Abstract
Green algae display a wide range of extracellular matrix (ECM) components that include various types of cell walls (CW), scales, crystalline glycoprotein coverings, hydrophobic compounds, and complex gels or mucilage. Recently, new information derived from genomic/transcriptomic screening, advanced biochemical analyses, immunocytochemical studies, and ecophysiology has significantly enhanced and refined our understanding of the green algal ECM. In the later diverging charophyte group of green algae, the CW and other ECM components provide insight into the evolution of plants and the ways the ECM modulates during environmental stress. Chlorophytes produce diverse ECM components, many of which have been exploited for various uses in medicine, food, and biofuel production. This review highlights major advances in ECM studies of green algae.
Advances Box.
Expanded sequencing and mining of genomes and transcriptomes of diverse green algal taxa has provided key insight into the biosynthesis and modulation of ECM components as well as evolutionary links to land plants.
Synthesis of field investigations and ecophysiology of specific taxa with molecular and biochemical studies has elucidated changes in ECM structure and dynamics during environmental stress.
Enhanced exploration of green algal ECM for applied uses in food, medicine, nutraceutical, and biofuel industries has catalyzed advanced biochemical analyses of the ECM.
Application of novel immunochemical and microscopy-based technologies has shed light on the complex structural designs and development of ECM components.
Outstanding Questions Box.
What is the composition of the CW proteome and what are the functions of proteins, including AGP, extensin, and expansin, of green algae?
What are the cytokinetic mechanisms involved in the ZCC clade of charophytes, including furrowing and cell plate/phragmoplast formation, and the CW polymers required for completion of cytokinesis?
What are the biosynthetic pathways, secretion dynamics, and functional roles of hydrophobic components of the ECM (e.g. algaenan, sporopollenin, phenolics, lipid coverings) of green algae?
How do the ECMs modulate in response to abiotic and biotic stress, and what are the roles of specific components such as sulfated polysaccharides and EPS gels?
What culturing, harvesting, and processing technologies will be optimal for efficacious use of green algae and their ECMs in the food, medicine, biofuels, and other industries?
Introduction
Green algae comprise a diverse assemblage of organisms that are part of the green lineage (Chloroplastida) of the Archaeplastida (i.e. green algae and plants; Adl et al. 2019; Bowles et al. 2022). These algae arose over 1 billion years ago, and their extant taxa now occupy most photic habitats of the planet, where they play major roles in the dynamics of the atmosphere, biogeochemistry, and food chains. Over 500 million years ago, an ancestor of the modern-day green algal group, the Zygnematophyceae (Charophyta or Streptophyta; Delwiche and Cooper 2015; Rensing 2020; Schreiber et al. 2022), successfully transitioned from a freshwater to a terrestrial habitat and ultimately gave rise to land plants, an event that profoundly changed the natural history of the planet (de Vries and Archiblad 2018; Fürst-Jansen et al. 2020; Permann et al. 2022). Green algae display a truly diverse range of phenotypes that include unicellular picoplankton (e.g. Ostreococcus), unicellular and colonial flagellates (e.g. Chlamydomonas, Volvox), filaments (e.g. Ulothrix, Spirogyra), coenocytic siphons (e.g. Acetabularia), and distinct 3-dimensional thalli (e.g. Ulva, Chara, Coleochaete). Green algae are major primary producers of marine, freshwater, and terrestrial habitats, may form spectacular blooms, are common residents of aquatic biofilms and may even form unique symbiotic associations with fungi (e.g. lichens) and various invertebrates. Central to their structure, development, physiology, and evolution is an extracellular matrix (ECM) that may manifest as a cell wall (CW), crystalline glycoprotein coverings, layers of scales, hydrophobic compounds, and/or a gel-like, extracellular polymeric substances (EPS) (Fig. 1). Green algal ECMs have long been studied by plant biologists (for reviews, see Popper et al. 2011; Domozych et al. 2012), and recently, detailed data acquired using molecular biology, immunology, high-resolution microscopy imaging, and ecophysiology have provided novel insight into our understanding of their structural architecture, biosynthesis, and stress-related modulations. Likewise, studies of green algae and their ECMs have been fueled by their multiple uses in human economy (e.g. food, medicines; Mišurcová et al. 2012; Roleda and Heesch 2021; Li et al. 2022) and, most recently, as sources of degradable biomass for production of biofuels (Soni et al. 2021). This update highlights recent major advances in ECM biology of the green algae.
Figure 1.
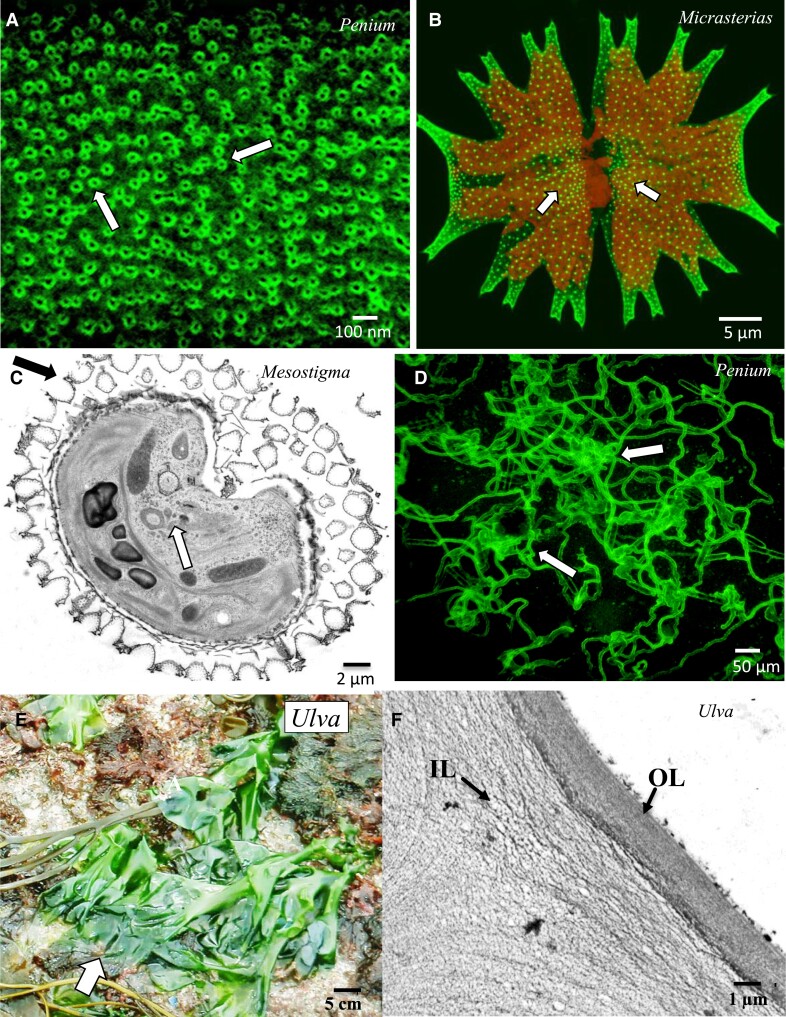
Examples of ECM diversity in green algae. A) The late-divergent charophyte Penium margaritaceum possesses a unique Ca2+complex pectin lattice (arrows) in its CW. Labeled with the HG-binding mAb JIM5. Fluorescence light microscopy (FLM). B) The CW of the late-divergent charophyte Micrasterias radiata possesses pores labeled with the AGP-specific mAb JIM13 (arrows, FLM). C) In the early-divergent charophyte Mesostigma viridie, multiple layers of scales cover the surface of motile cells (arrow). TEM image. D) The gliding trails of the EPS of P. margaritaceum (arrows). Labeled with 0.75 fluorescent beads (FLM). E) The Ulvophyte Ulva (arrow) is a common coastal seaweed whose CW polysaccharides have been extracted for multiple uses. F) The CW consists of an inner (IL) and outer (OL) layer and consists of cellulose, XyG, glucuronan, and the sulfated polysaccharide, ulvan (TEM).
The ECM of green algae: an overview
Green algae make up an estimated 22,000 extant species (Guiry 2012) that are currently classified in 3 phyla (Leliaert et al. 2012; Leliaert 2019; Li et al. 2020): (1) Streptophyta (also called Charophyta or basal Streptophyta; Mattox and Stewart 1984; Becker and Marin 2009); (2) Prasinodermophyta; and (3) Chlorophyta. It is important to note that our understanding of the taxonomy and phylogeny of green algae is continuously evolving as more information derived from genomic/transcriptomic sequencing and enhanced screening of more diverse assemblages of taxa have become available. For more up-to-date and detailed information, readers should consult Leliaert (2019), Li et al. (2021), Bachy et al. (2022), and Hess et al. (2022). Likewise, Table 1 lists those green algae whose genome/transcriptome have been investigated and may serve as valuable resources for elucidating multiple aspects of green algal biology.
Table 1.
Genome and transcriptome resources for ECM studies of green algae
aIndicates only transcriptome data available.
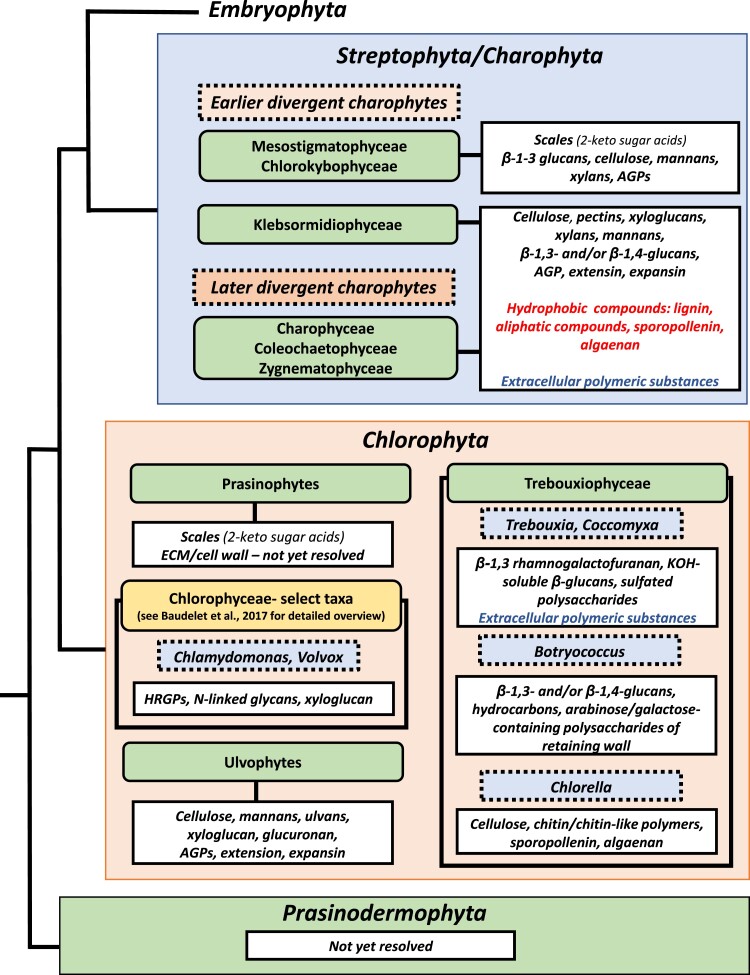
Much of what is known about the ECMs of green algae is based on biochemical, cellular, and molecular studies of small samples of select taxa. It is clear that we are only in an infancy stage of fully understanding this great diversity in ECM structures, their functions, and their development. Yet, what we do know paints a fascinating picture of how ECMs are critical to survival in vastly different ecosystems, modulations in response to environmental stress, and the evolution of modern-day green algae and plants. This review describes 5 major ECM types found in green algae based on both their structural/biochemical features (Text Box 1) and taxonomic/phylogenetic relationships (Fig. 2). These include scale coverings, glycoprotein coverings, hydrophobic components, EPS, and CWs. Although it is recognized that this grouping of ECM types is somewhat artificial, it provides a basis for our current understanding of the green algal ECM and a stimulus for future research.
Text Box 1.
The ECM components of the green algae
The ECM components of green algae can be loosely classified into 5 major types:
Scale coverings are commonly found on the plasma membrane/flagellar membranes of some unicellular taxa (e.g. prasinophytes and the charophyte, Mesostigma) and motile zoospores and sperm cells (e.g. the charophytes, Chara, Coleochaete).
Glycoprotein coverings that are highlighted by large amounts of the amino acid hydroxyproline are found in several chlorophyte taxa, most notably in unicellular and colonial flagellates including Chlamydomonas and Volvox.
Hydrophobic ECM components that include sporopollenin, algaenan, and lipid-like cuticle-like compounds have been identified throughout the chlorophytes and charophytes, including resting cell and spore coatings.
EPS consists of highly complex polysaccharide and protein components secreted beyond the CWs that often form gels/mucilages are found throughout the green algae.
CWs consisting of a framework of polysaccharide fibrils (e.g. cellulose, mannans, xylans) that are embedded in a matrix of polysaccharides and glycosylated proteins are exhibited in charophytes and chlorophytes.
Figure 2.
Summary of major ECM components found in the major groups of green algae.
Scales
Samples taken from the photic planktonic zones of most ocean waters will often contain prasinophytes. The Prasinophyceae comprise a paraphyletic assemblage of unicellular, mostly marine organisms that are believed to be the earliest divergent Chlorophyta (Bachy et al. 2022). Many taxa are distinguished by the presence of 1 or more layers of scales attached to the plasma membrane and/or flagellar membrane (Moestrup and Walne 1979; Perasso et al. 2000). Scale chemistry is highlighted by the 2-keto sugar acids, deoxy-5-O-methyl-manno-2-octulosonic acid and deoxy-lyxo-2-heptulosaric acid (Becker et al. 1994). The various shaped and sized scales are synthesized in the Golgi apparatus and are subsequently released onto the cell surface in a process that may include a vacuole-like scale reservoir. The 2-keto sugar acids found in scales are also found in the thecal covering of the chlorophyte, Tetraselmis (Becker et al. 1998; Kermanshahi-Pour et al. 2014) and in the spectacular basket scales of the early divergent charophyte Mesostigma viride (Domozych et al. 1991). Recently, biosynthesis and transport genes for KdO were identified in Mesostigma (Liang et al. 2019). It is also important to note that the zoospores and sperm cells of many green algae, including the charophytes Chlorokybus, Coleochaete, and Chara, contain scales, but their chemistry and functional roles are presently unknown.
A long-standing question concerning prasinophytes is whether they produce ECM components other than scales during different life cycle stages. Although no biochemical data are currently available, molecular studies have provided some initial insight (Worden et al. 2009; Blanc-Mathieu et al. 2014; Satjarak and Graham 2017). In a screening of the Pyramimonas parkeae genome for carbohydrate-active enzymes (CAZYmes), sequences that cluster with bacterial genes that encode cellulose synthases, including regions coding for domains of plant cellulose synthases, were identified (Satjarak and Graham 2017). Genes involved in pectin and xyloglucan (XyG) processing were also identified, including putative galacturonosyl-transferases, pectin acetyl esterases, pectin-degrading enzymes, and ß-galactosidase and xylosidase.
Glycoprotein coverings
Chlamydomonas, Volvox, and other related flagellates are commonly encountered chlorophytes of aquatic and terrestrial ecosystems. The ECMs of these organisms are comprised of distinct, hydroxyproline-rich glycoproteins (HRGP) that form a thin CW-like covering and, in colonial forms, an extensive matrix that holds the cells in place in the colony. The CW of the unicellular Chlamydomonas reinhardtii consists of an insoluble framework and about 20 hydroxyproline (Hyp)-containing, chaotrope-soluble glycoproteins (Lee et al. 2007; Voight et al. 2010; Barolo et al. 2022). A detailed, bioinformatics-based comparison of Chlamydomonas CW HRGPs with plant extensins may be found in Liu et al. (2016). Recently, a CW integrity–monitoring mechanism has been described in Chlamydomonas that senses osmotic stress and mechanical defects of its CWs and subsequently regulates CW-gene expression (Cronmiller et al. 2019). The colonial Volvox ECM consists of a diverse array of HRGPs (Sumper and Hallmann 1998). Its ECM matrix contains specific cell-type HRGPs, the pherophorins, that are central to the developmental dynamics of the colony (Hallmann 2006; von der Heyde and Hallmann 2022). In addition to the HRGP components of the ECM, Chlamydomonas also secretes complex N-linked glycans. Xylosyltransferases that heterogeneously xylosylate these glycans were recently described (Lucas et al. 2021). Elucidation of the biochemistry, genetics, and development of the ECMs of Chlamydomonas and Volvox has been critical in that they are central to understanding the biology of these valuable model organisms employed in studies of algal/plant stress biology, sexual reproduction, photosynthesis, organelle biosynthesis, circadian rhythms, and phototaxis (Matt and Umen 2016; Salomé and Merchant 2019; Umen 2020).
Hydrophobic components: Sporopollenin, algaenan, lignin, cuticles
The ECM of diverse taxa of the Chlorophyta and Charophyta possess hydrophobic components, including sporopollenin, algaenan, lignin, and other molecules. Sporopollenin is a highly resistant and complex heteropolymer partly composed of hydroxylated polyketides (Li et al. 2019) that is commonly found in the outer walls of pollen grains and land plant spores. Algaenan is a nonhydrolyzable aliphatic biopolymer composed of long-chain, even carbon–numbered, ω9-unsaturated ω-hydroxy fatty acid monomers varying in chain length from 30 to 34 carbon atoms (Rodrigues and de Silva Bon 2011). These are often ester-linked to form long, highly resistant polymers. Algaenan and sporopollenin have been identified in the spore walls of some charophytes (Niklas et al. 2017; de Vries and Archibald 2018; Permann et al. 2021a) and in the CWs of chlorophytes (Rodrigues and de Silva Bon 2011; Bachy et al. 2022). Polyketide synthase (PKS) and anther-specific chalcone synthase-like (ASCL) are fundamental players in the biosynthetic pathway of hydroxylated polyketides that serve as the precursors for sporopollenin (Suh and Ashton 2022). Screening of the genomes of the zygnematophytes Spirogloea muscicola, Mesotaenium endlicherianum (Cheng et al. 2019a, 2019b), and Penium margaritaceum (Jaio et al. 2020) has revealed putative PKSs, but they do not possess the recognized ASCL-specific active site residues (Suh and Ashton 2022).
Various phenolic compounds have been identified in charophytes (de Vries et al. 2017), including lignin in the CWs of later-divergent taxa (Sørensen et al. 2010, 2011). These and other phenolic compounds are products of the phenylpropanoid pathway (Rieseberg et al. 2023). Homologs of phenylalanine lyase, a key enzyme in phenylpropanoid synthesis, and other core phenylpropanoid biosynthetic genes have been identified in Klebsormidium nitens and Chara braunii (de Vries et al. 2017; Rieseberg et al. 2023) but were not found in P. margaritaceum (Jaio et al. 2020). This suggests a complex evolutionary history in the production of soluble lignin-like compounds, which are deposited into the CW of some charophytes.
Many phenolic compounds are employed in plants to enhance the structure and defense of the CW from abiotic and biotic stress, act as antioxidants, protect against UV radiation, and contribute to the regulation of water retention, for example, homoiohydry (Popper et al. 2011; de Vries and Archibald 2018; Kumar et al. 2020). For example, waterproof aromatic substances were detected by RAMAN spectroscopy in zygospores of some zygnematophytes (e.g. Permann et al. 2021a, 2021b; Permann et al. 2022). An array of phenolics (e.g. tannin-like compounds, flavonoids) have also been isolated and characterized in several charophytes (Aigner et al. 2013; Pichrtová et al. 2013; Permann et al. 2022). These studies have also described their importance in the ecophysiology of certain charophytes (e.g. Zygogonium) as well as in the overall health of glaciers (Dial et al. 2018; Davies et al. 2022). Most of these phenolics have been located in vacuoles or are released beyond the ECM. However, the discovery of an EPS-based “sunscreen” in the zygnematophyte Serritaenia (Busch and Hess (2022) that acts as a photoprotective and whose production is stimulated by UVB light suggests that some are closely associated with the ECM.
Recent interest has emerged as to the putative presence of aliphatic polymers in the ECM of some charophytes and their implications on the evolutionary origin of hydrophobic coverings in land plants (e.g. cutin, suberin), including the cuticle of epidermal cells and the endodermis of roots (Niklas et al. 2017; Philippe et al. 2020). Kondo et al. (2016) described a cuticle-like hydrophobic layer composed of lipids and glycoproteins in the ECM of Klebsormidium flaccidum. The lipid constituents of this layer, though, were notably different from the cutin polymers typically found in land plant cuticles. Screening of the P. margaritaceum genome and those of other late-divergent charophytes (Jiao et al. 2020) revealed homologs of genes for cuticle biosynthetic, transport, assembly frameworks (e.g. cutin synthase and BODYGUARD) that are known in Arabidopsis thaliana. However, other gene families involved in cuticle formation in land plants were not identified. It was concluded that phylogenetic distribution of these genes is consistent with stepwise expansion and neofunctionalization of ancient core lipid biosynthetic machinery to synthesize structural lipid precursors.
EPS: a future challenge for glycomics and proteomics
Many green algae produce an EPS that is secreted beyond their CWs (Boney 1981; Gerrath 1993; Domozych and Bagdan 2022). In many taxa, especially charophytes (e.g. zygnematophytes), the EPS manifests in distinct gel-like mucilages that surround the cell. EPS is produced by both planktonic and sessile green taxa and in certain cases, in very large amounts (e.g. massive amounts of EPS during spring ephemeral blooms by filamentous zygnematophytes; Domozych and Domozych 2007). EPS has multiple diverse functions in charophytes, including water retention, buoyancy, gliding motility, sexual reproduction, and contribution to the matrix of biofilms. In those charophytes whose EPS has been analyzed, highly-branched anionic polysaccharides with large amounts of fucose, galactose (Gal), and glucuronic acid (GlcA), with variable levels of sulfate and with associated proteins, have been described (Kiemle et al. 2007; Domozych and Bagdan 2022).
Many chlorophytes also secrete EPS beyond the cell. For example, some Chlorella species produce a large and complex ECM beyond the CW that provides herbicide and antibiotic resistance (Nazos et al. 2020). Spribille et al. (2020) described an “extracellular interaction matrix,” that is, a gelatinous-like zone beyond the CWs, that harbors nonphotosynthesizing organisms that are not traditionally recognized as lichen symbionts. EPS has also been examined as a possible remediation agent for microplastics deposited in marine and freshwater ecosystems (Cunha et al. 2019). Information concerning EPS biochemistry and function in chlorophytes is available elsewhere (Xiao and Zheng 2016; LaRoche 2022).
Much of what we know about EPS in green algae remains a major mystery, especially considering that EPS production requires large amounts of the alga's photosynthate and the subcellular secretory machinery. Clearly, detailed glycomics- and proteomics-based analyses will be required to understand the structural and functional roles of these ECM components.
CWs: Biochemical diversity and the origin of land plants
The CW represents the most well-studied type of ECM component of plants and green algae. Plant-like CWs are found in the later divergent charophytes and are considered to have been critical in the transition to terrestrial ecosystems and the evolution of land plants. Fibrillar matrix–based CWs as well as CWs with distinct polysaccharide and hydrophobic profiles are also found in the Chlorophyta. Here we present an update on recent discoveries in the CW biology of green algae.
The CWs of charophytes
The charophytes consists of 2 main paraphyletic clades: the earlier-diverging KCM-grade (Klebsormidiophyceae, Chlorokybophyceae, and Mesostigmatophyceae) and the later-diverging ZCC-grade (Zygnematophyceae, Coleochaetophyceae, and Charophyceae (de Vries et al. 2016). It is now widely accepted that an ancestor of the Zygnematophyceae emerged onto land approximately 500+ million years ago and gave rise to land plants (Delwiche and Cooper 2015; Hess et al. 2022; Permann et al. 2022). Previous immunochemical and biochemical studies (Domozych et al. 2007; Sørensen et al. 2010, 2011; Popper et al. 2011; Proseus and Boyer 2012; Mikkelsen et al. 2014) illustrated notable similarity in the CW constituents of many charophytes with the primary CWs of many land plants. This includes a scaffolding network of cellulose microfibrils that are embedded in a matrix of pectic polymers, xylan, XyG, mannan, mixed-linkage glucan, and proteins (e.g. arabinogalactan proteins [AGPs], extensin; Zhang et al. 2021). Recently, the sequencing of several charophyte genomes and transcriptomes (Table 1) along with correlative analyses that have synthesized biochemical and ecophysiological data have revealed new information about charophyte CWs/ECM that are refining our understanding of plant evolution, stress physiology, and cell biology (Fitzek et al. 2019; Leebens-Mack et al. 2019; Domozych and Bagdan 2022). Recently, a detailed gene family analysis by Feng et al. (2023) showed that the Zygnematophyceae share all the major enzymes with land plants for CW polysaccharide synthesis, degradation, and modifications. In fact, most of the enzymes for CW innovations that include polysaccharide backbone synthesis were gained more than 700 million years ago. The following represents a summary of highlights concerning recent discoveries of specific components of the CWs of charophytes.
Cellulose, the main load-bearing component of the plant CW, consists of β-D-glucose (Glc) linked by (1–4) glycosidic bonds that assemble to form higher-order fibers, microfibrils, with amorphous and crystalline regions (Nicolas et al. 2022). Cellulose microfibrils are synthesized by a complex of cellulose synthases A (CesA) on the plasma membrane of the cell (Wilson et al. 2021). Cellulose has been found in the late-diverging charophyte clades and in low amounts in the Klebsormidiophyceae and Chlorokybophyceae but not in the Mesostigmatophyceae (Domozych 2012). Fitzek et al. (2019) reported that land plant CesA orthologs exist only in Zygnematophyceae (i.e. the CesA clade) but not in Coleochaetophyceae and Charophyceae of the ZCC clade, nor in the KCM clade. However, cellulose synthaseD-like or CslD-like orthologs have been found in all charophytes analyzed (i.e. the ClsD clade, e.g. Coleochaetophyceae). The CslD-like orthologs were posited to be “charophyte-specific CesAs” that evolved in an ancestral charophyte line. Evolutionary divergence then occurred whereby branches in the CesA clade were only found in ancestral Zygnematophyceae and eventually became the ancestor of land plant CesAs, while another branch became the ancestor of all land plant CslDs (e.g. modern Coleochaetophyceae).
Callose, a β-1,3 glucan, is integral to a wide variety of physiological processes in plants, including rapid responses to abiotic and biotic stress, regulation of plasmodesmata-based transport, the growth of pollen tubes, and cell plate formation during cytokinesis (Schneider et al. 2016). During cytokinesis, callose hydrogels form a flexible and stabilizing matrix during the formation of the cell plate membrane network while also contributing to the radial expansion of the cell plate as it matures into a CW (Sinclair et al. 2022). Callose has been demonstrated in the cytokinetic apparatus of the charophytes Coleochaete and Chara (Scherp et al. 2001; Delwiche and Cooper 2015). Davis et al. (2020) recently described the role of callose in septum formation during cytokinesis in the unicellular zygnematophyte, P. margaritaceum, and posited that this polymer supports the cell plate membrane as it expands inward during new daughter cell pole formation. Putative callose synthase genes were also identified in this study.
Callose has been analyzed in the CW of the charophyte Klebsormidium, along with its role in enhancing CW regions experiencing biomechanical stress due to loss of turgor pressure (Herburger and Holzinger 2015, 2016; Steiner et al. 2020). The highly elastic properties of callose enhance/regulate CW flexibility that is necessary for life in habitats with fluctuating water availability, an ancient feature for CW support thought to be critical for dispersal and to have led to the widespread success of these algae.
Hemicelluloses represent a diverse array of CW heteropolysaccharides that possess a ß-(1–>4)–linked backbone and include XyGs, xylans, mannans and glucomannans, and ß-(1–>3,1–>4)-glucans or mixed linked glucans (MLGs) (Scheller and Ulvskov 2010). XyGs function in several ways, including increasing CW rigidity via the tethering of cellulose microfibrils, contributing to CW loosening during expansion, and aggregating soil particles in the rhizosphere around roots (Galloway et al. 2018; Zheng et al. 2018). The β-glucan backbone of XyGs is decorated with a variety of short side-chains interspersed with unsubstituted Glc units at regular intervals (Mikkelsen et al. 2021). Though not originally thought to be found in charophytes, XyGs have since been identified in many charophyte taxa (Sørensen et al. 2010, 2011; Herburger et al. 2018). In a screening of charophyte transcriptomes, Del-Bem (2018) demonstrated that XyG-processing enzymes were found in multiple charophyte taxa. This included the earlier diverging Klebsormidiophyceae (i.e. K. nitens) that contained homologs of most of the XyG-related genes first discovered in angiosperms, including all known enzymes required for XyG synthesis. One group of enzymes, the transglycosylases, are CW-remodeling enzymes that cleave off part of the backbone of a polysaccharide (donor) and graft it onto another (acceptor). These molecular rearrangements provide flexibility in the wall that allows for expansion. Transglycosylase activity (e.g. XyG endotransglucosylase (XET)) was demonstrated in the longitudinal CWs of young filaments of Klebsormidium, filaments of Zygnema (Zygnematophyceae), and the anticlinal and periclinal CWs of parenchymatous Chara (Charophyceae) thalli (Herburger et al. 2018). Heterotransglycanases are endotransglycosylases that catalyze transfers between XyGs and MLGs, xylans, or mannans and have also been identified in charophytes (Franková and Fry 2021). Mikkelsen et al. (2021) has also shown that orthologs to the biosynthetic enzymes associated with XyG fucosylation, long thought to be a late evolutionary elaboration, are present in the zygnematophyte Mesotaenium caldariorum. It has been postulated that XyGs likely evolved during land colonization by charophytes and contributed not only to cell expansion but also to processing of soils that were key to the conquest of land (Del-Bem 2018).
Mannans constitute another major group of hemicelluloses and contain a backbone of mannose (Man) linked in β-(1-4) configuration or a combination of Glc and Man that can be substituted by α-(1-6)–linked Gal (Moreira and Filho 2008; Scheller and Ulvskov 2010; Voiniciuc 2022). In land plants, mannans function as storage polysaccharides in seedlings and as structural components of the hemicellulose–cellulose network. Mannans have been identified in all groups of charophytes except Mesostigma (Sørensen et al. 2011; Permann et al. 2021a), but their location in the charophyte CW architecture and their functions are not yet known. In a recent analysis of transglycosylases, Franková and Fry (2021) noted a major difference in those found in charophytes vs those in land plants. High levels of trans-β-1,4-mannanase acting on mannan substrates were noted that, in turn, suggests that charophytes might prioritize mannan remodeling versus XyG remodeling, which is carried out by transglucanases (Verhage 2021).
Xylans consist of a β-(1-4)–linked D-xylose (Xyl) backbone that can be substituted by L-arabinose (Ara), D-Gal, GlcA, and acetyl groups (Scheller and Ulvskov 2010). Xylans function in growth and development and contribute to the structural integrity of the CW. Like mannans, they are found in most groups of charophytes (Domozych et al. 2010; Sørensen et al. 2011; Hsieh and Harris 2019). Genomic and transcriptomic mining of genes involved in xylan synthesis was performed for the charophyte K. flaccidum, which produces a highly substituted ß-1-4-xylan in its CW (Jensen et al. 2018). The protein K. flaccidum XYLAN SYNTHASE-1 was identified and shown to possess 1,4-β-xylan synthase activity. Also identified were the 1,4-β-xylan xylosyltransferases IRX9 and IRX14 required for xylan synthesis in planta and the glcosylase transfereases GT8, GT43, GT47, and DUF579 (domain of unknown function 579)/GXMT (glucuronoxylan 4-O-methyltransferase-like protein) that represent candidate genes implicated in β-1-4-xylan backbone formation, GlcA addition to the backbone, and subsequent GlcA methylation. C. brauniia, a deep-branching, highly diverged form of GT43, was identified as the most likely xylan synthase, providing the first hint that GT43 sequences are enzymatically involved in synthesizing xylan (Nishiyama et al. 2018).
Mixed linkage glucans or MLGs are composed of β-D (1-3) and β-D (1-4)–linked glucosyl residues and have also been identified in charophytes (Sørensen et al. 2011), but little is known about their function.
Pectins are galacturonic acid (GalA)-containing polymers of the plant CW that play critical roles in cell expansion and morphogenesis, ion uptake, and immune signaling pathways (Caffall and Mohnen 2009; Anderson and Keiber 2020; Haas et al. 2021; Cosgrove 2022). Pectins are ancient polymers, and 2 pectin subclasses—homogalacturonan (HG) and rhamnogalacturonan-I—have been found in many charophytes, especially taxa of the ZCC clade (Eder and Lütz-Meindl 2008, 2010; Sørensen et al. 2010, 2011; Domozych et al. 2014; Herburger et al. 2018). HG is a polymer made of (1,4)-GalA that can be substituted with methyl or acetyl groups. Rhamnogalacturonan-I is made of a repeating GalA-rhamnose (Rha) disaccharide that can be substituted with various side chains, such as galactan, arabinan, and type-I arabinogalactan (Caffall and Mohnen 2009). The structural and functional dynamics of charophyte pectins have been studied in detail in several charophytes, including Chara corallina and P. margaritaceum. In Chara, CW expansion encompasses a dynamic pectate cycle that coordinates with Ca2+ (calcium)-complexing of HG (Boyer 2016). In Penium, the demethylation of recently secreted HG at the central isthmus of the cell and subsequent Ca2+-complexing manifest in a bipolar expansion mechanism and the formation of the distinctive HG lattice on the outer CW layer (Domozych et al. 2014; Palacio-Lopez et al. 2020). Screening of the genome and transcriptome of P. margaritaceum (Jiao et al. 2020) demonstrated notable gene family expansion for CAZymes, including the classes of glycosyl hydrolase (GH), glycosyl transferase (GT), carbohydrate esterase, polysaccharide lyase, and carbohydrate binding modules. Interestingly, more GTs and polysaccharide lyases were found in Penium than in any of the green plant lineages. The precise roles of pectin in charophytes will require further detailed screening, but it is now apparent that these ancient macromolecules have multiple functions in cell expansion, cell differentiation, and the formation of the adhesive middle lamella of multicellular tissues.
Charophytes also possess structural and catalytic proteins in their ECMs. AGPs are one type of non-enzymatic Hyp-rich glycoproteins (Johnson et al. 2003, 2017) that are common CW/ECM components of many photosynthetic eukaryotes, including charophytes (Hervé et al. 2016; Knox 2016; Domozych and Bagdan 2022). These proteins are highly glycosylated (i.e. 90% of a plant's AGP may be carbohydrate), with inclusive arabinogalactan covalently linked to the protein moiety via Hyp (Silva et al. 2020). AGPs have various functions in plant cell expansion, development, and signal transduction across the plasma membrane (Pereira et al. 2015; Lin et al. 2022). In charophytes, putative AGPs have been identified in the CWs of nonconjugating Spirogyra filaments, especially in the area of transverse walls (Pfeifer et al. 2022), as well as in the CWs of zygospores of Spirogyra and Mougeotia (Permann et al. 2021a, 2021b). Biochemical analyses of the Spirogyra AGP showed that Ara was present only in small amounts, and the terminating sugars consisted predominantly of terminal and 1,3-linked Rha residues, leading to a new classification of “rhamnogalactan-protein” for this special AGP modification (Pfeifer et al. 2022). Palacio-Lopez et al. (2019) employed immunocytochemical methods to identify AGPs in the ECM surrounding Spirogyra rhizoids, Chlorokybus, Coleochaete, and Penium. It was shown that these AGPs were components of various adhesive mechanisms (e.g. cell-cell, cell-substrate).
Extensins are another type of Hyp-rich glycoproteins of plant CWs that act as scaffolds for the deposition of the main CW carbohydrate polymers and also contribute to plant defense (Møller et al. 2017; Castilleux et al. 2021). Extensins have been identified in CWs of a variety of charophytes (Sørensen et al. 2011; Permann et al. 2021a, 2021b). Little detail is known though about their location in the CW or their functions. Bioinformatic analyses reveal that green algae have no classical extensins but have a number of long chimeric extensins (Liu et al. 2016) that are not found in land plants. Glycosyltransferases associated with extension and AGP synthesis have also been identified in green algae (Ulvskov et al. 2013; Mikkelsen et al. 2021).
Using genome-wide transcript expression profiling, Vannerum et al. (2011) demonstrated expansin in the zygnematophyte Micrasterias denticulata. A GFP-tagged version of the expansin-resembling protein MdEXP2 localized to the CW and in Golgi-derived vesicles. Overexpression phenotypes altered lobe elongation and caused a loss of growth polarity and planarity. In a study of the transcriptome of Spirogyra, Van de Poel et al. (2016) displayed modification of the CW matrix by both expansins and XET/hydrolases.
ECMs of earlier diverging charophytes
The ECM of the earlier divergent taxa M. viride (Mesostigmatophyceae) and Chlorokybus atmophyticus (Chlorokybophyceae) are distinctly different from those found in the Klebsormidiophyceae and later-diverging charophytes (Sørensen et al. 2011). The motile stage of Mesostigma produces a covering consisting of layers of intricately shaped scales (see section on prasinophyte scales). The composition of the ECM of the nonmotile phase is unknown. The cells of nonmotile sarcinoid life cycle phase of Chlorokybus maintain a thin CW surrounded by an extensive extracellular sheath that contains AGP (Palacio-Lopez et al. 2019), and its motile zoospores are covered by scales. Recent screening of the Mesostigma and Chlorokybus genomes and transcriptomes (Demko et al. 2019; Liang et al. 2019; Wang et al. 2020; Irisarri et al. 2021) have provided novel insight into ECM dynamics in these 2 algae. Mesostigma possesses 81 CAZymes and 20 putative carbohydrate-binding modules (Wang et al. 2020). Three putative cellulose synthase-like (CSL) enzymes (CSLA/CSLC-like) were identified, but CESA was absent. The biosynthesis and transport genes for the 2-keto sugar acid, Kdo, a major component of its scales, was identified (Liang et al. 2019). Chlorokybus possesses 100 CAZymes and 22 carbohydrate-binding modules, including 3 CESA/CSLD-like homologues. Mesostigma possesses genes involved in CW polymer remodeling, including those involved in mannan and xylan metabolism (e.g. mannanases, mannosidase, and xylosidase), but Chlorokybus does not. XyG and xylan-degrading enzymes and pectin lyases are absent from these organisms.
Biosynthesis and secretion of the charophyte ECM
Due to their importance in the evolution of plants, charophytes and their subcellular machinery involved in ECM/CW synthesis and secretion have garnered much interest. The structure and dynamics of the endomembrane system and cytoskeletal network during CW/cell expansion and EPS production have been focal points of many of these studies, and detailed reviews may be found in Lütz-Meindl (2016) and Domozych and Bagdan (2022). The development of the CW/ECM during cytokinesis, including furrowing and phragmoplast/cell plate formation, the formation of the plasmodesmata, and morphogenetic events leading to multicellular thallus, has also attracted significant interest (Pickett-Heaps 1975; Buschmann and Zachgo 2016; Brunkard and Zambryski 2017; de Vries and Archibald 2018; Nishiyama et al. 2018; Buschmann 2020; Davis et al. 2020).
The CWs of chlorophytes
The Chlorophyta represent the largest and most diverse group of green algae. Several classes of chlorophytes have been identified (Leliaert 2019), most recently based on data from phylogenomic studies (Gulbrandsen et al. 2021; Li et al. 2021; Yamashita et al. 2021; Table 1). However, the very limited screening of the genomes/transcriptomes of vast assortment of chlorophyte taxa leaves many fundamental questions unresolved (Bachy et al. 2022). Many chlorophytes have been studied in detail, and several have become valuable model organisms for various biological investigations (e.g. Chlamydomonas, Volvox; Matt and Umen 2016; Craig et al. 2021). Others are recognized as keystone primary producers of the planet's oceans (e.g. Micromonas; Demory et al. 2019), and several are harvested and used for the medical, food, and biofuel applications (e.g. Ulva; Roleda and Heesch 2021). Chlorophyte phenotypes range widely from minute unicells to macroscopic unicellular siphons to large seaweeds. The ECMs of chlorophytes are very diverse. An excellent and thorough review of the structure and chemistry of their CWs and other extracellular components is presented in Baudelet et al. (2017). Three general types of ECMs have been recognized in the chlorophytes: Group 1 contains organisms with ECMs comprised of scaly or thecal coverings that contain deoxy-5-O-methyl-manno-2-octulosonic acid and deoxy-lyxo-2-heptulosaric acid (Becker et al. 1994; see section on prasinophyte scales); Group 2 taxa have glucan, mannan, and chitin-containing CWs with alagenan and some have crystalline glycoprotein CWs (see section on glycoprotein walls); and Group 3 organisms have glucan and mannan CWs that also contain sulfated or pyruvylated polysaccharides.
Recent highlights of select chlorophyte taxa
The Trebouxiophyceae consist of a diverse array of phenotypes commonly found in soil, and subaerial habitats and as the phycobionts of lichens. Several inclusive taxa have become important models for research and as economically important resources.
Chlorella: The unicellular chlorophyte Chlorella has been a popular organism in algal/plant physiology (e.g. the alga used by Melvin Calvin and co-workers in elucidating carbon fixation in photosynthesis; Nickelsen 2017). Today, Chlorella and related genera (e.g. Nannochloropsis gaditana) have shown much promise for yielding large CW-based biomass for conversion to biofuels (Rodrigues and de Silva Bon 2011; Taleb et al. 2015) as well as being employed in various medical and food supplement technologies (Bito et al. 2020). Species of this alga often have a bilayer or unilayer CW (Weber et al. 2022). The outermost layer of the bilayer wall often possesses a trilaminar structure and contains the highly resistant compound sporopllenin. Cellulose is found in the inner CW layer, and chitin or chitin-like polymers have been identified in the CW as well (Weber et al. 2022).
Botryococcus: One of more unique chlorophytes is the colonial Botryococcus. A total 86% of its dry weight can consist of long-chain hydrocarbons, including tri-terpine-based oils (e.g. botryococcenes), alkadienes, and alkatrienes. This and other features have brought Botryococcus into the limelight of applied technologies such as biofuels, food additives (e.g. carotenoids), cosmetics, nutraceuticals, feed stock, and antibiotic nanoparticle production (Cheng et al. 2019a, 2019b; Love 2022). The botryococcenes are produced in fenestrated cortical ER that directly delivers these hydrocarbons to the plasma membrane ,where they are deposited externally, exuded across the CW and into the ECM (Weiss et al. 2012). Botryococcus's ECM is structurally complex (Wang et al. 2022) and is made of 3 components: 1) a fibrillar, β-1, 4- and/or β-1, 3-glucan–containing CW; 2) an intracolonial space that consists of a cross-linked hydrocarbon network that is permeated with liquid hydrocarbons; and 3) a retaining wall that contains of a polysaccharide sheath of fibrils that sequester the ECM liquid hydrocarbons (Weiss et al. 2012). The hydrocarbon-ECM complex fills the interstices between cells and may act to stabilize the colony and facilitate gas exchange in the colony interior. The fibrils consist of 97% carbohydrate containing significant amounts of arabinose (42%) and Gal (39%).
Trebouxia, Coccomyxa: Trebouxia and Coccomyxa are 2 unicellular chlorophytes that may be free-living in terrestrial habitats or live as phycobionts of lichens (Centeno et al. 2016; González-Hourcade et al. 2020, 2021; Spribille et al. 2020, 2022). Lichens and their green microalgae are considered poikilohydric organisms, that is, they do not actively regulate their water content and must depend on the availability of water in their habitats. Many lichens live in habitats of extreme abiotic stress, especially with fluctuations in water availability, and recent work has shown that Trebouxia and Coccomyxa CW structural and chemical modulations are critical for their survival in the lichen complex (Baudelet et al. 2017). These lichen algae have flexible CWs that fold as the protoplast shrinks during dehydration and expand during rehydration. This flexibility is believed to prevent the mechanical stress occurring during dehydration in cells. β-3-Linked rhamnogalactofuranan, KOH-soluble β-glucans, and sulfated polysaccharides have been isolated from Trebouxia and Coccomyxa, and details of their CW chemistry and biochemical remodeling under stress can be obtained in González-Hourcade et al. (2021, 2020) and Gasulla et al. (2021).
Ulvophytes
The ulvophytes (Ulvophyceae) constitute a wide array of mostly macroscopic marine chlorophytes (i.e. green seaweeds) and some subaerial and freshwater taxa. Ulvophyte phenotypes range from unicells to macroscopic sheet-like thalli to giant-celled multinucleate siphonous and siphoncladous forms (Cocquyt et al. 2010). Ulva, a common and widely distributed green coastline seaweed, has been the focal point for many of ECM studies of ulvophytes (Lahaye and Robic 2007; Wichard et al. 2015; De Clerck et al. 2018; Wahlström et al. 2020a, 2020b; Kloareg et al. 2021; Wichard 2023). This seaweed grows in large amounts, has been harvested for a variety of human uses (Smetacek and Zingone 2013; Simon et al. 2022), and is also the cause of “green tides” (i.e. blooms caused by high nutrient run off in coastal zones; Hiraoka 2021). Four groups of polysaccharides have been characterized in the ECM of Ulva: semicrystalline cellulose, water-soluble ulvans, XyG, and a glucuronan (Kloareg et al. 2021). Ulvan, is a high–molecular weight (660,000–760,000 g/mol) sulfated polysaccharide that makes up to 30% of the alga's biomass and is composed of repeating units of sulfated disaccharides with Rha, Xyl, GlcA, and iduronic acid as the main building blocks (Kidgell et al. 2019, 2021; Wahlström et al. 2020a, 2020b; Kloareg et al. 2021). These include β-d-GlcA (1 → 4)-α-l-Rha-3-sulfate and α-l-iduronic acid (1 → 4)-α-l-Rha-3-sulfate. The iduronic acid or GlcA components may instead be a Xyl unit (sulfated or nonsulfated) forming the characteristic monomers β-d-Xyl (1 → 4)-α-l-Rha-3-sulfate and β-d-Xyl-2-sulfate(1 → 4)-α-l-Rha-3-sulfate. Sulfated polysaccharides like ulvan most likely serve as physiological adaptations to the high ionic strength of the saltwater habitats and contribute to moisture retention that enhances desiccation resistance. They also are important in cell-cell interactions, adhesion mechanisms, and as defense against pathogens (Ciancia et al. 2020). The development of an ulvan-specific monoclonal antibody has aided immunocytochemical studies (Rydahl et al. 2017). Sulfated polysaccharides from Ulva and other marine algae are gaining significant interest in that they are being screened for anti-cancer, anti-angiogenic, antioxidant, anti-inflammatory, immunostimulatory, hydrogel, and antiviral activities (Sulastri et al. 2021; López-Hortas et al. 2021; Meinita et al. 2022; Negreanu-Pirjol et al. 2022). Detailed comparative reviews of the complex sulfated polysaccharides found in other ulvophyte groups, including the Ulotrichales, the giant multinucleate (coenocytic) cells of the siphonous Bryopsidales and Dasycladales, and siphonocladous freshwater Cladophorales, are provided elsewhere (Fernández et al. 2015; Arata et al. 2017; Ciancia et al. 2020). Recently, pectins have been identified in Ulva (Holzinger et al. 2015), and the first identification of O-acetylation of pectin in green algae has been described in Bothwell et al. 2022. These results demonstrate that many CW polymers arose in green algae well before the emergence of land plants.
Cellulose is also a component of the CW of ulvophytes (Thygesen et al. 2021). Using atomic force microscopy, cellulose microfibril cross-linking and interactions with CW matrix components were demonstrated in Ventricaria ventricosa (Eslick et al. 2014). In another atomic force microscopy–based analysis of the CW of Valonia utricularis, a CW matrix component containing N-acetylgalactosamine was shown to be involved in the maintenance of the CW integrity through bonding of the neighboring CW layers (Mine and Sekida 2018). Linear β-1,4-mannans replace cellulose as the dominant structural polymer in some ulvophycean taxa, such as the green seaweed Codium vermilara (Fernández et al. 2015). This algal mannan is mostly fibrillar, with 2-linked sulfate on 23% of Man units resulting in partially soluble chains that could function in maintaining amorphous CW regions.
AGPs have also been identified in Ulva (Přerovská et al. 2021), and amino acid analysis showed a similarity between these and land plant AGPs. However, AGPs from Ulva revealed unique glycosylation patterns with relatively low amounts of arabinose and Gal but the presence of 3-O-methyl-hexose. These results likely show a specialized adaptation to the marine environment and add new insight to the evolution of CWs in ulvophytes and green algae in general.
Screening of the genomes of Ulva (Ulva mutabilis, De Clerck et al. 2018; Ulva compressa, Osorio et al. 2022) and Caulerpa lentillifera (Arimoto et al. 2019) have also contributed to our understanding of the ECM of ulvophytes. The Ulva species’ genomes contain multiple cellulose synthases, hydroxyprolyl O-arabinosyl transferases, glucomannan-mannosyl transferase, UDP-glucosamine transferase, UDP-rhamnose transporters, UDP-uronic acid transporters, pectin lyase, extensins, and expansin (Osorio et al. 2022). Interestingly, genes encoding for collagens and elastins have been detected in these 2 species as well. Comparative analyses of the biochemistry and genetics of ulvophytes and other seaweeds are provided in Kloareg et al. (2021) and Shao and Duan (2022).
The ECM of the Prasinodermophyta
The Prasinodermophyta is a newly recognized phylum of green algae thought to have diverged before the split of Chlorophyta and Charophyta 1 billion years ago. At present, the CW/ECM (Jouenne et al. 2011) has not been biochemically or structurally characterized. Mining of the Prasioderma coloniale genome revealed that genes responsible for the biosynthesis and remodeling of the major components of the plant primary CW (e.g. cellulose, mannans, xylans, XyGs) were not present (Li et al. 2020).
Concluding remarks
Over the last decade, an infusion of new information concerning the CWs/ECMs of green algae has significantly refined our understanding of green plant evolution; the CW and its role in expansion, development, and ecophysiology; and the applied uses of these cell coverings in multiple industries (see Advancements). Yet today, only a very small percentage of green algae, their CWs/ECM, and their biosynthetic/secretory pathways have been investigated in detail. Future studies will be needed to thoroughly elucidate the structure and function of both known polymers and the unique components of green algal ECMs. This information will yield critical insight into such areas as the architectural design and functional dynamics of specific polymers in the green plant CW, ECM modulations that occur during abiotic stress (e.g. low water stress, UV exposure), the functions of the complex EPS gels and hydrophobic molecules in cell coverings, as well as identify novel model organisms for investigating basic green plant life processes (Outstanding Questions Box). As important, these studies will directly contribute to our understanding of the causes and controls of algal blooms (e.g. green tides caused by Ulva), provide new ways to control atmospheric CO2 through mass algal growth, and reveal further uses of CW/ECM materials in food, medicine, and the development of clean biofuels.
Acknowledgments
We thank Lily Kozel, Kaylee Bagdan, Cormac Feeley, Tawn Tomasi, and Ruby Epstein (Skidmore College) for helpful discussions. We apologize to those colleagues whose work was not cited due to space constraints.
Contributor Information
David S Domozych, Department of Biology, Skidmore College, Saratoga Springs, NY 12866, USA.
Josephine G LoRicco, Department of Biology, Skidmore College, Saratoga Springs, NY 12866, USA.
Funding
This work was supported by National Science Foundation (USA) grant 2129443.
Data availability
All data is available upon request from the author, DSD.
References
- Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J Eukaryot Microbiol. 2019:66(1):4–119. 10.1111/jeu.12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner S, Remias D, Karsten U, Holzinger A. Unusual phenolic compounds contribute to ecophysiological performance in the purple-colored green alga Zygogonium ericetorum (Zygnematophyceae, Streptophyta) from a high-alpine habitat. J Phycol. 2013:49(4):648–660. 10.1111/jpy.12075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CT, Kieber JJ. Dynamic construction, perception, and remodeling of plant cell walls. Annu Rev Plant Biol. 2020:29(1):39–69. 10.1146/annurev-arplant-081519-035846 [DOI] [PubMed] [Google Scholar]
- Arata PX, Alberghina J, Confalonieri V, Errea MI, Estevez JM, Ciancia M. Sulfated polysaccharides in the freshwater green macroalga Cladophora surera not linked to salinity adaptation. Front Plant Sci. 2017:8:1927. 10.3389/fpls.2017.01927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto A, Nishitsuji K, Higa Y, Arakaki N, Hisata K, Shinzato C, Satoh N, Shoguchi E. A siphonous macroalgal genome suggests convergent functions of homeobox genes in algae and land plants. DNA Res. 2019:26(2):183–192. 10.1093/dnares/dsz002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriola MB, Velmurugan N, Zhang Y, Plunkett MH, Hondzo H, Barney BM. Genome sequences of Chlorella sorokiniana UTEX 1602 and Micractinium conductrix SAG 241.80: implications to maltose excretion by a green alga. Plant J. 2017:93(3):566–586. 10.1111/tpj.13789 [DOI] [PubMed] [Google Scholar]
- Bachy C, Wittmers F, Muschiol J, Hamilton M, Henrissat B, Worden AZ. The land-sea connection: insights into the plant lineage from a green algal perspective. Annu Rev Plant Biol. 2022:73(1):585–616. 10.1146/annurev-arplant-071921-100530 [DOI] [PubMed] [Google Scholar]
- Barolo L, Commault AS, Abbriano RM, Padula MP, Kim M, Kuzhiumparambil U, Ralph PJ, Pernice M. Unassembled cell wall proteins form aggregates in the extracellular space of Chlamydomonas reinhardtii strain UVM4. Appl Microbiol Biotechnol. 2022:106(11):4145–4156. 10.1007/s00253-022-11960-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudelet P-H, Ricochon G, Linder M, Muniglia L. A new insight in cell walls of Chlorophyta. Algal Res. 2017:25:333–317. 10.1016/j.algal.2017.04.008 [DOI] [Google Scholar]
- Becker B, Marin B. Streptophyte algae and the origin of embryophytes. Ann Bot. 2009:103(7):999–1004. 10.1093/aob/mcp044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Marin B, Melkonian M. Structure, composition, and biogenesis of prasinophyte cell coverings. Protoplasma. 1994:181(1–4):233–244. 10.1007/BF01666398 [DOI] [Google Scholar]
- Becker B, Melkonian M, Kamerling JP. The cell wall (theca) of Tetraselmis striata (Chlorophyta): macromolecular composition and structural elements of the complex polysaccharides. J Phycol. 1998:34(5):779–787. 10.1046/j.1529-8817.1998.340779.x [DOI] [Google Scholar]
- Bito T, Okumura E, Fujishima M, Watanabe F. Potential of Chlorella as a dietary supplement to promote human health. Nutrients. 2020:12(9):2524. 10.3390/nu12092524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, et al. The Chlorella variabilis NC64A genome reveals adaptations to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010:22(9):2943–2955. 10.1105/tpc.110.076406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Mathieu R, Verhelst B, Derelle E, Rombauts S, Bouget FY, Carré I, Château A, Eyre-Walker A, Grimsley N, Moreau H, et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genomics. 2014:15(1):1103. 10.1186/1471-2164-15-1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boney AD. Mucilage: a ubiquitous algal attribute. Br Phycol J. 1981:16(2):115–132. 10.1080/00071618100650101 [DOI] [Google Scholar]
- Bothwell JH, Goodridge AJ, Rapin M, Brennan PJ, López AT, Agrawal A, Fry SC, Campbell G, Blomme J.. Cell walls are dynamically O-acetylated in the green seaweed, Ulva compressa. bioRxiv 493306. bioRxiv. 2022. 10.1101/2022.05.24.493306, 25 May 2022, preprint: not peer reviewed. [DOI]
- Bowles AMC, Paps J, Bechtold U. Water-related innovations in land plants evolved by different patterns of gene cooption and novelty. New Phytol. 2022:235(2):732–742. 10.1111/nph.17981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JS. Enzyme-less growth in Chara and Terrestrial plants. Front Plant Sci. 2016:7:866. 10.3389/fpls.2016.00866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne DR, Jenkins J, Schmutz J, Shu S, Barry K, Grimwood J, Chiniquy J, Sharma A, Niehaus TD, Weiss TL, et al. Draft nuclear genome sequence of the liquid hydrocarbon-accumulating green microalga Botryococcus braunii race B (showa). Genome Announc. 2017:5(16):e00215–17. 10.1128/genomeA.00215-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkard JO, Zambryski PC. Plasmodesmata enable multicellularity: new insights into their evolution, biogenesis, and functions in development and immunity. Curr Opin Plant Biol. 2017:35:76–83. 10.1016/j.pbi.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Busch A, Hess S. Sunscreen mucilage: a photoprotective adaptation found in terrestrial green algae (Zygnematophyceae). Eur J Phycol. 2022:57(1):107–124. 10.1080/09670262.2021.1898677 [DOI] [Google Scholar]
- Buschmann H. Into another dimension: how streptophyte algae gained morphological complexity. J Exp Bot. 2020:71(11):3279–3286. 10.1093/jxb/eraa181 [DOI] [PubMed] [Google Scholar]
- Buschmann H, Zachgo S. The evolution of cell division: from streptophyte Algae to land plants. Trends Plant Sci. 2016:21(10):872–883. 10.1016/j.tplants.2016.07.004 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res. 2009:344(14):1879–1900. 10.1016/j.carres.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Castilleux R, Plancot B, Vicré M, Nguema-Ona E, Driouich A. Extensin, an underestimated key component of cell wall defence? Ann Bot. 2021:127(6):709–713. 10.1093/aob/mcab001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno DC, Hell AF, Braga MR, Del Campo EM, Casano LM. Contrasting strategies used by lichen microalgae to cope with desiccation-rehydration stress revealed by metabolite profiling and cell wall analysis. Environ Microbiol. 2016:18(5):1546–1560. 10.1111/1462-2920.13249 [DOI] [PubMed] [Google Scholar]
- Chen B-L, Mhuantong W, Ho S-H, Chang J-S, Zhao X-Q, Bai F-W. Genome sequencing, assembly, and annotation of the self-flocculating microalga Scenedesmus obliquus AS-6-11. BMC Genomics. 2020:21(1):743. 10.1186/s12864-020-07142-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Okada S, Zhou C, Chen P, Huo S, Li K, Addy M, Yan X, Ruan RR. High-value chemicals from Botryococcus braunii and their current applications—a review. Bioresour Technol. 2019a:291:121911. 10.1016/j.biortech.2019.121911 [DOI] [PubMed] [Google Scholar]
- Cheng S, Xian W, Fu Y, Marin B, Keller J, Wu T, Sun W, Li X, Xu Y, Zhang Y, et al. Genomes of subaerial zygnematophyceae provide insights into land plant evolution. Cell. 2019b:179(5):1057–1067.e14. 10.1016/j.cell.2019.10.019 [DOI] [PubMed] [Google Scholar]
- Ciancia M, Fernández PV, Leliaert F. Diversity of sulfated polysaccharides from cell walls of coenocytic green Algae and their structural relationships in view of green algal evolution. Front Plant Sci. 2020:11:554585. 10.3389/fpls.2020.554585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquyt E, Verbruggen H, Leliaert F, De Clerck O. Evolution and cytological diversification of the green seaweeds (Ulvophyceae). Mol Biol Evol. 2010:27(9):2052–2061. 10.1093/molbev/msq091 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Building an extensible cell wall. Plant Physiol. 2022:189(3):1246–1277. 10.1093/plphys/kiac184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig RJ, Hasan AR, Ness RW, Keightley PD. Comparative genomics of Chlamydomonas. Plant Cell. 2021:33(4):1016–1041. 10.1093/plcell/koab026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronmiller E, Toor D, Shao NC, Kariyawasam T, Wang MH, Lee JH. Cell wall integrity signaling regulates cell wall-related gene expression in Chlamydomonas reinhardtii. Sci Rep. 2019:9(1):12204. 10.1038/s41598-019-48523-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha C, Faria M, Nogueira N, Ferreira A, Cordeiro N. Marine vs freshwater microalgae exopolymers as biosolutions to microplastics pollution. Environ Pollut. 2019:249:372–380. 10.1016/j.envpol.2019.03.046 [DOI] [PubMed] [Google Scholar]
- Davies KM, Landi M, van Klink JW, Schwinn KE, Brummell DA, Albert NW, Chagné D, Jibran R, Kulshrestha S, Zhou Y, et al. Evolution and function of red pigmentation in land plants. Ann Bot. 2022:130(5):613–636. 10.1093/aob/mcac109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DJ, Wang M, Sørensen I, Rose JKC, Domozych DS, Drakakaki G. Callose deposition is essential for the completion of cytokinesis in the unicellular alga Penium margaritaceum. J Cell Sci. 2020:133(19):jcs249599. 10.1242/jcs.249599 [DOI] [PubMed] [Google Scholar]
- Del-Bem LE. Xyloglucan evolution and the terrestrialization of green plants. New Phytol. 2018:219(4):1150–1153. 10.1111/nph.15191 [DOI] [PubMed] [Google Scholar]
- De Clerck O, Kao SM, Bogaert KA, Blomme J, Foflonker F, Kwantes M, Vancaester E, Vanderstraeten L, Aydogdu E, Boesger J, et al. Insights into the evolution of multicellularity from the sea lettuce genome. Curr Biol. 2018:28(18):2921–2933.e5. 10.1016/j.cub.2018.08.015 [DOI] [PubMed] [Google Scholar]
- Delmont TO, Eren AM, Vineis JH, Post AF. Genome reconstructions indicate the partitioning of ecological functions inside a phytoplankton bloom in the Amundsen Sea, Antarctica. Front Microbiol. 2015:6:1090. 10.3389/fmicb.2015.01090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwiche CF, Cooper ED. The evolutionary origin of a terrestrial Flora. Curr Biol. 2015:25(19):R899–R910. 10.1016/j.cub.2015.08.029 [DOI] [PubMed] [Google Scholar]
- Demko V, Shen L, Han X, Zhang P, Gu X, Yu H. Mesostigma viride genome and transcriptome provide insights into the origin and evolution of streptophyta. Adv Sci (Weinh). 2019:7(1):1901850. 10.1002/advs.201901850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demory D, Baudoux AC, Monier A, Simon N, Six C, Ge P, Rigaut-Jalabert F, Marie D, Sciandra A, Bernard O, et al. Picoeukaryotes of the Micromonas genus: sentinels of a warming ocean. ISME J. 2019:13(1):132–146. 10.1038/s41396-018-0248-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Archibald JM. Plant evolution: landmarks on the path to terrestrial life. New Phytol. 2018:217(4):1428–1434. 10.1111/nph.14975 [DOI] [PubMed] [Google Scholar]
- de Vries J, De Vries S, Slamovits CH, Rose LE, Archibald JM. How embryophytic is the biosynthesis of phenylpropanoids and their derivatives in streptophyte algae? Plant Cell Physiol. 2017:58(5):934–945. 10.1093/pcp/pcx037 [DOI] [PubMed] [Google Scholar]
- de Vries J, Stanton A, Archibald JM, Gould SB. Streptophyte terrestrialization in light of plastid evolution. Trends Plant Sci. 2016:21(6):467–476. 10.1016/j.tplants.2016.01.021 [DOI] [PubMed] [Google Scholar]
- Dial RJ, Ganey GQ, Skiles SM. What color should glacier algae be? An ecological role for red carbon in the cryosphere. FEMS Microbiol Ecol. 2018:94(3). 10.1093/femsec/fiy007 [DOI] [PubMed] [Google Scholar]
- Domozych DS, Bagdan K. The cell biology of charophytes: exploring the past and models for the future. Plant Physiol. 2022:190(3):1588–1608. 10.1093/plphys/kiac390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domozych DS, Ciancia M, Fangel JU, Mikkelsen MD, Ulvskov P, Willats WG. The cell walls of green Algae: a journey through evolution and diversity. Front Plant Sci. 2012:3:82. 10.3389/fpls.2012.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domozych DS, Domozych CR. Desmids and biofilms of freshwater wetlands: development and microarchitecture. Microb Ecol. 2007:55(1):81–93. 10.1007/s00248-007-9253-y [DOI] [PubMed] [Google Scholar]
- Domozych D, Serfis A, Kiemle S, Gretz MR. The structure and biochemistry of charophycean cell walls: I. Pectins of Penium margaritaceum. Protoplasma. 2007:230(1–2):99–115. 10.1007/s00709-006-0197-8 [DOI] [PubMed] [Google Scholar]
- Domozych DS, Sørensen I, Pettolino FA, Bacic A, Willats WGT. The cell wall polymers of the charophycean green alga Chara corallina: immunobinding and biochemical screening. Int J Plant Sci. 2010:171(4):345–361. 10.1086/651227 [DOI] [Google Scholar]
- Domozych DS, Sørensen I, Popper ZA, Ochs J, Andreas A, Fangel JU, Pielach A, Sacks C, Brechka H, Ruisi-Besares P, et al. Pectin metabolism and assembly in the cell wall of the charophyte green alga Penium margaritaceum. Plant Physiol. 2014:165(1):105–118. 10.1104/pp.114.236257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domozych DS, Wells B, Shaw PJ. The basket scales of the green alga Mesostigma viride: chemistry, immunology and ultrastructure. J Cell Sci. 1991:100(2):397–407. 10.1242/jcs.100.2.397 [DOI] [Google Scholar]
- Eder M, Lütz-Meindl U. Pectin-like carbohydrates in the green alga Micrasterias characterized by cytochemical analysis and energy filtering TEM. J Microsc. 2008:231(2):201–214. 10.1111/j.1365-2818.2008.02036.x [DOI] [PubMed] [Google Scholar]
- Eder M, Lütz-Meindl U. Analyses and localization of pectin-like carbohydrates in cell wall and mucilage of the green alga Netrium digitus. Protoplasma. 2010:243(1–4):25–38. 10.1007/s00709-009-0040-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslick EM, Beilby MJ, Moon AR. A study of the native cell wall structures of the marine alga Ventricaria ventricosa (Siphonocladales, Chlorophyceae) using atomic force microscopy. Microscopy (Oxf). 2014:63(2):131–140. 10.1093/jmicro/dft083 [DOI] [PubMed] [Google Scholar]
- Feng X, Zheng J, Irisarri I, Yu H, Zheng B, Ali Z, de Vries S, Keller J, Fürst-Jansen JMR, Dadras A, et al. Chromosome-level genomes of multicellular algal sisters to land plants illuminate signaling network evolution. bioRxiv 2023:526407. 10.1101/2023.01.31.526407, 01 February 2023, preprint: not peer reviewed. [DOI] [Google Scholar]
- Fernández PV, Raffo MP, Alberghina J, Ciancia M. Polysaccharides from the green seaweed Codium decorticatum. Structure and cell wall distribution. Carbohydr Polym. 2015:117:836–844. 10.1016/j.carbpol.2014.10.039 [DOI] [PubMed] [Google Scholar]
- Fitzek E, Orton L, Entwistle S, Grayburn WS, Ausland C, Duvall MR, Yin Y. Cell wall enzymes in Zygnema circumcarinatum UTEX 1559 respond to osmotic stress in a plant-like fashion. Front Plant Sci. 2019:10:732. 10.3389/fpls.2019.00732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franková L, Fry SC. Hemicellulose-remodelling transglycanase activities from charophytes: towards the evolution of the land-plant cell wall. Plant J. 2021:108(1):7–28. 10.1111/tpj.15500 [DOI] [PubMed] [Google Scholar]
- Fürst-Jansen JMR, de Vries S, de Vries J. Evo-physio: on stress responses and the earliest land plants. J Exp Bot. 2020:71(11):3254–3269. 10.1093/jxb/eraa007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway AF, Pedersen MJ, Merry B, Marcus SE, Blacker J, Benning LG, Field KJ, Knox JP. Xyloglucan is released by plants and promotes soil particle aggregation. New Phytol. 2018:217(3):1128–1136. 10.1111/nph.14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasulla F, Del Campo EM, Casano LM, Guéra A. Advances in understanding of desiccation tolerance of lichens and lichen-forming Algae. Plants (Basel). 2021:10(4):807. 10.3390/plants10040807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrath JF. The biology of desmids: a decade of progress. Phycol Res. 1993:9:81–192. 10.1080/0028825X.1995.10410490 [DOI] [Google Scholar]
- González-Hourcade M, Del Campo EM, Braga MR, Salgado A, Casano LM. Disentangling the role of extracellular polysaccharides in desiccation tolerance in lichen-forming microalgae. First evidence of sulfated polysaccharides and ancient sulfotransferase genes. Environ Microbiol. 2020:22(8):3096–3111. 10.1111/1462-2920.15043 [DOI] [PubMed] [Google Scholar]
- González-Hourcade M, Del Campo EM, Casano LM. The under-explored extracellular proteome of aero-terrestrial microalgae provides clues on different mechanisms of desiccation tolerance in non-model organisms. Microb Ecol. 2021:81(2):437–453. 10.1007/s00248-020-01604-8 [DOI] [PubMed] [Google Scholar]
- Guiry MD. How many species of algae are there? J Phycol. 2012:48(5):1057–1063. 10.1111/j.1529-8817.2012.01222.x [DOI] [PubMed] [Google Scholar]
- Gulbrandsen ØS, Andresen IJ, Krabberød AK, Bråte J, Shalchian-Tabrizi K. Phylogenomic analysis restructures the ulvophyceae. J Phycol. 2021:57(4):1223–1233. 10.1111/jpy.13168 [DOI] [PubMed] [Google Scholar]
- Haas KT, Wightman R, Peaucelle A, Höfte H. The role of pectin phase separation in plant cell wall assembly and growth. Cell Surf. 2021:7:100054. 10.1016/j.tcsw.2021.100054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann A. The pherophorins: common, versatile building blocks in the evolution of extracellular matrix architecture in Volvocales. Plant J. 2006:45(2):292–307. 10.1111/j.1365-313X.2005.02627.x [DOI] [PubMed] [Google Scholar]
- He M, Yang Y, Shao Z, Zhang J, Feng C, Wang L, Mao W. Chemical structure and anticoagulant property of a novel sulfated polysaccharide from the green alga Cladophora oligoclada. Mar Drugs. 2021:19(10):554. 10.3390/md19100554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herburger K, Holzinger A. Localization and quantification of callose in the streptophyte green Algae Zygnema and Klebsormidium: correlation with desiccation tolerance. Plant Cell Physiol. 2015:56(11):2259–2270. 10.1093/pcp/pcv139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herburger K, Holzinger A. Aniline blue and Calcofluor white staining of callose and cellulose in the streptophyte green algae Zygnema and Klebsormidium. Bio Protoc. 2016:6(20):e1969. 10.21769/BioProtoc.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herburger K, Ryan LM, Popper ZA, Holzinger A. Localisation and substrate specificities of transglycanases in charophyte algae relate to development and morphology. J Cell Sci. 2018:131(2):jcs203208. 10.1242/jcs.203208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C, Siméon A, Jam M, Cassin A, Johnson KL, Salmeán AA, Willats WG, Doblin MS, Bacic A, Kloareg B. Arabinogalactan proteins have deep roots in eukaryotes: identification of genes and epitopes in brown algae and their role in fucus serratus embryo development. New Phytol. 2016:209(4):1428–1441. 10.1111/nph.13786 [DOI] [PubMed] [Google Scholar]
- Hess S, Williams SK, Busch A, Irisarri I, Delwiche CF, de Vries S, Darienko T, Roger AJ, Archibald JM, Buschmann H, et al. A phylogenomically informed five-order system for the closest relatives of land plants. Curr Biol. 2022:32(20):4473–4482.e7. 10.1016/j.cub.2022.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka M. Massive Ulva green tides caused by inhibition of biomass allocation to sporulation. Plants (Basel). 2021:10(11):2482. 10.3390/plants10112482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirooka S, Hirose Y, Kanesaki Y, Higuchi S, Fujiwara T, Onuma R, Era A, Ohbayashi R, Uzuka A, Nozaki H, et al. Acidophilic green algal genome provides insights into adaptation to an acidic environment. Proc Natl Acad Sci U S A. 2017:114(39):E8304–E8313. 10.1073/pnas.1707072114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A, Herburger K, Kaplan F, Lewis LA. Desiccation tolerance in the chlorophyte green alga Ulva compressa: does cell wall architecture contribute to ecological success? Planta. 2015:242(2):477–492. 10.1007/s00425-015-2292-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 2014:5(1):3978. 10.1038/ncomms4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YSY, Harris PJ. Xylans of red and green Algae: what is known about their structures and how they are synthesised? Polymers (Basel). 2019:11(2):354. 10.3390/polym11020354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iha C, Dougan KE, Varela JA, Avila V, Jackson CJ, Bogaert KA, Chen Y, Judd LM, Wick R, Holt KE, et al. Genomic adaptations to an endolithic lifestyle in the coral-associated alga Ostreobium. Curr Biol. 2021:31(7):1393–1402.e5. 10.1016/j.cub.2021.01.018 [DOI] [PubMed] [Google Scholar]
- Irisarri I, Darienko T, Pröschold T, Fürst-Jansen JMR, Jamy M, de Vries J. Unexpected cryptic species among streptophyte algae most distant to land plants. Proc Biol Sci. 2021:288(1963):2021–2168. 10.1098/rspb.2021.2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Busse-Wicher M, Poulsen CP, Fangel JU, Smith PJ, Yang JY, Peña MJ, Dinesen MH, Martens HJ, Melkonian M, et al. Identification of an algal xylan synthase indicates that there is functional orthology between algal and plant cell wall biosynthesis. New Phytol. 2018:218(3):1049–1060. 10.1111/nph.15050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao C, Sørensen I, Sun X, Sun H, Behar H, Alseekh S, Philippe G, Palacio Lopez K, Sun L, Reed R, et al. The Penium margaritaceum genome: hallmarks of the origins of land plants. Cell. 2020:181(5):1097–1111.e12. 10.1016/j.cell.2020.04.019 [DOI] [PubMed] [Google Scholar]
- Johnson KL, Cassin AM, Lonsdale A, Wong GK, Soltis DE, Miles NW, Melkonian M, Melkonian B, Deyholos MK, Leebens-Mack J, et al. Insights into the evolution of hydroxyproline-rich glycoproteins from 1000 plant transcriptomes. Plant Physiol. 2017:174(2):904–921. 10.1104/pp.17.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Schultz CJ, Bacic A. . Non-enzymic cell wall (glyco) proteins. In: Rose J, editor. The plant cell wall. Blackwell Publishing; 2003. p. 111–154. [Google Scholar]
- Jouenne F, Eikrem W, Le Gall F, Marie D, Johnsen G, Vaulot D. Prasinoderma singularis sp. nov. (prasinophyceae, chlorophyta), a solitary coccoid prasinophyte from the South-East Pacific Ocean. Protist. 2011:162(1):70–84. 10.1016/j.protis.2010.04.005 [DOI] [PubMed] [Google Scholar]
- Kermanshahi-Pour A, Sommer TJ, Anastas PT, Zimmerman JB. Enzymatic and acid hydrolysis of Tetraselmis suecica for polysaccharide characterization. Bioresour Technol. 2014:173:415–421. 10.1016/j.biortech.2014.09.048 [DOI] [PubMed] [Google Scholar]
- Kidgell JT, Carnachan SM, Magnusson M, Lawton RJ, Sims IM, Hinkley SFR, de Nys R, Glasson CRK. Are all ulvans equal? A comparative assessment of the chemical and gelling properties of ulvan from blade and filamentous Ulva. Carbohydr Polym. 2021:264:118010. 10.1016/j.carbpol.2021.118010 [DOI] [PubMed] [Google Scholar]
- Kidgell JT, Magnusson M, de Nys R, Glasson CRK. Ulvan: a systematic review of extraction, composition and function. Algal Res. 2019:39:101422. 10.1016/j.algal.2019.101422 [DOI] [Google Scholar]
- Kiemle SN, Domozych DS, Gretz MR. The extracellular polymeric substances of desmids (conjugatophyceae, streptophyta): chemistry, structural analyses and implications in wetland biofilms. Phycologia. 2007:46(6):617–627. 10.2216/06-97.1 [DOI] [Google Scholar]
- Kloareg B, Badis Y, Cock JM, Michel G. Role and evolution of the extracellular matrix in the acquisition of Complex multicellularity in eukaryotes: a macroalgal perspective. Genes (Basel). 2021:12(7):1059. 10.3390/genes12071059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox P. Delving in the deep for the origin of plant cell surface proteoglycans. New Phytol. 2016:209(4):1341–1343. 10.1111/nph.13862 [DOI] [PubMed] [Google Scholar]
- Kondo S, Hori K, Sasaki-Sekimoto Y, Kobayashi A, Kato T, Yuno-Ohta N, Nobusawa T, Ohtaka K, Shimojima M, Ohta H. Primitive extracellular lipid components on the surface of the charophytic alga Klebsormidium flaccidum and their possible biosynthetic pathways as deduced from the genome sequence. Front Plant Sci. 2016:7:952. 10.3389/fpls.2016.00952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Abedin MM, Singh AK, Das S. Role of phenolic compounds in plant-defensive mechanisms. In: Lone RShuab R, Kamili A, editors. Plant phenolics in sustainable agriculture. Singapore: Springer; 2020. p.517.–. [Google Scholar]
- Lahaye M, Robic A. Structure and functional properties of ulvan, a polysaccharide from green seaweeds. Biomacromolecules. 2007:8(6):1765–1774. 10.1021/bm061185q [DOI] [PubMed] [Google Scholar]
- Laroche C. Exopolysaccharides from microalgae and Cyanobacteria: diversity of strains, production strategies, and applications. Mar Drugs. 2022:20(5):336. 10.3390/md20050336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritano C, De Luca D, Amoroso M, Benfatto S, Maestri S, Racioppi C, Esposito F, Ianora A. New molecular insights on the response of the green alga Tetraselmis suecica to nitrogen starvation. Sci Rep. 2019:9(1):3336. 10.1038/s41598-019-39860-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Waffenschmidt S, Small L, Goodenough U. Between-species analysis of short-repeat modules in cell wall and sex-related hydroxyproline-rich glycoproteins of Chlamydomonas. Plant Physiol. 2007:144(4):1813–1826. 10.1104/pp.107.100891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leebens-Mack JH, Barker MS, Carpenter EJ, Deyholos MK, Gitzendanner MA, Graham SW, Grosse I, Li Z, Melkonian M, Mirarab S, et al. One thousand plant transcriptomes initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019:574(7780):679–685. 10.1038/s41586-019-1693-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F. Green algae: chlorophyta and streptophyta. In Thomas Schmidt ed. Encyclopedia of microbiology (4th ed). London: Academic Press; 2019. p. 457–468. [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O. Phylogeny and molecular evolution of the green Algae. Crit Rev Plant Sci. 2012:31(1):1–46. 10.1080/07352689.2011.615705 [DOI] [Google Scholar]
- Li J, He Z, Liang Y, Peng T, Hu Z. Insights into algal polysaccharides: a review of their structure, depolymerases, and metabolic pathways. J Agric Food Chem. 2022:70(6):1749–1765. 10.1021/acs.jafc.1c05365 [DOI] [PubMed] [Google Scholar]
- Li X, Hou Z, Xu C, Shi X, Yang L, Lewis LA, Zhong B. Large phylogenomic data sets reveal deep relationships and trait evolution in chlorophyte green Algae. Genome Biol Evol. 2021:13(7):evab101. 10.1093/gbe/evab101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FS, Phyo P, Jacobowitz J, Hong M, Weng JK. The molecular structure of plant sporopollenin. Nat Plants. 2019:5(1):41–46. 10.1038/s41477-018-0330-7 [DOI] [PubMed] [Google Scholar]
- Li L, Wang S, Wang H, Sahu SK, Marin B, Li H, Xu Y, Liang H, Li Z, Cheng S, et al. The genome of Prasinoderma coloniale unveils the existence of a third phylum within green plants. Nat Ecol Evol. 2020:4(9):1220–1231. 10.1038/s41559-020-1221-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Geng Y, Ji C, Du H, Wong CE, Zhang Q, Zhang Y, Zhang P, Riaz A, Chachar S, et al. Mesostigma viride genome and transcriptome provide insights into the origin and evolution of Streptophyta. Adv Sci (Weinh). 2019:7(1):1901850. 10.1002/advs.201901850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Miao Y, Huang H, Zhang Y, Huang L, Cao J. Arabinogalactan proteins: focus on the role in cellulose synthesis and deposition during plant cell wall biogenesis. Int J Mol Sci. 2022:23(12):6578. 10.3390/ijms23126578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wolfe R, Welch LR, Domozych DS, Popper ZA, Showalter AM. Bioinformatic identification and analysis of extensins in the plant kingdom. PLoS One. 2016:11(2):e0150177. 10.1371/journal.pone.0150177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Hortas L, Flórez-Fernández N, Torres MD, Ferreira-Anta T, Casas MP, Balboa EM, Falqué E, Domínguez H. Applying seaweed compounds in cosmetics. Cosmeceuticals and nutricosmetics. Marine Drug. 2021:19(10):552. 10.3390/md19100552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love J. Microbial pathways for advanced biofuel production. Biochem Soc Trans. 2022:50(2):987–1001. 10.1042/BST20210764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas PL, Mathieu-Rivet E, Song PCT, Oltmanns A, Loutelier-Bourhis C, Plasson C, Afonso C, Hippler M, Lerouge P, Mati-Baouche N, et al. Multiple xylosyltransferases heterogeneously xylosylate protein N-linked glycans in Chlamydomonas reinhardtii. Plant J. 2021:102(2):230–245. 10.1111/tpj.14620 [DOI] [PubMed] [Google Scholar]
- Lütz-Meindl U. Micrasterias as a model system in plant cell biology. Front Plant Sci. 2016:7:999. 10.3389/fpls.2016.00999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancipe NC, McLaughlin EM, Barney BM. Genomic analysis and characterization of Scenedesmus glucoliberatum PABB004:an unconventional sugar-secreting green alga. J Appl Microbiol. 2021:132(3):2004–2019. 10.1111/jam.15311 [DOI] [PubMed] [Google Scholar]
- Matt G, Umen J. Volvox: a simple algal model for embryogenesis, morphogenesis and cellular differentiation. Dev Biol. 2016:419(1):99–113. 10.1016/j.ydbio.2016.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattox KR, Stewart KD. Classification of the green algae: a concept based on comparative cytology. In: Irvine DEG, John DM, editors. Systematics of the green Algae. London: Academic Press; 1984. p. 29–72. [Google Scholar]
- Meinita MDN, Harwanto D, Choi JS. A concise review of the bioactivity and pharmacological properties of the genus Codium (Bryopsidales, Chlorophyta). J Appl Phycol. 2022:34(6):2827–2845. 10.1007/s10811-022-02842-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007:318(5848):245–250. 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Harholt J, Ulvskov P, Johansen IE, Fangel JU, Doblin MS, Bacic A, Willats WG. Evidence for land plant cell wall biosynthetic mechanisms in charophyte green algae. Ann Bot. 2014:114(6):1217–1236. 10.1093/aob/mcu171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Harholt J, Westereng B, Domozych D, Fry SC, Johansen IE, Fangel JU, Łężyk M, Feng T, Nancke L, et al. Ancient origin of fucosylated xyloglucan in charophycean green algae. Commun Biol. 2021:41(1):754. 10.1038/s42003-021-02277-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine I, Sekida S. Fibrous matrix component of cell wall in the giant-celled green alga Valonia utricularis observed by atomic force microscopy in liquid. Protoplasma. 2018:255(5):1575–1579. 10.1007/s00709-018-1251-z [DOI] [PubMed] [Google Scholar]
- Mišurcová L, Škrovánková S, Samek D, Ambrožová J, Machů L. Health benefits of algal polysaccharides in human nutrition. Adv Food Nutr Res. 2012:66:75–145. 10.1016/B978-0-12-394597-6.00003-3 [DOI] [PubMed] [Google Scholar]
- Moestrup O, Walne PL. Studies on scale morphogenesis in the Golgi apparatus of Pyramimonas tetrarhynchus (Prasinophyceae). J Cell Sci. 1979:36(1):437–459. 10.1242/jcs.36.1.437 [DOI] [PubMed] [Google Scholar]
- Møller SR, Yi X, Velásquez SM, Gille S, Hansen PLM, Poulsen CP, Olsen CE, Rejzek M, Parsons H, Yang Z, et al. Identification and evolution of a plant cell wall specific glycoprotein glycosyl transferase, ExAD. Sci Rep. 2017:7(1):45341. 10.1038/srep45341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau H, Verhelst B, Couloux A, Derelle E, Rombauts S, Grimsley N, Van Bel M, Poulain J, Katinka M, Hohmann-Marriot MF, et al. Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol. 2012:13(8):R74. 10.1186/gb-2012-13-8-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira LR, Filho EX. An overview of mannan structure and mannan-degrading enzyme systems. Appl Microbiol Biotechnol. 2008:79(2):165–178. 10.1007/s00253-008-1423-4 [DOI] [PubMed] [Google Scholar]
- Nazos TT, Kokarakis EJ, Valsami EA, Stratigakis NC, Poloniataki EG, Sfendourakis GP, Ghanotakis DF. Characterization of a novel herbicide and antibiotic-resistant Chlorella sp. with an extensive extracellular matrix. Photosynth Res. 2020:143(3):315–334. 10.1007/s11120-020-00710-5 [DOI] [PubMed] [Google Scholar]
- Negreanu-Pirjol BS, Negreanu-Pirjol T, Popoviciu DR, Anton RE, Prelipcean AM. Marine bioactive compounds derived from macroalgae as new potential players in drug delivery systems: a review. Pharmaceutics. 2022:14(9):1781. 10.3390/pharmaceutics14091781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen K. The organism strikes back: Chlorella algae and their impact on photosynthesis research, 1920s-1960s. Hist Philos Life Sci. 2017:39(2):9. 10.1007/s40656-017-0137-2 [DOI] [PubMed] [Google Scholar]
- Nicolas WJ, Fäßler F, Dutka P, Schur FKM, Jensen G, Meyerowitz E. Cryo-electron tomography of The Onion cell wall shows bimodally oriented cellulose fibers and reticulated homogalacturonan networks. Curr Biol. 2022:32(11):2375–2389.e6. 10.1016/j.cub.2022.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas KJ, Cobb ED, Matas AJ. The evolution of hydrophobic cell wall biopolymers: from algae to angiosperms. J Exp Bot. 2017:68(19):5261–5269. 10.1093/jxb/erx215 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Sakayama H, de Vries J, Buschmann H, Saint-Marcoux D, Ullrich KK, Haas FB, Vanderstraeten L, Becker D, Lang D, et al. The chara genome: secondary complexity and implications for plant terrestrialization. Cell. 2018:174(2):448–464.e24. 10.1016/j.cell.2018.06.033 [DOI] [PubMed] [Google Scholar]
- Osorio H, Tapia-Reyes P, Espinoza D, Laporte D, González A, Castro-Nallar E, Moenne A. The genome of the marine alga Ulva compressa (chlorophyta) reveals protein-coding genes with similarity to plants and green microalgae, but also to animal, bacterial, and fungal genes. Int J Mol Sci. 2022:23(13):7279. 10.3390/ijms23137279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacio-Lopez K, Sun L, Reed R, Kang E, Sørensen I, Rose JKC, Domozych DS. Experimental manipulation of pectin architecture in the cell wall of the unicellular Charophyte, Penium margaritaceum. Front Plant Sci. 2020:11:1032. 10.3389/fpls.2020.01032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacio-López K, Tinaz B, Holzinger A, Domozych DS. Arabinogalactan proteins and the extracellular matrix of charophytes: a sticky business. Front Plant Sci. 2019:10:447. 10.3389/fpls.2019.00447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenick B, Grimwood J, Aerts A, Grigoriev IV. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci U S A. 2007:104(18):7705–7710. 10.1073/pnas.0611046104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perasso L, Grunow A, Brüntrup IM, Bölinger B, Melkonian M, Becker B. The Golgi apparatus of the scaly green flagellate Scherffelia dubia: uncoupling of glycoprotein and polysaccharide synthesis during flagellar regeneration. Planta. 2000:210(4):51–62. 10.1007/s004250050044 [DOI] [PubMed] [Google Scholar]
- Pereira AM, Pereira LG, Coimbra S. Arabinogalactan proteins: rising attention from plant biologists. Plant Reprod. 2015:28(1):1–15. 10.1007/s00497-015-0254-6 [DOI] [PubMed] [Google Scholar]
- Permann C, Becker B, Holzinger A. Temperature- and light stress adaptations in Zygnematophyceae: the challenges of a semi-terrestrial lifestyle. Front Plant Sci. 2022:13:945394. 10.3389/fpls.2022.945394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permann C, Herburger K, Felhofer M, Gierlinger N, Lewis LA, Holzinger A. Induction of conjugation and zygospore cell wall characteristics in the alpine Spirogyra mirabilis (Zygnematophyceae, Charophyta): advantage under climate change scenarios? Plants (Basel). 2021a:10(8):1740. 10.3390/plants10081740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permann C, Herburger K, Niedermeier M, Felhofer M, Gierlinger N, Holzinger A. Cell wall characteristics during sexual reproduction of Mougeotia sp. (Zygnematophyceae) revealed by electron microscopy, glycan microarrays and RAMAN spectroscopy. Protoplasma. 2021b:258(6):1261–1275. 10.1007/s00709-021-01659-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer L, Utermöhlen J, Happ K, Permann C, Holzinger A, von Schwartzenberg K, Classen B. Search for evolutionary roots of land plant arabinogalactan-proteins in charophytes: presence of a rhamnogalactan-protein in Spirogyra pratensis (Zygnematophyceae). Plant J. 2022:109(3):568–584. 10.1111/tpj.15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe G, Sørensen I, Jiao C, Sun X, Fei Z, Domozych DS, Rose JK. Cutin and suberin: assembly and origins of specialized lipidic cell wall scaffolds. Curr Opin Plant Biol. 2020:55:11–20. 10.1016/j.pbi.2020.01.008 [DOI] [PubMed] [Google Scholar]
- Pichrtová M, Remias D, Lewis LA, Holzinger A. Changes in phenolic compounds and cellular ultrastructure of Arctic and antarctic strains of Zygnema (Zygnematophyceae, Streptophyta) after exposure to experimentally enhanced UV to PAR ratio. Microb Ecol. 2013:65(1):68–83. 10.1007/s00248-012-0096-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps JD. Green Algae: structure, reproduction and evolution in selected genera. Sunderland, MA: Sinauer Associates, Inc; 1975. [Google Scholar]
- Polle JE, Barry K, Cushman J, Schmutz J, Tran D, Hathwaik LT, Yim WC, Jenkins J, McKie-Krisberg Z, Prochnik S, et al. Draft nuclear genome sequence of the halophilic and Beta-carotene-accumulating green alga Dunaliella salina strain CCAP19/18. Genome Announc. 2017:5(43):e01105–17. 10.1128/genomeA.01105-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Hervé C, Domozych DS, Willats WG, Tuohy MG, Kloareg B, Stengel DB. Evolution and diversity of plant cell walls: from algae to flowering plants. Annu Rev Plant Biol. 2011:62(1):567–590. 10.1146/annurev-arplant-042110-103809 [DOI] [PubMed] [Google Scholar]
- Přerovská T, Henke S, Bleha R, Spiwok V, Gillarová S, Yvin JC, Ferrières V, Nguema-Ona E, Lipovová P. Arabinogalactan-like glycoproteins from ulva lactuca (chlorophyta) show unique features compared to land plants AGPs. J Phycol. 2021:57(2):619–635. 10.1111/jpy.13121 [DOI] [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010:329(5988):223–226. 10.1126/science.1188800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proseus TE, Boyer JS. Pectate chemistry links cell expansion to wall deposition in Chara corallina. Plant Signal Behav. 2012:7(11):1490–1492. 10.4161/psb.21777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Townsley BT, Ichihashi Y, Sinha NR, Chitwood DH. An intracellular atlas of the giant coenocyte Caulerpa taxifolia. PLoS Genet. 2015:11(1):e1004900. 10.1371/journal.pgen.1004900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA. How plants conquered land. Cell. 2020:181(5):964–966. 10.1016/j.cell.2020.05.011 [DOI] [PubMed] [Google Scholar]
- Rieseberg TP, Dadras A, Fürst-Jansen JMR, Dhabalia Ashok A, Darienko T, de Vries S, Irisarri I, de Vries J. Crossroads in the evolution of plant specialized metabolism. Semin Cell Dev Biol. 2023:134:37–58. 10.1016/j.semcdb.2022.03.004 [DOI] [PubMed] [Google Scholar]
- Rodrigues MA, da Silva Bon EP. Evaluation of chlorella (chlorophyta) as source of fermentable sugars via cell wall enzymatic hydrolysis. Enzyme Res. 2011:2011:405603. 10.4061/2011/405603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roleda MY, Heesch S. Chemical profiling of Ulva species for food applications: what is in a name? Food Chem. 2021:361:130084. 10.1016/j.foodchem.2021.130084 [DOI] [PubMed] [Google Scholar]
- Rydahl MG, Krac Un SK, Fangel JU, Michel G, Guillouzo A, Génicot S, Mravec J, Harholt J, Wilkens C, Motawia MS, et al. Development of novel monoclonal antibodies against starch and ulvan—implications for antibody production against polysaccharides with limited immunogenicity. Sci Rep. 2017:7(1):9326. 10.1038/s41598-017-04307-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Merchant SS. A series of fortunate events: introducing Chlamydomonas as a reference organism. Plant Cell. 2019:31(8):1682–1707. 10.1105/tpc.18.00952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satjarak A, Graham LE. Genome-wide analysis of carbohydrate-active enzymes in Pyramimonas parkeae (Prasinophyceae). J Phycol. 2017:53(5):1072–1086. 10.1111/jpy.12566 [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010:61(1):263–289. 10.1146/annurev-arplant-042809-112315 [DOI] [PubMed] [Google Scholar]
- Scherp P, Grotha R, Kutschera U. Occurrence and phylogenetic significance of cytokinesis-related callose in green algae, bryophytes, ferns and seed plants. Plant Cell Rep. 2001:20(2):143–149. 10.1007/s002990000301 [DOI] [PubMed] [Google Scholar]
- Schneider R, Hanak T, Persson S, Voigt CA. Cellulose and callose synthesis and organization in focus, what's New? Curr Opin Plant Biol. 2016:34:9–16. 10.1016/j.pbi.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Schreiber M, Rensing SA, Gould SB. The greening ashore. Trends Plant Sci. 2022:27(9):847–857. 10.1016/j.tplants.2022.05.005 [DOI] [PubMed] [Google Scholar]
- Shao Z, Duan D. The cell wall polysaccharides biosynthesis in seaweeds: a molecular perspective. Front Plant Sci. 2022:13:902823. 10.3389/fpls.2022.902823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Ferraz R, Dupree P, Showalter AM, Coimbra S. Three decades of advances in arabinogalactan-protein biosynthesis. Front Plant Sci. 2020:11:610377. 10.3389/fpls.2020.610377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, McHale M, Sulpice R. Applications of Ulva biomass and strategies to improve its yield and composition: a perspective for Ulva aquaculture. Biology (Basel). 2022:11(11):1593. 10.3390/biology11111593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair R, Hsu G, Davis D, Chang M, Rosquete M, Iwasa JH, Drakakaki G. Plant cytokinesis and the construction of new cell wall. FEBS Lett. 2022:596(17):2243–2255. 10.1002/1873-3468.14426 [DOI] [PubMed] [Google Scholar]
- Smetacek V, Zingone A. Green and golden seaweed tides on the rise. Nature. 2013:504(7478):84–88. 10.1038/nature12860 [DOI] [PubMed] [Google Scholar]
- Soni VK, Krishnapriya R, Sharma RK. Algae: biomass to biofuel. Methods Mol Biol. 2021:2290:31–51. 10.1007/978-1-0716-1323-8_3 [DOI] [PubMed] [Google Scholar]
- Soós V, Shetty P, Maróti IN, Badics E, Bálint ÖV, Balázs E. Biomolecule composition and draft genome of a novel, high-lipid producing Scenedesmaceae microalga. Algal Res. 2020:54:102181. 10.1016/j.algal.2020.102181 [DOI] [Google Scholar]
- Sørensen I, Domozych D, Willats WGT. How have plant cell walls evolved? Plant Physiol. 2010:153(2):366–372. 10.1104/pp.110.154427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen I, Pettolino FA, Bacic A, Ralph J, Lu F, O'Neill MA, Fei Z, Rose JK, Domozych DS, Willats WG. The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 2011:68(2):201–211. 10.1111/j.1365-313X.2011.04686.x [DOI] [PubMed] [Google Scholar]
- Spribille T, Resl P, Stanton DE, Tagirdzhanova G. Evolutionary biology of lichen symbioses. New Phytol. 2022:234(5):1566–1582. 10.1111/nph.18048 [DOI] [PubMed] [Google Scholar]
- Spribille T, Tagirdzhanova G, Goyette S, Tuovinen V, Case R, Zandberg WF. 3D Biofilms: in search of the polysaccharides holding together lichen symbioses. FEMS Microbiol Lett. 2020:367(5):fnaa023. 10.1093/femsle/fnaa023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkenburg SR, Polle JEW, Hovde B, Daligault HE, Davenport KW, Huang A, Neofotiz P, McKie-Krisberg Z. Draft nuclear genome, complete chloroplast genome, and complete mitochondrial genome for the biofuel/bioproduct feedstock Species scenedesmus obliquus strain DOE0152z. Genome Announc. 2017:5(32):e00617–17. 10.1128/genomeA.00617-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner P, Obwegeser S, Wanner G, Buchner O, Lütz-Meindl U, Holzinger A. Cell wall reinforcements accompany chilling and freezing stress in the streptophyte green alga Klebsormidium crenulatum. Front Plant Sci. 2020:11:873. 10.3389/fpls.2020.00873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh DY, Ashton NW. A sporopollenin definition for the genomics age. New Phytol. 2022:236(6):2009–2013. 10.1111/nph.18484 [DOI] [PubMed] [Google Scholar]
- Sulastri E, Lesmana R, Zubair MS, Elamin KM, Wathoni N. A comprehensive review on ulvan based hydrogel and its biomedical applications. Chem Pharm Bull (Tokyo). 2021:69(5):432–443. 10.1248/cpb.c20-00763 [DOI] [PubMed] [Google Scholar]
- Sumper M, Hallmann A. Biochemistry of the extracellular matrix of Volvox. Int Rev Cytol. 1998:180:51–85. 10.1016/S0074-7696(08)61770-2 [DOI] [PubMed] [Google Scholar]
- Taleb A, Pruvost J, Legrand J, Marec H, Le-Gouic B, Mirabella B, Legeret B, Bouvet S, Peltier G, Li-Beisson Y, et al. Development and validation of a screening procedure of microalgae for biodiesel production: application to the genus of marine microalgae Nannochloropsis. Bioresour Technol. 2015:177:224–232. 10.1016/j.biortech.2014.11.068 [DOI] [PubMed] [Google Scholar]
- Thygesen A, Fernando D, Ståhl K, Daniel G, Mensah M, Meyer AS. Cell wall configuration and ultrastructure of cellulose crystals in green seaweeds. Cellulose. 2021:28(5):2763–2778. 10.1007/s10570-021-03698-w [DOI] [Google Scholar]
- Ulvskov P, Paiva DS, Domozych D, Harholt J. Classification, naming and evolutionary history of glycosyltransferases from sequenced green and red algal genomes. PLoS One. 2013:8(10):e76511. 10.1371/journal.pone.0076511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG. Volvox and volvocine green algae. Evodevo. 2020:11(1):13. 10.1186/s13227-020-00158-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Poel B, Cooper ED, Van Der Straeten D, Chang C, Delwiche CF. Transcriptome profiling of the green alga Spirogyra pratensis (charophyta) suggests an ancestral role for ethylene in cell wall metabolism, photosynthesis, and abiotic stress responses. Plant Physiol. 2016:172(1):533–545. 10.1104/pp.16.00299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannerum K, Huysman MJ, De Rycke R, Vuylsteke M, Leliaert F, Pollier J, Lütz-Meindl U, Gillard J, De Veylder L, Goossens A, et al. Transcriptional analysis of cell growth and morphogenesis in the unicellular green alga Micrasterias (Streptophyta), with emphasis on the role of expansin. BMC Plant Biol. 2011:11(1):128. 10.1186/1471-2229-11-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage L. Tear down that wall—cell wall remodeling in charophyte algae. Plant J. 2021:108(1):5–6. 10.1111/tpj.15504 [DOI] [PubMed] [Google Scholar]
- Voigt J, Kiess M, Getzlaff R, Wöstemeyer J, Frank R. Generation of the heterodimeric precursor GP3 of the Chlamydomonas cell wall. Mol Microbiol. 2010:77(6):1512–1526. 10.1111/j.1365-2958.2010.07302.x [DOI] [PubMed] [Google Scholar]
- Voiniciuc C. Modern mannan: a hemicellulose's Journey. New Phytol. 2022:234(4):1175–1184. 10.1111/nph.18091 [DOI] [PubMed] [Google Scholar]
- von der Heyde B, Hallmann A. Cell type-specific pherophorins of Volvox carteri reveal interplay of both cell types in ECM biosynthesis. Cells. 2022:12(1):134. 10.3390/cells12010134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlström N, Edlund U, Pavia H, Toth G, Jaworski A, Pell AJ, Choong FX, Shirani H, Nilsson KP, Richter-Dahlfors A. Cellulose from the green macroalgae Ulva lactuca: isolation, characterization, optotracing, and production of cellulose nanofibrils. Cellulose. 2020a:27(7):3707–3725. 10.1007/s10570-020-03029-5 [DOI] [Google Scholar]
- Wahlström N, Nylander F, Malmhäll-Bah E, Sjövold K, Edlund U, Westman G, Albers E. Composition and structure of cell wall ulvans recovered from Ulva spp. along the Swedish west coast. Carbohydr Polym. 2020b:233:115852. 10.1016/j.carbpol.2020.115852 [DOI] [PubMed] [Google Scholar]
- Wang S, Li L, Li H, Sahu SK, Wang H, Xu Y, Xian W, Song B, Liang H, Cheng S, et al. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat Plants. 2020:6(2):95–106. 10.1038/s41477-019-0560-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WN, Li T, Li Y, Zhang Y, Wu HL, Xiang WZ, Li AF. Exopolysaccharides from the energy microalga strain Botryococcus braunii: purification, characterization, and antioxidant activity. Foods. 2022:11(1):110. 10.3390/foods11010110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S, Grande PM, Blank LM, Klose H. Insights into cell wall disintegration of Chlorella vulgaris. PLoS One. 2022:17(1):e0262500. 10.1371/journal.pone.0262500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss TL, Roth R, Goodson C, Vitha S, Black I, Azadi P, Rusch J, Holzenburg A, Devarenne TP, Goodenough U. Colony organization in the green alga Botryococcus braunii (Race B) is specified by a complex extracellular matrix. Eukaryot Cell. 2012:11(12):1424–1440. 10.1128/EC.00184-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichard T. From model organism to application: bacteria-induced growth and development of the green seaweed Ulva and the potential of microbe leveraging in algal aquaculture. Semin Cell Dev Biol. 2023:134:69–78. 10.1016/j.semcdb.2022.04.007 [DOI] [PubMed] [Google Scholar]
- Wichard T, Charrier B, Mineur F, Bothwell JH, Clerck OD, Coates JC. The green seaweed Ulva: a model system to study morphogenesis. Front Plant Sci. 2015:6:72. 10.3389/fpls.2015.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TH, Kumar M, Turner SR. The molecular basis of plant cellulose synthase complex organisation and assembly. Biochem Soc Trans. 2021:49(1):379–391. 10.1042/BST20200697 [DOI] [PubMed] [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009:324(5924):268–272. 10.1126/science.1167222 [DOI] [PubMed] [Google Scholar]
- Xiao R, Zheng Y. Overview of microalgal extracellular polymeric substances (EPS) and their applications. Biotechnol Adv. 2016:34(7):1225–1244. 10.1016/j.biotechadv.2016.08.004 [DOI] [PubMed] [Google Scholar]
- Yamashita S, Yamamoto K, Matsuzaki R, Suzuki S, Yamaguchi H, Hirooka S, Minakuchi Y, Miyagishima SY, Kawachi M, Toyoda A, et al. Genome sequencing of the multicellular alga astrephomene provides insights into convergent evolution of germ-soma differentiation. Sci Rep. 2021:11(1):22231. 10.1038/s41598-021-01521-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gao Y, Zhang L, Zhou Y. The plant cell wall: biosynthesis, construction, and functions. J Integr Plant Biol. 2021:63(1):251–272. 10.1111/jipb.13055 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wang X, Chen Y, Wagner E, Cosgrove DJ. Xyloglucan in the primary cell wall: assessment by FESEM, selective enzyme digestions and nanogold affinity tags. Plant J. 2018:93(2):211–226. 10.1111/tpj.13778 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data is available upon request from the author, DSD.