Abstract
Using interaction trap technology, we identified a putative extracytoplasmic-function (ECF) sigma factor (RpoE1) in Myxococcus xanthus, a bacterium which has a complex life cycle that includes fruiting body formation. The first domain of the response regulator protein FrzZ, a component of the Frz signal transduction system, was used as bait. Although the RpoE1 protein displayed no interactions with control proteins presented as bait, a weak interaction with a second M. xanthus response regulator (AsgA) was observed. While the specificity of the FrzZ-RpoE1 interaction therefore remains speculative, cloning and sequencing of the region surrounding rpoE1 localized it to a position downstream of the frzZ gene. A potential promoter site for binding of an ECF sigma factor was identified upstream of rpoE1, suggesting the gene may be autoregulated. However, primer extension studies suggested that transcription of rpoE1 occurs under both vegetative and developmental conditions from a ς70-like promoter. Dot blot analysis of RNA preparations confirmed the low-level, constitutive expression of rpoE1 during both stages of the life cycle. Analysis of an insertion mutant also indicated a role for RpoE1 under both vegetative and developmental conditions, since swarming was reduced on nutrient-rich agar and developmental aggregation was effected under starvation conditions, especially at high cell densities. An insertion mutation introduced into the gene directly downstream of rpoE1 (orf5) did not result in either swarming or developmental aggregation defects, even though the gene is transcribed as part of the same operon. Therefore, we propose that this new ECF sigma factor could play a role in the transcriptional regulation of genes involved in motility behavior during both stages of the complex M. xanthus life cycle.
Myxococcus xanthus moves via a surface-dependent motility mechanism called gliding, which, although still poorly understood, is known to involve many genetic loci (8). Hodgkin and Kaiser (9) first demonstrated that gliding motility involves two distinct systems: A (adventurous gliding), which allows movement of individual cells when isolated from other cells, and S (social gliding), which has recently been shown to be similar to twitching motility in Pseudomonas aeruginosa, since both involve type IV pili (25). Both A- and S-motility systems contribute to the wild-type gliding phenotype, and both systems appear to be under directional control from the Frz signal transduction system. Directed motility is of particular interest in M. xanthus since groups of cells can display complex motility behaviors, swarming under nutrient-rich conditions or, alternatively, aggregating into fruiting bodies when starved.
The Frz signal transduction system shows striking homology to the Che system of the enteric bacteria. However, chemotactic signal transduction in this slow-moving bacterium is still only partially understood (24). Whereas the enteric bacteria respond to defined chemical stimuli by using transmembrane receptor proteins (the methyl-accepting chemotaxis proteins [MCPs]), the MCP homologue in M. xanthus (FrzCD) has no membrane-spanning regions and must therefore either interact with membrane-associated sensors or detect stimuli within the cytoplasm. FrzCD is, however, methylated and demethylated during developmental aggregation (13) and vegetative swarming (17). This methylation and demethylation of FrzCD suggest that chemotaxis, which requires such adaptive modifications, may play a role in both fruiting body formation and colony dispersal, although chemokinesis (which does not require adaptation) has also been suggested to be involved in swarming behavior (23). The FrzE protein, which shows homology to both CheA and CheY, appears to act similarly to the enteric Che proteins; i.e., it is capable of autophosphorylation, and the phosphate group can be transferred from the histidine protein kinase (CheA domain) to the response regulator (CheY) domain (1). The Frz system is also known to involve several additional protein components, including FrzB and FrzZ. While little is known about the function of either protein, frzZ mutants show reduced swarming and developmental aggregation defects while retaining responses to repellent stimuli (21).
To date, no interactions between the motility-regulating Frz signal transduction system and the motility machinery of M. xanthus have been identified. We have used interaction trap methodology (6) to facilitate the identification of such interactions. Both the FrzE and FrzZ proteins have been cloned as bait into the two-hybrid system and used to screen a M. xanthus genomic DNA library for interacting proteins. These proteins were chosen as baits because they both encode CheY-like response regulator domains, which should presumably be involved in regulating output from the Frz system and might therefore be of use in the identification of motor proteins. The FrzZ protein is composed of two domains which both show homology to CheY but are quite distinct with respect to each other. These domains were therefore cloned separately into the two-hybrid system to determine if they interact with different proteins. In this study, we report an unexpected, potential interaction between the first domain of FrzZ and a putative extracytoplasmic-function (ECF) sigma factor involved in regulation of motility-associated behavior.
MATERIALS AND METHODS
Strains and culture conditions.
The strains used in this study are listed in Table 1. Plasmids are listed in Table 2. M. xanthus strains were grown in CYE medium (3), and developmental assays were performed on CF starvation media (7). Escherichia coli strains were grown in LB media. Yeast strains were grown in YPAD rich media or minimal SD media containing appropriate amino acids and 2% glucose (Stratagene).
TABLE 1.
Strains used in this study
| Strain | Genotype or relevant properties | Reference or source |
|---|---|---|
| M. xanthus | ||
| DZF1 | sglA (leaky) (A+ S−) | 3 |
| DZ2 | Wild type (A+ S+) | 4 |
| DZF4206 | rpoE1 sglA | This study |
| DZF4207 | orf5 sglA | This study |
| DZ24208 | rpoE1 | This study |
| DZ24209 | orf5 | This study |
| E. coli | ||
| XL1-Blue | General cloning strain | Stratagene |
| Top10 | Cloning strain of choice for pZErO; does not contain lacIq gene, allowing constitutive expression of ccdB | Invitrogen |
| S. cerevisiae YRG-2 | Reporter genes = lacZ and HIS3; transformation markers = LEU2 and TRP1 | Stratagene |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| pAD-GAL4 | Activation domain cloning vector; AmprLEU2 | Stratagene |
| pBD-GAL4 | Binding domain cloning vector; CmrTRP1 | Stratagene |
| p53 | Murine p53-binding domain fusion; AmprTRP1 | Stratagene |
| pSV40 | Simian virus 40 large-T-antigen–activation domain fusion; AmprLEU2 | Stratagene |
| pLaminC | Human lamin C-binding domain fusion; AmprTRP1 | Stratagene |
| pBD-Z1 | FrzZ first domain-binding domain fusion; CmrTRP1 | This study |
| pBD-Z2 | FrzZ second domain-binding domain fusion; CmrTRP1 | This study |
| pBD-E | FrzE-binding domain fusion; CmrTRP1 | This study |
| pBD-AsgA | AsgA-binding domain fusion; CmrTRP1 | Lynda Plamann, University of Missouri |
| pADLIB | Library of random fragments from M. xanthus DZF1-activation domain fusions; AmprLEU2 | This study |
| pAD-RpoE1 | RpoE1-activation domain fusion; AmprLEU2 | This study |
| pUC18 | General cloning vector; Ampr | 26 |
| pZErO-2 | Zero Background cloning vector; Kmr | Invitrogen |
| pZRpoE1 | Internal fragment of rpoE1 cloned in pZErO-2; Kmr | This study |
| pZOrf5 | Internal fragment of orf5 cloned in pZErO-2; Kmr | This study |
| pBS9 | 6,464-bp fragment containing the 3′ end of frzZ and downstream genes cloned in pBlueScript | This study |
HybriZAP two-hybrid system.
DNA-binding domain fusion constructs were prepared by PCR amplification of the DNA encoding the bait proteins. PCR was performed with the high-fidelity polymerase Pfu. Primers were designed with 5′ EcoRI restriction sites (underlined in the sequences below) to simplify cloning into the EcoRI site of pBD-GAL4. The following primer pairs were used: FrzZ1 (pBD-Z1), forward 5′-ATGAATTCACGATGTCGCGCGTACTGGTCATTGATGACAGCCCG and reverse 5′-ATGAATTCGATGCTCAGGGCGGGGGGGCCAATGAGACCCATGAC; FrzZ2 (pBD-Z2), forward 5′-ATGAATTCAAGCCGCGCATCCTCATCGTGGATGAC and reverse 5′-ATGAATTCCTACTCGTTACCGGTGGGCATCAGCTC; FrzE (pBD-E), forward 5′-ATGAATTCCGTACGCCGGCCATGGACACCGAGGCTCTC and reverse 5′-ATGAATTCTCAGGTCAGCCGGTCGATGGCCTGCGCGAG. The insertions within the resultant constructs (pBD-Z1, pBD-Z2, and pBD-E) were sequenced to ensure error-proof amplifications, and then the plasmids were transformed into Saccharomyces cerevisiae YRG-2. The transformants containing just the binding domain constructs were tested for reporter gene expression prior to retransformation with the activation domain library or test activation domain fusion constructs. No reporter gene activity was observed with any of the bait constructs transformed alone into YRG-2.
The activation domain library was constructed by using a sonicated sample of sheared M. xanthus genomic DNA (15a). Sonication sufficient to produce fragments of approximately 1 to 2 kb was performed. These fragments were blunt ended with Klenow enzyme, EcoRI and XhoI linkers were randomly ligated onto the free ends, and the fragments were cloned as EcoRI-XhoI fragments into predigested HybriZAP vector arms (as provided in the HybriZAP two-hybrid predigested vector/Gigapack cloning kit; Stratagene). The library was amplified, and pAD-LIB phagemid vector was excised in vivo. The excised library was again amplified prior to use.
Yeast transformations, the reporter gene assay for HIS3, and the filter lift assay for β-galactosidase activity were performed as stipulated in the Stratagene manual. Other procedures, including the isolation of plasmid DNA from yeast and the verification of specificity of protein-protein interactions, were also performed as specified in the Stratagene manual.
DNA manipulations and PCR.
All plasmids used in this study were prepared by using a QIAprep spin miniprep kit (Qiagen). Chromosomal DNA was prepared by a miniprep protocol using cetyltrimethylammonium bromide-NaCl and phenol-chloroform extraction (22). Restriction enzyme digests and modifying enzyme protocols were performed as specified by the manufacturers. PCR optimizations and cycling parameters were identified by the protocol of Kramer and Coen (11). In general, glycerol concentrations of 20% were required for high yields of pure products. Taq polymerase (Promega) and Pfu polymerase (Stratagene) were used in amplifications requiring low and high fidelity, respectively.
RNA preparation and analyses.
Total RNA of M. xanthus was isolated by using a modification of the hot phenol procedure of Oelmüller et al. (15). RNA from developing cells was prepared as follows. An exponentially growing culture of DZ2 was diluted to 107 CFU/ml in CYE. Thirteen sterile glass petri dishes were filled with 50 ml of this diluted culture and then incubated at 32°C for 24 h. After this time, the growing cells had adhered to the bottom of the dishes and the growth medium was carefully removed. The cells were washed once with water; then the water replaced with an aliquot (20 ml) of 1 mM CaCl2 in 10 mM morpholinepropanesulfonic acid (MOPS) buffer (pH 6.8). The dishes were further incubated at 32°C. Starvation-induced development was considered to start from the time point of growth medium removal. At 0.5, 1, 2, 4, 6, 8, 10, 12, 18, 28, 42, 63, and 80 h, the wash solution was removed and the cells were resuspended in a 5-ml aliquot of ice-cold AE buffer (20 mM sodium acetate [pH 5.5], 1 mM EDTA). A phenol solution (acid-phenol–chloroform, 5:1, pH 4.5; Ambion Inc.) containing 0.25% sodium dodecyl sulfate was prewarmed in a 60°C water bath and added to the cells. The liquid-cell mixture was placed in a tube and incubated for 10 min in a fast-shaking water bath (60°C). RNA from vegetative growing cells was isolated by taking 10 to 20 ml of the initial exponentially growing culture and quickly harvesting the sample in precooled vessels. Samples were then resuspended in 5 ml of ice-cold AE buffer and incubated in the hot phenol solution as described above. After phase separation by low-speed centrifugation (4,000 × g, 15 min, 4°C) the aqueous phase of all RNA preparations was adjusted to 0.25 M sodium acetate (pH 5.2) and extracted twice with phenol solution. After ethanol precipitation and washing and drying of the RNA pellet, the RNA was resuspended in DNase buffer (40 mM Tris-HCl [pH 7.5], 6 mM MgCl2) and treated with 100 U of DNase I (RNase free; Ambion). After another phenol extraction and ethanol precipitation, the RNA was resuspended in Tris-EDTA buffer at a concentration of 1 mg/ml. Per sample, 20 to 300 μg of total RNA was obtained.
Primer extension analysis was performed by a modification of the procedure of Kellmann et al. (10). Seventeen picomoles of the oligonucleotide 5′-GCGTCGCGCTCGTTCTTC, which lies downstream of the rpoE1 translational start, was radiolabeled with 10 U of polynucleotide kinase, using 10 μCi of γ-33P (Amersham) in a 10-μl reaction containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 10 mM dithioerythritol. Per primer extension reaction, 1 μl of the radiolabeled oligonucleotide was used. Annealing of the 33P-radiolabeled primer was performed in a total volume of 10 μl containing 5.5 μg of total RNA, 10 mM Tris-HCl (pH 7.9), 0.5 M KCl, and 12.5 U of RNase inhibitor (Ambion). After heating for 5 min at 80°C, the annealing reaction mixtures were incubated for 3 h at 45°C. For a 50-μl primer extension reaction, 10 μl of 5× reverse transcription (RT) buffer (250 mM Tris-HCl [pH 8.3], 125 mM KCl, 50 mM dithiothreitol, 15 mM MgCl2), 500 μM deoxynucleoside triphosphates, and 200 U of SuperScript II (Gibco/BRL) were added to the annealing reactions and incubated for 1 h at 37°C. The primer extension products were treated with RNase A (Ambion), phenol-chloroform extracted, ethanol precipitated, and analyzed on a polyacrylamide sequencing gel. The length of the primer extension reaction products was determined by running sequence reactions performed with the same primer on the same gel. Sequencing reactions for primer extension analysis were performed by the dideoxynucleotide chain termination method, using Sequenase (U.S. Biochemical Corp) and [α-35S]dATP (410 Ci mmol−1; Amersham) on double-stranded DNA. Each sample was run for 2 h on a 6% polyacrylamide gel prepared by using National Diagnostics Sequagel reagents.
For RNA dot blot preparation, 10 μg of RNA was incubated at 65°C for 5 min in 3 volumes of a solution containing 500 μl of formamide, 162 μl of formaldehyde (37%, vol/vol), 100 μl of 10× MOPS buffer (0.2 M MOPS 0.5 M sodium acetate [pH 7.0], 0.01 M Na2EDTA). After addition of 1 volume cold 20× SSC (3.0 M NaCl, 0.3 M sodium citrate), the RNA samples were spotted onto a Hybond-N nylon membrane (Amersham), prewet in 10× SSC. RNA was fixed onto the membranes by UV cross-linking. A radiolabeled, PCR-amplified internal fragment of the rpoE1 gene was used as the probe. Hybridization was performed under standard conditions, with high-stringency washes as specified for use with the Amersham Hybond-N membranes.
First-strand cDNA synthesis for RT-PCR was performed with SuperScript II (Gibco/BRL), using the manufacturer’ recommended conditions. The oligonucleotides used for template extension from within the orf5 and orf6 regions were 5′-GAGCATCCGGCGGATGTCGC and 5′-TGCAGTTCGGCCACCTTCGC, respectively. PCRs on the cDNA were then performed with Taq polymerase (Promega), again using the manufacturer’s recommended conditions. Thirty-five cycles of amplification were performed with the following oligonucleotides for amplification of an internal fragment of rpoE1: forward 5′-GCTCTATTCGGCGGCGCTGC and reverse 5′-AAGACGCCCTGGCCCTCGGC.
Production of mutants.
Mutants were constructed by cloning internal fragments of the specified genes into pZErO-2 (Invitrogen). These constructs were then electroporated into M. xanthus strains, and selection on CYE containing kanamycin (25 μg/ml) was used to identify mutants which had inserted the entire vector into the chromosome by homologous recombination. Internal fragments of the following genes were prepared by PCR using Taq polymerase (Promega). The following primer combinations were designed with 5′ EcoRI ends (underlined) to facilitate simple cloning into the EcoRI site of pZErO: rpoE1 (pZRpoE1), forward 5′-ATGAATTCGCTCTATTCGGCGGCGCTGC and reverse 5′-ATGAATTCAAGACGCCCTGGCCCTCGGC; orf5 (pZOrf5), forward 5′-ATGAATTCCCAGTTCGGCGTCTGGCTGC and reverse 5′-ATGAATTCGAGCATCCGGCGGATGTCGC.
Transformations were performed in E. coli Top10 cells, which allow Zero Background cloning (Invitrogen). Plasmids were then denatured with 1 N NaOH prior to electroporation into M. xanthus strains by the method described in reference 22. Chromosomal DNA was prepared from resultant strains, and the sites of insertion were confirmed by Southern blotting. Probes were constructed by PCR incorporating the hapten digoxigenin as digoxigenin-11-dUTP (Boehringer Mannheim), and detection was performed by enzyme immunoassay and an enzyme-catalyzed color reaction (Boehringer Mannheim).
Phenotypic analyses.
Carotenoid production was screened preliminarily by colony color after cells were left incubating in the light for 2 days. These cells were then extracted with methanol, and the extract was analyzed spectrophotometrically for absorption between wavelengths of 300 and 600 nm. Cells that had been incubated in the dark were used as control standards. Carotenoid production was considered positive if a peak at 480 nm was identified only in the light-grown cells.
Heat shock experiments were performed by growing cells in liquid media, at a nonrestrictive temperature, to Klett readings of 20 to 30. Duplicate flasks were then either returned to this temperature or transferred to 42°C. Klett readings of both cultures were taken from mutant and parent strains, and growth parameters were compared over an 8-h period.
Developmental defects were screened for on CF starvation agar either by plating 5 μl of cells at stated CFU-per-milliliter concentrations directly onto plates or by stabbing plates (thereby producing random inocula) and incubating them for 96 h. Aggregation patterns were photographed; then spore counts were performed by removing the cells from the agar and resuspending them in TM buffer (10 mM Tris-HCl, 8 mM MgSO4 [pH 7.6]). Spore clumps were dispersed by sonication, and appropriate dilutions were placed in a Petroff-Hausser chamber for counting under magnification.
Computer analysis.
Computer analyses of the sequenced genes, including GC coding predictions, identification of open reading frames (ORFs), translations, and sequence alignments, were performed with the program Gene Inspector (Textco Inc.).
Nucleotide sequence accession number.
The DNA sequence presented here have been submitted to the EMBL, GenBank, and DDBJ nucleotide sequence data libraries under accession no. AF049107.
RESULTS
Identification of a potential ECF sigma factor by using interaction trap technology.
A yeast two-hybrid screen, with the first domain of FrzZ (on pBD-Z1) used as bait, identified a construct, pAD-RpoE1, from a random M. xanthus genomic DNA library (pAD-LIB), which when present in combination with pBD-Z1 resulted in positive reporter gene assays in the system. Both plasmids were purified, and pAD-RpoE1 was retransformed into S. cerevisiae YRG-2 in combination with pBD-Z1 and other bait-encoding constructs (Table 3). While the same positive reporter gene expression was observed with the pAD-RpoE1–pBD-Z1 combination, the pAD-RpoE1 construct (one of the reporter systems in the Stratagene HybriZAP two-hybrid system) also produced a low level of β-galactosidase expression when transformed in combination with pBD-AsgA (kindly provided by L. Plamann, University of Missouri). Since asgA encodes a protein with two separate domains, a histidine protein kinase and a response regulator domain (16), we speculate that the protein encoded by pAD-RpoE1 may interact with a response regulator, but perhaps not specifically with FrzZ. However, the negative results produced by transforming YRG-2 cells with pAD-RpoE1 and other response regulator expression constructs (pBD-Z2 and pBD-E) suggest that the fusion protein expressed from pAD-RpoE1 does not interact nonspecifically with all response regulators. To extend our analysis of the gene cloned in pAD-RpoE1, the insert DNA was used as a probe to identify a large genomic fragment from within a cosmid library (kindly provided by T. Hartzell, University of Idaho). The identified clone (pBS9) contained the gene of interest and both upstream and downstream regions.
TABLE 3.
Two-hybrid reporter gene assays
| Plasmid combination
|
Reporter gene assay resulta
|
||
|---|---|---|---|
| Bait domain fusion | Activation domain fusion | SD-Leu-Trp-His | β-Galactosidase |
| pBD-Z1 | pAD-RpoE1 | ++ | + (5 h) |
| p53 | pAD-RpoE1 | + | − |
| pBD-Z2 | pAD-RpoE1 | + | − |
| pBD-E | pAD-RpoE1 | + | − |
| pBD-AsgA | pAD-RpoE1 | + | + (7 h) |
| p53 | pSV40 | +++++ | + (4 h) |
| pLaminC | pSV40 | + | − |
Various bait-domain fusions were tested against the activation domain-RpoE1 fusion for reporter gene activation in yeast. Expression of the HIS3 reporter gene was shown to be leaky under all conditions (+). Expression of β-galactosidase is reported as + for cells which turned blue on the filter paper and − for cells which showed no color change. The time when blue color was first observed on the filters is given in parentheses. The combination p53-pSV40 was used as the system positive control, while the combination pLaminC-pSV40 was used as the negative control.
The rpoE1 gene is positioned downstream of frzZ.
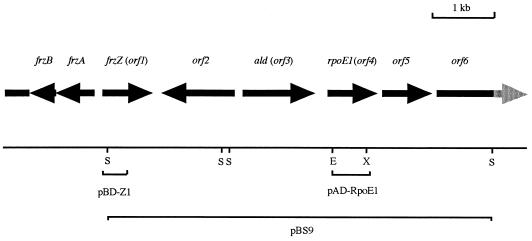
The 6,464-bp insert present on the construct pBS9 was sequenced and shown to encode six potential ORFs (Fig. 1), all with appropriate M. xanthus codon usage, a good indicator of translational potential (18). The first ORF spanned from the start of the sequenced region to a stop codon at bp 829 and was considered to be the 3′ end of a gene. A gapped BLAST analysis (2) of the translated protein identified it to be the C-terminal end of FrzZ (21), placing the gene encoding the potentially FrzZ-interacting protein downstream of the frzZ gene itself. The second ORF, present on the opposite DNA strand, spans from a potential ATG start codon at bp 2320 to a stop codon at bp 972. The orf2-encoded protein showed no homology to known proteins in a GenEMBL database search. The third ORF is transcribed divergently from orf2, with a potential GTG start codon at bp 2458 and a stop codon at bp 3580. A potential ribosome-binding (Shine-Dalgarno [SD]) site (GGAGG) was apparent 6 bp upstream of the proposed start codon. A gapped BLAST search showed orf3 to have significant similarity (57% identity) to the alanine dehydrogenase gene (ald) of Bacillus subtilis (19); orf3 has, therefore, been speculatively renamed ald. A potential terminator structure was identified downstream of the ald gene, suggesting it to be a discrete transcriptional unit. This structure could, however, play an alternative role, perhaps as a binding site for a DNA-binding regulatory protein.
FIG. 1.
Map of the rpoE1 region. The rpoE1 gene, which was shown to lie downstream of frzZ, was cloned and sequenced on pBS9 and various subclones (not shown). The EcoRI-XhoI fragment from pAD-RpoE1 which when expressed in yeast showed a potential interaction with the expressed insert in pBD-Z1, and was used as a probe to identify pBS9, is shown. The start of the large frz operon, which is transcribed divergently from frzZ, is also shown. Restriction enzyme cut sites: E, EcoRI; S, SacI; X, XhoI.
The fourth ORF spanned from a potential ATG start codon at bp 3825 to a stop codon at bp 4464 and contained the EcoRI-XhoI fragment cloned in pAD-RpoE1. Database searching suggested the translated product of orf4 to be a member of the ECF subfamily of transcriptional regulators. Figure 2 shows an alignment of the protein, named RpoE1, with the previously identified M. xanthus ECF sigma factor, CarQ (14), and E. coli RpoE (5). RpoE1 is 30% identical to CarQ and 34% identical to RpoE and shows several features distinctive of the ECF sigma factors (12), primarily a small size (estimated Mr of ∼25,000). It also lacks region 1 of ς70 but shows good homology in regions 2 and 4. Region 3 is truncated with respect to ς70 and not highly conserved. The RpoE1 protein is also seen to have a short C-terminal extension with respect to other RpoE proteins. A potential SD site (GGAAG) was identified 7 bp upstream of the start codon, but no obvious terminator structure was identified after the stop codon. The fifth ORF is proposed to have an ATG start codon at bp 4624 and a stop codon at bp 5372 of the sequenced region. The proposed translation product of the gene shows no homology to known proteins. A potential SD site (GGAGG) was identified upstream of the start codon, and a potential terminator structure was identified 28 to 44 bp after the stop codon. The final ORF presumably covers only the 5′ end of a gene, with a proposed ATG start codon at bp 5574 but with no stop codon in the sequenced fragment. A BLAST search suggests the first domain of the protein to be a “receiver module” of a response regulator protein from the two-component signal transduction family (20). A potential SD site (AGGA) was identified upstream of the start codon.
FIG. 2.
Alignment of the newly identified M. xanthus (M.x.) ECF sigma factor, RpoE1, with the previously identified M. xanthus CarQ sigma factor (14) and RpoE from E. coli (E.c.) (5).
Transcriptional regulation of rpoE1 expression.
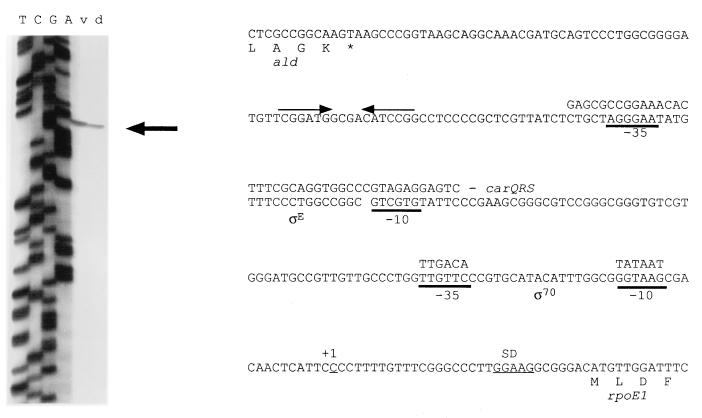
We found a potential terminator downstream of the 3′ end of the ald gene and a potential ECF sigma factor binding site upstream of the rpoE1 gene (by alignment with the carQRS promoter site [Fig. 3]). Although both structures were determined by sequence analysis alone, they were suggestive that the ald and rpoE1 genes could be transcriptionally separate. To determine if the rpoE1 gene could be transcribed from this putative ςE promoter, primer extension analysis, using a primer within the rpoE1 gene close to the ATG start codon, was performed on both vegetative and developmental RNA samples. A transcriptional start site was identified upstream of rpoE1 in both samples (Fig. 3), although it was shown to be positioned downstream of a potential ς70-type promoter rather than the potential ςE promoter. However, since primer extension studies identify only 5′ ends of RNA transcripts, further investigation will be required to confirm the promoter status of either site.
FIG. 3.
Primer extension analysis (left) used to identify the transcriptional start site for expression of the rpoE1 gene, within the ald-rpoE1 intergenic region (right). A sequencing reaction (TCGA), performed with the same primer, is shown next to the primer extension analysis on vegetative (v) and developmental (d) RNA templates. Potential ςE and ς70 promoters are underlined (bold), with alignments to the carQRS promoter region and the ς70 consensus, respectively. A second potential ς70 promoter (TGCATA-16 bp-GACAAC), closer to the proposed transcriptional start site, was also identified. The potential SD site upstream of rpoE1 is also underlined. The 3′ end of the ald gene is shown upstream of rpoE1, with the proposed transcriptional terminator identified by inverted arrows.
A dot blot analysis using RNA samples isolated from vegetative cells and from developing cells (taken at time points from 0.5 to 48 h poststarvation) was performed. The results suggested expression of the rpoE1 gene to be constitutive during both stages of the complex M. xanthus life cycle. No time point where expression was enhanced above a low level was identified (data not shown).
RT-PCR was performed on developmental, DNA-free RNA preparations to identify whether the genes downstream of rpoE1 are cotranscribed with rpoE1 or are transcriptionally separate, since the lack of a potential terminator structure after the rpoE1 gene suggests that rpoE1 and orf5 may be cotranscribed. cDNA was produced by using primers located within both orf5 and orf6 genes and then used as the template for PCR amplification of a fragment internal to the rpoE1 gene. PCR amplification of this region can, therefore, be obtained only if RNA transcripts provide a contiguous template for cDNA synthesis between orf5 and rpoE1 and between orf6 and rpoE1 (for the primers located in orf5 and orf6, respectively). Control PCRs were performed on non-reverse-transcribed RNA template to ensure that DNA contamination did not influence the results. This approach confirmed that both orf5 and orf6 genes are cotranscribed with rpoE1 (data not shown).
Mutational analyses.
A 500-bp, PCR-derived internal fragment of the rpoE1 gene was cloned into pZErO-2 to create pZRpoE1. This construct was electroporated into M. xanthus, and cells which had acquired the plasmid were selected for by antibiotic resistance. Since the vector is unable to replicate in M. xanthus, kanamycin-resistant electroporants should arise through the integration of the vector into the host genome at the rpoE1 gene by homologous recombination. A single crossover event using such an internal fragment of the rpoE1 gene would therefore result in two truncated copies of rpoE1 in the genome, separated by the kanamycin-resistant vector. Mutants produced by this technique were screened for, and shown to contain, correct insertion mutations by Southern analysis (data not shown).
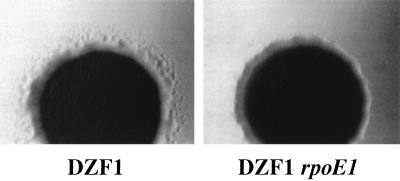
Since FrzZ effects motility and showed a potential interaction with RpoE1 in the two-hybrid analysis, motility behavior of the rpoE1 mutants was determined by analyzing swarming under vegetative conditions. Swarming in the DZF1 (sglA1) background was substantially reduced when observed after several days incubation (Fig. 4). In contrast, swarming in the fully motile (A+ S+) DZ2 background appeared only slightly reduced with respect to wild-type (not shown).
FIG. 4.
Swarming of DZF1 and DZF1 rpoE1 cells on CYE medium containing 1.5% agar. Cells were photographed after 3 days of incubation.
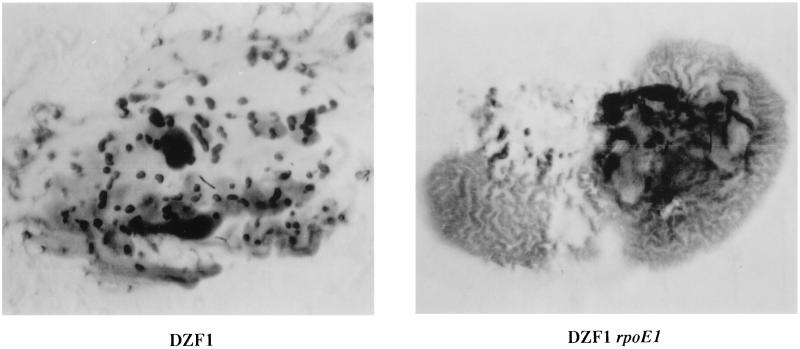
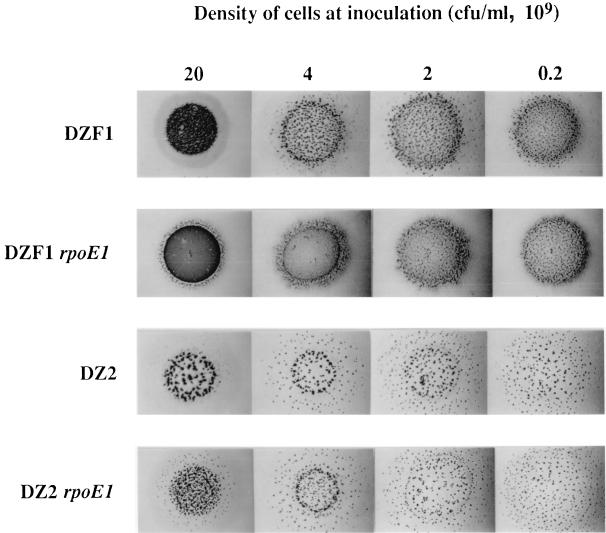
An analysis of developmental aggregation was initially performed in the DZF1 background. Cells were stabbed directly onto CF agar plates, and aggregation patterns were photographed at 24-h intervals. While the parent formed aggregates after 48 h of incubation, the DZF1 rpoE1 mutant was shown to produce a highly distinctive pattern, reminiscent of worm trails, rather than discrete aggregates, after just 24 h (Fig. 5). These trails darken to a solid mat after longer periods of incubation. However, areas of normal aggregation were identified within the rpoE1 mutant stabs (top left region), suggesting the occurrence of a density-dependent effect. To analyze this possibility further, mutants in both DZF1 and DZ2 backgrounds were spotted onto CF plates at high and low cell densities. In both genetic backgrounds, aggregation was shown to be effected at high cell density only (Fig. 6). This effect was most pronounced in the DZF1 background, where again close-packed trails of cells, rather than discrete fruiting bodies, were observed. In the DZ2 background, the mutants formed smaller and more closely packed aggregates. Spore counts performed on both DZF1 rpoE1 and DZ2 rpoE1 developmental cells showed that both strains produce normal levels of spores.
FIG. 5.
Developmental aggregation of DZF1 and DZF1 rpoE1 cells stabbed directly onto CF agar plates. The DZF1 rpoE1 aggregation phenotype was photographed after 24 h of incubation, since substantial darkening occurs after longer periods, obscuring the worm trail effect. The DZF1 aggregation pattern is shown after 48 h of incubation.
FIG. 6.
Density-dependent aggregation effect of the rpoE1 mutant shown in both DZF1 and DZ2 backgrounds. Cells were spotted directly onto CF plates and photographed after 96 h of incubation.
Since the previously identified ECF sigma factor, CarQ, from M. xanthus was shown to be involved with light-induced carotenoid biosynthesis, the production of carotenoids in the rpoE1 mutants was determined. After incubation in the light, the rpoE1 mutants turned orange and methanol extracts of whole-cell cultures showed a new peak on spectrophotometric analysis at 480 nm, similar to those seen in the parent strains but not present in cells grown in the dark. The ability to survive heat shock treatments was also analyzed in the rpoE1 mutants, since several ECF sigma factors have been shown to be induced under heat shock conditions. However, the M. xanthus rpoE1 mutants showed resistance to incubation at 42°C similar to that demonstrated by the parent strains.
A 500-bp, PCR-derived internal fragment of the orf5 gene was cloned into pZErO-2 to produce the construct pZOrf5, which was then introduced into M. xanthus DZF1 and DZ2 strains as described above. Insertion mutants with the desired genotype were confirmed by Southern analysis. Both vegetative swarming and developmental aggregation behaviors were analyzed, but mutants in either genetic background showed behavior patterns similar to that of the parent, suggesting that the mutant phenotypes identified in the rpoE1 mutants are not due to downstream effects.
DISCUSSION
In this study, we have identified a second homologue belonging to the ECF subfamily of sigma factors, RpoE1, in M. xanthus. The original identification, using interaction trap technology, with the first domain of the directed-motility-associated protein FrzZ as bait, suggested that RpoE1 could potentially interact with FrzZ. However, control interaction screens suggest that RpoE1 might also interact with a second response regulator protein, AsgA. While this second interaction was weaker than the RpoE1-FrzZ interaction, the possibility remains that both interactions are nonspecific. Therefore, the specificity of the RpoE1-FrzZ interaction remains speculative, and further studies, using alternative protein-protein analysis techniques, are required to convincingly characterize what would be an unusual and interesting interaction.
While a direct protein-protein interaction between RpoE1 and FrzZ remains unconfirmed, sequence analysis of the region surrounding the rpoE1 gene showed it to be genetically linked to the frzZ gene. Two transcriptionally divergent genes are positioned between the frzZ and rpoE1 genes. Neither gene has a known function in M. xanthus, although we presume that the ald gene may encode a functional alanine dehydrogenase due to the high degree of amino acid homology found in this protein. Mutational analyses performed on both genes (not reported in this study) suggest that neither is involved with vegetative swarming, developmental aggregation, or sporulation. The presence of a potential terminator structure between the ald and rpoE1 genes suggested that these genes could be transcriptionally separate.
The identification of a potential ECF sigma factor binding site upstream of the rpoE1 gene indicated that RpoE1, or another ECF sigma factor, might regulate transcription of rpoE1. However, primer extension studies performed on RNA samples extracted from both vegetatively growing and developmental cells suggested that the gene is transcribed under both conditions from a ς70-like promoter positioned downstream of the potential ςE-like promoter. A dot blot analysis confirmed the low-level, constitutive expression of the rpoE1 gene. While the possibility still exists that the proposed ςE promoter is functional under a specific set of conditions and might cause enhanced transcription of the gene, currently we have no indication of whether this region is actually a promoter or under what conditions it might be active.
The genetic organization of the ORFs downstream of rpoE1 suggested that rpoE1 and orf5 could be transcriptionally linked. The identification of a potential terminator structure at the end of the orf5 gene suggested that the orf6 gene might then be transcriptionally separate. RT-PCR was therefore used to analyze these possibilities and demonstrated that, in fact, both orf5 and orf6 are read on a single transcript with the rpoE1 gene. Since the currently available sequence covers only a portion of the orf6 gene, we are unable to speculate on the presence of further genes within this operon. However, it appears that rpoE1, orf5, and orf6 can be transcribed from a ς70-type promoter positioned upstream of the rpoE1 gene, although it remains possible that the downstream genes also have separate promoters.
ECF sigma factors have been proposed to respond to extracytoplasmic stimuli and regulate extracytoplasmic functions (12). Mutational analysis of cells with an insertion at the rpoE1 site suggest that rpoE1 may play a role in regulating motility behavior during both vegetative swarming and developmental aggregation. These phenotypes are unlikely to be associated with loss of function of the genes downstream of rpoE1, since cells with mutations in orf5 showed normal swarming and aggregation. However, rpoE1 mutants do still swarm and can form normal aggregates, particularly at lower cell densities, indicating that the role of RpoE1 may be cell density specific. The potential role of RpoE1 in regulating motility behavior during both vegetative swarming and developmental aggregation again draws analogy with the Frz signal transduction system. However, the aggregation phenotypes produced by frz mutants are highly distinctive, swirling patterns, disparate from the rpoE1 aggregation phenotype, and are not known to be cell density dependent.
In conclusion, we have identified a putative ECF sigma factor, RpoE1, in M. xanthus. The rpoE1 gene has been shown to be genetically linked to the frzZ gene. The RpoE1 protein also shows a potential (although currently speculative) interaction with the first domain of FrzZ in a two-hybrid analysis. In addition, RpoE1 appears to be involved in the regulation of motility behavior during both vegetative swarming and developmental aggregation. While the Frz system is known to regulate motility behavior under both of these situations, the RpoE1 developmental defect appears to be cell density specific and the resultant aggregation patterns are unique. Currently we are involved in expressing the RpoE1 protein to facilitate further protein-protein interaction studies (in order to confirm or refute an interaction between RpoE1 and the Frz signal transduction system) and to confirm functional sigma factor status for RpoE1. We are also interested in identifying an active ςE promoter and any genes which may be transcriptionally regulated by RpoE1.
ACKNOWLEDGMENTS
We particularly thank Bob Osborne for help in construction of the two-hybrid library. We also thank Lynda Plamann and Trish Hartzell for kindly providing other useful tools for this project. In addition, we are grateful to Stacia Hoover and Eric Bowman at the UC Davis Sequencing Facility for all of the sequence data published in this report.
Research in our laboratory is supported by Public Health Service grant GM20509 from the National Institutes of Health. A.T.-L. was funded by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Acuña G, Shi W, Trudeau K, Zusman D R. The ’CheA’ and ’CheY’ domains of Myxococcus xanthus FrzE function independently in vitro as an autokinase and a phosphate acceptor, respectively. FEBS Lett. 1995;358:31–33. doi: 10.1016/0014-5793(94)01389-i. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos J M, Geisselsoder J, Zusman D R. Isolation of bacteriophage Mx4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 4.Campos J M, Zusman D R. Regulation of development in Myxococcus xanthus: effect of 3′:5′ cyclic AMP, ADP and nutrition. Proc Natl Acad Sci USA. 1975;72:518–522. doi: 10.1073/pnas.72.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erikson J W, Gross C A. Identification of the ςE subunit of Escherichia coli RNA polymerase: a second alternate ς factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 6.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 7.Hagen D C, Bretscher A P, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 8.Hartzell P L, Youderian P. Genetics of gliding motility and development in Myxococcus xanthus. Arch Microbiol. 1995;164:309–323. doi: 10.1007/BF02529977. [DOI] [PubMed] [Google Scholar]
- 9.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 10.Kellmann J-W, Pichersky E, Piechulla B. Analysis of the diurnal expression patterns of the tomato chlorophyll a/b binding protein genes. Influence of light and characterization of the gene family. Photochem Photobiol. 1990;52:35–41. doi: 10.1111/j.1751-1097.1990.tb01752.x. [DOI] [PubMed] [Google Scholar]
- 11.Kramer M F, Coen D M. Enzymatic amplification of DNA by PCR: standard procedures and optimization, unit 15.1. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [DOI] [PubMed] [Google Scholar]
- 12.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase ς factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCleary W R, McBride M J, Zusman D R. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990;172:4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGowan S J, Gorham H C, Hodgson D A. Light induced carotenogenesis in Myxococcus xanthus: DNA sequence analysis of the carR region. Mol Microbiol. 1993;10:713–735. doi: 10.1111/j.1365-2958.1993.tb00943.x. [DOI] [PubMed] [Google Scholar]
- 15.Oelmüller U, Krüger N, Steinbüchel A, Friedrich C G. Isolation of prokaryotic RNA and detection of specific mRNA with biotinylated probes. J Microbiol Methods. 1990;11:73–84. [Google Scholar]
- 15a.Osborne, R. Personal communication.
- 16.Plamann L, Li Y, Cantwell B, Mayor J. The Myxococcus xanthus asgA gene encodes a novel signal transduction protein required for multicellular development. J Bacteriol. 1995;177:2014–2020. doi: 10.1128/jb.177.8.2014-2020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi W, Kohler T, Zusman D R. Chemotaxis plays a role in the social behaviour of Myxococcus xanthus. Mol Microbiol. 1993;9:601–611. doi: 10.1111/j.1365-2958.1993.tb01720.x. [DOI] [PubMed] [Google Scholar]
- 18.Shimkets L J. The myxobacterial genome. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: American Society for Microbiology; 1993. pp. 85–107. [Google Scholar]
- 19.Siranosian K J, Ireton K, Grossman A D. Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J Bacteriol. 1993;175:6789–6796. doi: 10.1128/jb.175.21.6789-6796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 25–51. [Google Scholar]
- 21.Trudeau K G, Ward M J, Zusman D R. Identification and characterization of FrzZ, a novel response regulator, necessary for swarming and fruiting-body formation in Myxococcus xanthus. Mol Microbiol. 1996;20:645–655. doi: 10.1046/j.1365-2958.1996.5521075.x. [DOI] [PubMed] [Google Scholar]
- 22.Ward M J, Mok K C, Astling D P, Lew H, Zusman D R. An ABC transporter plays a developmental aggregation role in Myxococcus xanthus. J Bacteriol. 1998;180:5697–5703. doi: 10.1128/jb.180.21.5697-5703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward M J, Mok K C, Zusman D R. Myxococcus xanthus displays Frz-dependent chemokinetic behavior during vegetative swarming. J Bacteriol. 1998;180:440–443. doi: 10.1128/jb.180.2.440-443.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward M J, Zusman D R. Regulation of directed motility in Myxococcus xanthus. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]
- 25.Wu S S, Kaiser D. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 26.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]