Abstract
Background and Objective:
Isocitrate dehydrogenase genes (IDH1 and IDH2) encode important enzymes that play pivotal role in cellular metabolism. Mutations in TET2 have been demonstrated to contribute to DNA hypermethylation, either expression of mutant IDH1/2 or TET2 resulted in poor cell differentiation and epigenetic alterations in hematopoietic cells, suggesting a sharing of the oncogenetic impact. In this study, we investigated the frequency of genetic alterations in IDH1/2 and TET2 genes in Egyptian cohort of adult patients with de novo AML, and the association of IDH1/2 and TET2 genetic Polymorphism with AML prognostic criteria and explore prognostic molecular markers with clinical outcome.
Methods:
The SNP assay for IDH1, IDH2 and TET2 genes polymorphism tested with RT-PCR included three polymorphisms that are rs121913500, rs121913503, and rs2454206 respectively, were tested on 141 adult Egyptian patients fulfilling the AML diagnostic criteria.
Result:
The incidence of IDH mutations is 11/141 (7.8%); 5/141 (3.5%) IDH1 mutant and 6/141 (4.3%) IDH2 mutant. And the incidence of TET2 mutations is 72/141 (51.1%); 15/141 (10.7%) homozygous mutation and 57/141 (40.4%) heterozygous mutations. IDH1, IDH2 and TET2 genes mutations with DFS and OS in AML patients were not significantly correlated.
Conclusions:
TET2 SNP is common in Egyptian AML patients. Further research on IDH, TET2 and their relationships to other hematological malignancies and leukemogenesis transformation is advised and a study of a larger number of cases is needed for potential statistical significance.
Key Words: Acute myeloid leukemia, isocitrate dehydrogenase, Ten-Eleven Translocation-2
Introduction
Acute myeloid leukemia (AML) is extremely heterogeneous and quickly progressing aggressive malignancy of the hematopoietic myeloid progenitor cells and the most frequent acute leukemia found in adults with an increasing incidence with age (Estey and Döhner, 2006). It is characterized by poor differentiation and abnormal clonal proliferation of immature myeloid precursor cells in the bone marrow which leads to accumulation of nonfunctional myeloblasts, abnormal hematopoiesis, bone marrow failure, and peripheral blood cytopenias, raising the risk of serious infections, anemia, bleeding, and other complications in patients (Döhner et al., 2015).
The identification of genetic mutations and epigenetic aberrations additional to classical molecular markers have been implicated in the diagnosis, prognosis and can also guide treatment of AML (Lagunas-Rangel et al., 2017)
IDH1 and IDH2 are two isoforms of Isocitrate dehydrogen gene (IDH) they encode for NADP-dependent isocitrate dehydrogenase, which is found in the cytosol and mitochondria, respectively. This enzyme is a key metabolic enzyme that catalyzes decarboxylation of isocitrate into alpha-ketoglutarate in Krebs cycle. TET proteins use this alpha-ketoglutarate during histone demethylation (Prada-Arismendy et al., 2017). IDH1/2 mutations in AML patients are heterozygous and often affect conserved arginine (R) residues found in the catalytic region of the enzyme (Ward et al., 2013). IDH1 and IDH2 genes located on chromosomes 2q33 and 16q26, respectively. IDH2 gene mutations are most common in AML, involving 8–19% of patients, and they are more common in older and intermediate-risk patient populations (Montalban-Bravo and DiNardo, 2018). IDH1 mutation occurs in 7− 14% of AML patients (Medeiros et al., 2017; Montalban-Bravo and DiNardo, 2018).
The detection of clinical and biological effects of isoforms 1 and 2 mutations of isocitrate dehydrogenases (IDH) result in the creation of a customized treatment strategy. Stimulating the differentiation and maturation of the cancerous clone targeting IDH is an emerging plan to enhance clinical responses in AML (Cerchione et al., 2021)
Ten–Eleven Translocation gene2 (TET-2) has pleiotropic function during hematopoiesis, including stem cells’ capacity for self-renewal, lineage commitment, and terminal of monocytes differentiation (Solary et al., 2014). It is located on the chromosome 4q24 region and have 11 exons (Albano et al., 2011) , Its proteins are ketoglutarate and Fe2+ dependent enzymes able to change DNA methylation state (Ko et al., 2011) . TET2 performs the conversion of 5-methyl-cytosine (5-mc) to 5-hydroxymethyl-cytosine (5-hmc), it is abundantly expressed in hematopoietic stem cells, thus, any mutation in the TET2 gene could cause the dysfunction of normal hematopoietic stem cells and disrupt the normal stem cell differentiation process through epigenetic modification (Feng et al., 2019 ; Wang et al., 2019) . TET2 gene was identified to be mutated in a numerous myeloid disorder (Weissmann et al., 2012). TET2 mutations are almost mutually exclusive with IDH1/2 mutations, however they can co-occur with NPM1, FLT3, JAK2, RUNX1, CEBPA, CBL, and KRAS gene mutations (Liu et al., 2014). About 7.7–27.4% of patients with AML have been reported with TET2 mutations (Gaidzik et al., 2012; Liu et al., 2014).
Our aim in this study is to investigate the frequency of genetic alterations in IDH1/2 and TET2 genes in Egyptian adult patients with de novo AML. Furthermore, the association of IDH1/2 and TET2 genetic Polymorphism with disease prognostic criteria and explore prognostic molecular markers with clinical outcome.
Materials and Methods
One hundred forty one adult Egyptian patients fulfilling the AML diagnostic criteria were the subject of the study. Patients’ ages varied from 18 to 74 years, with a median of 41 years, and they included 65 males (46.1% of patients) and 76 females (53.9% of patients). Between February 2019 and December 2021, they presented to the Medical Oncology clinics at Cairo University’s National Cancer Institute (NCI). The study was authorized by the Institutional Review Board (IRB) in accordance with the Helsinki declaration of studies involving human subjects after written informed permission was acquired from each patient.
Standard methods including complete blood picture, bone marrow aspirate, morphology, cytochemistry, immunophenotyping, cytogenetic analysis and molecular genetics were used to diagnose AML patients. Patients’ clinical characteristics are shown in Table 1 and routine molecular detection of NPM1, FLT3-ITD, FLT3-TKD and DNMT3A were performed. ELN 2017 classification of AML patients into low, intermediate and high risk groups was used to categorize the patients (Döhner et al., 2017). All patients received standard induction chemotherapy with 3+7 protocol (idarubicin as short infusion for 3 days with cytarabine 100mg/m2 continuous infusion for 7 days).
Table 1.
Descriptive Parameters of 141 Adult Patients with Acute Myeloid Leukemia
| Frequency (%) | |
|---|---|
| Age (n=141) | |
| ≤40 | 69 (48.9) |
| >40 | 72 (51.1) |
| Median (range) | 41.0 (18-74) |
| Gender (n=141) | |
| Female | 65 (46.1) |
| Male | 76 (53.9) |
| TLC (n=137) | |
| ≤100 X109/L | 97 (70.8) |
| >100x109/L | 40 (29.2) |
| Median (range) | 33 (1.38-452.9) |
| Hemoglobin (n=137) | |
| ≤10 g/dl | 123 (89.8) |
| >10 g/dl | 14 (10.2) |
| Median (range) | 7.3 (3.5-15.7) |
| Platelets (n=137) | |
| ≤100x109/L | 112 (81.8) |
| >100x109/L | 25 (18.2) |
| Median (range) | 45.0 (2.0-312.0) |
| Pb blast % (n=141) | |
| ≤50 | 37 (26.2) |
| >50 | 104 (73.8) |
| Median (range) | 70.0 (5.0-99.0) |
| BM blast% (n=141) | |
| ≤50 | 20 (14.2) |
| >50 | 121 (85.8) |
| Median (range) | 76.0 (20.0-98.0) |
| Cellularity (141) | |
| Hypercellular | 120 (85.1) |
| Normocellular | 13 (9.2) |
| Hypocellular | 8 (5.7) |
| FAB Classification (141) | |
| M0 | 4/141 (2.8) |
| M1 | 31/141 (22.0) |
| M2 | 41/141 (29.1) |
| M4 | 47/141 (33.3) |
| M5 | 15/141 (10.6) |
| M7 | 1/141 (0.7) |
| MPAL | 2 (1.4) |
| FAB Classification (134) | |
| M1&M2 | 72 (53.7) |
| M4&M5 | 62 (46.3) |
| Complete Remission (100) | |
| CR | 51 (51.0) |
| NO CR | 49 (49.0) |
| Lymphadenopathy (134) | |
| No | 78 (58.2) |
| Yes | 56 (41.8) |
| Organomegaly (134) | |
| No | 85 (63.4) |
| Yes | 49 (36.6) |
| Frequency (%) | |
| FLT3-ITD (141) | |
| Wild | 96 (68.1) |
| Mutant | 45 (31.9) |
| ITD ratio (n= 45) | |
| ≤0.525 | 20 (44.4) |
| >0.525 | 25 (55.6) |
| Median (range) | 0.58 (0.05-4.94) |
| FLT3-TKD (140) | |
| Wild | 134 (95.7) |
| Mutant | 6 (4.3) |
| NPM (128) | |
| Wild | 107 |
| Mutant | 21 |
| NPM&FLT3 (128) | |
| Favourable | 17 (13.3) |
| Unfavorable | 13 (10.2) |
| Intermediate | 98 (76.6) |
| DNMTA (139) | |
| Wild | 125 (89.9) |
| Mutant | 14 (10.1) |
| IDH1 (141) | |
| Wild | 136 (96.5) |
| Mutant | 5 (3.5) |
| IDH2 (141) | |
| Wild | 135 (95.7) |
| Mutant | 6 (4.3) |
| IDH1&IDH2 (141) | |
| Wild | 130 (92.2) |
| Mutant | 11 (7.8) |
| TET2 (141) | |
| Wild | 69 (48.9) |
| Mutant | 72 (51.1) |
| TET2 (141) | |
| Hetero | 57 (40.4) |
| Homo | 15 (10.7) |
| Wild | 69 (48.9) |
| CD34 (140) | |
| Positive | 69 (49.3) |
| Negative | 71 (50.7) |
| Dr (140) | |
| Positive | 100 (71.4) |
| Negative | 40 (28.6) |
| CD7 (140) | |
| Positive | 23 (16.4) |
| Negative | 117 (83.6) |
| OS time (months) median (range) | 2.07 (0.10-40.72) |
PB, peripheral blood; TLC, total leukocyte count; BM,bone marrow; CR, complete remission; OS, Overall survival; FLT3-ITD, FMS-like tyrosine kinase3internal tandem duplication; FLT3-TkD, FMS-like tyrosine kinase3 tyrosine kinase domain; IDH, Isocitrate dehydrogenase; TET2, Ten–Eleven Translocation gene2; NPM1, Nucleophosmin; DNMTA, DNA Methyltransferase
Isolation of DNA
Bone marrow samples or Whole blood samples were collected in the K2-EDTA tubes, Genomic DNA was isolated using QIAamp DNA blood Mini Kit (QIAGEN) (Cat no 51104). The quality of genomic DNA was assessed by using 2% agarose gel electrophoresis and the quantity of DNA was estimated by the spectrophotometer at absorbance of 260 nm.
The SNP assay for IDH1, IDH2 and TET2 gene polymorphisms was done using Thermo-Fisher predesigned SNP, USA, Quantstudio 3 Real-time PCR device included three polymorphisms that are C_167891677_20, rs121913500, C_163475619_10, rs121913503 and C_11566753_20, rs2454206 respectively.
Statistical Methods
IBM SPSS advanced statistics (Statistical Package for Social Sciences), version 24, was used to analyze the results (SPSS Inc., Chicago, IL). The median, range, or mean and standard deviation were used to express numerical data, whereas the number and percentage were used to describe qualitative data. The relationship between qualitative variables was investigated using the Pearson’s Chi-square (Fisher’s exact) test. Patients with AML were followed up using the Kaplan-Meier technique, and the log-rank test was used to compare two survival curves. To determine if statistically significant factors had independent prognostic effects at the univariate level, Cox regression analysis was used. The hazard ratio (HR) and its 95% confidence interval were then calculated (CI). All tests were two-tailed. A p-value of 0.05 or less was regarded as statistically significant.
Results
Demographic, clinical and laboratory data of the studied group are represented in Table 1. IDH mutations occurred in 11/141 (7.8%) patients. IDH1&2 were mutually exclusive, IDH1 mutation was found in 5/141 (3.5%) patients, while IDH2 mutation was found in 6/141 (4.3%) patients. TET2 mutation (Homozygous and heterozygous mutations) was positive in 72/141 (51.1%) patients, TET2 Homozygous mutation was positive in 15/141 (10.7) while TET2 heterozygous mutation was positive in 57/141(40.4%) patients. DNMTA mutation was positive in 14/140 (10.1%) patients. FLT3-TKD mutation was positive in 6/140 (4.3%) patients. FLT3-ITD mutation was positive in 45/140 (31.9%) patients while patients had FLT3-ITD with a high (>0.5) allelic ratio 25/45 (55.6%) patient. NPM1 mutation was positive in 21/128 (16.4%) patients.
There was no significant relationship between mutant and unmutated AML patients in terms of the IDH &TET2 mutations and the laboratory and clinical data as presented in Table 2. Statistical significance was encountered between TET2 mutation and aberrant expression of CD7 and MHC Class II (p value 0.018, 0.037 respectively). FLT-TKD gene showed a statistical association with mutant type TET2 (p=0.014) (Table 2).
Table 2.
Relationship between IDH1&IDH2 and TET2 Mutation and the Different Laboratory and Clinical Findings in Adult Acute Myeloid Leukemia Patients
| IDH1&IDH2 | TET2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Either IDH1 and or IDH2 mutant | both wild | P value | TET2 Mutant N=72 (51.1%) | TET2 Wild N=69 (48.9%) | p value | ||||||||
| Gende (n=141) | |||||||||||||
| Female | 4 (36.4) | 61 (46.9) | 0.5 | 37 (51.4) | 28 (40.6) | 0.198 | |||||||
| Male | 7 (63.6) | 69 (53.1) | 35 (48.6) | 41 (59.4) | |||||||||
| Age (n=141) | |||||||||||||
| ≤40 | 5 (45.5) | 64 (49.2) | 0.81 | 35 (48.6) | 34 (49.3) | 0.937 | |||||||
| >40 | 6 (54.5) | 66 (50.8) | 37 (51.4) | 35 (50.7) | |||||||||
| TLC X109/L (n=137) | |||||||||||||
| ≤100 | 9 (90.0% | 88 (69.3) | 0.166 | 51 (73.9) | 46 (67.6) | 0.42 | |||||||
| >100 | 1 (10.0) | 39 (30.7) | 18 (26.1) | 22 (32.4) | |||||||||
| Hb g/dl (n=137) | |||||||||||||
| ≤10 | 9 (90.0) | 114 (89.8) | 0.981 | 64 (92.8) | 59 (86.8) | 0.247 | |||||||
| >10 | 1 (10.0) | 13 (10.2) | 5 (7.2) | 9 (13.2) | |||||||||
| Plts X109/L (n=137) | |||||||||||||
| ≤100 | 9 (90.0) | 103 (81.1) | 0.483 | 55 (79.7) | 57 (83.8) | 0.533 | |||||||
| >100 | 1 (10.0) | 24 (18.9) | 14 (20.3) | 11 (16.2) | |||||||||
| PB blasts %(n=141) | |||||||||||||
| ≤ 50 | 3 (27.3) | 34 (26.2) | 0.935 | 19 (26.4) | 18 (26.1) | 0.968 | |||||||
| > 50 | 8 (72.7) | 96 (73.8) | 53 (73.6) | 51 (73.9) | |||||||||
| BMA blast%(n=141) | |||||||||||||
| ≤ 50 | 1 (9.1) | 19 (14.6) | 0.614 | 7 (9.7) | 13 (18.8) | 0.121 | |||||||
| > 50 | 10 (90.9) | 111 (85.4) | 65 (90.3) | 56 (81.2) | |||||||||
| FAB classification | |||||||||||||
| M1&M2 | 8 (72.7) | 64 (52.0) | 0.187 | 40 (57.1) | 32 (50.0) | 0.407 | |||||||
| M4&M5 | 3 (27.3) | 59 (48.0) | 30 (42.9) | 32 (50.0) | |||||||||
| LNS (n=134) | |||||||||||||
| No | 6 (54.5) | 72 (58.5) | 0.797 | 37 (54.4) | 41 (62.1) | 0.366 | |||||||
| Yes | 5 (45.5) | 51 (41.5) | 31 (45.6) | 25 (37.9) | |||||||||
| Organomegally(n=134) | |||||||||||||
| No | 8 (72.7) | 77 (62.6) | 0.504 | 41 (60.3) | 44 (66.7) | 0.444 | |||||||
| Yes | 3 (27.3) | 46 (37.4) | 27 (39.7) | 22 (33.3) | |||||||||
| CD34(n=140) | |||||||||||||
| No | 6 (54.5) | 65 (50.4) | 0.791 | 35 (48.6) | 36 (52.9) | 0.609 | |||||||
| Yes | 5 (45.5) | 64 (49.6) | 37 (51.4) | 32 (47.1) | |||||||||
| DR(n=140) | |||||||||||||
| No | 3 (27.3) | 37 (28.7) | 0.921 | 15 (20.8) | 25 (36.8) | 0.037 | |||||||
| Yes | 8 (72.7) | 92 (71.3) | 57 (79.2) | 43 (63.2) | |||||||||
| CD7(n=140) | |||||||||||||
| No | 11 (100.0) | 106 (82.2) | 0.126 | 55 (76.4) | 62 (91.2) | 0.018 | |||||||
| Yes | 0 (0.0) | 23 (17.8) | 17 (23.6) | 6 (8.8) | |||||||||
| FLT3-ITD (n=141) | |||||||||||||
| Mutant | 1 (9.1) | 44 (33.8) | 0.091 | 21 (29.2) | 24 (34.8) | 0.475 | |||||||
| Wild | 10 (90.9) | 86 (66.2) | 51 (70.8) | 45 (65.2) | |||||||||
| ITD Ratio (n=45) | |||||||||||||
| ≤0.525 | 0 (0.0) | 20 (45.5) | * | 11 (52.4) | 9 (37.5) | 0.316 | |||||||
| >0.525 | 1 (100.0) | 24 (54.5 | 10 (47.6) | 15 (62.5) | |||||||||
| IDH1&IDH2 | TET2 | ||||||||||||
| Either IDH1 and or IDH2 mutant | both wild | P value | TET2 Mutant N=72 (51.1%) | TET2 Wild N=69 (48.9%) | p value | ||||||||
| TKD (n=140) | |||||||||||||
| Mutant | 0 (0.0) | 6 (4.7) | 0.465 | 6 (8.5) | 0 (0.0) | 0.014 | |||||||
| Wild | 11 (100.0) | 123 (95.3) | 65 (91.5) | 69 (100.0) | |||||||||
| NPM (n=128) | |||||||||||||
| Mutant | 1 (10.0) | 20 (16.9) | 0.569 | 8 (12.5) | 13 (20.3) | 0.233 | |||||||
| Wild | 9 (90.0) | 98 (83.1) | 56 (87.5) | 51 (79.7) | |||||||||
| DNMTA(n=139) | |||||||||||||
| Mutant | 1 (9.1) | 13 (10.2) | 0.91 | 4 (5.7) | 10 (14.5) | 0.086 | |||||||
| Wild | 10 (90.9) | 115 (89.8) | 66 (94.3) | 59 (85.5) | |||||||||
| IDH1&IDH2 | |||||||||||||
| Either IDH1 and or IDH2 mutant | - | - | 6 (8.3) | 5 (7.2) | 0.81 | ||||||||
| Both wild | - | - | 66 (91.7) | 64 (92.8) | |||||||||
| TET2 gene | |||||||||||||
| mutant TET2 gene | 6 (54.5) | 66 (50.8) | 0.81 | - | - | ||||||||
| wild TET2 gene | 5 (45.5) | 64 (49.2) | - | - | |||||||||
| BMA cellularity | |||||||||||||
| Hypercellular | 7 (63.6) | 113 (86.9) | 0.083 | 59 (81.9) | 61 (88.4) | 0.388 | |||||||
| Hypocellular | 2 (18.2) | 6 (4.6) | 4 (5.6) | 4 (5.8) | |||||||||
| Normocellular | 2 (18.2) | 11 (8.5) | 9 (12.5) | 4 (5.8) | |||||||||
| combined FLT3&NPM (n=128) | |||||||||||||
| Favourable (n=17) | 1 (5.9) | 16 (94.1) | 0.847 | 4 (6.3) | 9 (14.1) | 0.097 | |||||||
| Unfavorable (n=13) | 0 (0.0) | 13 (100) | 50 (78.1) | 39 (60.9) | |||||||||
| Intermediate (n=98) | 9 (9.2) | 89 (90.8) | 10 (15.6) | 16 (25.0) | |||||||||
*, p value cannot be calculated because of small number within strata
According to the European Leukemia Net (ELN) 2017 risk classification, 17/128 (13.3%) were classified low risk (NPM1 positive and AR<0.5), 13/128(10.2%) high risk (NPM1 negative and AR>0.5) and 98/128 (76.6%) intermediate risk (NPM1 positive and AR>0.5) as shown in Table 3.
Table 3.
Relationship between Combined FLT3&NPM and the Different Laboratory and Clinical Findings in Adult Acute Myeloid Leukemia Patients
| Combined FLT3&NPM with ITD ratio | p value | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Favourable (n=17) | Unfavorable (n=13) | Intermediate (n=98) | |||||||
| Gender | |||||||||
| Female | 5a (29.4%) | 10b (76.9%) | 49a, b (50.0%) | 0.036 | |||||
| Male | 12a (70.6%) | 3b (23.1%) | 49a, b (50.0%) | ||||||
| Age (years) | |||||||||
| ≤40 | 8 (47.1%) | 9 (69.2%) | 46 (46.9%) | 0.314 | |||||
| >40 | 9 (52.9%) | 4 (30.8%) | 52 (53.1%) | ||||||
| TLC (n=124) | |||||||||
| ≤100 | 13 (76.5%) | 7 (58.3%) | 73 (76.8%) | 0.398 | |||||
| >100 | 4 (23.5%) | 5 (41.7%) | 22 (23.2%) | ||||||
| Hb (n=124) | |||||||||
| ≤10 | 15 (88.2%) | 12 (100.0%) | 83 (87.4%) | 0.541 | |||||
| >10 | 2 (11.8%) | 0 (0.0%) | 129 (12.6%) | ||||||
| PLTs (n=124) | |||||||||
| ≤100 | 14 (82.4%) | 12 (100.0%) | 75 (78.9%) | 0.221 | |||||
| >100 | 3 (17.6%) | 0 (0.0%) | 20 (21.1%) | ||||||
| PB blasts | |||||||||
| ≤ 50 | 3 (17.6%) | 2 (15.4%) | 32 (32.7%) | 0.308 | |||||
| > 50 | 14 (82.4%) | 11 (84.6%) | 66 (67.3%) | ||||||
| BMA blast | |||||||||
| ≤ 50 | 3 (17.6%) | 1 (7.7%) | 16 (16.3%) | 0.835 | |||||
| > 50 | 14 (82.4%) | 12 (92.3%) | 82 (83.7%) | ||||||
| BMA cellularity | |||||||||
| Hypercellular | 16 (94.1%) | 11 (84.6%) | 80 (81.6%) | 0.632 | |||||
| Hypocellular | 1 (5.9%) | 1 (7.7%) | 6 (6.1%) | ||||||
| Normocellular | 0 (0.0%) | 1 (7.7%) | 12 (12.2%) | ||||||
| FAB (n=122) | |||||||||
| M1&M2 | 7 (41.2%) | 7 (53.8%) | 49 (53.3%) | 0.648 | |||||
| M4&M5 | 10 (58.8%) | 6 (46.2%) | 43 (46.7%) | ||||||
| CR (n=90) | |||||||||
| No | 2a (18.2%) | 8b (72.7%) | 35a, b (51.5%) | 0.034 | |||||
| Yes | 9a (81.8%) | 3b (27.3%) | 33a, b (48.5%) | ||||||
| LNS (n=121) | |||||||||
| No | 12 (70.6%) | 7 (58.3%) | 51 (55.4%) | 0.509 | |||||
| Yes | 5 (29.4%) | 5 (41.7%) | 41 (44.6%) | ||||||
| Organomegally (n=121) | |||||||||
| No | 11 (64.7%) | 6 (50.0%) | 57 (62.0%) | 0.689 | |||||
| Yes | 6 (35.3%) | 6 (50.0%) | 35 (38.0%) | ||||||
| CD34 (n=127) | |||||||||
| No | 15a (88.2%) | 5b (38.5%) | 47b (48.5%) | 0.004 | |||||
| Yes | 2a (11.8%) | 8b (61.5%) | 50b (51.5%) | ||||||
| DR (n=127) | |||||||||
| No | 4 (23.5%) | 3 (23.1%) | 32 (33.0% | 0.676 | |||||
| Yes | 13 (76.5%) | 10 (76.9%) | 65 (67.0%) | ||||||
| CD7 (n=127) | |||||||||
| No | 17 (100.0%) | 11 (84.6%) | 78 (80.4%) | 0.135 | |||||
| Yes | 0 (0.0%) | 2 (15.4%) | 19 (19.6%) | ||||||
| Combined FLT3&NPM with ITD ratio | p value | ||||||||
| Favourable (n=17) | Unfavorable (n=13) | Intermediate (n=98) | |||||||
| TKD (n=127) | |||||||||
| Mutant | 1 (5.9%) | 0 (0.0%) | 5 (5.2%) | 1 | |||||
| Wild | 16 (94.1%) | 13 (100.0%) | 92 (94.8%) | ||||||
| DNMTA (n=126) | |||||||||
| Mutant | 2 (12.5%) | 1 (7.7%) | 11 (11.3%) | 1 | |||||
| Wild | 14 (87.5%) | 12 (92.3%) | 86 (88.7%) | ||||||
| IDH1&IDH2 | |||||||||
| Either IDH1 and or IDH2 mutant | 1 (5.9%) | 0 (0.0%) | 9 (9.2%) | 0.847 | |||||
| Both wild | 16 (94.1%) | 13 (100.0%) | 89 (90.8%) | ||||||
| TET2 gene | |||||||||
| mutant TET2 gene | 6a (35.3%) | 3a (23.1%) | 55a (56.1%) | 0.035 | |||||
| wild TET2 gene | 11a (64.7%) | 10a (76.9%) | 43a (43.9%) | ||||||
| BMA cellularity | |||||||||
| Hypercellular | 16 (94.1%) | 11 (84.6%) | 80 (81.6%) | 0.632 | |||||
| Hypocellular | 1 (5.9%) | 1 (7.7%) | 6 (6.1%) | ||||||
| Normocellular | 0 (0.0%) | 1 (7.7%) | 12 (12.2%) | ||||||
Hundred out of the 141 (70.9%) patients received induction chemotherapy, complete remission (CR) was achieved in 51 patients (51.0%), 8(8.0%) failed to achieve CR, and 41 (41.0%) died before reaching day 28. 41/141 (29.1%) patients with no available data as the patient refused therapy either due to treatment toxicity and/or his/her physical condition.
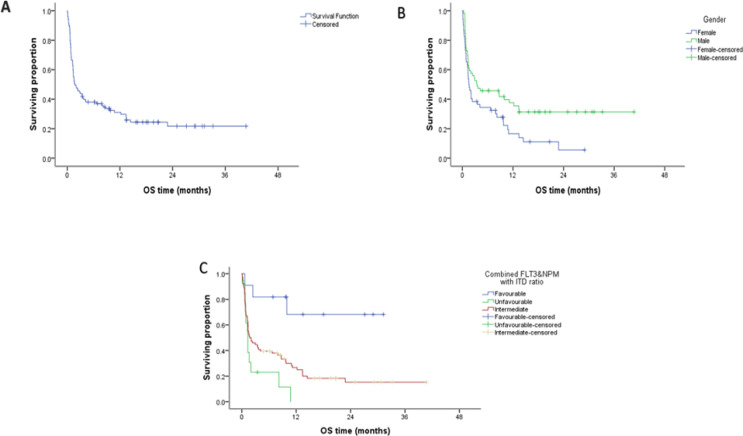
The overall survival (OS) was studied as regards the different parameters (Table 4). Patients with complete clinical data (111/141) were followed for a median of 2.07 months (range 0.10 - 40.72). The median overall survival was 2.07 months while the cumulative overall survival at 2 years was 0.198 %. Significant inferior survival was found in the female patients as compared to male group (p value 0. 014). Moreover, significant inferior overall survival was found in patients with FlT3-ITD as compared to the wild group, patients with wild NPM as compared to the mutant group (p value 0.035, 0.005 respectively) (Figure 1). Multivariate analysis using Cox regression hazard model to obviate the effect of confounders indicated that risk stratification according to ELN 2017 was the only independent prognostic factors for OS, being in the unfavorable category was associated with increased risk about eight times than being in favourable category, while being in the intermediate was associated with increased risk about 4.7 times than in favourable one, HR=8.2 , 4.7 respectively with 95% CI (2.2 →29.6 and 1.49 →15.2 , respectively ) P-value <0.001 and 0.008, respectively.
Table 4.
Effect of Different Variables on Overall Survival (OS) of AML Patients
| Total No. | No. of Events | Cumulative survival at 2year (%) |
Median survival time (months) | P value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Whole group | 111 | 83 | 0.198 | 2.07 | - | ||||||
| Gender | |||||||||||
| Female | 52 | 44 | 0.056 | 1.55 | 0.014 | ||||||
| Male | 59 | 39 | 0.313 | 3.45 | |||||||
| Age (years) | |||||||||||
| ≤40 | 57 | 41 | 0.183 | 4.14 | 0.216 | ||||||
| >40 | 54 | 42 | 0.2 | 1.41 | |||||||
| TLC (n=109) | |||||||||||
| ≤100 X109/L | 79 | 60 | 0.185 | 2.96 | 0.487 | ||||||
| >100 X109/L | 30 | 23 | 0.214 | 1.41 | |||||||
| Hemoglobin (n=109) | |||||||||||
| ≤10 g/dl | 104 | 80 | 0.176 | 2 | 0.34 | ||||||
| >10 g/dl | 5 | 3 | 0.400 | 12.2 | |||||||
| Platelets (n=109) | |||||||||||
| ≤100 X 109/L | 87 | 66 | 0.185 | 2 | 0.745 | ||||||
| >100 X 109/L | 22 | 17 | 0.205 | 3.42 | |||||||
| PB blasts% | |||||||||||
| ≤ 50 | 31 | 23 | 0.148 | 3.55 | 0.71 | ||||||
| > 50 | 80 | 60 | 0.221 | 2 | |||||||
| BMA blast % | |||||||||||
| ≤ 50 | 18 | 12 | 0.216 | 8.72 | 0.347 | ||||||
| > 50 | 93 | 71 | 0.202 | 2 | |||||||
| BMA cellularity | |||||||||||
| Hypercellular | 95 | 70 | 0.204 | 2.07 | 0.92 | ||||||
| Hypocellular | 7 | 6 | 10.76 | ||||||||
| Normocellular | 9 | 7 | 0.222 | 1.35 | |||||||
| Lymphadenopathy (n=105) | |||||||||||
| No | 62 | 44 | 0.245 | 2.07 | 0.618 | ||||||
| Yes | 43 | 34 | 0.142 | 2.34 | |||||||
| Organomegally (n=105) | |||||||||||
| No | 68 | 48 | 0.226 | 3.55 | 0.167 | ||||||
| Yes | 37 | 30 | 0.17 | 2 | |||||||
| FAB classification (n=105) | |||||||||||
| M1&M2 | 54 | 40 | 0.219 | 1.55 | 0.441 | ||||||
| M4&M5 | 51 | 37 | 0.219 | 3.68 | |||||||
| CD34 (n=110) | |||||||||||
| No | 53 | 38 | 0.24 | 2.43 | 0.674 | ||||||
| Yes | 57 | 44 | 0.162 | 2.07 | |||||||
| DR (n=110) | |||||||||||
| No | 29 | 23 | 0.162 | 2 | 0.656 | ||||||
| Yes | 81 | 59 | 0.228 | 2.43 | |||||||
| CD7 (n=110) | |||||||||||
| No | 91 | 69 | 0.204 | 2.07 | 0.387 | ||||||
| Yes | 19 | 13 | 0.167 | 4.14 | |||||||
| NPM (n=100) | |||||||||||
| Mutant | 14 | 5 | 0.595 | 0.005 | |||||||
| Wild | 86 | 70 | 0.133 | 1.61 | |||||||
| Total No. | No. of Events | Cumulative survival at 2year (%) |
Median survival time (months) | P value | |||||||
| FLT3 ITD | |||||||||||
| Mutant | 38 | 32 | 0.118 | 1.55 | 0.035 | ||||||
| Wild | 73 | 51 | 0.242 | 3.42 | |||||||
| Combined FLT3&NPM with ITD ratio (n=100) | |||||||||||
| Favourable (a) | 11 | 3 | 0.682 | NA | 0.002 | ||||||
| Unfavorable (b) | 13 | 12 | NA | 1.32 | |||||||
| Intermediate (b) | 76 | 60 | 0.153 | 1.74 | |||||||
| ITD Ratio (n=27) | |||||||||||
| ≤ 0.525 | 13 | 9 | 0.308 | 6.71 | 0.114 | ||||||
| >0.525 | 14 | 12 | NR | 1.32 | |||||||
| TKD (n=110) | |||||||||||
| Mutant | 5 | 3 | 0.3 | 7.89 | 0.489 | ||||||
| Wild | 105 | 79 | 0.194 | 2.07 | |||||||
| DNMTA (n=110) | |||||||||||
| Mutant | 11 | 8 | 0.227 | 2.96 | 0.755 | ||||||
| Wild | 99 | 74 | 0.198 | 2 | |||||||
| IDH1&IDH2 | |||||||||||
| Either IDH1 and or IDH2 mutant | 10 | 8 | 0.2 | 0.82 | 0.37 | ||||||
| Both wild | 101 | 75 | 0.197 | 2.43 | |||||||
| TET2 | |||||||||||
| Mutant | 58 | 43 | 0.189 | 2.83 | 0.645 | ||||||
| Wild | 53 | 40 | 0.207 | 1.55 | |||||||
| CR (n=98) | |||||||||||
| No | 47 | 46 | 0.021 | 0.79 | <0.001 | ||||||
| Yes | 51 | 24 | 0.413 | 14.44 | |||||||
Figure 1.
A. Kaplan–Meier plot showing the overall survivability (OS) of the entire patient group, B. OS according to gender, C. OS according to combined FLT3&NPM with ITD ratio
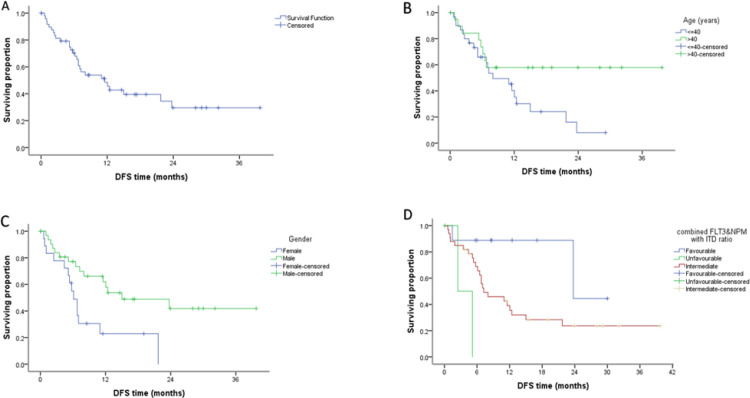
The median of Disease-free survival (DFS) was 11.55 months while the cumulative DFS at 1 year was 0.290 % (Table 5). The Disease-free survival of the studied group revealed also a significant inferior DFS in the female patients and patients > 40 years (p value 0.008, 0.048 respectively). Moreover, significant inferior DFS was found in the wild NPM patients as compared to Mutant group (p value 0.036) (Figure 2). Multivariate analysis using Cox regression hazard model to obviate the effect of confounders indicated that risk stratification according to ELN 2017 was the only independent prognostic factors for DFS, being in the unfavorable category was associated with increased risk about sixteen times than being in favourable category, while being in the intermediate was associated with increased risk about 3.7 times than in favourable one, HR=16.0 , 3.7 respectively with 95% CI (2.08 →123.56 and 0.88 →15.8 , respectively ) p-value 0.008 and 0.049, respectively.
Table 5.
Effect of Different Variables on Disease Free Survival (DFS) of AML Patients:
| Total No. | No. of Events | Cumulative survival at 1year (%) | Median survival time (months) | P value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Whole group | 51 | 29 | 0.29 | 11.55 | |||||||
| Gender | |||||||||||
| Female | 19 | 14 | NR | 6.12 | 0.008 | ||||||
| Male | 32 | 15 | 0.419 | 15 | |||||||
| Age (years) | |||||||||||
| ≤40 | 31 | 21 | 0.081 | 8.03 | 0.048 | ||||||
| >40 | 20 | 8 | 0.579 | NR | |||||||
| TLC | |||||||||||
| ≤100 x109/L | 38 | 21 | 0.309 | 10.99 | 0.762 | ||||||
| >100 x109/L | 11 | 6 | 0.36 | 12.04 | |||||||
| Hemoglobin | |||||||||||
| ≤10 g/dl | 46 | 26 | 0.28 | 10.99 | * | ||||||
| >10 g/dl | 3 | 1 | 0.667 | ||||||||
| Platelets | |||||||||||
| ≤100x109/L | 39 | 22 | 0.276 | 8.03 | 0.527 | ||||||
| >100 x109/L | 10 | 5 | 0.411 | 12.04 | |||||||
| PB blasts% | |||||||||||
| ≤ 50 | 13 | 8 | 0.186 | 7.24 | 0.862 | ||||||
| > 50 | 38 | 21 | 0.319 | 12.04 | |||||||
| BMA blast% | |||||||||||
| ≤ 50 | 8 | 5 | 0.219 | 6.78 | 0.96 | ||||||
| > 50 | 43 | 24 | 0.308 | 11.55 | |||||||
| BMA cellularity | |||||||||||
| Hypercellular | 44 | 25 | 0.273 | 10.99 | * | ||||||
| Hypocellular | 4 | 3 | 6.51 | ||||||||
| Normocellular | 3 | 1 | 0.667 | ||||||||
| FAB classification | |||||||||||
| M1&M2 | 23 | 13 | 0.385 | 6.78 | 0.448 | ||||||
| M4&M5 | 26 | 14 | 0.339 | 21.71 | |||||||
| LNS | |||||||||||
| No | 29 | 14 | 0.466 | 12.4 | 0.37 | ||||||
| Yes | 21 | 14 | 0.098 | 11.55 | |||||||
| Organomegally | |||||||||||
| No | 34 | 20 | 0.282 | 12.4 | 0.861 | ||||||
| Yes | 16 | 8 | 0.323 | 11.55 | |||||||
| CD34 | |||||||||||
| No | 23 | 13 | 0.306 | 10.99 | 0.942 | ||||||
| Yes | 28 | 16 | 0.275 | 11.55 | |||||||
| DR | |||||||||||
| No | 11 | 6 | 0.305 | 11.55 | 0.799 | ||||||
| Yes | 40 | 23 | 0.291 | 10.99 | |||||||
| CD7 | |||||||||||
| No | 40 | 21 | 0.335 | 12.04 | 0.579 | ||||||
| Yes | 11 | 8 | 0.17 | 10.99 | |||||||
| NPM | |||||||||||
| Mutant | 10 | 2 | 0.45 | 23.72 | 0.036 | ||||||
| Wild | 35 | 25 | 0.221 | 7.04 | |||||||
| FLT3 ITD | |||||||||||
| Mutant | 13 | 7 | 5.16 | 0.173 | |||||||
| Wild | 38 | 22 | 0.323 | 12.4 | |||||||
| ITD Ratio | |||||||||||
| ≤ 0.525 | 5 | 3 | NA | 12.04 | 0.171 | ||||||
| >0.525 | 5 | 2 | NA | 5.16 | |||||||
| TKD | |||||||||||
| Mutant | 3 | 2 | 0.333 | 6.78 | * | ||||||
| Wild | 47 | 26 | 0.285 | 11.55 | |||||||
| DNMTA | |||||||||||
| Mutant | 5 | 4 | 0.2 | 6.51 | 0.384 | ||||||
| Wild | 45 | 24 | 0.307 | 12.4 | |||||||
| Combined FLT3&NPM with ITD ratio | |||||||||||
| Favourable (a) | 9 | 2 | 0.889 | 23.72 | 0.011 | ||||||
| Unfavorable (b) | 3 | 2 | NA | 2.47 | |||||||
| Intermediate (b) | 33 | 23 | 0.391 | 7.24 | |||||||
| IDH1&IDH2 | |||||||||||
| Either IDH1 and or IDH2 mutant | 3 | 1 | 0.667 | * | |||||||
| Both wild | 48 | 28 | 0.262 | 10.99 | |||||||
| TET2 | |||||||||||
| Mutant | 31 | 20 | 0.323 | 7.04 | 0.079 | ||||||
| Wild | 20 | 9 | 0.71 | 21.71 | |||||||
Figure 2.
A. Kaplan–Meier plot showing the disease-free survival (DFS) of the entire patient group, B. DFS according to age, C. DFS according to gender, D. DFS according to combined FLT3&NPM with ITD ratio
Discussion
It has been demonstrated that better risk stratification may be achieved by identifying FLT3- ITD, NPM1, and CEBPA mutations and incorporating them into prognostic models, particularly in the vast population of patients with CN-AML (Dö et al., 2010). There is still a sizeable group of intermediate risk patients without FLT3-ITD, NPM1 and CEBPA mutations or other reliable prognostic markers, indicating the need for additional markers that could explain the differential outcome in this heterogeneous patient group (Estey, 2013). The most frequent mutations reported in AML are those that affect the genes that code for IDH1, IDH2, and TET2 (Abdel-Wahab and Levine, 2013).
In the current study, we investigated the prevalence and prognostic effects of the IDH1, IDH2 and TET2 SNPs 395G>T (rs121913500), 359G>A (rs121913503), 5284A>G (rs2454206) respectively. Studies have revealed an incidence of 2-14% for IDH mutations (Chotirat et al., 2012 ; Ahmad et al., 2014) and 8-19% for IDH2 (Mardis et al., 2009) in various patient groups. In our study cohort, the frequency of IDH1 mutations was 3.5% and the frequency of IDH2 mutations was 4.3%. However, in another study, IDH1/2 mutations were found in 5.5% and 4%, respectively (Raveendran et al., 2015) and IDH1 and 2 mutations were detected in 2.9% and 11.4%, respectively (ElNahass et al., 2020).
The correlation of IDH mutations with patient characteristics, various laboratory results and AML prognostic factors were studied. IDH mutations were more common in male, although the difference was not statistically significant and this result was in inconsistent with other report (Raveendran et al., 2015; Ali et al., 2018; ElNahass et al., 2020; Pastore et al.,2022). In agreement with another report (Patel et al., 2011), we did not find a significant correlation between IDH mutations and TLC, Hb concentration, or platelets count (p= 0.166 , 0.981 and 0.483 respectively).
In a previous study, it was shown that AML patients with IDH mutation shared several common clinical characteristics, including manifestation at an older age or higher platelets count at diagnosis (Marcucci et al., 2010). However, in the current study, IDH mutations were not significantly correlated with age or higher platelets count which consistent with a previous observation (Schnittger et al., 2010) .
Our findings demonstrated that the prevalence of IDH1 mutations did not significantly differ among the various FAB subtypes, in contrast to other studies that claimed IDH1 mutations were strongly related with FAB AML M1(Mardis et al., 2009 ; Schnittger et al., 2010).
FLT3-ITD was negative in 10/11 (90.9%) of IDH mutant patients with (p=0.091). This finding was in consistent with others who found that IDH mutations were not associated with FLT3-ITD mutations (Marcucci et al., 2010; Virijevic et al., 2016; ElNahass et al., 2020) However, these results are at odds with many other previous reports that found FLT3-ITD mutated AML associated with IDH mutations (DiNardo et al., 2016; Papaemmanuil et al., 2016; Boddu et al., 2017;) which could be explained by ethnic variations and other genetic markers interactions.
The prognostic significance of IDH mutations in AML has been extensively investigated but is still controversial. It varies greatly depending on the type of mutation that is found as well as whether or not other clinically significant genes have concurrent alterations. Despite the fact that certain research have revealed that individuals with CN-AML have a poorer prognosis (Marcucci et al., 2010; Yamaguchi et al., 2014;Virijevic et al., 2016; Xu et al., 2023), others have not found any prognostic effect (Thol et al., 2010; Wagner et al., 2010; Dinardo et al., 2015), which is in accordance with our findings. In our cohort of patients, we did not demonstrate any significant association of IDH mutations with EFS or OS, although it should be stressed that this finding is certainly limited by the small sample size as well as the low number of IDH-mutated patients.
The identification of the biological and clinical characteristics of mutated isoforms 1 and 2 of isocitrate dehydrogenases (IDH1/2) result in the creation of a customized treatment plan. Enhancing differentiation and maturation of the malignant clone that targets IDH is a strategy to enhance clinical outcomes in AML (Cerchione et al., 2021).
Several previous studies have reported that genetic variants in the TET2 gene are involved in the development of hematological malignancies including AML ( Weissmann et al., 2012; Feng et al., 2019; Zeng et al., 2019; Duployez et al., 2020).
In our study cohort, we found the frequency of wild 69/141(48.9%), homozygous 15/141(10.7%) and heterozygous 57/141 (40.4%) expression of TET2 gene polymorphism this mostly similar as Dammag et al., 2020 who reported that TET2 SNP was wild in AML in (46%), heterozygous in (44%) and with only 10% of patients being homozygous while Li et al. from Taiwan in 2011 reported that about78.6% of patients were presented with TET2 SNP, all SNPs were heterozygous, only 4 SNP were homozygous (Li et al., 2011).
In our study, TET2 mutation did not showed any statistical difference between polymorphism genotype and age, gender and FAB in AML patients this similar as reported by (Dammag et al., 2020) and we have not found a significant relation between TET2 mutations and CBC findings (including peripheral blast %) of patients This differed from that reported by (Metzeler et al., 2011; Wang et al., 2019)who reported that TET2 mutation were associated with higher pretreatment white blood cell counts and lower platelet count , and also differed from (Chehreghani et al., 2022) who said that patients harbored TET2 mut had higher Hb levels, lower Platelets counts and tended to have lower WBC counts ,while (Nibourel et al., 2010; Hamed et al., 2018) reported no relation between TET2 mutation and Hb, WBCs and platelets.
Contrary to previous studies (Hamed et al., 2018), which indicated that TET2 mutations were with high pattern associated with FAB AMLM1, M2 and M5 subtypes and (Wang et al., 2019) reported that the low differentiated subtype AML(M0/M1) had a significantly higher frequency of TET2 under-expression than the high differentiated subtypes AML(M2/M3). Our study suggested that the distribution of TET2 mutations didn’t show significant difference among the different FAB subtypes and this similar to Nibourel et al., 2010; Chehreghani et al., 2022.
Regarding our study, the TET2 mutation did not show any correlation with FLT3, NPM1 and DNMTA expression levels and this similar to Chehreghani et al., 2022.
Large cohort studies revealed that TET2 mutations did not impact the overall survival in AML patient (Nibourel et al., 2010; Kosmider et al., 2011) which similar with our cohort of patients, we did not found any significant association of TET2 mutation with EFS or OS, although it should be stressed that this finding is certainly limited by the small sample ,on the other side, some reports found that TET2 mutant AML(Abdel-Wahab et al., 2009; Metzeler et al., 2011; Weissmann et al., 2012; Liu et al., 2014; Wang et al., 2019; Xu et al., 2023) patients had worse outcomes compared to those without TET2 mutations.
TET2 mutations, as an early event in pathogenesis, could work in concert with other gene mutations, known as background mutations, to induce various hematological malignancies. For example, TET2 mutations, when harboring FLT3-ITD mutation, induced AML (Jan et al., 2012).
The National Comprehensive Cancer Network (NCCN) recommendations state that NPM1 mutation, in the absence of FLT3-ITD mutation, has a favourable prognostic effect in cytogenetically normal adult AML (Huang et al., 2019; Xu et al., 2020). However, in patients who have both a double NPM1 mutation and a FLT3-ITD mutation, the prognostic effect of the NPM1 mutation relies on the allelic ratio (AR) (Tsai et al., 2016; Döhner et al., 2017). Recent recommendations from the ELN for the risk categorization of AML include evaluating the prognostic impact of the FLT3-ITD in relation to the NPM1 mutation (Döhner et al., 2017). In contrast to the high risk group NPM1wild with high FLT3-ITD AR, patients in the low risk group NPM1mutant with low FLT3-ITD AR will not be candidates for post remission allogenic stem cell (Döhner et al., 2020) .
Multivariate analysis for OS in our AML group indicated that risk stratification according to ELN 2017 was the only independent prognostic factors for OS. In the unfavorable category OS was associated with increased risk about eight times than being in favourable category, while in the intermediate it was associated with increased risk about 4.7 times than in favourable one HR=8.2, 4.7 respectively with 95% CI (2.2-29.6 and 1.49-15.2, respectively) P-value <0.001 and 0.008, respectively. In agreement with our results Schneider et al., 2012 & Pratcorona et al., 2013 have confirmed the favorable impact effect of NPM1 mutation on patients with low level of FLT3-ITD mutation. However, other studies did not detect significant difference in the outcome between low risk group and the high risk group ( Sakaguchi et al., 2018; Feng et al., 2019; Shafik et al., 2021).
Author Contribution Statement
All authors contributed to the design and implementation of the research, to the analysis of the results and to the writing of the manuscript..
Acknowledgements
All authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, take responsibility for the integrity of the work and have given final approval to the version to be published. We acknowledge the patients and their families and all members of the National Cancer Institute, Cairo University, Egypt. The study was authorized by the Institutional Review Board (IRB) in accordance with the Helsinki declaration of studies involving human subjects after written informed permission was acquired from each patient.
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood. 2013;121:3563–72. doi: 10.1182/blood-2013-01-451781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Mullally A, Hedvat C, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–7. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F, Mohota R, Sanap S, Mandava S, Das BR. Molecular evaluation of DNMT3A and IDH1/2 gene mutation: Frequency, distribution pattern and associations with additional molecular markers in normal karyotype indian acute myeloid leukemia patients. Asian Pac J Cancer Prev. 2014;15:1247–53. doi: 10.7314/apjcp.2014.15.3.1247. [DOI] [PubMed] [Google Scholar]
- Albano F, Anelli L, Zagaria A, et al. Decreased TET2 gene expression during chronic myeloid leukemia progression. Leuk Res. 2011;35:220–2. doi: 10.1016/j.leukres.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Ali MAM, Ahmed EK, Assem MMA, Helwa R. The Synonymous Isocitrate Dehydrogenase 1 315C>T SNP Confers an Adverse Prognosis in Egyptian Adult Patients with NPM1-/CEBPA-Negative Acute Myeloid Leukemia. Indian J Hematol Blood Transfus. 2018;34:240–52. doi: 10.1007/s12288-017-0852-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddu P, Takahashi K, Pemmaraju N, et al. Influence of IDH on FLT3-ITD status in newly diagnosed AML. Leukemia. 2017;31:2526–9. doi: 10.1038/leu.2017.244. [DOI] [PubMed] [Google Scholar]
- Cerchione C, Romano A, Daver N, et al. IDH1/IDH2 Inhibition in Acute Myeloid Leukemia. Front Oncol. 2021;11:639387. doi: 10.3389/fonc.2021.639387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehreghani Z, Sadeghian MH, Ayatollahi H, et al. Detection of TET2 Mutation in Patients with De Novo Acute Myeloid Leukemia: A Mutation Analysis of 51 Iranian Patients. Asian Pac J Cancer Prev. 2022;23:803–6. doi: 10.31557/APJCP.2022.23.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotirat S, Thongnoppakhun W, Promsuwicha O, Boonthimat C, Auewarakul CU. Molecular alterations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) metabolic genes and additional genetic mutations in newly diagnosed acute myeloid leukemia patients. J Hematol Oncol. 2012;5:1–10. doi: 10.1186/1756-8722-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammag EA, Hamed NAM, Elhalawani NA, Kassem HS, Ayad MW. TET2 Single Nucleotide Polymorphism in Myeloid Neoplasms Among Egyptian Patients. Indian J Hematol Blood Transfus. 2020;36:91–6. doi: 10.1007/s12288-019-01172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo CD, Ravandi F, Agresta S, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol. 2015;90:732–6. doi: 10.1002/ajh.24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo CD, de Botton S, Stein EM, et al. Determination of IDH1 Mutational Burden and Clearance Via Next-Generation Sequencing in Patients with IDH1 Mutation-Positive Hematologic Malignancies Receiving AG-120, a First-in-Class Inhibitor of Mutant IDH1. Blood. 2016;128:1070. [Google Scholar]
- Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373:1136–52. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner K, Thiede C, Jahn N, et al. Impact of NPM1/FLT3-ITD genotypes defined by the 2017 European LeukemiaNet in patients with acute myeloid leukemia. Blood. 2020;135:371–80. doi: 10.1182/blood.2019002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duployez N, Goursaud L, Fenwarth L. Familial myeloid malignancies with germline TET2 mutation. Leukemia. 2020;34:1450–3. doi: 10.1038/s41375-019-0675-6. [DOI] [PubMed] [Google Scholar]
- ElNahass YH, Badawy RH, ElRefaey FA, et al. IDH mutations in AML patients; A higher association with intermediate risk cytogenetics. Asian Pac J Cancer Prev. 2020;21:721–5. doi: 10.31557/APJCP.2020.21.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013;88:317–27. doi: 10.1002/ajh.23404. [DOI] [PubMed] [Google Scholar]
- Feng S, Zhou L, Zhang X, et al. Impact of ELN risk stratification, induction chemotherapy regimens and hematopoietic stem cell transplantation on outcomes in hyperleukocytic acute myeloid leukemia with initial white blood cell count more than 100 × 109/L. Cancer Manag Res. 2019;11:9495–503. doi: 10.2147/CMAR.S225123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Li X, Cassady K, Zou Z, Zhang X. TET2 function in hematopoietic malignancies, immune regulation, and DNA repair. Front Oncol. 2019;9:210. doi: 10.3389/fonc.2019.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidzik VI, Paschka P, Späth D, et al. TET2 mutations in Acute Myeloid Leukemia (AML): Results from a comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol. 2012;30:1350–7. doi: 10.1200/JCO.2011.39.2886. [DOI] [PubMed] [Google Scholar]
- Hamed NAM, Nazer AA, Ayad MW, Sadiq TS. Ten Eleven Translocation Gene 2 (TET2) Polymorphism In Acute Myeloid Leukemia. Ann Adv Med. 2018;2:A49–53. [Google Scholar]
- Huang Y, Hu J, Lu T, et al. Acute myeloid leukemia patient with FLT3-ITD and NPM1 double mutation should undergo allogeneic hematopoietic stem cell transplantation in CR1 for better prognosis. Cancer Manag Res. 2019;11:4129–42. doi: 10.2147/CMAR.S194523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4:149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, et al. Ten-eleven-translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108:14566–71. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmider O, Delabesse E, de Mas VM, et al. TET2 mutations in secondary acute myeloid leukemias: A French retrospective study. Haematologica. 2011;96:1059–63. doi: 10.3324/haematol.2011.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunas-Rangel FA, Chávez-Valencia V, Gómez-Guijosa MÁ, Cortes-Penagos C. Acute Myeloid Leukemia-Genetic Alterations and Their Clinical Prognosis. Int J Hematol Oncol Stem Cell Res. 2017;11:328–39. [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Tan XH, Luo XP, et al. Prognostic significance of Tet methylcytosine dioxygenase 2 (TET2) gene mutations in adult patients with acute myeloid leukemia: A meta-analysis. Leuk Lymphoma. 2014;55:2691–8. doi: 10.3109/10428194.2014.893308. [DOI] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A cancer and leukemia group B study. J Clin Oncol. 2010;28:2348–55. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, et al. Recurring Mutations Found by Sequencing an Acute Myeloid Leukemia Genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros BC, Fathi AT, DiNardo CD, et al. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia. 2017;31:272–81. doi: 10.1038/leu.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng-Ju LI, Yang YL, Jou ST, et al. Prevalence & Prognosis Value of TET2 Gene Polymorphisms in Childhood Acute Myeloid Leukemia in Taiwan. Blood. 2011;118:1551. [Google Scholar]
- Metzeler KH, Maharry K, Radmacher MD, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: A cancer and leukemia group B study. J Clin Oncol. 2011;29:1373–81. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalban-Bravo G, DiNardo CD. The role of IDH mutations in acute myeloid leukemia. Future Oncol. 2018;14:979–93. doi: 10.2217/fon-2017-0523. [DOI] [PubMed] [Google Scholar]
- Nibourel O, Kosmider O, Cheok M, et al. Incidence and prognostic value of TET2 alterations in de novo acute myeloid leukemia achieving complete remission. Blood. 2010;116:1132–5. doi: 10.1182/blood-2009-07-234484. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore F, Pastore A, Rothenberg‐Thurley M, et al. Molecular profiling of patients with cytogenetically normal acute myeloid leukemia and hyperleukocytosis. Cancer. 2022;128:4213–22. doi: 10.1002/cncr.34495. [DOI] [PubMed] [Google Scholar]
- Patel KP, Ravandi F, Ma D, et al. Acute myeloid leukemia with IDH1 or IDH2 mutation: Frequency and clinicopathologic features. Am J Clin Pathol. 2011;135:35–45. doi: 10.1309/AJCPD7NR2RMNQDVF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada-Arismendy J, Arroyave JC, Röthlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017;31:63–76. doi: 10.1016/j.blre.2016.08.005. [DOI] [PubMed] [Google Scholar]
- Pratcorona M, Brunet S, Nomdedéu J, et al. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: relevance to post-remission therapy. Blood. 2013;121:2734–8. doi: 10.1182/blood-2012-06-431122. [DOI] [PubMed] [Google Scholar]
- Raveendran S, Sarojam S, Vijay S, et al. Mutation analysis of IDH1/2 genes in unselected de novo acute myeloid Leukaemia Patients in India - identification of a novel IDH2 mutation. Asian Pac J Cancer Prev. 2015;16:4095–4101. doi: 10.7314/apjcp.2015.16.9.4095. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M, Yamaguchi H, Najima Y, et al. Prognostic impact of low allelic ratio FLT3-ITD and NPM1 mutation in acute myeloid leukemia. Blood Adv. 2018;2:2744–54. doi: 10.1182/bloodadvances.2018020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider F, Hoster E, Unterhalt M, et al. The FLT3ITD mRNA level has high prognostic a impact in NPM1 mutated, butnot in NPM1 unmutated, AML witha normal karyotype. Blood. 2012;119:4383–6. doi: 10.1182/blood-2010-12-327072. [DOI] [PubMed] [Google Scholar]
- Schnittger S, Haferlach C, Ulke M, et al. IDH1 mutations are detected in 6 6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood. 2010;116:5486–96. doi: 10.1182/blood-2010-02-267955. [DOI] [PubMed] [Google Scholar]
- Shafik NF, Darwish AD, Allam RM, Elsayed GM. FLT3-ITD Allele Frequency Is an Independent Prognostic Factor for Poor Outcome in FLT3-ITD–Positive AML Patients. Clin Lymphoma Myeloma Leuk. 2021;21:676–85. doi: 10.1016/j.clml.2021.05.005. [DOI] [PubMed] [Google Scholar]
- Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W. The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia. 2014;28:485–96. doi: 10.1038/leu.2013.337. [DOI] [PubMed] [Google Scholar]
- Thol F, Damm F, Wagner K, et al. Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood. 2010;116:614–6. doi: 10.1182/blood-2010-03-272146. [DOI] [PubMed] [Google Scholar]
- Tsai CH, Hou HA, Tang JL, et al. Genetic Alterations and Their Clinical Implications in Older Patients with Acute Myeloid Leukemia. Leukemia. 2016;30:1485–92. doi: 10.1038/leu.2016.65. [DOI] [PubMed] [Google Scholar]
- Virijevic M, Karan-Djurasevic T, Marjanovic I, et al. Somatic mutations of isocitrate dehydrogenases 1 and 2 are prognostic and follow-up markers in patients with acute myeloid leukaemia with normal karyotype. Radiol Oncol. 2016;50:385–93. doi: 10.1515/raon-2016-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K, Damm F, Göhring G, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356–64. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- Wang R, Gao X, Yu L. The prognostic impact of tet oncogene family member 2 mutations in patients with acute myeloid leukemia: A systematic-review and meta-analysis. BMC Cancer. 2019;19:1–11. doi: 10.1186/s12885-019-5602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Lu C, Cross JR, et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem. 2013;288:3804–15. doi: 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann S, Alpermann T, Grossmann V, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26:934–42. doi: 10.1038/leu.2011.326. [DOI] [PubMed] [Google Scholar]
- Xu LH, Fang JP, Liu YC, Jones AI, Chai L. Nucleophosmin mutations confer an independent favorable prognostic impact in 869 pediatric patients with acute myeloid leukemia. Blood Cancer J. 2020:10. doi: 10.1038/s41408-019-0268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wang H, Han H, et al. Clinical characteristics and prognostic significance of DNA methylation regulatory gene mutations in acute myeloid leukemia. Clin Epigenetics. 2023;15:1–13. doi: 10.1186/s13148-023-01474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Iwanaga E, Tokunaga K, et al. IDH1 and IDH2 mutations confer an adverse effect in patients with acute myeloid leukemia lacking the NPM1 mutation. Eur J Haematol. 2014;92:471–7. doi: 10.1111/ejh.12271. [DOI] [PubMed] [Google Scholar]
- Zeng H, He H, Guo L, et al. Antibiotic treatment ameliorates Ten-eleven translocation 2 (TET2) loss-of-function associated hematological malignancies. Cancer Lett. 2019;467:1–8. doi: 10.1016/j.canlet.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]