang Z., Wang S., Jiang X., Cui H., Zhou T., He Z., Feng H., Guo Q., Song X., Cao L. ATM at the crossroads of reactive oxygen species and autophagy. Int. J. Biol. Sci. 2021;17:3080–3090. doi: 10.7150/ijbs.63963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cobley J.N., Fiorello M.L., Bailey D.M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol. 2018;15:490–503. doi: 10.1016/j.redox.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ovchinnikov D.A., Withey S.L., Leeson H.C., Lei U.W., Sundarrajan A., Junday K., Pewarchuk M., Yeo A.J., Kijas A.W., Lavin M.F., Wolvetang E.J. Correction of ATM mutations in iPS cells from two ataxia-telangiectasia patients restores DNA damage and oxidative stress responses. Hum. Mol. Genet. 2020;29:990–1001. doi: 10.1093/hmg/ddaa023. [DOI] [PubMed] [Google Scholar]
- 52.Shiloh Y., Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 53.Anderson P., Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 54.Saito M., Iestamantavicius V., Hess D., Matthias P. Monitoring acetylation of the RNA helicase DDX3X, a protein critical for formation of stress granules. Methods Mol. Biol. 2021;2209:217–234. doi: 10.1007/978-1-0716-0935-4_14. [DOI] [PubMed] [Google Scholar]
- 55.Valentin-Vega Y.A., Wang Y.D., Parker M., Patmore D.M., Kanagaraj A., Moore J., Rusch M., Finkelstein D., Ellison D.W., Gilbertson R.J., Zhang J., Kim H.J., Taylor J.P. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci. Rep. 2016;6 doi: 10.1038/srep25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Chalabi A., van den Berg L.H., Veldink J. Gene discovery in amyotrophic lateral sclerosis: implications for clinical management. Nat. Rev. Neurol. 2017;13:96–104. doi: 10.1038/nrneurol.2016.182. [DOI] [PubMed] [Google Scholar]
- 57.Baradaran-Heravi Y., Van Broeckhoven C., van der Zee J. Stress granule mediated protein aggregation and underlying gene defects in the FTD-ALS spectrum. Neurobiol. Dis. 2020;134 doi: 10.1016/j.nbd.2019.104639. [DOI] [PubMed] [Google Scholar]
- 58.Castelli L.M., Benson B.C., Huang W.P., Lin Y.H., Hautbergue G.M. RNA helicases in microsatellite repeat expansion disorders and neurodegeneration. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.886563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han J., Park H., Maharana C., Gwon A.R., Park J., Baek S.H., Bae H.G., Cho Y., Kim H.K., Sul J.H., Lee J., Kim E., Kim J., Cho Y., Park S., Palomera L.F., Arumugam T.V., Mattson M.P., Jo D.G. Alzheimer's disease-causing presenilin-1 mutations have deleterious effects on mitochondrial function. Theranostics. 2021;11:8855–8873. doi: 10.7150/thno.59776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cabal-Herrera A.M., Tassanakijpanich N., Salcedo-Arellano M.J., Hagerman R.J. Fragile X-associated tremor/ataxia syndrome (FXTAS): pathophysiology and clinical implications. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21124391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song G., Napoli E., Wong S., Hagerman R., Liu S., Tassone F., Giulivi C. Altered redox mitochondrial biology in the neurodegenerative disorder fragile X-tremor/ataxia syndrome: use of antioxidants in precision medicine. Mol. Med. 2016;22:548–559. doi: 10.2119/molmed.2016.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vandenberg G.G., Dawson N.J., Head A., Scott G.R., Scott A.L. Astrocyte-mediated disruption of ROS homeostasis in Fragile X mouse model. Neurochem. Int. 2021;146 doi: 10.1016/j.neuint.2021.105036. [DOI] [PubMed] [Google Scholar]
- 63.Elden A.C., Kim H.J., Hart M.P., Chen-Plotkin A.S., Johnson B.S., Fang X., Armakola M., Geser F., Greene R., Lu M.M., Padmanabhan A., Clay-Falcone D., McCluskey L., Elman L., Juhr D., Gruber P.J., Rub U., Auburger G., Trojanowski J.Q., Lee V.M., Van Deerlin V.M., Bonini N.M., Gitler A.D. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Didiot M.C., Subramanian M., Flatter E., Mandel J.L., Moine H. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol. Biol. Cell. 2009;20:428–437. doi: 10.1091/mbc.E08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lavin M.F., Gueven N., Bottle S., Gatti R.A. Current and potential therapeutic strategies for the treatment of ataxia-telangiectasia. Br. Med. Bull. 2007;81–82:129–147. doi: 10.1093/bmb/ldm012. [DOI] [PubMed] [Google Scholar]
- 66.Shiloh Y. The cerebellar degeneration in ataxia-telangiectasia: a case for genome instability. DNA Repair. 2020;95 doi: 10.1016/j.dnarep.2020.102950. [DOI] [PubMed] [Google Scholar]
- 67.Arsovic A., Halbach M.V., Canet-Pons J., Esen-Sehir D., Doring C., Freudenberg F., Czechowska N., Seidel K., Baader S.L., Gispert S., Sen N.E., Auburger G. Mouse ataxin-2 expansion downregulates CamKII and other calcium signaling factors, impairing granule-purkinje neuron synaptic strength. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21186673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scoles D.R., Pulst S.M. Spinocerebellar ataxia type 2. Adv. Exp. Med. Biol. 2018;1049:175–195. doi: 10.1007/978-3-319-71779-1_8. [DOI] [PubMed] [Google Scholar]
- 69.Ariza J., Rogers H., Monterrubio A., Reyes-Miranda A., Hagerman P.J., Martinez-Cerdeno V. A majority of FXTAS cases present with intranuclear inclusions within purkinje cells. Cerebellum. 2016;15:546–551. doi: 10.1007/s12311-016-0776-y. [DOI] [PubMed] [Google Scholar]
- 70.Hadano S., Benn S.C., Kakuta S., Otomo A., Sudo K., Kunita R., Suzuki-Utsunomiya K., Mizumura H., Shefner J.M., Cox G.A., Iwakura Y., Brown R.H., Jr., Ikeda J.E. Mice deficient in the Rab5 guanine nucleotide exchange factor ALS2/alsin exhibit age-dependent neurological deficits and altered endosome trafficking. Hum. Mol. Genet. 2006;15:233–250. doi: 10.1093/hmg/ddi440. [DOI] [PubMed] [Google Scholar]
- 71.Huber K.M. The fragile X-cerebellum connection. Trends Neurosci. 2006;29:183–185. doi: 10.1016/j.tins.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 72.Pernice H.F., Schieweck R., Jafari M., Straub T., Bilban M., Kiebler M.A., Popper B. Altered glutamate receptor ionotropic delta subunit 2 expression in stau2-deficient cerebellar purkinje cells in the adult brain. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20071797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chaudhari K., Wang L., Kruse J., Winters A., Sumien N., Shetty R., Prah J., Liu R., Shi J., Forster M., Yang S.H. Early loss of cerebellar Purkinje cells in human and a transgenic mouse model of Alzheimer's disease. Neurol. Res. 2021;43:570–581. doi: 10.1080/01616412.2021.1893566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Piccini A., Zanusso G., Borghi R., Noviello C., Monaco S., Russo R., Damonte G., Armirotti A., Gelati M., Giordano R., Zambenedetti P., Russo C., Ghetti B., Tabaton M. Association of a presenilin 1 S170F mutation with a novel Alzheimer disease molecular phenotype. Arch. Neurol. 2007;64:738–745. doi: 10.1001/archneur.64.5.738. [DOI] [PubMed] [Google Scholar]
- 75.Sepulveda-Falla D., Barrera-Ocampo A., Hagel C., Korwitz A., Vinueza-Veloz M.F., Zhou K., Schonewille M., Zhou H., Velazquez-Perez L., Rodriguez-Labrada R., Villegas A., Ferrer I., Lopera F., Langer T., De Zeeuw C.I., Glatzel M. Familial Alzheimer's disease-associated presenilin-1 alters cerebellar activity and calcium homeostasis. J. Clin. Invest. 2014;124:1552–1567. doi: 10.1172/JCI66407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chakrabarti L., Zahra R., Jackson S.M., Kazemi-Esfarjani P., Sopher B.L., Mason A.G., Toneff T., Ryu S., Shaffer S., Kansy J.W., Eng J., Merrihew G., MacCoss M.J., Murphy A., Goodlett D.R., Hook V., Bennett C.L., Pallanck L.J., La Spada A.R. Mitochondrial dysfunction in NnaD mutant flies and Purkinje cell degeneration mice reveals a role for Nna proteins in neuronal bioenergetics. Neuron. 2010;66:835–847. doi: 10.1016/j.neuron.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen P., Peng C., Luff J., Spring K., Watters D., Bottle S., Furuya S., Lavin M.F. Oxidative stress is responsible for deficient survival and dendritogenesis in purkinje neurons from ataxia-telangiectasia mutated mutant mice. J. Neurosci. 2003;23:11453–11460. doi: 10.1523/JNEUROSCI.23-36-11453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller L.C., Blandford V., McAdam R., Sanchez-Carbente M.R., Badeaux F., DesGroseillers L., Sossin W.S. Combinations of DEAD box proteins distinguish distinct types of RNA: protein complexes in neurons. Mol. Cell. Neurosci. 2009;40:485–495. doi: 10.1016/j.mcn.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 79.Nunomura A., Hofer T., Moreira P.I., Castellani R.J., Smith M.A., Perry G. RNA oxidation in Alzheimer disease and related neurodegenerative disorders. Acta Neuropathol. 2009;118:151–166. doi: 10.1007/s00401-009-0508-1. [DOI] [PubMed] [Google Scholar]
- 80.Abe T., Tohgi H., Isobe C., Murata T., Sato C. Remarkable increase in the concentration of 8-hydroxyguanosine in cerebrospinal fluid from patients with Alzheimer's disease. J. Neurosci. Res. 2002;70:447–450. doi: 10.1002/jnr.10349. [DOI] [PubMed] [Google Scholar]
- 81.Ding Q., Markesbery W.R., Cecarini V., Keller J.N. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer's disease. Neurochem. Res. 2006;31:705–710. doi: 10.1007/s11064-006-9071-5. [DOI] [PubMed] [Google Scholar]
- 82.Nunomura A., Perry G., Pappolla M.A., Wade R., Hirai K., Chiba S., Smith M.A. RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer's disease. J. Neurosci. 1999;19:1959–1964. doi: 10.1523/JNEUROSCI.19-06-01959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abe T., Isobe C., Murata T., Sato C., Tohgi H. Alteration of 8-hydroxyguanosine concentrations in the cerebrospinal fluid and serum from patients with Parkinson's disease. Neurosci. Lett. 2003;336:105–108. doi: 10.1016/s0304-3940(02)01259-4. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J., Perry G., Smith M.A., Robertson D., Olson S.J., Graham D.G., Montine T.J. Parkinson's disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am. J. Pathol. 1999;154:1423–1429. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang Y., Kong Q., Shan X., Tian G., Ilieva H., Cleveland D.W., Rothstein J.D., Borchelt D.R., Wong P.C., Lin C.L. Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PLoS One. 2008;3:e2849. doi: 10.1371/journal.pone.0002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Subramanian G.N., Yeo A.J., Gatei M.H., Coman D.J., Lavin M.F. 2022. Metabolic Stress and Mitochondrial Dysfunction in Ataxia-Telangiectasia. Antioxidants (Basel)11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Valentin-Vega Y.A., Maclean K.H., Tait-Mulder J., Milasta S., Steeves M., Dorsey F.C., Cleveland J.L., Green D.R., Kastan M.B. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119:1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han C., Liu Y., Wan G., Choi H.J., Zhao L., Ivan C., He X., Sood A.K., Zhang X., Lu X. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep. 2014;8:1447–1460. doi: 10.1016/j.celrep.2014.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elliott B.A., Ho H.T., Ranganathan S.V., Vangaveti S., Ilkayeva O., Abou Assi H., Choi A.K., Agris P.F., Holley C.L. Modification of messenger RNA by 2'-O-methylation regulates gene expression in vivo. Nat. Commun. 2019;10:3401. doi: 10.1038/s41467-019-11375-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Michel C.I., Holley C.L., Scruggs B.S., Sidhu R., Brookheart R.T., Listenberger L.L., Behlke M.A., Ory D.S., Schaffer J.E. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metabol. 2011;14:33–44. doi: 10.1016/j.cmet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee J., Harris A.N., Holley C.L., Mahadevan J., Pyles K.D., Lavagnino Z., Scherrer D.E., Fujiwara H., Sidhu R., Zhang J., Huang S.C., Piston D.W., Remedi M.S., Urano F., Ory D.S., Schaffer J.E. Rpl13a small nucleolar RNAs regulate systemic glucose metabolism. J. Clin. Invest. 2016;126:4616–4625. doi: 10.1172/JCI88069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang B., Chen Q. Cross-talk between oxidative stress and m(6)A RNA methylation in cancer. Oxid. Med. Cell. Longev. 2021;2021 doi: 10.1155/2021/6545728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang Y., Bai X., Guo Z., Dong H., Fu Y., Zhang H., Zhai G., Tian S., Wang Y., Zhang K. DNA-guided photoactivatable probe-based chemical proteomics reveals the reader protein of mRNA methylation. iScience. 2021;24 doi: 10.1016/j.isci.2021.103046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Youn J.Y., Dunham W.H., Hong S.J., Knight J.D.R., Bashkurov M., Chen G.I., Bagci H., Rathod B., MacLeod G., Eng S.W.M., Angers S., Morris Q., Fabian M., Cote J.F., Gingras A.C. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell. 2018;69:517–532 e511. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 95.Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., Cheng T., Gao M., Shu X., Ma H., Wang F., Wang X., Shen B., Wang Y., Feng X., He C., Liu J. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.