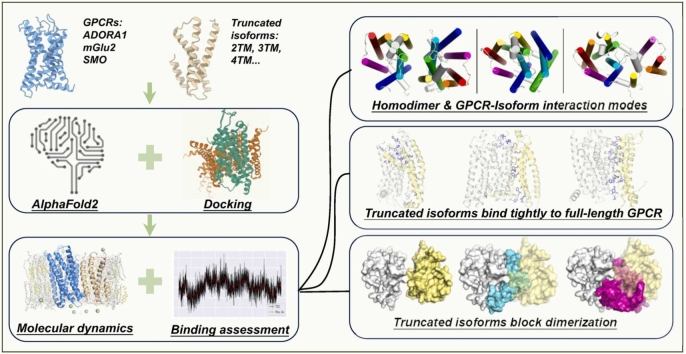
Abstract
G protein-coupled receptors (GPCRs) play a pivotal role in fundamental biological processes and disease development. GPCR isoforms, derived from alternative splicing, can exhibit distinct signaling patterns. Some highly-truncated isoforms can impact functional performance of full-length receptors, suggesting their intriguing regulatory roles. However, how these truncated isoforms interact with full-length counterparts remains largely unexplored. Here, we computationally investigated the interaction patterns of three human GPCRs from three different classes, ADORA1 (Class A), mGlu2 (Class C) and SMO (Class F) with their respective truncated isoforms because their homodimer structures have been experimentally determined, and they have truncated isoforms deposited and identified at protein level in Uniprot database. Combining the neural network-based AlphaFold2 and two physics-based protein-protein docking tools, we generated multiple complex structures and assessed the binding affinity in the context of atomistic molecular dynamics simulations. Our computational results suggested all the four studied truncated isoforms showed potent binding to their counterparts and overlapping interfaces with homodimers, indicating their strong potential to block homodimerization of their counterparts. Our study offers insights into functional significance of GPCR truncated isoforms and supports the ubiquity of their regulatory roles.
Keywords: AlphaFold2 prediction, Complex structure prediction, GPCR oligomerization, Molecular docking, Transmembrane truncation
Graphical abstract
1. Introduction
G protein-coupled receptors (GPCRs) transduce extracellular information into intracellular signals via G proteins, thereby activating signaling cascades and regulating physiological processes [1]. GPCRs represent one of the most abundant protein classes in the mammalian genome [2]. Among the 826 human GPCRs, approximately 350 non-olfactory members are considered druggable, with 165 validated as drug targets [3].
According to sequence homology and functional similarity, GPCRs can be generally categorized into six classes [4]: Class A (rhodopsin-like receptors), Class B (secretin receptors), Class C (metabotropic glutamate receptors), Class D (fungal mating pheromone receptors), Class E (cyclic AMP receptors) and Class F (Frizzled and Smoothened receptors) [5]. Class A is the largest class, involved in a wide range of physiological functions. They possess the conventional seven transmembrane helices and the eighth helix (Helix 8), with the ligand-binding site located within the extracellular region formed by the transmembrane domain. Class B GPCRs are characteristic of large extracellular domains (120–160 residues), which can bind to large peptides, such as hormones. The most notable characteristics of Class C GPCRs include: 1) a colossal extracellular domain (approximately 600 residues), including venus fly trap (VFT) domain and cysteine-rich domain (CRD); 2) obligated constitutive dimerization for receptor activation; 3) the presence of allosteric binding sites in the transmembrane domain, while orthosteric binding sites are located within the extracellular domain [1]. Class D and Class E are composed of non-mammalian GPCRs. Class F GPCRs share a conserved CRD in the extracellular region.
Protein isoforms are an important source of functional diversity, derived from tissue-specific alternative splicing [6]. Some GPCR isoforms were reported to exhibit distinct signaling patterns [7], [8], [9]. A systematic investigation [10] compiled diverse topological types of human GPCR isoforms and their functional characteristics and revealed distinct combinatorial expression patterns across different tissues. Interestingly, some GPCR isoforms are significantly truncated, yet they display unique functional roles [11]. Some of them can dimerize with their full-length counterparts, hampering cell surface trafficking and/or reducing ligand binding affinity [12], [13], [14], [15], [16], while some even show well-preserved individual functionality [17], [18].
Previously, we serendipitously discovered that a truncated GPCR variant could not only function as the negative regulator when co-expressed with the full-length counterpart, but also perform signaling individually [19]. It is reported that oligomerization of some GPCRs takes place in the endoplasmic reticulum [20], [21], [22] and tends to be essential for cell surface delivery of Class A and Class C GPCRs [23]. Thus, this might be a common regulatory strategy that, compared to normal homodimers, these truncated isoforms bind to their full-length counterparts with similar or higher affinity and overlapping interfaces, thereby inhibiting homodimerization. However, it remains largely unexplored how these truncated isoforms interact with full-length counterparts and whether such interaction is widespread.
Computational studies about GPCR dimerization interfaces have been emerging in last two decades. The methods used for interface prediction are mainly based on molecular docking, molecular modelling and coarse-grained molecular dynamics simulations [24], [25], [26]. Currently the most widely-used docking tools are typically based on Fast Fourier Transform, which efficiently generates geometrically complementary rigid-body docking poses [27]. Among these, ClusPro [28] is popular for its outstanding performance. Nevertheless, this approach cannot explicitly account for conformational flexibility [29]. Another docking method, LightDock [30], employs an artificial intelligence algorithm, Glowworm Swarm Optimization, capable of capturing multiple local and global energy minima in the docking energy landscape, accommodating multi-scale conformational flexibility. However, due to current computational capacity limitations and systematic error, using these physics-based approaches to accurately and comprehensively explore conformational space remains challenging.
The emergence of the neural network-based AlphaFold2 [31] offers an alternative approach for complex structure prediction, with its modified version, AlphaFold-Multimer [32], exhibiting exceptional prediction quality [33], [34], [35], [36], [37]. Recent research has shown that combining AlphaFold2 and the docking tool ClusPro through a nested-like structure can significantly enhance the success rate of complex structure prediction [38].
Here, we found that truncated isoforms occur in different classes of GPCRs, among which we select three GPCRs from three classes as the subjects in this study, including ADORA1 (Class A), mGlu2 (Class C) and SMO (Class F) because their homodimer structures have been experimentally determined, and they have truncated isoforms deposited and identified at protein level in Uniprot database. We studied their interaction patterns with their respective truncated isoforms. We generated the GPCR-isoform complex structure models by combining the neural network-based AlphaFold-Multimer and two physics-based protein-protein docking tools, ClusPro and LightDock. We assessed the binding affinities of the complexes using the Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) algorithm in the context of atomistic molecular dynamics (MD) simulations. Our results predict the presence of strong interactions between these GPCRs and their truncated isoforms, strongly indicating the potential of these truncated isoforms to inhibit homodimerization of the full-length receptors.
2. Methods
2.1. Complex structure model generation using AlphaFold-Multimer and structure processing
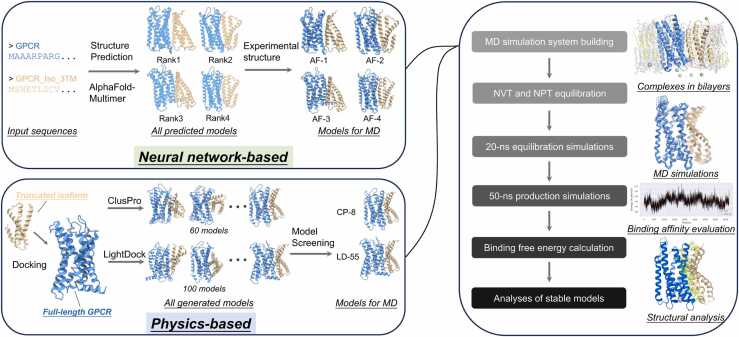
The structure prediction of all complexes was performed using AlphaFold-Multimer [32] via the ColabFold [39] pipeline applying mostly default parameters. Briefly, “alphafold2_multimer_v3” model type was applied; five structure models were output; all five structures were relaxed; “templates” mode was applied to maximize the similarity between predicted structures and experimental structures of protomers. The input sequences were from Uniprot database https://www.uniprot.org/ with entries summarized in Table S1. The alignment figures were made using Uniprot “align” tool https://www.uniprot.org/align; the topological figures were made using the TMHMM server [40] or Phobius server [41]. After the structure models were generated, we selected some of them and processed these models for MD simulations. The AlphaFold2 prediction confidence profiles of the models in Table 1 were summarized in Fig. S1. For each complex, we selected the models with unique interfaces. For each selected model, we aligned the protomer structure with its experimental structure and replaced the predicted structure by the experimental structure. Regarding truncated isoforms, we did not make modifications as no available experimental structures. For experimental structures, we used 5UEN (PDB entry) for ADORA1, 7EPA for mGlu2, and 4JKV for SMO and exported the GPCR structures for further use. The mutations made for crystallization and the missing loops connecting helices were repaired by inputting the original sequence and homogenous modelling using PyMod 3 [42]. For mGlu2, the N-terminal extracellular domain (residue 1–562) was removed. Steric clashes, if exist, were solved by rotating the clashing side chains, using PyMOL (version 2.5, Schrödinger). For the homodimer models, we aligned the processed monomer structure with the two protomers and replaced the original structure by the processed structure. The resultant structures were used as the inputs of MD simulations.
Table 1.
Summary of interface composition and binding free energy calculations of the most stable complex models.
| Complex (Subject-Partner) |
Model | Interface composition |
Binding free energy (kcal/mol) |
|
|---|---|---|---|---|
| Subject (left) | Partner (right) | |||
| ADORA1-ADORA1 | Noncry | TM3 (E) | Identical | 9.15 ± 0.24 |
| ADORA1-Iso_3TM | CP-6 | TM4, TM2 (E), TM3 (E) | TM2, TM1, TM3 (E) | -133.94 ± 0.25 |
| ADORA1-Iso_4TM | AF-1 | TM1, TM3 (E), TM4 (I) | TM4 | -111.64 ± 0.18 |
| AF-3 | TM1, TM2 | TM4 | -115.60 ± 0.39 | |
| mGlu2-mGlu2 | Inactive | TM4, TM3(E) | Identical | -61.06 ± 0.35 |
| Active | TM6, TM7 (E) | Identical | -43.55 ± 0.13 | |
| mGlu2-Iso_2TM | LD-15 | TM4, TM2, TM3 (E) | TM7, TM6 | -163.79 ± 0.26 |
| CP-H10 | TM7, TM6, TM1 | TM6, TM7 | -192.07 ± 0.32 | |
| SMO-SMO | TM4, TM5 | Identical | -69.66 ± 0.13 | |
| SMO-Iso_3TM | LD-34 | TM4, TM5 | TM4, TM5 | -95.90 ± 0.34 |
Note: The content of “Interface composition” is ordered by the contribution to the interface formation and only TM helices are shown. For example, “TM7, TM1, TM6” indicates TM7 > TM1 > TM6 in terms of contact contribution. E, the extracellular side-facing part of the helix; M, the middle part of the helix; I, the intracellular side-facing part of the helix. No parenthesis indicates that the majority of this helix is involved in the interface composition. “AF-1” denotes the Rank 1 model generated by AlphaFold-Multimer; “LD-12” denotes the No.12 model generated by LightDock; “CP-12” denotes the No.12 model generated by ClusPro with the default Balanced mode, and “CP-H12” denotes the No.12 model generated by ClusPro with the Hydrophobic-favored mode. Isoforms are displayed in the abbreviated forms without the prefix. For example, “mGlu2-Iso_2TM” denotes the complex formed by mGlu2 and its truncated isoform with two transmembrane helices. The numbering of transmembrane helices in truncated isoforms corresponds to their numbering in the full-length counterparts. Binding free energy values are shown as mean ± standard error of the mean (SEM).
2.2. Protein-protein docking using ClusPro and LightDock and structure processing
Protein-protein docking of complexes was performed using ClusPro server [28] applying all default parameters, or using LightDock [30] with the “flexible backbone” mode for the truncated isoform. The docking complexes include: ADORA1-Iso_3TM, ADORA1-Iso_4TM, mGlu2-Iso_2TM and SMO-Iso_3TM. Experimental structures of GPCRs and AlphaFold2 models of truncated isoforms (the PDB entries and structure processing same as abovementioned) were used as inputs of docking. For the resultant models of ClusPro, we selected “topology-compliant” models from 30 models obtained by the default “Balanced mode” and 30 models obtained by the “Hydrophobic-favored mode”. For the resultant models of LightDock, we selected “topology-compliant” models from all 100 models. “Topology-compliant” models are defined when the following rules are met: i) The orientation of both protomers is consistent with a typical GPCR orientation; ii) The putative membrane layers inferred from transmembrane regions of both protomers are basically spatially consistent. The selected models were used as the inputs of MD simulations.
2.3. Molecular dynamics simulations
The membrane-protein systems were built using the membrane builder [43] of the CHARMM-GUI web server [44]. The protein portion was centered in a rectangular box, with X/Y length of 12 nm for dimers and 11 nm for complexes containing truncated isoforms. The membrane consists of 70% 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) and 30% cholesterol. The system was solvated in TIP3P water with 150 mM KCl.
All MD simulations were performed using GROMACS 2022.3 [45]. The all-atom CHARMM36m force field was used. The system energy was minimized using the steepest descent method and the maximum forces were converged below 1000 kJ/mol/nm. Electrostatics were treated with Particle Mesh Ewald, and the cutoff for both Coulomb and van der Waals interactions was 1.2 nm. 125-ps equilibration simulations were performed using the standard six-step CHARMM-GUI protocol [46]. After NVT and NPT equilibration, a 20-ns further equilibration simulation was run to achieve stabilization of the complexes, which was assessed by the RMSD and trajectory of the complex. This was followed by a 50-ns production MD simulation, which next subjected to binding free energy calculations. These 70-ns simulations were performed under the following setting: 2-fs time step was used with the SHAKE algorithm. The van der Waals interactions were smoothly switched off at 10–12 Å by a force-switching function. Long-range electrostatic interactions were calculated using the particle-mesh Ewald method. The Parrinello-Rahman barostat was used with semi-isotropic coupling and the Nose-Hoover thermostat was used. Temperature was held at 310.15 K and pressure was held at 1 bar, respectively.
For conformational analysis, simulation frames were extracted every 250 ps and clustered (GROMACS built-in command “gmx cluster”) using the linkage method with a cut-off that produced the minimal number of clusters. The medoid structure of the largest cluster was exported and used for further analysis. The structure figures were made using PyMOL or ChimeraX [47].
2.4. Binding free energy calculation
The binding free energy of the complexes was calculated using Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) algorithm and performed using the gmx_MMPBSA tool [48]. For each model, all 5001 frames of the 50-ns simulation were subjected to calculations. The calculations were performed under the following setting, which is mostly consistent with the recommended setting for the protein-membrane systems prepared by CHARMM force fields https://valdes-tresanco-ms.github.io/gmx_MMPBSA/v1.5.6/examples/Protein_membrane_CHARMMff/: Poisson Boltzmann (PB) calculations were applied; the temperature was 310.15 K; the PB radii was “charmm_radii” (PBRadii = 7); a uniform membrane dielectric constant in a slab-like implicit membrane was used (memopt = 1); the membrane dielectric constant was 7.0; radii from the prmtop file for both the PB calculation and for the non-polar calculation were used (radiopt = 0); ionic strength was 0.15 M; the ratio between the longest dimension of the rectangular finite-difference grid and that of the solute was 1.25 (fillratio = 1.25); the total non-polar solvation free energy was modeled as a single term linearly proportional to the solvent accessible surface area (inp = 1); a geometric multigrid was used for iterative solvers (solvopt = 2); a classical geometric method was used for setting up a dielectric model for all numerical PB procedures (ipb = 1); periodic boundary condition was used (bcopt = 10); no focusing was used (nfocus = 1); computing total electrostatic energy and forces with the particle-particle particle-mesh (P3M) procedure outlined in Lu and Luo [49] was applied (eneopt = 1); atom-based cutoff distance to remove short-range finite-difference interactions and to add pairwise charge-based interactions was 7.0 nm (cutfd = 7.0); cutoff distance used for van der Waals interactions was 99.0 nm (cutnb = 99.0); using SASA to estimate cavity free energy was applied (use_sav = 0); number of dots used to store arc dots per atom was 15,000 (maxarcdot = 15,000); verbose mode is on (npbverb = 1); printing all residues within 5 Å between receptor and ligand was applied. For the analysis of binding-contributing resides, gmx_mmpbsa_ana tool was used. The binding free energy was calculated by summing Van der Waals contribution (ΔVDWAALS), Electrostatic contribution (ΔEEL) and Non-polar contribution to the solvation free energy (ΔENPOLAR). The detailed results of MMPBSA calculations were available in the Zenodo database: 10.5281/zenodo.10275624.
3. Results and discussion
3.1. Workflow of interface prediction and analysis
To investigate the potential of truncated isoforms to inhibit homodimerization of the full-length GPCRs, we selected the research subjects following two rules: their homodimer structures have been resolved, and they have truncated isoforms deposited and identified at protein level in Uniprot database. After we searched extensively, we found a total of three GPCRs that met our requirements: adenosine receptor A1 (ADORA1, Class A), metabotropic glutamate receptor 2 (mGlu2, Class C), and smoothened receptor (SMO, Class F) (Table S1).
We generated complex models combining approaches based on distinct mechanisms (Fig. 1). We employed AlphaFold-Multimer to generate five initial models, out of which those with unique interface compositions were selected. These models were then aligned and replaced with experimental structures, except for the truncated isoforms. Furthermore, we utilized the two docking tools, ClusPro and LightDock, to generate a total of 160 models, from which we selected those models that are topology-compliant. The topology-compliant models are defined when the following rules are met: i) The orientation of both protomers is consistent with a typical GPCR orientation; ii) The putative membrane layers inferred from transmembrane regions of both protomers are basically spatially consistent. The complex models were then used to construct protein-membrane systems, followed by all-atom MD simulations. After equilibration, 50-ns simulation trajectories were subjected to binding free energy (BFE) calculations using the MMPBSA algorithm. In our initial attempts, we found that most systems tended to maintain stabilization within around 50 ns after the pre-equilibration run of 20 ns, and did not show significant conformational deviations after we extended the simulations for another 50 ns (Fig. S2). For each complex, the most stable structure model, characterized by the lowest BFE, was selected for further analysis.
Fig. 1.
The workflow of our computational approach. Briefly, we generated initial complex models by combining neural network-based protein structure prediction tool AlphaFold2 and two physics-based protein-protein docking tools, ClusPro and LightDock. For complex structure prediction, we employed AlphaFold-Multimer to generate five models, out of which the models with unique interface composition were selected. These models were then aligned with experimental structures to replace predicted structures (except isoforms). For protein-protein docking, ClusPro generated 60 models and LightDock generated 100 models, from which topology-compliant models were chosen. Both sets of initial complex models were used as inputs to build protein complex-membrane systems for molecular dynamics (MD) simulations. After NVT and NPT equilibration, a 20-ns equilibration MD simulation was run to achieve stabilization of the complexes. This was followed by a 50-ns production MD simulation, subsequently subjected to binding free energy calculations based on MMPBSA algorithm. According to the results, the most stable complex models were selected for further analysis. The structure snapshots were made using PyMOL or ChimeraX.
The results of all complex models were summarized in Table S2-S4. For each GPCR-isoform complex, eight or nine models were input into MD simulations and BFE calculations, except for the ADORA1 isoform with three transmembrane helices (designated as ADORA1_Iso_4TM, or abbreviated as Iso_4TM when discussing the interaction with the full-length counterpart). We found that the most stable models (Table 1) could originate from AlphaFold-Multimer, ClusPro or LightDock, with no apparent preference, suggesting the advantage of our combinatorial strategy.
To our surprise, all truncated isoforms exhibited strong binding strengths, binding to the surfaces overlapping with the homodimerization interfaces of their full-length counterparts. In following sections, we will make a detailed analysis for each subject GPCR.
3.2. ADORA1: two different truncated isoforms both show the potential of blocking homodimerization
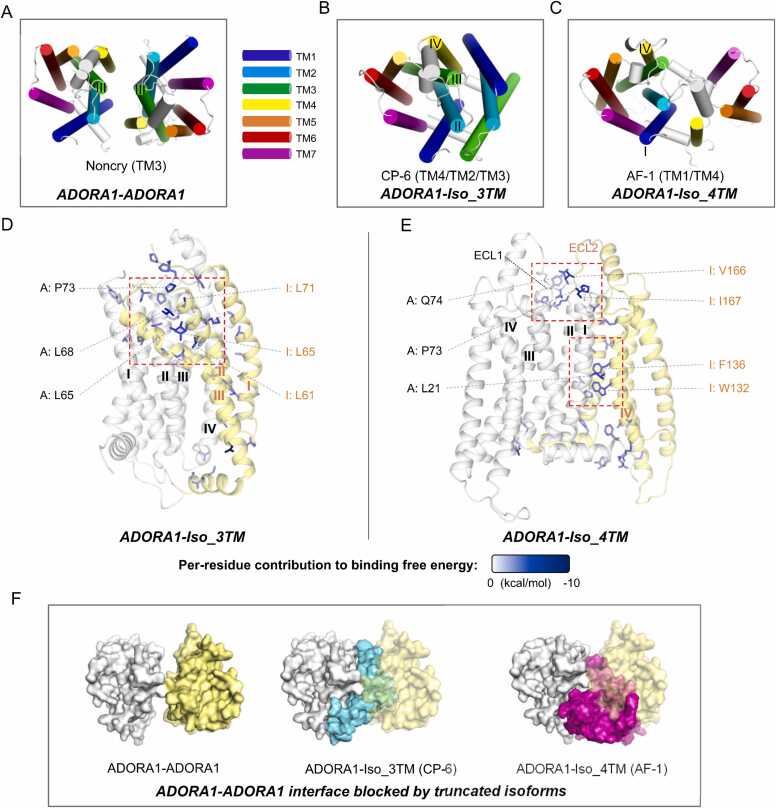
The homodimer structure of ADORA1 has previously been resolved. Glukhova et al. [50] demonstrated two types of interfaces: one is non-crystallographic, where only two pairs of aromatic residues on the extracellular side of TM3 contact; the other was crystallographic (between repeating units within a crystal), involving the extracellular side of TM4 and TM5. Although the latter interface was more extensive, it cannot be ruled out that it might be an artifact of crystallization. We conducted MD simulations and BFE calculations, revealing a weak binding for the non-crystallographic dimer (Fig. 2A) (BFE = 9.15 ± 0.24 kcal/mol).
Fig. 2.
Interaction patterns of ADORA1 homodimers and its complex with its truncated isoforms. (A-C) Top view of the conformations of ADORA1-ADORA1 (A) and the most stable complex models of ADORA1-Iso_3TM (B) and ADORA1-Iso_4TM (C), identified by binding free energy calculations. For every model, the designation and the information of the interface on ADORA1 are shown below the structure. Regarding the interface composition, the order of the helices is consistent with Table 1, i. e. determined by their contribution to the interface formation. The transmembrane helices are colored according to the scheme shown on the right side of A. Other parts in the structures are shown in white. (D-E) Side view of the interfaces of ADORA1-Iso_3TM (D) and ADORA1-Iso_4TM (E). The residues that contribute positively (per-residue contribution < −1 kcal/mol) to interface formation are shown as sticks, and in blue, with the color intensity determined by their contribution to binding free energy (as the bar shown below the diagram). ADORA1 backbones are shown in white and the isoform backbones are shown in yellow. Dash rectangles indicate the interface core regions. For the labels, “A” denotes ADORA1 and “I” denotes the isoform. The labeled residues are the residues that contribute the most to the formation of the interface core regions. The labels related to ADORA1 are shown in black with dashed connector, and those related to isoforms are shown in orange. The numbering of residues in truncated isoforms corresponds to their numbering in the full-length counterparts. (F) ADORA1 homodimerization interface could be blocked by truncated isoforms, shown as top view. The structures are shown as surfaces. To show the blocking effect, we aligned subject protein (here it is ADORA1) structure in the dimer model with that in the subject-blocker model, followed by hiding the subject protein structure in the subject-blocker model. All structure snapshots were from the medoid frame in the largest conformational cluster and made using PyMOL.
We identified two distinct truncated isoforms of ADORA1 in the Uniprot database. One is Iso_3TM (including residue 1–114 of ADORA1), comprising a complete N-terminus and the first three transmembrane helices; the other is Iso_4TM (including residue 117–326), encompassing the last four transmembrane helices and the full C-terminus (Fig. S3). For Iso_3TM, among all the ADORA1-Iso_3TM complex models, model CP-6 exhibited significantly higher binding affinity than the others (Table S2). In this model, TM2, TM3 and TM4 of ADORA1 interacted with TM1, TM2 and TM3 of Iso_3TM (Fig. 2B), with the interface core region on the extracellular-facing side. Notably, L65 and L68 in TM2, P73 in ECL1 of ADORA1, and L61, L65 and L71 in TM2 of Iso_3TM were major contributors to this region (Fig. 2D, and Fig. S4A for details). Regarding Iso_4TM, AlphaFold-Multimer generated five similar ADORA1-Iso_4TM models, and we selected two models with relatively large conformational differences. LightDock failed to produce topology-compliant models. Both AlphaFold models exhibited similarly strong binding affinities, significantly higher than other models (Table S2). Here, we chose the AF-1 model for structural analysis. The AF-1 model (Fig. 2C) revealed two interface core regions: one located in the middle of ADORA1's TM1 and Iso_4TM's TM4 (in this study, the numbering of components in truncated isoforms corresponds to that in the full-length counterparts), with L21 in TM1 of ADORA1, and W132 and F136 in TM4 of Iso_3TM as the major contributors (Fig. 2F). The other was located on the extracellular side, where P73 and Q74 in extracellular loop 1 (ECL1) of ADORA1 along with V166 and I167 in ECL2 of Iso_3TM serve as the major contributors (Fig. 2E, and Fig. S4B for details).
We found that both Iso_3TM and Iso_4TM have the potential to block ADORA1 homodimerization. That homodimerization could be blocked by the blocker is defined when the following rules are met: i) The homodimerization interface has significantly overlapped portion with the interface on the subject (here the subject is ADORA1) in the subject-blocker model; ii) The binding free energy of the subject-blocker model is lower or close compared to that of the homodimer model. In this case, the binding affinity of the homodimer is far lower than that of ADORA1-Iso_3TM (BFE = −133.94 ± 0.25 kcal/mol) and ADORA1-Iso_4TM (BFE = −111.64 ± 0.18 kcal/mol), and the small interface of the homodimer can be completely covered by the interfaces of Iso_3TM and Iso_4TM with ADORA1 (Fig. 2F). For the ADORA1-Iso_4TM AF-3 model, the situation is nearly identical to AF-1 (Fig. S5).
Our results suggest that the two different truncated isoforms of ADORA1 can stably bind to the full-length counterpart and exhibit the potential to block homodimerization.
3.3. mGlu2: its truncated isoform shows strong potential of blocking both two homodimerization patterns
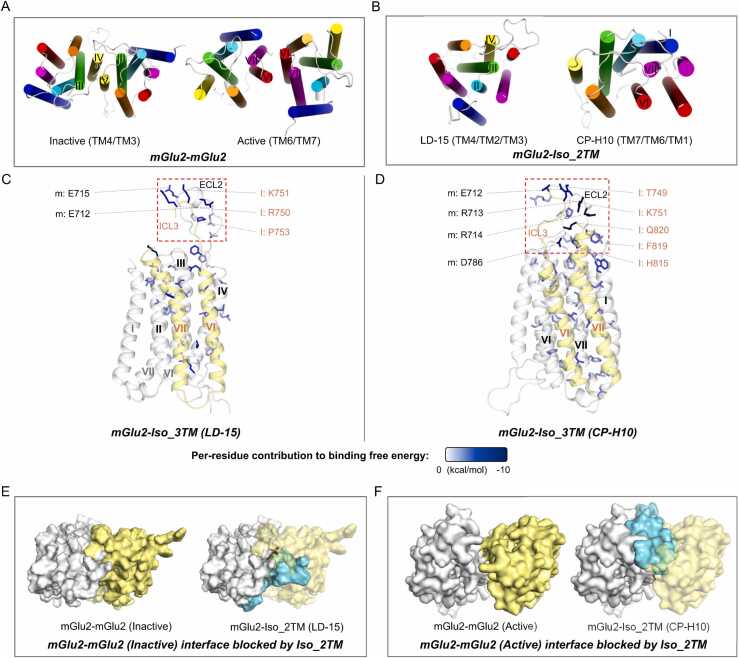
Du et al. [51] resolved two distinct dimeric structures of mGlu2, including an inactive apo structure and a structure bound simultaneously to an agonist and a positive allosteric modulator, the latter being indicated active. The dimeric interface of the inactive homodimer is composed of TM3 and TM4, while that of the active homodimer consists of TM6 and TM7 (Fig. 3A). We conducted MD simulations and BFE calculations for both homodimers, revealing moderate binding affinities in both cases (Inactive: −61.06 ± 0.35 kcal/mol; active: −43.55 ± 0.13 kcal/mol).
Fig. 3.
Interaction patterns of mGlu2 homodimers and its complex with its truncated isoform. (A-B) Top view of the conformations of mGlu2-mGlu2 (A) and the most stable complex models of mGlu2-Iso_2TM (B), identified by binding free energy calculations. The display style is the same as Fig. 2A. (C-D) Side view of the interfaces of and mGlu2-Iso_2TM Model LD-15 (C) and Model CP-H10 (D). Here the dash rectangles indicate the interface extracellular regions. The display style is the same as Fig. 2D. (E-F) mGlu2 homodimerization interface (E: Inactive; F: Active) could be blocked by Iso_2TM, shown as top view. The display style is the same as Fig. 2F.
A truncated isoform of mGlu2 with two transmembrane helices (Iso_2TM, including residue 1–470 and 749–872) was identified. It possesses a complete N-terminal VFT domain, TM6, TM7, and a full C-terminus, without the CRD and the first five helices of the transmembrane domain (Fig. S6). Two transmembrane topology prediction tools, TMHMM [40] and Phobius [41] both indicate that the VFT domain remains extracellular, yet TM6 and TM7 are inverted compared to the topology of full-length counterpart (Fig. S6C and D). We adopted this predicted topology as the correct form. Additionally, we only used the transmembrane domain for further investigation, without the extracellular portion, as we found that both AlphaFold2 and molecular docking were unable to generate models where simultaneously the both extracellular portions of the two protomers bound and the both transmembrane domains bound. We infer the main reason is that the linker between VFT domain and the transmembrane domain of Iso_2TM is too short and lacks CRD (cysteine rich domain) (Fig. S7). We suppose that the priority for binding lies with the transmembrane domains, as it is very unlikely that highly hydrophobic transmembrane domains can be entirely exposed to the extracellular environment.
For generated mGlu2-Iso_2TM models, both LD-15 (BFE = −163.79 ± 0.26 kcal/mol) and CP-H10 (BFE = −192.07 ± 0.32 kcal/mol) models exhibited remarkably strong binding affinities, while AlphaFold failed to produce a topology-compliant model. In the LD-15 model, TM2, TM3, and TM4 of mGlu2 contribute to interface formation, while the CP-H10 model, TM1, TM6, and TM7 of mGlu2 contribute to interface formation (Fig. 3B). We observed that both models exhibited strong interactions in the extracellular region. ECL2 of mGlu2 interacts with ICL3 of Iso_2TM mainly through electrostatic interactions (Fig. 3C and D). Particularly in the case of the CP-H10 model, the individual BFE contributions of several residues reached approximately − 10 kcal/mol. Within the transmembrane domain, both models displayed extensive interfaces with strong interactions spanning from the intracellular end to extracellular end. It is noteworthy that Iso_2TM in the two models utilized different sides to bind to mGlu2 (Fig. S8). We speculate that Iso_2TM, freed from the full-length framework, possesses higher structural flexibility and lower conformational constraints, which may explain the coexistence of two interaction patterns with the full-length counterpart.
Interestingly, we found that these two mGlu2-Iso_2TM models have the potential to block the two distinct patterns of homodimerization, respectively. In the LD-15 model, TM6 of Iso_2TM occupies the TM4 interface in the inactive homodimer (Fig. 3E). In model CP-H10, TM6 of Iso_2TM occupies the TM6 and TM7 interface in the active homodimer (Fig. 3F). Given the strong binding affinities observed in both models, we infer that Iso_2TM may have the capability to block interfaces in both active and inactive homodimers. This coincidence might imply that this could be a consequence of molecular evolution.
Our results suggest that the truncated isoform of mGlu2 can form highly stable complexes with the full-length counterpart in two distinct patterns and holds strong potential for blocking homodimerization.
3.4. SMO: SMO-Isoform interface highly overlaps with homodimeric interface
Wang et al. [52] experimentally determined the dimeric structure of SMO bound to an antagonist. In this homodimer, TM4 and TM5 form the interface (Fig. 4A). Through MD simulations and BFE calculations, we showed that this homodimeric structure exhibited moderate binding affinity (BFE = −69.66 ± 0.13 kcal/mol).
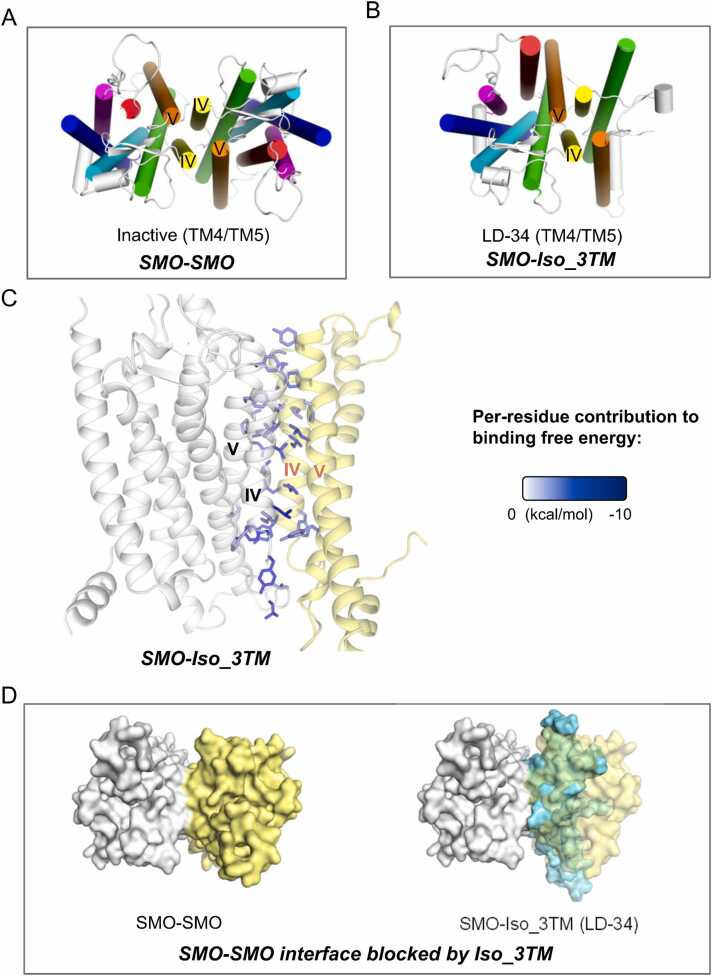
Fig. 4.
Interaction patterns of SMO homodimers and its complex with its truncated isoform. (A-B) Top view of the conformations of SMO-SMO (A) and the most stable complex model of SMO-Iso_3TM (B), identified by binding free energy calculations. The display style is the same as Fig. 2A. (C) Side view of the interfaces of and SMO-Iso_3TM. The display style is the same as Fig. 2D. (D) SMO homodimerization interface could be blocked by Iso_3TM, shown as top view. The display style is the same as Fig. 2F.
SMO has a truncated isoform that includes only the middle three transmembrane helices, namely TM3, TM4 and TM5 (including residue 308–452) (Fig. S9). In the generated SMO_Iso_3TM models, the LD-34 model exhibited the highest binding affinity (BFE = −95.90 ± 0.34 kcal/mol), with TM4 and TM5 from both protomers as the interface (Fig. 4B). In this model, interface residues were uniformly distributed, spanning from the intracellular end to the extracellular end (Fig. 4C). Interestingly, we observed that the interface in this model closely resembled that in the homodimer. Its higher binding affinity, compared to the homodimer, may be attributed to fine conformational tweaks, possibly enabled by increased structural flexibility and reduced conformational constraints. Consequently, this truncated isoform also demonstrates the potential to block homodimerization (Fig. 4D). It is noteworthy that this model originated from physics-based docking, instead of template-based methods such as AlphaFold. Furthermore, this truncated isoform encompasses TM4 and TM5, which are exactly required for homodimerization (TM3 could aid in maintaining the original conformation), suggesting that this may also be an outcome of molecular evolution.
4. Conclusions
Our computational study predicted the presence of strong interactions between three GPCRs and their truncated isoforms. Based on our computational results, truncated isoforms of GPCRs from three different classes (class A, class C and class F) consistently exhibited the potential to inhibit homodimerization of full-length counterparts, suggesting that their regulatory function through this mechanism might be widespread. Our findings not only could provide insights and guidance for further experimental research, but also the biologically functional roles of truncated isoforms. If the future experiments can verify our structural bioinformatic findings, these truncated isoforms could be developed as therapeutics, as interfering peptides targeting specific GPCR oligomerization has been emerging and considered as promising drug candidates for previously undruggable GPCRs [53], [54], [55].
Declaration of Generative AI and AI-assisted technologies in the writing process
This manuscript did not use any AI to generate the research data. All scientific results presented in this manuscript are carried out by us. We did not use AI, Chat GPT nor GPT4 to write this manuscript.
Declaration of Competing Interest
None.
Acknowledgements
This study is supported by the grant from National Natural Science Foundation of China (32170105). We thank Yanze Wang (Department of Chemistry, Massachusetts Institute of Technology) for his valuable advice on this study. The computations in this paper were run on the MIT SuperCloud (Lincoln Laboratory Supercomputing Center, Massachusetts Institute of Technology). M.L. acknowledge the student fellowship from Shanghai Jiao Tong University.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.12.008.
Appendix A. Supplementary material
Supplementary material
.
Data Avaiability
Data and code have been deposited in the Zenodo database, DOI: 10.5281/zenodo.10275624. Further information and requests for data should be directed to and will be fulfilled by S.Z., shuguang@mit.edu.
References
- 1.Odoemelam C.S., Percival B., Wallis H., Chang M.W., Ahmad Z., Scholey D., et al. G-Protein coupled receptors: structure and function in drug discovery. RSC Adv. 2020;10(60):36337–36348. doi: 10.1039/d0ra08003a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Congreve M., de Graaf C., Swain N.A., Tate C.G. Impact of GPCR structures on drug discovery. Cell. 2020;181(1):81–91. doi: 10.1016/j.cell.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Yang D., Zhou Q., Labroska V., Qin S., Darbalaei S., Wu Y., et al. G protein-coupled receptors: structure- and function-based drug discovery. Signal Transduct Target Ther. 2021;6(1) doi: 10.1038/s41392-020-00435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foord S.M., Bonner T.I., Neubig R.R., Rosser E.M., Pin J.P., Davenport A.P., et al. International union of pharmacology. XLVI. G protein-coupled receptor list. Pharm Rev. 2005;57(2):279–288. doi: 10.1124/pr.57.2.5. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh E., Kumari P., Jaiman D., Shukla A.K. Methodological advances: the unsung heroes of the GPCR structural revolution. Nat Rev Mol Cell Biol. 2015;16(2):69–81. doi: 10.1038/nrm3933. [DOI] [PubMed] [Google Scholar]
- 6.Buljan M., Chalancon G., Dunker A.K., Bateman A., Balaji S., Fuxreiter M., et al. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr Opin Struct Biol. 2013;23(3):443–450. doi: 10.1016/j.sbi.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Cong Z., Liang Y.L., Zhou Q., Darbalaei S., Zhao F., Feng W., et al. Structural perspective of class B1 GPCR signaling. Trends Pharm Sci. 2022;43(4):321–334. doi: 10.1016/j.tips.2022.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Kilpatrick G.J., Dautzenberg F.M., Martin G.R., Eglen R.M. 7TM receptors: the splicing on the cake. Trends Pharm Sci. 1999;20(7):294–301. doi: 10.1016/s0165-6147(99)01355-3. [DOI] [PubMed] [Google Scholar]
- 9.Markovic D., Challiss R.A. Alternative splicing of G protein-coupled receptors: physiology and pathophysiology. Cell Mol Life Sci. 2009;66(20):3337–3352. doi: 10.1007/s00018-009-0093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marti-Solano M., Crilly S.E., Malinverni D., Munk C., Harris M., Pearce A., et al. Combinatorial expression of GPCR isoforms affects signalling and drug responses. Nature. 2020;587(7835):650–656. doi: 10.1038/s41586-020-2888-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wise H. The roles played by highly truncated splice variants of G protein-coupled receptors. J Mol Signal. 2012;7(1):13. doi: 10.1186/1750-2187-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosse R., Schöneberg T., Schultz G., Gudermann T. Inhibition of gonadotropin-releasing hormone receptor signaling by expression of a splice variant of the human receptor. Mol Endocrinol. 1997;11(9):1305–1318. doi: 10.1210/mend.11.9.9966. [DOI] [PubMed] [Google Scholar]
- 13.Leung P.K., Chow K.B., Lau P.N., Chu K.M., Chan C.B., Cheng C.H., et al. The truncated ghrelin receptor polypeptide (GHS-R1b) acts as a dominant-negative mutant of the ghrelin receptor. Cell Signal. 2007;19(5):1011–1022. doi: 10.1016/j.cellsig.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 14.Cogé F., Guenin S.P., Renouard-Try A., Rique H., Ouvry C., Fabry N., et al. Truncated isoforms inhibit [3H]prazosin binding and cellular trafficking of native human alpha1A-adrenoceptors. Biochem J. 1999;343(Pt 1):231–239. doi: 10.1042/bj3430231. Pt 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez A., Borquez M., Trigo C.A., Brenet M., Sarmiento J.M., Figueroa C.D., et al. The splice variant of the V2 vasopressin receptor adopts alternative topologies. Biochemistry. 2011;50(22):4981–4986. doi: 10.1021/bi2001278. [DOI] [PubMed] [Google Scholar]
- 16.Martin C.B., Ramond F., Farrington D.T., Aguiar A.S., Jr, Chevarin C., Berthiau A.S., et al. RNA splicing and editing modulation of 5-HT(2C) receptor function: relevance to anxiety and aggression in VGV mice. Mol Psychiatry. 2013;18(6):656–665. doi: 10.1038/mp.2012.171. [DOI] [PubMed] [Google Scholar]
- 17.Ling K., Wang P., Zhao J., Wu Y.L., Cheng Z.J., Wu G.X., et al. Five-transmembrane domains appear sufficient for a G protein-coupled receptor: functional five-transmembrane domain chemokine receptors. Proc Natl Acad Sci U S A. 1999;96(14):7922–7927. doi: 10.1073/pnas.96.14.7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perron A., Sarret P., Gendron L., Stroh T., Beaudet A. Identification and functional characterization of a 5-transmembrane domain variant isoform of the NTS2 neurotensin receptor in rat central nervous system. J Biol Chem. 2005;280(11):10219–10227. doi: 10.1074/jbc.M410557200. [DOI] [PubMed] [Google Scholar]
- 19.Qing R., Tao F., Chatterjee P., Yang G., Han Q., Chung H., et al. Non-full-length water-soluble CXCR4(QTY) and CCR5(QTY) chemokine receptors: implication for overlooked truncated but functional membrane receptors. iScience. 2020;23(12) doi: 10.1016/j.isci.2020.101670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones K.A., Borowsky B., Tamm J.A., Craig D.A., Durkin M.M., Dai M., et al. GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature. 1998;396(6712):674–679. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- 21.White J.H., Wise A., Main M.J., Green A., Fraser N.J., Disney G.H., et al. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature. 1998;396(6712):679–682. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- 22.Wilson S., Wilkinson G., Milligan G. The CXCR1 and CXCR2 receptors form constitutive homo- and heterodimers selectively and with equal apparent affinities. J Biol Chem. 2005;280(31):28663–28674. doi: 10.1074/jbc.M413475200. [DOI] [PubMed] [Google Scholar]
- 23.Milligan G. The role of dimerisation in the cellular trafficking of G-protein-coupled receptors. Curr Opin Pharm. 2010;10(1):23–29. doi: 10.1016/j.coph.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Altwaijry N.A., Baron M., Wright D.W., Coveney P.V., Townsend-Nicholson A. An ensemble-based protocol for the computational prediction of helix-helix interactions in G protein-coupled receptors using coarse-grained molecular dynamics. J Chem Theory Comput. 2017;13(5):2254–2270. doi: 10.1021/acs.jctc.6b01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiedel A.C., Kose M., Barreto C., Bueschbell B., Morra G., Sensoy O., et al. Prediction and targeting of interaction interfaces in G-protein coupled receptor oligomers. Curr Top Med Chem. 2018;18(8):714–746. doi: 10.2174/1568026618666180604082610. [DOI] [PubMed] [Google Scholar]
- 26.Townsend-Nicholson A., Altwaijry N., Potterton A., Morao I., Heifetz A. Computational prediction of GPCR oligomerization. Curr Opin Struct Biol. 2019;55:178–184. doi: 10.1016/j.sbi.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Jiménez-García B., Roel-Touris J., Romero-Durana M., Vidal M., Jiménez-González D., Fernández-Recio J. LightDock: a new multi-scale approach to protein-protein docking. Bioinformatics. 2018;34(1):49–55. doi: 10.1093/bioinformatics/btx555. [DOI] [PubMed] [Google Scholar]
- 28.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., et al. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12(2):255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padhorny D., Kazennov A., Zerbe B.S., Porter K.A., Xia B., Mottarella S.E., et al. Protein-protein docking by fast generalized Fourier transforms on 5D rotational manifolds. Proc Natl Acad Sci U S A. 2016;113(30):E4286–E4293. doi: 10.1073/pnas.1603929113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiménez-García B., Roel-Touris J., Barradas-Bautista D. The LightDock server: artificial intelligence-powered modeling of macromolecular interactions. Nucleic Acids Res. 2023;51(W1) doi: 10.1093/nar/gkad327. W298-w304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richard E., Michael O.N., Alexander P., Natasha A., Andrew S., Tim G., et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv. 2021;2022 doi: 10.1101/2021.10.04.463034. 10.04.463034. [DOI] [Google Scholar]
- 33.Akdel M., Pires D.E.V., Pardo E.P., Jänes J., Zalevsky A.O., Mészáros B., et al. A structural biology community assessment of AlphaFold2 applications. Nat Struct Mol Biol. 2022;29(11):1056–1067. doi: 10.1038/s41594-022-00849-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibrahim T., Khandare V., Mirkin F.G., Tumtas Y., Bubeck D., Bozkurt T.O. AlphaFold2-multimer guided high-accuracy prediction of typical and atypical ATG8-binding motifs. PLoS Biol. 2023;21(2) doi: 10.1371/journal.pbio.3001962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu W., Shenoy A., Kundrotas P., Elofsson A. Evaluation of AlphaFold-Multimer prediction on multi-chain protein complexes. Bioinformatics. 2023;39(7) doi: 10.1093/bioinformatics/btad424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin R., Feng B.Y., Varshney A., Pierce B.G. Benchmarking AlphaFold for protein complex modeling reveals accuracy determinants. Protein Sci. 2022;31(8) doi: 10.1002/pro.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teufel F., Refsgaard J.C., Kasimova M.A., Deibler K., Madsen C.T., Stahlhut C., et al. Deorphanizing peptides using structure prediction. J Chem Inf Model. 2023;63(9):2651–2655. doi: 10.1021/acs.jcim.3c00378. [DOI] [PubMed] [Google Scholar]
- 38.Ghani U., Desta I., Jindal A., Khan O., Jones G., Hashemi N., et al. Improved docking of protein models by a combination of Alphafold2 and ClusPro. bioRxiv. 2021;2022 doi: 10.1101/2021.09.07.459290. 09.07.459290. [DOI] [Google Scholar]
- 39.Mirdita M., Schutze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19(6):679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 41.Käll L., Krogh A., Sonnhammer E.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35(Web Server issue):W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janson G., Paiardini A. PyMod 3: a complete suite for structural bioinformatics in PyMOL. Bioinformatics. 2021;37(10):1471–1472. doi: 10.1093/bioinformatics/btaa849. [DOI] [PubMed] [Google Scholar]
- 43.Wu E.L., Cheng X., Jo S., Rui H., Song K.C., Dávila-Contreras E.M., et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J Comput Chem. 2014;35(27):1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 45.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 46.Jo S., Lim J.B., Klauda J.B., Im W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys J. 2009;97(1):50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valdés-Tresanco M.S., Valdés-Tresanco M.E., Valiente P.A., Moreno E. gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J Chem Theory Comput. 2021;17(10):6281–6291. doi: 10.1021/acs.jctc.1c00645. [DOI] [PubMed] [Google Scholar]
- 49.Lu Q., Luo R. A Poisson–Boltzmann dynamics method with nonperiodic boundary condition. J Chem Phys. 2003;119(21):11035–11047. doi: 10.1063/1.1622376. [DOI] [Google Scholar]
- 50.Glukhova A., Thal D.M., Nguyen A.T., Vecchio E.A., Jörg M., Scammells P.J., et al. Structure of the Adenosine A(1) receptor reveals the basis for subtype selectivity. Cell. 2017;168(5):867–877. doi: 10.1016/j.cell.2017.01.042. e13. [DOI] [PubMed] [Google Scholar]
- 51.Du J., Wang D., Fan H., Xu C., Tai L., Lin S., et al. Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature. 2021;594(7864):589–593. doi: 10.1038/s41586-021-03641-w. [DOI] [PubMed] [Google Scholar]
- 52.Wang C., Wu H., Katritch V., Han G.W., Huang X.P., Liu W., et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497(7449):338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Botta J., Bibic L., Killoran P., McCormick P.J., Howell L.A. Design and development of stapled transmembrane peptides that disrupt the activity of G-protein-coupled receptor oligomers. J Biol Chem. 2019;294(45):16587–16603. doi: 10.1074/jbc.RA119.009160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lazim R., Suh D., Lee J.W., Vu T.N.L., Yoon S., Choi S. Structural characterization of receptor-receptor interactions in the allosteric modulation of G protein-coupled receptor (GPCR) Dimers. Int J Mol Sci. 2021;22(6) doi: 10.3390/ijms22063241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson G.P., Agwuegbo U., Jonas K.C. New insights into the functional impact of G protein–coupled receptor oligomerization. Curr Opin Endocr Metab Res. 2021;16:43–50. doi: 10.1016/j.coemr.2020.08.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data and code have been deposited in the Zenodo database, DOI: 10.5281/zenodo.10275624. Further information and requests for data should be directed to and will be fulfilled by S.Z., shuguang@mit.edu.