Abstract
The formation of crown gall tumors by Agrobacterium tumefaciens requires that the virulence (vir) genes be induced by chemical signals which consist of specific phenolic compounds and monosaccharides, synthesized at plant wound sites. Signal transduction in the activation of these genes is mediated by the VirA-VirG two-component regulatory system, together with ChvE, a glucose-galactose binding protein which interacts with VirA. We have previously presented genetic evidence that virA senses phenolic compounds directly (Y.-W. Lee, S. Jin, W.-S. Sim, and E. W. Nester, Proc. Natl. Acad. Sci. USA 92:12245–12249, 1995). The vir genes of strain KU12 can be induced by 4-hydroxyacetophenone, p-coumaric acid, and phenol, whereas these same phenolic compounds are weak inducers of the vir genes of strain A6. In this report, we show that a specific inducing sugar can broaden the specificity of the phenolic compound which VirA senses. 4-Hydroxyacetophenone and other related phenolic compounds function as inducing phenolic compounds with the virA gene of A6 if arabinose replaces glucose as the inducing sugar. We further demonstrate that this broadened specificity for phenolic inducers results from the increased level of ChvE through induction by arabinose via the regulatory protein GbpR. If high levels of ChvE are present, then poorly inducing phenolic compounds can induce the vir genes to high levels in combination with glucose. Comparing the induction response of the wild type and that of a VirA mutant with a mutation in its receiver domain revealed that the activity of the receiver domain is controlled by the periplasmic domain. We discuss these observations in terms of how VirA senses and transduces signals elicited by the two classes of plant signal molecules.
Agrobacterium tumefaciens infects a wide range of plants by transferring and integrating a piece (the T-DNA) of its tumor-inducing (Ti) plasmid into the plant genome, resulting in the formation of crown gall tumors. To initiate this process, a set of genes, the virulence (vir) genes, on the Ti plasmid must be activated by three kinds of environmental signals at the wound site of a plant. These signals include specific classes of plant phenolic compounds, monosaccharides, and an acidic pH. The signals are perceived by three proteins, the VirA-VirG two-component transduction system and ChvE, a periplasmic sugar binding protein. In response to these signals, the VirA sensor protein is autophosphorylated at His-474, and this phosphate is then transferred to Asp-52 of the response regulator, VirG. The activated VirG then induces the expression of all of the vir genes (for reviews, see references 15, 17, and 32).
The key molecule which allows Agrobacterium to sense environmental conditions favorable for T-DNA transfer is the VirA protein, which is anchored to the cytoplasmic membrane by two transmembrane domains. This protein contains four other domains: an amino-terminal periplasmic domain and three cytoplasmic domains. The latter domains include a linker, a kinase, and a carboxyl-terminal region termed the receiver because it contains a region that is homologous to the phosphorylatable receiver domain of VirG (5). The periplasmic domain is responsible for sensing a variety of monosaccharides involved in vir gene induction. The linker domain is necessary for perceiving phenolic compounds and acidity (5). The function of the receiver domain is less clear. Chang and Winans (5) reported that the A6 VirA receiver domain might play an inhibitory role in signal transduction, because when this domain was deleted, monosaccharides alone induced vir gene expression in the absence of phenolic compounds. Similar results were obtained by Gubba et al. (14). However, deleting the C-terminal receiver domain from the VirA protein of Agrobacterium rhizogenes resulted in a strain which showed poor vir gene induction and was nontumorigenic on Kalanchoe diagremontiana leaves (11). The A6 VirA receiver domain apparently restricts the recognition of phenolic inducers by VirA, since certain mutations in the domain or its removal widens the spectrum of inducers which VirA recognizes (6).
The chvE gene plays several roles, one in the uptake of specific sugars, another in chemotaxis to these sugars (4, 18), and a third in the VirA-VirG two-component signal transduction system. ChvE is homologous to two periplasmic proteins involved in sugar recognition and uptake in Escherichia coli (4, 18). The inducing monosaccharides, all of which are components of the plant cell wall, are sensed by the VirA protein via the ChvE protein (1, 4). The ChvE protein interacts with each of the various inducing monosaccharides, and the ChvE::monosaccharide complex in turn interacts with the periplasmic region of VirA (4, 26). The periplasmic domain of VirA may repress its function, and this repression is overcome by the binding of the ChvE::monosaccharide complex to the periplasmic domain of VirA (4, 15, 26). All Agrobacterium strains containing a defective chvE are defective in vir gene induction and have limited host ranges compared with those of wild-type strains (13, 18). In strain A348, mutants lacking the periplasmic domain of VirA have the same host range defects and inducing properties as chvE mutants. They are avirulent on some, but not all, plants (4). However, in strain C58, the loss of the ChvE protein results in the almost-complete loss of vir gene induction and an extremely limited host range (10). The chvE gene is regulated by the inducing monosaccharides through the gbpR gene product, GbpR, a member of the LysR family of transcriptional regulators (9). A subset of the monosaccharides which are involved in the induction of the vir genes induces the expression of ChvE a maximum of eightfold; in their absence, GbpR represses its expression (9).
In this paper, we present evidence that specific monosaccharides can broaden the spectrum of phenolic compounds which can serve as vir gene inducers. This effect is due to an increased level of the ChvE protein resulting from the induction of chvE gene expression by GbpR and the inducing sugar.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. A. tumefaciens strains were grown in MG/L or AB minimal medium at 28°C (21). E. coli strains were grown in Luria-Bertani medium (24) at 37°C. The following antibiotics were used at the indicated concentrations (in micrograms per milliliter): for A. tumefaciens, carbenicillin (100), kanamycin (100), gentamicin (100), spectinomycin (250), and tetracycline (5); and for E. coli, carbenicillin (100), kanamycin (100), gentamicin (5), and tetracycline (15). These concentrations were reduced by one-half for liquid media. For measuring the basal level of vir gene induction, glycerol induction broth (GIB) (2) was used. This medium contained AB mineral salts, 0.5 mM NaH2PO4, 50 mM MES [2-(N-morpholino)ethanesulfonic acid] (pH 5.5), and 0.5% (wt/vol) glycerol. As needed, different phenolic compounds and monosaccharides were added to GIB at the concentrations indicated. GIB medium also contained appropriate antibiotics to ensure plasmid maintenance.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Agrobacterium strains | ||
| C58 | Wild-type nopaline strain | 25 |
| A6 | Wild-type octopine strain | 25 |
| KU12 | Wild-type octopine strain | 20 |
| A136 | C58; Ti plasmid cured | 25 |
| A348 | A136(pTiA6) | 13 |
| A136MX1 | A136 chvE::Tn5 | 18 |
| C58ΔvirA | C58(ΔvirA) | 20 |
| At11058 | C58 chvE::Tn5 virA::Ω | T. Charles |
| A136gbpR | A136 gbpR | This work |
| C58ΔvirAgbpR | C58(ΔvirA) gbpR | This work |
| E. coli strains | ||
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 φ80d lacZ ΔM15 | Bethesda Research Laboratories |
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′[traD36 proAB+ lacIqlacZΔM15] | U.S. Biochemical Corp. |
| Plasmids | ||
| pGP159 | virA, virG, virB::lacZ of pTiA6 in IncP vector | 7 |
| pIB422 | pTiA6 virA virB::lacZ; IncP and pUC replion | 4 |
| pSM402 | virA, virG, virB::lacZ of pTiA6 in pVK102 | 28 |
| pSM243cd | virB::lacZ fusion of pTiA6 in pVK102 | 28 |
| pCH116 | virB::lacZ Plac-virG of pTiA6 in IncP vector | S. C. Winans (5) |
| pJZ105 | virA(I734N); IncW vector | S. C. Winans (6) |
| pPR1068 | pMAL-c2 derivative; NdeI at the start of MalE | New England Biolabs |
| pSL2L | pGC31; gbpR deletion at N terminus; nptII | 9 |
| pUFR047 | IncW broad-host-range vector | 12 |
| pWT120 | Ptac-chvE in pPR1068 | This work |
| pWT121 | Ptac-chvE lacIq in pUFR047 | This work |
| pWT122 | Ptac-chvE in pUFR047 | This work |
Enzymes and reagents.
Restriction enzymes, T4 DNA ligase, and Vent DNA polymerase were purchased from Bethesda Research Laboratories or New England Biolabs and used according to the supplier’s recommendations. Among the phenolic compounds used, acetosyringone (AS), acetovanillone, and 4-hydroxyacetophenone (HAP) were purchased from Aldrich Chemical Co.; syringaldehyde, syringic acid, ferulic acid, p-coumaric acid, and phenol were purchased from Sigma. The phenolic compounds were prepared in dimethyl sulfoxide as 1 M or 100 mM stock solutions. o-Nitrophenyl-β-d-galactoside (ONPG) and MES were purchased from Sigma. Isopropyl-β-d-thiogalactopyranoside (IPTG) was purchased from U.S. Biochemical. Antibiotics and other reagents were purchased from Sigma.
Overexpression of chvE.
To increase the expression level of chvE, we placed the chvE gene under the control of the Ptac promoter. Two primers, chvE-head, GGGAGAGTTAATGAAGTC, and chvE-tail, GCGAATTCTTATTTCAGCTGGTCTTC, were used to amplify the chvE coding region from A348 genomic DNA with high-fidelity Vent DNA polymerase. The primer chvE-tail has an EcoRI restriction site. The amplification reaction was performed in a total volume of 100 μl containing 1 μg of A348 genomic DNA, 20 nmol of each deoxynucleoside triphosphate, 20 pmol of each primer, and 2 U of Vent DNA polymerase. The reaction was performed as follows: 94°C for 5 min; 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min; followed by 72°C for 10 min. The 1-kb chvE PCR product (digested with MseI and EcoRI) was ligated with a 5.3-kb NdeI-EcoRI fragment of pPR1068 to create pWT120. A 2.6-kb MscI-HindIII fragment of pWT120 containing chvE and lacIq was cloned into SmaI- and HindIII-digested pUFR047 to create pWT121. A 1.7-kb EcoRV-HindIII fragment of pWT120 containing the chvE gene was cloned into pUFR047, which had been digested with SmaI and HindIII, to create pWT122.
Construction of gbpR mutants.
pSL2L was introduced into C58ΔvirA and A136 by electroporation, selecting for kanamycin-resistant colonies. These colonies were screened for gene replacement as judged by sensitivity to carbenicillin. Putative double-crossover events resulting in gene replacement were verified by Southern hybridization.
Western immunoblotting.
A. tumefaciens cultures (50 ml) were grown under the same growth conditions used for vir gene induction and then centrifuged, and the cell pellets were suspended in 2 ml of buffer containing 10 mM Tris (pH 8.0), 10 mM EDTA, 10% glycerol, and 1 mM phenylmethylsulfonyl fluoride. The cells were disrupted by two passages through a French press minicell at 1,000 lb/in2. The resulting crude cell lysates were centrifuged for 5 min, and the supernatants were recovered. Total protein concentrations were determined by using the Bio-Rad protein assay kit with bovine serum albumin as the standard. Equal amounts of protein (50 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide) and then transferred to polyvinylidene difluoride membranes (Millipore Corporation) with the Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad Laboratories). Antiserum which interacted with ChvE was raised in rabbits against the glutathione S-transferase–chvE fusion protein (27). Horseradish peroxidase-conjugated antibodies raised in goats against rabbit immunoglobulin G (Bio-Rad Laboratories) and the ECL Western blotting analysis system (Amersham Life Science) were used to visualize the ChvE protein.
vir gene induction assays.
Agrobacterium cells were grown in 25 ml of MG/L containing appropriate antibiotics to an optical density of about 0.4 at 600 nm. The cells were pelleted, resuspended in 1.0 ml of water containing 15% glycerol, and frozen at −70°C (31). Cells were thawed as needed and used to inoculate 2.0 ml of induction broth to an initial optical density at 600 nm of 0.1 in a 15-ml culture tube containing the phenolic and monosaccharide inducers indicated. Cultures were incubated with shaking at 28°C for 20 to 24 h. vir gene induction was measured as a function of β-galactosidase activity as assayed by the method described previously (22). All β-galactosidase activities reported are an average of three independent determinations.
RESULTS
The substrate specificity of VirAA6 can be broadened by using arabinose as the inducing sugar.
In a previous study (19), we demonstrated that different VirA proteins respond to different phenolic inducers. The vir genes of KU12 are induced by HAP, p-coumaric acid, and phenol, which are weak vir gene inducers in strains C58 and A6. In all of these studies, glucose was used as the inducing sugar. We extended our studies on the VirA-VirG–ChvE signal transduction system by testing other sugars in combination with these weak phenolic vir inducers.
Two wild-type A. tumefaciens strains, C58 and A6, and two genetically modified strains of C58 carrying either 1 to 2 or 10 to 15 copies of virA, virG, and the virB::lacZ translational fusion of strain A6 were tested for their responses to three weakly inducing phenolic compounds (HAP, p-coumaric acid, and phenol) (Table 2). When arabinose was used as the inducing sugar (Table 2, column 1), the vir genes of strain C58 were induced 16-fold by HAP. However, the wild-type strain A6 was not induced with these same inducers (Table 2, column 2). Interestingly, the VirAA6 from pSM402 was induced more than 12-fold by HAP with 10 mM arabinose when it was removed from its cognate Ti plasmid background and placed in a pTiC58 background (Table 2, column 3). This suggests that a pTiA6-encoded, VirAA6-specific inhibitor is present in strain A6. Evidence for such an inhibitor was also observed in studies of agroinfection (16).
TABLE 2.
Comparison of induction of virB::lacZ in A. tumefaciens strains with different sugars and phenolic compoundsa
| Inducing compounds | β-Galactosidase (U) obtained with A. tumefaciens strain:
|
|||
|---|---|---|---|---|
| C58 (pSM243cd) | A6 (pSM243cd) | C58ΔvirA (pSM402) | C58ΔvirA (pGP159) | |
| HAP | 3.5 ± 0.1 | 4.4 ± 0.6 | 3.4 ± 0.1 | 5.1 ± 0.5 |
| + Glucose | 15 ± 2.7 | 3.6 ± 1.0 | 9.6 ± 0.4 | 106 ± 3 |
| + Arabinose | 59 ± 2.8 | 7.0 ± 0.2 | 44 ± 0.2 | 376 ± 25 |
| p-Coumaric acid | 2.6 ± 0.6 | 11 ± 2.1 | 3.8 ± 0.4 | 4.8 ± 0.2 |
| + Glucose | 5.7 ± 1.6 | 4.7 ± 1.2 | 5.4 ± 0.5 | 11.6 ± 1.0 |
| + Arabinose | 53 ± 13 | 3.5 ± 0.1 | 17.3 ± 2.8 | 164 ± 11 |
| Phenol | 2.8 ± 0.1 | 6.1 ± 0.4 | 5.1 ± 0.3 | 5.4 ± 0.9 |
| + Glucose | 5 ± 0.5 | 3 ± 0.2 | 5.5 ± 0.1 | 12.7 ± 1.4 |
| + Arabinose | 30 ± 1.8 | 2 ± 0.1 | 10.6 ± 0.3 | 94 ± 16 |
a All strains were grown for 20 to 24 h in GIB induction medium containing phenolic compounds with or without monosaccharides at the indicated concentrations. The cultures were then assayed for β-galactosidase activity as described in the text. The data are means ± standard errors of the means for three samples. pSM243cd contains a virB::lacZ fusion. pSM402 contains pTiA6 virA, virG, and a virB::lacZ translational fusion in pVK102, a low-copy-number cosmid (1 to 2 copies/cell) (18a). pGP159 also contains pTiA6 virA, virG, and a virB::lacZ translation fusion in IncP vector pTJS75 (10 to 15 copies/cell) (8). β-Galactosidase activities of cultures lacking phenolic compounds were less than 2 U. HAP was used at 250 μM, while p-coumaric acid and phenol were used at 100 μM. Arabinose and glucose were used at 10 mM.
The data in Table 2 also indicate that the copy number of the virA gene can influence the level of induction by arabinose. When the copy number was increased approximately eightfold, the induction was increased another eightfold (Table 2, column 4 versus column 3). Even glucose allowed a 20-fold increase in induction with HAP in strains with elevated levels of virA (Table 2, column 4). The main difference between the two plasmids pSM402 and pGP159 is the copy number (7, 8, 28). As shown in Table 2, column 4, increasing the copy number of virA also led to a much higher level of induction by HAP, p-coumaric acid, and phenol.
Since pGP159 also contains 10 to 15 copies of virG, we explored the possibility that the extra copies of virG were responsible for the higher fold levels of induction by HAP and, to a lesser extent, p-coumaric acid and phenol. We compared the inducing activity of HAP in two derivatives of C58ΔvirA. One derivative contained pGP159 with multiple copies of virA and virG, while the other contained pIB422 with multiple copies of virA but no virG. The levels of induction were approximately the same (data not shown). We conclude that the extra copies of virA, and not virG, are responsible for the higher levels of induction by HAP.
Induction of vir genes in gbpR mutants by HAP.
Because a consistently higher level of induction was seen with arabinose than with glucose when weak phenolic inducers of the vir genes were present as inducers, we considered the possibility that higher levels of the ChvE protein might account for the differences observed. We have shown previously that arabinose induced the expression of ChvE about eightfold whereas glucose had no effect on the level of ChvE (9). Arabinose interacts with the GbpR protein, a member of the LysR family of regulatory proteins, to induce the synthesis of ChvE. In the absence of an inducing sugar, such as arabinose, GbpR represses chvE expression; in the presence of an inducing sugar, GbpR activates chvE expression. In the gbpR mutants, the level of chvE expression increased to a level which is intermediate between the basal and induced levels in the wild-type strain (9). Consequently, if the ChvE level determines vir gene induction by HAP, then in a gbpR mutant, the fold induction should be less than that in the wild type when arabinose is used as the inducing sugar while the fold induction should be more than that in the wild type when glucose is used. The results in Table 3 show that this is observed. A gbpR mutation in strain A136(pGP159) reduced the fold induction from 113 to 48 when arabinose was the inducing sugar, while it increased this fold induction from 14 to 29 with glucose. The same phenomenon was observed in the strain C58ΔvirA(pGP159). These data support the notion that the two sugars operate through the regulatory protein GbpR, presumably by controlling the level of the ChvE protein.
TABLE 3.
Effect of gbpR mutations on induction of virB::lacZ
| Strain | β-Galactosidase (U) obtained with inducing compoundsa:
|
||
|---|---|---|---|
| HAP | HAP + arabinose | HAP + glucose | |
| A136(pGP159) | 3.4 ± 0.2 | 388 ± 37 (113) | 48 ± 5.6 (14) |
| A136gbpR(pGP159) | 4.4 ± 0.6 | 213 ± 11 (48) | 127 ± 1.4 (29) |
| C58ΔvirA(pGP159) | 3.8 ± 0.2 | 348 ± 19 (90) | 102 ± 7.5 (26) |
| C58ΔvirAgbpR(pGP159) | 5.3 ± 1.0 | 189 ± 0.9 (36) | 204 ± 6.0 (39) |
a Data are means ± standard errors of the means for three samples. The numbers in parentheses are the fold increases in induction calculated from the values with HAP alone. HAP was used at 250 μM. Arabinose and glucose were used at 10 mM.
The level of ChvE is limiting for the induction of vir genes.
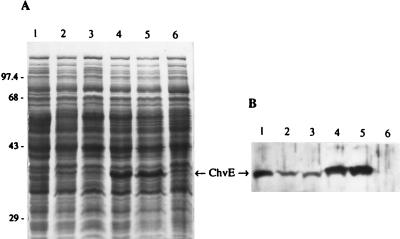
To demonstrate convincingly that the level of ChvE is the critical factor in determining what sugars influence vir gene induction by weak phenolic inducers, we put chvE under the control of the strong Ptac promoter, which functions independently of sugars. In such a construct we would expect that glucose and arabinose would be equally effective in inducing vir genes with HAP. The Ptac promoter has been used successfully to express Agrobacterium genes (6). We constructed two plasmids, one (pWT121) containing lacIq, which directs controlled expression of chvE upon addition of IPTG. The other plasmid (pWT122) does not contain lacIq, so chvE is expressed constitutively at high levels. From immunoblotting, we can clearly see the expected differences in the amounts of ChvE protein among the different bacterial cultures (Fig. 1). The ChvE protein concentration in Agrobacterium grown with 10 mM arabinose as the inducing sugar (Fig. 1, lane 1) is obviously much higher than what is observed with 10 mM glucose as the inducing sugar (Fig. 1, lane 2). As expected, we observed considerably higher levels of chvE expression when using the Ptac promoter (Fig. 1, lanes 4 and 5).
FIG. 1.
Western immunoblotting assay for the expression of ChvE protein. Cells were incubated on induction medium with different monosaccharides and other additives. (A) Protein profiles of different A. tumefaciens cultures as analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. (B) Western immunoblotting. Lanes: 1, C58ΔvirA(pGP159) grown in GIB plus 10 mM arabinose; 2, C58ΔvirA(pGP159) grown in GIB plus 10 mM glucose; 3, C58ΔvirA(pGP159, pWT121) grown in GIB plus 10 mM glucose; 4, C58ΔvirA(pGP159, pWT121) grown in GIB plus 10 mM glucose plus 1 mM IPTG; 5, At11058(pGP159, pWT122) grown in GIB plus 10 mM glucose; 6, At11058(pGP159) grown in GIB plus 10 mM arabinose. The numbers on the left are molecular masses.
In order to observe the effect of the increased ChvE level, we introduced the constitutive expression construct pWT122 into two chvE mutants, A136MX1(pGP159) and At11058(pGP159). As shown in Table 4, the induction by HAP with glucose indeed increased, from 2 to 28 for A136(pGP159) and from 7 to 58 for C58ΔVirA(pGP159), presumably as a result of the increased levels of ChvE. Most importantly, glucose was just as good an inducer as arabinose when ChvE was highly expressed.
TABLE 4.
Effect of ChvE concentration on induction of virB::lacZ
| Straina | β-Galactosidase (U) obtained with inducing compoundsb:
|
||
|---|---|---|---|
| HAP | HAP + arabinose | HAP + glucose | |
| A136(pGP159) | 7 ± 1 | 238 ± 10 (33) | 18 ± 1.5 (2) |
| A136MX1(pGP159, pWT122) | 8.7 ± 1.2 | 206 ± 6 (24) | 243 ± 0.5 (28) |
| C58ΔvirA(pGP159) | 8.6 ± 1.4 | 363 ± 21 (42) | 60 ± 3.7 (7) |
| At11058(pGP159, pWT122) | 6.7 ± 1.0 | 384 ± 8 (57) | 385 ± 3.6 (58) |
a A136MX1 is A136 with a Tn5 insertion in the chvE gene. At11058 is C58 with mutations in chvE and virA. pWT122 directs the constitutive high expression of the chvE gene.
b Data are means ± standard errors of the means for three samples. The numbers in parentheses are the fold increases in induction calculated from the values with HAP alone. HAP was used at 250 μM. Arabinose and glucose were used at 10 mM.
In order to observe the effects of differing ChvE concentrations in the same strain, we introduced the controlled expression construct pWT121, which contains lacIq, into the chvE wild-type strain C58ΔvirA(pGP159). As shown in Table 5, in the absence of IPTG, the induction level increased only 10-fold when glucose was present as the inducing sugar, but it increased 50-fold when IPTG was added to the induction medium. Significantly, glucose was as effective an inducer as arabinose when chvE was under the control of the Ptac promoter and IPTG was added.
TABLE 5.
Effect of ChvE level on induction of virB::lacZ
| Inducing compound | β-Galactosidase (U) obtained with straina:
|
Fold induction |
|---|---|---|
| C58ΔvirA(pGP159, pWT121) | ||
| HAP | 4.4 ± 1.5 | |
| + Arabinose | 167 ± 13 | 38 |
| + Glucose | 46 ± 4 | 10 |
| HAP + IPTG | 6.0 ± 1.4 | |
| + Arabinose | 278 ± 8 | 47 |
| + Glucose | 295 ± 18 | 50 |
a pWT121 directs the controlled high expression of chvE upon addition of IPTG. The data are means ± standard errors of the means for three independent samples. HAP was used at 250 μM. Arabinose and glucose were used at 10 mM. IPTG was used at 1 mM.
Induction of vir genes by HAP with galacturonic acid.
Ankenbauer and Nester (1) found that induction of the vir genes of A6 is much more sensitive to galacturonic acid than to the other 11 inducing sugars, all of which function through ChvE. However, galacturonic acid does not induce the expression of chvE, indicating that the galacturonic acid-ChvE complex is especially efficient in vir gene induction (9). We used HAP in combination with galacturonic acid to determine whether the ChvE-sugar complex is important for activation of VirA by HAP. The vir genes were not induced by HAP with galacturonic acid (data not shown). This observation indicates that the different ChvE-sugar complexes are not the reason for induction by HAP, but it is consistent with the notion that an increased level of ChvE is the important end result of arabinose being in the medium.
The activity of the VirA receiver domain is controlled by the periplasmic domain.
Chang et al. (6) have shown that a strain with a point mutation in the receiver domain of VirA, VirA(I734N), and a VirA mutant lacking the entire receiver domain recognize a broader set of phenolic compounds than the parent strain does. We wanted to compare the effects of different sugars on the induction responses of wild-type VirA and VirA(I734N). As shown in Table 6, even the VirA(I734N) strain requires monosaccharides for vir gene induction, but HAP serves as a good inducer with either glucose or arabinose present. Apparently, the mutation I734N allows VirA to be induced to high levels with low levels of the ChvE protein. As a result, glucose as the inducing sugar can provide the level of ChvE needed to achieve a high induction level. In contrast, to achieve a high induction level, the wild-type VirA protein requires a high level of the ChvE protein, which glucose cannot provide. The mutation of the VirA receiver domain makes it much more sensitive to induction by phenolic compounds. It seems that the receiver domain restricts the ability of VirAA6 to respond to these normally weak vir gene inducers, and the conformational change of the receiver domain resulting from the mutation is favorable for vir gene induction. The binding of the ChvE::monosaccharide complex to the VirA protein must also change the conformation of the receiver domain.
TABLE 6.
Responses of wild-type VirA and VirA(I734N) to induction by HAP with glucose and arabinose
| Straina | β-Galactosidase (U) obtained with inducing compoundsb:
|
||
|---|---|---|---|
| HAP | HAP + arabinose | HAP + glucose | |
| A136(pGP159) | 5.4 ± 0.2 | 388 ± 16 (72) | 27.4 ± 1.2 (5) |
| A136(pCH116, pJZ105) | 10.6 ± 1.6 | 982 ± 34 (93) | 570 ± 7 (54) |
a pJZ105 contains VirA(I734N). pCH116 contains a Plac-virG promoter fusion to permit constitutive expression of virG and a virB::lacZ gene fusion.
b Data are means ± standard errors of the means for three samples. The numbers in parentheses are the fold increases in induction calculated from the values with HAP alone. HAP was used at 250 μM. Arabinose and glucose were used at 10 mM.
Effects of glucose and arabinose on the inducing abilities of phenolic inducers in C58 and KU12.
We determined if sugars influenced the inducing activities of a variety of phenolic compounds previously shown to induce vir genes to various levels. We studied the wild-type strains C58 and KU12 induced with phenolic compounds containing one methoxy group (acetovanillone and ferulic acid) or two methoxy groups (AS, syringaldehyde, and syringic acid) and phenolic compounds lacking both methoxy groups (HAP, p-coumaric acid, and phenol). The data in Table 7 reveal that only the compounds with the weakest inducing activity (HAP, p-coumaric acid, and phenol) showed an increased inducing activity when arabinose replaced glucose as the inducing sugar. Stronger inducers showed only a slight but consistent increase. Furthermore, this increase in inducing activity was only seen in strain C58.
TABLE 7.
Induction of virB::lacZ in A. tumefaciens strains by specific phenolic compounds
| Inducing compoundsa | β-Galactosidase (U)b
|
|
|---|---|---|
| C58(pSM243cd) | KU12(pSM243cd) | |
| Acetosyringone | 109 ± 5 | 92 ± 1 |
| + Arabinose | 535 ± 8 | 145 ± 5 |
| + Glucose | 499 ± 22 | 133 ± 2 |
| Syringaldehyde | 42 ± 0.2 | 76 ± 2 |
| + Arabinose | 185 ± 6 | 107 ± 7 |
| + Glucose | 132 ± 3 | 92 ± 1 |
| Syringic acid | 9.1 ± 1.7 | 7.3 ± 1.6 |
| + Arabinose | 96 ± 5 | 126 ± 6 |
| + Glucose | 103 ± 1 | 92 ± 5 |
| Acetovanillone | 6.0 ± 1.5 | 6.9 ± 2.1 |
| + Arabinose | 192 ± 5 | 229 ± 5 |
| + Glucose | 163 ± 3 | 207 ± 2 |
| Ferulic acid | 14.7 ± 4 | 1.5 ± 0.1 |
| + Arabinose | 260 ± 4 | 242 ± 3 |
| + Glucose | 216 ± 4 | 211 ± 3 |
| 4-Hydroxyacetophenone | 2.7 ± 0.1 | 13.7 ± 0.1 |
| + Arabinose | 88 ± 5 | 371 ± 7 |
| + Glucose | 25 ± 1 | 354 ± 8 |
| p-Coumaric acid | 2.6 ± 0.5 | 7.1 ± 1.3 |
| + Arabinose | 54 ± 13 | 347 ± 19 |
| + Glucose | 5.7 ± 1.5 | 368 ± 8 |
| Phenol | 2.8 ± 0.1 | 3.8 ± 0.7 |
| + Arabinose | 30 ± 2 | 242 ± 15 |
| + Glucose | 5.0 ± 0.5 | 195 ± 12 |
a All phenolic compounds were used at 100 μM except for 4-hydroxyacetophenone, which was used at 250 μM. Arabinose and glucose were used at 10 mM.
b Data are means ± standard errors of the means for three samples. β-Galactosidase activities of cultures lacking phenolic compounds were less than 2 U.
An unexpected finding was the observation that strain KU12 was induced to a significant extent by AS and other compounds with two methoxy groups. This contrasts with a previous report that these compounds were very poor inducers (19). The reason for these differences is not certain, but it may reflect differences in culture conditions used in vir gene induction. In the present studies, the precultures used for the assay of vir gene induction were always in the logarithmic phase of growth. This was not carefully controlled in the previous experiments. These new observations help to explain why strain KU12, which is induced by all of the phenolic compounds tested here, has an expanded host range compared with that of strain A348 (20).
DISCUSSION
The data in the present paper demonstrate that the VirA protein in a variety of strains of Agrobacterium can recognize a broad variety of phenolic compounds for vir gene induction. Any differences that exist between different strains and different inducers are quantitative and not qualitative. This conclusion is obvious when arabinose is substituted for glucose and the number of copies of VirA is increased in the course of vir gene induction.
The data in this paper can be considered in terms of the model of vir gene regulation proposed by Heath et al. (15). These investigators proposed that VirA has three states of activity: “off,” “standby,” and “on.” The equilibrium which exists among these three states is driven by the two classes of inducing compounds, monosaccharides and phenolic compounds. In this model, the VirA sensor proteins of cells grown in medium at neutral pH is not autophosphorylated, but it becomes autophosphorylated in response to acidic conditions independently of monosaccharides, ChvE, or phenolic inducers. In the absence of monosaccharides, the VirA remains in the off state, with a small proportion of the molecules in the standby state, because of the equilibrium. In the off state, VirA is in a conformation in which the receiver and kinase domains interact. This intramolecular interaction between the receiver and kinase domains may prevent VirA from binding the phenolic inducers, perhaps by blocking access to the linker domain, the site at which the phenolic inducers bind. ChvE, with its bound monosaccharide, interacts with the C-terminal half of the periplasmic domain of VirA (4, 15, 26), thereby converting the VirA protein to the standby state. The interaction between ChvE and the periplasmic domain of VirA alters the conformation of the receiver domain so that it no longer interacts with the kinase domain. This binding of ChvE to the periplasmic domain may also relieve a poorly defined repressive or inhibitory function of the periplasmic domain for signal transduction (3, 4, 15, 26). In the standby mode, the VirA molecules are able to interact with specific phenolic compounds, most likely in the linker domain, and thereby become converted to the on state. This state allows the transfer of the phosphate molecule from VirA to VirG, which then transcriptionally activates all of the vir genes by binding to the vir box in the promoter region of each of the genes. The genes activated include virA in most, but not all, strains that have been studied.
The present data emphasize the importance of the concentrations of the VirA and ChvE proteins in vir gene induction. Only a few molecules of the VirA protein are ordinarily present in the inner membrane. In strain A6 this protein can be induced up to eightfold by AS, and it is induced in nopaline strains such as C58 as well (23, 30, 33). If this induction does not occur, then the induction of other vir genes will be limited, even by ordinarily effective vir gene inducers such as AS. This was demonstrated by Turk et al. (30), who showed that replacing the promoter region of a virA gene which was not inducible by AS because it did not have a vir box (strain Ag162) with a promoter region that contained a vir box resulted in virB gene induction by AS. Thus, in order for vir genes to be induced, it appears that a threshold level of VirA protein must be present. In order for HAP, p-coumaric acid, and phenol, which are weak inducers of VirAA6, to function more effectively, the level of VirA protein in the standby state must be increased. This can be achieved to some extent by increasing the number of copies of the virA gene (Table 2). Once the concentration is increased, the binding of the ChvE::monosaccharide complex to VirA increases the level of VirA in the standby form. Elevated levels of this form can then react with poor vir gene inducers and convert the standby form to the on form. In strain C58, apparently the threshold level of VirA is present and extra copies do not have to be introduced in order for the induction by weak inducers to be observed.
The equilibrium between the off and standby forms of VirA can also be shifted in favor of the standby form by increasing the concentration of the other reactant in the equation, the ChvE protein. This protein is also present in a low concentration for vir gene induction. This is not surprising, since ChvE, which is chromosomally encoded, performs several functions in the physiology of Agrobacterium in addition to its role in vir gene induction. These include a role in the uptake of monosaccharides as well as in chemotaxis towards these molecules. ChvE becomes limiting for VirA activation when the level of VirA is increased by increasing the number of copies of virA. Therefore, increasing the level of ChvE results in additional molecules of VirA being converted to the standby mode and an increased level of vir gene induction by weak phenolic inducers.
The binding of the ChvE protein to the periplasmic domain of VirA must affect the conformation of the receiver domain and in some way allows the VirA molecule to interact with normally poor inducers, such as HAP, p-coumaric acid, and phenol. The change in conformation of the receiver domain can result either through mutation, such as I734N, or through the binding of ChvE to the periplasmic domain. However, the mutation I734N merely sensitizes the VirA protein so that it becomes more amenable to induction by HAP. This induction still requires ChvE, although not in as high a concentration as is required by the wild-type VirA, since glucose and arabinose are equally good inducing sugars for VirA(I734N). This suggests that the activity of the receiver domain is controlled by the periplasmic domain, a conclusion that was also reached by Turk (29).
The effect of the increased levels of ChvE is most pronounced when the weakest phenolic inducers are used. Compounds like acetovanillone and ferulic acid are equally good inducers of strain C58 when either glucose or arabinose is the inducing sugar (Table 7). In the case of KU12, which is induced to significant levels by all vir gene inducers tested here, no significant difference is seen between the inducing properties of arabinose and those of glucose.
These studies expand our knowledge of the role of sugars in the interaction of Agrobacterium with its host cells and illustrate the interaction between chromosomally and plasmid-encoded gene products. Previous studies have shown that sugars synthesized by wounded plant cells could serve as attractants as well as carbon sources for Agrobacterium through pathways involving ChvE (4). Sugars also play a critical role by binding to ChvE, which increases the affinity of ChvE for VirA (27) and thereby increases the concentration of VirA in the standby state. Previous studies also demonstrated that specific sugars determine the concentration of ChvE through an induction process involving the transcriptional regulator, GbpR. However in these earlier studies, no physiological importance could be attributed to GbpR, since mutations in the locus encoding this protein had no observable effect on any physiological characters tested. These included chemotaxis, sugar utilization, and tumor formation. The present observations demonstrate that GbpR, through its differential interaction with specific sugars, plays a significant role in expanding the inducing capabilities of weak phenolic inducers. By this interaction, the presence or absence of certain sugars determines the breadth of inducing capabilities of specific phenolic compounds and, in a subtle way, the host range of Agrobacterium.
ACKNOWLEDGMENTS
We thank Y. Machida (Nagoya University) for the gift of anti-ChvE serum and S. C. Winans for the gifts of pJZ105 and pCH116. We thank W. Deng for helpful suggestions and members of this laboratory for many stimulating discussions. Lin Lee provided important technical assistance. We also thank S. L. Doty for critical reading of the manuscript.
This work was supported by Public Health Service grant GM32618 from the National Institutes of Health to E. W. Nester.
REFERENCES
- 1.Ankenbauer R G, Nester E W. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J Bacteriol. 1990;172:6442–6446. doi: 10.1128/jb.172.11.6442-6446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ankenbauer R G, Best E A, Palanca C A, Nester E W. Mutants of the Agrobacterium tumefaciens virA gene exhibiting acetosyringone-independent expression of the vir regulon. Mol Plant-Microbe Interact. 1991;4:400–406. doi: 10.1094/mpmi-4-400. [DOI] [PubMed] [Google Scholar]
- 3.Banta L M, Joerger R D, Howitz V R, Campbell A M, Binns A N. Glu-225 outside the predicted ChvE binding site in VirA is crucial for sugar enhancement of acetosyringone perception by Agrobacterium tumefaciens. J Bacteriol. 1994;176:3242–3249. doi: 10.1128/jb.176.11.3242-3249.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cangelosi G A, Ankenbauer R G, Nester E W. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci USA. 1990;87:6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang C H, Winans S C. Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein. J Bacteriol. 1992;174:7033–7039. doi: 10.1128/jb.174.21.7033-7039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C H, Zhu J, Winans S C. Pleiotropic phenotypes caused by genetic ablation of the receiver module of the Agrobacterium tumefaciens VirA protein. J Bacteriol. 1996;178:4710–4716. doi: 10.1128/jb.178.15.4710-4716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das A, Pazour G J. Delineation of the regulatory region sequences of Agrobacterium tumefaciens virB operon. Nucleic Acids Res. 1989;17:4541–4550. doi: 10.1093/nar/17.12.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das A, Xie Y-H. Replication of the broad-host-range plasmid RK2: isolation and characterization of a spontaneous deletion mutant that can replicate in Agrobacterium tumefaciens but not in Escherichia coli. Mol Gen Genet. 1995;246:309–315. doi: 10.1007/BF00288603. [DOI] [PubMed] [Google Scholar]
- 9.Doty S L, Ming C, Nester E W. The chromosomal virulence gene, chvE, of Agrobacterium tumefaciens is regulated by a LysR family member. J Bacteriol. 1993;175:7880–7886. doi: 10.1128/jb.175.24.7880-7886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doty S L, Yu M C, Lundin J I, Heath J D, Nester E W. Mutational analysis of the input domain of the VirA protein of Agrobacterium tumefaciens. J Bacteriol. 1996;178:961–970. doi: 10.1128/jb.178.4.961-970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endoh H, Oka A. Functional analysis of the VirG-like domain contained in the Agrobacterium VirA protein that senses plant factors. Plant Cell Physiol. 1993;34:227–235. [Google Scholar]
- 12.Feyter R D, Yang Y, Gabriel D W. Gene for gene interactions between cotton R genes and Xanthomonas campestris pv malvacearum avr genes. Mol Plant-Microbe Interact. 1993;6:225–237. doi: 10.1094/mpmi-6-225. [DOI] [PubMed] [Google Scholar]
- 13.Garfinkel D J, Nester E W. Agrobacterium tumefaciens mutants affected in crown gall tumorigenesis and octopine catabolism. J Bacteriol. 1980;144:732–743. doi: 10.1128/jb.144.2.732-743.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gubba S, Xie Y-H, Das A. Regulation of Agrobacterium tumefaciens virulence gene expression: isolation of a mutation that restores virGD52E function. Mol Plant-Microbe Interact. 1995;8:788–791. doi: 10.1094/mpmi-8-0788. [DOI] [PubMed] [Google Scholar]
- 15.Heath J D, Charles T C, Nester E W. Ti plasmid and chromosomally encoded two-component systems important in plant cell transformation by Agrobacterium species. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 367–385. [Google Scholar]
- 16.Heath J D, Boulton M I, Rainer D M, Doty S L, Mushegian A R, Charles T C, Davies J W, Nester E W. Discrete regions of the sensor protein VirA determine the strain-specific ability of Agrobacterium to agroinfect maize. Mol Plant-Microbe Interact. 1997;10:221–227. doi: 10.1094/MPMI.1997.10.2.221. [DOI] [PubMed] [Google Scholar]
- 17.Hooykaas P J J, Beijersbergen A G M. The virulence system of Agrobacterium tumefaciens. Annu Rev Phytopathol. 1994;32:157–179. [Google Scholar]
- 18.Huang M-L, Cangelosi G A, Halperin W, Nester E W. A chromosomal Agrobacterium tumefaciens gene required for effective plant signal transduction. J Bacteriol. 1990;172:1814–1822. doi: 10.1128/jb.172.4.1814-1822.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Knauf V C, Nester Eugene W. Wild host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y-W, Jin S, Sim W-S, Nester E W. Genetic evidence for the direct sensing of phenolic compounds by the VirA protein of Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 1995;92:12245–12249. doi: 10.1073/pnas.92.26.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y-W, Jin S, Sim W-S, Nester E W. The sensing of plant signal molecules by Agrobacterium: genetic evidence for the direct recognition of phenolic inducers by the VirA protein. Gene. 1996;179:83–88. doi: 10.1016/s0378-1119(96)00328-9. [DOI] [PubMed] [Google Scholar]
- 21.Lichtenstein C P, Draper J. Genetic engineering of plants. In: Glover D M, editor. DNA cloning: a practical approach. II. Washington, D.C: IRL Press; 1985. pp. 67–119. [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 23.Rogowsky P M, Close T J, Chimera J A, Shaw J J, Kado C I. Regulation of the vir genes of Agrobacterium tumefaciens plasmid pTiC58. J Bacteriol. 1987;169:5101–5112. doi: 10.1128/jb.169.11.5101-5112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Sciaky D, Montoya A L, Chilton M-D. Fingerprints of Agrobacterium Ti plasmids. Plasmid. 1978;1:238–253. doi: 10.1016/0147-619x(78)90042-2. [DOI] [PubMed] [Google Scholar]
- 26.Shimoda N, Toyoda Y A, Nagamine J, Usami S, Katayama M, Sakagami Y, Machida Y. Control of expression of Agrobacterium vir genes by synergistic actions of phenolic signal molecules and monosaccharides. Proc Natl Acad Sci USA. 1990;87:6684–6688. doi: 10.1073/pnas.87.17.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimoda N, Toyoda Y A, Aoki S, Machida Y. Genetic evidence for an interaction between the VirA sensor protein and the ChvE sugar-binding protein of Agrobacterium. J Biol Chem. 1993;268:26552–26558. [PubMed] [Google Scholar]
- 28.Stachel S E, Zambryski P C. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell. 1986;46:325–333. doi: 10.1016/0092-8674(86)90653-7. [DOI] [PubMed] [Google Scholar]
- 29.Turk S C H J. Ph.D. thesis. The Netherlands: Leiden University; 1993. [Google Scholar]
- 30.Turk S C H J, Nester E W, Hooykaas P J J. The virA promoter is a host-range determinant in Agrobacterium tumefaciens. Mol Microbiol. 1993;7:719–724. doi: 10.1111/j.1365-2958.1993.tb01162.x. [DOI] [PubMed] [Google Scholar]
- 31.Winans S C. Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media. J Bacteriol. 1990;172:2433–2438. doi: 10.1128/jb.172.5.2433-2438.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winans S C. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol Rev. 1992;56:12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winans S C, Kerstetter R A, Nester E W. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J Bacteriol. 1988;170:4047–4054. doi: 10.1128/jb.170.9.4047-4054.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]