Abstract
SurA is a periplasmic peptidyl-prolyl isomerase required for the efficient folding of extracytoplasmic proteins. Although the surA gene had been identified in a screen for mutants that failed to survive in stationary phase, the role played by SurA in stationary-phase survival remained unknown. The results presented here demonstrate that the survival defect of surA mutants is due to their inability to grow at elevated pH in the absence of ςS. When cultures of Escherichia coli were grown in peptide-rich Luria-Bertani medium, the majority of the cells lost viability during the first two to three days of incubation in stationary phase as the pH rose to pH 9. At this time the surviving cells resumed growth. In cultures of surA rpoS double mutants the survivors lysed as they attempted to resume growth at the elevated pH. Cells lacking penicillin binding protein 3 and ςS had a survival defect similar to that of surA rpoS double mutants, suggesting that SurA foldase activity is important for the proper assembly of the cell wall-synthesizing apparatus.
Bacteria have evolved a multitude of starvation-inducible mechanisms that lead to profound changes in cellular morphology and physiology (25, 40, 42). Induction of these changes results in cells with a greatly increased ability to survive many different environmental stresses, including starvation itself (26). Another survival strategy that appears to be quite widespread in the microbial world is a phenomenon we have termed GASP (for growth advantage in stationary phase) (45–47). When bacterial cultures are incubated for prolonged periods in stationary phase under many different conditions, there can be a dramatic loss in the number of CFU. Surprisingly, the survivors almost invariably are mutants able to grow at the time that the majority of the cells lose viability. The GASP phenomenon is easily observed when cultures of Escherichia coli K-12 are incubated in Luria-Bertani (LB) medium at 37°C. After reaching an initial titer of approximately 5 × 109 CFU/ml after overnight incubation, titers drop during the first week and then stabilize at approximately 108 CFU/ml (47). The surviving cells are invariably mutants expressing the GASP phenotype, which can be assessed by their ability to take over a wild-type strain in mixed cultures during stationary-phase incubation.
In an effort to understand more about the surviving cells of E. coli cultures kept in stationary phase for long periods, we designed a screen for mutants unable to survive under these conditions and in this way discovered surA (43). Mutants that lacked SurA grew normally but completely lost viability after 3 to 5 days of incubation in rich medium. The nucleotide sequence of surA, determined later, indicated that SurA was similar in amino acid sequence to parvulin, a peptidyl-prolyl isomerase (PPIase) (37). Subsequently, it was determined that SurA was a periplasmic protein required for the proper assembly of several outer membrane proteins (22) and that it possessed PPIase activity (29, 36).
We wondered how the loss of a periplasmic PPIase could result in a survival defect during stationary phase. E. coli possesses three periplasmic PPIases: PpiA (23), FkpA (16), and SurA (29, 36, 37). Mutants that lack PpiA or FkpA have no obvious phenotype (39). Therefore, the phenotype associated with the lack of SurA was very unusual. This difference in phenotype suggested that the functions of these PPIases are not entirely redundant. The differential, but interconnected, regulation of the three activities also suggested nonredundant functions. Both ppiA and fkpA are induced in response to periplasmic stresses (e.g., unfolded proteins or elevated pH) and are members of the CpxRA and ςE regulons, respectively (5, 6, 31). The surA gene does not appear to be a member of either of these regulons, but its loss induces the expression of the ςE regulon (5, 29, 31, 36). In an effort to understand why loss of this particular PPIase should cause cells to die in stationary phase, we further analyzed the properties of surA cells. The results of these analyses are presented here.
MATERIALS AND METHODS
Strains and growth conditions.
Our standard laboratory wild-type strain of E. coli is K-12 ZK126 (W3110 ΔlacU169 tna-2) (4). Mutations were transduced into this strain with bacteriophage P1vir (28), and the resulting strains are described in Table 1. Cells were grown in LB medium and incubated at 37°C with aeration for the time periods indicated in the figures. Cultures were supplemented with kanamycin (50 μg/ml) or tetracycline (15 μg/ml) where appropriate.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| ZK126 | W3110 ΔlacU169 tna-2 | 4 |
| ZK1000 | ZK126 rpoS::Km | 3 |
| ZK1171 | ZK126 rpoS::Tn10 | 24 |
| SL238 | ZK126 rpoS::Tn10 | This work |
| ZK819 | ZK126 rpoS819 | 47 |
| ZK880 | ZK819 surA1::mini-Tn10(Km) | 43 |
| SL133 | ZK126 ΔsurA3::Km rpoS+ | 22 |
| SL239 | SL238 ΔsurA3::Km | This work |
| EC295 | ftsI23 leu::Tn10 | 11 |
| SL268 | ZK126 ftsI23 leu::Tn10 | This work |
| SL270 | ZK1000 ftsI23 leu::Tn10 | This work |
Microscopy.
Samples from cultures were spotted onto microscope slides coated with 0.8% agar and then viewed with a Nikon Optiphot-2 phase-contrast microscope at ×60 magnification. Images were collected with a charge-coupled device camera linked to a Macintosh 8600 computer with Scion Image software for image processing.
Immunoblot detection of proteins.
Detection of SurA was performed as previously described, (10) with rabbit polyclonal antiserum raised against SurA. This antiserum was obtained under our contract with Immuno-Dynamics Inc. (La Jolla, Calif.) with purified SurA (22). Log-phase cells were at an optical density at 600 nm (OD600) of 0.6, and stationary-phase cultures grew overnight.
pH sensitivity assay.
To determine the effect of pH on growth, exponentially growing cultures (OD600, 0.1) were resuspended into fresh prewarmed LB medium that had been either untreated or adjusted to pH 9 with NaOH. Growth was monitored by observing OD600.
High-pressure liquid chromatography (HPLC).
The composition of the murein sacculus was determined by following the protocol of Glauner (8). Cells from mid-log-phase cultures (OD600, 0.7) were used for analysis.
Penicillin binding protein (PBP) assay.
Cells were grown at 37°C in 10 ml of LB medium to an approximate OD600 of 1. Culture volumes were adjusted with LB medium as necessary to an OD600 of 1. Five milliliters of cells was pelleted and resuspended in 125 μl of spheroplast buffer (50 mM Tris [pH 8], 5 mM EDTA, 500 mM sucrose). Forty microliters of this suspension was transferred to a fresh tube and incubated for 10 min at 4°C. Then 2.6 μl of 300 μM fluorescein-hexanoic acid-6-amino penicillanic acid (Flu-Apa) (gift of D. Weiss) was added, and the mixture was incubated at 30°C for 30 min. An equal volume of 2× loading buffer (1) was added, and cells were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were visualized and quantitated with a Fluorimager (Molecular Dynamics).
RESULTS
Characterization of surA mutant cells.
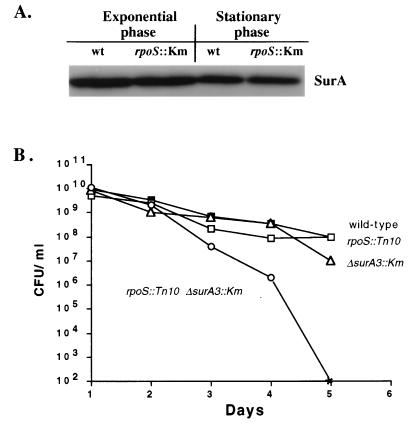
SurA appeared to be specifically required for survival during stationary phase (43). Thus, it made sense to investigate whether surA expression was growth phase regulated. To determine whether SurA was specifically induced in stationary phase and whether it was regulated by ςS, the stationary-phase-specific sigma factor, we quantitated the abundance of SurA in a strain lacking ςS (20). The results of Western blot analysis are shown in Fig. 1A. The amount of SurA was unchanged in an rpoS::Km strain relative to that in the wild type and did not change significantly when the cells were in stationary phase.
FIG. 1.
(A) SurA levels are independent of ςS and growth phase. The levels of SurA in wild-type (wt) and rpoS::Km cells were detected by immunoblot analysis in exponential-phase (OD600, 0.6) and stationary-phase (overnight) cells. (B) Mutations in both rpoS and surA are required to express the Sur− phenotype. Levels of viability of the wild type (filled squares), rpoS::Tn10 mutant (open squares), ΔsurA3::Km mutant (open triangles), and rpoS::Tn10 ΔsurA3::Km double mutant (open circles) in LB medium are shown. The asterisk represents <102 CFU/ml. This experiment was repeated more than three times, and representative data are shown.
While ςS does not control the expression of surA, we serendipitously discovered that the allelic state of rpoS greatly affects the expression of the survival (Sur−) phenotype. When the original surA1::mini-Tn10(Km) allele was transduced into a wild-type strain, we observed that the Sur− phenotype was lost, i.e., transductants survived in stationary phase like the wild type. Since it has been reported that many laboratory wild-type E. coli K-12 strains harbor mutant rpoS alleles (17, 18, 47), we tested the original surA1::mini-Tn10(Km) strain for the activity of ςS. We determined that the original strain in which the surA1::mini-Tn10(Km) mutation was isolated did indeed contain a mutation in rpoS (rpoS819) and that this mutation was also required for the loss of viability in stationary phase. To further test the notion that in order to express the Sur− phenotype a strain needs mutations in both surA and rpoS, we constructed strains containing combinations of null alleles of these genes (ΔsurA3::Km and rpoS::Tn10). Strains carrying null alleles of either rpoS or surA alone did not have noticeable defects in stationary-phase survival (Fig. 1B); only when mutant alleles of both genes were present did all the cells of the culture lose viability in stationary phase (Fig. 1B).
Effect of pH on the survival of surA rpoS double mutants.
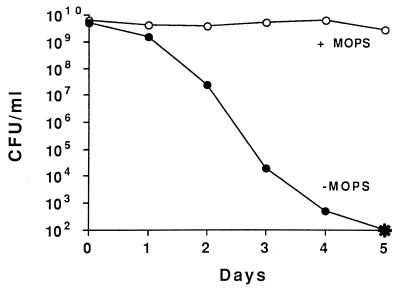
SurA is found in growing and nongrowing cells but is essential only during stationary phase. This finding suggests that particular changes in the environment during stationary phase are responsible for rendering SurA essential only during this phase. In order to understand why SurA function is required in the absence of ςS, environmental changes that occur during stationary phase in LB medium cultures had to be determined. Tormo et al. (43) observed that, unlike cells in LB medium, surA1::mini-Tn10(Km) rpoS819 cells grown in minimal glucose medium were able to survive for a long time (20 days) during incubation in stationary phase. Since the main carbon source in LB medium is peptides and amino acids, these cultures become alkaline in stationary phase due to the release of excess amine-containing compounds. In contrast, the pH of M63-glucose-grown culture supernatants remains near neutrality (pH 6.5 to 7). To test whether the increase in pH in LB medium cultures was responsible for the loss of viability of surA1::mini-Tn10(Km) rpoS819 mutants, 40 mM MOPS (morpholinepropanesulfonic acid; pH 7.4) was added to buffer the LB medium cultures. In this medium, surA1::mini-Tn10(Km) rpoS819 cells did not lose viability (Fig. 2). That the Sur− phenotype is observed only in cells carrying both mutations indicates that a wild-type allele of either surA or rpoS is sufficient for cell survival in stationary phase at elevated pH.
FIG. 2.
Growth of the surA1::mini-Tn10(Km) rpoS819 double mutant in LB medium (filled circles) or LB medium plus 40 mM MOPS (pH 7.4) (open circles). The asterisk represents <102 CFU/ml. This experiment was repeated three times, and representative data are shown.
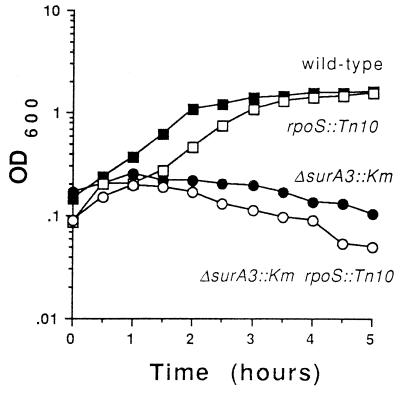
Although SurA is present in exponentially growing cells, little if any ςS is present during exponential phase when cells are grown in LB medium at 37°C. We therefore hypothesized that loss of SurA should also increase the sensitivity of exponential-phase cells to alkaline pH. To test this, exponentially growing cells of four strains, the wild type, a ΔsurA3::Km strain, an rpoS::Tn10 strain, and a ΔsurA3::Km rpoS::Tn10 strain, were resuspended in fresh LB medium adjusted to pH 9 with sodium hydroxide and incubated at 37°C (Fig. 3). For the first 30 min of incubation, all four strains continued to grow exponentially. After that time, the wild-type strain continued to grow, the rpoS::Tn10 strain lagged for a bit and then resumed growth to a level similar to that of the wild type, and the two strains lacking SurA stopped growing and began to lyse (Fig. 3). Lysis was confirmed by microscopic observation (Fig. 4). These results demonstrate that SurA is required for growth at pH 9.
FIG. 3.
SurA is required for growth at pH 9. Exponentially growing cells (OD600, 0.1) were resuspended into LB medium (pH 9) and incubated for 5 h at 37°C with aeration. Growth was monitored by observing OD. Shown are results for the wild type (filled squares), the rpoS::Tn10 mutant (open squares), the ΔsurA3::Km mutant (filled circles), and the rpoS::Tn10 ΔsurA3::Km double mutant (open circles). This experiment was repeated more than three times, and representative data are shown.
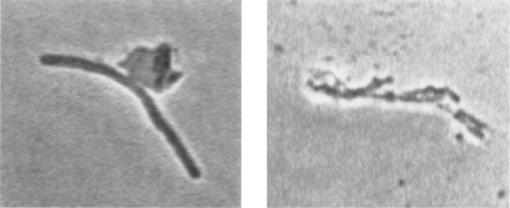
FIG. 4.
Typical morphology of surA1::mini-Tn10(Km) rpoS819 cells in the process of lysing after transfer to pH 9. Cultures of surA1::mini-Tn10(Km) rpoS819 cells growing exponentially (OD600, 0.1) in LB medium were transferred to LB medium at pH 9. After 30 min at 37°C cells were observed microscopically with phase contrast.
Both the ΔsurA3::Km and the ΔsurA3::Km rpoS::Tn10 strain lysed in the experiment shown in Fig. 3, whereas in stationary phase the ΔsurA3::Km strain survived and only the double mutant died. During exponential growth the ΔsurA3::Km mutant strain should not contain much ςS, since ςS is not highly expressed during exponential phase in LB medium at 37°C. As cell growth slows in early-stationary-phase cultures, the ςS regulon is induced, before the pH of the medium becomes alkaline. This suggests that ςS expression can compensate for the loss of SurA.
The results shown in Fig. 3 demonstrate that SurA is required during exponential growth at elevated pH but do not answer the question of why both SurA and ςS are required for long-term survival in stationary phase. The answer may lie in the fact that stationary-phase LB medium cultures are highly dynamic. As most of the cells died, GASP mutants took over the culture. For the first two days of incubation in stationary phase, the ΔsurA3::Km rpoS::Tn10 strain lost viability, with kinetics similar to those of the wild type. The difference in levels of viability between the two cultures became apparent on the third to fourth days of incubation (Fig. 1B), the same period during which the GASP mutants grew and took over the culture (47). This suggests that either SurA or ςS activity was specifically required for growth of the GASP mutants at elevated pH in these stationary-phase cultures. Consistent with the idea that SurA is required only for growth is the observation that after overnight incubation the nongrowing cells were completely viable although the medium was at pH 9 (Fig. 1B).
Morphological changes in a ΔsurA3::Km rpoS::Tn10 mutant.
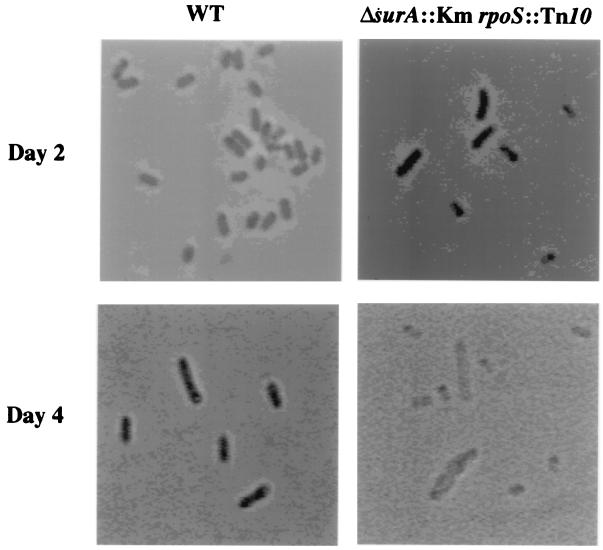
Microscopic analysis of mutant cells in stationary phase was performed to determine whether the mutant cells had altered morphology. These experiments indicated that ΔsurA3::Km rpoS::Tn10 mutant cells indeed had aberrant morphology (Fig. 5). The ΔsurA3::Km rpoS+ cells appeared similar to the wild-type cells in overnight cultures (data not shown). Two-day-old cultures of the wild-type and ΔsurA3::Km rpoS::Tn10 strains also appeared similar (Fig. 5). After 4 days of incubation, the cell morphology became drastically altered. Many of the double-mutation cells were misshapen, having bulges in their middle sections. Other cells appeared extremely small (Fig. 5). Experiments with the vital stain acridine orange suggested that the large bulging cells were the cells that were attempting to resume growth and that the small cells had lost viability (data not shown). Decrease in the OD of the culture and accumulation of cellular debris in the culture suggested that the cells lysed as they attempted to grow and that lysis was the ultimate reason for the death of the surA rpoS double mutants in stationary phase.
FIG. 5.
Morphology of ΔsurA3::Km rpoS::Tn10 cells in stationary phase. The morphologies of wild-type (WT) and ΔsurA3::Km rpoS::Tn10 cells at days 2 and 4 of incubation in LB medium are shown.
Cell shape is determined by the murein sacculus, which is composed of short, cross-linked strands of peptidoglycan (14). The altered morphology of ΔsurA3::Km rpoS::Tn10 cells suggested that changes in peptidoglycan composition occurred in these mutant cells. At least 15 enzymes act in concert to synthesize the sacculus (9, 14). Although disruption of any one of these enzymes need not be lethal to the cell, morphological defects can occur (14). The concurrent disruption of multiple peptidoglycan-synthesizing enzymes can cause cell lysis, as evidenced by the action of β-lactam antibiotics (38). The misshapen morphology of ΔsurA3::Km rpoS::Tn10 cells suggested that the relative activities of these sacculus-synthesizing enzymes were no longer in balance, resulting in a sacculus with structural defects.
The fine structure of a sacculus can be determined by isolating the peptidoglycan from the cell and then digesting it into its component muropeptides (8). At least 80 different muropeptides are generated from this digestion, and these can be separated by reverse-phase HPLC (8). Changes in a chromatogram profile reflect changes in relative enzyme activities in vivo. This technique has been used extensively to monitor the changes that a sacculus undergoes under a variety of conditions, including the changes seen upon the entry of cells into stationary phase (27, 30, 44). The peptidoglycans of the wild type and the surA1::mini-Tn10(Km) rpoS819 mutant were analyzed by this technique. The results indicated that the mutant cells had an approximately twofold enrichment of muropeptides with 1,6-anhydrous moieties. These moieties are the signature activities of the lytic transglycosylases, enzymes that break the β-1,4-acetylglucosaminic links between N-acetyl-muramic acid and N-acetyl-glucosamine residues of the peptidoglycan backbone (15). This activity is analogous to that of lysozyme, so it is not surprising that increasing the amount of it can result in cell lysis.
At least three lytic transglycosylases are present in E. coli, the predominant one being the 70-kDa soluble lytic transglycosylase (Slt70) (35). These three enzymes are required for cellular growth, since links within a sacculus must be broken in order for new strands of peptidoglycan to be inserted. Little is known about the regulation of any of the peptidoglycan-metabolizing enzymes. For instance, overexpression of Slt70 is insufficient to lyse cells unless the stringent response is inactivated, but the mechanism(s) by which the stringent response is involved is still unknown (2). This lack of cell lysis under these conditions suggests that although the relative amount of lytic transglycosylase activity is probably increased in the surA1::mini-Tn10(Km) rpoS819 strain, some other activities must also be altered in order for lysis to occur.
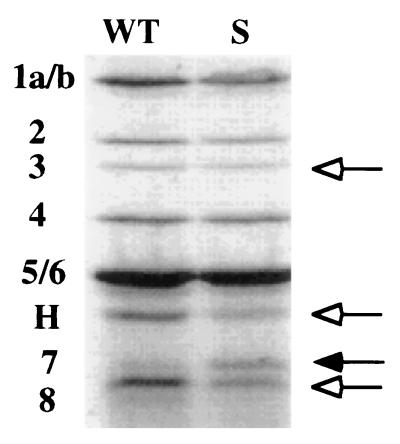
To investigate what other cell wall-synthesizing activities might have been altered in the ΔsurA3::Km rpoS::Tn10 double mutant, the relative amounts of nine PBPs were measured. These enzymes catalyze a variety of functions required for the growth of the sacculus. A penicillin-fluorescein conjugate, Flu-Apa, was used to visualize and quantitate the PBPs (32). Wild-type and ΔsurA3::Km cells were incubated with Flu-Apa, and the proteins were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 6). During growth at neutral pH the amounts of several PBPs were altered in the ΔsurA3::Km mutant. Most notably, PBP8 and AmpH decreased and PBP7 increased. The amounts of PBP1, -2, -3, and -8 and AmpH decreased between 15 and 40% relative to amounts in the wild type, while the amount of PBP4 was approximately the same. In overnight cultures few differences were observed (data not shown). Since cultures of stationary-phase cells contain mixtures of viable and nonviable cells, the PBPs were not measured during extended incubation.
FIG. 6.
The pattern of PBPs is altered in a ΔsurA3::Km mutant. Cells from exponential-phase cultures (OD600, 0.1) grown in LB medium at 37°C were labeled with Flu-Apa (see the text). Shown are bands that decreased (open arrows) or increased (filled arrow) in intensity in the mutant relative to the intensities of corresponding bands in the wild type. WT, wild type; S, ΔsurA3::Km mutant; H, AmpH. Numbers indicate designations of PBPs.
PBP7 is an endopeptidase that cleaves the dd-diaminopimelate-d-alanine bonds in intact cell walls, the same bond that PBP3 creates (33). PBP7 is processed into PBP8, a smaller but still active form (12, 33). AmpH is similar to the class C β-lactamases but does not possess β-lactamase activity (13). Cells lacking AmpH as well as PBP1a and PBP5 have aberrant morphology during exponential growth, though they have no noticeable defect when AmpH alone is missing (13).
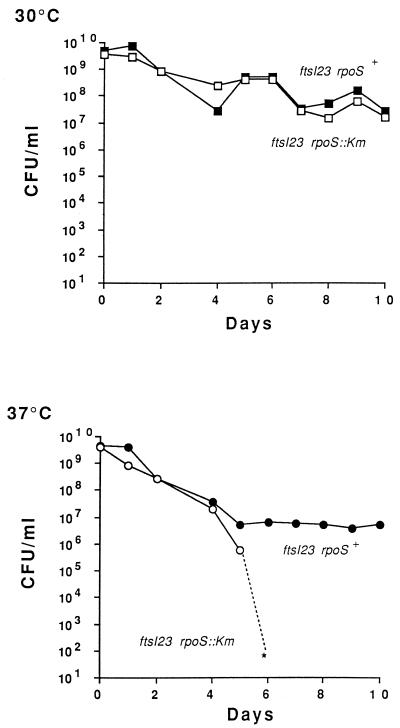
It has been reported that PBP3, PBP7/8, and Slt70 might function together in a complex and that the activity of Slt70 might be modulated by the complex (34). PBP3, encoded by ftsI, forms the cross-links between muropeptide side chains that create the septa of dividing cells (14). Inactivation of PBP3 does not cause cell lysis but rather inhibits cell division, leading to filament formation (14). Since the HPLC data suggested that Slt activity might be increased and since the PBP profile data indicated that the amounts of PBP7/8 were altered, we decided to test whether a mutation in one of the genes of this complex, ftsI, would confer a Sur− phenotype upon the cell. To do this, we tested whether cells lacking ςS and PBP3 lysed during stationary phase. Although loss of PBP3 results in filamentation in growing cells, the effect of losing this enzyme in stationary-phase cultures was unknown. PBP3 is essential for growth, so mutations in the gene encoding PBP3, ftsI, are conditional mutations. These cultures must be maintained at the permissive temperature, 30°C, and switched to 37°C to deplete PBP3 from the cells. The ftsI23 mutation (11) was transduced into our wild-type strain, as well as into the isogenic rpoS::Km mutant. Cultures were grown at 30°C for 15 h, and then the temperature was shifted to 37°C for long-term incubation of the cultures in stationary phase (Fig. 7). Control cell cultures maintained at 30°C displayed normal stationary-phase survival characteristics, similar to what occurred with the wild type (Fig. 7A). Cells in cultures whose temperature was shifted to 37°C behaved differently, mimicking the Sur− phenotype. The ftsI12 rpoS::Km double mutant completely lost viability within 6 to 7 days, similar to what occurred with a surA rpoS mutant (Fig. 7B). No decrease in viability was seen in an rpoS+ background, again similar to what occurred in a surA rpoS+ background (Fig. 7B).
FIG. 7.
Survival of ftsI mutants in stationary phase. E. coli ftsI23 strains containing rpoS (filled symbols) or rpoS::Km (open symbols) were grown at 30°C. The cultures were either maintained at 30°C (top) or switched to 37°C (bottom) after 15 h of incubation. The asterisk represents <102 CFU/ml. This experiment was repeated three times, and representative data are shown.
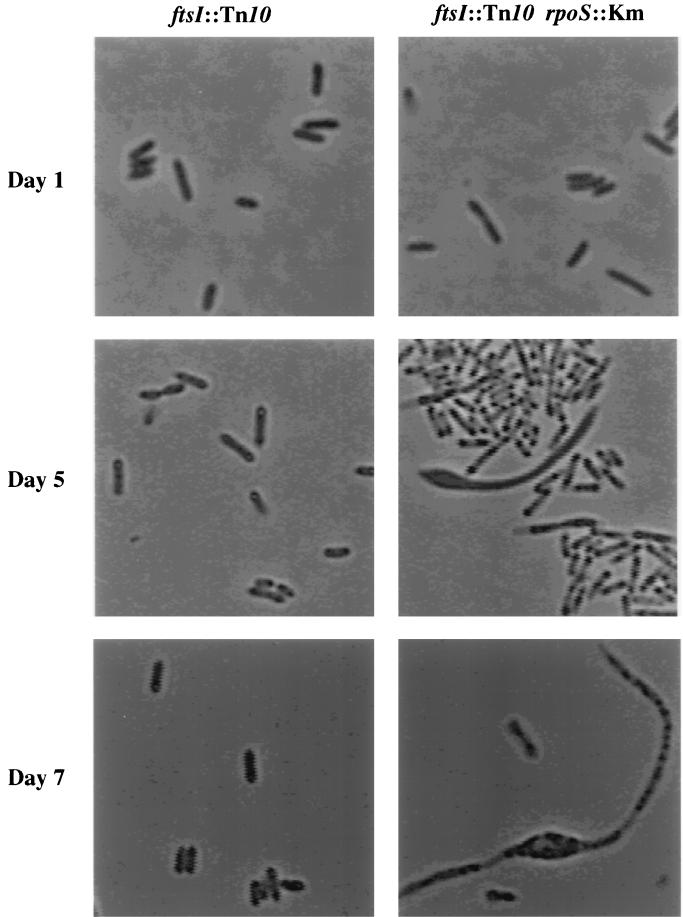
The morphology of the ftsI23 mutants in stationary phase was also similar to that of surA mutants. After the switch to 37°C, in an otherwise wild-type background cells carrying the ftsI23 allele appeared normal (Fig. 8). However, cells from overnight cultures of the double mutant appeared long and thin, reminiscent of the morphology of the ΔsurA3::Km rpoS::Tn10 mutant (Fig. 8). As incubation continued and viability decreased, the cells bulged and lysed, which is again very similar to what occurred with the morphology of the ΔsurA3::Km rpoS::Tn10 mutant (Fig. 8).
FIG. 8.
Morphology of cells lacking ftsI in stationary phase. The morphologies of cells carrying ftsI23 or ftsI23 rpoS::Km at days 1, 5, and 7 of incubation are shown. The cultures were grown at 30°C for 15 h and then switched to 37°C.
DISCUSSION
We have investigated the requirement for SurA in stationary-phase cultures. SurA is expressed in both exponentially growing and nongrowing cells and is not regulated by ςS. In the absence of SurA, some outer membrane proteins do not fold properly (22), resulting in pleiotropic defects, including increased sensitivity to antibiotics and large hydrophobic compounds (21). These phenotypes are observed in both rpoS+ and rpoS mutant cells. Cells lacking SurA grow in LB medium under standard laboratory conditions without noticeable defects. However, cells growing at pH 9 require SurA, presumably to allow nascent outer membrane proteins to fold. The pH of LB medium stationary-phase cultures slowly increases to pH 9, causing the majority of cells in the culture to lose viability. Presumably the death of the majority of cells provides nutrients for the GASP mutants to grow. For wild-type cells or cells lacking either SurA or ςS, this is not a problem. However, loss of both of these proteins prevents the cell from growing at pH 9 by causing cell lysis.
The results presented here indicate that lytic transglycosylase activity is increased in the surA1::mini-Tn10(Km) rpoS819 mutant relative to that in the wild type and that the levels of PBP7/8 and AmpH are altered in the ΔsurA3::Km mutant. Since Slt is believed to be in a complex with PBP7/8 and PBP3, we speculate that in cells lacking SurA this complex is altered, perhaps destabilized, resulting in a disruption of the regulatory interactions between the enzymes. Presumably at high pH this complex or some other enzyme involved in sacculus integrity is lost, resulting in lysis.
The reason for the altered levels of PBPs is unclear. Four enzymes, namely, Slt70, endopeptidases 30 and 49, and PBP3, were tested for the ability to fold in the absence of SurA at neutral pH. These proteins do not require SurA function to fold properly (22; data not shown). Presumably, some other protein which does require SurA to fold alters the stability of the PBP3–PBP7/8–Slt70 complex. Alternatively, these enzymes might require SurA function to fold only at elevated pH.
The changes in peptidoglycan and the PBPs caused by loss of SurA are all observed in exponentially growing cells, at a time when cellular growth and morphology appears normal (data not shown). The morphology and growth defects are observed only in cells growing at alkaline pH in the absence of ςS. Several enzymes involved in cell shape and structure are regulated by ςS, namely, bolA, ftsQAZ, and dacA (3, 7). Mutant cells lacking rpoS have been reported to have altered shape in overnight cultures (19). The combined loss of SurA and ςS presumably results in the altered expression of numerous proteins required for maintaining the cell structure, and loss of these enzymes results in a compromised structure that fails to maintain cell integrity at elevated pH. The lack of ςS-dependent functions that mediate extreme alkali resistance (41) might also contribute to cell lysis of the mutants in stationary phase. Alternatively, ςS may regulate some other folding chaperone that can partially supplem
l2;5qent SurA function.
ACKNOWLEDGMENTS
We thank Elaine Tuomanen for guidance and interpretation of HPLC results, David Weiss for Flu-Apa, the Beckwith lab for strains, and K. Young for strains and sharing information prior to publication. We also thank S. Finkel for critical reading of the manuscript.
This work was supported by a grant from the National Science Foundation.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1992. [Google Scholar]
- 2.Betzner A S, Ferreira L C S, Höltje J-V, Keck W. Control of the activity of the soluble lytic transglycosylase by the stringent response in Escherichia coli. FEMS Microbiol Lett. 1990;67:161–164. doi: 10.1016/0378-1097(90)90187-u. [DOI] [PubMed] [Google Scholar]
- 3.Bohannon D E, Connell N, Keener J, Tormo A, Espinosa-Urgel M, Zambrano M M, Kolter R. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and the role of ς70. Jstrain Bacteriol. 1991;173:4482–4492. doi: 10.1128/jb.173.14.4482-4492.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connell N, Han Z, Moreno F, Kolter R. An E. coli promoter induced by the cessation of growth. Mol Microbiol. 1987;1:195–201. doi: 10.1111/j.1365-2958.1987.tb00512.x. [DOI] [PubMed] [Google Scholar]
- 5.Danese P N, Silhavy T J. The ςE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 6.Danese P N, Snyder W B, Cosma C L, Davis L J B, Silhavy T J. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 7.Dougherty T J, Pucci M J. Penicillin-binding proteins are regulated by rpoS during transitions in growth states of Escherichia coli. Antimicrob Agents Chemother. 1994;38:205–210. doi: 10.1128/aac.38.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glauner B. Separation and quantification of muropeptides with high performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 9.Glauner B, Höltje J V. Growth pattern of the murein sacculus of Escherichia coli. J Biol Chem. 1990;265:18988–18996. [PubMed] [Google Scholar]
- 10.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 11.Hedge P J, Spratt B G. Resistance to β-lactam antibiotics by remodeling the active site of an Escherichia coli penicillin-binding protein. Nature. 1985;318:478–480. doi: 10.1038/318478a0. [DOI] [PubMed] [Google Scholar]
- 12.Henderson T A, Templin M, Young K D. Identification and cloning of the gene encoding penicillin-binding protein 7 of Escherichia coli. J Bacteriol. 1995;177:2074–2079. doi: 10.1128/jb.177.8.2074-2079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson T A, Young K D, Denome S A, Elf P K. AmpC and AmpH, proteins related to the class C beta-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J Bacteriol. 1997;179:6112–6121. doi: 10.1128/jb.179.19.6112-6121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Höltje J-V. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch Microbiol. 1995;164:243–254. doi: 10.1007/BF02529958. [DOI] [PubMed] [Google Scholar]
- 15.Höltje J-V, Mirelman D, Sharon N, Schwarz U. Novel type of murein transglycosylase in Escherichia coli. J Bacteriol. 1975;24:1067–1076. doi: 10.1128/jb.124.3.1067-1076.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horne S M, Young K D. Escherichia coli and other species of the Enterobacteriaceae encode a protein similar to the family of Mip-like FK506-binding proteins. Arch Microbiol. 1995;163:357–365. doi: 10.1007/BF00404209. [DOI] [PubMed] [Google Scholar]
- 17.Ivanova A, Renshaw M, Guntaka R V, Eisenstark A. DNA base sequence variability in katF (putative sigma factor) gene of Escherichia coli. Nucleic Acids Res. 1992;20:5479–5480. doi: 10.1093/nar/20.20.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jishage M, Ishihama A. Variation in RNA polymerase sigma subunit composition with different stocks of Escherichia coli W3110. J Bacteriol. 1997;179:959–963. doi: 10.1128/jb.179.3.959-963.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor ςS (rpoS) J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary phase gene expression in E. coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 21.Lazar S W. Survival of Escherichia coli in stationary phase. Ph.D. thesis. Cambridge, Mass: Harvard University; 1996. [Google Scholar]
- 22.Lazar S W, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Walsh C T. Peptidyl-prolyl cis-trans-isomerase from Escherichia coli: a periplasmic homolog of cyclophilin that is not inhibited by cyclosporin A. Proc Natl Acad Sci USA. 1990;87:4028–4032. doi: 10.1073/pnas.87.11.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loewen P C. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J Bacteriol. 1984;157:622–626. doi: 10.1128/jb.157.2.622-626.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Losick R, Shapiro L. Microbial development. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 26.Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991;5:3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 27.Mengin-Lecreulx D, Van Heijenoort J. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J Bacteriol. 1985;163:208–212. doi: 10.1128/jb.163.1.208-212.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 29.Missiakas D, Betton J-M, Raina S. New components of protein folding in the extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. EMBO J. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 30.Pisabarro A G, de Pedro M A, Vazquez D. Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. J Bacteriol. 1985;161:238–242. doi: 10.1128/jb.161.1.238-242.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pogliano J, Lynch A S, Belin D, Lin E C C, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 32.Popham D L, Setlow P. Phenotypes of Bacillus subtilis mutants lacking multiple class A high molecular weight penicillin-binding-proteins. J Bacteriol. 1996;178:2079–2085. doi: 10.1128/jb.178.7.2079-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romeis T, Höltje J V. Pennicillin-binding protein 7/8 of Escherichia coli is a DD-endopeptidase. Eur J Biochem. 1994;224:597–604. doi: 10.1111/j.1432-1033.1994.00597.x. [DOI] [PubMed] [Google Scholar]
- 34.Romeis T, Höltje J V. Specific interaction of penicillin-binding proteins 3 and 7/8 with soluble lytic transglycosylase in Escherichia coli. J Biol Chem. 1994;269:21603–21607. [PubMed] [Google Scholar]
- 35.Romeis T, Vollmer W, Höltje J-V. Characterization of three different lytic transglycosylases in Escherichia coli. FEMS Microbiol Lett. 1993;111:141–146. doi: 10.1111/j.1574-6968.1993.tb06376.x. [DOI] [PubMed] [Google Scholar]
- 36.Rouviere P E, Gross C A. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- 37.Rudd K E, Sofia H J, Koonin E V, Plunkett III G, Lazar S, Rouviere P E. A new family of peptidyl-prolyl isomerases. Trends Biochem Sci. 1995;20:12–14. doi: 10.1016/s0968-0004(00)88940-9. [DOI] [PubMed] [Google Scholar]
- 38.Satta G, Cornaglia G, Mazzariol A, Golini G, Valisena S, Fontana R. Target for bacteriostatic and bacteriocidal activities of beta-lactam antibiotics against Escherichia coli resides in different penicillin-binding proteins. Antimicrob Agents Chemother. 1995;39:812–818. doi: 10.1128/aac.39.4.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmid F X. Prolyl-isomerases join the fold. Curr Biol. 1995;5:993–994. doi: 10.1016/s0960-9822(95)00197-7. [DOI] [PubMed] [Google Scholar]
- 40.Shapiro L, Kaiser D, Losick R. Development and behavior in bacteria. Cell. 1993;73:835–836. doi: 10.1016/0092-8674(93)90264-q. [DOI] [PubMed] [Google Scholar]
- 41.Small P, Blankerhorn D, Welty D, Zinser E, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith I, Slepecky R A, Setlow P. Regulation of prokaryotic development. Structural and functional analysis of bacterial sporulation and germination. Washington, D.C: American Society for Microbiology; 1989. [Google Scholar]
- 43.Tormo A, Almirón M, Kolter R. surA, an Escherichia coli gene essential for survival in stationary phase. J Bacteriol. 1990;172:4339–4347. doi: 10.1128/jb.172.8.4339-4347.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuomanen E, Cozens R. Changes in peptidoglycan composition and penicillin-binding proteins in slowly growing Escherichia coli. J Bacteriol. 1987;169:5308–5310. doi: 10.1128/jb.169.11.5308-5310.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zambrano M M, Kolter R. Escherichia coli mutants lacking NADH dehydrogenase I have a competitive disadvantage in stationary phase. J Bacteriol. 1993;175:5642–5647. doi: 10.1128/jb.175.17.5642-5647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zambrano M M, Kolter R. GASPing for life in stationary phase. Cell. 1996;86:181–184. doi: 10.1016/s0092-8674(00)80089-6. [DOI] [PubMed] [Google Scholar]
- 47.Zambrano M M, Siegele D A, Almirón M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]