Abstract
The ability of Rhodobacter sphaeroides 2.4.1T to respire anaerobically with the alternative electron acceptor dimethyl sulfoxide (DMSO) or trimethylamine N-oxide (TMAO) is manifested by the molybdoenzyme DMSO reductase, which is encoded by genes of the dor locus. Previously, we have demonstrated that dor expression is regulated in response to lowered oxygen tensions and the presence of DMSO or TMAO in the growth medium. Several regulatory proteins have been identified as key players in this regulatory cascade: FnrL, DorS-DorR, and DorX-DorY. To further examine the role of redox potentiation in the regulation of dor expression, we measured DMSO reductase synthesis and β-galactosidase activity from dor::lacZ fusions in strains containing mutations in the redox-active proteins CcoP and RdxB, which have previously been implicated in the generation of a redox signal affecting photosynthesis gene expression. Unlike the wild-type strain, both mutants were able to synthesize DMSO reductase under strictly aerobic conditions, even in the absence of DMSO. When cells were grown photoheterotrophically, dorC::lacZ expression was stimulated by increasing light intensity in the CcoP mutant, whereas it is normally repressed in the wild-type strain under such conditions. Furthermore, the expression of genes encoding the DorS sensor kinase and DorR response regulator proteins was also affected by the ccoP mutation. By using CcoP-DorR and CcoP-DorY double mutants, it was shown that the DorR protein is strictly required for altered dor expression in CcoP mutants. These results further demonstrate a role for redox-generated responses in the expression of genes encoding DMSO reductase in R. sphaeroides and identify the DorS-DorR proteins as a redox-dependent regulatory system controlling dor expression.
The ability of the facultative phototrophic bacterium Rhodobacter sphaeroides to respire anaerobically using dimethyl sulfoxide (DMSO) or trimethylamine-N-oxide (TMAO) as an alternative electron acceptor has been well documented (15, 32). A single enzyme, DMSO reductase (DMSOR), is responsible for the reduction of both compounds and has been characterized at the structural and biochemical levels, as well as at the genetic level (13, 16, 23, 24, 27, 30).
We demonstrated previously that the structural components of DMSOR of R. sphaeroides 2.4.1T are encoded by the dorCBA operon, whose expression is dependent on the presence of DMSO (or TMAO) and the absence of oxygen (Fig. 1) (16). Upstream of this operon are genes encoding regulators of dorCBA expression (16). The dorS and dorR genes, respectively, encode the sensor kinase and response regulator components of a two-component sensory-transduction system, which was previously suggested to be involved in DMSO-dependent dor gene regulation (Fig. 1) (19, 28). The dorX and dorY genes appear to encode a novel regulatory system which is required for the activation of dorCBA expression (Fig. 1) (17). The DorX protein appears to possess a DNA-binding domain at its carboxy terminus and a transmembrane domain at its amino terminus, suggesting an interaction with the cytoplasmic membrane. We have also demonstrated a requirement for the FnrL protein, a homolog of the Escherichia coli Fnr protein that serves as a global regulator of gene expression in response to anaerobiosis, in the expression of the dorS gene, which in turn affects the synthesis of DMSOR (19, 33). Thus, the expression of the dor genes is subject to regulation by multiple signals through multiple regulatory pathways.
FIG. 1.
Physical map of the dor locus of R. sphaeroides 2.4.1T. The large arrows show the genes and the directions of transcription. The functions of the gene products are shown below the genes. The small arrows indicate upstream regulatory sequence (URS) regions, and the stem-loop structures show putative rho-independent transcriptional terminators.
In our laboratory, a number of regulatory components which mediate oxygen-dependent control of photosynthesis gene expression in R. sphaeroides have been identified. These include the Prr regulatory system, the AppA-PpsR system, the TspO protein, and the FnrL protein (7, 8, 10, 31, 34). In addition to these elements, mutations in the ccoNOQP gene cluster, encoding the cbb3 oxidase, or in the rdxB gene, encoding a membrane-bound ferredoxin-like protein, result in the oxygen-insensitive formation of photosynthetic membranes and altered carotenoid accumulation (21, 35). It has been proposed that these membrane protein complexes generate a redox signal or intermediate which normally inhibits photosynthesis gene expression under high oxygen tensions. When the oxygen concentration is lowered, it is presumed that there is a decrease in or loss of this signal, which allows the expression of photosynthesis genes. The ability for this signal to be translated into altered photosynthesis gene expression has been shown to occur through the Prr system and possibly also via the FnrL protein (20, 35).
Since dor expression is also dependent on the absence of oxygen and is affected by light intensity, we wished to determine whether mutations in the ccoNOQP gene cluster and rdxB led to any effect on dor expression in response to the signal generated through their gene products. As we demonstrate here, the lack of ccoP or rdxB resulted in altered dor expression, suggesting that the Cco-Rdx-generated signal is required for normal dor expression. Furthermore, we can now redefine the DorSR regulatory system as being responsive to redox and not to DMSO, as we previously suggested (19). The results reported here also demonstrate that redox-dependent signalling via the Cco-Rdx proteins is not restricted solely to photosynthesis gene expression but appears to be a global signalling system regulating anaerobic gene expression in R. sphaeroides 2.4.1T.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions.
Bacterial strains and plasmids used or constructed in this work are listed in Table 1. E. coli strains were grown at 37°C in Luria-Bertani medium, and R. sphaeroides strains were grown at 30°C in Sistrom’s minimal medium A containing succinate as the carbon source (3, 29). Where appropriate, DMSO was added at a final concentration of 60 mM. Cells were grown anaerobically in sealed glass tubes and were incubated in the dark for anaerobic-dark growth or in front of a 10-W m−2 light source for photoheterotrophic growth, except in the experiments involving different light intensities. Aerobic cultures were grown by continuous sparging with a mixture of 30% O2–69% N2–1% CO2. Semiaerobic growth was achieved by growing cells on a rotary shaker in glass tubes. Antibiotics were used as follows to maintain selection for plasmids or to select for recombinant strains: ampicillin, 100 μg ml−1 (E. coli); kanamycin, 25 μg ml−1 (R. sphaeroides and E. coli); spectinomycin, 25 μg ml−1 (R. sphaeroides and E. coli); streptomycin, 25 μg ml−1 (R. sphaeroides and E. coli); tetracycline, 1 μg ml−1 (R. sphaeroides) and 10 μg ml−1 (E. coli); and trimethoprim, 50 μg ml−1 (R. sphaeroides).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype and/or characteristic(s) | Reference or source |
|---|---|---|
| R. sphaeroides | ||
| 2.4.1T | Wild-type | 29 |
| CCOP1 | ccoP::ΩTpr | 21 |
| RDXB1 | rdxB::ΩTpr | 21 |
| NM16 | dorR::ΩStr/Spr | 16 |
| NM22 | dorY::ΩStr/Spr | 17 |
| NM23 | ccoP::ΩTprdorY::ΩStr/Spr | This study |
| NM24 | ccoP::ΩTprdorR::ΩStr/Spr | This study |
| E. coli | ||
| DH5αphe | F− φ80dlacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17(rK− mK+) supE44 λ− thi-1 gyrA relA1 phe::Tn10dCm | 7 |
| HB101 | F− Δ(gpt-proA)62 leuB6 supE44 ara-14 glaK2 lacYI Δ(mcrC-mrr) rpsL20 (Str) xyl-5 mtl-1 recA13 | 1 |
| Plasmids | ||
| pRK2013 | Conjugative helper plasmid | 9 |
| pML5 | Promoterless lacZ transcriptional fusion vector, Tcr | 14 |
| pNMT40 | pSUP202 containing 3.0-kb XhoI frag-ment containing dorR::Ω Str/Spr | 16 |
| pNMT77 | pML5 containing dorS::lacZ | 19 |
| pNMT78 | pML5 containing dorC::lacZ | 19 |
| pNMT91 | pSUP202 containing dorY::Ω Str/Spr | 17 |
| pNMT94 | pML5 containing dorR::lacZ | 19 |
| pNMT114 | pML5 containing dorX::lacZ | 17 |
DNA manipulations.
The integration sites for the antibiotic resistance cassettes in the R. sphaeroides genome were confirmed by nonradioactive Southern hybridizations of restriction digests of genomic DNA probed with appropriate DNA fragments, as described previously (22). Labeling of DNA probes and detection of hybridized sequences by chemiluminescence were performed using a NEBlot Phototope kit (New England Biolabs Inc., Beverly, Mass.) according to the manufacturer’s instructions.
Conjugation techniques.
The dorR::Ω and dorY::Ω insertion or insertion-deletion mutation pSUP202-derived plasmids were conjugated into R. sphaeroides 2.4.1T by using triparental matings with the suicide vector pRK2013, as described previously (5). The plasmid-borne mutated genes were integrated into the R. sphaeroides genome by homologous recombination.
Cell extract preparation and assays of β-galactosidase activity.
Preparation of crude cell extracts and determination of β-galactosidase activities were performed as described previously (25), using reagent-grade o-nitrophenyl-β-d-galactopyranoside (Sigma Chemical Co., St. Louis, Mo.) as the substrate.
Determination of protein concentration.
Cell extract protein concentrations were determined with the Pierce bicinchoninic acid protein assay reagent (Pierce, Rockford, Ill.), using bovine serum albumin as a reference standard.
Immunoblotting.
Crude cell extracts separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12.5% polyacrylamide gels were transferred to nitrocellulose membranes by wet electrotransfer in 50 mM Tris–380 mM glycine–0.1% SDS–20% methanol buffer. The DMSOR (DorA) polypeptide was detected on the protein blots by the alkaline phosphatase color detection system (Promega Corp., Madison, Wis) with polyclonal rabbit antiserum against purified R. capsulatus DorA protein (1:2,500 dilution) and secondary goat anti-rabbit alkaline phosphatase-linked immunoglobulin (1:25,000 dilution).
Heme staining.
Heme staining of polypeptides electrophoresed on 15% polyacrylamide gels was performed with 3,3′,5,5′-tetramethylbenzidine according to the previously described protocol of Thomas et al. (26).
RESULTS
CcoP and RdxB mutants exhibit altered DMSOR protein production.
Previous studies demonstrated that strains possessing mutations in either the ccoNOQP gene cluster or in the rdxB gene appear to have altered expression of genes regulated by anaerobiosis (20, 21, 35). Since genes for DMSOR are also known to be under anaerobic control, we wondered if cco and rdxB mutations likewise affected dor expression. Our initial experiments focused on whether CcoP or RdxB mutants had altered expression of c-type cytochromes. R. sphaeroides cells were grown semiaerobically, and protein extracts from mid-log-phase cultures were analyzed for c-type cytochromes by staining SDS-polyacrylamide gels of electrophoresed protein extracts with 3,3′,5,5′-tetramethylbenzidine, as described previously (16). In both the presence and absence of DMSO, the CcoP and RdxB mutants synthesized the 44-kDa c-type cytochrome which is associated with DMSOR and which we previously showed to be encoded by the dorC gene (data not shown) (16). This is in contrast to the wild-type strain, in which DorC was synthesized only in the presence of DMSO. It should be noted here that the synthesis of other c-type cytochromes synthesized by R. sphaeroides was unaffected, except in the CcoP mutant, in which the ccoP-encoded cytochrome was absent.
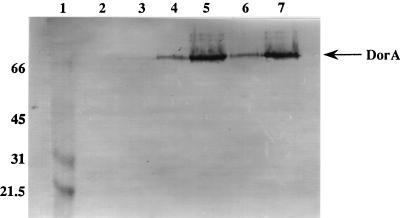
To more specifically examine the effects of the ccoP and rdxB mutations on DMSOR protein production, we performed an immunoblotting experiment using anti-DorA antiserum with cells grown with 30% O2 in the presence or absence of DMSO. Immunoblot analysis of crude protein extracts from the wild-type strain revealed that under these conditions, DMSOR was not produced (Fig. 2, lanes 2 and 3). These results differ from those of the heme staining experiment since the cells were grown here with 30% O2 (fully aerobic) and not semiaerobically. In contrast, both the CcoP and RdxB mutants synthesized detectable levels of DMSOR, even in the absence of DMSO (Fig. 2, lanes 4 to 7). However, DMSOR production was stimulated by DMSO in these strains, indicating that DMSO-dependent regulation was still apparent in the CcoP and RdxB mutants. These results suggested that the Cco-Rdx-generated redox signal is required for normal regulation of DMSOR synthesis. Since the effects on DorA and DorC production were comparable for both the CcoP and RdxB mutants and since the effects on photosynthesis gene expression have been shown to be identical for both mutants, the following experiments were performed only with the CcoP mutant (or derivatives thereof) (21).
FIG. 2.
Synthesis of DorA by wild-type and CcoP and RdxB mutant strains of R. sphaeroides. Fifty micrograms of whole-cell proteins were subjected to SDS-PAGE and blotted onto nitrocellulose membranes. The presence of DorA was detected with polyclonal antiserum to DorA and visualized by the AP detection system (Promega). Lanes: 1, molecular weight markers (in thousands) (Bio-Rad); 2, 2.4.1T aerobic; 3, 2.4.1T aerobic plus DMSO; 4, CCOP1 aerobic; 5, CCOP1 aerobic plus DMSO; 6, RDXB1 aerobic; 7, RDXB1 aerobic plus DMSO.
dorC::lacZ expression is affected in the CcoP mutant.
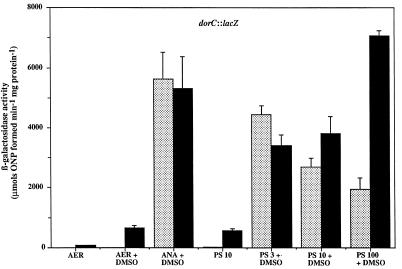
To determine whether the effects on DMSOR protein synthesis were due to altered transcription of the dorCBA operon, which encodes the structural components of DMSOR, we measured β-galactosidase activity from a dorC::lacZ transcriptional fusion which was introduced into wild-type and CcoP mutant strains on a low-copy-number plasmid. It was previously shown that the copy number of plasmids with an RSF1010 replicon, such as pML5, is four to six in R. sphaeroides 2.4.1T and does not vary with growth condition, and thus, our interpretation of these results is not likely to be affected by plasmid copy number (25). Cells were grown under different environmental conditions, and their β-galactosidase activities were assayed. As we showed previously, the wild-type strain, 2.4.1T, exhibited extremely low levels of activity after growth under strictly aerobic (30% O2) conditions, even when DMSO was present (Fig. 3) (19). In contrast, the CcoP mutant was able to expressdorC::lacZ at much higher levels than the wild type in both the presence and absence of DMSO, although an approximately sevenfold increase in dorC::lacZ expression was observed in the presence of DMSO compared to levels of expression in the absence of DMSO. An approximately 50-fold increase in β-galactosidase activity was observed for the CcoP mutant, compared to the wild-type strain, after photosynthetic growth in the absence of DMSO. However, DMSO-dependent induction of expression was still observed. As we demonstrated previously, dorC::lacZ expression in the wild-type strain is repressed with increasing light intensity in the presence of DMSO (19). Here, however, dorC::lacZ expression in the CcoP mutant increased with increasing light intensity, although slightly decreased levels of dorC::lacZ expression were observed under low and medium light intensities when compared to the levels observed under anaerobic-dark growth conditions in the presence of DMSO. At the high light intensity (100 W m−2) dorC::lacZ expression was approximately 3.5-fold higher in the CcoP mutant than in the wild-type strain. It should also be noted here that in the absence of ccoP the expression of dorC::lacZ was unaffected when cells were grown under anaerobic-dark conditions in the presence of DMSO. These results suggest that the transcription of dorC is indeed affected by the ccoP mutation but that DMSO-dependent regulation still exists. Furthermore, it appears that the Cco-generated signal is required for the normal regulation of dorC expression in response to increasing light intensity under photosynthetic growth conditions.
FIG. 3.
β-Galactosidase activities from cell extracts of R. sphaeroides strains containing the dorC::lacZ transcriptional fusion plasmid pNMT78. Strains are 2.4.1T (░⃞) and CCOP1 (■). Growth conditions are as follows: AER, aerobically; ANA, anaerobically in the dark; PS 3, photosynthetically at a light intensity of 3 W m−2; PS 10, photosynthetically at 100 W m−2; PS 100, photosynthetically at 100 W m−2. Where indicated cultures were supplemented with DMSO to a final concentration of 60 mM. Results are the mean values from triplicate assays of at least three independent cultures and are corrected for activity from the vector alone under the same conditions (pML5, <35 μmol of o-nitrophenol [ONP] formed min−1 mg of protein−1). Vertical bars represent the standard deviations from the means.
Effects of the ccoP mutation on expression of genes encoding regulatory proteins.
Since the expression of dorC has been shown to be dependent on the regulatory proteins DorS, DorR, and DorXY, we determined whether the expression of the genes encoding these regulators was also affected in the CcoP mutant background (17, 19). Plasmids containing dorS::lacZ, dorR::lacZ, and dorX::lacZ transcriptional fusions were introduced into the CcoP mutant, and as for the dorC::lacZ expression studies, β-galactosidase activities were assayed after growth under different conditions.
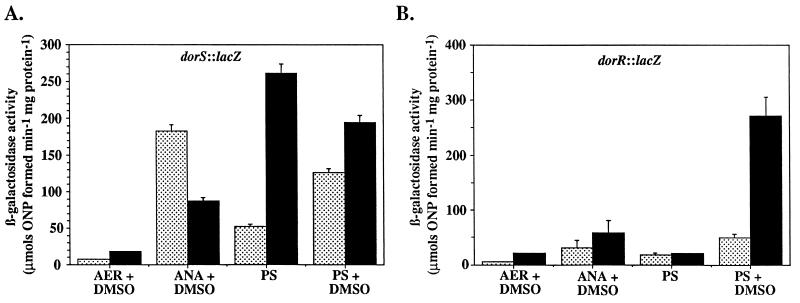
Measurement of β-galactosidase activities from the dorS::lacZ and dorR::lacZ fusions revealed altered patterns of expression for both fusions in the CcoP mutant when compared to expression in the wild-type strain. dorS::lacZ expression was increased approximately 1.5 to 2-fold in the CcoP mutant when cells were grown aerobically or photosynthetically with DMSO and decreased approximately twofold when cells were grown under anaerobic-dark conditions with DMSO compared to expression in the wild-type strain under similar conditions (Fig. 4A). The most significant effect was an approximately fivefold increase in dorS::lacZ expression in the CcoP mutant, over that in the wild type, after photosynthetic growth in the absence of DMSO.
FIG. 4.
(A) β-Galactosidase activities from cell extracts of R. sphaeroides strains containing the dorS::lacZ transcriptional fusion plasmid pNMT77. (B) β-Galactosidase activities from cell extracts of R. sphaeroides strains containing the dorR::lacZ transcriptional fusion plasmid pNMT94. Strains are 2.4.1T (░⃞) and CCOP1 (■). Growth conditions are as follows: AER, aerobically; ANA, anaerobically in the dark; PS, photosynthetically at a light intensity of 10 W m−2. Where indicated cultures were supplemented with DMSO to a final concentration of 60 mM. Results are the mean values from triplicate assays of at least three independent cultures and are corrected for activity from the vector alone under the same conditions (pML5, <35 μmol of o-nitrophenol [ONP] formed min−1 mg of protein−1). Vertical bars represent the standard deviations from the means.
In contrast to dorS::lacZ expression, the ccoP mutation had no effect on dorR::lacZ expression when cells were grown photosynthetically without DMSO but resulted in a sixfold increase in expression in the presence of DMSO when compared to levels of β-galactosidase activity for the wild-type strain (Fig. 4B). Approximately twofold increases in dorR::lacZ expression were observed for the CcoP mutant when cells were grown aerobically or anaerobically in the dark with DMSO compared to those for the wild-type strain.
We have previously demonstrated that dorX::lacZ expression appears to be constitutive and essentially unregulated (17). In contrast to the expression of dorS and dorR, the ccoP mutation did not affect normal levels of dorX::lacZ expression under any growth condition (data not shown). The results indicate that the expression of the dorS and dorR gene, but not that of the dorX gene, are normally responsive to the Cco-generated signal.
The DorSR system is required for CcoP-mediated effects on dorC::lacZ expression.
In order to determine which, if either, of the DorXY and DorSR regulatory systems was responsive to the redox changes manifested by the ccoP mutation, we constructed DorY-CcoP and DorR-CcoP double mutants, as described above. The dorC::lacZ fusion plasmid, pNMT78, and the vector plasmid, pML5, were both introduced into these double-mutant strains, and β-galactosidase activities were assayed after photosynthetic growth with medium light intensity (10 W m−2) in the presence or absence of DMSO. In the wild-type background, the presence of either the dorY or dorR mutation resulted in extremely low levels of dorC::lacZ expression, even in the presence of DMSO (Table 2). In contrast, in the CcoP-DorY double mutant, strain NM23, there was dorC::lacZ expression in the presence of DMSO but at a level approximately 20% of that in the CcoP single mutant. Very low expression was observed for strain NM23 in the absence of DMSO, comparable to that of the DorY mutant alone, but at a level 2% of that in the CcoP single mutant. The CcoP-DorR double mutant, strain NM24, exhibited very low levels of dorC::lacZ expression in either the presence or absence of DMSO after photosynthetic growth (Table 2). These results suggest that the DorSR system, but not DorXY, is obligatory for the CcoP-mediated effects on dorC::lacZ expression but that both systems play significant roles in dorC expression.
TABLE 2.
Effects of dorY, dorR, and ccoP mutations on dorC::lacZ expression
| Strain | β-Galactosidase activity after
growtha
|
|
|---|---|---|
| Photoheterotrophic | Photoheterotrophic + DMSO | |
| 2.4.1T (wild type) | 12 ± 7 | 2,683 ± 297 |
| NM16 (DorR) | 17 ± 5 | 39 ± 5 |
| NM22 (DorY) | 19 ± 7 | 23 ± 14 |
| CCOP1 (CcoP) | 567 ± 70 | 3,813 ± 569 |
| NM23 (DorY CcoP) | 12 ± 10 | 783 ± 60 |
| NM24 (DorR CcoP) | 2 ± 1 | 7 ± 3 |
Cultures were grown to mid-log phase under photosynthetic conditions at a light intensity of 10 W m−2 in the presence or absence of 60 mM DMSO. Cell extracts were assayed for β-galactosidase activity. Units of activity are micromoles of o-nitrophenol (ONP) formed minute−1 milligram of protein−1. Values represent the mean values ± the standard deviations from triplicate assays of at least three independent growths and are corrected for activity from the vector alone under the same conditions (pML5, <35 μmol of ONP formed min−1 mg of protein−1).
DMSOR activity is required for dorC::lacZ expression.
We were interested to know whether redox flux brought about by the activity of the DMSOR enzyme itself affected dorC::lacZ expression. In order to determine whether the absence of DMSOR affects dorC expression, we introduced plasmids pML5 (vector) and pNMT78 (dorC::lacZ) into strain NM19, which contains an insertion of the ΩSt/Spr cassette in dorA and thus is unable to make DMSOR or exhibit DMSOR specific activity (16). Intriguingly, in strain NM19 dorC::lacZ expression appeared to be insensitive to the presence or absence of DMSO when cells were grown photosynthetically (Table 3). Furthermore, the DorA mutant was able to express dorC::lacZ in the absence of DMSO, unlike the wild-type strain. However, dorC::lacZ expression in the presence of DMSO for strain NM19 was approximately 60% less than that for the wild-type strain. Under aerobic growth conditions, the effects of the DorA mutation are more reduced, and only a small, but reproducible, increase in dorC::lacZ expression was observed when cells were grown in the presence of DMSO (Table 3). The results suggest that the functioning of DMSOR is required for normal dorC regulation.
TABLE 3.
Effects of a dorC mutation on dorC::lacZ expression
| Strain | β-galactosidase activity after
growtha:
|
|||
|---|---|---|---|---|
| Aerobic | Aerobic + DMSO | Photohetero-trophic | Photoheterotrophic + DMSO | |
| 2.4.1T (wild type) | 1 ± 1 | 4 ± 3 | 12 ± 7 | 2,683 ± 297 |
| NM19 (DorA) | 4 ± 3 | 30 ± 4 | 932 ± 53 | 1,048 ± 172 |
Cultures were grown to mid-log phase under the conditions indicated and assayed for β-galactosidase activity. Photosynthetic growth occurred at a light intensity of 10 W m−2. Units of activity are micromoles of o-nitrophenol (ONP) formed minute−1 milligram of protein−1. Values represent the mean values ± the standard deviations from triplicate assays of at least three independent growths and are corrected for activity from the vector alone under the same conditions (pML5, <35 μmol of ONP formed min−1 mg of protein−1).
DISCUSSION
Our previous work demonstrated that the regulation of genes encoding components of DMSOR in R. sphaeroides 2.4.1T was complex and required several regulatory factors (17, 19, 33). We showed that the expression of the dorCBA operon was regulated by the presence of DMSO and the absence of oxygen and required the regulatory proteins FnrL, DorSR, and DorXY. Further, dorC::lacZ expression was shown to be modulated by varying light intensity, which we believe to be representative of differing redox balances or intermediates in the cell during photosynthetic growth conditions (19). In this study, we extended our observations on dor gene regulation to include effects mediated by the Cco and Rdx systems, which have previously been suggested to be responsible for the generation of a redox-dependent signal or intermediate which is inhibitory for photosynthesis gene expression under high oxygen tensions (20, 21, 35).
Our initial experiments focused on the ability of strains possessing mutations in either the ccoP or rdxB gene to synthesize components of DMSOR. Analysis of crude protein extracts by heme staining and immunoblotting with anti-DorA antiserum revealed that the CcoP and RdxB mutants were equally able to synthesize the DorC c-type cytochrome and the DorA DMSOR protein under aerobic growth conditions (Fig. 2 and data not shown). This is in contrast to the wild-type strain, which is unable to produce the DMSOR components under aerobic growth conditions. These results indicated to us that the Cco-Rdx-generated signal is important in the regulation of DMSOR synthesis.
Analysis of the expression of dor::lacZ transcriptional fusions in the CcoP mutant demonstrated that the effects of the ccoP mutation occurred at the transcriptional level. The CcoP mutant exhibited altered dorC::lacZ expression under both aerobic and photosynthetic growth conditions when compared to that of the wild-type strain (Fig. 3). The most striking example was the increase in dorC::lacZ expression observed when cells were grown with increasing light intensity. Normally in the wild-type strain dorC::lacZ expression is repressed by increasing light intensity (19). We believe that this difference is due to the loss of or alteration in a redox signal or intermediate caused by the lack of the CcoP protein and that this signal or intermediate is normally derived from light-mediated reactions (see below).
The expression of genes encoding the regulatory proteins DorS and DorR was also affected by the ccoP mutation, albeit the effects on the two genes were different from each other (Fig. 4). We previously demonstrated that the expression of dorS and dorR is autoregulated by the DorS and DorR proteins themselves (19). Furthermore, the expression of dorS was shown to be induced by the FnrL protein in response to lowering oxygen tension. It was previously demonstrated that the Cco-Rdx pathway affects the expression of the FnrL-regulated hemA gene, which encodes 5-aminolevulinic acid synthase (35). Whether the effects of the Cco-Rdx pathway are exerted directly on the activity of FnrL remains to be addressed. Recent evidence from our laboratory suggests that the CcoP mutation has little effect on fnrL::lacZ expression (6). Since dorS expression is regulated by FnrL, the ccoP mutation may affect not only the DorSR system (see below) but also FnrL, and this may account for the differences in expression between the dorS and dorR genes, since we believe that dorR expression is not regulated directly by FnrL (19).
We have previously demonstrated the requirement for two specific regulatory systems in the activation of dorC expression, DorXY and DorSR (17, 19). In order to determine which of these systems may be affected by the ccoP mutation, we determined dorC::lacZ expression in CcoP-DorR and CcoP-DorY double mutants (Table 2). In contrast to the CcoP-DorR double mutant, which expresses dorC::lacZ at extremely low levels under all conditions, the CcoP-DorY double mutant is able to express dorC::lacZ, albeit at levels lower than those expressed by the wild-type strain. The results suggest that the ccoP mutation still results in a partial phenotype in the DorY background but not in the DorR background, indicating that CcoP normally functions with the DorSR proteins to control dorC expression. This suggests that the ccoP mutation increases the activity of DorS, leading to high levels of phosphorylated DorR, which is able to activate dor transcription in the absence of DorY. However, the CcoP-DorY double mutant does exhibit reduced levels of dorC::lacZ expression compared to levels for the CcoP single mutant, demonstrating the requirement for DorY for maximal expression. The results also suggest that DMSO stimulates the DorSR system, allowing for expression of dorC::lacZ in the CcoP-DorY double mutant in the presence of DMSO.
In these respects, the DorSR system resembles the Prr system, which communicates with the CcoP and RdxB proteins to regulate gene expression in response to redox (4, 7, 20). Furthermore, both systems resemble the ArcBA system of E. coli, which functions as a regulator of gene expression in response to decreasing oxygen tensions through either sensing the redox state of a component of the electron transport chain or by sensing the levels of some compound associated with redox poise (11, 12). The functional homology between DorSR and ArcBA is an interesting one, especially in light of the fact that ArcB has been identified as a member of a family of proteins which possess S (sensory) boxes that have been suggested to form a common fold required for proteins that sense redox or related stimuli (2). Comparison of the residues which form the S boxes of ArcB and the N-terminal signaling domain of DorS reveals that many of these residues are conserved between both proteins, indicating that DorS belongs to this family of proteins (18). It would therefore be of interest to perform a detailed biochemical study of DorS.
Since the DMSOR enzyme itself is a redox-active protein, requiring electrons for the reduction of DMSO to dimethyl sulfide, we were interested in determining whether loss of this reductase protein (or activity) affected dor expression. Measurement of dorC::lacZ expression in a DorA mutant revealed that indeed a functional DMSOR is required for normal dorC expression. Thus, it appears that activity of DMSOR plays a key role in the dor regulatory pathway, possibly by monitoring electron flux through the DorCBA proteins via the DorSR and/or DorXY regulatory systems. It is thus speculative to suggest that the cell senses the presence of DMSO, not directly by sensing the DMSO molecule itself, but rather by sensing the activity of the DMSOR enzyme, which utilizes DMSO as a substrate. Further experiments are clearly required to investigate this hypothesis.
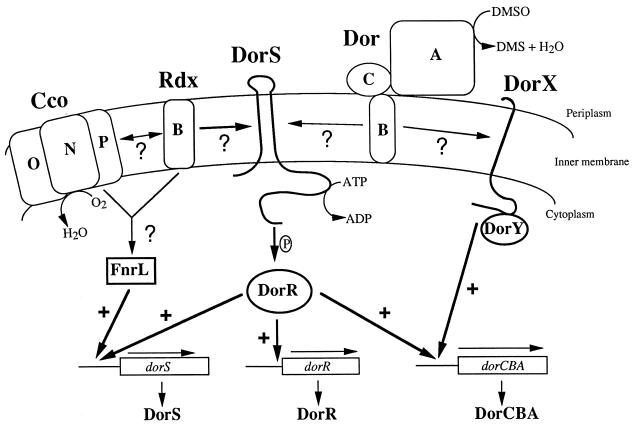
Taking these results together with those of our previous studies, we propose a revised and more extensive model for the regulation of DMSOR (dor) gene expression in R. sphaeroides 2.4.1T. Under aerobic growth conditions the Cco oxidase and the RdxB protein either transmit a redox signal or allow accumulation of a redox intermediate which directly or indirectly keeps FnrL in an inactive state (Fig. 5). Consequently, expression of the dorS, dorR, and dorCBA genes is low and DMSOR is not synthesized. As the oxygen concentration is lowered, the Cco-Rdx-mediated redox signal is either diminished, abolished, or modified such that FnrL is able to activate dorS expression. DorS is also responsive to the Cco-Rdx-generated redox signal, and as a result, DorS becomes autophosphorylated and is able to phosphorylate its cognate regulator, DorR. Phosphorylated DorR is able to activate transcription of the dorR and dorS genes in the absence of DMSO. When DMSO is present, the dorCBA operon is expressed via DorR and the DorXY system and functional DMSOR is produced. Furthermore, since DMSOR is required for normal dorCBA expression, we propose that a redox signal is transmitted from DMSOR itself to either or both of the DorXY and DorSR systems to modulate dor expression. Under photosynthetic conditions in the presence of DMSO, increasing light intensity results in an alteration of the Cco-Rdx-generated redox signal and dorCBA expression is repressed. Clearly, many further studies are required to validate this model, but we believe that this model provides the framework for future experiments.
FIG. 5.
Model for regulation of DMSO reductase (dor) gene expression in R. sphaeroides 2.4.1T. Question marks indicate unknown regulatory signals. Plus signs indicate activating roles for the various regulatory proteins. See the text for further details.
ACKNOWLEDGMENTS
We thank A. G. McEwan for generously providing antiserum to the R. capsulatus DorA protein. We also thank Jesus Eraso for helpful discussions and for critical reading of the manuscript.
This work was supported by U.S. Public Health Service grant GM15590 to S.K.
REFERENCES
- 1.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 2.Capaldi R A. Structure and function of cytochrome coxidase. Annu Rev Biochem. 1990;59:569–596. doi: 10.1146/annurev.bi.59.070190.003033. [DOI] [PubMed] [Google Scholar]
- 3.Cohen-Bazire G, Sistrom W R, Stanier R Y. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Comp Physiol. 1956;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 4.Cramer W A, Knaff D B. Energy transduction in biological membranes. Berlin, Germany: Springer-Verlag KG; 1990. [Google Scholar]
- 5.Davis J, Donohue T J, Kaplan S. Construction, characterization, and complementation of a puf mutant of Rhodobacter sphaeroides. J Bacteriol. 1988;170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eraso, J. M., and S. Kaplan. Unpublished observations.
- 7.Eraso J M, Kaplan S. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eraso J M, Kaplan S. Oxygen-insensitive synthesis of the photosynthesis membranes of Rhodobacter sphaeroides: a mutant histidine kinase. J Bacteriol. 1995;177:2695–2706. doi: 10.1128/jb.177.10.2695-2706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figurski D H, Helsinki D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomelsky M, Kaplan S. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides2.4.1. J Bacteriol. 1997;179:128–134. doi: 10.1128/jb.179.1.128-134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iuchi S, Chepuri V, Fu H-A, Gennis R B, Lin E C C. Requirement for terminal cytochromes in generation of the aerobic signal for the arc regulatory system in Escherichia coli: study utilizing deletions and lac fusions of cyo and cyd. J Bacteriol. 1990;172:6020–6025. doi: 10.1128/jb.172.10.6020-6025.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iuchi S, Matsuda Z, Fujiwara T, Lin E C C. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arcmodulon. Mol Microbiol. 1990;4:715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- 13.Kisker C, Schindelin H, Rees D C. Molybdenum-cofactor-containing enzymes: structure and mechanism. Annu Rev Biochem. 1997;66:233–267. doi: 10.1146/annurev.biochem.66.1.233. [DOI] [PubMed] [Google Scholar]
- 14.Labes M, Pühler A, Simon R. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene. 1990;89:37–46. doi: 10.1016/0378-1119(90)90203-4. [DOI] [PubMed] [Google Scholar]
- 15.McEwan A G. Photosynthetic electron transport and anaerobic metabolism in purple non-sulfur bacteria. Antonie van Leeuwenhoek. 1994;66:151–164. doi: 10.1007/BF00871637. [DOI] [PubMed] [Google Scholar]
- 16.Mouncey N J, Choudhary M, Kaplan S. Characterization of genes encoding dimethylsulfoxide reductase of Rhodobacter sphaeroides 2.4.1T: an essential metabolic gene function encoded on chromosome II. J Bacteriol. 1997;179:7617–7624. doi: 10.1128/jb.179.24.7617-7624.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mouncey, N. J., and S. Kaplan. Unpublished observations.
- 18.Mouncey, N. J., and S. Kaplan. Unpublished observations.
- 19.Mouncey N J, Kaplan S. Cascade regulation of DMSO reductase (dor) gene expression in the facultative phototroph Rhodobacter sphaeroides 2.4.1T. J Bacteriol. 1998;180:2924–2930. doi: 10.1128/jb.180.11.2924-2930.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Gara J P, Eraso J M, Kaplan S. A redox-responsive pathway for aerobic regulation of photosynthesis gene expression in Rhodobacter sphaeroides2.4.1. J Bacteriol. 1998;180:4044–4050. doi: 10.1128/jb.180.16.4044-4050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Gara J P, Kaplan S. Evidence for the role of redox carriers in photosynthesis gene expression and carotenoid biosynthesis in Rhodobacter sphaeroides2.4.1. J Bacteriol. 1997;179:1951–1961. doi: 10.1128/jb.179.6.1951-1961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Satoh T, Kurihara F N. Purification and properties of dimethylsulfoxide reductase containing a molybdenum cofactor from a photodenitrifier, Rhodopseudomonas sphaeroides f.s.p. denitrificans. J Biochem. 1987;102:191–197. doi: 10.1093/oxfordjournals.jbchem.a122032. [DOI] [PubMed] [Google Scholar]
- 24.Schindelin H, Kisker C, Hilton J, Rajagopalan K V, Rees D C. Crystal structure of DMSO reductase: redox-linked changes in molybdopterin coordination. Science. 1996;272:1615–1621. doi: 10.1126/science.272.5268.1615. [DOI] [PubMed] [Google Scholar]
- 25.Tai T-N, Havelka W A, Kaplan S. A broad host-range vector system for cloning and translational lacZfusion analysis. Plasmid. 1988;19:175–188. doi: 10.1016/0147-619x(88)90037-6. [DOI] [PubMed] [Google Scholar]
- 26.Thomas P E, Ryan D, Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 27.Ujiiye T, Nakama H, Okubo A, Yamazaki S, Satoh T. Nucleotide sequence of the genes, encoding the pentaheme cytochrome (dmsC) and the transmembrane protein (dmsB), involved in dimethyl sulfoxide respiration from Rhodobacter sphaeroides f. sp. denitrificans. Biochim Biophys Acta. 1996;1277:1–5. doi: 10.1016/s0005-2728(96)00101-6. [DOI] [PubMed] [Google Scholar]
- 28.Ujiiye T, Yamamoto I, Satoh T. The dmsR gene encoding a dimethyl sulfoxide-responsive regulator for expression of dmsCBA (dimethyl sulfoxide respiration genes) in Rhodobacter sphaeroides f. sp. denitrificans. Biochim Biophys Acta. 1997;1353:84–92. doi: 10.1016/s0167-4781(97)00062-6. [DOI] [PubMed] [Google Scholar]
- 29.van Neil C B. The culture, general physiology, morphology, and classification of the non-sulfur purple and brown bacteria. Bacteriol Rev. 1944;8:1–118. doi: 10.1128/br.8.1.1-118.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto I, Wada N, Ujiiye T, Tachibana M, Matsuzaki M, Kajiwara H, Watanabe Y, Hirano H, Okubo A, Satoh T, Yamazaki S. Cloning and nucleotide sequence of the gene encoding dimethyl sulfoxide reductase from Rhodobacter sphaeroides f. sp. denitrificans. Biochim Biotechnol Biochem. 1995;59:1850–1855. doi: 10.1271/bbb.59.1850. [DOI] [PubMed] [Google Scholar]
- 31.Yeliseev A A, Kaplan S. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides2.4.1. J Biol Chem. 1995;270:21167–21175. doi: 10.1074/jbc.270.36.21167. [DOI] [PubMed] [Google Scholar]
- 32.Yen H C, Marrs B L. Growth of Rhodopseudomonas capsulatusunder dark anaerobic conditions with dimethylsulphoxide. Arch Biochem Biophys. 1977;181:411–418. doi: 10.1016/0003-9861(77)90246-6. [DOI] [PubMed] [Google Scholar]
- 33.Zeilstra-Ryalls J H, Gabbert K, Mouncey N J, Kaplan S, Kranz R G. Analysis of the fnrL gene and its function in Rhodobacter capsulatus. J Bacteriol. 1997;179:7264–7273. doi: 10.1128/jb.179.23.7264-7273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeilstra-Ryalls J H, Kaplan S. Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrLgene. J Bacteriol. 1995;177:6422–6431. doi: 10.1128/jb.177.22.6422-6431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeilstra-Ryalls J H, Kaplan S. Control of hemA expression in Rhodobacter sphaeroides2.4.1: regulation through alterations in the cellular redox state. J Bacteriol. 1996;178:985–993. doi: 10.1128/jb.178.4.985-993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]