Summary
CRISPR-Cas genome engineering in the unicellular green algal model Chlamydomonas reinhardtii has until recently suffered from low integration efficiencies despite traditional genetics being well established. Here, we present a protocol for efficient homology-directed knockin mutagenesis in all commonly used strains of Chlamydomonas. We describe steps for scarless integration of fusion tags and sequence modifications of almost all proteins without the need for a preceding mutant line. We further empower this genetic-editing approach by efficient crossing and highly robust screening protocols.
For complete details on the use and execution of this protocol, please refer to Nievergelt et al. (2023).1
Subject areas: Cell Biology, CRISPR, Genetics, Molecular Biology
Graphical abstract

Highlights
-
•
CRISPR-Cas allows efficient gene disruption and endogenous protein tagging
-
•
Small-volume electroporation reduces reagent waste and improves cell viability
-
•
A highly robust (q)-PCR-based screening procedure reduces variability
-
•
FACS is used to increase the speed of crossing experiments
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
CRISPR-Cas genome engineering in the unicellular green algal model Chlamydomonas reinhardtii has until recently suffered from low integration efficiencies despite traditional genetics being well established. Here, we present a protocol for efficient homology-directed knockin mutagenesis in all commonly used strains of Chlamydomonas. We describe steps for scarless integration of fusion tags and sequence modifications of almost all proteins without the need for a preceding mutant line. We further empower this genetic-editing approach by efficient crossing and highly robust screening protocols.
Before you begin
This protocol describes an efficient pipeline for precisely editing the nuclear genome of Chlamydomonas to generate knock-outs as well as endogenous knock-in tags. Moreover, we developed streamlined protocols for crossing, screening, and verifying mutants with the goal of producing the exact mutant line needed for a particular project in minimal time and manual work by using equipment widely available in most laboratories.
New mutants can be created from a variety of backgrounds, such as commonly available wild type strains, insertional mutants such as the CLiP library or rescue mutants. We use quantitative PCR (qPCR) to genotype or screen candidate cells with minimal human effort before verification of the selected cells with Sanger sequencing followed ideally by next-generation whole genome sequencing (WGS).
Cell culture
Timing: 3–10 days
-
1.
Dissolve a streak of the desired source cell line in TAP medium and plate on TAP in 1.5% agar.
Note: Plating is best achieved with a spreading tool made from a glass Pasteur pipette bent into a triangular shape with the help of a Bunsen burner.
Note: In our hands, basically any Chlamydomonas strain can be used as a source cell line, as long as there is no overlap between an already present antibiotic resistance and the resistance of the intended CRISPR insertion.
-
2.
Let cells grow in a 14 h light / 10 h dark cycle. Cells are usually ready for use after 3 days and can be used for a week thereafter.
Autolysin preparation
Timing: 1 week
Autolysin is the supernatant of a mating reaction of Chlamydomonas gametes which contains the cell wall digesting enzyme MMP1 (previously called gamete lytic enzyme Cre17.g718500_4532.1). This protocol is an adapted version of the Waffenschmidt protocol.2
-
3.
Inoculate 10 × 100 mm TAP agar (1.5%) plates each of CC-620 (mt+) and CC-621 (mt-) cells either from dense liquid culture (100 μL/plate) or scraped from a plate and dissolved in TAP medium, while aiming for an even spread.
-
4.
Grow plates in constant light for 5–7 days until cells are stationary.
-
5.Flood plates in the morning with 5–10 mL TAP-N.
-
a.Scrape off cells with an inoculation loop, collecting plus and minus cells separately
-
b.Centrifuge each at 600 RCF for 10 min.
-
c.Discard supernatant to remove cell debris.
-
d.Resuspend with ∼10× pellet volume.
-
e.Pour the cells into separate 2 L beakers on a light board or under strong lights.
-
a.
Note: The cell suspension when harvesting cells from plates can be moved from plate to plate to reduce the total volume.
Note: 10 × 10 mm plates typically result in about 2–3 mL of pellet volume and can be resuspended in ∼25 mL TAP-N.
-
6.
Periodically mix 20 μL of the two mating types and check under a microscope for mating activity. Mating can generally be observed after 5 h post washing, but full mating competence is usually reached about 9 h post wash (See also Methods video S1).
Note: Mating competence can be assessed directly after combining opposite mating types by the presence of cells aggregated into mating clusters. The few cells are freely swimming the higher the mating efficiency is likely to be. Additionally, the mating efficiency can be confirmed 20–30 min after initiating mating by adding ∼2 μL of the mating mixture to a drop of ∼10 μL of TAP + 2 mM EGTA on a microscope slide which makes cells adhere. The fraction of cells with four cilia can be easily estimated this way using a phase microscope and should be above 75%.
-
7.When mating is at full efficiency, combine all cells in one beaker.
-
a.Allow mating to proceed for 20–30 min.
-
b.Remove cells by centrifugation at 5000 RCF for 10 min.Note: Autolysin prepared this way usually has a slight red tinge.
-
c.Collect supernatant and filter sterilize through a 0.22 μm filter.
-
d.Freeze sterile autolysin in aliquots at −20°C for short-term (few months) or −80°C for long-term (years) storage.Note: The produced autolysin can be tested by adding it to a small pellet of a non-hatching Chlamydomonas strain grown in liquid culture. The cells of such strains are found in clusters of multiple cells enclosed by parental cell walls (palmelloids) which dissolve within about a minute of adding autolysin to release single cells. We use CC-4375 IFT46::NIT for this but any aciliate strain should work just as well.
-
a.
Clusters of cells, stuck by their cilia can be observed in many places and freshly shed cell walls appear as dark shells. Related to preparation step Autolysin preparation and Crossing by FACS.
Guide sequence selection
Timing: 10 min
As the variable element of a CRISPR-Cas experiment, the guide RNA is crucial and needs to be carefully selected by trading off three main properties in order of importance: specificity, location and activity. Specificity ensures that the chosen guide RNA is sufficiently unique across the whole genome to minimize off-target cuts. Neglecting to check a guide for off-targets can in the best-case result in erroneous integration and in the worst case cause chromosomal recombination and consequently cross-lethal lines. The high GC content of Chlamydomonas is for once advantageous, since the probability of finding the protospacer motif NGG is very high.
The location of the guide defines the place of genomic cleavage and subsequently the integration of the donor DNA.
For knock-out lines, any guide cutting one of the first few exons of a gene is usually sufficient.
When targeting a knock-in tag, the cut site should be chosen close to the targeted terminus, ensuring the cut site is in the coding sequence (see Figure 1, options 2–5). Importantly, for any chosen cut site, the surrounding region should be verified a priori by Sanger sequencing for every individual source cell line, as any nucleotide differences with respect to the reference will result in a significant reduction of transformation efficiency.
Note: Activity scoring can improve the efficiency of cutting, resulting in higher transformation efficiency. We use the “Doench 2016” algorithm included in Geneious Prime.3
Note: We use Geneious prime which includes off-target and activity scoring by default. Alternatives tools that could be used for guide RNA design are: Benchling (https://www.benchling.com/), CasOFFfinder (http://www.rgenome.net/cas-offinder/), CHOPCHOP (http://chopchop.cbu.uib.no/) or CRISPOR (http://crispor.tefor.net/).
Note: Guides without off-targets and a Doench activity score >0.4 usually work well in our hands. For a positive control, the ARMC2 co-targeting source vectors (available at CRC, see Data S1 for maps) can be used directly after digestion with BspQI in conjunction with the gRNA CTGACGACCGTAGTCCGCTGGGG. This combination should result in >75% of colonies with paralyzed cilia.
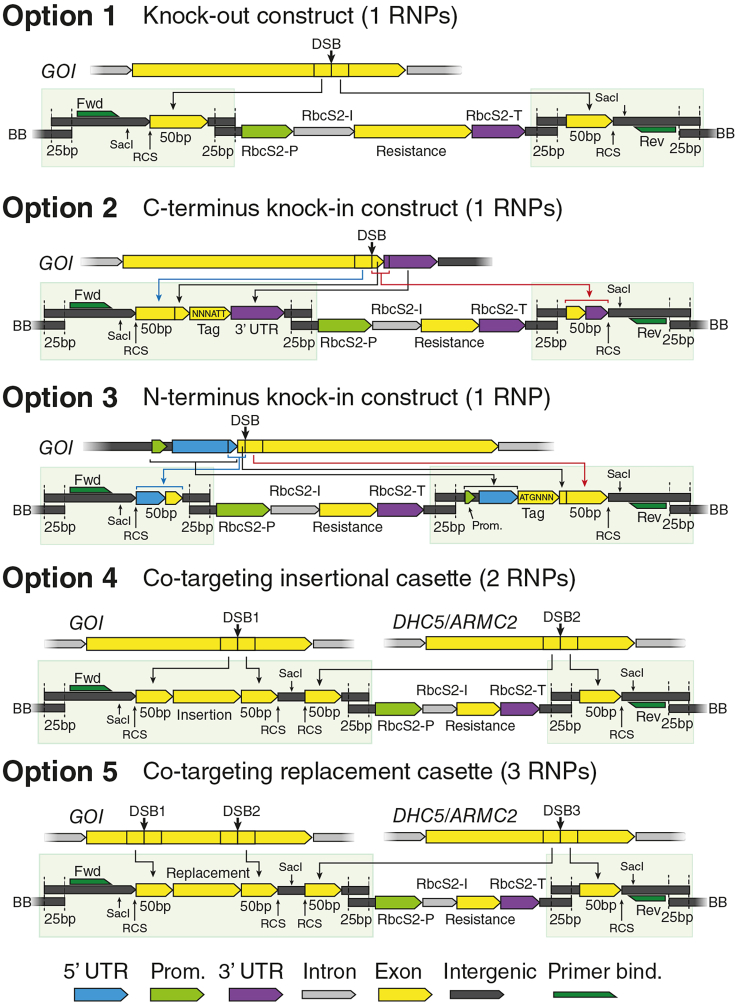
Figure 1.
Design options for Donor DNA design and corresponding guide RNA placement
Donor DNA design
Timing: 1 h
While the exact makeup of the donor DNA is dependent on the intended genetic modification, a resistance cassette is generally included, consisting of a promoter, resistance CDS and a terminator. For this reason, we have designed vectors containing resistance cassettes for Paromomycin (AphVIII),4 Nourseothricin (NAT),5 Spectinomycin (AadA)6 and Blasticidin S (BSD)7 under the RbcS2 promoter-terminator pair. We flanked the cassette with unique EcoRI and NheI sites to split the cassette from the backbone and allow for easy cloning of homology arms by ligation to synthetic linkers by Gibson assembly. For any new construct, linkers have to be designed upstream and downstream of the cassette. Apart from the homology arms flanking each insert, the design of the inserts is limited mainly by creativity, but should follow general Chlamydomonas genetic lore and care should be taken to design for appropriate codon bias, introns and UTRs. A design that does not work as an insertional rescue is unlikely to work in an endogenous edit either.
Non-coding linker upstream of resistance (Linker 1):
| Element | Length (bp) |
|---|---|
| Homology to the bacterial backbone for Gibson assembly | 18–25 |
| Artificial primer binding site | 18–30 |
| Diagnostic SacI site | 6 |
| Type IIs restriction site (BspQI or BtsI) | 6-8 |
| Genomic sequence upstream of the CRISPR cut site | 50 |
| Homology to the resistance cassette for Gibson assembly | 18-25 |
Non-coding linker downstream of resistance (Linker 2):
| Element | Length (bp) |
|---|---|
| Homology to resistance cassette for Gibson assembly | 18-25 |
| Genomic sequence downstream of the CRISPR cut site | 50 |
| Type IIs restriction site (BspQI or BtsI) | 6-8 |
| Diagnostic SacI site | 6 |
| Artificial primer binding site | 18-30 |
| Homology to the bacterial backbone for Gibson assembly | 18-25 |
Protein-coding linker upstream of resistance (Linker 3):
| Element | Length (bp) |
|---|---|
| Homology to the bacterial backbone for Gibson assembly | 18–25 |
| Artificial primer binding site | 18–30 |
| Diagnostic SacI site | 6 |
| Type IIs restriction site (BspQI or BtsI) | 6–8 |
| Genomic sequence upstream of the CRISPR cut site | 50 |
| Genomic sequence between the CRISPR cut site and up to the endogenous stop codon, mutated to not include the gRNA sequence. | Variable |
| Linker peptide and/or epitope tag (Optional but recommended) | Variable |
| The coding sequence of the desired fusion tag, followed by a stop codon | Variable |
| The complete 3′ UTR of the endogenous gene | Variable |
| Homology to the resistance cassette for Gibson assembly | 18–25 |
Protein coding linker downstream of resistance (Linker 4):
| Element | Length (bp) |
|---|---|
| Homology to resistance cassette for Gibson assembly | 18–25 |
| The complete 5′ UTR of the endogenous gene, plus ∼300 bp extra on the 5′ end to include remote promoters. Alternatively, a different, known promoter can be used. | Variable |
| A start codon followed by the coding sequence of the desired fusion tag. Adding introns is highly recommended. | Variable |
| Linker peptide and/or epitope tag (Optional but recommended) | Variable |
| The genomic sequence from the endogenous start codon to the CRISPR cut site, mutated to not include the gRNA sequence | Variable |
| Genomic sequence downstream of the CRISPR cut site | 50 |
| Type IIs restriction site (BspQI or BtsI) | 6–8 |
| Diagnostic SacI site | 6 |
| Artificial primer binding site | 18–30 |
| Homology to the bacterial backbone for Gibson assembly | 18–25 |
Co-targeting linker upstream of resistance (Linker 5):
| Element | Length (bp) |
|---|---|
| Homology to resistance cassette for Gibson assembly | 18–25 |
| Type IIs restriction site (BspQI or BtsI) | 6–8 |
| Genomic sequence upstream of the primary CRISPR cut site | 50 |
| The desired insert sequence, usually matched to the frame of the homology arms | Variable |
| Genomic sequence upstream of the primary CRISPR cut site | 50 |
| Type IIs restriction site (BspQI or BtsI) directed upstream | 6–8 |
| Diagnostic SacI site | 6 |
| Type IIs restriction site (BspQI or BtsI) directed downstream | 6–8 |
| Genomic sequence upstream of the secondary CRISPR cut site | 50 |
| Homology to the bacterial backbone for Gibson assembly | 18–25 |
-
8.Chose a design option (see tables above for linker constituents).
-
a.For a gene disruption, construct linkers 1 and 2. See Figure 1, option 1.
-
b.For a c-terminal knock-in construct linkers 3 and 2. See Figure 1, option 2.
-
c.For an n-terminal knock-in construct linkers 1 and 4. See Figure 1, option 3.
-
d.For a co-targeting design construct linker 5 and linker 1 targeted to a secondary gene with a known phenotype. See Figure 1, option 4 or option 5.
-
a.
Note: This option results in a multi-fragment cassette that is co-targeted to two places in the genome by two to three different RNPs. This option results in a line where the selectable marker is inserted in a known secondary locus on a different chromosome (e.g., DRC4, ARMC2 or other gene that results in an observable phenotype when disrupted). The selectable marker can then be crossed out in a second step (see Crossing by FACS).
Note: Construction of this type of donor can be simplified using the pAPN_noAB-ARMC2-∗ vectors that already encode the secondary insertion. A primary insertion can easily be cloned in by replacing the NruI flanked ccdB cassette with a homology-flanked insertion sequence.
CRITICAL: This design results in significantly fewer correct clones (usually in the few percent) and higher screening effort is to be expected.
-
9.In a DNA editor, verify correct assembly of the designed linkers with the virtually digested source plasmid by virtual homology cloning.
-
a.Verify that all guide sequences used are mutated to prevent RNPs from degrading correct insertions.
-
b.Verify that the resulting insert does not contain the chosen type IIs site, except the intentional ones flanking the insert. Change type IIs if necessary.
-
a.
Note: Restriction sites in the backbone portion of the resulting plasmids are usually unproblematic and can usually be safely ignored.
Cloning
Timing: 3–4 days
Note: For fully annotated working example sequences for the different options, see the “Examples” folder in Data S1.
The designed fragments can be purchased as full synthetic pieces or split into a mix of multiple smaller fragments and purchased or amplified by PCR (Figure 2).
-
10.Mix all components to roughly 200 fmol in a total of 5 μL. See Data S1 for examples.Note: PCR products generally don’t need to be purified and 0.2–0.5 μL can be used directly for most fragments.
-
a.Add 5 μL of NEBuilder HiFi master mix.Alternatives: a different Gibson assembly master mix can be used if so desired.
-
b.Incubate for 30 min at 50°C.
-
c.Transform chemically competent E. coli with 2 μL of the assembly reaction, diluted with sterile water to 10 μL by heat shocking for 45 s at 42°C on a thermoblock.
-
d.Recover cells by adding 1 mL of SOC medium.
-
e.Plate 20 μL and 100 μL immediately or after 5–15 min of recovery at 37°C on selective LB agar plates.Note: This procedure usually results in ∼50–200 colonies for the 20 μL plate after 14–18 h at 37°C.
-
a.
-
11.Mini-prep 3–6 colonies.Optional: Constructs can easily be pre-screened by restriction digestion with SacI.
 CRITICAL: CRISPR-Cas donor plasmids tend to contain repeat regions that in seldom cases will result in highly efficient recombination in E. coli, which results in exclusively failed assemblies. While uncommon, in case cloning by Dh5α or other common cloning strains repeatedly fails with mutations that aren’t attributable to PCR or Gibson assembly, use recombination deficient strains such as NEB Stable or Invitrogen Stbl4.
CRITICAL: CRISPR-Cas donor plasmids tend to contain repeat regions that in seldom cases will result in highly efficient recombination in E. coli, which results in exclusively failed assemblies. While uncommon, in case cloning by Dh5α or other common cloning strains repeatedly fails with mutations that aren’t attributable to PCR or Gibson assembly, use recombination deficient strains such as NEB Stable or Invitrogen Stbl4.-
a.Verify the resulting DNA by Sanger or NGS (whole plasmid) sequencing.
-
b.Midi-prep with a high purity kit such as ZymoPure II Midiprep.Note: Midi-preps can be done by re-transforming the verified DNA or using retained bacterial stocks from before mini-prepping.
-
a.
-
12.To prepare the final construct for transformation into Chlamydomonas, digest 20–30 μg with the chosen type IIs enzyme (BspQI/BtsI).
-
a.Column purify using a Zymo Clean and Concentrator-25 kit or equivalent.
 CRITICAL: Avoid ethanol precipitation or columns where droplets of residual buffer can get trapped.Note: Eluted DNA should be > 300 ng/μL. Every transformation will need about 1.5 μg of digested donor DNA.
CRITICAL: Avoid ethanol precipitation or columns where droplets of residual buffer can get trapped.Note: Eluted DNA should be > 300 ng/μL. Every transformation will need about 1.5 μg of digested donor DNA. -
b.Freeze eluted DNA for 12 h at -20°C to increase sterility.
 Pause point: Frozen donor DNA can be stored for years without loss of activity.
Pause point: Frozen donor DNA can be stored for years without loss of activity.
-
a.
Figure 2.
Molecular cloning vector using modular resistance vectors
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli DH5α | NEB | Cat#C2987H |
| E. coli GB06 | Lab stock | N/A |
| E. coli DB3.1 | Lab stock | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| K2HPO4 | Sigma-Aldrich | P3786 |
| KH2PO4 | Merck | 1.04873 |
| MgSO4·7H2O | Sigma-Aldrich | 3138 |
| NH4Cl | Sigma-Aldrich | A9434 |
| CaCl2·2H2O | Merck | C7902 |
| KCl | Merck | 1.04936 |
| Tris base | Sigma-Aldrich | T1503 |
| Acetic acid, glacial | Merck | 1.00063 |
| Agar | Fisher Scientific | BD 214010 |
| Sucrose | Alfa Aesar | 36508 |
| Alt-R S.p. Cas9 Nuclease V3 | IDT | Cat#1081058 |
| Platinum SuperFi II PCR master mix (2×) | Thermo Fisher Scientific | Cat#12368010 |
| Platinum II Hot-Start PCR master mix (2×) | Thermo Fisher Scientific | Cat#14000013 |
| Paromomycin | TCI | P2092 |
| Nourseothricin | Jena Bioscience | AB-102XL |
| Blasticidin S | Carl Roth | CP14.2 |
| Spectinomycin | Merck | S4014-5G |
| EvaGreen Plus | Biotium | 31077-T |
| GelGreen | Biotium | 41005 |
| Betaine | Merck | 61962 |
| NEBuilder HiFi assembly master mix | NEB | E2621L |
| BspQI | NEB | R0712L |
| BtsI-v2 | NEB | R0667L |
| SacI-HF | NEB | R3156L |
| EcoRI-HF | NEB | R3101L |
| NheI-HF | NEB | R3131S |
| Hutner’s trace elements | Chlamydomonas Resource Center | N/A |
| IGEPAL CA-630 | Merck | I8896 |
| Difco Bacto Agar | BD | BD 214010 |
| Nuclease-free duplex buffer | IDT | 11-05-01-12 |
| QuickExtract solution | Lucigen | QE09050 |
| Critical commercial assays | ||
| ZymoPURE II Plasmid Midiprep Kit | Zymo Research | D4200 |
| ZymoPURE II Plasmid Miniprep Kit | Zymo Research | D4211 |
| DNA Clean & Concentrator-25 | Zymo Research | D4033 |
| Neon electroporator | Thermo Fisher Scientific | MPK5000 |
| 10 μL Neon transfection kit | Thermo Fisher Scientific | MPK1025 |
| QuantStudio 7 Pro | Thermo Fisher Scientific | A43185 |
| Light board | Artograph | LightPad 940LX |
| FACS sorter | Sony | MA-900 |
| Experimental models: Organisms/strains | ||
| Chlamydomonas WT (+) | Chlamydomonas Resource Center | CC-620 |
| Chlamydomonas WT (-) | Chlamydomonas Resource Center | CC-621 |
| Chlamydomonas WT (-) | Dutcher Lab | CC124 32M |
| Chlamydomonas WT (+) | Chlamydomonas Resource Center | CC-125 |
| Oligonucleotides | ||
| Alt-R CRISPR-Cas9 tracrRNA, 20 nmol | IDT | Cat#1072533 |
| CRISPR-Cas9 crRNAs, 2 nmol | IDT | N/A |
| crMTM_q_1F | IDT | TGCTAGCTCAATCTGACTGAAGGT |
| crMTM_q_1R | IDT | GGGACATGGTGCTCGGGG |
| crMTP_q_1F | IDT | AGTATCACAGTACGGAATGCCC |
| crMTP_q_1R | IDT | TCATTGTTTGCTAGGGGTGC |
| Recombinant DNA | ||
| pAB262_NAT-R | This study | N/A |
| pAPN_BSD | This study | N/A |
| pAPN_AphVIII | This study | N/A |
| pAPN_AadA | This study | N/A |
| pAPN_noAB-ARMC2-BSD-ccdb | This study | N/A |
| pAPN_noAB-ARMC2-PAR-ccdb | This study | N/A |
| pAPN_noAB-ARMC2-NAT-ccdb | This study | N/A |
| Custom synthetic fragments | Twist Bioscience | N/A |
| Custom synthetic fragments | IDT | N/A |
| Custom synthetic fragments | Eurofins Genomics | N/A |
| Software and algorithms | ||
| Geneious Prime | Biomatters | 2021-2023.0.1 |
| Design and Analysis | Thermo Fisher Scientific | 2.6.0 |
| Fiji | Fiji | https://imagej.net/software/fiji/ |
| iQ3 | Andor | https://andor.oxinst.com/products/iq-live-cell-imaging-software/ |
Materials and equipment
Phosphate solution:
| Reagent | Final concentration | Amount |
|---|---|---|
| H2O | N/A | 80 mL |
| K2HPO4 | 1.65 M | 28.8 g |
| KH2PO4 | 1.06 M | 14.4 g |
| H2O | N/A | to 100 mL |
| Total | N/A | 100 mL |
Autoclave solution for 20 min at 140°C and store at 4°C. The solution is stable for at least 5 years if sealed properly.
TAP salts:
| Reagent | Final concentration | Amount |
|---|---|---|
| H2O | 800 mL | |
| NH4Cl | 280 mM | 15 g |
| MgSO4·7H2O | 16.2 mM | 4 g |
| CaCl2·2H2O | 13.6 mM | 2 g |
| H2O | to 1 L | |
| Total | N/A | 1 L |
Dissolve in 800 mL and adjust final volume to 1 L. Autoclave solution for 20 min at 140°C and store at 4°C. The solution is stable for at least 5 years.
TAP-N salts:
| Reagent | Final concentration | Amount |
|---|---|---|
| H2O | N/A | 800 mL |
| KCl | 282 mM | 21 g |
| MgSO4·7H2O | 16.2 mM | 4 g |
| CaCl2·2H2O | 13.6 mM | 2 g |
| H2O | to 1 L | |
| Total | N/A | 1 L |
Autoclave solution for 20 min at 140°C and store at 4°C. The solution is stable for at least 5 years
TAP Medium:
| Reagent | Final concentration | Amount |
|---|---|---|
| H2O | N/A | 850 mL |
| Tris base (H2NC(CH2OH)3) | 20 mM | 2.42 g |
| TAP salts | N/A | 25 mL |
| Phosphate solution | N/A | 1 mL |
| Hutner’s trace elements | N/A | 1 mL |
| Acetic acid, glacial | to pH 7 | ∼1.8 mL |
| H2O | N/A | to 1 L |
| Total | N/A | 1 L |
Add components in order and mix completely between additions. Autoclave solution for 20 min at 140°C and store at RT. The medium is stable for at least one year.
TAP-N Medium for gametogenesis:
| Reagent | Final concentration | Amount |
|---|---|---|
| H2O | N/A | 850 mL |
| Tris base (H2NC(CH2OH)3) | 20 mM | 2.42 g |
| TAP-N salts | N/A | 25 mL |
| Phosphate solution | N/A | 1 mL |
| Hutner’s trace elements | N/A | 1 mL |
| Acetic acid, glacial | to pH 7 | ∼1.8 mL |
| H2O | N/A | to 1 L |
| Total | N/A | 1 L |
Add components in order and mix completely between additions. Autoclave solution for 20 min at 140°C and store at RT. The medium is stable for at least one year.
-
•
1.5% TAP Agar: Add 4.5 g of Difco Agar to 300 mL of TAP medium in a 500 mL bottle and autoclave at 121°C for 20 min. Store at 4°C–30°C and melt in a microwave when pouring plates. Properly sealed bottles are stable for at least one year.
-
•
10× TAPS Medium for electroporation and recovery: Add 13.7 g of sucrose to 100 mL of TAP medium to a final concentration of 400 mM. Sterilize by passing through a 0.22 μm syringe filter and store in 10 mL aliquots at 4°C–30°C. Dilute to 1× TAPS by diluting with TAP. Aliquots are stable for at least one year.
Note: Sucrose can be used at up to 50 mM final without a notable change in survivability.
The electroporation part of the protocol relies on the Invitrogen Neon electroporation system. Unlike conventional electroporators this system does not use conventional vertical cuvettes and . We used a first generation Neon electroporator with 10 μL tips. The newer Neon NxT has been reported to work with this protocol, but consumables may be incompatible between systems. The required Buffer E to fill the pedestal tube of the Neon electroporator is included with the electroporation tips.
Most screening procedures in this protocol make heavy use of a qPCR machine. We have successfully used Roche Lightcycler 96 and Invitrogen QuantStudio 7 Pro systems in our work and we assume that most other qPCR instruments can be used with no or minimal changes, as long as the device supports the acquisition of high-resolution melting traces. If available, we recommend to use a 384 well capable cycler for screening procedures in order to save time and reagents.
We have generated a set of different resistance plasmids that can be used as a base for constructing new CRISPR donor DNA. The individual vectors have compatible ends and can be used interchangeably when following this protocol to select a resistance of choice:
| Plasmid name | Resistance (work conc.) | Restriction sites | Purpose |
|---|---|---|---|
| pAB262-NAR-R | Nourseothricin (7.5 μL/mL) | NheI, EcoRI | Monolithic insert |
| pAPN_AphVIII | Paromomycin (10 μL/mL) | NheI, EcoRI | Monolithic insert |
| pAPN_crAadA | Spectinomycin (100 μL/mL ) | NheI, EcoRI | Monolithic insert |
| pAPN_crBSDr | Blasticidin S (50 μL/mL) | NheI, EcoRI | Monolithic insert |
| pAPN_noAB-ARMC2-NAT-ccdB | Nourseothricin (7.5 μL/mL) | NruI | Cotargeting insert |
| pAPN_noAB-ARMC2-PAR-ccdB | Paromomycin (10 μL/mL) | NruI | Cotargeting insert |
| pAPN_noAB-ARMC2-BSD-ccdB | Blasticidin S (50 μL/mL) | NruI | Cotargeting insert |
The first set of vectors is used for general monolithic inserts with a fused antibiotic resistance. The second set allows for co-targeting constructs that insert the resistance for selection in the ARMC2 gene which results in paralyzed cilia and can later be crossed out.
CRITICAL: The co-targeting vectors are constructed with an excisable ccdB cassette that virtually eliminates cloning background, but requires ccdB resistant cells such as DB3.1 or ccdB survival cells to propagate.
Step-by-step method details
RNP assembly
Timing: 20 min
This step prepares the Cas9 ribonucleoprotein (RNP) complexes required for transformation.
The CRISPR-Cas system requires a guide RNA (gRNA) bound to the Cas9 enzyme, known together as the RNP. The gRNA is usually hybridized from a variable CRISPR RNA (crRNA) and an invariable trans-activating crRNA (tracrRNA). Optionally these two parts can be combined during synthesis and are commercially available as single-guide RNA (sgRNA).
Note: We used AltR Cas9, crRNA and tracrRNA from IDR for all of our work, but we expect RNA from other suppliers such as STEMCELL Technologies, Thermo Fisher or GenScript to work as well.
CRITICAL: crRNA and tracrRNA must be sourced from the same supplier, as suppliers may use proprietary hybridization regions which may not be compatible.
-
1.Dissolve crRNA and tracrRNA to 100 μM each in duplex buffer. If using sgRNA, dissolve to 50 μM and skip sub-step a.
-
a.Mix 5 μL of crRNA with 5 μL of tracrRNA.
-
b.Heat to 95°C for 2 min then let cool on the bench.Note: This results in 50 μM gRNA.
-
c.Mix 16 μL of duplex buffer with 2 μL of (s)gRNA and 1.6 μL of Cas9 enzyme (62 μM glycerol stock) in 1.5 mL tube
-
d.Flick the tube briefly or pulse vortex to mix.Note: This results in a 5 μM RNP solution.Note: In our hands, left-over gRNA can be frozen at −20°C for months without any notable decrease in efficiency.Note: We generally avoid storing assembled RNPs and instead prefer to scale down the amount of freshly prepared to match the number of needed reactions in order to reduce losses. One electroporation of 10 μL needs 1 μL of RNP solution and we generally do four repeat reactions.
-
a.
Transformation
Timing: 2 days
In this step, Chlamydomonas cells are transformed with donor DNA and RNPs to induce precise genetic changes.
Since transformation efficiency is cell cycle dependent, it is critical to perform this step at the proper time. Given a start of the light cycle at 8:00 o’clock, electroporation should generally be performed between 12:00 and 14:00. Perform all steps in a strictly sterile fashion.
In our hands, this transformation procedure results in a transformation efficiency of about 200 colonies per 10 μL electroporation reaction. Of picked colonies, on average 65%–90% have an insertion genotype sufficient to knock out the gene of interest and about 20% of colonies have an error-free scarless integration sufficient for endogenous tagging.
-
2.Harvest.
-
a.Scrape cells from the culture plate into TAP medium in a 1.5 mL tube.
-
b.Centrifuge at 600 RCF for 1 min and aspirate supernatant to remove debris.
-
c.Resuspend pellet in 100 μL TAP medium.
-
d.Measure new total volume to determine total pellet volume. For each planned mutant line, dispense the equivalent of 15 μL of pellet ( cells) into a new 1.5 mL tube.
-
a.
Note: Each aliquot from this step serves for transforming two different constructs.
-
3.Remove cell walls.
-
a.Fill up with autolysin to 1.2 mL.Note: For fully cell wall deficient lines such as CW92 autolysin can be replaced by TAP-N, however many cw strains still have a partial wall and should undergo the full autolysin treatment.
-
b.Using a higher concentration of cells will result in cell death after heat shock, which drastically reduces transformation efficiency. Incubate for 40–60 min.
-
c.Centrifuge cells at 500 RCF for 3 min.
-
d.Carefully aspirate supernatant.
-
e.Resuspend in fresh autolysin.
-
f.Incubate for 40–60 min at 20°C–30°C on a light board.
 CRITICAL: Cells without cell walls are much more delicate and can lyse when centrifuged too hard.
CRITICAL: Cells without cell walls are much more delicate and can lyse when centrifuged too hard. -
g.Mix a 20 μL sample with 1 μL of 10% IGEPAL CA-630 (0.5% final) and observe cell lysis under a phase contrast microscope. Lysing cells should dissolve smoothly and not leave behind remnants of the cell wall.
-
h.If any cell walls remain, repeat from step c.Note: Typically 2–3 cycles of autolysin treatment are needed to fully strip all cell walls.
- i.
-
a.
-
4.Set up electroporation environment.
-
a.Fill a clean, sterile Neon tube with 3 mL of Neon Buffer E.
-
b.Place the Neon tube in the Neon pedestal by clicking it in place.
-
c.Prepare a 24 well plate by filling the wells with 1 mL TAPS medium.Note: We usually repeat the electroporation up to 4 times. With 4 repeats a single 24 well plate can accommodate up to 6 different constructs.
-
d.Pick up a new Neon tip.
-
i.Fully pushing the plunger of the Neon Pipette
-
ii.Press the pipette vertically onto a fresh tip (Figures 3A and 3B) until the tip clicks into place.
-
iii.Release the plunger.Note: Each new construct should be electroporated with a fresh tip, but multiple repeats can be performed with the same tip.
-
i.
-
a.
-
5.Wash cells three times in TAPS.
 CRITICAL: MMP1, the cell wall removing enzyme in autolysin is highly cytotoxic when electroporated into cells and will result in cells dying if not removed thoroughly.
CRITICAL: MMP1, the cell wall removing enzyme in autolysin is highly cytotoxic when electroporated into cells and will result in cells dying if not removed thoroughly.-
a.Centrifuge at 500 RCF for 2 min.
-
b.Resuspend gently in 1.2 mL TAPS.
-
c.Repeat.
-
a.
-
6.Prepare electroporation reactions.
-
a.Centrifuge at 500 RCF for 2 min.
-
b.Resuspend cells in 50–90 μL TAPS to a final cell density of cells/mL.
-
c.Re-aliquot 50 μL of this solution per into fresh 1.5 mL tubes at one tube per intended construct.
-
d.Add 5 μL RNP solution to a tube.
-
e.Add 1.5 μg of cut donor DNA matching the RNPs to the same tube and flick tube to mix.
-
a.
-
7.Perform the electroporation.
-
a.Draw 10 μL of the final mixture into the Neon pipette, making sure there are no bubbles in the pipette (Figure 3C).Note: Unlike most electroporators, the Neon system can diagnose bad contacts and will alert the operator to the presence of bubbles or bad contacts.
-
b.Push pipette with filled pipette tip into the filled Neon tube placed in the pedestal until a click is felt (Figures 3D and 3E).
-
c.Electroporate for 3 pulses at 2300 V for 12 ms by setting the parameters on the Neon touch screen and pressing the large round “Start” button.Note: Some versions of the Neon system might fail at this step with a warning that the chosen parameters are over the power limit. Lowering the Voltage slightly to 2200 V should resolve the problem in these cases without significantly impacting the transformation result.
-
d.Discharge pipette into 1 mL of TAP + 50 mM sucrose in a 24 well plate.Note: Repeat this step up to 5 times per pipette (Figure 3F), one well per transformation. Pooling reactions is not recommended and should generally not be necessary.
 CRITICAL: When transforming multiple constructs using background strains which are already resistant to any of the resistance vectors used, transform resistant cells last or exchange the Neon cuvette to avoid contamination colonies.
CRITICAL: When transforming multiple constructs using background strains which are already resistant to any of the resistance vectors used, transform resistant cells last or exchange the Neon cuvette to avoid contamination colonies.
-
a.
-
8.Recover and select transformants.
-
a.Let the cells recover 16–24 h in the light.
 CRITICAL: Do not skip recovery as transformation efficiency will drastically decrease especially for Nourseothricin or Blasticidin S.
CRITICAL: Do not skip recovery as transformation efficiency will drastically decrease especially for Nourseothricin or Blasticidin S. -
b.Plate one well per plate on selective 1.5% TAP agar plates (Figure 3G).Note: An ethanol-sterilized glass spreader made from a long glass Pasteur pipette works best in our hands for achieving an even spread, but other spreading tools such as disposable plastic loops can be used.Note: In our hands, 2 plates per construct are almost always sufficient. On a single plate, typically between 100 and 800 colonies will form.Note: Concentrating the transformation mixture before plating by centrifugation at 500 RCF for 2 min is recommended to reduce the plating volume to ∼100 μL.
-
c.Wait for colonies to develop.
-
d.Pick 24–48 colonies for knock-outs and 48–96 colonies for knock-ins into a 96 well plate filled with 200 μL TAP per well.
-
e.Proceed to section Screening by qPCR.
-
a.
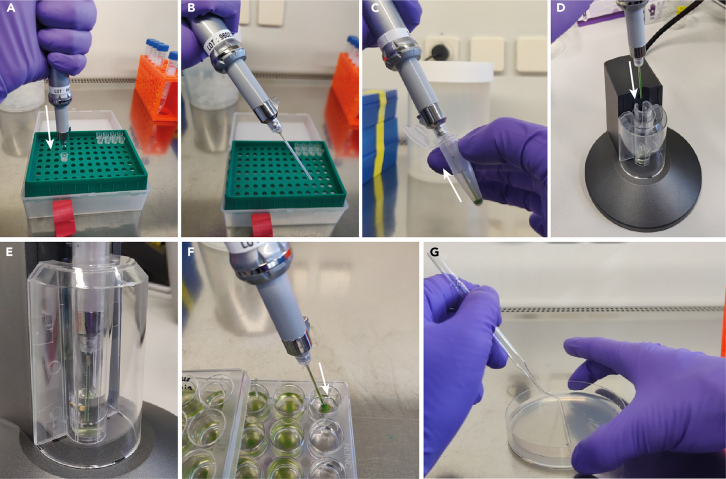
Figure 3.
Chlamydomonas electroporation procedure using the Neon electroporator
(A) Push the plunger of the positive displacement pipette to the end and push onto 10 μL Neon tip.
(B) Correctly attached Neon tip.
(C) Fill the Neon tip with cell/DNA/RNP mixture by slowly releasing the pressed plunger of the pipette, making sure no bubbles are aspirated.
(D) Insert the filled pipette into the electroporation pedestal.
(E) Fully inserted pipette with filled tip ready for electroporation by 3 pulses at 2300 V for 12 ms (set and started by Neon touchscreen).
(F) Release successfully electroporated reactions slowly into 1 mL TAPS buffer in a 24-well plate.
(G) After overnight recovery, spread one reaction per plate on 1.5% TAP-Agar plates with the correct antibiotic added. Use a homemade glass or disposable plastic spreading loop to distribute cells evenly across the plate.
Screening by qPCR
Timing: 3–4 h
Due to inefficiencies in the mutagenesis process, the picked colonies need to be screened for correct insertion. Here, a qPCR screen is used to identify candidates that have undergone the expected homology directed repair.
-
9.Prepare crude extracts.
-
a.Transfer 5–20 μL of cells of interest into a 96-well PCR plate.
-
b.Add the same volume of QuickExtract solution to each well and seal the plate with PCR foil.
-
c.Quickly vortex the plate to mix.
-
d.Centrifuge for a few seconds to collect all liquid in the bottom of the wells.
-
e.In a thermocycler, heat the plate to 65°C for 6 min, then to 98°C for 2 min to lyse cells.
-
f.Briefly vortex again.
-
a.
-
10.
Assemble 96-well or 384-well qPCR plate with 6 μL reactions with primers either specific to the desired genotype or a mix of the two primer pairs for each mating type.
The final reactions should consist of:
| Reagent | Amount |
|---|---|
| DNA template (cell extract) | 0.5 μL |
| Platinum 2 HS Master Mix | 3 μL |
| Primer 1 (20 mM) | 0.06 μL |
| Primer 2 (20 mM) | 0.06 μL |
| 2 M Betaine | 2.32 μL |
| EvaGreen (Plus) 20× in Water | 0.06 μL |
CRITICAL: Most PCR master mixes, especially those marketed for qPCR fail to amplify reliably when crude Chlamydomonas lysate is used. When using other products than suggested here, the reaction products should be verified for specificity before the master mix is used for large scale screening.
Note: It is highly recommended to premix all ingredients except the template (cell lysate) in bulk and dispense to reduce pipetting errors.
CRITICAL: Most PCR master mixes, especially those marketed for qPCR fail to amplify reliably when crude Chlamydomonas lysate is used. When using other products than suggested here, the reaction products should be verified for specificity before the master mix is used for large scale screening.
Note: It is highly recommended to premix all ingredients except the template (cell lysate) in bulk and dispense to reduce pipetting errors.
Note: When using a 384 well format, the total reaction can be halved to 3 μL per well, but care should be taken to work quickly to prevent wells from evaporating. We recommend electronic repeater pipettes for dispensing the PCR mix into the wells.
Note: When determining mating type by qPCR, two primer pairs (crMTM_q_1F, crMTM_q_1R, crMTP_q_1F and crMTP_q_1R) need to be used. We keep these primers at double the normal concentration for ease of use. These four primer pairs can be used as a reliable positive control and will result in two easily distinguishable peaks for the two mating types in high resolution melting analysis.
-
11.Run the qPCR reaction.
-
a.Seal plate with qPCR foil.
-
b.Briefly vortex to mix.
-
c.Centrifuge at 1000 RCF for 1 min to remove any air bubbles.
-
d.Cycle reactions in a qPCR machine with the following conditions:
-
a.
| Step | Temperature | Time | Cycles | Record |
|---|---|---|---|---|
| Initial Denaturation | 94°C | 2 min | 1 | No |
| Denaturation | 94°C | 15 s | 42 cycles | No |
| Annealing | 60°C | 20 s | No | |
| Extension | 68°C | 15 s/kB | Yes | |
| Denaturation | 95°C | 15 s | 1 | No |
| Annealing | 60°C | 1 min | 1 | No |
| High resolution melting | 60°C–98°C | 0.075 °C/s | 1 | Yes |
| Hold | 24°C | forever | No | |
-
12.
Analyze resulting melting curves for quantitation cycle Cq values per gene of interest (see Figure 4 for example).
Note: Successful amplifications generally have a Cq of 18-28, with higher values indicating the absence of the selected amplicon and subsequent non-specific amplification.
Note: Cq values for the same gene of interest should clearly cluster, but the average Cq can vary somewhat. Combined mating type primers should always amplify, and mating type can be directly determined from the high resolution melting profile of the finished reaction.
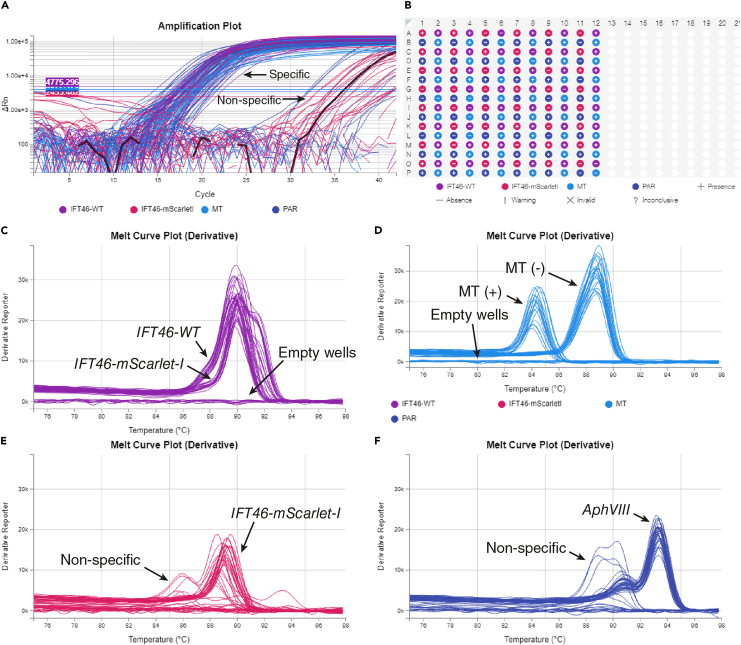
Figure 4.
qPCR analysis of FACS sorted progeny of back-crossing IFT46-mScarlet drc4::PAR x CC-125
(A) Amplification plot showing difference between positive amplifications (Cq<25) and nonspecific amplifications (Cq>25).
(B) Presence-absence plate map for all probed targets.
(C) High-resolution melting (HRM) of IFT-46 c-terminus amplification, showing two similar but distinct profiles for IFT46-mScarletI and wild-type IFT46.
(D) HRM based mating type analysis unambiguously distinguishing MT(-), MT(+) and empty wells.
(E) HRM of amplification specific to c-terminal mScarlet fusion of IFT46 with specific and non-specific curves.
(F) HRM of amplification specific to paromomycin resistance gene (AphVIII) with specific and non-specific curves.
Selection and verification
Timing: 2 days
Among the candidates identified in Screening by qPCR, only a subset will have integrated the donor DNA without additional mutations. The most common defects encountered are micro-homology driven fragment exchanges in the donor DNA. This step selects for mutants that have integrated the full donor DNA without additional unwanted changes.
Nuclear transformation in Chlamydomonas however suffers from unwanted side effects, ranging from commonly encountered non-specific integration of donor DNA, donor template recombination or Cas9 off-target activity, to rarer but highly problematic events such as interchromosomal crossover. In the context of creating engineered Chlamydomonas cell lines, genetic insertions should be unconditionally verified by sanger sequencing to confirm the integrity of the desired mutation.
Note: For cell lines that result from a crossing experiment, the results of the qPCR screening in combination with phenotypic controls is generally sufficient to select a progeny line for long term propagation or storage.
-
13.Select 5–10 colonies that were identified to have correct homology directed repair flanks in Screening by qPCR.
-
a.Select primers suitable to amplify the functional part of the wanted insertion.
-
b.Assemble 12 μL PCR reactions with the following composition:
-
a.
| Reagent | Amount |
|---|---|
| DNA template from Screening by qPCR | 0.5 μL |
| Platinum SuperFi2 Master Mix | 6 μL |
| Primer 1 (20 mM) | 0.3 μL |
| Primer 2 (20 mM) | 0.3 μL |
| 2 M Betaine | 4.9 μL |
Note: If extremely high GC content is present in the amplified regions (>30 bp islands over 90% GC) the added Betaine concentration can be increased up to 5 M (2 M final). This can slightly decrease amplification efficiency for amplicons with less GC content.
-
14.
Cycle using the following conditions:
| Step | Temperature | Time | Cycles |
|---|---|---|---|
| Initial Denaturation | 98°C | 2 min | 1 |
| Denaturation | 98°C | 15 s | 34 cycles |
| Annealing | 60°C | 20 s | |
| Extension | 72°C | 25 s/kB | |
| Hold | 24°C | forever | |
-
15.Select reactions with the expected size band.
-
a.Run 0.5 μL of the product in loading buffer on an agarose gel.
-
b.Identify bands with approximately the expected size.Note: The correct band is generally the consensus size on the gel.
-
c.Purify selected reactions.
-
d.Mix products with sequencing primers and send for Sanger sequencing.
-
a.
-
16.
Verify the absence of indels and SNPs in resulting sequences.
Crossing by FACS
Timing: 2 weeks
Here, the genome of two Chlamydomonas lines of opposite mating type is combined via meiosis in a rapid process that utilizes the ability of modern FACS sorters to isolate single cells directly from a liquid suspension.
Despite the general ease of introducing exogenous DNA into the genome of Chlamydomonas when following this protocol, it is still frequently desirable to cross different lines. Some common reasons for wanting to do so include:
Combining or transferring insertions: Crossing allows for rapid combination of multiple desired traits without the need for re-sequencing the loci of interest. Additionally, traits can generally be combined even if they express the same antibiotic resistances.
Detecting and removing unwanted insertions: Since Chlamydomonas cells tend to integrate exogenous DNA by random insertion, this can lead to undesired secondary inserts during CRISPR-Cas mediated mutagenesis. Back-crossing to a wild-type by this protocol offers simultaneously an easy readout for multiple insertions as well as a means of selecting for single-copy insertions. When screening the crossing progeny for the endogenous edit (by primers flanking the insertion) as well as for the used resistance (both primers on the resistance cassette), ideally only endogenous edit positives are also resistance positive. If endogenous edit negative but antibiotic resistance positive wells are detected, secondary insertions are likely present and re-crossing progeny with high Cq(resistance)/Cq(endogenous insert) is advised.
Changing mating type: If opposite mating types are required for downstream experiments, crossing naturally produces progeny of both mating types which can be identified purely based only on qPCR analysis. This is also a direct result of the back-crossing workflow above.
Conversely, there are cases in which crossing should be avoided:
Subtle phenotypes: If a gene of interest causes a very mild phenotype with no specific readout available, crossing can introduce phenotypic differences that outweigh the studied one, especially when very disparate source lines are used.
Genes in close proximity: If genes are in too close proximity on the same chromosome (<200 kbp) recombination frequency becomes prohibitively low and CRISPR-Cas editing should be used instead.
-
17.Derp Prepare mating competent cells.
-
a.Dissolve streaks of source cells of opposite mating type in TAP medium and plate on TAP 1.5% agar. Place plates in constant light for 5–7 days.
-
b.Scrape cells off both plates and dissolve in TAP-N medium in separate 1.5 mL tubes.
-
c.Wash cells once by centrifuging at 600 RCF for 1 min to remove debris and resuspend in at least 1 mL of TAP-N.
-
d.Leave cells for 6–8 h in high light. Ensure that cells develop cilia.
-
e.Mix a small aliquot of both mating types and check for mating under a microscope.
-
a.
-
18.Mate cells when sufficient competency is reached.
-
a.Combine both tubes into one.
-
b.Check for mated cells after 20–30 min in a phase microscope.Note: High efficiency mating like for autolysin preparation is not required for crossing.
-
c.Spread mating mixture on TAP 1.5% agar plates and place in constant light for about 18 h.
-
a.
-
19.
Wrap plates in aluminum foil and place in the dark for at least 5 days.
Pause point: Plates can be kept in the dark for weeks to months, but hatching efficiency will decrease over time.
-
20.Isolate mating progeny.
-
a.Remove aluminum foil.
-
b.Wash plates thoroughly by pipetting to remove all unmated cells.Note: A dense layer of highly adherent zygospores should be visible as a yellow sheen on the agar surface.
 CRITICAL: Continue washing until barely any swimming cells can be observed in a cell culture microscope.
CRITICAL: Continue washing until barely any swimming cells can be observed in a cell culture microscope. -
c.Place the plate in constant light and wait for zygospores to hatch (1–2 days).
-
d.Scrape some of the hatched cells off the plate with an inoculation loop and dissolve in 200–2000 μL of TAP medium in a FACS tube.
-
e.Load FACS tube into FACS machine.
-
f.Record forward and side scatter profile and sort the peak as single cells into a 96-well plate filled with TAP medium.
 CRITICAL: If palmelloids (multiple cells held together by a parental cell wall) are present in the solution, add 0.1 V autolysin to the tube and briefly vortex in order to sort only single cells.Note: sorting only the peak reduces the probability of sorting debris or cell clumps.
CRITICAL: If palmelloids (multiple cells held together by a parental cell wall) are present in the solution, add 0.1 V autolysin to the tube and briefly vortex in order to sort only single cells.Note: sorting only the peak reduces the probability of sorting debris or cell clumps.
-
a.
-
21.
Wait for cells to grow (typically ∼5–7 days), optionally re-plate by desired phenotype and proceed to Screening by qPCR.
Expected outcomes
The procedures described in this protocol enable the construction of genetically edited lines of arbitrary Chlamydomonas strains. The protocol is optimized towards robustness and minimizing operator work time. As such, the majority of process time is spent for cell growth. Our approach enables highly efficient tagging of proteins at either terminus by using a fused cassette which cannot be crossed out. Alternatively, the presented transformation and screening procedure is robust and efficient enough to co-target multiple insertions, which can be used to place a selective marker in a phenotypically scorable gene. Using the described crossing protocol, the selective marker can then be easily removed.
Limitations
While the presented method allows targeting and editing almost any gene of Chlamydomonas, modifications can have unintended effects. For example, some genes are lethal or cause severe growth defects when knocked out. The present method also does not inherently prevent unintended secondary insertions of vector fragments that are typical for Chlamydomonas genetics and back-crossing and sequencing strains for functional studies is highly recommended.
Troubleshooting
Problem 1
Low mating efficiency after mixing cells during Autolysin preparation.
Potential solution
-
•Ensure cells are healthy and ciliated:
-
○If few cells with cilia are present after several hours in TAP-N, start over.
-
○If cilia are present in >95% of cells, resuspend cells in fresh TAP-N and leave in high light for another 2–16 h.
-
○
Problem 2
Cloning of donor DNA only results in wrongly assembled colonies in section Cloning.
Potential solution
In some cases, common E.coli cloning strains will recombine inserts due to the repeats present in UTRs and homology arms. If there is evidence of recombination of fragments, use a recombination deficient strain such as NEB Stable or Stbl3 cells.
Problem 3
No or very few colonies form after transformation with Cas9 RNPs and donor DNA during Transformation.
Potential solution
-
•
When starting autolysin treatment, ensure cells are healthy and free of contamination.
-
•
Ensure the right antibiotic is used at the right concentration.
-
•
Some genes, when edited, are toxic or even lethal. Promoter replacement can be used to knock-down instead of knock-out a gene.
Problem 4
Sequenced inserts have scars close to the insertion site in Selection and verification.
Potential solution
-
•
Ensure the gRNA sequence is mutated in the donor DNA so that the donor DNA and the repaired genomic region will not be recut.
-
•
Make sure homology arms are centered 3 bp upstream of the PAM.
Problem 5
After crossing, spores don’t hatch or hatch at extremely low efficiency in step Crossing by FACS.
Potential solution
This likely indicates chromosomal translocations in one of the parental strains. Use a different clone for crossing and/or verify by long read sequencing.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gaia Pigino (gaia.pigino@fht.org).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Adrian Nievergelt (adrian@nievergelt-merz.ch).
Materials availability
-
•
Strains and plasmids generated in this study will be deposited to the Chlamydomonas Resource Center at time of publication.
Data and code availability
-
•
This protocol does not report new datasets.
-
•
This protocol does not report original code.
-
•
Any additional information required to reanalyze the data reported in this protocol is available from the lead contact upon request.
Acknowledgments
The authors would like to acknowledge M. Sarov, I. Reichardt-Gomez, and J. Koellner from the Genome Engineering Facility at MPI-CBG as well as the sequencing facility and the FACS Facility at MPI-CBG. We would also like to thank C. Peano and N. Alfonso from the Genomics Facility of Human Technopole. We would like to acknowledge support from the MPI-CBG Light Microscopy Facility. A.P.N. was supported by an EMBO Long-term Fellowship under ALTF number 891–2018 as well as by an HFSP Cross-disciplinary Fellowship with reference number LT000515/2019. We would like to acknowledge the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 819826) and the DFG grant GACR-DFG Cooperation 2019 no. PI1218/3-1 to G.P. T.B. is supported by DFG (INST 269/768-1).
Author contributions
Conceptualization, A.P.N. and G.P.; methodology, A.P.N. and D.R.D.; investigation, A.P.N., D.R.D., and A.B.; data curation, A.P.N.; visualization, A.P.N.; formal analysis, A.P.N.; sequencing analysis, T.B.; writing – original draft, A.P.N.; writing – review and editing, A.P.N., D.R.D., and G.P.; funding acquisition, A.P.N. and G.P.; resources, G.P.; supervision, G.P.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102774.
Contributor Information
Adrian Pascal Nievergelt, Email: adrian@nievergelt-merz.ch.
Gaia Pigino, Email: gaia.pigino@fht.org.
Supplemental information
Related to steps Guide sequence selection, Donor DNA design and Cloning, as well as Figures 1 and 2.
References
- 1.Nievergelt A.P., Diener D.R., Bogdanova A., Brown T., Pigino G. Efficient precision editing of endogenous Chlamydomonas reinhardtii genes with CRISPR-Cas. Cell Reports Methods. 2023;3 doi: 10.1016/j.crmeth.2023.100562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waffenschmidt S. 2005. Preparation of Gamete Autolysin-Improved Protocol - Chlamydomonas Resource Center.https://www.chlamycollection.org/methods/preparation-of-gamete-autolysin-improved-protocol/ [Google Scholar]
- 3.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sizova I., Fuhrmann M., Hegemann P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene. 2001;277:221–229. doi: 10.1016/s0378-1119(01)00616-3. [DOI] [PubMed] [Google Scholar]
- 5.Yang X., Peng J., Pan J. Nourseothricin N-acetyl transferase (NAT), a new selectable marker for nuclear gene expression in Chlamydomonas. Plant Methods. 2019;15:140. doi: 10.1186/s13007-019-0526-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meslet-Cladière L., Vallon O. Novel shuttle markers for nuclear transformation of the green alga chlamydomonas reinhardtii. Eukaryot. Cell. 2011;10:1670–1678. doi: 10.1128/EC.05043-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Carpentier F., Le Peillet J., Boisset N.D., Crozet P., Lemaire S.D., Danon A. Blasticidin S Deaminase: A New Efficient Selectable Marker for Chlamydomonas reinhardtii. Front. Plant Sci. 2020;11:242. doi: 10.3389/fpls.2020.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greiner A., Kelterborn S., Evers H., Kreimer G., Sizova I., Hegemann P. Targeting of Photoreceptor Genes in Chlamydomonas reinhardtii via Zinc-Finger Nucleases and CRISPR/Cas9. Plant Cell. 2017;29:2498–2518. doi: 10.1105/tpc.17.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Picariello T., Hou Y., Kubo T., McNeill N.A., Yanagisawa H., Oda T., Witman G.B. TIM, a targeted insertional mutagenesis method utilizing CRISPR/Cas9 in Chlamydomonas reinhardtii. PLoS One. 2020;15 doi: 10.1371/journal.pone.0232594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clusters of cells, stuck by their cilia can be observed in many places and freshly shed cell walls appear as dark shells. Related to preparation step Autolysin preparation and Crossing by FACS.
Related to steps Guide sequence selection, Donor DNA design and Cloning, as well as Figures 1 and 2.
Data Availability Statement
-
•
This protocol does not report new datasets.
-
•
This protocol does not report original code.
-
•
Any additional information required to reanalyze the data reported in this protocol is available from the lead contact upon request.