Abstract
Essential trace metals play key roles in the survival, replication, and virulence of bacterial pathogens. Helicobacter pylori (H. pylori), the main bacterial cause of gastric ulcers, requires Ni(ii) to colonize and persist in the acidic environment inside the stomach, exploiting the nickel-containing enzyme urease to catalyze the hydrolysis of urea to ammonia and bicarbonate and create a pH-buffered microenvironment. Urease utilizes Ni(ii) as a catalytic cofactor for its activity. In ureolytic bacteria, unique transmembrane (TM) transporters evolved to guarantee the selective uptake and efflux of Ni(ii) across cellular membranes to meet the cellular requirements. NixA is an essential Ni(ii) transporter expressed by H. pylori when the extracellular environment experiences a drop in pH. This Class I nickel–cobalt transporter of the NiCoT family catalyzes the uptake of Ni(ii) across the inner membrane from the periplasm. In this study, we characterized NixA using a platform whereby, for the first time on a NiCoT transporter, recombinantly expressed and purified NixA and key mutants in the translocation pathway have been reconstituted in artificial lipid bilayer vesicles (proteoliposomes). Fluorescent sensors responsive to Ni(ii) transport (Fluozin-3-Zn(ii)), luminal pH changes (pyranine), and membrane potential (oxonol VI) were encapsulated in the proteoliposomes lumen to monitor, in real-time, NixA transport properties and translocation mechanism. Kinetic transport analysis revealed that NixA is highly selective for Ni(ii) with no substrate promiscuity towards Co(ii), the other putative metal substrate of the NiCoT family, nor Zn(ii). NixA-mediated Ni(ii) transport exhibited a Michaelis–Menten-type saturable substrate concentration dependence, with an experimental KM, Ni(ii) = 31.0 ± 1.2 μM. Ni(ii) transport by NixA was demonstrated to be electrogenic, and metal translocation did not require a proton motive force, resulting in the generation of a positive-inside transmembrane potential in the proteoliposome lumen. Mutation analysis characterized key transmembrane residues for substrate recognition, binding, and/or transport, suggesting the presence of a three-step transmembrane translocation conduit. Taken together, these investigations reveal that NixA is a Ni(ii)-selective Class I NiCoT electrogenic uniporter. The work also provides an in vitro approach to characterize the transport properties of metal transporters responsible for Ni(ii) acquisition and extrusion in prokaryotes.
Helicobacter pylori NixA is a Ni(ii)-selective Class I NiCoT transmembrane electrogenic importer.
Introduction
Nickel is an essential trace metal present predominantly in prokaryotic (and plant) enzymes.1–9 Ni(ii) participates in the maturation and as catalytic cofactor in metalloenzymes that catalyze challenging chemical reactions, such as in the cases of CODH (carbon monoxide dehydrogenase) and [NiFe] hydrogenase.1–9 CODH enzymatic activity produces low potential electrons and drive the reversible and selective interconversion of carbon monoxide to carbon dioxide. As a result, CODH activity is central for carbon fixation processes and the survival of organisms possessing CODH.1–9 [NiFe] hydrogenase catalyzes the reversible conversion of molecular hydrogen to protons and electrons that are needed in critical metabolic pathways.1–9 To guarantee that the necessary Ni(ii) cellular “quotas” are met, bacteria requiring Ni(ii) evolved a network of nickel binding proteins that allows selective metal ion uptake, extrusion, delivery, storage, and chaperoning to target metalloenzymes, so that cellular Ni(ii) concentrations are tightly controlled to meet the physiological needs without reaching toxic levels.3–12
Helicobacter pylori (H. pylori), a Gram-negative bacterium, has been identified as the causative agent of human gastric and duodenal ulcers, both of which can eventually lead to gastric cancer.4,13 The extremely low pH environment in the stomach, where H. pylori replicates, presents a challenge to its survival and replication. H. pylori utilizes another key cytosolic nickel-containing metalloenzyme, urease, to cope and persist in this harsh environment. Urease is matured via insertion of two Ni(ii) ions as cofactors in its catalytic site, to catalyze the hydrolysis of urea to ammonia and carbon dioxide.14 The produced ammonium and bicarbonate generate a pH-buffered microenvironment that allow H. pylori to survive in the acidic milieu of the gastric tissue.4,11,14 Thus, Ni(ii) uptake across its cellular membranes, is crucial for H. pylori survival and pathogenicity.2,4,11,15–17
In H. pylori, ABC (ATP Binding Cassette) transporters and nickel–cobalt permeases (NiCoTs) have been identified to be the prominent transmembrane Ni(ii) transporters for nickel acquisition across its inner membrane.10,17–25In vivo studies on the activity of nickel–cobalt permeases have proven their importance in Ni(ii) uptake and urease maturation,10,11,17–25 but the knowledge regarding their transport properties, such as translocation mechanism, substrate selectivity bias, and transport modality remains to be investigated, as biochemical and biophysical investigations on purified NiCoT transporters are lacking. The Ni(ii) uptake pathway to the cytoplasm involves FrpB4, an outer membrane protein supported by the TonB protein, and FecA3, which translocates Ni(ii) from the outer membrane to the periplasm.11,26–28 Subsequently, Ni(ii) is transported from the periplasm to the cytoplasm through NixA or the Ni(ii) ABC transporter NiuBDE.17,29,30 Once in the cytoplasm, a complex network of metallochaperones interact with Ni(ii) to store this metal ion and mediate nickel-enzymes maturation, as widely investigated for urease and hydrogenase.3–12
NixA is known to be a high-affinity nickel transporting protein, facilitating the import of Ni(ii) across the inner membrane in H. pylori.17,29,30 NixA is a member of the NiCoT importer family.10,17–25,29,30 These permeases feature several subclasses based on their substrate preference towards the metal substrates Ni(ii) and Co(ii). The family is postulated to consist of at least three transporter groups, where Class I appear to be exclusively selective for Ni(ii). Classes II and III appear to be instead promiscuous, and catalyze the translocation of both Ni(ii) and Co(ii), but with a preferential selectivity bias for Co(ii) or Ni(ii), respectively.21,31 NixA is classified as a Class I NiCoT, and expected to exclusively transport Ni(ii) based on in vivo evidence.20,31
Existing studies have determined NixA activity through indirect in vivo methods such as metal accumulation analysis (liquid scintillation) and urease activation assays upon cellular exposure to different metals.30,32 NixA has been observed to selectively transport Ni(ii) while discriminating against other divalent cations such as Zn(ii), Co(ii), and Cu(ii).6 Eight putative transmembrane helices (TM) are conserved across all proteins in the NiCoT family and it has been proposed that TMs II and III are responsible for the transport activity of NixA. Mutation of amino acids located in TMs II (H44, D47, D49, D50, D55) and III (H79), believed to be critical for NixA activity, to isoleucine, reduce Ni(ii) uptake and urease activity in vivo.29,30,32 Despite the crucial role of NixA in the survival of H. pylori, the molecular mechanism by which the protein facilitates Ni(ii) transport remain to be fully elucidated.
Biochemical and biophysical characterization of the transport properties on purified NiCoT systems, including NixA, are currently lacking due to the limitations of methodologies to express and purify NiCoTs in active forms, and shortage of molecular tools that allow direct measurement of substrate translocation, co-/counter-ion transport, and electrogenicity with transporter reconstituted in native-like membranes.
This work describes an effort to fill this gap, through the development of a platform to recombinantly express and purify NixA for reconstitution in artificial lipid bilayers (small unilamellar vesicles (SUVs), also called proteoliposomes). Through this platform, which mimics the biophysical properties of cellular membranes, we determined key NixA transport properties. Three liposome-compatible and impermeable fluorescent probes were encapsulated in the proteoliposomes' lumen to characterize NixA metal translocation mechanism and kinetics: (1) a Ni(ii)-sensitive fluorescent probe, Fluozin-3-Zn(ii) (FZ-3-Zn),33–36 (2) the pH sensitive probe pyranine,37 (3) and the membrane potential sensor oxonol VI.38 Based on transport assays, we determine that NixA is a highly-selective, high-affinity Ni(ii) uniporter, and that metal transport is electrogenic. By mutational analysis, coupled to metal binding determinations and transport assays, we also identified and dissected key residues in the putative NixA transmembrane translocation pathway that control Ni(ii) recognition, selection and transport across the lipid bilayer, thus providing novel molecular insight in the transport properties of NixA. While the use of proteoliposomes with encapsulated fluorescent probes responsive to diverse stimuli in the lumen has been utilized to the study of other metal transporters, in this work we developed a novel methodology that allows these investigations to study Ni(ii) transmembrane transporters, opening the venue for the characterization of the translocation properties in real-time of other Ni(ii)/Co(ii) transporter families upon their reconstitution in native-like artificial lipid bilayers.
Materials and methods
Expression and purification of wild-type NixA and inactive mutants
Synthetic DNA encoding codon-optimized wild-type NixA (wtNixA Uniprot: Q48262) from H. pylori was synthetized and cloned in a pET-52b(+) vector between the SmaI and SacI restriction sites (Genscript Inc.), generating a construct encoding a thrombin cleavage site followed by a His-10-tag at the protein C-terminus. Single-point and combination mutants were commercially generated by Genscript Inc. following a proprietary mutagenesis strategy. Mutant's constructs were verified by sequencing using primers listed in ESI Table S1.† Plasmids were transformed in E. coli BL21 (DE3) GOLD (Agilent Technologies) and clones selected on Luria Broth agar (LB-agar) plates after O/N growth at 37 °C. Precultures were prepared in Terrific Broth (TB) media by O/N growth at 37 °C. The main cultures, in TB-media supplemented with 1% glycerol, were inoculated with precultures at 1% (v/v). Cells were grown in the presence of 50 μg mL−1 ampicillin, under constant shaking (140 rpm), at 37 °C. The OD600 was recorded regularly, and the temperature was decreased to 23 °C when OD600 reached a value of 2, both for the expression of both wild-type NixA (wtNixA) and related mutants. After 1 h, 0.1 mM isopropyl thiogalactopyranoside (IPTG) was added to the cultures and the target proteins were expressed at 23 °C for 16 h, at 140 rpm.
Cells were harvested by centrifugation, at 4000×g for 25 min at 4 °C (Thermo Scientific Sorvall LYNX 6000 centrifuge). The cells were resuspended in lysis buffer (20 mM Tris/HCl, pH 8, 100 mM NaCl, 5 mM MgCl2, 30 μg mL−1 deoxyribonuclease I from bovine pancreas (Sigma-Aldrich) and 2× EDTA-free protease inhibitor cocktail tablets (Thermo Scientific)). The cells were lysed at a low temperature (∼4 °C) using a microfluidizer (Microfluidics M-110P) at 20 000 psi by three passages through a Z-shaped diamond chamber cooled on ice. The lysate was centrifuged at 17 000×g for 25 min at 4 °C (Thermo Scientific Sorvall LYNX 6000 centrifuge) to remove cell debris. The supernatant was collected and subjected to ultracentrifugation (37 000 rpm, 1 h, 4 °C) to isolate the cell membrane fraction that contained wtNixA or its mutants. The membrane pellet was resuspended in storing buffer (20 mM Tris/HCl, pH 8, 500 mM NaCl, 1% glycerol (w/v), 1× EDTA-free protease inhibitor cocktail tablets (Thermo Scientific)) to a final membrane concentration of 1.0 g of cells per 1.0 mL of buffer. The membrane suspension was subsequently frozen in liquid N2 and stored at −80 °C until protein purification.
For purification, 10 mL of the membrane suspensions were mixed with 40 mL of membrane resuspension buffer (20 mM Tris/HCl pH 8.0, 500 mM NaCl, 1× EDTA-free protease inhibitor cocktail tablets (Thermo Scientific), 25 mM imidazole, 5 mM β-mercaptoethanol). To solubilize the membrane proteins, n-tetradecylphosphocholine (Fos-choline-14) was added to the membrane suspension to a final concentration of 1% (w/v), and the mixture stirred for 1 h, at 4 °C, to guarantee wtNixA/mutants extraction. The suspension was subsequently subjected to ultracentrifugation (37 000 rpm, 1 h, 4 °C; Beckman Optima XPN80) to separate the solubilized membrane protein fraction from the residual membrane debris. Afterwards, the supernatant was loaded to a Ni-NTA HisTrap FF column pre-equilibrated with binding buffer (20 mM Tris/HCl pH 8.0, 500 mM NaCl, 25 mM imidazole, 1 mM tris(2-carboxyethyl) phosphine (TCEP), 0.05% (w/v) Fos-choline-14), using an AKTA Pure FPLC system (Cytiva). The column was washed with 30 column volumes (CV) of wash buffer (20 mM Tris/HCl pH 8.0, 500 mM NaCl, 25 mM imidazole, 1 mM TCEP, 0.05% (w/v) Fos-choline-14), to remove unbound proteins and impurities. The bound proteins were eluted with a 5 CV linear imidazole gradient (0–100%) generated by mixing wash buffer and elution buffer (20 mM Tris/HCl pH 8.0, 500 mM NaCl, 500 mM imidazole, 1 mM tris(2-carboxyethyl) phosphine (TCEP), 0.05% (w/v) 7-cyclohexyl-1-heptyl-β-d-maltoside (CYMAL-7)). The fractions containing NixA were collected and concentrated to ∼5–10 mg mL−1 using 30 000 Da MWCO cutoff filter (Sartorius VIVASPIN 20) via centrifugation (4000×g, 4 °C). The protein was further purified through a size exclusion chromatographic (SEC) step, by loading the protein solution onto a Superdex 200 Increase 10/300 column equilibrated with 20 mM MOPS/NaOH buffer, pH 7.0, 500 mM NaCl, 1 mM TCEP and 0.05% (w/v) CYMAL-7. The SEC fractions containing wtNixA/mutants were eluted using the same buffer, collected and concentrated to ∼1–5 mg mL−1.
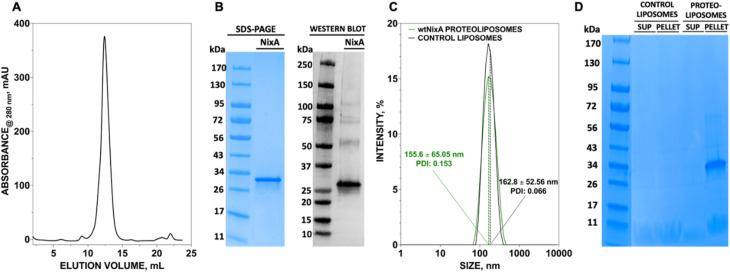
The purity of the obtained protein was verified by SDS-PAGE (4–15% Tris-Glycine Mini-PROTEAN gels, BioRad) and western blot analysis utilizing an antibody recognizing the His10-tag present at the C-terminus (6x-His Tag Monoclonal Antibody, HIS.H8, Invitrogen). The protein concentration was determined from the absorbance at 280 nm measured with a Nanodrop spectrophotometer (Thermo Scientific Nanodrop One), given the extinction coefficient of ε280 = 43 890 M−1 cm−1 for wild-type NixA. The protein was frozen in liquid N2 and stored at −80 °C until further use. All mutants were expressed and purified following the same protocol of wtNixA.
Small-scale detergent screening
A small-scale detergent optimization was performed to identify the optimal detergent to be used for membrane protein extraction. 200 μL of isolated membranes were mixed with 800 μL of membrane resuspension buffer (20 mM Tris/HCl pH 8.0, 500 mM NaCl, 1× EDTA-free protease inhibitor cocktail tablets (Thermo Scientific), 25 mM imidazole, 5 mM β-mercaptoethanol). Detergent stock solutions (DMNG: decyl maltose neopentyl glycol; FC-14: Fos-choline-14; CYMAL-7: 7-cyclohexyl-1-heptyl-β-d-maltoside; LDAO: lauryldimethylamine-N-oxide; OG: n-octyl-β-d-glucopyranoside; DDM: n-dodecyl-β-d-maltopyranoside) were added to individual membrane mixtures at a final 1% (w/v). The mixtures were homogenized using a Benchmark D1000 homogenizer. The suspension was incubated for 1 h, at 4 °C, while tilting. The membrane solutions were then subjected to ultracentrifugation at 150 000×g for 30 min at 4 °C (Sorvall mX120+ Micro-Ultracentrifuge) to separate the solubilized supernatant and the insoluble membrane pellet. The supernatant and pellet were then analyzed via western blot and SDS-PAGE.
His10-tag removal for Ni2+-binding stoichiometry determination
The histidine-tag was removed from the purified wtNixA by addition of bovine Thrombin (20 units per mg NixA; EMD Millipore Crop) to a final 1 : 5 protein : thrombin (v/v) ratio. The cleavage was carried out at 21 °C for 10 h, under constant shaking. Thrombin and other impurities were removed by SEC, using a pre-equilibrated Superdex 200 Increase 10/300 column, and eluted with 20 mM MOPS/NaOH buffer, pH 7.0, 500 mM NaCl, 1 mM TCEP, 0.05% (w/v) CYMAL-7.
NixA and mutants reconstitution in liposomes
Liposomes were prepared by mixing l-α-phosphatidylcholine from chicken egg (Avanti Polar Lipids) and E. coli polar lipids extract (Avanti Polar Lipids) at a 1 : 3 ratio (w/w). The lipid mixture, dissolved in chloroform, was dried using a stream of nitrogen gas under rotation and residual solvent was removed overnight in a vacuum desiccator.
The dried lipid cake was hydrated and resuspended using a Chelex®-treated (BioRad) solution of 1 mM TCEP in MilliQ H2O. The lipid suspension was buffered to final 20 mM MOPS/NaOH, pH 7, 100 mM NaCl, 1 mM TCEP, using a Chelex®-treated 10× buffer stock (200 mM MOPS/NaOH, pH 7, 1 M NaCl, 1 mM TCEP). The lipids suspensions were prepared to a final total lipid concentration of 25 mg mL−1.
Liposomes were prepared by subjecting the lipid suspension to three freeze–thaw cycles in liquid nitrogen, followed by sequential 11 extrusions through polycarbonate (PC) membranes of decreasing pore sizes (1 μm, 0.4 μm then 0.2 μm filters), using a 1 mL airtight syringe system (Avanti, Polar Lipids, Inc.) to produce unilamellar vesicles. The liposomes suspensions were collected and subsequently destabilized by addition of CYMAL-7 detergent at a final concentration of 0.02% (w/v) and incubation for 1 h, at room temperature, under tilting, prior to protein incorporation. Purified wtNixA and mutants (∼1–5 mg mL−1) were reconstituted into the liposomes by adding purified proteins stocks to a final 1 : 25 (w/w) protein : lipid ratio. Similarly, control liposomes were prepared by buffer addition to the detergent destabilized liposomes.
The liposome and protein mixtures, as well as the control liposomes, were tilted for 1 h at 4 °C. The CYMAL-7 detergent was removed using Bio-Beads (SM-2; BioRad). The Bio-Beads were activated via consecutive washings with methanol, ethanol, and water, followed by drying through vacuum filtration. The activated Bio-Beads were added to the proteoliposomes and the control liposome to a final concentration of 60 mg mL; then, the mixtures were tilted for 1 h at 4 °C for the first Bio-Bead exchange prior to subsequent BioBead resin exchanges carried out for 12, 14, and 16 h.
The prepared liposomes were collected by ultracentrifugation at 180 000×g for 30 min at 4 °C (Sorvall mX120+ Micro-Ultracentrifuge). The pellets were collected and resuspended in 20 mM MOPS/NaOH, pH 7, 100 mM NaCl, 1 mM TCEP (Chelex®-treated), to a final total lipid concentration of 25 mg mL−1 (wtNixA/mutants = 1 mg mL−1). In order to assess the level of protein incorporation in the proteoliposomes, SDS-PAGE (4–15% Tris-Glycine Mini-PROTEAN gels, BioRad) densitometric analysis was performed on the supernatant and the resuspended proteoliposomes.
The proteoliposomes and the control liposomes were frozen in liquid N2 and stored at −80 °C.
The number of NixA molecules incorporated in each proteoliposome particle was estimated by determining the SUV diameter by dynamic light scattering and knowing the lipid concentration, under the assumption that the SUVs are spherical particles.35 The total lipid head group area (STOT) can be calculated as the sum of the proteoliposome outer layer surface area (Sout) and the inner layer surface area (Sin). The bilayer thickness is approximately 3 nm (h = 3 nm), and STOT can be calculated as:STOT = Sout + Sin = 4πr2 + 4π(r − h)2
The total number of lipid molecules per proteoliposome (NTOT) can be estimated from STOT and the average polar head surface (SL 0.7 nm2 per phospholipid):NTOT = STOT/SL
Assuming that the average molecular weight of phospholipids is 750 g mol−1, the approximate molecular weight of single proteoliposome was obtained. The number of NixA molecules per SUV was subsequently calculated knowing the concentrations of lipids and protein in the proteoliposome preparations.
Determination of SUV's size distribution by dynamic light scattering (DLS)
The size distribution of the NixA proteoliposomes and control liposomes were obtained by UV-Vis Dynamic Light Scattering (DLS) with a Zeta Sizer Nano ZS DLS instrument (Malvern Panalytical). The samples were placed in a polystyrene cuvette and the measurements conducted at 25 °C, at a scattering angle of 175°, with a laser wavelength of 633 nm (refractive index of sample medium = 1.33 and refractive index of material = 1.45).35 The polydispersity index (PDI) for control liposome and NixA proteoliposomes were 0.066 and 0.153 respectively.
Fluozin-3-Zn(ii) probe encapsulation in SUVs and in vitro Ni(ii) transport assays
Control liposomes and proteoliposomes carrying wtNixA and its mutants were diluted to a final concentration of 12.5 mg mL−1 in the transport buffer (20 mM MOPS/NaOH, pH 7, 100 mM NaCl, 1 mM TCEP). Equimolar amounts of Fluozin-3 (final concentration = 10 μM) and Zn(ii) (10 μM) were added to the suspensions, and the Fluozin-3-Zn(ii) complex was encapsulated in the lumen by membrane freeze-fracture upon three freeze–thaw cycles in liquid nitrogen. Liposomes were subsequently sequentially extruded through 1 μm and 0.2 μm PC membrane filters using an airtight syringe system. The unencapsulated Fluozin-3-Zn(ii) complex was removed by ultracentrifugation at 160 000×g for 45 min at 4 °C in a Sorvall mX120+ Micro-Ultracentrifuge. The pellets were collected and washed with the transport buffer (1 mL), collected via a subsequent ultracentrifugation step, and subsequently recollected and resuspended in the transport assay buffer (20 mM MOPS/NaOH, pH 7, 100 mM NaCl, 1 mM TCEP). The control liposomes were subjected to the same protocol.
Buffer solutions (20 mM MOPS/NaOH, pH 7, 100 mM NaCl, 1 mM TCEP) and Chelex®-treated MilliQ water were made oxygen-free on a Schlenk-line by at least four vacuum/nitrogen gas cycles. Stock solutions of Ni(ii) were freshly prepared in an aerobic glove box with constant nitrogen gas flow. The Ni(ii) stock solutions were prepared from a 0.1 M NiCl2 stock solution. 100× Ni(ii) stocks (100 μM–10 mM) for transport assay were prepared via dilution in oxygen-free transport buffer. The stock solutions were stored in a sealed tube prior to use.
The Ni(ii) transport assays were conducted at 25 °C using a spectrofluorometer (Horiba Scientific Fluoromax-4) at excitation (λexc) and emission (λexc) wavelengths of 480 nm (slit widths: 2.5 nm) and 515 nm (slit widths: 2.5 nm), respectively. NixA proteoliposomes (120 μL), with the encapsulated Fluozin-3-Zn(ii) probe complex, were placed in a sub-micro quartz cuvette and the baseline fluorescence was collected for 50 s prior to the addition of Ni2+.
Ni(ii) transport was triggered by the addition of 1.2 μL Ni(ii) stock solutions and the fluorescence signal change monitored for 500 s at 0.1 s intervals. Ni(ii) transport was determined by plotting the ΔF/F0versus time, where ΔF is the difference between the fluorescence measured at time t (Ft) and the fluorescence immediately before Ni(ii) addition (F0). The same protocol was utilized for the characterization of control liposomes and NixA mutants.
Ni(ii)/H+ co-/counter-ion transport monitored by pH sensitive probe pyranine
Control liposomes and proteoliposomes carrying wtNixA and its mutants were diluted (1 : 2 (v/v)) in the transport buffer (4 mM MOPS/NaOH, pH 7, 100 mM NaCl, 1 mM TCEP). Subsequently, a 20 mM pyranine stock solution in H2O (Alfa Aesar) was added to a final concentration of 1 mM. The pH sensitive probe was encapsulated through three freeze–thaw cycles, followed by consecutive extrusion through 1 μm, 0.4 μm, and 0.2 μm PC membrane filters using gas-tight syringes. The proteoliposome and control liposome pellets were collected by ultracentrifugation (160 000×g, 45 min, 4 °C, Sorvall mX120+ Micro-Ultracentrifuge), and the supernatant containing excess pyranine removed. The proteoliposome pellets and control liposomes were washed using the transport buffer (1 mL) to further remove any unencapsulated pyranine excess. Pellets were collected by ultracentrifugation (180 000×g, 30 min, 4 °C; Sorvall mX120+ Micro-Ultracentrifuge) and resuspended in the same transport buffer for H+ counter-ion transport assays.
The H+ co-/counter-ion transport assays were conducted as described for the Ni(ii) transport assays. The reaction was triggered by addition of 1.2 μL of Ni(ii) stocks (100×; 2.5 mM) to NixA proteoliposomes and control liposomes. The fluorescence signal change was monitored for 500 s at 0.1 s intervals (λexc: 480 nm, slit width: 1 nm; λem: 515 nm, slit width: 1 nm). Kinetics were reported by plotting ΔF/F0 as a function of time.
Ni(ii) transport electrogenicity monitored with the membrane potential probe oxonol VI
Control liposomes and proteoliposomes carrying wtNixA and its mutants were diluted to a final lipid concentration of 12 mg mL−1 in the transport buffer (20 mM MOPS/NaOH pH 7, 100 mM NaCl, 1 mM TCEP). The membrane potential fluorescent probe oxonol VI, which can detect transmembrane potential changes, was added to the suspension to a final concentration of 5 μM. Oxonol VI was encapsulated into the proteoliposomes and control liposomes by three freeze–thaw cycles in liquid N2, followed by sequential extrusion through 1 μm, 0.4 μm, and 0.2 μm PC membrane filters using gas-tight syringes. The suspension was centrifuged (160 000×g, 45 min, 4 °C, Sorvall mX120+ Micro-Ultracentrifuge) to collect proteoliposome/liposome pellets and remove the unencapsulated oxonol VI in the supernatant. The pellets were further washed with the transport buffer (1 mL) to remove the excess probe. The pellets were collected by ultracentrifugation (160 000×g, 45 min, 4 °C, Sorvall mX120+ Micro-Ultracentrifuge)) and subsequently resuspended in the same buffer.
The membrane potential transport assays were conducted as described for the Ni(ii) transport assays. Proteoliposomes or control liposomes (120 μL) were placed in a sub-micro quartz cuvette and equilibrated at 25 °C. Subsequently, 1.2 μL Ni(ii) stock solutions (100×; 2.5 mM) were added to trigger substrate transport. The fluorescence signal was monitored for 500 s at 0.1 s intervals (λexc: 580 nm, slit width: 4 nm; λEm: 660 nm; slit width: 5 nm). Kinetics of transmembrane potential changes were reported by plotting ΔF/F0 as a function of time.
wtNixA and mutants Ni(ii)-binding stoichiometry determination
Prior to the binding experiments, purified wtNixA and its mutants were incubated with EDTA to a final concentration of 1 mM. Subsequently, EDTA was removed by injecting the protein samples on a SEC column (Superdex 200 10/300 Increase) and eluted using a Chelex®-treated SEC buffer (20 mM MOPS/NaOH, pH 7.0, 500 mM NaCl, 1 mM TCEP, 0.05% (w/v) CYMAL-7). Subsequently, wtNixA and its mutants were concentrated to at least 10 μM by centrifugation using 30 000 Da MWCO cutoff filter concentrators (Sartorius VIVASPIN 20; 4000×g, 4 °C). The concentration of the protein was determined by absorbance at 280 nm on a Nanodrop spectrometer (Thermo Scientific Nanodrop One). Three Ni(ii) equivalents (NiCl2) dissolved in Chelex®-treated SEC buffer (20 mM MOPS/NaOH, pH 7.0, 500 mM NaCl, 1 mM tris(2-carboxyethyl) phosphine (TCEP), 0.05% (w/v) 7-cyclohexyl-1-heptyl-β-d-maltoside (CYMAL-7)), were added to the metal-free protein sample (500 μL; 10–20 μM). The mixture was incubated for 10 min at room temperature under shaking. Protein samples were injected in a SEC column (Superdex 200 10/300 Increase) to remove the unbound Ni(ii). The protein eluate (1.4 mL) was collected and analyzed by SDS-PAGE (4–15% Tris-Glycine Mini-PROTEAN gels, BioRad) to confirm the presence of protein. To determine the binding stoichiometry (metal : protein ratio), the metal concentration was determined by Inductively Coupled Plasma Mass Spectrometry (ICP-MS) and the protein concentration by a Bradford assay.
The protein eluate aliquots (100 μL) were digested in concentrated HNO3 overnight. The mixture was diluted to 2 mL using Chelex®-treated water. The Ni(ii) concentration was determined using ICP-MS on a 7800 Inductively Coupled Plasma Mass Spectrometer (Agilent), equipped with an autosampler. Prior to the Ni(ii) analysis, Ni(ii) standards (Nickel Standard for ICP-MS by Sigma Aldrich) were prepared in 2% HNO3 to generate a calibration curve.
To determine the protein concentration, protein samples (4 μL) were mixed with of 1.0 M NaOH (4 μL) and Bradford reagent (192 μL). The absorbance at 595 nm was determined using a plate reader (TECAN Spark Multimode Microplate Reader), and protein concentrations calculated using a standard calibration curve generated using a purified NixA stock with a known concentration.
Isothermal titration calorimetry
The thermodynamics of Ni(II) binding to wtNixA was investigated using isothermal titration calorimetry (ITC). Titrations were performed using a high-sensitivity VP-ITC microcalorimeter (MicroCal LLC, MA, USA.), at 25 °C. The reference cell of the microcalorimeter was filled with MilliQ water. A 20 μM wtNixA solution in Chelex®-treated buffer containing 20 mM MOPS buffer, 500 mM NaCl, 1 mM TCEP, 0.05% CYMAL-7, at pH 7.0, loaded into the sample cell (1.4093 mL), was titrated by performing forty 5 μL injections of a 0.4 mM NiCl2 solution, dissolved in the same buffer, using a computer-controlled microsyringe. A control experiment was carried out, under identical conditions, by titrating the NiCl2 solution into buffer alone, to exclude any significant heat of dilution. The integrated heat data obtained for the titration were fitted using the AFFINImeter software,39 carrying out a nonlinear least-squares minimization of two alternative models, namely (i) a stoichiometric equilibria (SE) model, or (ii) an independent sites (IS) model. The SE model is based on stoichiometric (or macroscopic) equilibrium constants and is typically applied when the different binding sites are coupled to each other; the IS model is based on site-specific (or microscopic) equilibrium constants relative to different binding sites that do not influence each other.39 The molar enthalpy change (ΔH, cal mol−1) and binding constant (Ka, M−1) were the floating thermodynamic parameters in the fit.39 The parameters rM (scaling parameter for the protein concentration) and Qdil (heat of dilution, cal mol−1) were also adjusted as fitting parameters. The molar entropy change was calculated using the relationships ΔG = −RT ln Ka (R = 1.9872 cal mol−1 K−1, T = 298 K) and ΔG = ΔH − TΔS. The statistical goodness of fit (GoF) parameter was used to assess the best fit.
Results and discussion
Recombinant NixA expression in E. coli, purification, and reconstitution in SUVs
We developed a methodology to express NixA in E. coli recombinant cells using a codon-optimized construct encoding for the amino acid sequence from H. pylori (Uniprot: Q48262). Protein expression was maximized in BL21(DE3)-Gold E. coli cells. The membranes from the lysed cells expressing NixA were isolated by ultracentrifugation, followed by solubilization of membrane proteins using a biocompatible detergent. Prior to develop a purification strategy, a small-scale detergent screen, utilizing six detergents classes with different polar head and/or hydrophobic tails, was conducted to identify an ideal molecule to guarantee the highest NixA membrane extraction efficiency (ESI Fig. S1†). The zwitterionic Fos-choline-14 was selected as the most efficient detergent for extraction, as demonstrated by SDS-PAGE analysis of the solubilized fraction vs. the insolubilized NixA in the membrane pellet isolated by ultracentrifugation. The NixA construct was designed to encode a C-terminal poly-histidine tag, and the first purification step was performed by Immobilized Metal Affinity Chromatography (IMAC). IMAC purification yielded 5–10 mg NixA per liter of culture, with >90% purity as determined by SDS-PAGE analysis (ESI Fig. S2†). The protein eluate was subsequently further purified by Size Exclusion Chromatography (SEC), allowing to switch the detergent to cyclohexyl-1-heptyl-β-d-maltoside (CYMAL-7), a milder polar non-charged maltoside detergent. This exchange proved to be critical in minimizing protein aggregation. SEC analysis revealed monodispersed NixA-CYMAL-7 micelle complexes with no significant aggregation (Fig. 1A). This protocol allowed micellar NixA purification in monomeric form to >95% purity, as demonstrated by SDS-PAGE and western blot analysis (Fig. 1B).
Fig. 1. Purification of NixA and reconstitution in proteoliposomes. (A) Size exclusion chromatogram profile of NixA purified in CYMAL-7 micelles, and (B) SDS-PAGE of purified NixA after SEC, and corresponding and western blot analysis utilizing an Anti-His6 tag monoclonal antibody. (C) Dynamic light scattering (DLS) analysis of control liposomes and NixA proteoliposomes showing the monodisperse size of SUVs and corresponding polydispersity index (PDI). (D) SDS-PAGE analysis of NixA reconstitution in the proteoliposome fraction isolated by ultracentrifugation (pellet) compared to non-incorporated soluble NixA (supernatant, SUP), and corresponding control liposomes.
To generate NixA proteoliposomes and control liposomes, artificial lipid bilayer SUV vesicles were generated by extrusion, and the obtained liposomes destabilized by CYMAL-7 addition, followed by the incorporation of NixA at a 1 : 25 protein/lipid (mg mL−1) ratio. Leakage-free proteoliposomes were obtained upon detergent removal with BioBeads SM2 resin. Characterization of control liposomes and NixA proteoliposomes by Dynamic Light Scattering (DLS) revealed monodisperse size distributions, with average diameters of 163 ± 53 nm and 156 ± 65 nm, respectively (Fig. 1C). The amount of reconstituted NixA in the proteoliposomes was determined by SDS-PAGE densitometric analysis, upon ultracentrifugation to remove non-reconstituted proteins. NixA quantification in both the soluble and proteoliposome pellet fractions (Fig. 1D) revealed successful reconstitution, with >95% NixA incorporation in the lipid bilayer. Further, based on the average size of the NixA proteoliposomes and the ratio of protein-to-lipid reconstitution, the number of NixA molecules incorporated in each SUV was estimated as approximately 150 molecule per vesicle.
Ni(ii) transport monitored by FZ-3-Zn(ii) probe complex
The biochemical and biophysical transport properties of NixA, such as its translocation mechanism, substrate selectivity, and kinetics, have not yet been fully established due to the absence of an appropriate platform to study substrate translocation events in purified Ni(ii)-transporters. Previous studies attempting to address its transport mechanism were based on in vivo studies of membrane extracts with radiolabeled metals, and in vivo urease assays to indirectly quantify the NixA activity.29,30,32 Analysis on crude membrane extracts and indirect urease activity assays remain of challenging interpretation as a result of the activity of other promiscuous metal protein transporters for the same metal substrate, resulting in possible interferences. Moreover, real-time kinetics determinations of Ni(ii) transport are lacking in literature due to the scarcity of highly-sensitive, lipid-compatible, and impermeant fluorescent probes that can be encapsulated in small unilamellar vesicles to detect Ni(ii) translocation.
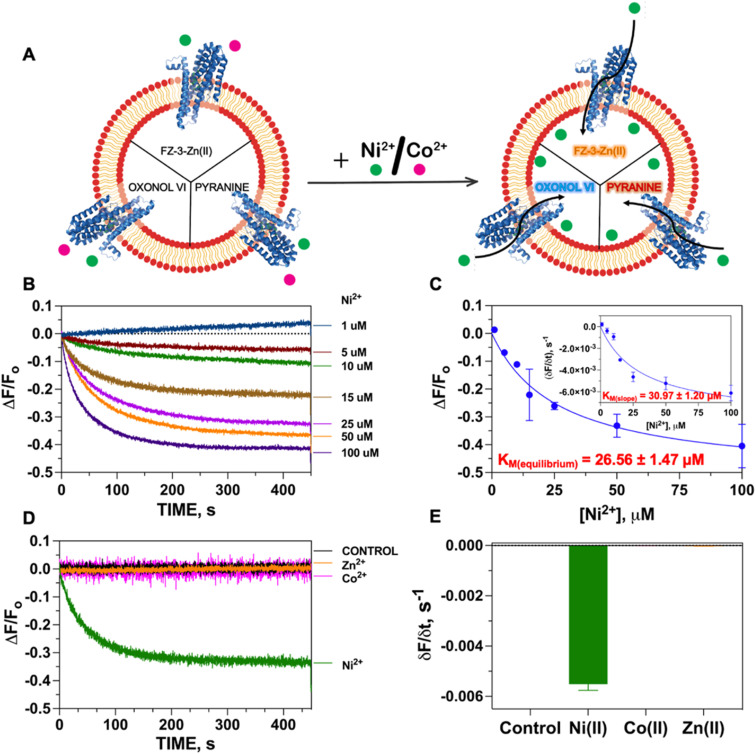
We thus established a fluorescent-probe screening procedure to identify a suitable fluorescent probe that could detect the desired metal substrate when encapsulated inside the liposomal lumen. The turn-on fluorescent probe Fluozin-3 was initially developed to detect Zn(ii) in cellular systems.33 Application of FZ-3 in monitoring other transition metals revealed a turn-on fluorescent response with other substrates such as Fe(ii), Co(ii), Ni(ii) and Cd(ii) in the presence and absence of lipids.35,36 However, in the putative application of FZ-3 to detect Ni(ii) transport activity by NixA in proteoliposomes potential problems were encountered related to: (i) residual Zn(ii) contamination in lipids isolated from native sources, and (ii) the FZ-3 significantly higher sensitivity for Zn(ii) compared to Ni(ii), which would affect batch-to-batch reproducibility. To circumvent these potential problems, we decided to exploit the comparable affinities of FZ-3 for Zn(ii) and Ni(ii) (or Co(ii)), and the significantly lower fluorescent turn-on response (fluorescence yield) for FZ-3-Ni(ii)/Co(ii) complexes (compared to FZ-3-Zn(ii)).34 We thus mixed Zn(ii) with FZ-3 (1 : 1 stoichiometric ratio) to generate FZ-3-Zn(ii), and encapsulated it in the proteoliposome lumen. We hypothesized that encapsulation of FZ-3-Zn(ii) would result in a significant fluorescence quenching response in case of Ni(ii) translocation, resulting from the metal-swapping of Zn(ii) with Ni(ii) in the FZ-3-metal complex, together with potential collisional quenching contributions. Control experiments conducted by titrating FZ-3-Zn(ii) with Ni(ii) and Co(ii) in lipid environments confirmed a concentration-dependent significant fluorescence quenching upon exposure of FZ-3-Zn(ii) to Ni(ii) and Co(ii) (ESI Fig. S3†). These data substantiate the use of FZ3-Zn(ii) as a probe to detect Ni(ii) or Co(ii) translocation events by NixA, when encapsulated in proteoliposomes (Fig. 2A).
Fig. 2. Transport activity of NixA monitored by FZ-3-Zn(ii) complex. (A) Overall scheme for the characterization of NixA transport properties utilizing fluorescent probes responsive to diverse stimuli. (B) Fluorescence quenching transport traces indicative of Ni(ii) transport by NixA in proteoliposomes, as monitored by FZ-3-Zn(ii) encapsulated in the SUV lumen, as a function of increasing Ni(ii) concentrations (1–100 μM; λexc = 480 nm; λem = 515 nm) (n = 3). Traces are plotted as differential fluorescence at time t (Ft − F0; ΔF) normalized to the fluorescence prior to the addition of Ni(ii) (F0), with traces corrected by subtracting the signals obtained with control liposomes. (C) Michaelis–Menten-type fitting of the maximum F change (ΔF/F0) as a function of Ni(ii) concentrations (KM, Ni(ii) = 26.6 ± 1.5 μM and (ΔF/F0)MAX = −0.52 ± −0.01 s−1) inset: maximal initial Ni(ii) transport rates in NixA proteoliposomes and corresponding fit with a Michaelis–Menten-like equation: (δF/δt) = (δF/δt)MAX × [Ni(ii)]/(KM + [Ni(ii)]); KM, Ni(ii) = 31.0 ± 1.2 μM and (δF/δt)MAX = −0.0089 ± −0.0004 s−1. (D) Fluorescence quenching transport traces upon addition of Ni(ii) and Co(ii) (25 μM) to NixA proteoliposomes, monitored by FZ-3-Zn(ii) probe complex, and Zn(ii) transport traces monitored by metal-free FZ-3. Signals were corrected by subtracting the background signal of control liposomes (n = 3). (E) Maximal initial transport rates (initial slope) obtained for control (−1.22 ± 0.05 × 10−5 s−1), Ni(ii) (−5.51 ± 0.20 × 10−3 s−1), and Co(ii) (−4.35 ± 0.24 × 10−6 s−1) (25 μM) in NixA proteoliposomes monitored by FZ-3-Zn(ii) probe, and Zn(ii) (−2.73 ± 0.20 × 10−5) (25 μM) in NixA proteoliposomes using FZ-3. Signals were corrected by subtracting the background signal of control liposomes (n = 3).
The Ni(ii) translocation in NixA proteoliposomes was monitored by recording the fluorescence immediately after the addition of the metal substrate to the proteoliposome solutions at the emission maximum of the FZ-3-Ni(ii) complex (λexc = 480 nm; λem = 515 nm). The kinetic traces obtained from NixA proteoliposomes revealed a time-dependent development of the fluorescence quenching signal, with a signal equilibration reached approximately after 5–8 min (Fig. 2B). The quenching fluorescence signals were also corrected by subtracting the almost negligible background signals of the corresponding control liposomes. These kinetic traces are indicative of a real-time detection of NixA-mediated Ni(ii) transport events across the proteoliposome lipid bilayer. The ΔF/F0 traces as a function of time showed increasing and faster quenching response upon exposure of the proteoliposomes to increasing extravesicular Ni(ii) concentrations, ranging from 1 to 100 μM (Fig. 2B). Exponential fitting of the Ni(ii) transport traces was applied to establish apparent initial transport rates (δF/δt) (Fig. 2C inset). The apparent initial velocities obtained revealed a saturable hyperbolic dependency resulting from NixA-dependent Ni(ii) transport. The slopes from the kinetic traces at different Ni(ii) concentrations were fitted with a Michaelis–Menten-like equation, resulting in a KM, Ni(ii) = 31.0 ± 1.2 μM and a (δF/δt)MAX = −0.0089 ± −0.0004 s−1 (Fig. 2C). The equilibrium fluorescence quenching response values (ΔF/F0) were also analyzed and fitted with a Michaelis–Menten-like equation, which revealed a similar KM, Ni(ii) = 26.6 ± 1.5 μM and a (ΔF/F0)MAX = −0.52 ± −0.01 s−1 (Fig. 2C).
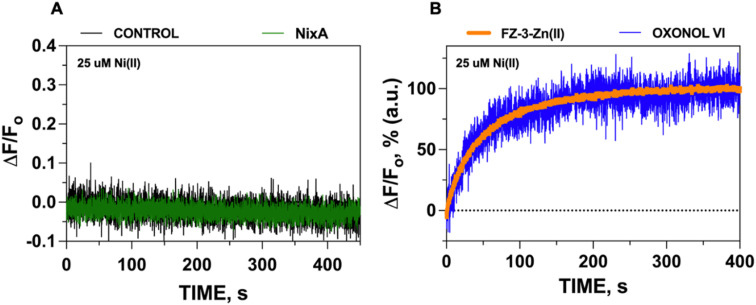
Based on previous in vivo metal accumulation assays, NixA was tentatively classified as a Class I NiCoT protein, suggesting that NixA might selectively transport only Ni(ii) but not Co(ii), despite belonging to the NiCoT family.20,21,31 We thus investigated the metal selectivity of NixA in our proteoliposome system by monitoring the transport of Ni(ii) and Co(ii) in control liposomes and NixA proteoliposomes. In parallel, we also investigated the transport properties of NixA towards Zn(ii), a more abundant non-substrate essential transition metal ion, via encapsulation of metal-free FZ-3, which possesses high zinc affinity (Kd ∼15 nM) and dramatic turn-on fluorescent response (>50-fold). Transport assays were conducted by supplementing the metal substrates, and by recording the proteoliposome fluorescence traces corrected for response of control liposomes, as described for the Ni(ii) transport assays. The fluorescence traces (ΔF/F0) as a function of time, resulting from the NixA-mediated transport, showed a significant exponential quenching response only upon addition of Ni(ii), and the absence of signals upon the addition of Co(ii) or Zn(ii) indicated the lack of translocation activity towards these two substrates. Therefore, this investigation unambiguously confirm that NixA is a high-affinity, Ni(ii)-selective Class I NiCoT transporter with low substrate promiscuity (Fig. 2D and E).
Putative Ni(ii)/H+ co-/counter-transport monitored with the pH sensitive probe pyranine
Given the acidic environment (pH 4.5–6.5) of H. pylori periplasm, it is hypothesized that NixA could co-transport H+ while acquiring the metal substrate.22 To determine if NixA strictly requires and exploit the proton motive force to catalyze Ni(ii) translocation, we encapsulated the fluorescent pH sensitive probe pyranine in the proteoliposome lumen. This aryl sulfonate derivative (8-hydroxy-1,3,6-pyrenetrisulfonate) is a water-soluble, membrane-impermeant pH-sensitive probe, with a fluorescent emission centered at 515 nm. The fluorescent emission yield of pyranine depends on the protonation state of the 8-hydroxyl group (pKa 7.2).37 These pyranine properties makes it suitable as a membrane-compatible sensor to report pH changes, as it does not bind with the lipid membranes because of the repulsion between negative charges of pyranine and the polar head of the lipids. When pyranine is deprotonated, the increase in pH results in a turn-on fluorescence response. Therefore, encapsulation of pyranine in the liposomal lumen can allow for the monitoring of proton influx or efflux through a decrease or increase in the time-dependent fluorescence signal, respectively.40 Pyranine was encapsulated in the lumen of control liposomes and NixA proteoliposomes by freeze–thaw membrane fracture. Ni(ii) transport was triggered by rapid mixing with buffered solutions containing Ni(ii), and the fluorescence traces were recorded at the pyranine emission maxima (λexc = 480 nm: λem = 515 nm), utilizing identical time frames used to study Ni(ii) transport with the FZ-3-Zn(ii) probe. Analysis of the kinetic traces revealed no detectable change in pyranine fluorescence in NixA proteoliposomes, with identical traces compared to control liposomes. This analysis indicates that, under the condition used, no H+ transport occurs upon Ni(ii) translocation (Fig. 3A). Thus, the presence of a proton motive force is not a strict requirement for NixA-mediated Ni(ii) translocation, even though the role of the proton gradient in promoting Ni(ii) acquisition in vivo remains to be investigated in detail. Regardless, our observation reveals a novel mechanistic feature of NixA translocation, as no NiCoT permease has been characterized using pyranine in previous studies.
Fig. 3. Determination of H+ transport and electrogenicity in NixA proteoliposomes. (A) Determination of H+ translocation in NixA proteoliposomes upon Ni(ii) transport (Ni(ii) = 25 μM) monitored by tracking changes in luminal pH as a function of time, utilizing the pH indicator pyranine (n = 3). (B) Superimposition of kinetic traces of Ni(ii) transport monitored by FZ-3-Zn(ii) and transmembrane potential changes monitored by oxonol VI. The kinetic traces were normalized and corrected with corresponding the control background signals (n = 3). The close correspondence of the two traces are consistent with a positive-inside transmembrane potential generated by NixA-mediated Ni(ii) transport.
Electrogenic Ni(ii) transport monitored by the membrane potential probe oxonol VI
The results from the metal transport assays with FZ-3-Zn(ii) and H+ proton translocation with pyranine revealed that NixA potentially allows the selective translocation of Ni(ii) across without a net proton co-/counter-transport. Therefore, we postulated that Ni(ii) translocation by NixA could be electrogenic, resulting in the accumulation of net positive charges in the proteoliposomes, if no other ion (e.g. Na+) is co-transported.
In order to determine Ni(ii) transport electrogenicity, oxonol VI (bis-(3-propyl-5-oxoisoxazol-4-yl) pentamethine oxonol) was used as a probe to monitor the changes in transmembrane membrane potential (Ψ). Oxonols are known to be relatively slow-response fluorescent probes that detects transmembrane membrane potential by differential partitioning between the aqueous phase and lipid bilayers. This change in partitioning as a function of membrane potential results in a modification of the fluorescence response. Compared to other oxonol compounds, oxonol VI has the fastest response to changes in potential.38,41 These characteristics make oxonol VI suitable for the real-time monitoring of transmembrane potential in liposomes and in cells, allowing the study of ion channels and electrogenic pumps.38,41
To test if the transport of Ni(ii) is electrogenic, the same methodologies were employed for oxonol VI reconstitution in NixA proteoliposomes, as described for the metal transport assays. The fluorescence kinetic traces were analyzed at the oxonol VI emission maximum (λexc = 580 nm: λem = 660 nm), upon mixing the proteoliposomes with Ni(ii) stock solutions to trigger substrate translocation. Analysis of the fluorescence kinetic traces indicated that, as Ni(ii) is translocated by NixA inside the proteoliposome lumen, a transmembrane potential is generated as a result of an accumulation of net positive charges, resulting in an increase in the oxonol VI fluorescence response. Thus, at least under the condition used, the transport the NixA-mediated Ni(ii) transport is electrogenic, creating a net transfer of positive charges from the extravesicular environment into the proteoliposome lumen.
To further substantiate that the positive-inside membrane potential generated in the proteoliposome lumen (detected by oxonol VI) is a result of Ni(ii) transport (as monitored by FZ-3-Zn(ii)), we normalized the signals obtained from these transport assays and overlapped their kinetic traces. The almost perfect superimposition of the traces confirms that the net translocation of the positive charges stems from the translocation of Ni(ii) (Fig. 3B). Together, this analysis also substantiates that the Ni(ii) translocation monitored by FZ-3-Zn(ii) reports the metal translocation events in real-time.
In conclusion, our developed platform based on fluorescent reporter probes sensitive to different stimuli (metal substrates, pH, transmembrane potential) provides a new tool to study the translocation properties and mechanism of NixA, and could potentially be applied to other Ni(ii) transporters. Based on the analysis, we discovered that H. pylori NixA is a non-promiscuous transmembrane electrogenic uniporter strictly selective for Ni(ii).
Mutation studies of NixA Ni(ii) translocation pathway
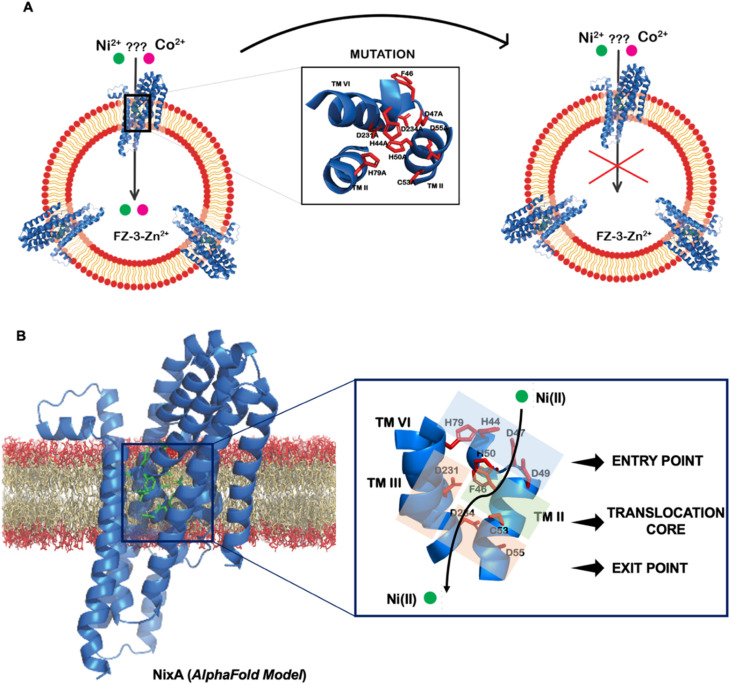
Topology prediction of NixA analyzed with TOPCONS, indicated that the protein contains eight transmembrane (TM) helices with a long hydrophilic loop connecting TM IV and TM V.21 Previous in vivo studies have shown that important conserved motifs (also known as recognition sequences), potentially responsible for substrate recognition and translocation, are present in TM II and TM III. These conserved motifs in TM II and TM III are (HA(F/V)DADH(I/L)) and (G(Xaa)5GHSSVV), respectively.21,23 As we developed a methodology to monitor the transport properties of purified NixA reconstituted in proteoliposomes, we proceeded to further dissect the critical residues responsible for substrate recognition and transport via mutation studies. We introduced selected single and combination mutations in NixA based on in vivo studies which analyzed membrane extracts and quantified the protein activity through cellular metal accumulation assays and urease activity.21,29,30,32 The structural model of NixA (H. pylori) generated by AlphaFold as previously described (Fig. 4A)11 was used to identify and locate the relative positions of the amino acids in these recognition sequences. Based on the model, the NixA conserved motifs (HA(F/V)DADH(I/L) and G(Xaa)5GHSSVV) were located in the innermost part of the transmembrane protein scaffold that spans from the periplasm to the cytoplasm (Fig. 4B).30,32In vivo studies identified that mutations in TM II (H44I, D47I, D49I, H50I, D55I), TM III (H79I) and TM VI (D23I, D234I) caused reduced metal accumulation and/or reduced/abolished urease activity.30 We assessed the relative positions of these conserved amino acid sequences in the structural model and noticed that these residues were arranged in a transmembrane pathway-like structure which consisted of an entry point, translocation core, and exit point (Fig. 4B).
Fig. 4. Structural model of NixA (H. pylori). (A) Schematic depicting the AlphaFold model for NixA and approach to study key transmembrane residues essential for transport activity. (B) Relative position of conserved NiCoT permeases recognition motifs and the putative translocation pathway based on the AlphaFold model.
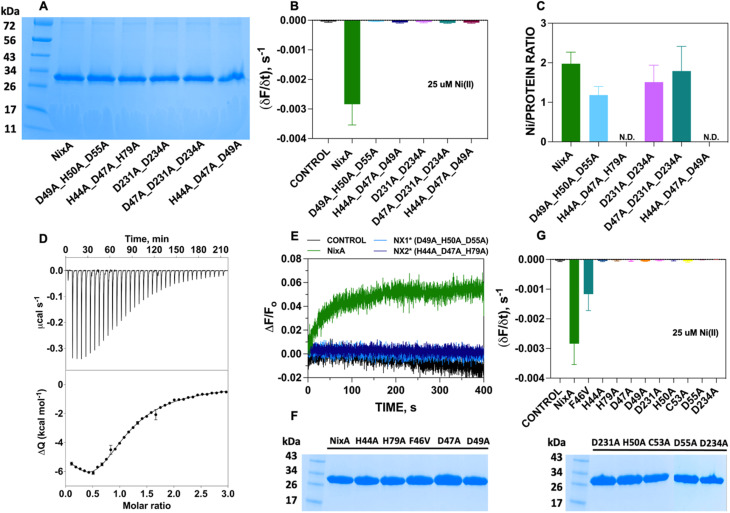
Five NixA combination mutants were initially designed, based on potential metal-coordinating amino acid residues clustered in each position in the translocation pathway, which were replaced with a non-coordinating alanine (Table 1). These mutant proteins were expressed, purified in stable forms (ESI Fig. S4†), and reconstituted utilizing the same protocols described for wild-type NixA. After purification, the monodisperse proteins were successfully incorporated into proteoliposomes with the same reconstitution efficiencies as compared to wild-type NixA (Fig. 5A), and the effect of the mutations in the transport activity – whether they changed the substrate recognition and/or terminated the protein activity – were investigated. The transport assays of these mutants were carried out as described for wtNixA. Analysis of the transport traces and their respective apparent initial transport velocities revealed that the Ni(ii) transport activities of all these combination mutants were completely abolished, substantiating key roles of these residues in the NixA transmembrane translocation pathway (Fig. 5B, ESI S5, and Table S2†).
NixA mutants and relative positions of the mutations based on the AlphaFold model.
| Mutant name | Transmembrane helix | Amino acid mutations | Position in the translocation pathway |
|---|---|---|---|
| H44A_D47A_H79A | TM2 | H44A | Entry point |
| D47A | |||
| TM3 | H79A | ||
| H44A_D47A_D49A | TM2 | H44A | Entry point |
| D47A | |||
| D49A | |||
| D49A_D50A_D55A | TM2 | D49A | Translocation core |
| H50A | |||
| D55A | |||
| D231A_D234A | TM6 | D231A | Translocation core |
| D234A | |||
| D47A_D231A_D234A | TM2 | D47A | Exit point |
| TM6 | D231A | ||
| D234A |
Fig. 5. NixA mutation studies. (A) SDS-PAGE analysis of NixA and NixA combination mutants reconstituted in proteoliposomes utilized for the Ni(ii) transport assays. (B) Maximal initial Ni(ii) transport rates by NixA and combination mutants monitored in proteoliposomes with FZ-3-Zn(ii) encapsulated in the SUV lumen (25 μM; λexc = 480 nm; λem = 515 nm) (n = 3). (C) NixA and NixA mutants Ni(ii) binding stoichiometry measured by ICP-MS (Ni(ii) concentration) and Bradford assays (protein concentration) (n = 3). (D) Ni(ii) titration of HpNixA followed by ITC. Heat response for injections of 400 μM Ni(ii) into 20 μM HpNixA (top); integrated heat data of the titration as a function of Ni(ii)/NixA molar ratio. The continuous line represents the best fit (goodness of fit, GoF = 78.4%) obtained with a stoichiometric equilibria model which involves two sets of metal-binding sites (bottom). (E) Real-time traces of transmembrane potential generation by NixA, NX1, and NX2 mutants in proteoliposomes upon incubation with Ni(ii) (25 μM), monitored by encapsulating oxonol VI in the proteoliposome lumen (n = 3). (F) SDS-PAGE analysis of NixA and single-point NixA mutants reconstituted in proteoliposomes utilized for Ni(ii) transport assays. (G) Maximal initial Ni(ii) transport rates in NixA and NixA point-mutants monitored in proteoliposomes with FZ-3-Zn(ii) encapsulated in the SUV lumen (Ni(ii) = 25 μM; λexc = 480 nm; λem = 515 nm) (n = 3).
To further understand the role of each combination mutations in recognition and Ni(ii) binding, we performed metal-binding determinations. Samples were prepared by mixing the purified NixA and mutants in detergent micelles with metal substrate in a 1 : 3 molar ratio; then, weakly bound and unbound metals were removed by SEC. Protein concentrations were determined via a modified Bradford assays, and Ni(ii) concentrations were determined by ICP-MS. Analysis of the Ni(ii) stoichiometry ratios revealed that wtNixA features two high-affinity transmembrane metal binding sites (Ni(ii):NixA = 2.0 ± 0.3 ion mol−1; Fig. 5C).
To estimate the binding affinity of these two Ni(ii) binding sites, Isothermal Titration Calorimetry (ITC) determinations were conducted on wtNixA samples. Ni(ii) titration of NixA produced an exothermic reaction, as indicated by negative peaks following each injection of Ni(ii), with a biphasic behavior, revealing the presence of two distinct binding events. This observation, together with the results of the metal analysis discussed above, clearly hints to the presence of two binding sites on wtNixA, each containing a single Ni(ii) ion (Fig. 5D, upper panel). The integrated heat data were fitted using either a stoichiometric equilibria (SE) model or an independent sites (IS) model (both assuming two sets of single-metal-binding sites) (Fig. 5D, lower panel). The goodness-of-fit parameters obtained for the two models are identical (see ESI Table 3† for a complete list of the fitted parameters), so that a clear distinction cannot be made. The detailed speciation and heat contribution to the binding isotherms for the fits of the two models are provided in ESI Fig. S6–S9.†
The dissociation constants are in the low μM range for both models (KD1 (SE) = 1.3 ± 0.4 μM, KD1 (IS) = 1.6 ± 0.4 μM, and KD2 (SE) = 9.2 ± 3.9 μM, KD2 (IS) = 7.7 ± 2.4 μM), confirming the characteristic of tight binding for both sites. The estimated sub-stoichiometric active protein concentration (rMca. 44% for both models) might result from residual metal copurification in the proteins samples (approx. 15%, as determined by ICP-OES), together with a potential equilibrium with one or multiple conformational states inaccessible to metal binding, and/or partial loss of protein activity due to sample partial aggregation or degradation. Nevertheless, in agreement with the presence of two distinct binding sites in NixA, the two binding events showed different thermodynamic properties. The first binding event is both enthalpically and entropically driven (ESI Table S3†), while the second binding event is enthalpically driven while possessing a negative entropic contribution (ESI Table S3†).
While the stoichiometry of Ni(ii) binding to D231A/D234A NixA (1.5 ± 0.4 mol mol−1) and D47A/D231A/D234A NixA (1.8 ± 0.6 mol mol−1) was not drastically affected, the two mutants H44A/D47A/H79A and H44A/D47A/D49A showed completely abolished Ni(ii) binding abilities. Contrarily, the D49A/H50A/D55A mutant showed decreased binding (1.2 ± 0.2 mol mol−1) consistent with the alteration on only one of the two high affinity binding sites (Fig. 5C). Based on this analysis, and consistent with the AlphaFold structural model, we conclude that the amino acid residues H44, D47, D49, H79, which are situated in the entry point of the translocation pathway, are centrally involved not only in translocation but also in the first step of metal recognition and binding.
To further support and validate our findings, we performed oxonol VI experiments on two representative mutants: D49A/H50A/D55A (capable of binding one Ni(ii) instead of two, with no transport) and H44A/D47A/H79A (incapable of Ni(ii) binding, with no transport). The working hypothesis were as follows: if these mutants eliminated the ability of NixA to bind and/or transport Ni(ii), no response from oxonol VI should be observed, as no Ni(ii) substrate can be bound and/or transported. Kinetic traces from the oxonol VI electrogenicity assay showed complete absence of turn-on fluorescence signal, indicating that no electrogenic Ni(ii) transport occurred upon mutation of these key residues (Fig. 5E).
Given the results obtained on the combination mutants, single-point mutants in the translocation pathway were generated to further identify and dissect the contribution of each amino acid residue to NixA transport activity. These mutants were expressed, purified (ESI Fig. 4†), and reconstituted into liposomes. The SDS-PAGE analysis of the corresponding proteoliposomes pellets showed identical incorporation efficiency in proteoliposomes as observed for wtNixA (Fig. 5F). Transport assays on proteoliposomes in the presence of Ni(ii), monitored by FZ-3-Zn(ii), were conducted as described for the wtNixA to determine the relative transport activity of each mutant. Transport traces collected on H44A, H79A, D47A, D49A, H50A, C53A, D55A, D231A, and D234A NixA single-site mutants showed absence of quenching response, indicating the total abolishment of Ni(ii) transport activity, with the exception of F46V which displayed reduced transport activity compared to NixA (∼33%, Fig. 5G, ESI 5, and Table S2†). The amino acid (F/V) in the conserved motif at TM II (HAF/VDADHIA) in NiCoTs has been postulated to be potentially responsible for the selectivity towards Ni(ii) vs. Co(ii) substrates.21 Our data suggest that phenylalanine indeed contributes, but is not critical, for the transport of Ni(ii). The role of this phenylalanine in modulating substrate selectivity can potentially arise from putative cation–π interactions in the selectivity filter.
Based on this data, we can conclude that H44, H79, D47, D49, H50, C53, D55, D231, and D234 are critical residues in Ni(ii) translocation by NixA, further supporting the structural model which indicate the presence of a three-stage translocation pathway. These findings, moreover, validate in vivo observations on some of these mutants investigated via Ni(ii) accumulation assays or urease activation.21,29,30,32 Overall, our work further supports that NixA evolved chemically and structurally to exclusively recognize and transport Ni(ii), owing to the presence of a characteristic transmembrane recognition and translocation pathway characterized by an arrangement of key and conserved metal coordinating residues which allow non-promiscuous Ni(ii) substrate recognition and transport.
Conclusions
In summary, this work established a platform to recombinantly express and purify transmembrane NiCoT transporter proteins in active form, which allowed us to reconstitute them in artificial lipid bilayer vesicles to study Ni(ii) translocation events, transport mechanism, and kinetics. This study also revealed that H. pylori NixA is a Class I NiCoT permease with no substrate promiscuity. The transport assays in proteoliposomes substantiate that NixA is a high affinity, high-capacity Ni(ii) transporter with KM, Ni(ii) = 31.0 ± 1.2 μM, allowing for the uptake of Ni(ii) at physiologically relevant concentrations with fast translocation rates. Unprecedented co-transport and electrogenicity measurements in proteoliposomes carrying a purified NiCoT transporter revealed that NixA can act as an electrogenic Ni(ii) uniporter. Mutational analysis identified key transmembrane residues (H44, D47, H79, and D49) that are critical in high-affinity Ni(ii) binding, which together with a number of additional transmembrane (possibly transient) coordinating residues (H50, C53, D55, D231, and D234) create a potential three-stage transmembrane translocation pathway to allow for the selective translocation of Ni(ii) across the lipid bilayer. A detail comparison of NixA transport properties with the other Ni(ii) importer present in H. pylori, the Ni(ii) ABC transporter NiuBDE, is limited by the lack of detailed molecular-level structural and functional characterizations of the transmembrane component of Ni(ii) ABC transporter systems. While the NikABCDE protein complex found in the E. coli inner membrane is the canonical and best characterized nickel-responsive ABC transporter, only its periplasmic substrate binding protein (SBP), NikA, is well-characterized. Apo- and holo- NikA crystal structures featuring a classical SBP topology and coordination of one Ni(ii) ion (with aquo or l-histidine ligands) have been reported, and a Ni(ii) dissociation constant (KD) of approx. 10 μM has been determined by ITC, showing a similar affinity regime as determined for NixA in this study.42,43 Studies on NikA homologues have suggested that NikA and other Ni-SBPs are highly promiscuous, binding to different nickel forms (complexes) during uptake.42–44 However, the Ni(ii) affinities and transport kinetics of the transmembrane transporter component, responsible for transmembrane Ni(ii) translocation across the lipid bilayer, remain to be established. Thus, this study, carried out for the first time on NixA, opens new venues to investigate and understand the transport properties of other NiCoT transporter subclasses as well as other Ni(ii) transporter families, which will allow for a detailed comparative investigations of their diverse Ni(ii) translocation properties.
Data availability
Data for this paper are available upon request to the corresponding author.
Author contributions
Jayoh A. Hernandez: investigation, formal analysis, methodology, writing – original draft; Paul S. Micus, Sean Alec Lois Sunga: investigation, formal analysis, methodology, writing – review & editing; Luca Mazzei, Stefano Ciurli: investigation, formal analysis, methodology, writing, resources; Gabriele Meloni: conceptualization, supervision, investigation, formal analysis, methodology, writing – original draft, resources.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
The work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM128704 (to G. M.) and by the Robert A. Welch Foundation (AT-2073-20210327 to G. M). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. L. M. and S. C. received financial support from the University of Bologna and the Consorzio Interuniversitario di Risonanze Magnetiche di Metallo-Proteine (CIRMMP).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc05135h
References
- Gray H. B., Stiefel E. I., Valentine J. S., and Bertini I., Biological Inorganic Chemistry Structure and Reactivity, University Science Books, Sausalito, Californie, 1st edn, 2007 [Google Scholar]
- Palmer L. D. Skaar E. P. Transition Metals and Virulence in Bacteria. Annu. Rev. Genet. 2016;50:67–91. doi: 10.1146/annurev-genet-120215-035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney M. J. Ciurli S. Nonredox nickel enzymes. Chem. Rev. 2014;114:4206–4228. doi: 10.1021/cr4004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney M. J. Ciurli S. Nickel as a virulence factor in the Class I bacterial carcinogen, Helicobacter pylori. Semin. Cancer Biol. 2021;76:143–155. doi: 10.1016/j.semcancer.2021.04.009. [DOI] [PubMed] [Google Scholar]
- Leach M. R. Zamble D. B. Metallocenter assembly of the hydrogenase enzymes. Curr. Opin. Chem. Biol. 2007;11:159–165. doi: 10.1016/j.cbpa.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Li Y. Zamble D. B. Nickel homeostasis and nickel regulation: an overview. Chem. Rev. 2009;109:4617–4643. doi: 10.1021/cr900010n. [DOI] [PubMed] [Google Scholar]
- Kaluarachchi H. Chan Chung K. C. Zamble D. B. Microbial nickel proteins. Nat. Prod. Rep. 2010;27:681–694. doi: 10.1039/B906688H. [DOI] [PubMed] [Google Scholar]
- Sydor A. M. Zamble D. B. Nickel metallomics: general themes guiding nickel homeostasis. Met. Ions Life Sci. 2013;12:375–416. doi: 10.1007/978-94-007-5561-1_11. [DOI] [PubMed] [Google Scholar]
- Zeer-Wanklyn C. J. Zamble D. B. Microbial nickel: cellular uptake and delivery to enzyme centers. Curr. Opin. Chem. Biol. 2017;37:80–88. doi: 10.1016/j.cbpa.2017.01.014. [DOI] [PubMed] [Google Scholar]
- Eitinger T. Mandrand-Berthelot M. A. Nickel transport systems in microorganisms. Arch. Microbiol. 2000;173:1–9. doi: 10.1007/s002030050001. [DOI] [PubMed] [Google Scholar]
- Camporesi G. Minzoni A. Morasso L. Ciurli S. Musiani F. Nickel import and export in the human pathogen Helicobacter pylori, perspectives from molecular modelling. Metallomics. 2021;13(12):mfab066. doi: 10.1093/mtomcs/mfab066. [DOI] [PubMed] [Google Scholar]
- Musiani F. Zambelli B. Bazzani M. Mazzei L. Ciurli S. Nickel-responsive transcriptional regulators. Metallomics. 2015;7:1305–1318. doi: 10.1039/C5MT00072F. [DOI] [PubMed] [Google Scholar]
- Ruggiero P. Helicobacter pylori and inflammation. Curr. Pharm. Des. 2010;16:4225–4236. doi: 10.2174/138161210794519075. [DOI] [PubMed] [Google Scholar]
- Mazzei L. Musiani F. Ciurli S. The structure-based reaction mechanism of urease, a nickel dependent enzyme: tale of a long debate. J. Biol. Inorg. Chem. 2020;25:829–845. doi: 10.1007/s00775-020-01808-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney S. B. Hausinger R. P. Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 2003;27:239–261. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- Hood M. I. Skaar E. P. Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F. Robbe-Saule M. Turlin E. Mancuso F. Michel V. Richaud P. Veyrier F. J. De Reuse H. Vinella D. Characterization in Helicobacter pylori of a Nickel Transporter Essential for Colonization That Was Acquired during Evolution by Gastric Helicobacter Species. PLoS Pathog. 2016;12:e1006018. doi: 10.1371/journal.ppat.1006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov D. A. Hebbeln P. Gelfand M. S. Eitinger T. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 2006;188:317–327. doi: 10.1128/JB.188.1.317-327.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitinger T. Suhr J. Moore L. Smith J. A. C. Secondary transporters for nickel and cobalt ions: theme and variations. BioMetals. 2005;18:399–405. doi: 10.1007/s10534-005-3714-x. [DOI] [PubMed] [Google Scholar]
- Hebbeln P. Eitinger T. Heterologous production and characterization of bacterial nickel/cobalt permeases. Fems Microbiol. Lett. 2004;230:129–135. doi: 10.1016/S0378-1097(03)00885-1. [DOI] [PubMed] [Google Scholar]
- Degen O. Eitinger T. Substrate specificity of nickel/cobalt permeases: Insights from mutants altered in transmembrane domains I and II. J. Bacteriol. 2002;184:3569–3577. doi: 10.1128/JB.184.13.3569-3577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degen O. Kobayashi M. Shimizu S. Eitinger T. Selective transport of divalent cations by transition metal permeases: the Alcaligenes eutrophus HoxN and the Rhodococcus rhodochrous NhlF. Arch. Microbiol. 1999;171:139–145. doi: 10.1007/s002030050691. [DOI] [PubMed] [Google Scholar]
- Eitinger T. Wolfram L. Degen O. Anthon C. A Ni2+ binding motif is the basis of high affinity transport of the Alcaligenes eutrophus nickel permease. J. Biol. Chem. 1997;272:17139–17144. doi: 10.1074/jbc.272.27.17139. [DOI] [PubMed] [Google Scholar]
- Eitinger T. Friedrich B. A Topological Model for the High-Affinity Nickel Transporter of Alcaligenes Eutrophus. Mol. Microbiol. 1994;12:1025–1032. doi: 10.1111/j.1365-2958.1994.tb01090.x. [DOI] [PubMed] [Google Scholar]
- Eitinger T. Friedrich B. Cloning, Nucleotide-Sequence, and Heterologous Expression of a High-Affinity Nickel Transport Gene from Alcaligenes-Eutrophus. J. Biol. Chem. 1991;266:3222–3227. doi: 10.1016/S0021-9258(18)49977-2. [DOI] [PubMed] [Google Scholar]
- Noinaj N. Guillier M. Barnard T. J. Buchanan S. K. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet A. H. Stoof J. Vlasblom R. Wainwright S. A. Hughes N. J. Kelly D. J. Bereswill S. Bijlsma J. J. Hoogenboezem T. Vandenbroucke-Grauls C. M. Kist M. Kuipers E. J. Kusters J. G. The role of the Ferric Uptake Regulator (Fur) in regulation of Helicobacter pylori iron uptake. Helicobacter. 2002;7:237–244. doi: 10.1046/j.1523-5378.2002.00088.x. [DOI] [PubMed] [Google Scholar]
- Postle K. TonB system, in vivo assays and characterization. Methods Enzymol. 2007;422:245–269. doi: 10.1016/S0076-6879(06)22012-3. [DOI] [PubMed] [Google Scholar]
- Fulkerson Jr J. F. Mobley H. L. Membrane topology of the NixA nickel transporter of Helicobacter pylori: two nickel transport-specific motifs within transmembrane helices II and III. J. Bacteriol. 2000;182:1722–1730. doi: 10.1128/JB.182.6.1722-1730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulkerson Jr J. F. Garner R. M. Mobley H. L. Conserved residues and motifs in the NixA protein of Helicobacter pylori are critical for the high affinity transport of nickel ions. J. Biol. Chem. 1998;273:235–241. doi: 10.1074/jbc.273.1.235. [DOI] [PubMed] [Google Scholar]
- Deng X. He J. M. He N. Comparative study on Ni2+-affinity transport of nickel/cobalt permeases (NiCoTs) and the potential of recombinant Escherichia coli for Ni2+ bioaccumulation. Bioresour. Technol. 2013;130:69–74. doi: 10.1016/j.biortech.2012.11.133. [DOI] [PubMed] [Google Scholar]
- Wolfram L. Bauerfeind P. Conserved low-affinity nickel-binding amino acids are essential for the function of the nickel permease NixA of Helicobacter pylori. J. Bacteriol. 2002;184:1438–1443. doi: 10.1128/JB.184.5.1438-1443.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee K. R. Zhou Z. L. Qian W. J. Kennedy R. Detection and imaging of zinc secretion from pancreatic beta-cells using a new fluorescent zinc indicator. J. Am. Chem. Soc. 2002;124:776–778. doi: 10.1021/ja011774y. [DOI] [PubMed] [Google Scholar]
- Zhao J. Bertoglio B. A. Devinney Jr M. J. Dineley K. E. Kay A. R. The interaction of biological and noxious transition metals with the zinc probes FluoZin-3 and Newport Green. Anal. Biochem. 2009;384:34–41. doi: 10.1016/j.ab.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeyrathna S. S. Abeyrathna N. S. Thai N. K. Sarkar P. D'Arcy S. Meloni G. IroT/MavN Is a Legionella Transmembrane Fe(II) Transporter: Metal Selectivity and Translocation Kinetics Revealed by in Vitro Real-Time Transport. Biochemistry. 2019;58:4337–4342. doi: 10.1021/acs.biochem.9b00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeyrathna S. S. Abeyrathna N. S. Basak P. Irvine G. W. Zhang L. Meloni G. Plastic recognition and electrogenic uniport translocation of 1st-, 2nd-, and 3rd-row transition and post-transition metals by primary-active transmembrane P1B-2-type ATPase pumps. Chem. Sci. 2023;14:6059–6078. doi: 10.1039/D3SC00347G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement N. R. Gould J. M. Pyranine (8-hydroxy-1,3,6-pyrenetrisulfonate) as a probe of internal aqueous hydrogen ion concentration in phospholipid vesicles. Biochemistry. 1981;20:1534–1538. doi: 10.1021/bi00509a019. [DOI] [PubMed] [Google Scholar]
- Apell H. J. Bersch B. Oxonol VI as an optical indicator for membrane potentials in lipid vesicles. Biochim. Biophys. Acta. 1987;903:480–494. doi: 10.1016/0005-2736(87)90055-1. [DOI] [PubMed] [Google Scholar]
- Pineiro A. Munoz E. Sabin J. Costas M. Bastos M. Velazquez-Campoy A. Garrido P. F. Dumas P. Ennifar E. Garcia-Rio L. Rial J. Perez D. Fraga P. Rodriguez A. Cotelo C. AFFINImeter: A software to analyze molecular recognition processes from experimental data. Anal. Biochem. 2019;577:117–134. doi: 10.1016/j.ab.2019.02.031. [DOI] [PubMed] [Google Scholar]
- Wang K. Sitsel O. Meloni G. Autzen H. E. Andersson M. Klymchuk T. Nielsen A. M. Rees D. C. Nissen P. Gourdon P. Structure and mechanism of Zn2+-transporting P-type ATPases. Nature. 2014;514:518–522. doi: 10.1038/nature13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R. J. Apell H. J. A stopped-flow kinetic study of the interaction of potential-sensitive oxonol dyes with lipid vesicles. Biophys. Chem. 1989;34:225–237. doi: 10.1016/0301-4622(89)80061-4. [DOI] [PubMed] [Google Scholar]
- Heddle J. Scott D. J. Unzai S. Park S. Y. Tame J. R. H. Crystal Structures of the Liganded and Unliganded Nickel-Binding Protein NikA from Escherichia Coli. J. Biol. Chem. 2003;278:50322–50329. doi: 10.1074/jbc.M307941200. [DOI] [PubMed] [Google Scholar]
- Lebrette H. Iannello M. Fontecilla-Camps J. C. Cavazza C. The Binding Mode of Ni-(L-His)2 in NikA Revealed by X-Ray Crystallography. J. Inorg. Biochem. 2013;121:16–18. doi: 10.1016/j.jinorgbio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Lebrette H. Brochier-armanet C. Zambelli B. de Reuse H. Borezée-Durant E. Ciurli S. Cavazza C. Promiscuous Nickel Import in Human Pathogens: Structure, Thermodynamics, and Evolution of Extracytoplasmic Nickel-Binding Proteins. Structure. 2014;22:1421–1432. doi: 10.1016/j.str.2014.07.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for this paper are available upon request to the corresponding author.