Abstract
Long non-coding RNAs (lncRNAs) constitute the largest and most diverse class of non-coding RNAs. They localize to the nucleus, cytoplasm, or both compartments, and regulate gene expression through various mechanisms at multiple levels. LncRNAs tend to evolve faster and present higher tissue- and developmental stage-specific expression than protein-coding genes. Initially considered byproducts of erroneous transcription without biological function, lncRNAs are now recognized for their involvement in numerous biological processes, such as immune response, apoptosis, pluripotency, reprogramming, and differentiation. In this study, we focused on Heart Brake lncRNA 1 (HBL1), a lncRNA recently reported to modulate the process of pluripotent stem cell differentiation toward cardiomyocytes. We employed RT-qPCR and high-resolution RNA FISH to monitor the expression and localization of HBL1 during the differentiation progression. Our findings indicate a significant increase in HBL1 expression during mesodermal and cardiac mesodermal stages, preceding an anticipated decrease in differentiated cells. We detected the RNA in discrete foci in both the nucleus and in the cytoplasm. In the latter compartment, we observed colocalization of HBL1 with Y-box binding protein 1 (YB-1), which likely results from an interaction between the RNA and the protein, as the two were found to be coimmunoprecipitated in RNP-IP experiments. Finally, we provide evidence that HBL1, initially reported as an independent lncRNA gene, is part of the LINC00458 (also known as lncRNA-ES3 or ES3) gene, forming the last exon of some LINC00458 splice isoforms.
Subject terms: Cell biology, Stem cells
Introduction
Long non-coding RNAs are defined as transcripts that have no protein-coding function and are longer than 200 nucleotides, which distinguishes them from the other classes of non-coding RNAs, such as microRNAs (miRNAs), small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs). Localized in intergenic regions (lincRNAs) or overlapping with protein-coding genes, they are predominantly transcribed by RNA polymerase II and undergo capping, splicing, and polyadenylation. When compared with protein-coding genes, lncRNAs are more numerous, less conserved evolutionarily, generally exhibit lower expression levels and are more developmental stage- and tissue-specific1–4. They are gaining increasing recognition as important regulators of various cellular processes, such as proliferation, apoptosis, stress response, pluripotency maintenance, and differentiation, through the control of gene expression at both the transcriptional and posttranscriptional levels2,5. LncRNAs are able to fold into complex three-dimensional structures, and to bind RNA, DNA and proteins, offering platforms for complex molecular assemblies. LncRNAs can localize within the nucleus, cytoplasm, or both, where they act by distinct mechanisms6.
In the nucleus, lncRNAs are involved in nuclear organization by contributing to nuclear body formation and chromatin organization. Nuclear lncRNAs act at the epigenetic level to modulate chromatin structure and function, at the transcriptional level to either suppress or activate the transcription of neighboring or distant genes, and at the posttranscriptional level to affect RNA splicing, stability or transport7. In the cytoplasm, lncRNAs can negatively regulate gene expression by inhibiting translation and mediating RNA degradation. They can also enhance gene expression by competing with miRNA binding sites on messenger RNAs or by sequestering and inactivating miRNAs acting as miRNA molecular sponges1,6,8. The latter ones, termed competitive endogenous RNAs (ceRNAs), illustrate an interesting interplay between lncRNAs and miRNAs. Apart from their physiological functions, lncRNAs are also involved in disease development, including cancer, cardiovascular disease and neurological disorders9.
Pluripotent stem cells (PSCs) are characterized by their exceptional capacity for self-renewal and the ability to differentiate into cells across all three germ layers. This makes them widely used in the stem cell field in the areas of disease modeling, drug discovery, and regenerative medicine10. Intricate mechanisms, which are in part contingent on long non-coding RNAs, underpin the pluripotency of these stem cells6. This is particularly relevant considering that many physiological pathways in stem cells are upregulated in cancer cells, thereby contributing to their tumorigenic properties. Hence, a comprehensive understanding of lncRNA functions within stem cell biology, covering both the preservation of pluripotency and the regulation of differentiation, is crucial not only for deciphering fundamental cellular mechanisms but also for exploring potential therapeutic applications and advancing our knowledge of cancer cell biology.
Genome-wide screens in pluripotent stem cells have identified many lncRNAs as essential components for maintaining pluripotency and driving differentiation. A recently reported lncRNA, Heart Brake lncRNA 1 (HBL1), is involved in the regulation of the differentiation of human pluripotent stem cells toward cardiomyocytes (CMs) through counteracting microRNA-1 (miR-1) and guiding Polycomb Repressive Complex 2 (PRC2) chromatin occupancy11,12.
We set out our research to examine the progression of HBL1 expression and its nuclear-cytoplasmic distribution pattern during the differentiation of human induced pluripotent stem cells (hiPSCs) into cardiomyocytes. First, our findings revealed that during CM differentiation, HBL1 levels increased significantly during the mesoderm and cardiac mesoderm stages and subsequently decreased as the cells transitioned into cardiac progenitors. Second, utilizing RNA FISH, we showed that HBL1 accumulated in discrete foci within the nucleus and cytoplasm, and that in the latter compartment it colocalized with the YB-1 protein. We observed that the number of HBL1 foci initially increased and then decreased over the course of differentiation. Finally, we present evidence that HBL1 is expressed as the 3′ end exon of LINC00458 (also known as lncRNA-ES3 or ES3), resulting from intricate alternative splicing occurring at this particular genomic location.
Results
HBL1 expression peaks during the mesodermal stage of hiPSC differentiation to CMs
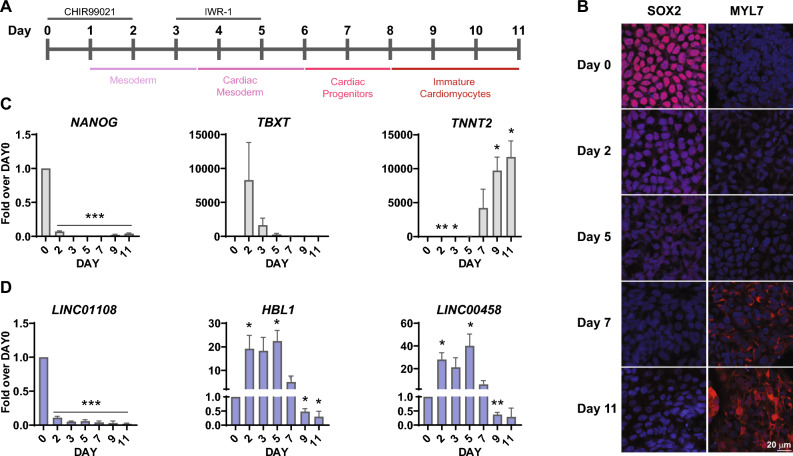
HBL1, a recently described lncRNA implicated in cardiogenesis, is present in human pluripotent stem cells but not in differentiated CMs (Day 20)11. We showed that its expression is indeed predominant in pluripotent stem cells compared with human umbilical cord-derived mesenchymal stem cells (UC-MSCs), human bone osteosarcoma epithelial cells (U2OS), and human embryonic kidney cells (HEK 293 T) (Supplementary Fig. S1) and wished to carefully monitor its exact temporal expression profile during hiPSC differentiation to CMs. We therefore applied a small molecule-based approach to differentiate cardiomyocytes from hiPSCs13 (Fig. 1A). The efficiency of differentiation was evaluated using immunofluorescence and flow cytometry. The pluripotency marker SOX2 decreased after differentiation initiation, while the expression of cardiomyocyte markers (cardiac troponin T (TNNT2) and atrial myosin light chain 2 (MYL7)) appeared on day 7, and their levels subsequently increased (Fig. 1B and Supplementary Fig. S2A). As assessed by flow cytometry, cells positive for both TNNT2 and MYL7 constituted 80% of the cell population on day 12 of differentiation (Supplementary Fig. S2B). Furthermore, RT-qPCR was applied to monitor the levels of selected differentiation markers and lncRNAs (Fig. 1C and D). NANOG was present in undifferentiated hiPSCs, and its levels decreased significantly at early differentiation timepoints (Day 2). The expression of TBXT, an early mesodermal marker, peaked on the second and third days of differentiation at the mesodermal stage. Finally, TNNT2 appeared in cardiac progenitor cells, subsequently increasing its expression level (Fig. 1C). Having confirmed the efficiency of our differentiation protocol, we further applied RT-qPCR to monitor HBL1 expression. We also included two other lncRNAs that are reported to be specifically expressed in pluripotent cells: LINC01108 (lncRNA-ES1; ES1) and LINC0045814. We expected a steep decrease in their expression levels, similar to the pluripotency-specific expression of NANOG (Fig. 1C). The anticipated pattern was true only for LINC01108, while both HBL1 and LINC00458 significantly increased at the mesodermal stage of the differentiation process and decreased only when cells committed to the cardiac differentiation pathway (Fig. 1D). This decrease is clearly correlated with increased expression of cardiac-specific genes, e.g., TNNT2 (Fig. 1C). To rule out the possibility that the observed expression is specific for the hiPSC line in use (established from endothelial progenitor cells, derived from peripheral blood), we subjected another hiPSC line (reprogrammed from peripheral blood mononuclear cells) to the same differentiation protocol. Despite the fact that differentiation efficiency appeared weaker in this cell line, as judged by TBXT and TNNT2 expression levels, HBL1 did exhibit a comparable expression profile in both cell lines (Supplementary Fig. S3). Thus, the peak of HBL1 expression between days 2 and 5 of hiPSC differentiation to CMs is not an artifact of one specific hiPSC line. The exact biological significance and function, as well as mechanisms controlling this increased expression at the mesodermal stage prior to cardiomyocyte commitment, remain to be addressed in the future. It can be hypothesized that this expression pattern is related to the role of HBL1 in guiding and maintaining PRC2 binding to cardiogenic genes, as the onset of cardiac gene expression (e.g., TNNT2) is concomitant with a decrease in HBL1 levels (Fig. 1D), thus being in line with the proposed involvement of HBL1 in cellular mechanisms controlling the maintenance (high HBL1 expression) and subsequent derepression (low HBL1 expression) of PRC2-mediated gene silencing of cardiogenesis-related factors12.
Figure 1.
Expression profile of HBL1 during hiPSC differentiation to cardiomyocytes. (A) Schematic representation of the small molecule-based differentiation protocol. Cells were incubated with the Wnt pathway activator CHIR99021 and, subsequently, the Wnt pathway inhibitor IWR-1. Beating cells started to appear around days 10–11 of differentiation. (B) Cells at selected points of differentiation were fixed and permeabilized, and immunofluorescence staining was performed to visualize selected markers of pluripotency (SOX2) and cardiomyocytes (MYL7). Scale bar = 20 µm. (C,D) RNA was purified from cells at the indicated timepoints and subjected to RT-qPCR analysis with primers amplifying (C) markers of pluripotency (NANOG), mesoderm (TBXT), and cardiomyocytes (TNNT2), as well as (D) selected lncRNAs. Data are shown as the mean ± standard deviation (n = 3). * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, Student’s t-test, two-tailed, paired.
Subcellular localization of HBL1 in hiPSCs
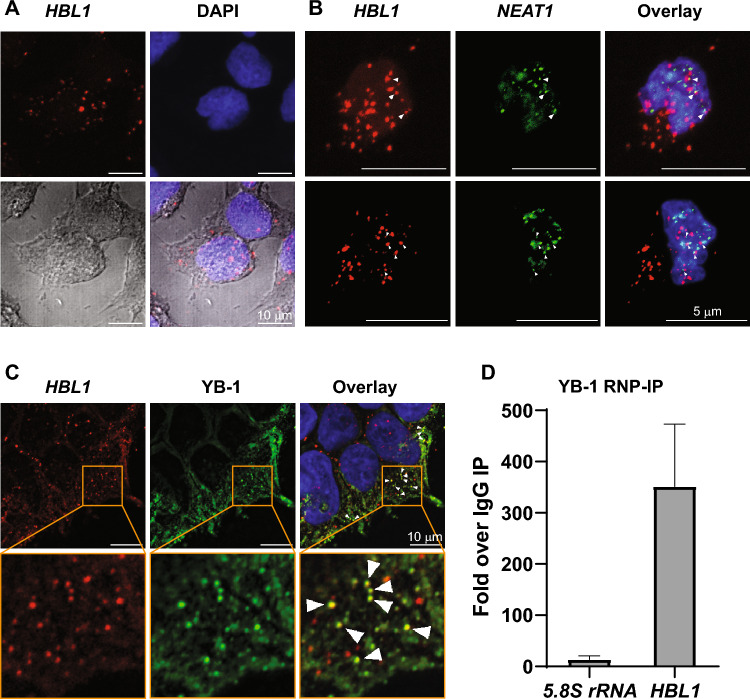
HBL1 is present in both the nucleus and the cytoplasm of human pluripotent stem cells, as reported by Liu et al. using nuclear and cytoplasmic biochemical fractionation protocol11. We wished to assess its subcellular localization at the single-cell level in human induced pluripotent stem cells by taking advantage of RNA Fluorescence In Situ Hybridization assay (RNA FISH) and confocal microscopy. First, we optimized a protocol for HBL1 lncRNA detection in human pluripotent stem cells using 20 sequence-specific probes recognizing HBL1. As shown in Fig. 2A, HBL1 lncRNA exhibits punctate localization, forming foci that are scarcely distributed in the nucleus and the cytoplasm. The specificity of the FISH probes was validated in hiPSC-derived endothelial cells that do not express HBL1 (Supplementary Fig. S4). The formation of HBL1 foci may be driven by phase separation, as in the case of other membrane-free intracellular condensates composed of nucleic acids and proteins observed in both nuclei and the cytoplasm15. A similar localization pattern consisting of larger and smaller nuclear and cytoplasmic foci was also described for other lncRNAs, e.g., ANCR or GAS516.
Figure 2.
Subcellular localization of HBL1. (A) Human induced pluripotent stem cells were subjected to RNA FISH to detect HBL1 lncRNA (shown in red). Nuclei were stained with DAPI (blue). Cells were visualized with transmitted light to appreciate the nuclear and cytoplasmic localization of HBL1. Scale bar = 10 µm. (B) On day 5 of differentiation toward cardiomyocytes, cells were subjected to double RNA FISH with probes targeting HBL1 and NEAT-1 lncRNAs. HBL1 is shown in red, NEAT-1 is shown in green, and DAPI-stained nuclei are shown in blue. Adjacent localization is shown by white triangles. Scale bar = 5 µm. (C) Human induced pluripotent stem cells were subjected to HBL1 and YB-1 immuno-RNA FISH. HBL1 is shown in red, YB-1 is shown in green, and DAPI-stained nuclei are shown in blue. Colocalization is shown by white triangles. Scale bar = 10 µm. (D) HiPSCs were collected and subjected to RNP-IP protocol using a Magna RIP® RNA-Binding Protein Immunoprecipitation Kit according to the manufacturer’s instructions. A nonspecific IgG control was included in the experiment. Data are shown as the mean ± standard deviation (n = 3). P-values relative to IgG IP are: 0.1181 for 5.8S rRNA and 0.0553 for HBL1, Student’s t-test, two-tailed, paired.
To further enhance our understanding of HBL1’s cellular functions and operational mechanisms, we aimed to explore whether there could be any overlap or interaction between HBL1 and other cellular structures. As intracellular condensates are usually composed of nucleic acids and proteins, we focused on cellular bodies that contain proteins found among proteins interacting with HBL1 in HBL1 pulldown followed by mass spectrometry, i.e., splicing factor, proline- and glutamine-rich (SFPQ) and Y box binding protein 1 (YB-1)12. We thus first focused our attention on paraspeckles, nuclear bodies that comprise, among others, the SFPQ protein and their structural component NEAT1 lncRNA. Our interest in paraspeckles also stems from the fact that HBL1 function is related to chromosome modifications, and paraspeckles are reported to associate with genomic regions through NEAT1 lncRNA12,17. To this end, we developed a double RNA FISH protocol to visualize both HBL1 and NEAT1 lncRNAs. We stained cells on day 5 of differentiation, as we expected to detect both HBL1 foci, based on the high expression level of the transcript at this timepoint (Fig. 1D), and NEAT1-containing paraspeckles that form once cells exit the pluripotency state18. As shown in Fig. 2B, HBL1 lncRNA appeared in both cellular compartments, while paraspeckles were exclusively nuclear, as expected. The majority of the nuclear HBL1 lncRNA foci did not colocalize with NEAT1 paraspeckles; however, we observed adjacent localizations between HBL1 and NEAT1 in a few cases. Further studies are needed to determine whether these RNAs bind overlapping chromatin sites within the nucleus. Moreover, it remains to be established whether any other nuclear bodies overlap with HBL1 nuclear foci and to determine which other proteins or RNAs reside in HBL1 nuclear foci. The first obvious candidates would be the PRC2 complex components.
Subsequently, we investigated the potential colocalization of HBL1 with YB-1, a multifunctional RNA- and DNA-binding protein19,20. YB-1 is mainly localized in the cytoplasm and detected in small cytoplasmic mRNP granules, as well as in P-bodies (PBs) in unstressed cells and upon stress conditions in stress granules (SGs)21–23. YB-1 regulates apoptosis, cell proliferation, differentiation and stress responses and has been reported to be involved in several functional interactions with lncRNAs, such as CAR10, GAS5, TMEM92-AS1, BASP-AS1, HOXC-AS3 and HOTAIR, some of which have been implicated in tumorigenesis24–28. Thus, given the presence of YB-1 in cytoplasmic bodies and reported functional interactions with lncRNAs, we wished to assess its potential interplay with HBL1. To this end, we used immuno-RNA FISH to visualize both YB-1 protein and HBL1 lncRNA in pluripotent cells. As shown in Fig. 2C, YB-1 exhibited mainly cytoplasmic localization with some punctate staining in the cytoplasm. Interestingly, we observed colocalization between HBL1 and YB-1 in the cytoplasm (Fig. 2C) in a subset of foci. We further wished to determine whether HBL1 and YB-1 form ribonucleoprotein particles (RNPs), which we assessed by RNA immunoprecipitation (RIP) approach with anti-YB-1 antibodies. We detected an enrichment of HBL1 in YB-1 IP compared with nonspecific IgG IP (Fig. 2D). The binding of YB-1 to highly abundant rRNAs was much lower, indicating a specific interaction between YB-1 and HBL1. While initially, the literature indicated a preference of YB-1 for A/C-rich regions29,30, the latest publications that applied CLIP and RNAcompete methods to study RNA–protein interactions indicate that YB-1 binds to a CU box (C/GCUC/GC/UC/GA/U) and a consensus motif CUGCG, respectively31,32. Nine CU box motifs can be found within the sequence of HBL1 (Supplementary Fig. S5). To summarize, HBL1 foci are distinct from paraspeckles and colocalize with YB-1-containing foci in the cytoplasm. It can be hypothesized that this colocalization is related to known HBL1 interactions with miRNA-1 (miR-1) and Argonaute 2 (AGO2), the latter residing in P-bodies, sites of mRNA decay, where YB-1 is also detected in unstressed cells11,22. The functional significance of the HBL1 interaction and colocalization with YB-1 will be pursued in the future.
Visualization of HBL1 lncRNA during hiPSC differentiation to cardiomyocytes
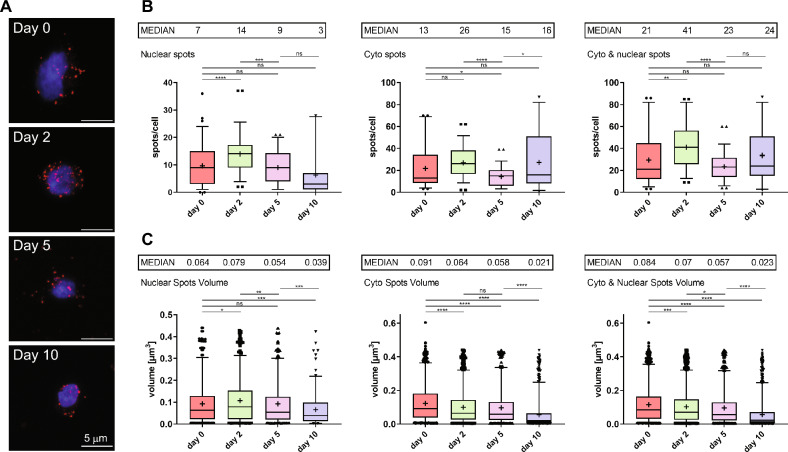
In parallel to cellular HBL1 level measurement during differentiation using the RT-qPCR approach, the subcellular distribution of HBL1 foci was addressed over time using our RNA FISH approach. Cells were collected at days 0, 2, 5 and 10, fixed, subjected to an RNA FISH protocol to detect HBL1 lncRNA, and the number and volume of nuclear and cytoplasmic HBL1 lncRNA foci were calculated (Fig. 3A). On day 0, we observed a median number of 7 condensates in nuclei and 13 in the cytoplasm (Fig. 3B). On day 2, we observed a twofold increase in the number of nuclear and cytoplasmic HBL1 foci as compared to day 0 (median of 7 on day 0 and 14 on day 2 nuclear spots/cell and median of 13 on day 0 and 26 on day 2 cytoplasmic spots/cell). On day 5, the number of HBL1 foci began to decrease when compared to day 2 (median of 9 nuclear spots/cell and of 15 cytoplasmic spots/cell). On day 10, we observed a further decrease in the number of nuclear spots (median of 3 nuclear spots/cell) and no changes in the number of cytoplasmic spots (median of 16 cytoplasmic spots/cell). Next, the volume of the nuclear and cytoplasmic HBL1 foci was measured, which in general slightly decreased on day 2 and day 5 when compared to day 0, except for nuclear foci on day 2, where we observed an increase (Fig. 3C). On day 10, the volume of nuclear and cytoplasmic foci strongly decreased – we observed a 15- and 4.5-fold decrease compared to day 0, respectively. Altogether, we observed that the expression of HBL1 peaked on day 2 of differentiation, as shown by the increase in the number of nuclear and cytoplasmic HBL1 foci, and began to fall starting on day 5, with a strong decrease on day 10, as reflected by the drop in the number and volume of nuclear and cytoplasmic spots. Of note, cytoplasmic HBL1 condensates did not change on day 10 compared to day 5, which suggests different stabilities of nuclear and cytoplasmic foci. In summary, the increase in HBL1 lncRNA expression at the mesodermal stage (days 2 and 3) of differentiation correlates with increases in the number of nuclear and cytoplasmic foci (day 2), and the lower RNA abundance in cardiomyocytes (day 11) corresponds to a decrease in the number of nuclear foci and to a decrease in the volume of nuclear and cytoplasmic foci on day 10. Thus, the number and size of foci in general positively correlate with cellular levels of HBL1.
Figure 3.
HBL1 nuclear and cytoplasmic localization during hiPSC differentiation toward cardiomyocytes by RNA FISH. Human induced pluripotent stem cells were differentiated into cardiomyocytes over 10 days. On days 0, 2, 5, and 10, cells were collected and subjected to RNA FISH targeting HBL1 lncRNA and analyzed using confocal microscopy. (A) Representative images of cells on days 0, 2, 5, and 10 as maximal projections reconstructed from confocal image stacks. HBL1 RNA is shown in red, and DAPI-stained nuclei are shown in blue. Scale bar = 5 µm. The number (B) and volume [µm3] (C) of nuclear, cytoplasmic, and total (nuclear + cytoplasmic) HBL1 lncRNA foci. Images of single cells were acquired with a confocal microscope, and HBL1 RNA foci were quantified inside DAPI-stained nuclei and in the cytoplasm in z-stacks from 10 images/sample in three independent differentiations (n = 30). The results are presented as the mean values (depicted as “ + ”) ± s.d. Median values are shown as bars and indicated above each graph. Statistics were performed using the Welch two-sample t-test. Statistical comparisons are indicated if p < 0.05.
Relationship of the HBL1 gene with the LINC00458 gene
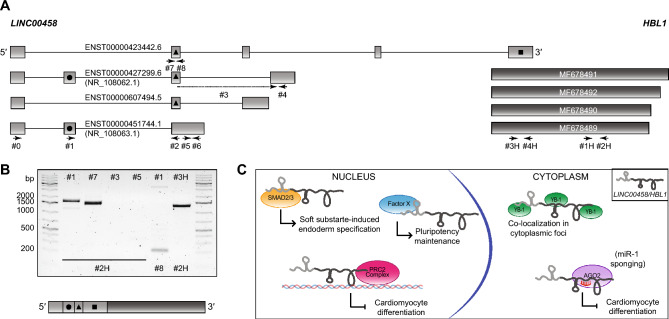
Finally, our attention was drawn by the similar expression profiles of HBL1 and LINC00458 observed during differentiation (Fig. 2D). LINC00458 precedes HBL1 on chromosome 13 and has multiple alternative splicing (AS) isoforms, some of which overlap with HBL1 (Fig. 4A). With each new release of GENCODE annotations, the completeness and complexity of this locus increases (Supplementary Fig. S6A). This reflects a general trend for lncRNA annotation, resulting from a current drive in the GENCODE project to identify new transcripts and extend existing models based on the targeted incorporation of long-read datasets. HBL1 was first described as an independent gene with 4 transcript variants differing in the length of their 3′ ends based on 5′ and 3′ RACE and PCR analysis with primers targeting selected sequences within HBL1 and LINC0045811. However, through personal communication with Dr. Jonathan Mudge and Dr. Sílvia Carbonell Sala from the GENCODE Project, we established that long-read transcriptomics data clearly support HBL1 being part of the same gene as LINC00458, constituting the last exon of some LINC00458 AS isoforms. A subset of these longer isoforms is currently reported in the newest GENCODE release, V43 (Supplementary Fig. S6A). To further confirm these data, we amplified selected regions within LINC00458 and HBL1, as well as amplicons starting within the LINC00458 sequence and finishing at the HBL1 3′ end (Fig. 4B). We obtained PCR products for two amplicons encompassing exons located in the 5′ end of LINC00458 and HBL1 (primers #1 and #7 combined with primer #2H). No PCR products were detected for putative amplicons containing 3′ end sequences present in annotated AS isoforms of LINC00458 (primers #3 and #5 combined with primer #2H). As positive controls, we amplified regions within LINC00458 and HBL1 (primers #1 combined with #8 and #3H combined with #2H, respectively). Additionally, we sequenced PCR products amplified by primers #0 and #1 combined with primer #2H (Fig. 4B and Supplementary Fig. S6B) and detected sequences of both LINC00458 and HBL1, as depicted in Fig. 4B. The sequencing results were in agreement with the DNA molecule sizes estimated from the electrophoresis run. We thus conclude that the extremely complex LINC00458 locus yields a variety of alternatively spliced isoforms, a subset of which encompasses the sequence of HBL1. These data do not completely rule out the possibility that HBL1 exists as a separate gene. However, this possibility seems unlikely based on FANTOM5 CAGE data that do not support the presence of a transcription start site (TSS) for HBL1 yet show a very clear TSS for LINC00458 (Supplementary Fig. S7). In summary, based on the presented results and long-read transcriptomics data (from WTC-11 iPSC line), we postulate that LINC00458 and HBL1 constitute a single gene driven by the promoter region located immediately upstream of LINC00458.
Figure 4.
Relationship of the HBL1 gene with the LINC00458 gene. (A) Selected LINC00458 isoforms according to GENCODE V39 12/2022 (light gray) and HBL1 mRNAs from GenBank (dark gray). GENCODE transcript numbers and NCBI RefSeq numbers are indicated. The localization of primers is depicted by arrows and single digit numbers for LINC00458 and H followed by single digit numbers for HBL1. Exons are represented as boxes, and introns are represented as lines. (B) RT-PCR products amplified with indicated primers selected such as to detect the presence of amplicons that encompass both LINC00458 and HBL1 sequences were separated by electrophoresis (upper panel). Forward primers are indicated at the top of the gel and reverse primers at the bottom. See Fig. 4A for the localization of the primers within the transcripts. PCR products amplified using primer #1 (as well as #0) combined with #2H were purified from the gel and subjected to Sanger sequencing, and the resulting sequence corresponded to the indicated LINC00458 exons and HBL1 (lower panel). (bp = base pairs). The original raw agarose gel image is presented in Supplementary Fig. S8A. (C) Model of LINC00458/HBL1 cellular function proposed based on published data and our work. We propose that LINC00458/HBL1 interacts with its binding partners and fulfils its diverse nuclear and cytoplasmic functions through separate functional domains encompassed within its sequence. The sequence corresponding to the 5′ exons of LINC00458 is shown in light gray, and the 3′ end containing the HBL1 sequence is shown in dark gray.
Discussion
In this study, we examined the expression and subcellular localization patterns of Heart Brake lncRNA 1 during the differentiation of human-induced pluripotent stem cells into cardiomyocytes. Our findings can be summarized into the following key observations. First, the cellular levels of HBL1 markedly increased during the mesoderm and cardiac mesoderm stages of differentiation, followed by a decrease at later stages of cardiac differentiation. Future research will investigate the functional implications of this expression pattern and the mechanisms driving its regulation. Second, we discovered that HBL1 is localized in foci within both the nucleus and cytoplasm. The nuclear foci are distinct from paraspeckles, but adjacent localization is observed in some instances. This proximity may suggest an association with specific genomic regions necessary for gene expression control during development, a feature often associated with many lncRNAs33. We found that HBL1 cytoplasmic condensates colocalize with YB-1, a multifunctional protein known to interact functionally with other lncRNAs, such as HOTAIR25. This colocalization might be related to HBL1 interaction with miR1 and AGO211, all of which localize to cytoplasmic P bodies, or could be linked to others yet to be discovered cellular processes. Third, we uncovered that HBL1, initially characterized as a separate gene11, appears to be expressed as the last exon of LINC00458. We support this observation with several lines of evidence: (1) identical expression profiles of the two transcripts during differentiation, (2) detection of PCR amplicons spanning the sequences of both transcripts, (3) the presence of LINC00458 isoforms that encompass the HBL1 sequence in current GENCODE long-read transcriptomic data, and (4) FANTOM5 CAGE data indicating a transcription start site that clearly precedes the transcribed sequence of LINC00458 but not HBL1. This said, we cannot fully exclude low levels of independent HBL1 transcription from a minor transcription start site.
The observation that LINC00458 and HBL1 are part of the same gene raises the question how the distinct functions reported for the two transcripts, i.e., pluripotency maintenance, as well as endodermal lineage specification for LINC00458, and control of cardiac differentiation for HBL1, can be reconciled. We propose that it could be achieved through the presence of distinct structural and functional domains within the same RNA, each serving a separate function. This concept aligns with the RNA modular code hypothesis, which suggests that lncRNAs possess multiple structural domains, each with its own unique function facilitated by specific interactions with RNA, DNA sequences, or protein complexes34.
In this context, the 5′ end of the transcript, which corresponds to the initially reported LINC00458 sequence, might be involved in pluripotency maintenance interactions, although the mechanisms remain undefined14. Conversely, the 3′ terminal sequence of HBL1 could regulate differentiation toward cardiomyocytes by counteracting miR-1 and directing the chromatin occupancy of PRC11,12. Additionally, an expected but unidentified SMAD2/3 interaction site might relate to LINC00458’s role in substrate-specific endodermal lineage specification35. While the impact of LINC00458 knockdown by siRNA or LNA GapmeRs on PSC differentiation into cardiomyocytes was not assessed14,35, the specific effect of HBL1 on cardiomyocyte development, but not on stem cell pluripotency, was evaluated using CRISPR/Cas9 knockout of HBL1. This was achieved by targeting the sequence of HBL1, likely preserving the expression of shorter LINC00458 isoforms11. Consequently, experimental data gathered in currently available literature do not rule out the possibility that LINC00458/HBL1 might integrate the reported functions in pluripotency maintenance and modulation of differentiation by spatially separating interaction sites with specific binding factors within its sequence (Fig. 4C). This potential multifunctionality underscores the complexity and versatility of lncRNAs.
Methods
Cell culture and differentiation
A human iPSC line established from endothelial progenitor cells (derived from peripheral blood) was purchased from Reprocell and cultured on truncated recombinant human vitronectin (rhVTN-N) in Essential 8™ Flex medium. Cells were controlled for pluripotency level by flow cytometry analysis of Oct3/4 and Sox2 expression in cells and for mycoplasma by using the Venor GeM qEP kit. hiPSCs were differentiated toward cardiomyocytes based on the protocol published by Ref.13. Briefly, hiPSCs at 90% confluency were treated with 3 μM CHIR99021 in CDM3 medium (RPMI 1640 supplemented with 500 μg/mL recombinant human albumin and 213 μg/mL L-ascorbic acid 2-phosphate) for 48 h. After 24 h in CDM3 medium, the cells were treated for 48 h with 4 μM IWR-1 in CDM3 medium. Following 48 h in CDM3 medium, the cells were cultured in RPMI medium with B-27 supplement. Additionally, an alternative hiPSC line reprogrammed from peripheral blood mononuclear cells (PBMCs) was cultured and differentiated under the same conditions.
RNA isolation, RT, qPCR, PCR
Total RNA was isolated using TRIzol™ Reagent, DNase-treated, and reverse-transcribed with a mixture of oligo(dT) reverse primer and random hexamers using an NG dART RT kit. qPCR was performed using SG qPCR Master Mix on a Quantstudio 6 Flex real-time PCR system (Thermo Fisher). The reactions were performed according to the manufacturer’s protocols. To calculate fold changes in mRNA levels, the following equation was used: 2ΔCt(target)/2ΔCt(norm), where ΔCt = Ct (control sample) – Ct (experimental sample). Ct stands for threshold cycle value, “target” indicates the gene of interest, and “norm” indicates RNA for normalization. The cDNA amplified by conventional PCR with selected primers complementary to HBL1 and LINC00458 sequences was subjected to agarose gel analysis. Digital images of agarose gels were acquired using a ChemiDoc™ MP Imaging System (Bio-Rad) with Image Lab Touch Software (Bio-Rad), and bands selected for Sanger sequencing were cut out of the gel and purified using a GeneJET Gel Extraction Kit. See Supplementary Table S1 for primer sequences.
Flow cytometry
Cells were harvested and dissociated using TrypLE™ Select Enzyme, fixed with 1% PFA for 20 min, permeabilized with 90% methanol for 10 min, and stained using 1:40 mouse monoclonal IgG1 TNNT2 and 1:200 mouse monoclonal IgG2b MLC2A (MYL7) for 1 h at RT. Mouse IgG1 and mouse IgG2b isotype controls were used. Secondary staining was performed with 1:1000 Alexa Fluor 488 goat anti-mouse IgG1 and 1:1000 Alexa Fluor 647 goat anti-mouse IgG2b for 30 min at RT. Cells were analyzed using a Navios (Beckman Coulter) flow cytometer. Data were analyzed using Kaluza 2.2 (Beckman Coulter) software.
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde/PIPES for 10 min, permeabilized for 10 min with 0.5% Triton X-100, blocked in serum, and incubated with appropriate primary antibodies for 1 h at RT and fluorescent dye-conjugated secondary antibodies for 30 min at RT. Finally, coverslips were mounted in ProLong™ Gold Antifade Mountant with DAPI. Images were collected using an Axio Observer Z1 inverted microscope (Carl Zeiss, Jena, Germany) equipped with the LSM 880 AiryScan confocal module with ZEN 2012 SP1 black edition software and processed in ImageJ Fiji (National Institutes of Health, Bethesda, MD). A complete list of antibodies used is presented in Supplementary Table S2.
RNA fluorescence in situ hybridization (RNA FISH) and confocal microscopy
hiPSCs and hiPSC-CMs on selected days of differentiation (days 0, 2, 5 and 10) were harvested, dissociated using TrypLE™ Select Enzyme, resuspended in an appropriate medium and plated on laminin-coated coverslips for 60 min at 37 °C. Next, the cells were fixed with 3.7% paraformaldehyde in 1 × PHEM (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 4 mM MgSO4·7H20) buffer for 10 min at RT. Next, cells were subjected to an RNA-FISH protocol using hybridization chain reaction (HCR) technology from Molecular Instruments, Inc. as previously described with modifications36. Briefly, cells were permeabilized with 0.1% Tween-20 in PBS for 10 min followed by incubation with a set of 20 DNA HCR v3.0 probes complementary to HBL1 lncRNA (see Supplementary Table S3 for probe sequences) for 12 h at 37 °C. In the case of double RNA-FISH, cells were incubated simultaneously with two sets of probes complementary to HBL1 ncRNA and NEAT1 lncRNA. Next, cells were extensively washed and hybridized with HCR amplifiers labeled with Alexa 647 dye (for HBL1) and, in the case of double RNA-FISH, with additional amplifiers labeled with Alexa 488 dye (for NEAT1) for 12 h at RT in the dark and washed again. Finally, the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (D1306; Thermo Fisher Scientific), and the cells were mounted on slides with Prolong Diamond Antifade Mounting Medium (P36970; Invitrogen). Fluorescence images were acquired using an Axio Observer Z1 inverted microscope (Carl Zeiss, Jena, Germany) equipped with the LSM 880 AiryScan confocal module with ZEN 2012 SP1 black edition software and processed in ImageJ Fiji (National Institutes of Health, Bethesda, MD). Extrapolated data were analyzed and plotted with R [R Core Team (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/ and Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. ISBN 978-3-319-24277-4, https://ggplot2.tidyverse.org].
Immuno-RNA FISH
HiPSCs were fixed using 3.7% PFA, washed thoroughly with PBS and permeabilized for 10 min using 0.1% Tween-20 in PBS (PBST) solution. The cells were then incubated for 30 min at 37 °C in probe hybridization buffer. Next, coverslips were put into a hand-made humidity chamber to incubate with HBL1 lncRNA-specific probes overnight at 37 °C. Cells were washed 4 times with probe wash buffer preheated to 37 °C and 2 times with 5 × SSCT buffer. Next, the cells were incubated with snap-cooled Alexa 647-labeled hairpin amplificators overnight at room temperature in the dark. Cells were then washed 5 times with 5 × SSCT buffer, blocked in 5% BSA in PBST solution for 2 h at room temperature and subjected to rabbit α-YB-1 (1:200) antibody incubation in 1% BSA and 0.02% Tween-20 in PBS at 4 °C overnight. The next day, the cells were washed using PBST buffer 3 times, incubated with goat α-rabbit Alexa 594 antibodies (1:400) in PBS for 2 h at RT and then washed 3 times using PBST buffer. Finally, the nuclei were stained with DAPI in PBS for 10 min at room temperature and subjected to confocal microscopy analysis. See Supplementary Table S2 for antibody information.
Immunoprecipitation of ribonucleoproteins (RNP-IP)
The assay was performed using a Magna RIP® RNA-Binding Protein Immunoprecipitation Kit according to the manufacturer’s instructions. The isolated RNA was DNase-treated and subjected to reverse transcription. Fold enrichment of a specific antibody-precipitated RNA over nonimmune IgG IP was determined by quantitative real-time PCR (qPCR). The following equation was used: 2(Ct(IgG)—Ct(Spec)), where Ct stands for threshold cycle value, IgG for nonimmune IgG IP and Spec for specific antibody IP.
Supplementary Information
Acknowledgements
We thank Dr. Jonathan Mudge and Dr. Sílvia Carbonell Sala from the GENCODE Project for input regarding the results of long-read sequencing data and helpful discussion, Prof. Ewa Zuba-Surma (Department of Cell Biology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Krakow, Poland) for sharing hiPSCs reprogrammed from endothelial progenitor cells, Dr. Neli Kachamakova-Trojanowska (Malopolska Centre of Biotechnology, Jagiellonian University, Krakow, Poland) for sharing hiPSCs reprogrammed from PBMCs, and Dr. Sylwia Noga (Malopolska Centre of Biotechnology, Jagiellonian University, Krakow, Poland) for providing hUC-MSC cell pellets for RNA isolation.
Author contributions
M.P. and A.K.P. envisioned the project. M.P., A.K.P. and K.P. designed the experimental concepts. P.Mac., F.A. and M.P. performed the differentiation, RT-qPCR, ICC and RNP-IP experiments; A.S. and J.W. ran the RNA FISH and immune-RNA FISH experiments; P.Mai. quantified the number and volume of HBL1 foci during hiPSC differentiation to cardiomyocytes; M.B. analyzed protein expression levels by flow cytometry. M.P. and A.K.P. prepared the figures and wrote the manuscript, with input from all other authors.
Funding
This work was supported by the OPUS 17 grant (2019/33/B/NZ3/02837 to MP) from the National Science Centre (NCN). AKP and AS acknowledge funding from the National Science Centre, Poland (Sonata Bis Grant UMO2018/30/E/NZ1/00874). AKP and JW acknowledge funding from the National Science Centre, Poland (OPUS Grant UMO-2022/45/B/NZ3/03890).
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Patrycja Maciak and Agnieszka Suder.
Contributor Information
Anna Kula-Pacurar, Email: anna.kula-pacurar@uj.edu.pl.
Marta Pabis, Email: marta.pabis@uj.edu.pl.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-49753-3.
References
- 1.Charles Richard JL, Eichhorn PJA. Platforms for investigating LncRNA functions. SLAS Technol. 2018;23:493–506. doi: 10.1177/2472630318780639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabili M, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong C, Popp MWL, Maquat LE. Biochemical analysis of long non-coding RNA-containing ribonucleoprotein complexes. Methods. 2012;58:88–93. doi: 10.1016/j.ymeth.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson EK, Covarrubias S, Carpenter S. The how and why of lncRNA function: An innate immune perspective. Biochim. et Biophys. Acta Gene Regul. Mech. 2020;1863:194419. doi: 10.1016/j.bbagrm.2019.194419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fico A, Fiorenzano A, Pascale E, Patriarca EJ, Minchiotti G. Long non-coding RNA in stem cell pluripotency and lineage commitment: Functions and evolutionary conservation. Cell. Mol. Life Sci. 2019;76:1459–1471. doi: 10.1007/s00018-018-3000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chujo T, Hirose T. Nuclear bodies built on architectural long noncoding RNAs: Unifying principles of their construction and function. Mol. Cells. 2017;40:889–896. doi: 10.14348/molcells.2017.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592:2884–2900. doi: 10.1002/1873-3468.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimbrel EA, Lanza R. Pluripotent stem cells: The last 10 years. Regen. Med. 2016;11:831–847. doi: 10.2217/rme-2016-0117. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Li Y, Lin B, Sheng Y, Yang L. HBL1 is a human long noncoding RNA that modulates cardiomyocyte development from pluripotent stem cells by counteracting MIR1. Dev. Cell. 2017;42:333–348.e5. doi: 10.1016/j.devcel.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, J. et al. LncRNA HBL1 is Required for Genome-Wide PRC2 Occupancy and Function in Cardiogenesis from Human Pluripotent Stem Cells. (2021). [DOI] [PMC free article] [PubMed]
- 13.Burridge PW, et al. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:6357. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 16.Cabili MN, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015 doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosch M, Ittermann S, Shaposhnikov D, Drukker M. Chromatin-associated membraneless organelles in regulation of cellular differentiation. Stem Cell Rep. 2020;15:1220–1232. doi: 10.1016/j.stemcr.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosch M, et al. Nucleus size and DNA accessibility are linked to the regulation of paraspeckle formation in cellular differentiation. BMC Biol. 2020 doi: 10.1186/s12915-020-00770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- 20.Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Moscow) 2011;76:1402–1433. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- 21.Mateu-Regué À, et al. Single mRNP analysis reveals that small cytoplasmic mRNP granules represent mRNA singletons. Cell Rep. 2019;29:736–748.e4. doi: 10.1016/j.celrep.2019.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 23.Somasekharan SP, et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J. Cell Biol. 2015;208:913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suresh PS, Tsutsumi R, Venkatesh T. YBX1 at the crossroads of non-coding transcriptome, exosomal, and cytoplasmic granular signaling. Eur. J. Cell Biol. 2018;97:163–167. doi: 10.1016/j.ejcb.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Li S, et al. Long noncoding RNA HOTAIR interacts with Y-Box Protein-1 (YBX1) to regulate cell proliferation. Life Sci. Alliance. 2021;4:e202101139. doi: 10.26508/lsa.202101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song S, et al. A novel long noncoding RNA, TMEM92-AS1, promotes gastric cancer progression by binding to YBX1 to mediate CCL5. Mol. Oncol. 2021;15:1256–1273. doi: 10.1002/1878-0261.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li YL, et al. LncRNA BASP1-AS1 interacts with YBX1 to regulate Notch transcription and drives the malignancy of melanoma. Cancer Sci. 2021;112:4526–4542. doi: 10.1111/cas.15140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang E, et al. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018 doi: 10.1186/s13059-018-1523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stickeler E, et al. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 2001;20:3821–3830. doi: 10.1093/emboj/20.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skabkina OV, Lyabin DN, Skabkin MA, Ovchinnikov LP. YB-1 Autoregulates translation of its own mRNA at or prior to the step of 40S ribosomal subunit joining. Mol. Cell. Biol. 2005;25:3317–3323. doi: 10.1128/MCB.25.8.3317-3323.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodarzi H, et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray D, et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009;27:667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 33.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006 doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 34.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, Y.-F. et al. Control of matrix stiffness promotes endodermal lineage specification by regulating SMAD2/3 via lncRNA LINC00458. Sci. Adv. vol. 6. http://advances.sciencemag.org/ (2020). [DOI] [PMC free article] [PubMed]
- 36.Kula-Pacurar A, et al. Visualization of SARS-CoV-2 using immuno RNA-fluorescence in situ hybridization. J. Visual. Exp. 2020 doi: 10.3791/62067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.