The parasitic plant Phtheirospermum japonicum modifies the pectin methylesterification status in its haustoria in a dynamic and tissue-specific manner to allow efficient infection of the host plant.
Abstract
Parasitic plants are globally prevalent pathogens with important ecological functions but also potentially devastating agricultural consequences. Common to all parasites is the formation of the haustorium which requires parasite organ development and tissue invasion into the host. Both processes involve cell wall modifications. Here, we investigated a role for pectins during haustorium development in the facultative parasitic plant Phtheirospermum japonicum. Using transcriptomics data from infected Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), we identified genes for multiple P. japonicum pectin methylesterases (PMEs) and their inhibitors (PMEIs) whose expression was upregulated by haustoria formation. Changes in PME and PMEI expression were associated with tissue-specific modifications in pectin methylesterification. While de-methylesterified pectins were present in outer haustorial cells, highly methylesterified pectins were present in inner vascular tissues, including the xylem bridge that connects parasite to host. Specifically blocking xylem bridge formation in the haustoria inhibited several PME and PMEI genes from activating. Similarly, inhibiting PME activity using chemicals or by overexpressing PMEI genes delayed haustoria development. Our results suggest a dynamic and tissue-specific regulation of pectin contributes to haustoria initiation and to the establishment of xylem connections between parasite and host.
Introduction
Parasitic plants, which constitute around 1% of angiosperm species (Westwood et al. 2010), are important contributors to ecological systems but also include devastating pests that cause major agricultural losses each year (Rodenburg et al. 2016). Parasitism has evolved independently at least 12 times (Nickrent 2020), and despite these diverse origins, all parasitic plants form an invasive structure, the haustorium, which penetrates the host and allows the uptake of nutrients, hormones and signalling molecules (Birschwilks et al. 2006; Spallek et al. 2017; Shahid et al. 2018; Liu et al. 2020). The development of the haustorium starts with the perception of a suitable host through haustorium inducing factors (HIFs). Treatment with 2,6-Dimethoxybenzoquinone (DMBQ), the first discovered HIF, is sufficient to induce the formation of pre-haustoria in the parasitic plant family Orobanchaceae, even in the absence of a host (Chang et al. 1986). Other HIFs include hormones like cytokinin and lignin-related compounds (Goyet et al. 2017; Cui et al. 2018; Aoki et al. 2022). In the facultative root parasite Phtheirospermum japonicum the perception of HIFs is mediated by leucine-rich-repeat receptor-like kinases (Laohavisit et al. 2020) and increases auxin polar transport and auxin biosynthesis. The auxin signalling peak promotes cell expansion and division, leading to the formation of a swelling called the pre-haustorium (Ishida et al. 2016; Wakatake et al. 2020). Penetration of the host by the pre-haustorium is thought to depend on haustorium-secreted cell-wall-modifying enzymes such as expansins and peroxidases that loosen the host cell walls (Losner-Goshen et al. 1998; Veronesi et al. 2007; Honaas et al. 2013; Olsen et al. 2016). The invasion of the host tissues is then mediated by the intrusive cells, which differentiate from epidermal cells at the parasite-host interface and drive haustorial growth towards the host vasculature (Heide-Jørgensen and Kuijt 1995; Hood et al. 1998). Finally, a vascular connection develops between the parasite and the host. All parasitic plants form a xylem connection, which begins its differentiation from the cambium-like tissue at the centre of the haustorium (Wakatake et al. 2018). A mass of xylem tissue then develops close to the parasite vasculature (plate xylem) before strands of xylem (xylem bridges) differentiate to connect the xylem of the parasite to the xylem of the host.
Despite recent advances in our understanding of haustorium development, the mechanisms regulating haustoria initiation and host invasion remain largely unknown, but likely rely in part on cell wall modifications. In plants, lateral organ development relies on the fine tuning of cell wall modifications which are required for cell expansion, division and differentiation. These processes all require the modification of cell-to-cell adhesion. The main mediator of cell adhesion in plants is pectin, a jelly-like matrix composed of homogalacturonan, rhamnogalacturonan I and rhamnogalacturonan II (Pelloux et al. 2007; Daher and Braybrook 2015). Homogalacturonan is secreted to the cell wall in a highly methylesterified state and is then modified in the cell wall by different families of pectin modifying enzymes including pectin methylesterases (PMEs), PME inhibitors (PMEIs), polygalacturonases (PGs) and pectate lyases (PLs). Highly methylesterified pectin forms a tight matrix with less elastic properties. During developmental processes such as tissue expansion and lateral organ emergence, homogalacturonans undergo de-methylesterification by PMEs to become looser (Pelloux et al. 2007; Daher and Braybrook 2015). In addition to the fundamental roles of PMEs and PMEIs in plant development, many plant pathogens hijack plant pectin modification mechanisms to allow tissue intrusion (Hewezi et al. 2008; Raiola et al. 2011). For example, the cyst nematode Heterodera schachtii secretes into Arabidopsis (Arabidopsis thaliana) a cellulose binding protein (CBP) that activates PME3, facilitating the entry of the nematode (Hewezi et al. 2008). A. thaliana pme3 mutants are less susceptible to nematode infection (Hewezi et al. 2008). Furthermore, some nematodes and fungi can directly secrete PMEs that mimic plant PMEs and facilitate host tissue invasion (Valette-Collet et al. 2003; Vicente et al. 2019). Parasitic plants are thought to secrete cell-wall modifying enzymes (CWMEs) to promote growth of the haustorium, adhesion to the host and loosening of host tissues to allow for invasion (Veronesi et al. 2007; Honaas et al. 2013; Yang et al. 2015; Kurotani et al. 2020). The secretion of CWMEs has been partially investigated in some parasitic plant species. For example, different species from the Orobanche genus secrete PMEs, PGs, PLs and peroxidases close to the site of infection to modify the host's cell wall (Ben-Hod et al. 1993; Losner-Goshen et al. 1998; Veronesi et al. 2007), while the shoot parasite Cuscuta upregulates PMEI transcription important for host penetration and increases pectin degrading enzyme activity during infection (Nagar et al. 1984; Jhu et al. 2022).
Even though cell wall modifications are suggested to be crucial for haustorium development, this aspect remains largely unexplored. Here, we use a combination of transcriptomic and genetic approaches to identify PMEs and PMEIs relevant for haustoria formation. We go on to describe dynamic and tissue specific changes in pectin methylesterification and show the effects of perturbing host and parasite PME-related enzymes. Together, this study describes the role of pectin during parasitic plant infection and reveals the importance of pectin methylesterification-related genes for haustoria development.
Results
PjPMEs and PjPMEIs are differentially expressed during haustorium development
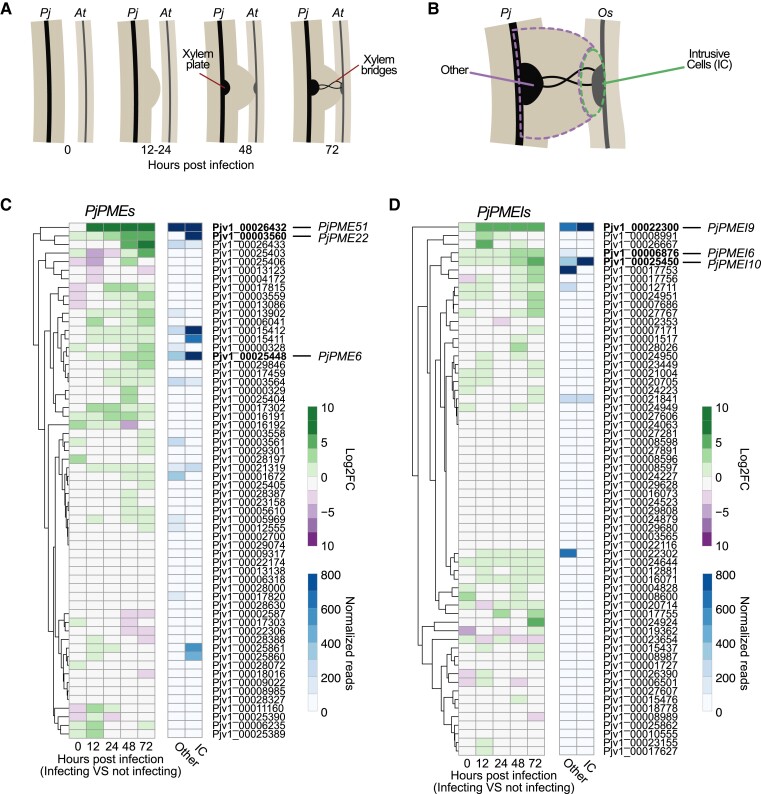
To study the role of pectin in P. japonicum, we focused on modifications of pectin by the pectin methylesterase (PME) and PME-inhibitor (PMEI) enzyme families. We performed a Hidden-Markov-Model search on the P. japonicum (Pj) proteome (Cui et al. 2020) and identified 73 putative PjPMEs and 62 putative PjPMEIs (Supplemental Table S1). We further filtered PjPMEs based on the presence of at least three of the five conserved catalytic amino acids (Johansson et al. 2002; Markovic and Janecek 2004) and retained 60 PjPMEs for downstream analyses (Supplemental Table S1). We aligned PjPMEs and PjPMEIs with A. thaliana PMEs and PMEIs, respectively, and built two Maximum-Likelihood phylogenetic trees. The trees showed co-clustering of A. thaliana and P. japonicum sequences, suggesting conservation in PMEs and PMEIs between parasite and host (Supplemental Fig. S1, A and B). We then looked at the expression of PjPMEs and PjPMEIs in two different published P. japonicum transcriptomic datasets. The first dataset sampled tissues at the site of haustorium development during a time-course infection of A. thaliana (Fig. 1A, Kokla et al. 2022), while the second dataset sampled intrusive cells (ICs) and non-ICs in mature haustoria infecting rice (Oryza sativa) (Fig. 1B, Ogawa et al. 2021). RNA levels of several PjPME and PjPMEI genes increased during haustorium formation, particularly at 48 and 72 h post infection (hpi), while very few genes showed decreased RNA levels (Fig. 1, C and D). Many PjPME and PjPMEI genes were also highly expressed during rice infection, including several in common with A. thaliana infection. We observed a greater number of highly expressed PjPMEs (>400 reads) in ICs compared to non-IC tissues, while several PjPMEIs were highly expressed in both IC and non-IC tissues (Fig. 1, C and D). In the A. thaliana host dataset, few AtPMEs and AtPMEIs changed expression during haustorium development with AT1G23200 (PME) and AT2G01610 (PMEI) showing the most consistent pattern of increased expression over multiple time points (Supplemental Fig. S1, C and D).
Figure 1.
PjPMEs and PjPMEIs are differentially expressed during haustorium development. A) Illustration of P. japonicum (Pj) haustorium development during infection of A. thaliana (At) corresponding to the time points selected for the time course RNA-Seq (Kokla et al. 2022). B) Illustration of the sampling for the P. japonicum intrusive cells (IC) or rest of the haustorial tissues (other) during O. sativa (Os) infection for the RNA-Seq dataset presented in Ogawa et al. 2021. C-D) Heatmaps of the expression of candidate P. japonicum PMEs (PjPMEs) or PMEIs (PjPMEIs): log2 fold change (FC) between P. japonicum infecting and not infecting over five time points during infection, and normalized reads in intrusive cells and other haustorial tissue. The genes are clustered by expression according to the time-course dataset.
PME activity increases during haustorium development and is higher in intrusive cells
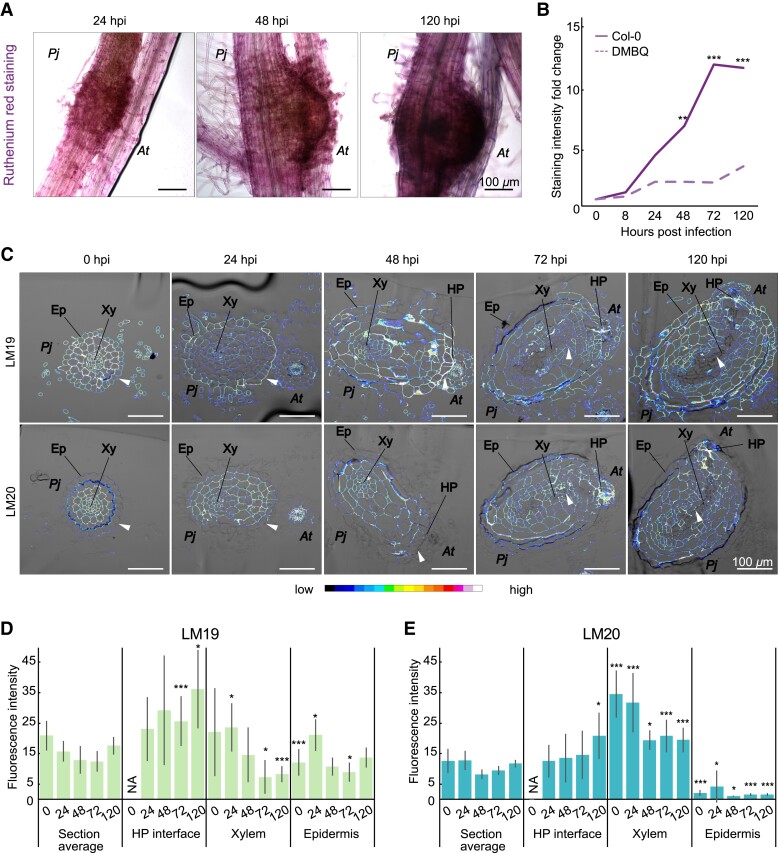
Since we observed differential expression of PjPMEs and PjPMEIs during haustorium development, we tested whether pectin methylesterification levels could also be affected. We first performed ruthenium red staining on whole roots during an infection time course (Fig. 2A). Ruthenium red binds with higher affinity to de-methylesterified homogalacturonan, and therefore a higher staining often corresponds with higher PME activity (Downie et al. 1998). We observed an increase in ruthenium red staining by 24 hpi at the interface between P. japonicum and A. thaliana. The staining intensity increased further in later stages corresponding to host invasion (48 hpi) and xylem differentiation (72 and 120 hpi) (Fig. 2, A and B, Supplemental Fig. S2A), but was not observed when inducing haustoria using DMBQ (Fig. 2B, Supplemental Fig. S2B), which does not induce xylem bridge formation. We also used LM19 and LM20 antibodies specific for de-methylesterified and highly methylesterified pectin (Verhertbruggen et al. 2009), respectively, to measure pectin modifications from 0 to 120 hpi (Fig. 2, C, D and E, Supplemental Fig. S2C). LM19 staining was mostly localised to the outer epidermis and at the host-parasite interface, especially in later time points (72 and 120 hpi), whereas staining intensity was reduced in xylem tissues during infection (Fig. 2, C and D). LM20 staining was nearly absent in epidermal tissues and staining was instead mainly focused to xylem tissues and at the host-parasite interface (Fig. 2, C and E). These results showed that pectin methylesterification was modified dynamically during haustorium development in a tissue-specific manner.
Figure 2.
PME activity increases during haustorium development and is higher in intrusive cells. A) Ruthenium red staining of the developing haustoria at 24, 48 and 120 h post infection (hpi). Scale bars 100 μm. B) Quantification of staining intensity in infecting haustoria (solid line) or pre-haustoria formed on DMBQ (dashed line) during a time course from 0 to 120 hpi. Staining intensity normalized to 0 hpi. Asterisks indicate significant difference in staining intensity between haustoria and pre-haustoria at the same time point. (** P < 0.01, *** P < 0.001, Wilcoxon test, n = 11 to 20 roots, 1 replicate). C) Fluorescence images of antibody staining with LM19 (unmethylesterified homogalacturonan) and LM20 (highly methylesterified homogalacturonan) on P. japonicum (Pj) haustoria cross sections at 0, 24, 48, 72 and 120 hpi of A. thaliana (At). Arrowheads denote areas of differential staining between LM19 and LM20. Ep = epidermis, Xy = xylem, HP = host-parasite interface. D, E) Fluorescence quantification in whole P. japonicum sections (section average), host-parasite (HP) interface, xylem (root xylem and xylem bridge) and epidermis tissues for LM19 and LM20 antibodies. Asterisks indicate significant difference between a specific tissue and the section average at the same time point (Student's t-test, P-value corrected for multiple testing), bars represent standard deviation. Scale bars 100 μm; NA = tissue not present at the time point. * for P < 0.05, ** for P < 0.01, *** for P < 0.001, n = 3 to 15 sections.
Differentially expressed PjPMEs and PjPMEIs activate in intrusive cells and cambium-like tissue
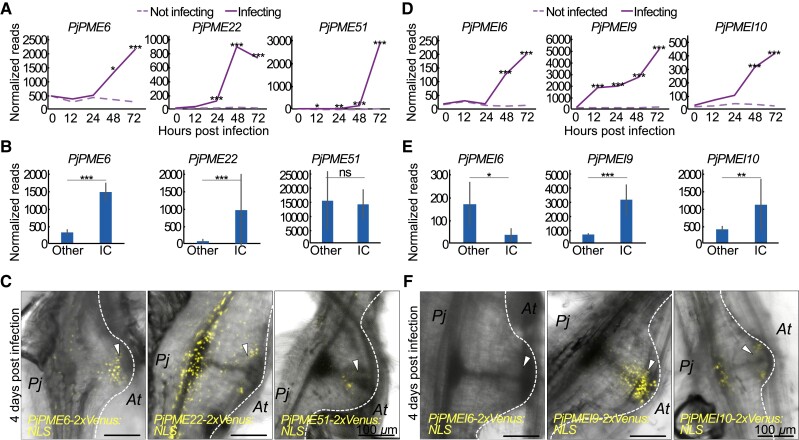
Since the pectin degree of methylesterification (DM) in the developing haustorium was different in specific tissues, we investigated candidate PjPMEs and PjPMEIs to understand if their expression pattern corresponded with the pattern of pectin DM. We chose three PjPME genes with increased expression in the time course dataset and renamed them based on the A. thaliana homolog (Fig. 1C, Fig. 3A). All three genes were also upregulated during P. japonicum-O. sativa infection. PjPME6 (Pjv1_00025448) and PjPME22 (Pjv1_00003560) expression levels increased in ICs compared to other tissues, while PjPME51 (Pjv1_00026432) was highly expressed in both IC and non-IC tissues (Fig. 3B). We made and transformed transcriptional reporters in P. japonicum hairy roots and found PjPME6 and PjPME51 reporters showed signal mainly in ICs, while the PjPME22 reporter was mostly expressed in P. japonicum vasculature (Fig. 3C). We then selected three PjPMEI genes upregulated during haustorium development in A. thaliana and O. sativa infections (Fig. 1D and 3D). In the P. japonicum-O. sativa dataset, PjPMEI9 (Pjv1_00022300) and PjPMEI10 (Pjv1_00025450) showed increased expression in ICs, while PjPMEI6 (Pjv1_00006876) was more expressed in non-IC tissues (Fig. 3E). Our transcriptional reporters for these genes showed expression in ICs and cambium-like tissues for PjPMEI9, and plate xylem and ICs for PjPMEI10 (Fig. 3F). The PjPMEI6 reporter showed no fluorescence in 4 dpi haustoria (Fig. 3F). All the PjPMEs and PjPMEIs we investigated were not expressed in the primary root tip of the hairy roots, and showed little or no fluorescence at the lateral root emergence sites of the hairy roots, suggesting that upregulation of these genes was specific to haustorium development (Supplemental Fig. S3).
Figure 3.
Upregulated PjPMEs and PjPMEIs are primarily expressed in intrusive cells and cambium-like tissue. A, D) Normalized reads of PjPME6, PjPME22, PjPME51, PjPMEI6, PjPMEI9 and PjPMEI10 over five time points during infection for P. japonicum infecting and not infecting. Asterisks indicate a significant difference between infecting and not infecting (Wald test with Benjamini-Hochberg correction for multiple testing, n = 3 libraries). B, E) Normalized reads of PjPME6, PjPME22, PjPME51, PjPMEI6, PjPMEI9 and PjPMEI10 in intrusive cells and non-IC (other) tissues. Asterisks indicate a significant difference between IC and other tissues (Student's t-test, n = 3 libraries). Bars represent standard deviation. C, F) Images of infecting transgenic hairy roots expressing PjPME6, PjPME22, PjPME51, PjPMEI6, PjPMEI9 and PjPMEI10 nuclear-localized (NLS) transcriptional reporters in fully developed haustoria (4 dpi). Arrowheads denote intrusive cells. Scale bars 100 μm. For all panels * for P < 0.05, ** for P < 0.01, *** for P < 0.001.
Inhibition of PME activity impairs haustoria induction and development
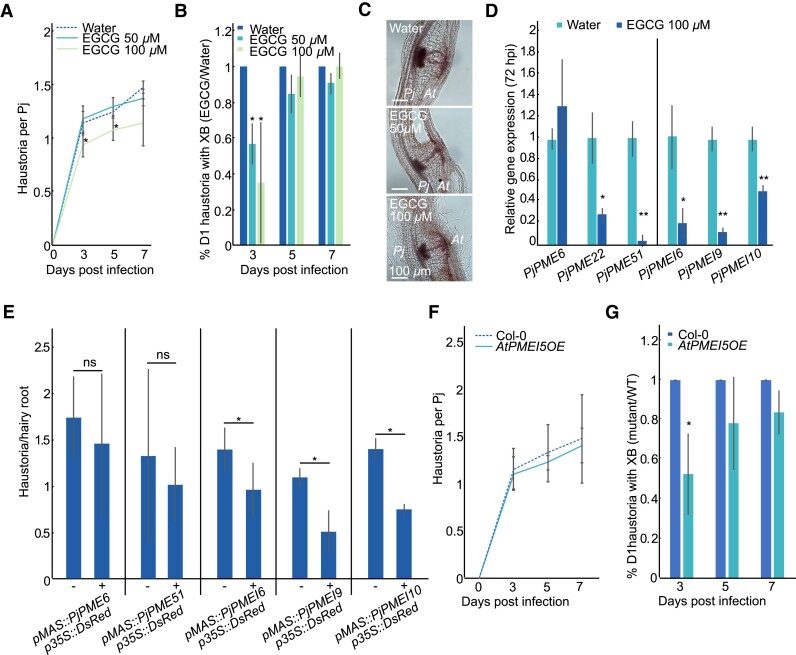
To determine if PME activity is necessary for haustorium development, we treated infecting P. japonicum with 50 µM or 100 µM of epigallocatechin gallate (EGCG), a chemical inhibitor of PME enzymes (Lewis et al. 2008). Treatment reduced the number of haustoria (Fig. 4A) and delayed the formation of xylem bridge connections to the host (Fig. 4B), although it did not substantially affect plate xylem area and number of xylem bridge connections at 7 dpi (Fig. 4C, Supplemental Fig. S4, A and B). EGCG treatment also reduced PjPME22, PjPME51 and PjPMEI10 expression at 72 hpi (Fig. 4D), and reduced PjPMEI6 and PjPMEI9 expression at both 0 and 72 hpi, suggesting that chemical inhibition of PMEs affected both haustoria development and the transcriptional regulation of endogenous PMEs and PMEIs (Fig. 4D, Supplemental Fig. S4C). We next overexpressed PjPME6 and PjPME51 in P. japonicum hairy roots (Supplemental Fig. S4, D and E) but did not observe defects in haustorium induction or development (Fig. 4E, Supplemental Fig. S4, F and G). However, overexpression of PjPMEI6, PjPMEI9 and PjPMEI10 significantly inhibited haustoria induction (Fig. 4E) but did not affect xylem connections (Supplemental Fig. 4, F and G). Finally, we tested whether modifying pectin status in the host could affect infection by using the PMEI5-overexpressing A. thaliana line AtPMEI5OE, which is characterised by highly methylesterified pectin (Wolf et al. 2012; Jonsson et al. 2021). Haustoria induction was not affected in the mutant compared to wild-type Col-0 (Fig. 4F), however, xylem bridge formation was delayed during infection of AtPMEI5OE (Fig. 4G). Taken together, these results suggest that parasitic PME activity is important for efficient induction and development of haustoria.
Figure 4.
Inhibition of PME activity impairs haustorium induction and development. A) Number of haustoria per P. japonicum plant at four time points during treatment with 50 μM ECGC, 100 μM EGCG or water control. Asterisks indicate significance compared to control (Student's t-test, n = 3 replicates). B) Ratio of the percentage of day one (D1) haustoria with a xylem bridge (XB) formed during treatment with 50 or 100 μM EGCG over water at three time points. Asterisks indicate significance compared to control (Student's t-test, n = 3 replicates). C) Images of 7 dpi haustoria formed on water, 50 μM EGCG and 100 μM EGCG. D) Relative gene expression of selected PjPMEs and PjPMEIs at 72 hpi in P. japonicum haustoria treated with 100 μM EGCG, normalised to water. Asterisks indicate significance compared to control (Student's t-test, n = 3 replicates). E) Numbers of haustoria per hairy root transformed with PjPME and PjPMEI overexpression constructs. Non-transgenic hairy roots are marked as “-” and transgenic roots are marked as “+”. Asterisks indicate significance compared to control (Fisher's exact test; n = 2 to 4 replicates, 12 to 53 total roots per sample). F) Number of haustoria per P. japonicum plant at four time points during infection of AtPMEI5OE mutant or Col-0 as control (n = 3 replicates). G) Ratio of the percentage of D1 haustoria with a XB formed during infection of AtPMEI5OE over Col-0 at three time points. Asterisks indicate significance compared to control (Student's t-test, n = 3 replicates). For all panels * for P < 0.05, ** for P < 0.01, bars represent standard deviation.
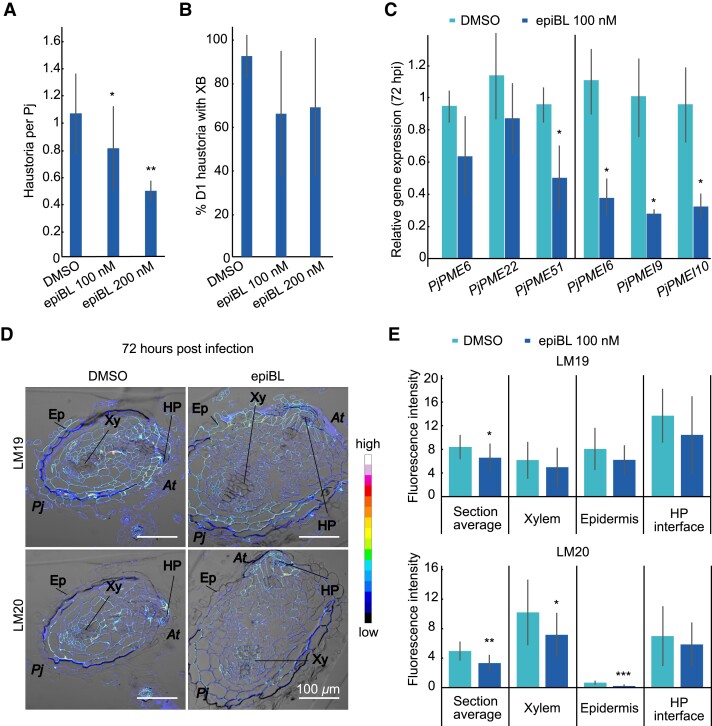
Brassinosteroid treatment reduces PjPME and PjPMEI expression and delays haustorium development
Brassinosteroid (BR) signalling mediates cell wall biosynthesis and remodelling, and has been implicated in feedback mechanisms with PME and PMEI activity and EGCG treatments (Wolf et al. 2012). To test the effect of BRs, we applied 100 nM or 200 nM of epibrassinolide (epiBL) during P. japonicum infection and found it reduced the number of haustoria per P. japonicum (Fig. 5, A and B, Supplemental Fig. S5A), similar to EGCG treatment (Fig. 4, A and , B). We also tested the expression of PjPMEs and PjPMEIs in haustoria following epiBL treatment. PjPME51, PjPMEI6, PjPMEI9 and PjPMEI10 were downregulated in haustoria treated with epiBL at 72 hpi but not at 0 hpi (Fig. 5C, Supplemental Fig. S5B), suggesting transcriptional control of pectin methylesterification by BR signalling during haustorium development. To determine if BR treatment could modify pectin methylesterification levels, we performed antibody staining using LM19 and LM20 on cross sections of 0 hpi and 72 hpi haustoria untreated or treated with epiBL (Fig. 5D, Supplemental Fig. S5C). The haustoria treated with epiBL had lower levels of both unmethylesterified pectin (LM19) and highly methylesterified pectin (LM20) compared to the control (Fig. 5E), suggesting overall pectin levels were reduced following epiBL treatment. In particular, staining for highly methylesterified pectin (LM20) was significantly lower in xylem and epidermis tissues treated with epiBL (Fig. 5E), corresponding with the reduced PjPMEI gene expression previously observed (Fig. 5C). Finally, we infected the A. thaliana BRI1-EMS-SUPPRESSOR 1 mutants with modified BR signalling, bes1-2 and bes1-D, to test the role of host BR signalling during infection. P. japonicum could efficiently infect both mutants and establish xylem connections (Supplemental Fig. S5, D and E), suggesting host BR signalling is not crucial for haustoria development and instead BR signalling might be important for parasite cell wall modifications during infection.
Figure 5.
BR treatment reduces PjPME and PjPMEI expression and inhibits haustorium development. A) Number of haustoria per P. japonicum plant at 7 dpi during treatment with 100 nM epiBL, 200 nM epiBL or DMSO control (n = 4 to 6 replicates). B) Percentage of day one (D1) haustoria with a xylem bridge (XB) formed during treatment with 100 nM epiBL, 200 nM epiBL or DMSO at 7 dpi (n = 4 to 6 replicates). C) Relative gene expression of selected PjPMEs and PjPMEIs at 72 h post infection in P. japonicum haustoria treated with 100 nM epiBL, normalised to DMSO (n = 3 replicates). D) Fluorescence images of antibody staining using LM19 (unmethylesterified homogalacturonan) and LM20 (highly methylesterified homogalacturonan) on cross sections of 72 hpi haustoria formed on DMSO control or 100 nM epiBL. Scale bars 100 μm. Ep = epidermis, Xy = xylem, HP = host-parasite interface. At = A. thaliana, Pj = P. japonicum. E) Fluorescence quantification in whole P. japonicum sections (section average), host-parasite (HP) interface, xylem (root xylem and xylem bridge) and epidermis tissues for LM19 and LM20 antibodies (n = 7 to 14 sections). For all panels, asterisks indicate significance compared to DMSO (Student's t-test): * for P < 0.05, ** for P < 0.01, bars represent standard deviation.
PjPMEs and PjPMEIs expression associates with xylem bridge development
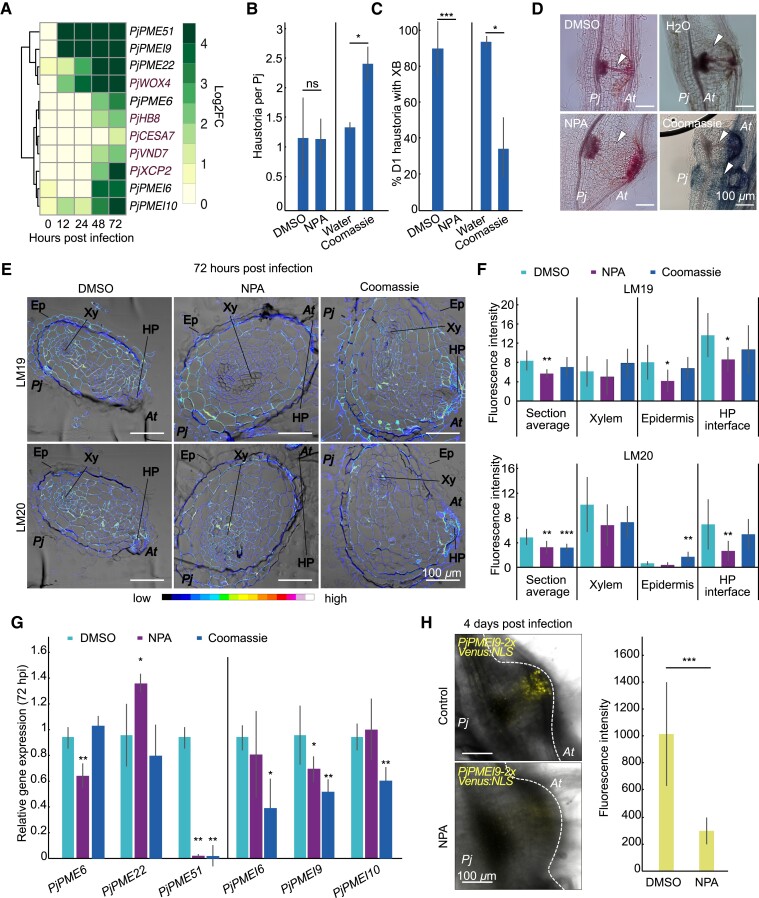
Since some PjPMEs and PjPMEIs are expressed in the cambium and xylem-like tissues during haustorium development (Fig. 3C-F) and inhibiting PME activity delays xylem formation (Fig. 4B), we investigated the role of PjPMEs and PjPMEIs during xylem-bridge formation. Looking at the expression pattern of cambium and xylem marker genes, we found the cambium marker WUSCHEL RELATED HOMEOBOX 4 (PjWOX4) (Wakatake et al. 2018) co-expressed with PjPME22, PjPME51 and PjPMEI9 (Fig. 6A). The putative procambium marker HOMEOBOX GENE 8 (PjHB8) co-expressed with PjPME6. The xylem-markers CELLULOSE SYNTHASE A 7 (PjCESA7) (Wakatake et al. 2018), VASCULAR RELATED NAC-DOMAIN PROTEIN 7 (PjVND7) (identified through BLAST using AtVND7 as a query) and XYLEM CYSTEINE PEPTIDASE 2 (PjXCP2) (Kokla et al. 2022) co-expressed with PjPMEI6 and PjPMEI10 (Fig. 6A, Supplemental Fig. 6A). To investigate if pectin methylesterification levels change in response to xylem-bridge development, we chemically inhibited xylem bridge formation by treatment with the auxin transport inhibitor N-1-naphthylphthalamidic acid (NPA). NPA treatment did not affect haustoria numbers, yet inhibited xylem bridge connection (Wakatake et al. 2020, Fig. 6, B and D). Treatment with the synthetic auxin 1-naphthaleneacetic acid (NAA) did not affect haustoria numbers or xylem bridge formation (Supplemental Fig. S6B, C and D). We also found that the commonly used dye Coomassie Brilliant Blue, an inhibitor of xyloglucan endotransglucosylase/hydrolase (XTH) activity (Olsen and Krause 2017), increased haustoria numbers, yet reduced xylem bridge formation by approximately 60% (Fig. 6B-D). LM19 and LM20 antibody staining of 72 hpi sections showed both de-methylesterified pectin and highly methylesterified pectin were reduced following NPA treatment, mostly in the host-parasite interface (Fig. 6, E and F, Supplemental Fig. S6E). Staining with LM20 was also reduced following Coomassie treatment (Fig. 6, E andF, Supplemental Fig. S6E). Expression levels of several PMEs and PMEIs were significantly affected by NPA and Coomassie treatments (Fig. 6G) including PjPME51 and PjPMEI9 that were decreased by both treatments at 72 hpi (Fig. 6g) but not at 0 hpi (Supplemental Fig. S6F). This decreased expression of PjPME51 and PjPMEI9 appeared specific to xylem bridge inhibition and suggested a role for these genes in xylem bridge formation. We then infected hairy roots expressing PjPMEI9-2xVenus:NLS, which showed fluorescence in cambium-like tissues (Fig. 3F, Supplemental Video S1). At 4 days post infection we observed a marked decrease in fluorescence when hairy roots expressing PjPMEI9-2xVenus:NLS were treated with NPA compared to the control, consistent with the RT-qPCR data (Fig. 6H) and demonstrating that xylem bridge formation is important for PMEI9 expression.
Figure 6.
PjPMEs and PjPMEIs expression associates with xylem bridge development. A) Expression heatmap of P. japonicum cambium and xylem marker genes (red) and selected PjPMEs and PjPMEIs: log2 fold change between P. japonicum infecting and not infecting over five time points during infection, clustered by expression. B) Number of haustoria per P. japonicum plant at 7 dpi during treatment with DMSO, 5 μM NPA (n = 2 replicates), water or 0.05 mM Coomassie (n = 3 replicates). C) Percentage of day one (D1) haustoria with a xylem bridge (XB) formed during treatment with DMSO, 5 μM NPA (n = 2 replicates), water or 0.05 mM Coomassie (n = 3 replicates). D) Images of 7 dpi haustoria formed on DMSO, 5 μM NPA, water or 0.05 mM Coomassie. White arrowheads denote the location of XB development. E) Fluorescence images of antibody staining using LM19 and LM20 on cross sections of 72 hpi haustoria developed on DMSO, 5 μM NPA or 0.05 mM Coomassie + DMSO. Ep = epidermis, Xy = xylem, HP = host-parasite interface. At = A. thaliana, Pj = P. japonicum. F) Fluorescence quantification in whole P. japonicum sections (section average), host-parasite (HP) interface, xylem (root xylem and xylem bridge) and epidermis tissues for LM19 and LM20 antibodies. Asterisks indicate significance compared to DMSO (Student's t-test, P-value corrected for multiple testing, n = 8 to 14 sections). G) Relative gene expression of select PjPMEs and PjPMEIs at 72 hpi in P. japonicum haustoria treated with 5 μM NPA or 0.05 mM Coomassie + DMSO normalised to DMSO (n = 3 replicates). H) Images of haustoria developed on hairy roots expressing the PjPMEI9 nuclear-localised (NLS) transcriptional reporter at 4 dpi on DMSO or 5 μM NPA, and quantification of fluorescence intensity (n = 7 to 10 haustoria). Scale bars 100 μm. For all panels, asterisks indicate significance compared to control (Student's t-test): * for P < 0.05, ** for P < 0.01, *** for P < 0.001, bars represent standard deviation.
Discussion
Here, we investigated the role of PME-mediated pectin modifications during haustorium development in P. japonicum and identified multiple PjPMEs and PjPMEIs upregulated during A. thaliana and O. sativa infections. Induction dynamics varied with some genes highly activated during early stages of infection whereas others peaked late when xylem bridges formed (Fig. 1C-D and 3, A and D), suggesting they had different developmental roles as infection progressed. The reporters we generated showed PME-related gene expression in intrusive cells (Fig. 3B-C) and these outer tissues also showed low methylesterification (Fig. 2C) suggesting cell wall loosening was relevant for expansion of outer tissues and interaction with the host. Suppressing PME activity by overexpressing PMEIs or by chemical treatments with EGCG also reduced the ability for haustoria to form (Fig. 4) consistent with a role for PMEs and pectin loosening in organogenesis and haustoria expansion. Recently, it was found that pectin methylesterification levels are also crucial for lateral root initiation (Wachsman et al. 2020). Our findings suggested aspects of lateral root formation and haustoria emergence are conserved in P. japonicum as it has been previously suggested for Cuscuta and Thesium parasites (Ichihashi et al. 2018; Jhu et al. 2021).
We also observed strong induction of PMEI-related gene expression including in the vascular tissues of the haustoria (Fig. 1D and 3D-F). Antibody staining revealed high levels of methylesterified pectin in the inner haustoria tissues and xylem bridges (Fig. 2C-D) that could provide structural support to these tissues. Thus, the apparent co-expression of both esterase (PME) and inhibitor (PMEI) could be explained in part by differences in spatial expression and the degree of methylesterification in different tissues. Asymmetric pectin methylesterification is required in several plant developmental processes, including apical hook formation during seedling emergence and leaf patterning (Jonsson et al. 2021; Peng et al. 2022). Furthermore, differences in pectin methylesterification between lateral roots and its progenitor tissues might allow the emergence of the lateral root while preventing its own digestion (Laskowski et al. 2006). We propose that in haustoria high PME activity in intrusive cells drives host cell wall loosening and penetration, while high PMEI activity maintains inner haustorial tissue integrity and helps these structures push towards the host.
Our investigations also revealed a tight association between pectins and xylem bridge formation. DMBQ induces pre-haustoria that lack xylem bridges (Cui et al. 2016) and we observed little ruthenium red staining of DMBQ treatment samples (Fig. 2B, Supplemental Fig. S2B) suggesting that pectins were not highly de-methylesterified during pre-haustoria formation. Similarly, EGCG, NPA and Coomassie Brilliant Blue treatments all delayed or inhibited xylem bridge formation (Fig. 4B and 6C) and reduced the expression of PME51 and PMEI9 (Fig. 4D and 6G) suggesting a close relation between xylem bridge formation and PME51 and PMEI9 activation. In alfalfa (Medicago sativa), xylem cell walls contain about 4% pectin, compared to 25% pectin in other tissues (Grabber et al. 2002), suggesting pectin might be degraded during xylem differentiation. In A. thaliana, five PMEs are expressed in xylem tissues (Pelloux et al. 2007), and the demethylesterification of pectin might be important for lignification (Lairez et al. 2005; Pelloux et al. 2007). PME activity including from PjPME51 might therefore be required in the first stage of xylem bridge differentiation to allow pectin degradation, followed by lignification.
In the A. thaliana host, our data suggest the role of pectin modifications during infection is less important. Only AT1G23200 (PME) and AT2G01610 (PMEI) showed clear increases in expression during infection (Supplemental Fig. S1, C and D), suggesting these genes might either be involved in a defence response to the parasite or are activated by P. japonicum to facilitate parasitism. Notably, the A. thaliana overexpressor AtPMEI5OE (Wolf et al. 2012), which has high methylesterification levels (Wolf et al. 2012; Jonsson et al. 2021), delayed xylem bridge connections (Fig. 4, F and G) indicating that pectin modifications by the host could influence parasite development. Our EGCG treatments likely inhibited both parasite and host PMEs so our finding that EGCG reduces xylem bridge formation could be explained in part due to inhibition of host PMEs. Thus, cell wall modifications by both host and parasite appear relevant for successful parasitic plant infection and deserve further attention. By better understanding and modifying the host cell wall response, it may be possible to achieve durable resistance to parasites.
Materials and methods
Plant materials and growth conditions
Phtheirospermum japonicum and Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized by washing with 70% v/v ethanol for 20 min, followed by 95% v/v ethanol for 5 min, and sown on 1⁄2 MS medium with 1% w/v sucrose and 0.8% w/v bactoagar. After stratification at 4 °C in darkness for 1 or 2 days for P. japonicum and A. thaliana respectively, the plates were moved to a growth cabinet at 25 °C in long day conditions (16 h light/8 h darkness), 100 μmol m−2 s−1 light. The A. thaliana Col-0 accession was used unless otherwise stated. The AtPMEI5OE line has been previously published (Wolf et al. 2012). The BR-signalling mutants bes1-2 and bes1-D have been previously published (Yin et al. 2002; Lachowiec et al. 2013).
PjPMEs and PjPMEIs identification and phylogenetic analyses
A. thaliana PME (Louvet et al. 2006) and PMEI (Wang et al. 2013) sequences were downloaded from the Phytozome database (Goodstein et al. 2012). P. japonicum putative PMEs and PMEIs were identified by searching the HMM profiles (PF01095 and PF04043 respectively) on a P. japonicum proteome obtained from the published genome (Cui et al. 2020) using the HMMER3 software (Finn et al. 2011). The putative PjPMEs were aligned using Clustal W in MEGAX (Tamura Stecher and Kumar 2021), and the sequences lacking more than one of the five conserved catalytic amino acids (Johansson et al. 2002; Markovic and Janecek 2004) were removed from downstream analyses. P. japonicum and A. thaliana PME and PMEI sequences were aligned using ClustalW. Maximum-Likelihood phylogenetic trees were built using MEGAX with 100 bootstraps.
In vitro infection assays with Phtheirospermum japonicum
Infection assays were performed according to Kokla et al. 2022. Briefly, five days after germination P. japonicum seedlings were moved from nutrient medium to nutrient-free medium (water agar) for starvation. After three days, a six-day old A. thaliana seedling was aligned root-to-root to each P. japonicum seedling to allow infection. 50 μM or 100 μM EGCG, 100 nM or 200 nM epiBL, 5 μM NPA, 0.05 mM Coomassie Brilliant Blue and 0 to 500 nM NAA were applied directly in the nutrient-free medium and left until the end of the infection period. For measuring the plate xylem area and the number of xylem bridges, 7 days post infection (dpi) haustoria were stained with Safranin-O following the method in Spallek et al. 2017. Pictures were taken using an Axioscope A1 microscope and analysed in Fiji (Schindelin et al. 2012; Rueden et al. 2017).
Immunohistochemical staining of pectin residues
P. japonicum infecting A. thaliana was harvested at 0, 24, 48, 72 and 120 hpi for infections on water. For infections on DMSO, 100 nM epiBL (dissolved in DMSO), 0.05 mM Coomassie (plus DMSO) and 5 μM NPA (dissolved in DMSO) treated samples were harvested and at 0 and 72 hpi. The seedlings were fixed in a 1% v/v glutaraldehyde, 4% w/v formaldehyde, 0.05 M NaPi aqueous solution by vacuuming twice for 20 min, followed by overnight incubation at 4 °C. The samples were then dehydrated with an ethanol gradient (30 min in each of 10%, 30%, 50%, 70%, 96%, 100%, 100% v/v ethanol) and incubated overnight in a 1:1 solution of 100% ethanol:Historesin solution (Leica). The solution was exchanged with Historesin and the samples incubated again overnight at 4 °C. The seedlings were then oriented in molds following the method in (Scheres et al. 1994), aligning the haustoria of different seedlings. The shoot was removed, and a 14:1 solution of Historesin and hardener was added to form a hard resin sheet. The haustoria were cross-sectioned at 8 μm thickness using a Microm HM355 S microtome. The sections were rehydrated in PBS, incubated in 0.05 M glycine in PBS for 20 min, and blocked in 2% w/v BSA in PBS (blocking buffer) for 30 min. Three consecutive slides were stained in 1:20 dilutions of LM19, LM20 in PBS or just PBS for the negative control and incubated for 2 h. After rinsing with blocking buffer three times, the sections were incubated for 1 h in a 1:100 dilution of Goat anti-Rat IgG Alexa Fluor 647 secondary antibody. After rinsing three times with PBS, the sections were mounted in PBS and immediately imaged on a Zeiss LSM-780 confocal microscope with 633 nm excitation, 0.5% laser power, 650 gain and 633 to 695 nm detection. Fluorescence was quantified in Fiji using the mean gray value measurement after selecting the area corresponding to the desired tissue on the brightfield channel. Three to fifteen different haustoria were imaged and quantified for each time point and treatment. The same DMSO control was used for epiBL presented in Fig. 5 and Coomassie and NPA presented in Fig. 6. Representative images were processed equally for each panel using the 16-colors LUT to allow easier visualization of fluorescence intensity.
Ruthenium red staining
Ruthenium red staining was performed by dipping infecting roots at 0, 24, 48, 72 and 120 hpi in 0.05% w/v ruthenium red in deionised water for 5 min, followed by rinsing 2 times with deionised water and mounting on 20% v/v glycerol. Pictures were taken using an Axioscope A1 microscope and analysed in Fiji (Schindelin et al. 2012; Rueden et al. 2017).
RNA-Seq datasets and gene accession numbers
The RNA-seq dataset used for gene expression analyses of the infection time course in P. japonicum and A. thaliana is presented in Kokla et al. 2022. The dataset used for gene expression analyses in intrusive and non-intrusive haustorial cells is presented in Ogawa et al. 2021. The heatmaps were generated using the “pheatmap” function in RStudio on the log2 fold change (time-course dataset) or normalised reads (IC vs non-IC dataset) of the genes indicated in each heatmap. The heatmaps were clustered by expression in the time course dataset. Genes IDs and accession numbers for the P. japonicum genes mentioned in the text are available in Supplemental Table S2.
Gene expression analyses
Forty 5-day-old P. japonicum seedlings per biological replicate per treatment were transferred to the starvation medium with 100 μM EGCG, 100 nM epiBL, 5 μM NPA, 0.05 mM Coomassie, or 1 μM NAA for three days or control DMSO. After infecting A. thaliana, 2 mm of root around the haustorium was collected at 0 or 72 h post infection. RNA was extracted using the ROTIPrep RNA MINI kit (Carl Roth, 8485) following the manufacturer's instructions. cDNA was synthesised with the Maxima First Strand cDNA Synthesis Kit for RT-qPCR (ThermoFisher, K1642) following the manufacturer's instructions. RT-qPCR was performed using the Maxima SYBR Green/ROX qPCR Master Mix 2x (Thermo Scientific, K022). P. japonicum SERINE/THREONINE PROTEIN PHOSPHATASE 2A (PjPP2A) was used as normalisation control (Serivichyaswat et al. 2022). For each experiment, three biological replicates and at least two technical replicates were used. The relative gene expression was calculated using the Pfaffl method. The primers used are available in Supplemental Table S3.
Cloning of PjPMEs and PjPMEIs and plasmid construction
All cloning was based on the Greengate cloning method following the standard protocols (Lampropoulos et al. 2013). Greengate plasmids used for cloning have been previously published (Lampropoulos et al. 2013). All primers used for GreenGate cloning are listed in Supplemental Table S3. Digestion and ligation reactions were performed using the BsaI-HFv2 (NEB #R3733) and T4 DNA Ligase (NEB M0202) enzymes respectively. For the overexpression constructs, the CDS of PjPME6, PjPME51, PjPMEI9, PjPMEI6 and PjPMEI10 were amplified using the CloneAmp HiFi PCR Premix (TakaraBio) from the cDNA of P. japonicum and inserted into the entry vector pGGC000 (Addgene plasmid # 48858) to yield pGGC-CDS vectors. The ligated plasmids were amplified in chemically competent Escherichia coli DH5α and confirmed by Sanger sequencing. The final binary vector assembly was performed using pGGA-pMAS, pGGB003 (Addgene plasmid # 48821), pGGC-CDS, pGGD002 (Addgene plasmid # 48834), pGGE-terMAS, pGGF-DsRed and pGGZ001 (Addgene plasmid # 48868). pGGA-pMAS, pGGE-terMAS and pGGF-DsRed were previously published (Kokla et al. 2022). For the reporter constructs, a sequence of ∼3 kb upstream the starting codon was cloned as the promoter of the genes of interest (pGOI). PjPME6 (3087 bp), PjPME51 (2876 bp), PjPME22 (3044 bp), PjPMEI9 (3021 bp), PjPMEI10 (3071 bp) and PjPMEI6 (2988 bp) promoters were amplified using the CloneAmp HiFi PCR Premix (TakaraBio) from the gDNA of P. japonicum and inserted into the entry vector pGGA000 (Addgene plasmid # 48856) to yield pGGA-pGOI vectors. A 2xVenus-NLS sequence was cloned from a previously published GoldenGate vector backbone (Cui et al. 2016) and inserted in the pGGC000 entry vector to create pGGC-2xVenus-NLS. The ligated plasmids were amplified in chemically competent E. coli DH5α and confirmed by Sanger sequencing. The final binary vector assembly was performed using pGGA-pGOI, pGGB003, pGGC-2xVenus-NLS, pGGD002, pGGE001 (Addgene plasmid # 48839), pGGF-DsRed and pGGZ001. The final overexpression and reporter plasmids were co-transformed in electrocompetent Agrobacterium rhizogenes AR1193 with the pSoup plasmid (Addgene plasmid # 165419), and the bacteria cultured in LB broth with 50 μg/ml spectinomycin and 50 μg/ml rifampicin.
P. japonicum hairy root transformation
P. japonicum transformation was performed according to Ishida et al. 2011. Seven-day-old P. japonicum seedlings were sonicated for 10 s and vacuum-infiltrated for 5 min in a solution of AR1193 carrying the construct of interest. The seedlings were then moved to solid B5 medium supplemented with 1% w/v sucrose and 450 μM acetosyringone and kept at 22 °C in the dark for 2 days. Seedlings were then moved to B5 medium containing 300 μg/ml cefotaxime and grown at 25 °C in long day conditions until formation of hairy roots. Transgenic hairy roots were identified through red fluorescence using a Leica M205 FA stereo microscope and placed on starvation medium for 4 days before addition of A. thaliana. Non-fluorescent hairy roots from the same transformation experiment were used as a control for each construct. Counting of haustoria and safranin-O staining were performed at 7 dpi for overexpression constructs. Imaging of transcriptional reporters was performed on 4 dpi haustoria using a Zeiss LSM780 confocal microscope with 514 nm excitation, 2.8% laser power, 950 gain and 519 to 550 nm detection.
Statistics
All experiments were replicated at least three times unless otherwise stated. For infection assays each biological replicate consisted of the average of results from at least 15 plants, and one-tailed Student's t-tests on means were used for single comparisons. For assays with transformed hairy roots overexpressing PjPMEs or PjPMEIs, the data from the biological replicates were pooled and divided in categories of 0, 1 or ≥2 haustoria per hairy root. A Fisher exact test was then used to calculate significance. For RT-qPCR data, one-tailed Student's t-tests on biological replicates were used for single comparisons of treatment vs control. For ruthenium red staining, one replicate was performed with 10 to 20 plants per time point and treatment. For antibody staining assays, two replicates were performed for each time point and treatment. The quantifications from each replicate were pooled together and one-tailed Student's t-tests were used for single or multiple comparisons. The p-values for multiple comparisons were adjusted using the Bonferroni correction.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers listed in Supplemental Table S2.
Supplementary Material
Acknowledgments
We thank Sebastian Wolf for providing the AtPMEI5OE seeds, Thomas Spallek for providing the GoldenGate plasmid containing the 3xVenus-NLS sequence and Judith Lundberg-Felten for kindly donating the LM19 and LM20 antibodies.
Contributor Information
Martina Leso, Department of Plant Biology, Linnean Center for Plant Biology, Swedish University of Agricultural Sciences, Almas allé 5, 756 51 Uppsala, Sweden.
Anna Kokla, Department of Plant Biology, Linnean Center for Plant Biology, Swedish University of Agricultural Sciences, Almas allé 5, 756 51 Uppsala, Sweden.
Ming Feng, Department of Plant Biology, Linnean Center for Plant Biology, Swedish University of Agricultural Sciences, Almas allé 5, 756 51 Uppsala, Sweden.
Charles W Melnyk, Department of Plant Biology, Linnean Center for Plant Biology, Swedish University of Agricultural Sciences, Almas allé 5, 756 51 Uppsala, Sweden.
Author contributions
ML and CWM conceived the experiments. ML, AK and MF performed the experiments. ML and CWM wrote the manuscript. All authors revised the final manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. A. thaliana PMEs and PMEIs are differentially expressed during haustorium development
Supplemental Figure S2. PME activity is increased during haustorium development
Supplemental Figure S3. PjPMEs and PjPMEIs are specific to haustoria
Supplemental Figure S4. PjPME and PjPMEI overexpression does not affect xylem connection to the host
Supplemental Figure S5. A. thaliana BR signalling mutants do not affect parasitism efficiency
Supplemental Figure S6. NAA treatment does not affect parasitism efficiency
Supplemental Table S1. List of identified PjPMEs and PjPMEIs
Supplemental Table S2. P. japonicum gene accession numbers and IDs
Supplemental Table S3. Primers used in the study
Supplemental Video S1. Confocal Z-stack of a haustorium expressing PjPMEI9-2xVenus::NLS
Funding
M.L., M.F., and C.W.M. were supported by a European Research Council starting grant (GRASP-805094). A.K. and C.W.M were supported by a Wallenberg Academy Fellowship from the Knut och Alice Wallenbergs Stiftelse (2016-0274). M.F. and C.W.M were supported by a Svenska Forskningsrådet Formas Grant (2018-00533).
References
- Aoki N, Cui SK, Yoshida S. Cytokinins induce prehaustoria coordinately with quinone signals in the parasitic plant striga hermonthica. Plant Cell Physiol. 2022:63(10):1446–1456. 10.1093/pcp/pcac130 [DOI] [PubMed] [Google Scholar]
- Ben-Hod G, Losner D, Joel DM, Mayer AM. Pectin methylesterase in calli and germinating seeds of orobanche aegyptiaca. Phytochemistry. 1993:32(6):1399–1402. 10.1016/0031-9422(93)85145-H [DOI] [Google Scholar]
- Birschwilks M, Haupt S, Hofius D, Neumann S. Transfer of phloem-mobile substances from the host plants to the holoparasite Cuscuta sp. J Exp Bot. 2006:57(4):911–921. 10.1093/jxb/erj076 [DOI] [PubMed] [Google Scholar]
- Chang M, Lynn DG, Netzly DH, Butler LG. Chemical regulation of distance: characterization of the first natural host germination stimulant for striga asiatica. J Am Chem Soc. 1986:108(24):7858–7860. 10.1021/ja00284a074 [DOI] [PubMed] [Google Scholar]
- Cui S, Kubota T, Nishiyama T, Ishida JK, Shigenobu S, Shibata TF, Toyoda A, Hasebe M, Shirasu K, Yoshida S. Ethylene signaling mediates host invasion by parasitic plants. Sci Adv. 2020:6(44):eabc2385. 10.1126/sciadv.abc2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Wada S, Tobimatsu Y, Takeda Y, Saucet SB, Takano T, Umezawa T, Shirasu K, Yoshida S. Host lignin composition affects haustorium induction in the parasitic plants phtheirospermum japonicum and striga hermonthica. New Phytol. 2018:218(2):710–723. 10.1111/nph.15033 [DOI] [PubMed] [Google Scholar]
- Cui S, Wakatake T, Hashimoto K, Saucet SB, Toyooka K, Yoshida S, Shirasu K. Haustorial hairs are specialized root hairs that support parasitism in the facultative parasitic plant, phtheirospermum japonicum. Plant Physiol. 2016:170(3):1492–1503. 10.1104/pp.15.01786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher FB, Braybrook SA. How to let go: pectin and plant cell adhesion. Front Plant Sci. 2015:6:523. 10.3389/fpls.2015.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie B, Dirk LMA, Hadfield KA, Wilkins TA, Bennett AB, Bradford KJ. A gel diffusion assay for quantification of pectin methylesterase activity. Analy Biochem. 1998:264(2):149–157. 10.1006/abio.1998.2847 [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER Web server: interactive sequence similarity searching. Nucleic Acids Res. 2011:39(suppl):29–37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012:40(D1):1178–1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyet V, Billard E, Pouvreau JB, Lechat MM, Pelletier S, Bahut M, Monteau F, Spíchal L, Delavault P, Montiel G, et al. Haustorium initiation in the obligate parasitic plant phelipanche ramosa involves a host-exudated cytokinin signal. J Exp Bot. 2017:68(20):5539–5552. 10.1093/jxb/erx359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabber JH, Panciera MT, Hatfield RD. Chemical composition and enzymatic degradability of xylem and nonxylem walls isolated from alfalfa internodes. J Agric Food Chem. 2002:50(9):2595–2600. 10.1021/jf011598c [DOI] [PubMed] [Google Scholar]
- Heide-Jørgensen HS, Kuijt J. The haustorium of the root parasite triphysaria (Scrophulariaceae), with special reference to xylem bridge ultrastructure. Am J Bot. 1995:82(6):782–797. 10.1002/j.1537-2197.1995.tb15691.x [DOI] [Google Scholar]
- Hewezi T, Howe P, Maier TR, Hussey RS, Mitchum MG, Davis EL, Baum TJ. Cellulose binding protein from the parasitic nematode heterodera Schachtii interacts with arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism. Plant Cell. 2008:20(11):3080–3093. 10.1105/tpc.108.063065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honaas LA, Wafula EK, Yang Z, Der JP, Wickett NJ, Altman NS, Taylor CG, Yoder JI, Timko MP, Westwood JH, et al. Functional genomics of a generalist parasitic plant: laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol. 2013:13(1):1–19. 10.1186/1471-2229-13-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME, Condon JM, Timko MP, Riopel JL. Primary haustorial development of striga asiatica on host and nonhost species. Phytopathology. 1998:88(1):70–75. 10.1094/PHYTO.1998.88.1.70 [DOI] [PubMed] [Google Scholar]
- Ichihashi Y, Kusano M, Kobayashi M, Suetsugu K, Yoshida S, Wakatake T, Kumaishi K, Shibata A, Saito K, Shirasu K. Transcriptomic and metabolomic reprogramming from roots to haustoria in the parasitic plant, Thesium chinense. Plant Cell Physiol. 2018:59(4):724–733. 10.1093/pcp/pcx200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida JK, Wakatake T, Yoshida S, Takebayashi Y, Namba S, Wafula E, DePamphilis CW, Kasahara H, Shirasu K. Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant phtheirospermum japonicum. Plant Cell. 2016:28(8):1795–1814. 10.1105/tpc.16.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida JK, Yoshida S, Ito M, Namba S, Shirasu K. Agrobacterium Rhizogenes-mediated transformation of the parasitic plant phtheirospermum japonicum. PLoS One. 2011:6(10):1–8. 10.1371/journal.pone.0025802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhu MY, Farhi M, Wang L, Zumstein K, Sinha NR. Investigating host and parasitic plant interaction by tissue-specific gene analyses on tomato and Cuscuta campestris interface at three haustorial developmental stages’. Front Plant Sci. 2022:12:764843. 10.3389/fpls.2021.764843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhu MY, Ichihashi Y, Farhi M, Wong C, Sinha NR. LATERAL ORGAN BOUNDARIES DOMAIN 25 functions as a key regulator of haustorium development in dodders. Plant Physiol. 2021:186(4):2093–2110. 10.1093/plphys/kiab231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson K, El-Ahmad M, Friemann R, Jörnvall H, Markovič O, Eklund H. Crystal structure of plant pectin methylesterase. FEBS Lett. 2002:514(2-3):243–249. 10.1016/S0014-5793(02)02372-4 [DOI] [PubMed] [Google Scholar]
- Jonsson K, Lathe RS, Kierzkowski D, Routier-Kierzkowska AL, Hamant O, Bhalerao RP. Mechanochemical feedback mediates tissue bending required for seedling emergence. Curr Biol. 2021:31(6):1154–1164.e3. 10.1016/j.cub.2020.12.016 [DOI] [PubMed] [Google Scholar]
- Kokla A, Leso M, Zhang X, Simura J, Serivichyaswat PT, Cui SK, Ljung K, Yoshida S, Melnyk CW. Nitrogen represses haustoria formation through abscisic acid in the parasitic plant phtheirospermum japonicum. Nat Commun. 2022:13(1):2976. 10.1038/s41467-022-30550-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotani K, Wakatake T, Ichihashi Y, Okayasu K, Yu SW, Ogawa S, Cui SK, Suzuki T, Shirasu K, Notaguchi M. Host-parasite tissue adhesion by a secreted type of beta-1,4-glucanase in the parasitic plant phtheirospermum japonicum. Commun Biol. 2020:3(1):407. 10.1038/s42003-020-01143-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachowiec J, Lemus T, Thomas JH, Murphy PJM, Nemhauser JL, Queitsch C. The protein chaperone HSP90 can facilitate the divergence of gene duplicates. Genetics. 2013:193(4):1269. 10.1534/genetics.112.148098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lairez D, Cathala B, Monties B, Bedos-Belval F, Duran H, Gorrichon L. Aggregation during coniferyl alcohol polymerization in pectin solution: a biomimetic approach of the first steps of lignification. Biomacromolecules. 2005:6(2):763–774. 10.1021/bm049390y [DOI] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J. GreenGate - A novel, versatile, and efficient cloning system for plant transgenesis. PLoS One. 2013:8(12);e83043. 10.1371/journal.pone.0083043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laohavisit A, Wakatake T, Ishihama N, Mulvey H, Takizawa K, Suzuki T, Shirasu K. Quinone perception in plants via leucine-rich-repeat receptor-like kinases. Nature. 2020:587(7832):92–97. 10.1038/s41586-020-2655-4 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Biller S, Stanley K, Kajstura T, Prusty R. Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 2006:47(6):788–792. 10.1093/pcp/pcj043 [DOI] [PubMed] [Google Scholar]
- Lewis KC, Selzer T, Shahar C, Udi Y, Tworowski D, Sagi I. Inhibition of pectin methyl esterase activity by green tea catechins. Phytochemistry. 2008:69(14):2586–2592. 10.1016/j.phytochem.2008.08.012 [DOI] [PubMed] [Google Scholar]
- Liu N, Shen G, Xu Y, Liu H, Zhang J, Li S, Li J, Zhang C, Qi J, Wang L, et al. Extensive inter-plant protein transfer between Cuscuta parasites and their host plants. Mol Plant. 2020:13(4):573–585. 10.1016/j.molp.2019.12.002 [DOI] [PubMed] [Google Scholar]
- Losner-Goshen D, Portnoy VH, Mayer AM, Joel DM. Pectolytic activity by the haustorium of the parasitic plant orobanche L. (Orobanchaceae) in host roots. Ann Bot. 1998:81(2):319–326. 10.1006/anbo.1997.0563 [DOI] [Google Scholar]
- Louvet R, Cavel E, Gutierrez L, Guénin S, Roger D, Gillet F, Guerineau F, Pelloux J. Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta. 2006:224(4):782–791. 10.1007/s00425-006-0261-9 [DOI] [PubMed] [Google Scholar]
- Markovic O, Janecek S. Pectin methylesterases: sequence-structural features and phylogenetic relationships. Carbohydr Res. 2004:339:2281–2295. 10.1016/j.carres.2004.06.023 [DOI] [PubMed] [Google Scholar]
- Nagar R, Singh M, Sanwal GG. Cell-Wall degrading enzymes in Cuscuta-Reflexa and its hosts’. J Exp Bot. 1984:35(8):1104–1112. 10.1093/jxb/35.8.1104 [DOI] [Google Scholar]
- Nickrent DL. Parasitic angiosperms: how often and how many? Taxon. 2020:69(1):5–27. 10.1002/tax.12195 [DOI] [Google Scholar]
- Ogawa S, Wakatake T, Spallek T, Ishida JK, Sano R, Kurata T, Demura T, Yoshida S, Ichihashi Y, Schaller A, et al. Subtilase activity in intrusive cells mediates haustorium maturation in parasitic plants. Plant Physiol. 2021:185(4):1381–1394. 10.1093/plphys/kiaa001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen S, Krause K. Activity of xyloglucan endotransglucosylases/hydrolases suggests a role during host invasion by the parasitic plant Cuscuta reflexa. PLoS One. 2017:12(4):e0176754. 10.1371/journal.pone.0176754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen S, Striberny B, Hollmann J, Schwacke R, Popper Z, Krause K. Getting ready for host invasion: elevated expression and action of xyloglucan endotransglucosylases/hydrolases in developing haustoria of the holoparasitic angiosperm Cuscuta. J Exp Bot. 2016:67(3):695–708. 10.1093/jxb/erv482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloux J, Rustérucci C, Mellerowicz EJ. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007:12(6):267–277. 10.1016/j.tplants.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Peng ZY, Alique D, Xiong YY, Hu JR, Cao XW, Lu SQ, Long M, Wang Y, Wabnik K, Jiao YL. Differential growth dynamics control aerial organ geometry. Curr Biol. 2022:32(22):4854. -+. 10.1016/j.cub.2022.09.055 [DOI] [PubMed] [Google Scholar]
- Raiola A, Lionetti V, Elmaghraby I, Immerzeel P, Mellerowicz EJ, Salvi G, Cervone F, Bellincampi D. Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol Plant Microbe Interact. 2011:24(4):432–440. 10.1094/MPMI-07-10-0157 [DOI] [PubMed] [Google Scholar]
- Rodenburg J, Demont M, Zwart SJ, Bastiaans L. Parasitic weed incidence and related economic losses in rice in Africa. Agric Ecosyst Environ. 2016:235:306–317. 10.1016/j.agee.2016.10.020 [DOI] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. Imagej2: imageJ for the next generation of scientific image data. BMC Bioinformatics. 2017:18(1):529. 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P. Embryonic origin of the Arabidopsis primary root and root-meristem initials. Development. 1994:120(9):2475–2487. 10.1242/dev.120.9.2475 [DOI] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012:9(7):676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serivichyaswat PT, Bartusch K, Leso M, Musseau C, Iwase A, Chen Y, Sugimoto K, Quint M, Melnyk CW. High temperature perception in leaves promotes vascular regeneration and graft formation in distant tissues. Development. 2022:149(5):dev200079. 10.1242/dev.200079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid S, Kim G, Johnson NR, Wafula E, Wang F, Coruh C, Bernal-Galeano V, Phifer T, Depamphilis CW, Westwood JH, et al. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature. 2018:553(7686):82–85. 10.1038/nature25027 [DOI] [PubMed] [Google Scholar]
- Spallek T, Melnyk CW, Wakatake T, Zhang J, Sakamoto Y, Kiba T, Yoshida S, Matsunaga S, Sakakibara H, Shirasu K. Interspecies hormonal control of host root morphology by parasitic plants. Proc Natal Acad Sci U S A. 2017:114(20):5283–5288. 10.1073/pnas.1619078114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA11 Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021:38(7):3022–3027. 10.1093/molbev/msab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valette-Collet O, Cimerman A, Reignault P, Levis C, Boccara M, Umr PV, National I, Paris A, De Biochimie L. Disruption of Botrytis cinerea pectin methylesterase gene bcpme1 reduces virulence on several host plants. Mol Plant Microbe Interact. 2003:16(4):360–367. 10.1094/MPMI.2003.16.4.360 [DOI] [PubMed] [Google Scholar]
- Verhertbruggen Y, Marcus SE, Haeger A, Ordaz-Ortiz JJ, Knox JP. An extended set of monoclonal antibodies to pectic homogalacturonan’. Carbohydrate Res. 2009:344(14):1858–1862. 10.1016/j.carres.2008.11.010 [DOI] [PubMed] [Google Scholar]
- Veronesi C, Bonnin E, Calvez S, Thalouarn P, Simier P. Activity of secreted cell wall-modifying enzymes and expression of peroxidase-encoding gene following germination of orobanche ramosa. Biologia Plantarum. 2007:51(2):391–394. 10.1007/s10535-007-0084-y [DOI] [Google Scholar]
- Vicente CSL, Nemchinov LG, Mota M, Eisenback JD, Kamo K, Vieira P. Identification and characterization of the first pectin methylesterase gene discovered in the root lesion nematode pratylenchus penetrans. PLoS One. 2019:14(2):e0212540. 10.1371/journal.pone.0212540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsman G, Zhang JY, Moreno-Risueno MA, Anderson CT, Benfey PN. Cell wall remodeling and vesicle trafficking mediate the root clock in Arabidopsis. Science. 2020:370(6518):819. 10.1126/science.abb7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatake T, Ogawa S, Yoshida S, Shirasu K. Auxin transport network underlies xylem bridge formation between the hemi-parasitic plant phtheirospermum japonicum and host Arabidopsis. Development. 2020:147(14):dev187781. 10.1242/dev.187781 [DOI] [PubMed] [Google Scholar]
- Wakatake T, Yoshida S, Shirasu K. Induced cell fate transitions at multiple cell layers configure haustorium development in parasitic plants. Development. 2018:145:dev164848. 10.1242/dev.164848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yuan D, Gao W, Li Y, Tan J, Zhang X. A comparative genome analysis of PME and PMEI families reveals the evolution of pectin metabolism in plant cell walls. PLoS One. 2013:8:e72082. 10.1371/journal.pone.0072082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010:15(4):227–235. 10.1016/j.tplants.2010.01.004 [DOI] [PubMed] [Google Scholar]
- Wolf S, Mravec J, Greiner S, Mouille G, Höfte H. Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol. 2012:22(18):1732–1737. 10.1016/j.cub.2012.07.036 [DOI] [PubMed] [Google Scholar]
- Yang Z, Wafula EK, Honaas LA, Zhang H, Das M, Fernandez-Aparicio M, Huang K, Bandaranayake PCG, Wu B, Der JP, et al. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol Biol Evol. 2015:32(3):767–790. 10.1093/molbev/msu343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YH, Wang ZY, Mora-Garcia S, Li JM, Yoshida S, Asami T, Chory J. BES1 Accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002:109(2):181–191. 10.1016/S0092-8674(02)00721-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.