Abstract
Resistance to β-lactam antibiotics in Streptococcus pneumoniae is due to alteration of penicillin-binding proteins (PBPs). S. pneumoniae PBP 1a belongs to the class A high-molecular-mass PBPs, which harbor transpeptidase (TP) and glycosyltransferase (GT) activities. The GT active site represents a new potential target for the generation of novel nonpenicillin antibiotics. The 683-amino-acid extracellular region of PBP 1a (PBP 1a*) was expressed in Escherichia coli as a GST fusion protein. The GST-PBP 1a* soluble protein was purified, and its domain organization was revealed by limited proteolysis. A protease-resistant fragment spanning Ser 264 to Arg 653 exhibited a reactivity profile against both β-lactams and substrate analogues similar to that of the parent protein. This protein fragment represents the TP domain. The GT domain (Ser 37 to Lys 263) was expressed as a recombinant GST fusion protein. Protection by moenomycin of the GT domain against trypsin degradation was interpreted as an interaction between the GT domain and the moenomycin.
The synthesis of the bacterial cell wall requires cytoplasmic and periplasmic enzymes. The final steps of peptidoglycan biosynthesis occur outside the cytoplasmic membrane, and they are catalyzed by membrane-bound penicillin-binding proteins (PBPs). PBPs play essential roles in cell division and morphology (6, 20, 31). Based upon their molecular sizes and amino acid sequence similarities, PBPs can be classified into two groups (6): low-molecular-weight (low-Mr) PBPs, which act as d,d-carboxypeptidases, and high-molecular-weight (high-Mr) PBPs, which carry transpeptidase (TP) and glycosyltransferase (GT) activities. The high-Mr group can be further divided into bifunctional enzymes with TP and GT activities (class A) and monofunctional TP enzymes (class B).
β-Lactam antibiotics bind with high affinity specifically to d,d-carboxypeptidase and TP domains because of their structural similarity to the natural substrates, the stem peptides. This binding results in the formation of a covalent acyl-PBP enzyme complex, leading to the inactivation of PBPs.
High-Mr PBPs are multidomain proteins (6). The three-dimensional structure of Streptococcus pneumoniae PBP 2x (class B high-Mr PBP) illustrates this domain organization (25). The only non-penicillin-binding domain of known function is the GT domain, corresponding to the N-terminal region of class A PBPs. This GT activity, clearly identified in Escherichia coli PBP 1b, is difficult to measure (23, 29, 31–35). It is insensitive to penicillin but sensitive to moenomycin, an antibiotic which is not used for human therapy (23, 29, 32, 33).
S. pneumoniae is one of the major human pathogens of the upper respiratory tract, causing pneumonia, meningitis, and ear infections. Six PBPs have been identified in S. pneumoniae: high-Mr PBPs 1a, 1b, 2a, 2x, and 2b and low-Mr PBP 3 (8). PBPs 1a, 1b, and 2a belong to class A, while PBPs 2x and 2b are monofunctional class B proteins. Deletion of pbp2x and pbp2b in S. pneumoniae is lethal for the bacteria, while the deletion of pbp1a is tolerated (11), probably due to compensation by PBP 1b. This has been observed for E. coli class A PBP 1a, whose deletion can be compensated for by PBP 1b (36). In clinical isolates of resistant pneumococci, pbp1a, pbp2x, and pbp2b genes were shown to present a mosaic organization, encoding PBPs with reduced affinity for β-lactam antibiotics (2, 5, 15, 18). The specific resistance to ceftriaxone and cefotaxime of S. pneumoniae from the hospital environment is mediated by modification of PBP 2x and PBP 1a (22). Furthermore, gene transfer of pbp1a, pbp2x, and pbp2b from resistant strains conferred penicillin resistance on sensitive S. pneumoniae strains under laboratory conditions (2–4, 14, 15, 27, 30).
The effort to overcome resistance to antibiotics in S. pneumoniae might therefore benefit from a detailed understanding of the molecular basis of TP and GT activities. The GT domain represents a new potential target for novel nonpenicillin antibiotics. Here, we delineate the GT and TP domains of S. pneumoniae PBP 1a* (a water-soluble form of PBP 1a) by limited proteolytic digestion and expression of recombinant domains. The TP activity of PBP 1a* and that of the isolated TP domain were compared. We also present evidence for an interaction between the isolated GT domain and moenomycin.
MATERIALS AND METHODS
Materials.
The thioester pseudosubstrates S2a (N-benzoyl-glycylmercaptoacetic acid) and S2d (N-benzoyl-d-alanylmercaptoacetic acid) (10) were obtained from L. Christiaens (Université de Liège, Liège, Belgium). 4,4′-Dithiopyridine, trypsin, chymotrypsin, glutathione, penicillin G, and cefotaxime were purchased from Sigma. Moenomycin was a gift from Hoechst (Frankfurt/Main, Germany).
Bacterial strains and culture conditions.
Cloning and protein expression were performed in E. coli MC1061 [F− araD139Δ(ara-leu)7636 galE15 galK16 Δ(lac)X74 rpsL mcrA mcrB1 hsdR2 (rK− mK+)]. Cultures were grown in Luria-Bertani (LB) medium (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl per liter) (Life Technologies) supplemented with 100 μg of ampicillin per ml when necessary.
Construction of the expression systems.
pbp1a is the DNA sequence, produced by PCR amplification, that codes for PBP 1a*, a water-soluble form of PBP 1a lacking the putative transmembrane domain at its N terminus. Genomic preparation of S. pneumoniae R6 DNA was performed as described previously (37). The primers used were 5′ATCGCAGCCATTGTCTTAGGGGATCCCAT ATGATCGAAGGTCGTAGCAAGGCTCCTAGCCTATC (upstream) and 5′GAAAAAATCACCCAGGGATCCCTCGAGGCGGCCGCTTATGGT TGTGCTGGTTG (downstream).
The upstream primer contains an NdeI site (underlined characters), an ATG start codon beginning at Ser 37 (following the putative transmembrane region), and a BamHI site (bold characters). The downstream primer includes the TAA stop codon and NotI (underlined characters) and BamHI (bold characters) sites to facilitate the cloning. The amplification of a 2.1-kb DNA fragment was performed with the Tth polymerase according to the manufacturer’s instructions (Clontech, Palo Alto, Calif.). The PCR product was washed in a microconcentrator, digested with BamHI, and ligated into pGEX-2T (Pharmacia, Piscataway, N.J.) with T4 DNA ligase from GIBCO BRL (Grand Island, N.Y.). The ligation reaction product was transformed into competent DH5α cells, and identification of plasmid DNA carrying the insert in the desired orientation within the vector resulted in pJAH143. DNA sequencing was used to verify the sequence (19).
The GT domain was expressed independently by site-directed mutagenesis (12) following replacement of the Ser 264 codon in pJAH143 with a TAA stop codon (bold characters) by using the mutagenic primer 5′-AGTCTCAAATAATCGATTAATTAC-3′. The mutation was checked by restriction analysis (ClaI site, underlined) and DNA sequencing.
Protein purification.
E. coli MC1061(pJAH143) was first grown overnight with vigorous shaking at 37°C in LB medium supplemented with 100 μg of ampicillin per ml. The preculture (20 ml) was diluted into 1 liter of fresh LB medium containing 100 μg of ampicillin per ml and grown at 37°C until the optical density at 600 nm reached 1. The culture was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) overnight at 22°C. Cells were harvested by centrifugation at 11,300 × g for 15 min and resuspended in 40 ml of a solution of 50 mM Tris-HCl (pH 8.0)–200 mM KCl containing one tablet of Complete, a protease inhibitor cocktail (Boehringer Mannheim). The resulting suspension was disrupted by a 2-min sonication step. The lysate was centrifuged at 31,000 × g for 20 min at 4°C. The supernatant was loaded onto a 3.5-ml glutathione-Sepharose column (Pharmacia). The column was equilibrated and washed with 50 mM Tris-HCl (pH 8.0)–200 mM KCl. The fusion protein was either eluted with 10 mM glutathione in 50 mM Tris-HCl (pH 8.0)–200 mM KCl or cleaved while bound to the column in the presence of 50 U of thrombin (Sigma) for 1 h at room temperature. The thrombin activity was then inhibited by 1 mM phenylmethylsulfonyl fluoride. Both enzyme preparations were adjusted to 20% glycerol (vol/vol) and stored at −20°C. All the purification steps were performed at 4°C except when specified.
The TP domain was prepared by trypsin digestion of PBP 1a* (the trypsin/PBP 1a* ratio was 1:20 [wt/wt]) for 30 min at 37°C. A solution of about 5 to 10 mg, resulting from PBP 1a* digestion, was applied to a gel filtration column (Superdex 75, preparation-grade HiLoad 26/60; Pharmacia) at a flow rate of 40 ml/h in order to eliminate the small peptides generated in the proteolysis step.
Production and purification of glutathione S-transferase (GST)–GT followed the same protocol as for GST-PBP 1a*, except that the culture was incubated at 37°C for 3 h during the induction step.
Proteolytic digestion and protein sequencing.
GST-PBP 1a* (about 0.5 mg/ml) in 50 mM Tris-HCl (pH 8.0)–200 mM KCl was incubated at 37°C with either trypsin or chymotrypsin (protease/fusion protein ratio of 1:2 to 1:8, wt/wt). Aliquots were withdrawn at various times, and the reaction was stopped by the addition of sodium dodecyl sulfate (SDS) loading buffer. The samples were boiled for 5 min at 100°C and then loaded onto an SDS–12.5% polyacrylamide gel for electrophoresis. The proteins were either stained with a Coomassie blue solution or transferred to a Problot membrane (Applied Biosystems) and stained according to the supplier’s instructions. The protein bands were cut from the membrane and sequenced by automated Edman degradation on an Applied Biosystems gas-phase sequencer, model 477A, with on-line analysis of the phenylthiohydantion derivatives.
Limited proteolysis of GST-GT and PBP 2x (about 0.3 and 1 mg/ml, respectively) was performed by using a trypsin/GST-GT fusion protein ratio from 1:1,500 to 1:15 (wt/wt) and trypsin/PBP 2x protein ratios of 1:100 and 1:10 (wt/wt) in the absence and presence of 5 mM moenomycin. The incubation time was 10 min, and the reaction was performed at 37°C.
Determination of kinetic parameters.
High-Mr PBPs interact with β-lactam antibiotics according to the following three-step scheme: k1 k2 k3 E + I ⇄ EI → EI* → E + P k−1
where K is equal to k−1/k1, E is the PBP enzyme, I is the β-lactam antibiotic, EI is the Michaelis-Menten complex, EI* is the acyl-enzyme covalent complex, and P is the product of the reaction (degraded β-lactam antibiotic).
The k2/K parameter, accounting for the efficiency of the acylation step, was determined by monitoring the decrease of the intrinsic fluorescence of the protein in the presence of antibiotic at 37°C by using spectrofluorometric measurements coupled with a Biologic SFM3 stopped-flow apparatus. The excitation wavelength for tryptophan was 280 nm, and the emission was measured in the 305- to 360-nm range. PBP 1a* and derivatives at a concentration of 0.6 μM in 10 mM sodium phosphate (pH 7.0)–200 mM KCl were incubated with concentrations of β-lactam antibiotic ranging from 200 μM to 1 mM.
Transpeptidase activity of PBP, i.e., hydrolysis activity, was measured by using S2d, a synthetic thioester analog of cell wall stem peptide, according to the method described by Zhao et al. (37). Assays were performed at 37°C. Protein concentrations were determined with the bicinchoninic acid detection kit (Pierce) with bovine serum albumin as a standard.
The titration of functional PBP and the determination of k3 were performed with [3H]benzylpenicillin (20 Ci/mmol, 1 mCi/ml; Amersham). Purified PBP (2 μM) in 50 mM Tris-HCl (pH 8.0)–200 mM KCl was incubated at 37°C with different concentrations of [3H]benzylpenicillin for 15 min, at which time the reaction was completed. The samples were then subjected to SDS-polyacrylamide gel electrophoresis (PAGE). [3H]benzylpenicillin bound to proteins was monitored by two different procedures. The gel was stained with Coomassie blue, destained, incubated with Amplify (Amersham), dried, and either exposed to a film for 16 h or cut around the protein bands. In the latter case, the radioactivity of the gel slice was measured with a liquid scintillation analyzer (Packard model 2100TR) after mixing with 10 ml of liquid scintillation counting cocktail (Picofluor 15; Packard).
The deacylation reaction obeys the following equation: −k3t = ln[EI*]t/[EI*]0, where [EI*]0 is the initial concentration of acyl-enzyme and [EI*]t is its concentration at time t. Determination of k3 was performed as follows: 2 μM purified PBP was labelled with 1 μM [3H]benzylpenicillin during 15 min at 37°C. Cold benzylpenicillin (15 mM) was added, and the incubation was continued at 37°C. Samples were removed at various times and loaded onto an SDS-polyacrylamide gel, and the amount of radioactivity was measured in the protein bands as mentioned above.
RESULTS
Molecular organization of class A PBPs and MGT.
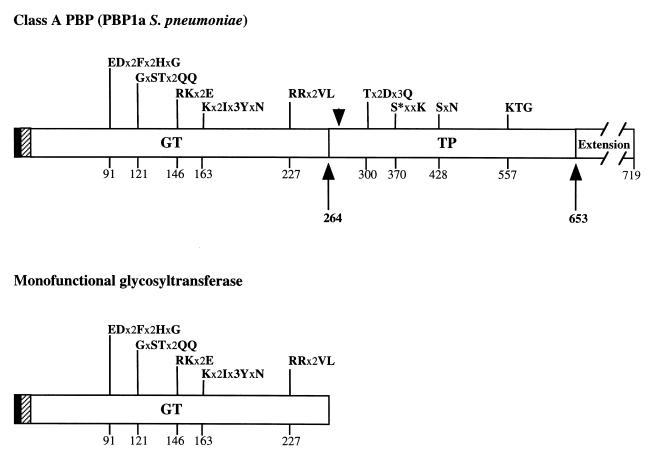
Monofunctional glycosyltransferases (MGTs) are open reading frames possessing a high degree of similarity with the N-terminal region of class A PBPs, i.e., the GT domain. They were found in E. coli (1, 9, 28), Haemophilus influenzae, Klebsiella pneumoniae, Neisseria gonorrhoeae (28), and Ralstonia eutropha (24). MGTs from E. coli and R. eutropha were expressed, with or without their transmembrane anchors (1, 24). The R. eutropha MGT is expressed in the bacteria under normal growth conditions, and a peptidoglycan-synthesizing activity has been found in E. coli MGT (1, 9). These results indicate that MGT proteins are functional. Figure 1 underlines the sequence similarity between the S. pneumoniae PBP 1a GT domain and the MGT protein.
FIG. 1.
Comparison of the molecular organization of class A PBP and MGT. The filled and hatched boxes represent the N-terminal cytoplasmic region and the membrane anchor, respectively. The C-terminal Ser- and Asn-rich extension is also depicted in PBP 1a. GT and TP domains are represented together with their conserved motifs; active serine 370 is indicated by an asterisk. x represents any amino acid. The amino acid numbering is that of S. pneumoniae PBP 1a (19). The arrows delineate the proteolytic PBP 1a* TP domain. The arrowhead refers to the permissive site for insertion mutations in E. coli PBP 1b as described by Lefèvre et al. (16).
Purification of soluble GST-PBP 1a* and PBP 1a*.
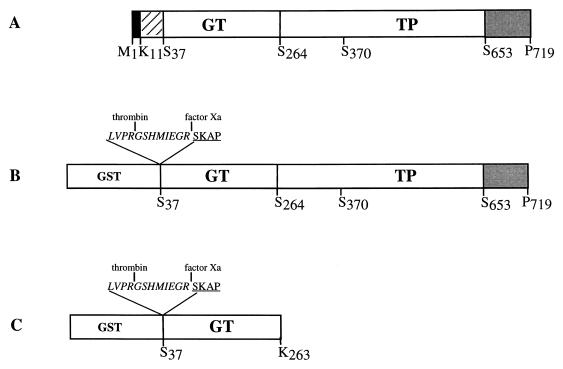
PBP 1a is a 719-amino-acid protein which includes an N-terminal transmembrane region and a short cytoplasmic portion (Fig. 1 and 2A). To study the domain organization and the enzymatic characteristics of S. pneumoniae PBP 1a, we expressed the periplasmic region of PBP 1a fused to the GST moiety (Fig. 2A). Cleavage sites specific for thrombin and factor Xa have been added between GST and the N terminus of PBP 1a* (Fig. 2B).
FIG. 2.
Schematic diagrams of the PBP 1a-derived constructs. (A) Organization of the native PBP 1a protein. Filled, hatched, and shaded boxes indicate the N-terminal cytoplasmic region, the membrane anchor, and the Ser- and Asn-rich C-terminal extension, respectively. (B) Construction of the GST-PBP 1a* fusion protein. (C) Construction of the GST-GT fusion protein. The active-site serine 370 is indicated. Peptide at the GST-GT junctions includes sequences specific for thrombin and factor Xa (italic characters) and N-terminal amino acids of GT (underlined characters).
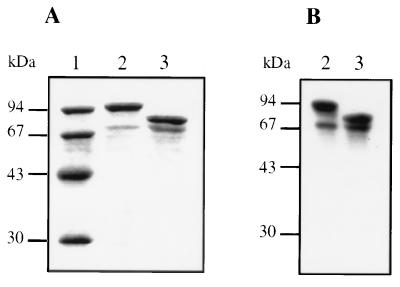
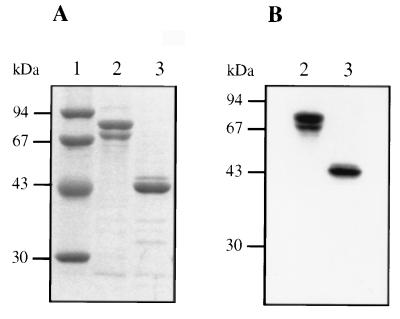
The fusion protein was purified by affinity chromatography. This single-step procedure results in a major species with an apparent molecular mass of about 100 kDa (Fig. 3A, lane 2). This fusion protein is functional since it can be labelled with [3H]benzylpenicillin (Fig. 3B, lane 2). PBP 1a* was released from the fusion protein by cleavage with thrombin. The apparent molecular mass of PBP 1a* is 75 kDa, in good agreement with the value calculated from its amino acid sequence (76.1 kDa) (Fig. 3A, lane 3). N-terminal sequencing confirms the expected sequence, Gly-Ser-His-Met (Fig. 2B). Specific labelling with [3H]benzylpenicillin shows that purified PBP 1a* is functional (Fig. 3B, lane 3). A species of about 70 kDa is present in GST-PBP 1a* and PBP 1a* preparations (Fig. 3A, lanes 2 and 3). This species, which represents 25 to 30% of the PBP 1a* quantity (Fig. 3A, lane 3), is competent for [3H]benzylpenicillin binding (Fig. 3B, lanes 2 and 3). N-terminal sequencing of this species revealed a secondary thrombin cleavage upstream of Asn 113. Further purification steps, including anion exchange and gel filtration, were not successful in removing this minor contaminant. The purification yields for the fusion protein and PBP 1a* are approximately 8 and 10 mg/liter, respectively.
FIG. 3.
Analysis of purified GST-PBP 1a* and PBP 1a*. (A) Proteins were separated by SDS–12.5% PAGE and stained with Coomassie blue. (B) Autoradiogram of a dried SDS–12.5% PAGE of proteins (2 μg) labelled with [3H]benzylpenicillin prior to the migration. Numbers at the left indicate sizes of standard molecular mass markers. Lanes: 1, molecular mass markers; 2, GST-PBP 1a*; 3, PBP 1a*.
Limited proteolysis of GST-PBP 1a*.
A series of proteases with different specificities were used in an attempt to identify structural and functional domains of PBP 1a*. We used GST-PBP 1a* since it had no previous contact with a protease, unlike PBP 1a* preparations. GST-PBP 1a* was incubated for various times in the presence of different amounts of trypsin or chymotrypsin (Fig. 4). Proteolytic products were analyzed by SDS-PAGE.
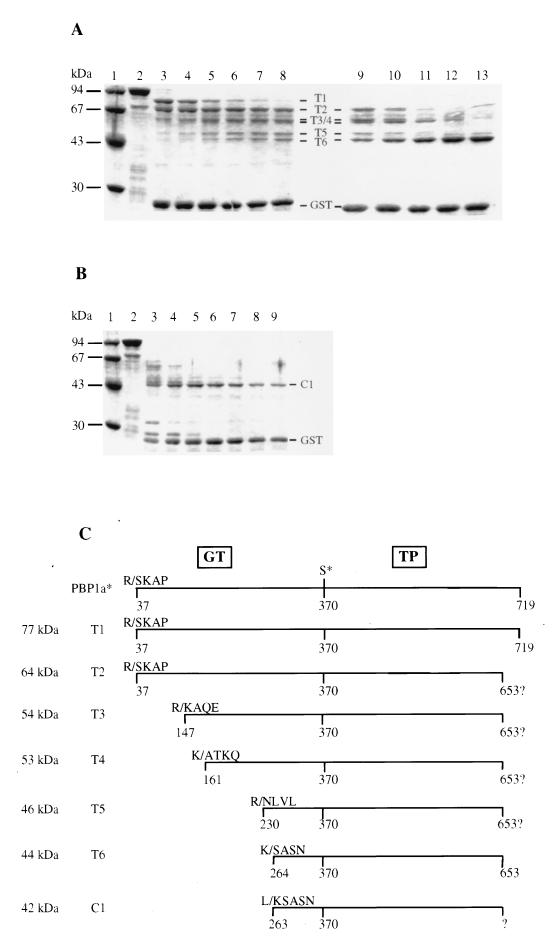
FIG. 4.
Characterization of peptides derived from limited proteolysis of GST-PBP 1a*. (A) Trypsin digestion. Lanes: 1, standard molecular mass markers; 2 to 8, incubation times of 15, 30, 60, 90, 120, and 150 min, respectively, at a trypsin/GST-PBP 1a* ratio of 1:8 (wt/wt); 9 to 13, incubation times of 30, 60, 120, 180, and 240 min, respectively, at a trypsin/GST-PBP 1a* ratio of 1:4 (wt/wt). (B) Chymotrypsin digestion. Lanes: 1, standard molecular mass markers; 2 to 9, incubation time of 5, 15, 30, 60, 90, 120, and 150 min, respectively, at a chymotrypsin/GST-PBP 1a* ratio of 1:8 (wt/wt). Numbers at the left indicate sizes of standard molecular mass markers. Protein fragments and GST are identified. (C) Schematic representation of the proteolytic fragments. The molecular mass of each fragment was measured by SDS-PAGE. The N-terminal sequence of the fragments was experimentally determined. The putative C termini of T2, T3, T4, and T5 are shown. The C terminus of T6 is derived from mass spectrometry measurements. The positions of the active-site serine 370 and the GT and TP domains are indicated. K/ and R/, trypsin digestion sites; L/, chymotrypsin site (the number corresponds to the N-terminal residue freed after digestion).
Incubation of GST-PBP 1a* with trypsin at a protease/fusion protein ratio of 1:8 (wt/wt) generates a pattern of protein bands designated T1 (77 kDa), T2 (64 kDa), T3 (53 kDa), T4 (53 kDa), T5 (46 kDa), and T6 (44 kDa), according to their decreasing apparent molecular mass (Fig. 4A). The 26-kDa GST moiety is released early. Fifteen minutes after the beginning of the reaction, two major species, T1 and T2, are produced concomitantly (Fig. 4A, lane 3). T1 disappears rapidly; T2, however, is still present after incubation with trypsin for 150 min (Fig. 4A, lane 8). As T2 intensity progressively decreases, smaller fragments (T3, T4, T5, and T6) appear, suggesting that these proteins are produced by further digestion of T2 (Fig. 4A, lane 8). A twofold increase in the relative concentration of trypsin (1:4) along with a prolonged incubation generates T6 as the dominant species (Fig. 4A, lane 13), indicating its extreme resistance to trypsin cleavage.
A similar experiment with chymotrypsin provides a very different pattern (Fig. 4B): the GST portion is released 5 min after the beginning of the incubation, and most of the fusion protein is converted into a single major 42-kDa band (C1) (Fig. 4B, lane 9).
Peptide mapping of GST-PBP 1a* proteolytic fragments.
We used N-terminal sequencing and molecular mass determination, along with protease cleavage specificity, to identify the proteolytic products. The peptide mapping results are shown in Fig. 4C. The most accessible trypsin cleavage sites lead to the liberation of the GST, T1, and T2. The latter two protein fragments start at Ser 37, which is the first amino acid of recombinant soluble PBP 1a*. Given the lower molecular mass of T2 than of T1, and the potential trypsin cleavage sites, we deduce the presence of an exposed trypsin site at the C terminus of the molecule. T2 cannot extend beyond position Arg 653, since that residue is the most C-terminal putative trypsin site. T1 is likely to correspond to the complete PBP 1a*, since both proteins comigrate on SDS-PAGE. N-terminal amino acid sequencing of T3, T4, T5, and T6 revealed that these fragments begin at Lys 147, Ala 161, Asn 230, and Ser 264, respectively. The differences in molecular mass of the various protein fragments are consistent with the calculated N-terminal deletions (Fig. 4C). They indicate that protein fragments T3 to T6 have not been further cleaved at their C termini and that they are likely to extend up to position Arg 653.
The N-terminal sequence of the chymotrypsin C1 product reveals that this fragment arises from a C-terminal cleavage to Leu 262 (Fig. 4C). C1 and T6 have neighboring N termini and very similar molecular masses, indicating that they have similar C termini.
Purification and characterization of the T6 (TP) tryptic fragment of PBP 1a*.
Extensive proteolysis of purified PBP 1a* with trypsin resulted in the production of T6 (Fig. 5A). Small peptide fragments were removed by gel filtration. The T6 product eluted as a single peak with an apparent molecular mass of approximately 50 kDa, providing clear evidence of the monomeric and soluble properties of the T6 fragment. This peak includes the unique T6 species of 44 kDa, as shown by SDS-PAGE analysis (Fig. 5A, lane 3). This fragment is able to bind [3H]benzylpenicillin (Fig. 5B, lane 3), as is intact PBP 1a* (Fig. 5B, lane 2). The removal of the N-terminal portion of PBP 1a* does not affect the binding of β-lactam antibiotics significantly. In fact, we used the quantitative binding of [3H]benzylpenicillin to PBP 1a* and T6 to perform active-site titration. Protein concentrations measured by the bicinchoninic acid method are identical to those determined by active-site titration (data not shown). This shows that both protein preparations are fully functional.
FIG. 5.
Analysis of purified PBP 1a* and TP. (A) Proteins were separated by SDS–12.5% PAGE and stained with Coomassie blue. (B) Autoradiogram of a dried SDS–12.5% PAGE of proteins (2 μg) labelled with [3H]benzylpenicillin prior to the migration. Numbers at the left indicate sizes of standard molecular mass markers. Lanes: 1, standard molecular mass markers; 2, PBP 1a*; 3, TP.
The molecular mass of T6, obtained by electrospray ionization-mass spectrometry, is 43,574 ± 1.6 Da. Since T6 starts at Ser 264, the measured molecular mass confirms Arg 653 as its C-terminal amino acid (calculated mass of 43,572.3 Da). Thus, the T6 portion of recombinant PBP 1a* did not undergo posttranslational modification. T6 is the most stable proteolytic fragment of PBP 1a* able to bind benzylpenicillin and is referred to herein as the TP domain.
Functional studies of GST-PBP 1a*, PBP 1a*, and TP.
Kinetic properties of purified GST-PBP 1a*, PBP 1a*, and TP were determined by using the cephalosporin and penicillin classes of antibiotics and two substrate analogues (Table 1). The second-order rate constants of acylation (k2/K) of PBP 1a* and TP by both β-lactams are comparable with differences for the average values of about 30%. The k2/K of GST-PBP 1a* is about 30% higher than that of TP for both antibiotics. The rate of acylation by benzylpenicillin is approximately two- to threefold that of cefotaxime. kcat/Km values for hydrolysis by GST-PBP 1a*, PBP 1a*, and TP are in the same range of values for both the S2d and S2a thioester pseudosubstrates (Table 1). The efficiency of hydrolysis of S2d is about 10 times that of S2a. The most significant variations are between GST-PBP 1a* and PBP 1a* (more than threefold) for both S2d and S2a. The k3 parameter for benzylpenicillin for the three proteins is comparable (Table 1) and corresponds to a half-life ranging from 14 h (GST-PBP 1a*) to 18 h (PBP 1a*). Taken together, these results show that isolation of the TP domain from PBP 1a* has not modified its enzymatic characteristics significantly.
TABLE 1.
Kinetic parameters of GST-PBP 1a*, PBP 1a*, and TP by using antibiotics and two substrate analoguesa
| Protein | Cefotaxime k2/K (M−1 s−1) | Benzylpenicillin
|
Substrate analogue kcat/Km (M−1 s−1)
|
||
|---|---|---|---|---|---|
| k2/K (M−1 s−1) | k3 (s−1) | S2d | S2a | ||
| GST-PBP 1a* | 25,840 ± 2,008 | 70,054 ± 3,923 | 1.3 × 10−5 ± 1 × 10−6 | 415 ± 39 | 32.8 ± 4 |
| PBP 1a* | 11,933 ± 2,309 | 34,200 ± 1,555 | 1.0 × 10−5 ± 1 × 10−6 | 128 ± 12 | 5.7 ± 0.4 |
| TP | 18,110 ± 1,173 | 47,700 ± 424 | 1.2 × 10−5 ± 1 × 10−6 | 195 ± 27 | 9.1 ± 1.6 |
Values are means ± standard deviations.
Production, purification, and characterization of GST-GT.
The identification of the boundaries of the functional TP domain within PBP 1a provided an opportunity to propose a location for the GT domain. We expressed GST fused to the Ser 37-to-Lys 263 region of PBP 1a (Fig. 2C). GST-GT recombinant protein was purified by affinity chromatography with a yield of 1 to 2 mg/liter of E. coli culture. N-terminal sequencing of the GT moiety following cleavage by thrombin confirmed the expected sequence (Fig. 2C).
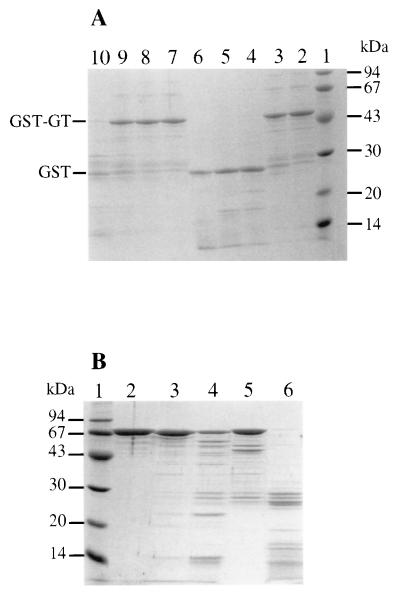
The antibiotic moenomycin has been proposed to interact with the GT domain of class A PBPs, presumably in a competitive manner (12, 34). In this study, we used moenomycin as a functional probe for GT. Preliminary experiments have shown that the migration on native gels of GST-PBP 1a* is modified in a moenomycin concentration-dependent manner (data not shown). This indicates that moenomycin interacts with PBP 1a* and might therefore protect the GT domain of PBP 1a* from trypsin cleavage. The trypsin digestion pattern of GST-GT is clearly affected by the presence of moenomycin (Fig. 6A). A 1:1,500 (wt/wt) trypsin/GST-GT ratio has little effect on GST-GT (Fig. 6A, lane 3). With a ≥10-fold relative increase of trypsin, the GT domain is fully degraded and GST is released (Fig. 6A, lanes 4 to 6). A completely different result was obtained when the proteolysis was carried out in the presence of an excess of moenomycin. The antibiotic completely prevents GST-GT degradation by trypsin up to a protease/fusion protein ratio of 1:75 (Fig. 6A, lanes 7 to 9). This is not due to an inhibition of the protease by moenomycin, since a further increase in relative concentration of trypsin (1:15) leads to a full degradation of the fusion protein (Fig. 6A, lane 10). This is further supported by the fact that class B S. pneumoniae PBP 2x is degraded by trypsin in the presence of moenomycin (Fig. 6B). The latter experiment also demonstrates the specificity of the effect of moenomycin on PBP 1a.
FIG. 6.
(A) Protection of GST-GT from trypsin hydrolysis by moenomycin. Lanes: 1, standard molecular mass markers; 2, no trypsin; 3 and 7, trypsin/GST-GT ratio of 1:1,500 (wt/wt); 4 and 8, trypsin/GST-GT ratio of 1:150 (wt/wt); 5 and 9, trypsin/GST-GT ratio of 1:75 (wt/wt); 6 and 10, trypsin/GST-GT ratio of 1:15 (wt/wt). Lanes 7 to 10, hydrolysis in the presence of 5 mM moenomycin. Numbers at the left indicate sizes of standard molecular mass markers. (B) Trypsin digestion of PBP 2x. Lanes: 1, molecular mass standards; 2, no trypsin; 3 and 5, trypsin/PBP 2x ratio of 1:100 (wt/wt) 4 and 6, trypsin/PBP 2x ratio of 1:10 (wt/wt). Lanes 5 and 6, hydrolysis in the presence of 5 mM moenomycin.
DISCUSSION
Class A PBPs are essential for cell survival. Disruption of both E. coli PBP 1a and PBP 1b is lethal for the cell, while the disruption of either gene is compensated for by the other (36). Similar behavior of class A PBPs has also been reported for S. pneumoniae (11). S. pneumoniae resistance to β-lactams originates from the selection of PBPs (1a, 2x, and 2b) with mosaic structures, which have a low affinity for the antibiotic (2, 5, 15, 18). Given the clinical relevance of class A PBPs, the GT activity, which is specific to this class, represents a novel nonpenicillin drug target.
Class A PBPs are poorly characterized from a structural and functional point of view. Recent sequencing of complete bacterial genomes provides close to 20 sequences of class A PBPs. Some of them have been overexpressed in E. coli: PBP 1 from Mycobacterium leprae (17) and PBP 1 from N. gonorrhoeae and Neisseria meningitidis (26). Kinetic parameters of their transmembrane forms were determined either from total membrane extracts or from detergent-solubilized purified proteins. MGTs were found in E. coli (9, 28), H. influenzae, K. pneumoniae, N. gonorrhoeae (28), and R. eutropha (24). MGTs from E. coli and from R. eutropha were expressed, with or without their transmembrane anchors, in E. coli (1, 24).
Sequence comparison revealed that the GT domain of S. pneumoniae PBP 1a is homologous to the corresponding domains of E. coli PBPs 1a and 1b (19). However, the TP sequence of S. pneumoniae PBP 1a has a low sequence similarity with TP domains of class B PBPs, low-Mr PBPs, or class A β-lactamases, though it does contain the same catalytic motifs: SxxK, SxN, and KTG (19). Based on this observation, the tridimensional structure of the PBP 1a* TP domain is likely to have the characteristic fold of penicilloyl serine transferases (6, 25).
Here, we report the production of a soluble class A PBP, PBP 1a* from S. pneumoniae deprived of its cytoplasmic and transmembrane regions. This is the first successful attempt to solubilize a functional high-Mr class A PBP. We were also interested in studying the multidomain organization of PBP 1a*. Therefore, we undertook the production, purification, and characterization of soluble GT and TP domains from S. pneumoniae PBP 1a* which were defined by limited proteolysis experiments.
The proteolytic experiments showed that the GT domain is more sensitive to trypsin and chymotrypsin than the TP domain. The resistance of the TP domain indicates a compact structure. When subjected to trypsin digestion under similar conditions, PBP 2x from S. pneumoniae is totally degraded (data not shown). Since trypsin and chymotrypsin proteolysis of PBP 1a* generates TP domains whose N-terminal residues are Ser 264 and Lys 263, respectively, this zone site is likely to belong to an exposed hinge region located between the GT and TP domains. Lefèvre et al. (16) identified a permissive insertion site in E. coli PBP 1b (Gly 427 to Lys 431) which is within 15 amino acid positions downstream from the equivalent location in PBP 1a (Fig. 1). Another indication that this hinge region may link the two functional domains is that the recombinant PBP 1a* GT domain (residues 37 to 263) has a size similar to that of MGT (Fig. 1). In conclusion, PBP 1a* cleavage at the 264 position leads to the release of the TP functional domain and of the MGT equivalent GT domain.
Kinetic parameters of TP activities for GST-PBP 1a*, PBP 1a*, and TP were very similar. GST-PBP 1a* values were consistently higher than for the other two proteins; this might be the consequence of GST dimerization (data not shown). The substrate kcat/Km values and the k2/K acylation rates for antibiotics of PBP 1a* and the isolated TP domain were similar. This implies that the GT domain and the highly hydrophilic Ser- and Asn-rich C-terminal extension do not influence the TP activity significantly (Fig. 1 and 2). Thus, the C-terminal region might constitute an independent structural domain. Efforts to express functional TP domain individually in E. coli have failed (data not shown). This situation is reminiscent of the failure to express the functional TP domain of class B PBPs (E. coli PBP 3 and Enterococcus hirae PBP 5) following extensive modification of their N-terminal regions (7, 21). A large discrepancy exists between S. pneumoniae PBP 1a* TP kinetic parameters and those of M. leprae class A TP (17). Such differences were also noticeable when S. pneumoniae PBP 1a* (Table 1) and PBP 2x* (10) were compared. For instance, the PBP 2x* k2/k value for cefotaxime is about 25-fold higher than that of PBP 1a*, and the S2d kcat/Km value is around 10 times higher.
The PBP 1a* N-terminal region was expressed as a soluble GST-GT fusion protein. Measurement of the glycosyltransferase activity with E. coli lipid II as a substrate is difficult (32–35). Indeed, to our knowledge, this assay has been successful only when used with membrane-bound PBPs of E. coli origin (24). Moenomycin (flavomycin) inhibits peptidoglycan elongation (23), a property which may be explained by its analogy to the structure of lipid II. Moenomycin was efficient in protecting GST-GT (Fig. 6A) or PBP 1a* from trypsin proteolysis (data not shown) but not PBP 2x* (Fig. 6B). The specificity of this effect was confirmed by the fact that the migration of PBP 1a* in native gels was modified in the presence of moenomycin while that of PBP 2x* and GST was not modified (data not shown). Moenomycin is a phospholipid with detergent properties and is known to form high-molecular-mass aggregates (13). It is unlikely that these micelles are responsible for the migration shift of PBP 1a*, since other detergents, including dodecyl maltoside, octyl glucoside, Triton X-100, and CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-3-propanesulfa-nate}, used at concentrations above their critical micelle concentration did not cause a similar effect. The substrate-specific trypsin protection or migration shift is indicative of an interaction between moenomycin and the GT domain, triggering a conformation change of the GT domain.
The availability of large quantities of purified soluble PBP 1a* and the possibility of obtaining isolated GT and TP domains allow us to undertake attempts to determine the tridimensional structure of this class A PBP.
ACKNOWLEDGMENTS
This work was supported by a grant from Eli Lilly.
We thank Y. Pétillot for mass spectrometry analysis.
Footnotes
Publication 536 of the Institut de Biologie Structurale Jean-Pierre Ebel.
REFERENCES
- 1.Di Berardino M, Dijkstra A, Stüber D, Keck W, Gubler M. The monofunctional glycosyltransferase of Escherichia coli is a member of a new class of peptidoglycan-synthesising enzymes. Overexpression and determination of the glycan-polymerisation activity. FEBS Lett. 1996;392:184–188. doi: 10.1016/0014-5793(96)00809-5. [DOI] [PubMed] [Google Scholar]
- 2.Dowson C G, Hutchison A, Brannigan J A, George R C, Hansman D, Linares J, Tomasz A, Maynard Smith J, Spratt B G. Horizontal transfer of penicillin-binding protein genes in penicillin resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowson C G, Hutchison A, Woodford N, Johnson A P, George R C, Spratt B G. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1990;87:5858–5862. doi: 10.1073/pnas.87.15.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowson C G, Coffey T J, Spratt B G. Origin and molecular epidemiology of penicillin-binding protein-mediated resistance to β-lactam antibiotics. Trends Microbiol. 1994;2:361–366. doi: 10.1016/0966-842x(94)90612-2. [DOI] [PubMed] [Google Scholar]
- 5.Georgopapadakou N H. Penicillin-binding proteins and bacterial resistance to β-lactams. Antimicrob Agents Chemother. 1993;37:2045–2053. doi: 10.1128/aac.37.10.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghuysen J-M. Molecular structures of penicillin-binding proteins and β-lactamases. Trends Microbiol. 1994;10:372–380. doi: 10.1016/0966-842x(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 7.Goffin C, Fraipont C, Ayala J, Terrak M, Nguyen-Distèche M, Ghuysen J-M. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J Bacteriol. 1996;178:5402–5409. doi: 10.1128/jb.178.18.5402-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakenbeck R, Briese T, Ellerbrok H, Laible G, Martin C, Metelmann C, Schier H-M, Tornette S. Targets of β-lactams in Streptococcus pneumoniae. In: Actor P, Daneo-Moore L, Higgins M L, Salton M R J, Shockman G D, editors. Antibiotic inhibition of bacterial cell surface assembly and function. Washington, D.C: American Society for Microbiology; 1988. pp. 390–399. [Google Scholar]
- 9.Hara H, Suzuki H. A novel glycan polymerase that synthesizes uncross-linked peptidoglycan in Escherichia coli. FEBS Lett. 1984;168:155–160. doi: 10.1016/0014-5793(84)80226-4. [DOI] [PubMed] [Google Scholar]
- 10.Jamin M, Damblon C, Millier S, Hakenbeck R, Frère J-M. Penicillin-binding protein 2x of Streptococcus pneumoniae: enzymatic activities and interactions with β-lactams. Biochem J. 1993;292:735–741. doi: 10.1042/bj2920735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kell C M, Sharma U K, Dowson C G, Town C, Balganesh T S, Spratt B G. Deletion analysis of the essentiality of penicillin-binding proteins 1a, 2b and 2x of Streptococcus pneumoniae. FEMS Microbiol Lett. 1993;106:171–176. doi: 10.1111/j.1574-6968.1993.tb05954.x. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurtz M, Guba W, Vertezy L. Three-dimensional structure of moenomycin A. A potent inhibitor of penicillin-binding protein 1b. Eur J Biochem. 1998;252:500–507. doi: 10.1046/j.1432-1327.1998.2520500.x. [DOI] [PubMed] [Google Scholar]
- 14.Laible G, Hakenbeck R, Sicard M A, Joris B, Ghuysen J-M. Nucleotide sequences of the pbpx genes encoding the penicillin-binding proteins 2x from Streptococcus pneumoniae R6 and a cefotaxime-resistant mutant, C506. Mol Microbiol. 1989;3:1337–1348. doi: 10.1111/j.1365-2958.1989.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 15.Laible G, Spratt B G, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP2x in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 16.Lefèvre F, Rémy M-H, Masson J-M. Topographical and functional investigation of Escherichia coli penicillin-binding protein 1b by alanine stretch scanning mutagenesis. J Bacteriol. 1997;179:4761–4767. doi: 10.1128/jb.179.15.4761-4767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lepage S, Dubois P, Ghosh T K, Joris B, Mahapatra S, Kundu M, Basu J, Chakrabarti P, Cole S T, Nguyen-Distèche M, Ghuysen J-M. Dual multimodular class A penicillin-binding proteins in Mycobacterium leprae. J Bacteriol. 1997;179:4627–4630. doi: 10.1128/jb.179.14.4627-4630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin C, Sibold C, Hakenbeck R. Relatedness of penicillin-binding protein 1a genes from different clones of penicillin-resistant Streptococcus pneumoniae isolated in South Africa and Spain. EMBO J. 1992;11:3831–3836. doi: 10.1002/j.1460-2075.1992.tb05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin C, Briese T, Hakenbeck R. Nucleotide sequences of genes encoding penicillin-binding proteins from Streptococcus pneumoniae and Streptococcus oralis with high homology to Escherichia coli penicillin-binding proteins 1a and 1b. J Bacteriol. 1992;174:4517–4523. doi: 10.1128/jb.174.13.4517-4523.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuhashi M. Utilization of the lipid-linked precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanism of their regulation. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 55–72. [Google Scholar]
- 21.Moolerach M E, Partoune P, Coyette J, Ghuysen J-M. Importance of the E46-D160 polypeptide segment of the non-penicillin binding module for the folding of the low affinity, multimodular class B penicillin-binding protein 5 of Enterococcus hirae. J Bacteriol. 1996;178:1774–1775. doi: 10.1128/jb.178.6.1774-1775.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munoz R, Dowson C G, Daniels M, Coffey T J, Martin C, Hakenbeck R, Spratt B G. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1992;6:2461–2465. doi: 10.1111/j.1365-2958.1992.tb01422.x. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa J, Tamaki S, Tomioka S, Matsuhashi M. Functional biosynthesis of cell wall peptidoglycan by polymorphic bifunctional polypeptides. J Biol Chem. 1984;259:13937–13946. [PubMed] [Google Scholar]
- 24.Paik J, Jendrossek D, Hakenbeck R. A putative monofunctional glycosyltransferase is expressed in Ralstonia eutropha. J Bacteriol. 1997;179:4061–4065. doi: 10.1128/jb.179.12.4061-4065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pares S, Mouz N, Pétillot Y, Hakenbeck R, Dideberg O. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat Struct Biol. 1996;3:284–289. doi: 10.1038/nsb0396-284. [DOI] [PubMed] [Google Scholar]
- 26.Ropp P A, Nicholas R A. Cloning and characterization of the ponA gene encoding penicillin-binding protein 1 from Neisseria gonorrhoeae and Neisseria meningitidis. J Bacteriol. 1997;179:2783–2787. doi: 10.1128/jb.179.8.2783-2787.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spratt B G. Resistance to β-lactam antibiotics. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 517–534. [Google Scholar]
- 28.Spratt B G, Zhou J, Taylor M, Merrick M J. Monofunctional biosynthetic peptidoglycan transglycosylases. Mol Microbiol. 1996;3:639–647. doi: 10.1046/j.1365-2958.1996.442924.x. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki H, van Heijenoort Y, Tamura T, Mizoguchi J, Hirota Y, van Heijenoort J. In vitro peptidoglycan polymerization catalysed by penicillin-binding protein 1b of Escherichia coli K12. FEBS Lett. 1980;110:245–249. doi: 10.1016/0014-5793(80)80083-4. [DOI] [PubMed] [Google Scholar]
- 30.Tomasz A, Munoz R. β-Lactam antibiotic resistance in Gram-positive bacterial pathogens of the upper respiratory tract: a brief overview of mechanisms. Microb Drug Resist. 1995;1:103–109. doi: 10.1089/mdr.1995.1.103. [DOI] [PubMed] [Google Scholar]
- 31.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1025–1034. [Google Scholar]
- 32.van Heijenoort Y, Derrien M, van Heijenoort J. Polymerisation by transglycosylation in the biosynthesis of the peptidoglycan of Escherichia coli K12 and its inhibition by antibiotics. FEBS Lett. 1978;89:141–144. doi: 10.1016/0014-5793(78)80540-7. [DOI] [PubMed] [Google Scholar]
- 33.van Heijenoort Y, van Heijenoort J. Biosynthesis of the peptidoglycan of Escherichia coli K12. Properties of the in vitro polymerisation by transglycosylation. FEBS Lett. 1980;110:241–244. doi: 10.1016/0014-5793(80)80082-2. [DOI] [PubMed] [Google Scholar]
- 34.van Heijenoort Y, Leduc M, Singer H, van Heijenoort J. Effects of moenomycin on Escherichia coli. J Gen Microbiol. 1987;133:667–674. doi: 10.1099/00221287-133-3-667. [DOI] [PubMed] [Google Scholar]
- 35.van Heijenoort Y, Gómez M, Derrien M, Ayala J, van Heijenoort J. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J Bacteriol. 1992;174:3549–3557. doi: 10.1128/jb.174.11.3549-3557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youssif S, Broome-Smith J-K, Spratt B G. Lysis of Escherichia coli by β-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1a and 1b. J Gen Microbiol. 1985;131:2839–2845. doi: 10.1099/00221287-131-10-2839. [DOI] [PubMed] [Google Scholar]
- 37.Zhao G, Yeh W-K, Carnahan R H, Flokowitsch J, Meier T I, Alborn W E, Jr, Becker G W, Jaskunas S R. Biochemical characterization of penicillin-resistant and -sensitive penicillin-binding protein 2x transpeptidase activities of Streptococcus pneumoniae and mechanistic implications in bacterial resistance to β-lactam antibiotics. J Bacteriol. 1997;179:4901–4908. doi: 10.1128/jb.179.15.4901-4908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]