Abstract
Both the APOE ε4 and TOMM40 rs10524523 (‘523’) genes have been associated with risk for Alzheimer’s disease (AD) and neuroimaging biomarkers of AD. No studies have investigated the relationship of TOMM40’523-APOE ε4 on the structural complexity of the brain in AD individuals. We used a comprehensive approach to quantify brain morphology and multiple cortical attributes in individuals with mild cognitive impairment (MCI) and AD (n=94), then tested whether APOE ε4 or TOMM40 poly-T genotypes were related to AD morphological biomarkers in cognitively unimpaired (CU, n=148) and MCI/AD individuals. We identified several AD-specific phenotypes in brain morphology and found that TOMM40 poly-T short alleles are associated with early, AD-specific brain morphological differences in healthy aging. We observed decreased cortical thickness, sulcal depth and fractal dimension in CU individuals with the poly-T short alleles. Moreover, in MCI/AD participants, the APOE ε4 (TOMM40 L) individuals had a higher rate of gene-related morphological markers indicative of AD. Our data suggest that TOMM40’523 is associated with early brain structure variations in the precuneus, temporal and limbic cortices.
Keywords: TOMM40, APOE, Poly-T, Cortical Thickness, Alzheimer’s Disease, gray matter volume, neuroimage, mitochondria
1. Introduction
Late-onset Alzheimer’s disease (LOAD) is a complex and progressive neurodegenerative disease, and large genome-wide association studies (GWAS) have identified more than 20 independent loci associated with AD (Bellenguez et al., 2020; Kunkle et al., 2019). The ε4 allele of the Apolipoprotein E gene (APOE) is located on chromosome 19 and is the most known genetic risk factor for LOAD (Coon et al., 2007). TOMM40, or Translocase of the Mitochondrial Membrane 40, is a close neighbor to APOE (2104 base pairs away) and in linkage disequilibrium with APOE (Lyall et al., 2013). TOMM40 is one of several genes on chromosome 19 (APOE, TOMM40, APOC1, APOC1PA) that are in high linkage disequilibrium and have interactive effects on AD risk (Kulminski et al., 2022b; Ortega-Rojas et al., 2022). The haplotype of TOMM40, APOE, and APOC1 has been described as a longevity hub (Torres et al., 2022), yet there is little data to help understand how these genes interact independently or together in relation to AD risk.
Sequence variants in both TOMM40 and APOE have been associated with cognitive aging, longevity, aging-related brain structure and function biomarkers, as well as possible genetic contribution to the “mitochondrial cascade hypothesis” (Swerdlow and Khan, 2004, 2009). The mitochondrial cascade hypothesis suggests that multiple interacting factors impact baseline and age-related decline in mitochondrial function (Swerdlow, 2012). APOE’s mechanism contributing to risk for AD most likely involves mediation of cell and tissue lipid transport, however the APOE ε4 allele is not always a determinant for AD risk. TOMM40’s mechanism contributing to risk for AD is most likely a disruption of cellular bioenergetics in the mitochondria, but it is unclear if this contribution is independent of APOE.
A common polymorphism of TOMM40 is the TOMM40 variable length homopolymeric T variation, or poly-T polymorphism, at rs10524523 (“TOMM40’523”). The alleles on TOMM40’523 are grouped into short (S), long (L), and very long (VL) depending on the T length (Roses et al., 2010). A number of studies have used neuroimaging and cognitive measures in cognitively unimpaired (CU) individuals to tease out independent effects of TOMM40’523 markers of aging and risk for AD (reviewed in (Chiba-Falek et al., 2018)). TOMM40 poly-T allelic variations have been related to the acceleration of Alzheimer’s disease (Yu et al., 2017a; Yu et al., 2017b) and the VL allele has been associated with hippocampal thickness (Burggren et al., 2017), gray matter volume(Johnson et al., 2011), and cognitive decline (Caselli et al., 2012; Greenbaum et al., 2014; Hayden et al., 2012; Johnson et al., 2011) in APOE ε3 carriers. To complicate things, other studies have shown that the VL variant is associated with lower risk for AD (Cruchaga et al., 2011; Jun et al., 2012). Most studies have limited their sample to APOE ε3 carriers due to the linkage equilibrium between the L and the APOE ε4 in Caucasian samples, and most have focused on hippocampal volumetrics or diffusion tensor imaging of white matter with mixed results. Given the evidence from transcriptomic data that TOMM40 may functionally impact temporal cortex thickness (Varathan et al., 2022), a goal of this study was to more comprehensively assess effects of the APOE-“523” haplotype on regional complexity of cortical shape and volume in cognitively unimpaired and MCI/AD individuals.
Structural MRI of the brain is a widely used biomarker of neurodegeneration in AD, and there a growing number of additional features of structural MRI that may provide more sensitive phenotypes to smaller genetic effects in the brain. For instance, quantification of local fractal dimension measures (FD) that use spherical harmonic reconstructions provide more detailed information about the complexity of cortical folding (Yotter et al., 2011a; Yotter et al., 2011b). Fractal dimension can detect significant variations in structural complexity of gray matter in healthy aging and neurological disease, with the complexity decreasing with age (King et al., 2010; Liu et al., 2020; Marzi et al., 2020). Moreover, measurements of cortical thinning have identified distinct regional patterns in AD (Choi et al., 2019; Du et al., 2007), and are present in early stages of cognitive decline (Kalin et al., 2017). Gyrification index quantitatively measures a ratio of total pial surface area to superficial cortical surface area, providing a measure of cortical folding and subsequent changes during atrophy. Sulcal depth measures the Euclidean distance between pial and outer surfaces (Yun et al., 2013) and may be sensitive to the detection of MCI (Im et al., 2008). No previous study has investigated the relationship of APOE and TOMM40 on the structural complexity of the brain using fractal dimension, gyrification index, or sulcal depth, for which there may be subtle differences in mitochondrial-related gene changes. Moreover, no previous study has compared the impact of TOMM40’523-APOE ε4 genotypes on brain imaging phenotypes in people with MCI/AD.
We aimed to use whole brain voxel-based (VBM) and surface-based morphology (SBM) methods test whether APOE ε4 or TOMM40 genetic variation differentially impacted AD-related measures of volume, cortical thickness, sulcal depth, fractal dimension, and gyrification index. Based on structural imaging studies on APOE ε4 and TOMM40 to date, we hypothesized that presence of an APOE ε4 and/or APOE ε4− TOMM40 VL allele, regardless of diagnosis, would be related to reduced volume, thinner cortex, shallower sulcal depth, reduced fractal dimension, and lower gyrification index in AD-related temporal and parietal regions compared to TOMM40 S-carriers.
2. Materials and Methods
2.1. Standard Protocol Approvals, Registrations, and Patient Consents
Study procedures were approved by the University of Kansas School of Medicine Institutional Review Board and were in accordance with U.S. federal regulations. All participants provided written informed consent.
2.2. Participants
Participants were recruited as part of intervention and observational studies at the University of Kansas Alzheimer’s Disease Center (KU ADC) and were part of the Clinical Cohort. We have previously reported results from these investigations (Morris et al., 2017; Morris et al., 2020; Vidoni et al., 2015). The KUADC collects longitudinal data on a clinical cohort that includes over 400 individuals. The cohort includes participants with cognitive impairment as well as healthy cognition. Individuals who were cognitively unimpaired (CU) were included at age 60 and older, while individuals with AD were included regardless of age. The Uniform Data Set (UDS) was created in 2005 to collect standard clinical data on participants from the National Institute on Aging (NIA)-supported Alzheimer’s Disease Centers (ADCs). The UDS is administered to ADC Clinical Cohort participants on an approximately annual basis.
The KU ADRC is part of the U.S. network of Alzheimer’s Disease Centers of Excellence that support research into brain aging and dementia. The KU ADRC has established an infrastructure for the identification, recruitment, and characterization of older adults both with and without dementia. Beginning in 2004, we developed a registry of individuals who have consented to be contacted regarding research studies, details of which have been published elsewhere (Vidoni et al., 2012). 242 participants underwent brain imaging as part of these ongoing observational and intervention-based studies (pre-intervention timepoint only) on fitness, exercise, aging and risk for AD (ClinicalTrials.gov: NCT01129115, NCT02000583, NCT00267124).
All participants also underwent a standard examination, which includes a thorough clinical and cognitive evaluation with a clinician at the KU ADC. This clinical evaluation includes a semi-structured interview (Clinical Dementia Rating, CDR) with the participant and study partner (Morris, 1993), as well as a physical and neurological examination. Clinical evaluation results were used to determine dementia status, which were reviewed along with psychometric battery results and finalized at a consensus diagnostic conference attended by clinicians and psychometricians using the NINCDS-ADRDA criteria as well as the McKann NIA-AA workgroup diagnostic guidelines (Beach et al., 2012; McKhann et al., 2011). Diagnostic criteria for AD require the gradual onset and progression of impairment in memory and in at least one other cognitive and functional domain on the CDR. MCI was diagnosed by a clinician and verified with medical records. Individuals were excluded from participating if they had other neurological disorders that could impair cognition, evidence of bleeding disorders during screening, clinically significant disease, psychiatric disorder, systemic illness, stroke, or myocardial infarction.
A psychometrician administered a standard psychometric battery as described in previous publication (Weintraub et al., 2009). The cognitive test battery included logical memory IA and IIA, selective reminding test, digit span forward and backward, letter number sequencing, trailmaking B, category fluency, the digit symbol substitution test, and the interference condition from the Stroop test. As published previously, we used Mplus to combine test scores into cognitive domain specific factor scores using confirmatory factor analysis, and specific tests were organized by whether they measured attention, verbal memory, or executive function(Watts et al., 2019). Tests of attention included digits forward, digits backward, and letter-number sequencing. Tests of verbal memory included immediate and delayed logical memory, as well as the sum of three selective reminding trials. Tests of executive function assessed set maintenance and set shifting and included category fluency (sum of animal and vegetable categories), Stroop color word interference, trailmaking test B, and the digit symbol substitution test. We included domain-specific factor scores as variables in our demographics analysis. Other measured covariates included the Geriatric Depression Scale (GDS) and Mini-Mental State Examination (MMSE). We characterized ‘Age at first decline’ as the age identified by the clinician when the decline first began, and is not the same as age of disease onset. The decline may have occurred in cognition, behavior or motor skills.
2.3. Family History and Genotyping Procedures
Participants completed thorough family history examinations using a standard family history questionnaire, as has been described elsewhere(Honea et al.; Xiong et al., 2011). Briefly, a family history of dementia included at least one first-degree relative whose dementia onset was between the ages of 60 and 80 years. Participants (or their caregivers or study partners in the case of those with MCI and AD) self-reported relationships, dates of birth, age at death, age at onset of disease, and clinical information of affected and unaffected family members. Participants family history data were not included if both of their parents had not lived to the average age at risk of Late-onset AD (i.e., 60 years) and a true code could not be assigned.
Determination of APOE genotype was performed by the National Cell Repository for Alzheimer’s Disease (NCRAD), with independent verification of selected samples by the KUADC Mitochondrial Genomics and Metabolism Core using a previously described allelic discrimination assay(Wilkins et al., 2017). TOMM40’523 genotyping was performed by Polymorphic DNA technologies (PDT) with independent verification performed as with APOE. Poly-T length reproducibility between the PDT and KUADC measurements was uniformly within 1 T. Blood samples for genotyping were available for 210 of the 242 participants.
For TOMM40, the lengths of the poly-T length polymorphisms at “523” were classified as short (14–20 T residues; i.e. ‘S’), long (21–29 T residues, i.e., ‘L’) or very long (>29 T residues, i.e., ‘VL’), as has been done in the literature(Roses et al., 2010). Examination of poly-T length by APOE allele in this sample has been published already(Watts et al., 2019). For comparisons of APOE genotype, participants were categorized into two groups: those carrying at least one ε4 allele (ε4+) and those without any ε4 allele (ε4-). The TOMM40 L variant is almost exclusively linked to the APOE ε4 allele while the VL and S variants are in strong linkage disequilibrium with APOE ε3, participants were further classified into the following 4 groups: those homozygous for S (TOMM40 S/S), homozygous for VL (TOMM40 VL/VL, excluding those with an APOE ε4 allele), heterozygous (TOMM40 S/VL), and APOE ε4 carriers (primarily made up of TOMM40 L variant carriers).
2.4. Structural Brain Imaging Acquisition
All participants coming through neuroimaging studies at the KUADC underwent magnetic resonance imaging (MRI) of the brain in either a Siemens 3.0 Tesla Allegra or Skyra scanner. We obtained a high resolution T1-weighted image (MP-RAGE; 1×1×1mm voxels; TR=2500ms, TE=4.38ms, TI=1100, FOV=256X256 with 18% oversample, 1mm slice thickness, flip angle 8deg) for detailed anatomy with high gray-white matter contrast. We did cortical surface-based (estimation of cortical thickness, complexity of cortical folding based on fractal dimension (FD), gyrification index, and sulcal depth) analyses along with VBM and region of interest analyses. Every scan was checked for image artifacts and gross anatomical abnormalities. 242 individuals with MPRAGE scans passed quality control. The overall time between the cognitive assessment and brain scans ranged from 4–6 weeks. There were no statistical differences in scanner type between TOMM40’523-APOE ε4 groups, thus we did not include it as a variable of interest in our analysis.
2.5. Voxel-Based and Surface-Based Morphometry
For VBM and SBM analysis and pre-processing of T1-weighted images, we used the Computational Anatomical Toolbox 12 (CAT12 Version 12.6, C. Gaser, Structural Brain Mapping Group, Jena University Hospital, Jena, Germany; http://dbm.neuro.uni-jena.de/cat/) through Statistical Parametric Mapping version 12 (SPM12; Wellcome Trust Centre for Neuroimaging, London, UK; http://www.fil.ion.ucl.ac.uk/spm/software/spm12/)) that operate under Matlab (R2019b) (the Mathworks, Natick, MA) on Mac. This was used for brain volume (VBM), and surface-based measures such as cortical thickness (CT), sulcal density (SD), GI (gyrification index) and fractal dimension (FD). All the SBM procedures (http://www.neuro.uni-jena.de/cat12/CAT12-Manual.pdf) were conducted using default settings.
T1 images were corrected for bias-field inhomogeneities, registered using linear (12-parameter affine) and non-linear transformations, spatially normalized using the high-dimensional DARTEL algorithm into MNI space(Ashburner, 2007), and segmented into gray matter (GM), white matter (WM), cerebrospinal fluid (CSF) and white matter hyperintensity (WMH). We calculated total intracranial volume (TIV) using total gray, white, and CSF volumes. We used TIV, and TIV-adjusted GM, WM, and WMH in our comparison between diagnostic groups and TOMM40’523-APOE ε4 haplogroups. The amount of volume changes were scaled, in order to retain the original local volumes (modulating the segmentations) (Good et al., 2001). The modulated gray matter segmentations were smoothed using a 10 × 10 × 10 mm full-width at half-maximum Gaussian kernel prior to group level voxel-wise analysis.
2.5.1. VBM- Statistical Analysis
For all analyses, voxels are reported with reference to the MNI standard space within SPM12. To avoid possible edge effects at the border between GM and WM and to include only relatively homogeneous voxels, we used an absolute threshold masking of 0.10 for each analysis. In order to investigate associations between TOMM40’523-APOE ε4 groups and gray matter volume differences we included age, sex, and total intracranial volume (TIV), as variables of no interest in our full factorial model. Statistics were done in imaging space across all voxels. A full-factorial analysis was done comparing 1) CU and CI groups, 2) the 4 genotype groups within CU individuals and 3) the 4 genotype groups within CI individuals. The statistical threshold was set at Pheight < 0.05, family wise error (FWE) corrected, with a minimum cluster size of 100 voxels (k>100) for all analyses. Anatomical labeling from the Wakeforest Pickatlas AAL atlas was used to identify peak coordinate regions.
2.5.2. SBM- Statistical Analysis
We followed the workflow specified in CAT12 (Dahnke et al., 2013). This workflow comprises tissue segmentation to estimate the white matter distance from gray matter, which is then used to project the local maxima to other gray matter voxels. Resampled surface data for cortical thickness (CT), fractal dimension (FD) and sulcal depth (SD) were smoothed using a 15mm FWHM kernel, and data for gyrification were smoothed using a 20mm FWHM kernel, prior to 2nd level analyses. In 2nd level between-group t-tests we computed vertex-wise analyses across each hemisphere. A full-factorial analysis was done comparing 1) CU and CI groups, 2) the 4 genotype groups within CU individuals and 3) the 4 genotype groups within CI individuals. Statistical significance was based on FWE corrected cluster-level and/or peak level threshold of p<.05 with a cluster extent threshold of 50 vertices. Clusters surviving the uncorrected peak threshold at p<.001 were also included in our results given the exploratory nature of this analysis. The Desikan-Killiany (Desikan et al., 2006) atlas was used for SBM to extract mean regional values from the processed images after voxel-wise analysis.
2.6. Statistical Analyses- Demographic and Sample Characteristics
SPSS 22.0 (IBM Corp., Armonk, NY) was used for the statistical analyses performed outside of imaging space. Continuous demographic, cognitive, and volumetric imaging variables (dependent variables) were compared first between the diagnostic groups (CU and MCI/AD combined), then between the TOMM40’523-APOE ε4 groups within the CU sample (the independent variables) using the one-way multivariate analysis of covariance (MANCOVA) for the descriptive statistics. A chi-square analysis was used to compare categorical demographic variables between groups. Cohen’s κ coefficients (Cohen, 1960) evaluated the concordance between APOE ε4 and TOMM40’523-L genotypes. We included participants’ age, sex, and years of education as covariates in the MANCOVA when testing factor cognitive scores and brain volumes. Raw p-values <0.05 were considered to be nominally significant.
3. Results
3.1. Demographics and Group Tables
Diagnostic groups were not significantly different in mean age, education, and geriatric depression scale scores (GDS). Diagnostic groups were significantly different in MMSE, CDR, sex, cognitive factor scores in verbal, attention, and executive function domains, and overall normalized brain volumes (Table 1). There was no significant difference between family history positivity between the groups.
Table 1.
Sample Participant Characteristics by Diagnosis
| CU | MCI/AD | p-value | Effect size | |
|---|---|---|---|---|
|
|
||||
| N=242 | 148 | 94 | ||
|
| ||||
| Years of Age (SD) | 74.5 (6.1) | 74.1 (7.4) | 0.688 | 0.001 |
| Education (years) | 16.4 (2.9) | 15.88 (3.2) | 0.206 | 0.007 |
| Sex (M/F) | 58/90 | 57/37 | 0.001 | 0.209 |
| MMSE | 29.21 (.985) | 24.82 (3.7) | <.001 | 0.431 |
| CDR (0/.5/1.0/2.0) | 148/0/0/0 | 12/50/29/2 | <.001 | 0.898 |
| GDS Score | .973 (1.6) | 1.04 (1.6) | 0.743 | 0.000 |
| Verbal Memory Factor | 1.04 (.89) | −1.51 (1.3) | <.001 | 0.577 |
| Attention Factor | .264 (.38) | −.37 (.63) | <.001 | 0.291 |
| Executive Function Factor | .551 (.57) | −.92 (.96) | <.001 | 0.483 |
| Total Intracranial Volume (TIV, mm3) | 1383.46 (146) | 1397.14 (150.8) | 0.485 | 0.002 |
| Gray Matter Volume, TIV adjusted (mm3) | .404 (.03) | .373 (.03) | <.001 | 0.245 |
| White Matter Volume, TIV adjusted (mm3) | .338 (.02) | .327 (.03) | <.001 | 0.048 |
| White Matter Hyperintensity (mm3) | 5.19 (7.9) | 7.6 (8.7) | 0.023 | 0.021 |
| FH (−/FHm/FHp/FHboth) (N=124) | 28/29/11/12 | 18/15/6/5 | 0.901 | 0.068 |
| FH (−/+) (N=124) | 29/52 | 18/26 | 0.515 | 0.059 |
| TOMM40/APOE Haplotype (N=210) | ||||
| APOEε4 Carrier (−/+) | 105/43 | 44/50 | <.001 | 0.242 |
| ε4− S/S: ε4− S/VL: ε4− VL/VL: ε4+ Carrier | 29/40/17/43 | 7/16/8/50 | <.001 | 0.288 |
Demographic, neuropsychological, and MRI characteristics of the overall diagnostic groups. Values are mean (SD (standard deviation)) except for sex and age range. Effect sizes indicating differences among the groups were calculated partial eta2 for ANOVA, and Cramer’s V for Pearson Chi-Square which was used for categorical variables. R; Right, L; Left, FH; family history of dementia, +; positive family history of dementia, −; negative family history of dementia, M; male, F; female, TIV; Total Intracranial Volume, mm; millimeter, MMSE; Mini-Mental Status Exam, GDS; Geriatric Depression Score, N; number, FHm; maternal family history of dementia, FHp; paternal family history of dementia, FHboth; maternal and paternal family history of dementia. For p indicating the level of significance compared with CU group are
p< 0.05;
p < 0.01,
p<0.001.
In the total sample across all diagnostic groups, APOE had genotype frequencies of: ε2/ε2 = 3 (1.2%), ε2/ε3 = 24 (9.9%), ε3/ε3 = 121 (50%), ε3/ε4 = 74 (30.6%), ε2/ε4 = 3 (1.2%), ε4/ε4 = 17 (7%) (total = 242). TOMM40’523 had genotype frequencies of S/S = 38 (15.7%), S/L = 34 (14%), S/VL = 57 (23.6%), L/L = 13 (5.4%), L/VL = 24 (9.9%), VL/VL = 28 (11.6%) (Histogram of distribution shown in Figure 1). The frequencies of APOE and TOMM40’523 genotypes for individuals with both genotypes, broken down by APOE ε4, are shown on Table 2. Consistent with linkage between ε4 and ‘523-L, very few of the ε4-non carriers had a ‘523-L (1.7%), whereas 89.6% of the ε4 carriers had an ‘523-L. The classification was highly concordant (Cohen’s κ = .899, standard error =.031). As expected, there was a significantly higher (p<.001) proportion of ε4-Carriers in individuals with MCI/AD (53%) than in individuals who were CU (29%).
Figure 1.
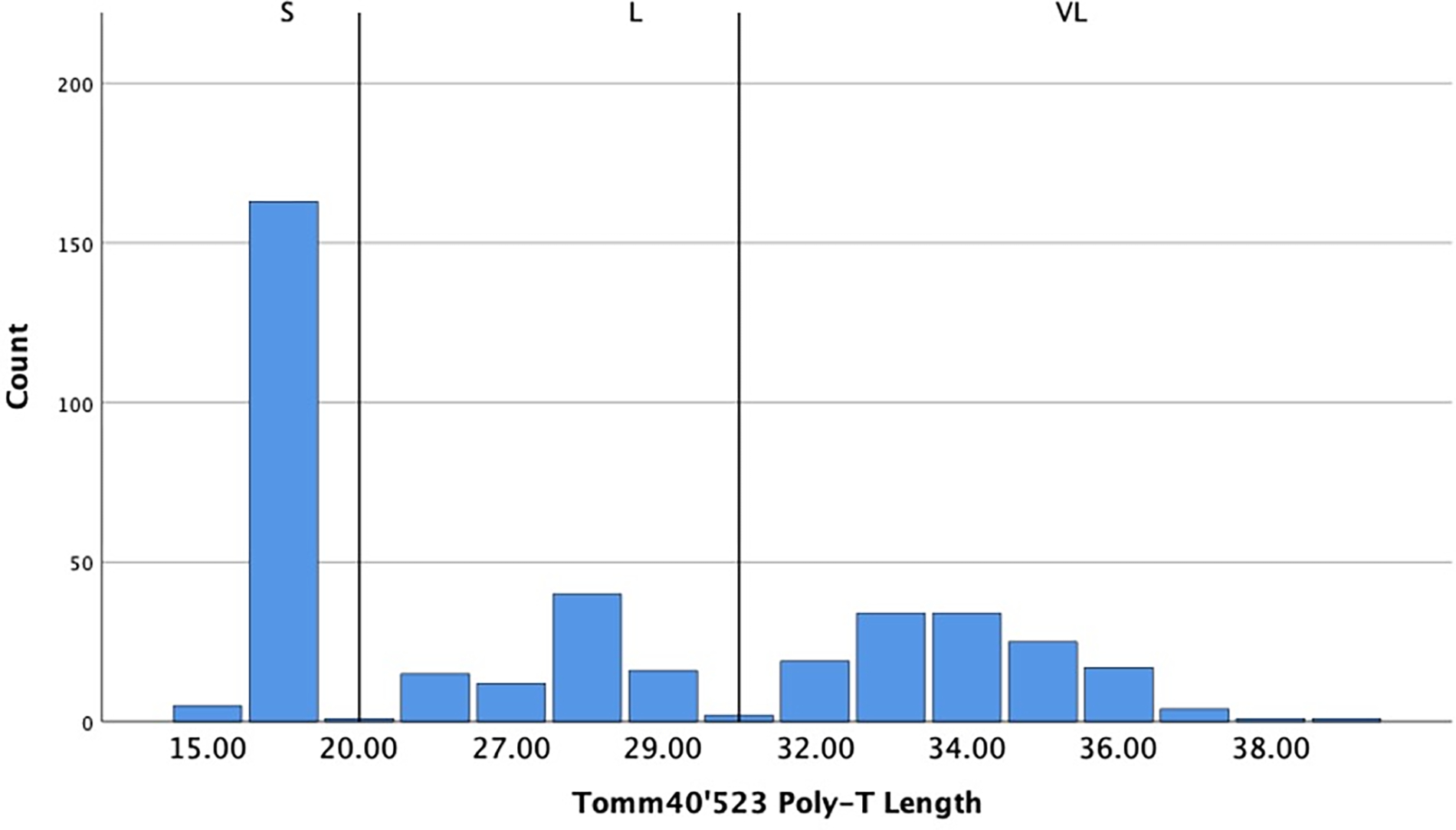
T-repeat distribution of TOMM40’523 poly-T lengths counts classified into short ((14–20 T residues; i.e. ‘S’), long (21–29 T residues, i.e., ‘L’) or very long (>29 T residues, i.e., ‘VL’).
Table 2.
Sample distribution counts of TOMM40’523 poly-T lengths and APOE ε4 genotypes by APOE ε4 across whole sample
| APOE Genotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| Non ε4 carriers | ε4 carriers | ||||||||||
|
|
|
||||||||||
| TOMM40’523 Genotype | 22 | 32 | 33 | Frequency | % | 34 | 42 | 44 | Frequency | % | Total |
| S/S | 0 | 7 | 28 | 35 | 29.9 | 1 | 1 | 1 | 3 | 3.9 | 38 |
| S/L | 0 | 0 | 0 | 0 | 0 | 33 | 1 | 0 | 34 | 44.2 | 34 |
| L/L | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 13 | 16.8 | 13 |
| L/VL | 0 | 0 | 2 | 2 | 1.7 | 19 | 1 | 2 | 22 | 28.6 | 24 |
| S/VL | 2 | 11 | 42 | 55 | 47 | 2 | 0 | 0 | 2 | 2.6 | 57 |
| VL/VL | 0 | 3 | 22 | 25 | 21.4 | 3 | 0 | 0 | 3 | 3.9 | 28 |
|
|
|
||||||||||
| Total | 2 | 21 | 94 | 117 | 100 | 58 | 3 | 16 | 77 | 100 | 194 |
In the CU sample used for VBM and SBM analysis, TOMM40’523-APOE ε4 genotype groups were significantly different in age (ε4− S/S older than ε4+ group, p=.01) and education (ε4+ group had fewer years of education than the VL/VL group, p=.032, Table 3). Genotype groups were not significantly different in sex, MMSE, GDS score, family history, verbal memory, attention, and executive function factor score. There were no significant differences in overall brain volumes except for TIV-adjusted white matter volume (ε4− VL/VL group had significantly less white matter volume than the ε4− S/VL group, p=.049).
Table 3.
Demographic variables for CU TOMM40’523-APOE genotype groups
| CU Individuals from VBM and SBM analysis | ε4− S/S | ε4−S/VL | ε4− VL/VL | ε4+ | p value | Effect Size |
|---|---|---|---|---|---|---|
|
| ||||||
| N=129 | 29 | 40 | 17 | 43 | ||
|
| ||||||
| Years of Age (SD) | 76.7 (5.9) | 73.6 (5.5) | 76.2 (7.5) | 73.2 (4.9) | 0.032 | 0.068 |
| Education (years) | 16.5 (3.3) | 16.5 (2.5) | 17.9 (3.1) | 15.2 (2.8) | 0.01 | 0.086 |
| Sex (M/F) | 13/16 | 14/26 | 12-May | 15/28 | 0.723 | 0.101 |
| MMSE | 29.17 (.928) | 29.50 (.751) | 29.06 (1.1) | 29.14 (.99) | 0.99 | 0.034 |
| GDS Score | .82 (1.3) | 1.2 (2.1) | .47 (.72) | 1.0 (1.5) | 0.233 | 0.034 |
| Verbal Memory Factor | .95 (.95) | 1.29 (.86) | .83 (1.03) | 1.03 (.79) | 0.296 | 0.03 |
| Attention Factor | .235 (.33) | .229 (.39) | .379 (.48) | .286 (.37) | 0.408 | 0.023 |
| Executive Function Factor | .66 (.55) | .49 (.62) | .63 (.68) | .57 (.51) | 0.198 | 0.037 |
| Total Intracranial Volume (mm3) | 1403 (160) | 1364 (157) | 1402 (139) | 1367 (136) | 0.566 | 0.016 |
| Gray Matter Volume, TIV adjusted (mm3) | .397 (.02) | .408 (.03) | .401 (.02) | .210 (.03) | 0.746 | 0.01 |
| White Matter Volume, TIV adjusted (mm3) | .336 (.02) | .346 (.02) | .327 (.02) | .338 (.02) | 0.049 | 0.062 |
| White Matter Hyperintensity (mm3) | 7.1 (10.2) | 3.5 (2.9) | 7.2 (11.4) | 3.6 (3.6) | 0.273 | 0.031 |
| FH (FH+/FH-) | 9-Jun | 13/9 | 4-Aug | 21/5 | 0.066 | .031 |
Demographic, neuropsychological, and MRI characteristics of the CU individuals from the VBM and SBM analysis. Values are mean (SD (standard deviation)) except for sex and age range. Effect sizes indicating differences among the groups were calculated partial eta2 for ANOVA, and Cramer’s V for Pearson Chi-Square which was used for categorical variables, and covariates included age, sex, and education for univariate analysis. R; Right, L; Left, FH; family history of dementia, FH+; positive family history of dementia, FH−; negative family history of dementia, M; male, F; female, TIV; Total Intracranial Volume, mm; millimeter, MMSE; Mini-Mental Status Exam, GDS; Geriatric Depression Score, N; number. Significant values in bold.
In the MCI/AD sample used for VBM and SBM analysis, TOMM40’523-APOE ε4 genotype groups were not significantly different in age, education, sex, or GDS Score. There was a significant difference between the groups on MMSE score (E4 group had lower MMSE than the ε4− S/VL group, p=.049 Bonferroni corrected, Table 4). Genotype groups were not significantly different on Attention and Executive Function factor scores, however there was a significant (p=.049) difference in Verbal Memory Factor with the ε4+ group having the lowest scores. There were no significant differences in age at first cognitive decline, however notably the ε4− S/S group had a younger age of decline (65.6) than the other groups. There were no significant differences in overall brain volumes between the groups.
Table 4.
Demographic variables for Cognitively Impaired (MCI/AD) TOMM40’523-APOE genotype groups
| MCI/AD Individuals from VBM and SBM analysis | ε4− S/S | ε4− S/VL | ε4− VL/VL | ε4−+ | p value | Effect Size |
|---|---|---|---|---|---|---|
|
| ||||||
| N=81 | 7 | 16 | 8 | 50 | ||
|
| ||||||
| Years of Age (SD) | 74.4 (6.5) | 76.4 (8.6) | 76.7 (5.6) | 73.2 (7.3) | 0.306 | 0.047 |
| Education (years) | 17.1 (1.2) | 15.3 (3.2) | 16.3 (4.5) | 15.5 (3.1) | 0.642 | 0.022 |
| Sex (M/F) | 5/2 | 11/5 | 7/1 | 26/24 | 0.077 | 0.242 |
| MMSE | 26.71 (2.7) | 26.56 (2.5) | 25.63 (3.5) | 23.64 (4.1) | 0.026 | 0.116 |
| GDS Score | 2.14 (2.9) | 1.37 (1.8) | 1.62 (2.7) | .82 (1.1) | 0.116 | 0.065 |
| Verbal Memory Factor | −.63 (1.5) | −1.32 (1.4) | −1.1 (1.4) | −1.9 (1.1) | 0.049 | 0.099 |
| Attention Factor | −.16 (.81) | −.32 (.47) | −.26 (.64) | −.50 (.66) | 0.469 | 0.033 |
| Executive Function Factor | −.31 (.86) | −.72 (1.1) | −.74 (.87) | −1.21 (.89) | 0.105 | 0.079 |
| Age at first cognitive Decline~ | 65.6 (3.5) | 71.3 (9.8) | 70.7 (6.6) | 68.8 (7.8) | 0.513 | 0.033 |
| Total Intracranial Volume (mm3) | 1496 (129) | 1426 (141) | 1440 (125) | 1381 (161) | 0.485 | 0.032 |
| Gray Matter Volume, TIV adjusted (mm3) | .375 (.02) | .366 (.03) | .373 (.04) | .370 (.03) | 0.717 | 0.018 |
| White Matter Volume, TIV adjusted (mm3) | .325 (.03) | .328 (.03) | .321 (.03) | .328 (.03) | 0.761 | 0.016 |
| White Matter Hyperintensity (mm3) | 7.1 (7.6) | 11.8 (12.0) | 6.2 (4.9) | 7.5 (8.6) | 0.408 | 0.038 |
| FH (FH+/FH-) | 0/1 | 4/4 | 4/2 | 13/7 | 0.55 | 0.23 |
Demographic, neuropsychological, and MRI characteristics of the MCI/AD individuals from the VBM and SBM analysis. Values are mean (SD) except for sex and age range. Effect sizes indicating differences among the groups were calculated partial eta2 for ANOVA, and Cramer’s V for Pearson Chi-Square which was used for categorical variables, and covariates included age, sex, and education for univariate analysis. For age at first decline, n= (6, 11, 6, 50) respectively. R; Right, L; Left, FH; family history of dementia, +; positive family history of dementia, −; negative family history of dementia, M; male, F; female, TIV; Total Intracranial Volume, mm; millimeter, MMSE; Mini-Mental Status Exam, GDS; Geriatric Depression Score, N; number. Significant values in bold.
3.2. Voxel and Surface Based Morphometry between Diagnostic groups
In the voxel-based analysis of gray matter volume across all diagnostic groups, we found that individuals with MCI/ AD (Table 5) had significantly decreased volume in the medial temporal complex, specifically the left parahippocampal gyrus and hippocampus, the bilateral temporal cortex and bilateral middle frontal cortex (Table 5, Figure 2A; p<.05 FWE corrected). The most powerful surface-based phenotype for discriminating between participant groups based on number of clusters and T-score in our sample was cortical thickness (CT), which was decreased in people with AD in the bilateral parahippocampal gyrus, bilateral precuneus, superior temporal and inferior temporal cortices, precentral and paracentral gyrus, and the middle frontal gyrus (Table 5, Figure 2E; p<.05 FWE corrected). Individuals with MCI/AD had decreases of sulcal depth (SD) in primarily the left and right parahippocampal gyrus, right superior temporal gyrus, and insula (Table 5, Figure 2D; p<.05 FWE corrected). Individuals with MCI/AD had a significantly smaller gyrification index in the insula, but a larger gyrification index in the left parahippocampal gyrus than CU (Table 5, Figure 2C; p<.05 FWE corrected). Individuals with AD had less fractal dimension (or less cortical complexity) in the right rectal gyrus, the posterior cingulate, the bilateral insula, and right middle frontal gyrus (Table 5, Figure 2B; p<.05 FWE corrected).
Table 5.
Voxel Based and Surface Based Morphometry results between Diagnosis Groups in MCI/AD compared to CU, as well as increased (red, bilateral insula) in MCI/AD compared to CU. L; left, R; right. Color to be used in print.
| Comparison | Size (vertexes) | Peak t value | Peak-level p value (FWE Corrected) | Coordinates (mm mm mm) | Brain Region |
|---|---|---|---|---|---|
|
| |||||
| Volume | |||||
|
| |||||
| CU > MCI/AD | 149895 | 14.19 | <.001 | −28 –22 -20 | Left Parahippocampal Gyrus/Hippocampus/Medial Temporal Cortex |
| 1911 | 6.26 | <.001 | 28 –6 48 | Right Precentral Gyrus | |
| 714 | 5.74 | <.001 | −28 50 12 | Left Middle Frontal Gyrus | |
| 384 | 5.53 | <.001 | 26 38 27 | Right Middle Frontal Gyrus | |
| 694 | 5.2 | .002 | −27 –3 52 | Left Middle Frontal Gyrus | |
| 1294 | 5.18 | .002 | −28 –76 -52 | Right Cerebelum | |
| 144 | 4.75 | .013 | −34 8 38 | Left Middle Frontal Gyrus | |
|
| |||||
| Cortical Thickness | |||||
|
| |||||
| CU > MCI/AD | 4062 | 11.5 | <.001 | −25 –5 -29 | Left Parahippocampal Gyrus |
| 4539 | 10.67 | <.001 | 25 –7 -30 | Right Parahippocampal Gyrus | |
| 1229 | 7.7 | <.001 | 18 –72 35 | Right Precuneus | |
| 1088 | 6.46 | <.001 | 50 –62 32 | Right Angular Gyrus | |
| 1284 | 6.14 | <.001 | −15 –69 33 | Left Precuneus | |
| 417 | 6.11 | <.001 | 4 –33 62 | Right Paracentral Lobe | |
| 566 | 5.67 | <.001 | −34 –18 46 | Left Precentral Gyrus | |
| 134 | 5.54 | <.001 | −18 –67 -12 | Left Lingual Gyrus | |
| 598 | 5.53 | <.001 | −40 –56 38 | Left Inferior Parietal, Angular | |
| 114 | 5.33 | <.001 | 23 32 –16 | Right Middle Frontal Gyrus | |
| 88 | 5.32 | <.001 | 58 –54 -18 | Right Inferior Temporal Gyrus | |
| 159 | 5.25 | .001 | −44 –53 20 | Left Superior Temporal Gyrus | |
| 100 | 5.01 | .002 | −39 3 38 | Left Precentral Gyrus | |
| 71 | 4.76 | .005 | −24 –43 -10 | Left Parahippocampal Gyrus | |
| 57 | 4.59 | .010 | 52 8 31 | Right Inferior Frontal Gyrus | |
| 69 | 4.58 | .010 | −5 –35 58 | Left Paracentral Gyrus | |
|
| |||||
| Sulcal Depth | |||||
|
| |||||
| CU > MCI/AD | 79 | 7.41 | <.001 | −31 –2 -22 | Left Parahippocampal Gyrus/ Amygdala |
| 1403 | 6.39 | <.001 | −51 –30 6 | Left Superior Temporal Gyrus | |
| 386 | 5.55 | <.001 | 34 –47 -8 | Right Parahippocampal Gyrus | |
| 358 | 5.53 | <.001 | 49 –34 13 | Right Superior Temporal Gyrus | |
| 53 | 4.73 | .007 | 32 –5 45 | Right Precentral Gyrus | |
| 89 | 4.64 | .011 | 44 –10 14 | Right Insula | |
|
| |||||
| Gyrification Index | |||||
|
| |||||
| CU > MCI/AD | 539 | 5.86 | <.001 | −37 –15 -7 | Left Insula |
| 255 | 5.46 | <.001 | 34 –30 17 | Right Insula | |
| MCI/AD > CU | 212 | 7.39 | <.001 | 24 –8 -29 | Right Parahippocampal Gyrus |
| 213 | 6.51 | <.001 | −25 –5 -29 | Left Parahippocampal Gyrus | |
|
| |||||
| Fractal Dimension | |||||
|
| |||||
| CU > MCI/AD | 231 | 6.92 | <.001 | 8 27 –24 | Right Rectal Gyrus |
| 51 | 5.45 | <.001 | −4 11 –8 | Left Anterior Cingulate/Caudate | |
| 106 | 5.22 | 0.001 | −16 –50 0 | Left Lingual Gyrus | |
| 122 | 5.2 | 0.001 | 8 –24 41 | Right Cingulate Gyrus | |
| 61 | 4.99 | 0.002 | 27 4 51 | Right Middle Frontal Gyrus | |
| 51 | 4.84 | 0.004 | 34 11 11 | Right Insula | |
| 69 | 4.59 | 0.011 | −34 –22 17 | Left Insula | |
Figure 2.
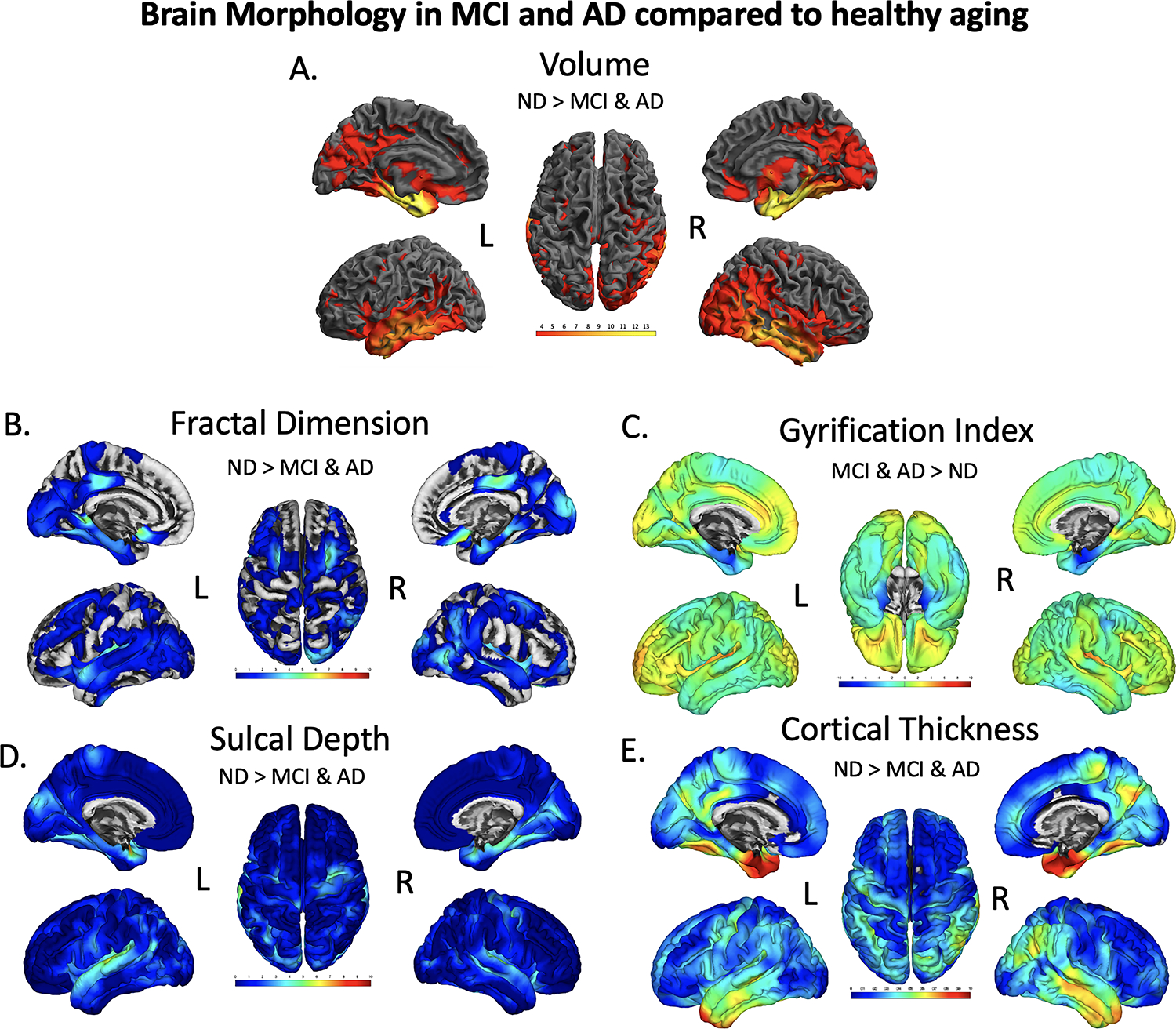
Brain regions showing significant differences in volume (A), fractal dimension (B), gyrification index (C), sulcal depth (D), and cortical thickness (E), between CU and dementia groups. The results were FWE-corrected p<.05 with a cluster extent threshold of 100 voxels or vertices, controlling for age, sex, education and TICV (TICV was only used as a covariate in the Volume (A) analysis, not for surfaces). For Volume (A), yellow color indicates regions with a significant reduction volume in MCI/AD compared with CU, with a T-value of 3 being red and yellow being a 14 on the color bar. For fractal dimension (B), sulcal depth (D), and cortical thickness (E) a turquoise to yellow to red indicates the level of significance for a decrease in fractal dimension, sulcal depth, and cortical thickness in MCI/AD compared to CU. Gyrification index measures were found to be both significantly decreased (blue, bilateral parahippocampal gyrus)
3.3. Voxel and Surface Based Morphometry between TOMM40’523- APOE ε4 groups in CU individuals
In the voxel-based analysis of gray matter volume there were no significant differences between TOMM40’523-APOE ε4 groups (Figure 3A, Table 6). In our analysis of fractal dimension (or cortical complexity), S/S individuals had significantly decreased fractal dimension in the right superior parietal cortex (AD phenotype) compared to VL-carriers (p=.046, peak T=3.46 k=274) (Figure 3B and Table 6). TOMM40’523 S-carriers had significantly decreased sulcal depth in the right precentral right precentral gyrus compared to VL/VL individuals (p=.048, peak T=3.42, k=297). The most significant finding was decreased sulcal depth in the left posterior cingulate at in TOMM40’523 S/S individuals (p<.000, peak T=3.77, k=823) (Figure 3D, Table 6). TOMM40’523 S-carriers had a significant decrease in cortical thickness compared to APOE ε4+ and VL/VL carriers in the left fusiform gyrus at a cluster level FWE corrected significance (p=.019, peak T=3.80, k=426), like the AD phenotype (Figure 3E, Table 6). There were no significant differences in gyrification index between TOMM40’523-APOE ε4 groups.
Figure 3.
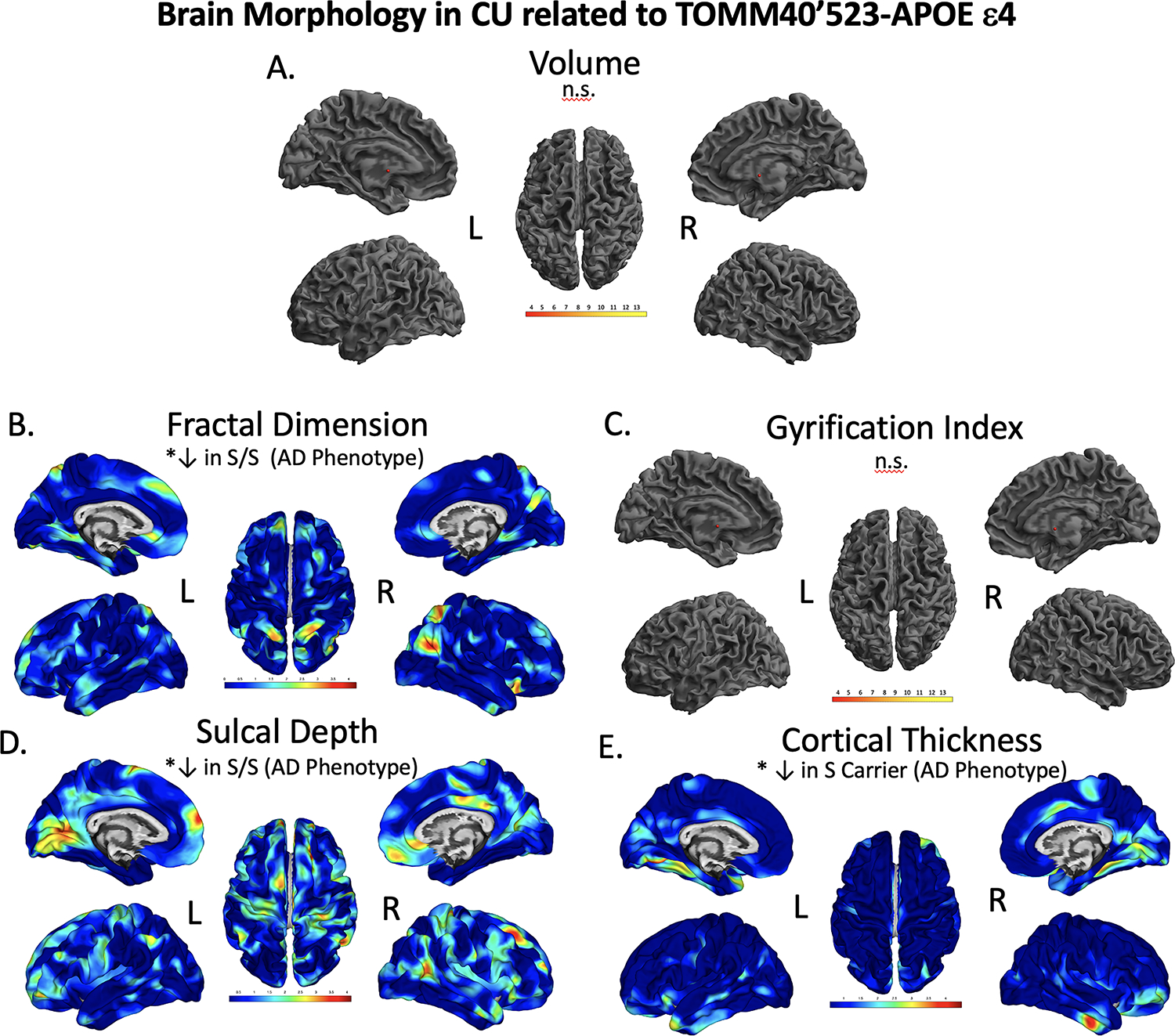
Brain regions showing significant differences in volume (A), fractal dimension (B), gyrification index (C), sulcal depth (D), and cortical thickness (E), between CU TOMM40’523-APOE ε4 groups. The results were FWE-corrected p<.05 with a cluster extent threshold of 100 voxels or vertexes, controlling for age, sex, and education. Yellow to red indicate significant decrease in morphology in fractal dimension (B), sulcal depth (D), and cortical thickness (E). We found significantly reduced cortical thickness, sulcal depth, and fractal dimension in individuals with an S/S compared to VL/VL, and APOEε4 groups. L; left, R; right, n.s.; not significant at our threshold. Color to be used in print.
Table 6. Voxel Based and Surface Based Morphometry results between TOMM40’523 and APOE groups in CU Individuals.
Corrected significant values in bold. L; left, R; right, n.s.; not significant.
| Comparison | Size (vertexes) | Peak t value | Cluster level p value (FWE) corrected | Peak-level p value (FWE Corrected) | uncorrected p value | Coordinates (mm mm mm) | Brain Region |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Volume | |||||||
|
| |||||||
| n.s. | |||||||
|
| |||||||
| Cortical Thickness | |||||||
|
| |||||||
| VL/VL and ε4+ > S Carrier | 426 | 3.80 | .019 | .216 | <.000 | −18 –69 -12 | L Fusiform Gyrus |
|
| |||||||
| Sulcal Depth | |||||||
|
| |||||||
| VL/VL > SS | 823 | 3.77 | 0.000 | 0.559 | <.000 | −15 –69 7 | L Posterior Cingulate |
| VL/VL > S Carrier | 297 | 3.42 | 0.048 | 0.602 | <.000 | 29 –30 69 | R Precentral Gyrus |
|
| |||||||
| Gyrification Index | |||||||
|
| |||||||
| n.s. | |||||||
| Fractal Dimension | |||||||
|
| |||||||
| VL Carrier > SS | 274 | 3.46 | .047 | .605 | <.000 | 25 –57 57 | R Superior Parietal |
3.4. Voxel and Surface Based Morphometry between TOMM40’523- APOE ε4 groups in MCI/AD
For all voxel and surface-based morphometry analyses, statistics were done in imaging space across all voxels and vertices, controlling for age, sex, education, and in the volume-based analysis for TICV as well. A full-factorial analysis was done comparing each genotype group within MCI/AD individuals, only FWE-corrected significant results are reported. In the voxel-based analysis of gray matter volume APOE ε4 positive individuals had significantly decreased volume in the right superior parietal cortex (p=.031, peak T=4.79, k=134), a larger cluster centering around the right superior temporal gyrus and medial temporal cortex (p=.003, peak T=4.54, k=3351), and a large cluster in the precuneus cortex (p=.011, peak T=4.42, k=3369) compared to TOMM40’523 S/S individuals. APOE ε4+ individuals also had significantly lower volume in the right angular gyrus compared to TOMM40’523 S-carriers (p=.001, peak T=5.06, k=3992) (Figure 4A, Table 7).
Figure 4.
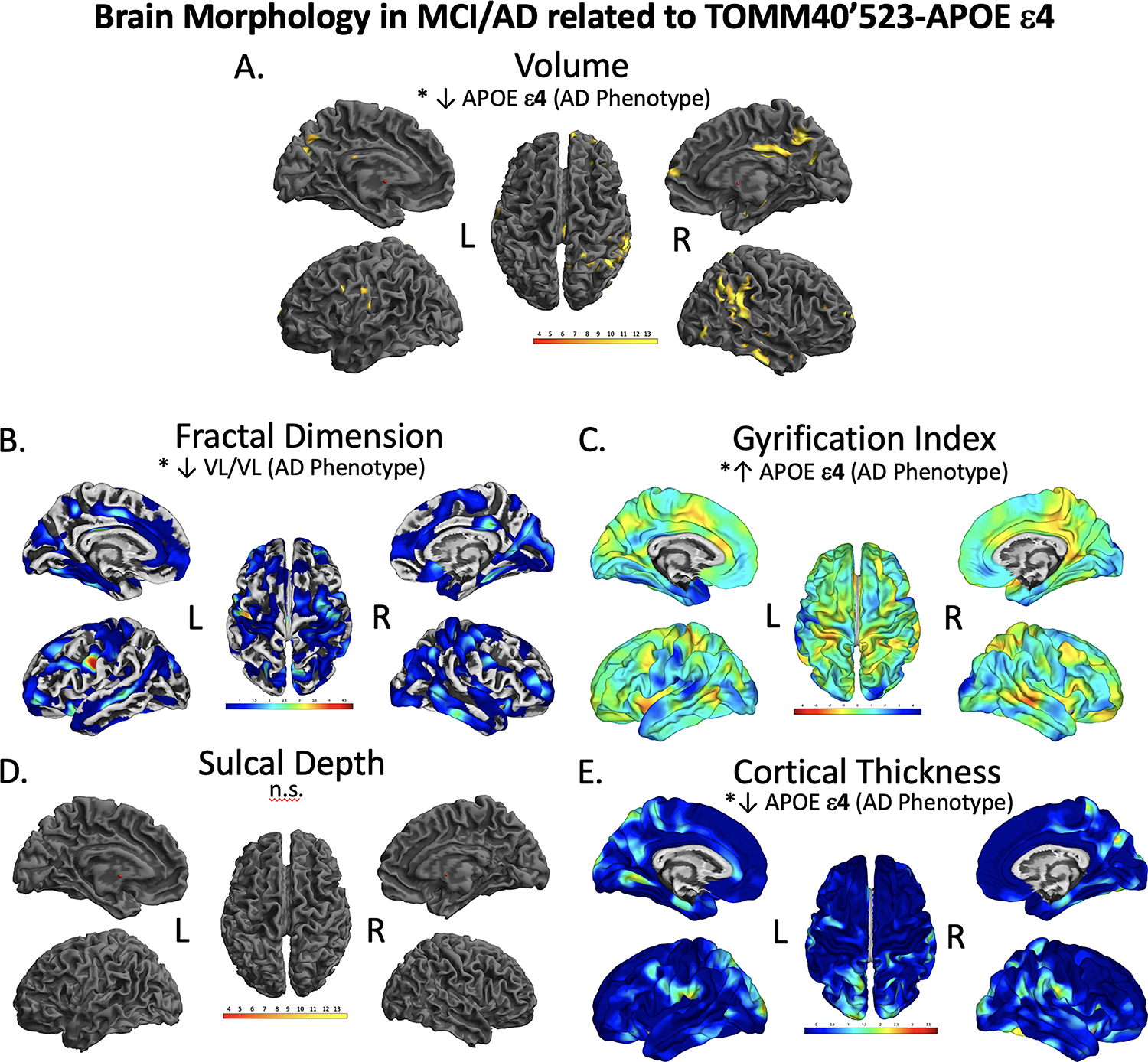
Brain regions showing significant differences in volume (A), fractal dimension (B), gyrification index (C), sulcal depth (D), and cortical thickness (E), between MCI/AD TOMM40’523-APOE ε4 groups. The results were FWE-corrected p<.05 with a cluster extent threshold of 100 voxels or vertexes, controlling for age, sex, and education. Yellow color indicates regions with a significant reduction in volume (A), Red indicated significant decrease in morphology in fractal dimension (B), sulcal depth (D), and cortical thickness (E), and blue indicates a significant increase in gyrification (C). Volume in the precuneus, gyrification in the temporal cortex and cortical thickness in the precuneus were significantly lower in the APOEε4 group. Individuals with a VL/VL had significantly decreased fractal dimension. L; left, R; right. n.s.; not significant at our threshold. Color to be used in print.
Table 7.
Voxel Based and Surface Based Morphometry results between TOMM40’523 and APOE groups in MCI/AD Individuals
| Comparison | Size (vertexes) | Peak t value | Cluster level p value (FWE) corrected | Peak-level p value (FWE Corrected) | uncorrected p value | Coordinates (mm mm mm) | Brain Region |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Volume | |||||||
|
| |||||||
| SS > ε4+ | 134 | 4.79 | .700 | .031 | <.000 | 24 –51 72 | R Superior Parietal Cortex |
| 3351 | 4.54 | .003 | .068 | <.000 | 64 –40 9 | R Superior Temporal Gyrus | |
| 3369 | 4.42 | .011 | .098 | <.000 | −16 –76 21 | LPrecuneus | |
| S Carrier > ε4+ | 3992 | 5.06 | .001 | .013 | <.000 | 46 –56 30 | R Angular Gyrus (Parietal) |
| 213 | 4.60 | .593 | .057* | <.000 | −16 –76 21 | LPrecuneus | |
|
| |||||||
| Cortical Thickness | |||||||
|
| |||||||
| ε4− > ε4+ | 171 | 4.19 | .035 | .076 | <.000 | −22 –37 61 | LPostcentral Gyrus |
| 516 | 4.12 | <.000 | .096 | <.000 | 17 –40 68 | R Postcentral Gyrus | |
| 172 | 4.11 | .034 | .099 | <.000 | 56 –38 47 | R Inferior Parietal Cortex | |
| VL VL > ε4+ | 97 | 4.38 | .155 | .044 | <.000 | 30 –49 44 | R Inferior Parietal Cortex |
|
| |||||||
| Sulcal Depth | |||||||
|
| |||||||
| n.s. | |||||||
|
| |||||||
| Gyrification Index | |||||||
|
| |||||||
| ε4+ > ε4− SS and VL | 464 | 4.11 | .027 | .097 | <.000 | −50 10 –15 | L Superior Temporal Gyrus |
| ε4+ > S-Carrier | 416 | 4.18 | .048 | .079 | <.000 | −42 12 25 | L Inferior Frontal Gyrus |
| 641 | 3.94 | .003 | .156 | <.000 | −66 –33 -10 | L Middle Temporal Gyrus | |
|
| |||||||
| Fractal Dimension | |||||||
|
| |||||||
| ε4+ > ε4− SS and VL | 96 | 3.88 | .076 | .281 | <.000 | 52 –61 6 | R Middle Temporal Gyrus |
| SS > VLVL | 141 | 3.82 | .025 | .328 | <.000 | −43 –18 33 | L Postcentral gyrus |
| S Carrier > VLVL | 160 | 4.35 | .009 | .076 | <.000 | −54 –5 29 | L Precentral Gyrus |
L; left, R; right, n.s.; not significant.
indicates trend
In our analysis of cortical thickness, APOE ε4+ MCI/AD individuals had significantly thinner cortex at the left postcentral gyrus (p=.035, peak T=4.19, k=171), the right postcentral gyrus (p<.000, peak T=4.12, k=516), and the right inferior parietal cortex (p=.034, peak T=4.11, k=172) compared to all other groups (Figure 4E, Table 7). In our analysis of gyrification index, APOE ε4+ MCI/AD individuals had significantly increased gyrification in the left superior temporal gyrus compared to APOE ε4− S/S and VL/VL individuals (p=.027, peak T=4.11, k=464), and also increased gyrification in the left inferior frontal gyrus (p=.048, peak T=4.18, k=416) and left middle temporal gyrus (p=.003, peak T=3.94, k=641) compared to TOMM40’523 S-carriers. In our analysis of fractal dimension, there were no significant decreases in the APOE ε4+ group, however TOMM40’523 VL/VL individuals had significantly decreased fractal dimension in the left precentral (p=.025, peak T=3.82, k=141) and left postcentral (p=.009, peak T=4.35, k=160) gyri compared to S-carriers. There were no significant differences in sulcal depth between TOMM40’523-APOE ε4 groups.
4. Discussion
In this study, we sought to characterize 5 morphological biomarkers of cortical complexity in a large sample, then investigate the relationship of APOE and TOMM40’523 on these biomarkers in CU and CI individuals. We identified several AD-specific phenotypes in brain morphology and found that healthy aging individuals with TOMM40’523 poly-T S alleles have more AD-related biomarkers of cortical complexity than those with APOE ε4 and TOMM40 VL alleles. While APOE ε4 was not significantly associated with brain structure in CU individuals, we did find that APOEε4+ MCI/AD individuals had significantly decreased temporal and parietal volume, cortical thickness, and fractal dimension as well as increased gyrification index in the frontal and temporal cortices compared to ε4 negative TOMM40’523 S and VL carriers.
Multiple processed data types from MPRAGE have become standard AD biomarkers, namely gray matter volume and thickness, as regional variations in atrophy mark progression of the disease (Choi et al., 2019; Du et al., 2007). Fewer studies have quantified cortical complexity in MCI and AD using gyrification index, fractal dimension, and sulcal depth. A recent study (Choi et al., 2019) compared thickness and gyrification index metrices across a spectrum of AD individuals and found that cortical thickness may be preferable to gyrification index in characterizing structural change in AD-related cognitive decline. In our analysis, voxel-wise cortical thickness measurements captured more significant differences in temporal and limbic regions between diagnosis groups, with gyrification index identifying diagnostic differences in smaller clusters in the insula and parahippocampal gyrus. Our data fit with previous studies showing regional patterns of cortical thinning in AD across the medial temporal, superior temporal, and parietal cortices (Kalin et al., 2017). We identified significant widening measured by gyrification index in the parahippocampal gyrus in AD compared to CU individuals, which was also found in Ruiz et al., 2017 (Ruiz de Miras et al., 2017). Individuals with AD also had decreased fractal dimension compared to CU individuals in the cingulate, insula, rectal gyrus, and middle frontal cortex, the same regions of fractal dimension differences as a recent report (Nicastro et al., 2020). Fractal dimension may be more sensitive to structural differences in AD than gyrification index because it is scale-free (Chen et al., 2020; Madan and Kensinger, 2016), and the complexity and folding of the brain decreases with age (King et al., 2010; Liu et al., 2020; Marzi et al., 2020).
Our data contribute to a growing literature supporting the role of TOMM40 variant repeat length in cortical complexity of the brain in limbic and temporal, and precuneus cortices in the preclinical phase. Our results are supported by other studies showing an association of rs10524523 and risk of LOAD in individuals with a short TOMM40 Poly-T allele (Cruchaga et al., 2011). There have also been a number of studies in cognitively unimpaired individuals assessing the relationship of TOMM40’523 Poly-T alleles to CSF, imaging, cognitive, and mitochondrial function. Chiba-Falek et al., reviewed effects of TOMM40’523 Poly-T alleles on Alzheimer’s imaging phenotypes in healthy individuals and found that the VL allele may effect brain regions affected early in AD (Chiba-Falek et al., 2018). However, the two studies they reference had a younger sample than ours (mean age 55 (Burggren et al., 2017) and 63 (Johnson et al., 2011)), and no studies have yet used metrics of shape complexity, which may capture more subtle, age and disease related variations in the brain (Ziukelis et al., 2022). A recent study found significant gene-AD association in several SNPs of TOMM40 with cortical thickness in the temporal lobe, adding functional support for the role of TOMM40 in cortical morphometry changes leading to AD (Varathan et al., 2022). We also found decreased cortical thickness in the temporal cortex in CU TOMM40 S/S individuals. Moreover, a study using PET tau and amyloid binding found that S/VL individuals had greater binding in the medial temporal lobe (MTL), which may point towards a preclinical change (Siddarth et al., 2018). Our analysis of sulcal depth and fractal dimension both identified decreases in the metabolic hub of the parietal cortex and posterior cingulate in APOE ε3 individuals with a TOMM40 S allele. SNPs in the TOMM40 gene associated with AD have also been shown to mediate gene expression and DNA methylation in the prefrontal cortex (Marioni et al., 2018). Decreases in fractal dimension have been reported in the parietal and posterior cingulate associated with risk for AD, as well as with cognitive decline (Kinno et al., 2017; Ruiz de Miras et al., 2017).
Imaging results from this current study support our previous association between TOMM40 S alleles with worse cognitive performance in the absence of APOE ε4, and regardless of clinical status (Watts et al., 2019). Presence of the TOMM40’523 S in APOE ε3 individuals has also been shown to impact rate of cognitive decline in Parkinson’s disease and Parkinson’s disease dementia (Bakeberg et al., 2021). Finally, we did not find a significant difference in brain morphometry between APOE ε4 and APOE ε3 CU individuals. Our current findings are in line with other studies have shown that APOE ε4 may not impact limbic volume in cognitively unimpaired individuals (Morra et al., 2009; Protas et al., 2013).
No studies have investigated the relationship of TOMM40’523 on the structural complexity of the brain in MCI/AD individuals, however there is a large amount of data supporting the data that APOE ε4 impacts brain structure and is associated with multiple facets of AD pathology (Yamazaki et al., 2019). In our analysis, MCI/AD individuals with an APOE ε4 had more brain degeneration across multiple measures of cortical complexity. APOE ε4 carriers with cognitive impairment had significantly decreased volume cortical thickness, and increased gyrification index in the superior and middle temporal, inferior parietal, precuneus and postcentral gyri. Our results point to APOE-related differences in these regions that are beginning to change cortically after cognitive decline has occurred, and possibly further along in a temporal trajectory. It is also important to consider that the analysis of genetic effects in MCI and AD groups may also be confounded by the effects of disease pathology, stage and progression, and APOE-related differences presented here may also just reflect degree of progression differences. That said, a recent large database study characterized the temporal evolution of AD biomarkers across the lifespan (18–103) and between APOE ε4 groups, and their findings demonstrate a variety of temporal trajectories of decline across biomarkers and ε4− and ε4+ groups, primarily showing that the presence of an ε4+ may increase the rates of brain atrophy (Luo et al., 2022).
Ours and others’ analyses of the APOE ε3-TOMM40’523 cis-haplotypes argue for the relevance of these two adjacent genes to the clinical expression of AD. There is growing evidence that APOE and TOMM40 genes work interactively on Chromosome 19 to impact downstream mitochondrial metabolic function in aging (Caselli et al., 2012). TOMM40 and APOC1 genes modulate the effect of the APOE ε4, and this interplay of genes may explain the differing roles of Aβ and Tau in the pathology of AD, as well as the age of onset (Kulminski et al., 2022a; Lutz et al., 2010). Our analysis in aging individuals shows that TOMM40 S (or APOE ε3) has an impact on the brain, perhaps at a developmental level or during physiologic brain aging, that this does not necessarily result in increased risk for AD. While our data is not longitudinal and does not show conversion rates to AD, we do present data showing that once an individual is cognitively impaired, having an APOE ε4 negatively impacts neurodegeneration. What will be key to tease out will be the epigenetic mechanisms, or perhaps promotor-enhancer relationships with nearby genes on chromosome 19, through which the toxicity that APOE ε4 expression becomes an active part of neurodegeneration.
4.1. Study Limitations
We are limited by the cross-sectional nature of the design of this observational study and cannot infer causality or longitudinal risk based on these results. There were a higher than normal (global prevalence 8%, http://www.alzgene.org/meta.asp?geneID=83) proportion of APOE ε2/ε2 (n=2) and APOE ε3/ε2 (n=7) in our cognitively unimpaired sample who had a TOMM40’523 S/VL genotype, making up 22% of this TOMM40’523 group. Both the APOE ε3/ε2 and ε2/ε2 are uncommon genotypes in Caucasian AD patients (4%) (http://www.alzgene.org/meta.asp?geneID=83). This might have contributed to a more protective phenotype, a.k.a. healthier brain morphometry with less AD-related change. However, these proportions of the ε3/ε2 and ε2/ε2 genotypes within an aging/AD cohort sample are similar to another TOMM40 published study from the ADNI dataset in which, among 365 APOE ε4− individuals, they had 60 APOE ε2/ε2 or ε3/ε2, making up 16% of their sample (Willette et al., 2017). In fact, individuals with an APOE ε2 allele have been included in the majority of brain imaging studies on the TOMM40’523APOEε4 haplotype (Bruno et al., 2012a; Bruno et al., 2012b; Burggren et al., 2017; Lyall et al., 2014a, b; Lyall et al., 2015; Lyall et al., 2013; Yu et al., 2017b). While the presence of an APOE ε2 allele has been shown to possibly increase the age of onset of AD, studies that have directly looked for effects of the ε2 allele have not found effects on hippocampal volume, white matter tract integrity, or change in cognition in aging (Lyall et al., 2014a; Lyall et al., 2013; Payton et al., 2016; Willette et al., 2017). While the data here are too small to investigate ε2 allele groups separately, a future study in a larger dataset of participants with neuroimaging and TOMM40 data would be a useful analysis.
While one of the main strengths of our study is our well-characterized cohort of patients across a spectrum of cognitive decline, we had a small sample of ε4− MCI/AD individuals with a TOMM40’523 S/S or VL/VL genotype. We tried to overcome this limitation by adjusting for several confounding factors and applying multiple test corrections, however given the complete lack of imaging studies of the TOMM40’523APOEε4 haplotype in MCI/AD individuals we felt it important to include these analyses. We chose to combine MCI and AD individuals to increase our power in this unique analysis, however in doing so we may have added heterogeneity of cognitive decline into the analysis. That said, our center uses strict criteria for diagnosis of MCI and AD (in Methods), and when looking at the larger picture of our MCI individuals longitudinal data, we know that 66% of our MCI sample have since converted to AD (12 out of 18), possibly more will, and thus the majority of our MCI individuals are in a state of early AD. That said, an additional limitation to our study is the lack of diagnostic confirmation with CSF or PET biomarkers.
The KUADRC Clinical cohort is primarily Caucasian who typically show tight linkage disequilibrium between APOE ε4 and TOMM40’523 L alleles, which is evidenced in this sample. Therefore, we could not test the independent contributions of the TOMM40’523 long allele and APOE ε4, however another study in an African American cohort with less linkage disequilibrium between the TOMM40’523 long allele and APOE ε4 has reported unique varying levels of haplotype contributions incident dementia(Yu et al., 2017b).
5. Conclusion
Our study is the first to use comprehensive morphological analysis techniques to show varying levels of impact of TOMM40’523 and APOE ε4 genotypes on AD-related brain phenotypes across the spectrum of disease. As expected, we found that APOEε4+ MCI/AD (TOMM40 L) individuals had a higher rate of gene-related significant morphological markers indicative of AD. We also found that TOMM40’523 short variants were related to reduced cortical thickness, sulcal depth, gyrification, and fractal dimension in the precuneus, limbic regions, and temporal cortex in cognitively unimpaired individuals negative for an APOE ε4 genotype. This is especially interesting as the precuneus has been identified in other studies, and it may be particularly sensitive to dysfunction in bioenergetic changes associated with faulty mitochondria. This adds to a growing literature implicating TOMM40’523 and mitochondrial function in late-onset Alzheimer’s disease progression. Identifying APOE ε3 TOMM40 S-carrying individuals in those at risk for AD is key for understanding the complexities of genetic risk outside of the primary focus of APOE ε4.
Acknowledgments
Portions of this work were supported by the following grants: R03AG026374, R21AG029615, R01AG034614, R01AG033673, R01AG062548, R21AG061548, R00AG050490, from the National Institutes on Aging, K23NS058252 from the National Institute on Neurological Disorders and Stroke, and the Alzheimer’s Association Park the Cloud Grant. The University of Kansas Alzheimer’s Disease Research Center Cohort is supported grant P30AG035982 and P30AG072973 (Cohort). The Hoglund Biomedical Imaging Center is supported by grants C76 HF00201, S10 RR29577, UL1 TR000001. Much of the study data were collected and managed using REDCap electronic data capture tools hosted at University of Kansas Medical Center(Harris et al., 2019; Harris et al., 2009). REDCap (Research Electronic Data Capture) is a secure, web-based software platform designed to support data capture for research studies and is provided for by CTSA Award # UL1TR002366.The authors thank the members of the KU ADRC team for their assistance with data collection and study support, and the participants at the KU ADRC for their generosity of time and spirit, which makes this research possible.
Footnotes
The authors have no competing interests to declare
References
- Ashburner J, 2007. A fast diffeomorphic image registration algorithm. Neuroimage 38(1), 95–113. https://doi.org:S1053-8119(07)00584-8 [pii] 10.1016/j.neuroimage.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Bakeberg MC, Gorecki AM, Pfaff AL, Hoes ME, Koks S, Akkari PA, Mastaglia FL, Anderton RS, 2021. TOMM40 ‘523’ poly-T repeat length is a determinant of longitudinal cognitive decline in Parkinson’s disease. NPJ Parkinsons Dis 7(1), 56. 10.1038/s41531-021-00200-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach TG, Monsell SE, Phillips LE, Kukull W, 2012. Accuracy of the clinical diagnosis of Alzheimer disease at National Institute on Aging Alzheimer Disease Centers, 2005–2010. Journal of neuropathology and experimental neurology 71(4), 266–273. 10.1097/NEN.0b013e31824b211b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellenguez C, Grenier-Boley B, Lambert JC, 2020. Genetics of Alzheimer’s disease: where we are, and where we are going. Current Opinion in Neurobiology 61, 40–48. 10.1016/j.conb.2019.11.024 [DOI] [PubMed] [Google Scholar]
- Bruno D, Nierenberg JJ, Ritchie JC, Lutz MW, Pomara N, 2012a. Cerebrospinal fluid cortisol concentrations in healthy elderly are affected by both APOE and TOMM40 variants. Psychoneuroendocrinology 37(3), 366–371. 10.1016/j.psyneuen.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno D, Pomara N, Nierenberg J, Ritchie JC, Lutz MW, Zetterberg H, Blennow K, 2012b. Levels of cerebrospinal fluid neurofilament light protein in healthy elderly vary as a function of TOMM40 variants. Exp Gerontol 47(5), 347–352. 10.1016/j.exger.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggren AC, Mahmood Z, Harrison TM, Siddarth P, Miller KJ, Small GW, Merrill DA, Bookheimer SY, 2017. Hippocampal thinning linked to longer TOMM40 poly-T variant lengths in the absence of the APOE epsilon4 variant. Alzheimers Dement 13(7), 739–748. 10.1016/j.jalz.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli RJ, Dueck AC, Huentelman MJ, Lutz MW, Saunders AM, Reiman EM, Roses AD, 2012. Longitudinal modeling of cognitive aging and the TOMM40 effect. Alzheimers Dement 8(6), 490–495. 10.1016/j.jalz.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Huang NX, Zou TX, Chen HJ, 2020. Brain Cortical Complexity Alteration in Amyotrophic Lateral Sclerosis: A Preliminary Fractal Dimensionality Study. BioMed research international 2020, 1521679. 10.1155/2020/1521679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba-Falek O, Gottschalk WK, Lutz MW, 2018. The effects of the TOMM40 poly-T alleles on Alzheimer’s disease phenotypes. Alzheimers Dement 14(5), 692–698. 10.1016/j.jalz.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Youn H, Kim D, Lee S, Suh S, Seong JK, Jeong HG, Han CE, 2019. Comparison of neurodegenerative types using different brain MRI analysis metrics in older adults with normal cognition, mild cognitive impairment, and Alzheimer’s dementia. PloS one 14(8), e0220739. 10.1371/journal.pone.0220739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JA, 1960. A coefficient of agreement for nominal scales. Educ Psychol Meas 20 37–46. [Google Scholar]
- Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Walker DG, Ravid R, Heward CB, Rogers J, Papassotiropoulos A, Reiman EM, Hardy J, Stephan DA, 2007. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J Clin Psychiatry 68(4), 613–618. 10.4088/jcp.v68n0419 [DOI] [PubMed] [Google Scholar]
- Cruchaga C, Nowotny P, Kauwe JS, Ridge PG, Mayo K, Bertelsen S, Hinrichs A, Fagan AM, Holtzman DM, Morris JC, Goate AM, Alzheimer’s Disease Neuroimaging I, 2011. Association and expression analyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease. Arch Neurol 68(8), 1013–1019. 10.1001/archneurol.2011.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahnke R, Yotter RA, Gaser C, 2013. Cortical thickness and central surface estimation. Neuroimage 65, 336–348. 10.1016/j.neuroimage.2012.09.050 [DOI] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ, 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31(3), 968–980. 10.1016/j.neuroimage.2006.01.021 [DOI] [PubMed] [Google Scholar]
- Du AT, Schuff N, Kramer JH, Rosen HJ, Gorno-Tempini ML, Rankin K, Miller BL, Weiner MW, 2007. Different regional patterns of cortical thinning in Alzheimer’s disease and frontotemporal dementia. Brain 130(Pt 4), 1159–1166. 10.1093/brain/awm016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good CD, Ashburner J, Frackowiak RS, 2001. Computational neuroanatomy: new perspectives for neuroradiology. Rev Neurol (Paris) 157(8–9 Pt 1), 797–806. [PubMed] [Google Scholar]
- Greenbaum L, Springer RR, Lutz MW, Heymann A, Lubitz I, Cooper I, Kravitz E, Sano M, Roses AD, Silverman JM, Saunders AM, Beeri MS, 2014. The TOMM40 poly-T rs10524523 variant is associated with cognitive performance among non-demented elderly with type 2 diabetes. European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology 24(9), 1492–1499. 10.1016/j.euroneuro.2014.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, McLeod L, Delacqua G, Delacqua F, Kirby J, Duda SN, Consortium RE, 2019. The REDCap consortium: Building an international community of software platform partners. J Biomed Inform 95, 103208. 10.1016/j.jbi.2019.103208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG, 2009. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42(2), 377–381. https://doi.org:S1532-0464(08)00122-6 [pii] 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden KM, McEvoy JM, Linnertz C, Attix D, Kuchibhatla M, Saunders AM, Lutz MW, Welsh-Bohmer KA, Roses AD, Chiba-Falek O, 2012. A homopolymer polymorphism in the TOMM40 gene contributes to cognitive performance in aging. Alzheimers Dement 8(5), 381–388. 10.1016/j.jalz.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honea RA, Swerdlow RH, Vidoni ED, Goodwin J, Burns JM, Reduced gray matter volume in normal adults with a maternal family history of Alzheimer disease. Neurology 74(2), 113–120. https://doi.org:74/2/113 [pii] 10.1212/WNL.0b013e3181c918cb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im K, Lee JM, Seo SW, Hyung Kim S, Kim SI, Na DL, 2008. Sulcal morphology changes and their relationship with cortical thickness and gyral white matter volume in mild cognitive impairment and Alzheimer’s disease. Neuroimage 43(1), 103–113. 10.1016/j.neuroimage.2008.07.016 [DOI] [PubMed] [Google Scholar]
- Johnson SC, La Rue A, Hermann BP, Xu G, Koscik RL, Jonaitis EM, Bendlin BB, Hogan KJ, Roses AD, Saunders AM, Lutz MW, Asthana S, Green RC, Sager MA, 2011. The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOE epsilon3/epsilon3 genotype. Alzheimers Dement 7(4), 456–465. 10.1016/j.jalz.2010.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Vardarajan BN, Buros J, Yu CE, Hawk MV, Dombroski BA, Crane PK, Larson EB, Alzheimer’s Disease Genetics C, Mayeux R, Haines JL, Lunetta KL, Pericak-Vance MA, Schellenberg GD, Farrer LA, 2012. Comprehensive search for Alzheimer disease susceptibility loci in the APOE region. Arch Neurol 69(10), 1270–1279. 10.1001/archneurol.2012.2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalin AM, Park MT, Chakravarty MM, Lerch JP, Michels L, Schroeder C, Broicher SD, Kollias S, Nitsch RM, Gietl AF, Unschuld PG, Hock C, Leh SE, 2017. Subcortical Shape Changes, Hippocampal Atrophy and Cortical Thinning in Future Alzheimer’s Disease Patients. Frontiers in aging neuroscience 9, 38. 10.3389/fnagi.2017.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RD, Brown B, Hwang M, Jeon T, George AT, Alzheimer’s Disease Neuroimaging I, 2010. Fractal dimension analysis of the cortical ribbon in mild Alzheimer’s disease. Neuroimage 53(2), 471–479. 10.1016/j.neuroimage.2010.06.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinno R, Shiromaru A, Mori Y, Futamura A, Kuroda T, Yano S, Murakami H, Ono K, 2017. Differential Effects of the Factor Structure of the Wechsler Memory Scale-Revised on the Cortical Thickness and Complexity of Patients Aged Over 75 Years in a Memory Clinic Setting. Frontiers in aging neuroscience 9, 405. 10.3389/fnagi.2017.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulminski AM, Jain-Washburn E, Loiko E, Loika Y, Feng F, Culminskaya I, Alzheimer’s Disease Neuroimaging I, 2022a. Associations of the APOE epsilon2 and epsilon4 alleles and polygenic profiles comprising APOE-TOMM40-APOC1 variants with Alzheimer’s disease biomarkers. Aging (Albany NY) 14(24), 9782–9804. 10.18632/aging.204384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulminski AM, Philipp I, Shu L, Culminskaya I, 2022b. Definitive roles of TOMM40-APOE-APOC1 variants in the Alzheimer’s risk. Neurobiology of aging 110, 122–131. 10.1016/j.neurobiolaging.2021.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, Bellenguez C, Frizatti A, Chouraki V, Martin ER, Sleegers K, Badarinarayan N, Jakobsdottir J, Hamilton-Nelson KL, Moreno-Grau S, Olaso R, Raybould R, Chen YN, Kuzma AB, Hiltunen M, Morgan T, Ahmad S, Vardarajan BN, Epelbaum J, Hoffmann P, Boada M, Beecham GW, Garnier JG, Harold D, Fitzpatrick AL, Valladares O, Moutet ML, Gerrish A, Smith AV, Qu LM, Bacq D, Denning N, Jian XQ, Zhao Y, Del Zompo M, Fox NC, Choi SH, Mateo I, Hughes JT, Adams HH, Malamon J, Sanchez-Garcia F, Patel Y, Brody JA, Dombroski BA, Naranjo MCD, Daniilidou M, Eiriksdottir G, Mukherjee S, Wallon D, Uphill J, Aspelund T, Cantwell LB, Garzia F, Galimberti D, Hofer E, Butkiewicz M, Fin B, Scarpini E, Sarnowski C, Bush WS, Meslage S, Kornhuber J, White CC, Song Y, Barber RC, Engelborghs S, Sordon S, Voijnovic D, Adams PM, Vandenberghe R, Mayhaus M, Cupples LA, Albert MS, De Deyn PP, Gu W, Himali JJ, Beekly D, Squassina A, Hartmann AM, Orellana A, Blacker D, Rodriguez-Rodriguez E, Lovestone S, Garcia ME, Doody RS, Munoz-Fernadez C, Sussams R, Lin HH, Fairchild TJ, Benito YA, Holmes C, Karamujic-Comic H, Frosch MP, Thonberg H, Maier W, Roshchupkin G, Ghetti B, Giedraitis V, Kawalia A, Li S, Huebinger RM, Kilander L, Moebus S, Hernandez I, Kamboh MI, Brundin R, Turton J, Yang Q, Katz MJ, Concari L, Lord J, Beiser AS, Keene CD, Helisalmi S, Kloszewska I, Kukull WA, Koivisto AM, Lynch A, Tarraga L, Larson EB, Haapasalo A, Lawlor B, Mosley TH, Lipton RB, Solfrizzi V, Gill M, Longstreth WT, Montine TJ, Frisardi V, Diez-Fairen M, Rivadeneira F, Petersen RC, Deramecourt V, Alvarez I, Salani F, Ciaramella A, Boerwinkle E, Reiman EM, Fievet N, Rotter JI, Reisch JS, Hanon O, Cupidi C, Uitterlinden AGA, Royall DR, Dufouil C, Maletta RG, de Rojas I, Sano M, Brice A, Cecchetti R, St George-Hyslop P, Ritchie K, Tsolaki M, Tsuang DW, Dubois B, Craig D, Wu CK, Soininen H, Avramidou D, Albin RL, Fratiglioni L, Germanou A, Apostolova LG, Keller L, Koutroumani M, Arnold SE, Panza F, Gkatzima O, Asthana S, Hannequin D, Whitehead P, Atwood CS, Caffarra P, Hampel H, Quintela I, Carracedo A, Lannfelt L, Rubinsztein DC, Barnes LL, Pasquier F, Frolich L, Barral S, McGuinness B, Beach TG, Johnston JA, Becker JT, Passmore P, Bigio EH, Schott JM, Bird TD, Warren JD, Boeve BF, Lupton MK, Bowen JD, Proitsi P, Boxer A, Powell JF, Burke JR, Kauwe JSK, Burns JM, Mancuso M, Buxbaum JD, Bonuccelli U, Cairns NJ, McQuillin A, Cao CH, Livingston G, Carlson CS, Bass NJ, Carlsson CM, Hardy J, Carney RM, Bras J, Carrasquillo MM, Guerreiro R, Allen M, Chui HC, Fisher E, Masullo C, Crocco EA, DeCarli C, Bisceglio G, Dick M, Ma L, Duara R, Graff-Radford NR, Evans DA, Hodges A, Faber KM, Scherer M, Fallon KB, Riemenschneider M, Fardo DW, Heun R, Farlow MR, Kolsch H, Ferris S, Leber M, Foroud TM, Heuser I, Galasko DR, Giegling I, Gearing M, Hull M, Geschwind DH, Gilbert JR, Morris J, Green RC, Mayo K, Growdon JH, Feulner T, Hamilton RL, Harrell LE, Drichel D, Honig LS, Cushion TD, Huentelman MJ, Hollingworth P, Hulette CM, Hyman BT, Marshall R, Jarvik GP, Meggy A, Abner E, Menzies GE, Jin LW, Leonenko G, Real LM, Jun GR, Baldwin CT, Grozeva D, Karydas A, Russo G, Kaye JA, Kim R, Jessen F, Kowall NW, Vellas B, Kramer JH, Vardy E, LaFerla FM, Jockel KH, Lah JJ, Dichgans M, Leverenz JB, Mann D, Levey AI, Pickering-Brown S, Lieberman AP, Klopp N, Lunetta KL, Wichmann HE, Lyketsos CG, Morgan K, Marson DC, Brown K, Martiniuk F, Medway C, Mash DC, Nothen MM, Masliah E, Hooper NM, McCormick WC, Daniele A, McCurry SM, Bayer A, McDavid AN, Gallacher J, Mckee AC, van den Bussche H, Mesulam M, Brayne C, Miller BL, Riedel-Heller S, Miller CA, Miller JW, Al-Chalabi A, Morris JC, Shaw CE, Myers AJ, Wiltfang J, O’Bryant S, Olichney JM, Alvarez V, Parisi JE, Singleton AB, Paulson HL, Collinge J, Perry WR, Mead S, Peskind E, Cribbs DH, Rossor M, Pierce A, Ryan NS, Poon WW, Nacmias B, Potter H, Sorbi S, Quinn JF, Sacchinelli E, Raj A, Spalletta G, Raskind M, Caltagirone C, Bossu P, Orfei MD, Reisberg B, Clarke R, Reitz C, Smith AD, Ringman JM, Warden D, Roberson ED, Wilcock G, Rogaeva E, Bruni AC, Rosen HJ, Gallo M, Rosenberg RN, Ben-Shlomo Y, Sager MA, Mecocci P, Saykin AJ, Pastor P, Cuccaro ML, Vance JM, Schneider JA, Schneider LS, Slifer S, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tang M, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu CE, Yu L, Saba Y, Pilotto A, Bullido MJ, Peters O, Crane PK, Bennett D, Bosco P, Coto E, Boccardi V, De Jager PL, Lleo A, Warner N, Lopez OL, Ingelsson M, Deloukas P, Cruchaga C, Graff C, Gwilliam R, Fornage M, Goate AM, Sanchez-Juan P, Kehoe PG, Amin N, Ertekin-Taner N, Berr C, Debette S, Love S, Launer LJ, Younkin SG, Dartigues JF, Corcoran C, Ikram MA, Dickson DW, Nicolas G, Campion D, Tschanz J, Schmidt H, Hakonarson H, Clarimon J, Munger R, Schmidt R, Farrer LA, Van Broeckhoven C, O’Donovan MC, DeStefano AL, Jones L, Haines JL, Deleuze JF, Owen MJ, Gudnason V, Mayeux R, Escott-Price V, Psaty BM, Ramirez A, Wang LS, Ruiz A, van Duijn CM, Holmans PA, Seshadri S, Williams J, Amouyel P, Schellenberg GD, Lambert JC, Pericak-Vance MA, ADGC EADI, Genomic CHAR, Defining GERA, 2019. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates A beta, tau, immunity and lipid processing (vol 51, pg 414, 2019). Nat Genet 51(9), 1423–1424. 10.1038/s41588-019-0495-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu T, Jiang J, Cheng J, Liu Y, Li D, Dong C, Niu H, Li S, Zhang J, Brodaty H, Sachdev P, Wen W, 2020. Differential longitudinal changes in structural complexity and volumetric measures in community-dwelling older individuals. Neurobiology of aging 91, 26–35. 10.1016/j.neurobiolaging.2020.02.023 [DOI] [PubMed] [Google Scholar]
- Luo JQ, Agboola F, Grant E, Morris JC, Masters CL, Albert MS, Johnson SC, McDade EM, Fagan AM, Benzinger TLS, Hassenstab J, Bateman RJ, Perrin RJ, Wang GQ, Li Y, Gordon B, Cruchaga C, Day GS, Levin J, Voglein J, Ikeuchi T, Suzuki K, Allegri RF, Xiong CJ, Alzheimer DI, 2022. Accelerated longitudinal changes and ordering of Alzheimer disease biomarkers across the adult lifespan. Brain 145(12), 4459–4473. 10.1093/brain/awac238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz MW, Crenshaw DG, Saunders AM, Roses AD, 2010. Genetic variation at a single locus and age of onset for Alzheimer’s disease. Alzheimers Dement 6(2), 125–131. 10.1016/j.jalz.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall DM, Harris SE, Bastin ME, Munoz Maniega S, Murray C, Lutz MW, Saunders AM, Roses AD, Valdes Hernandez Mdel C, Royle NA, Starr JM, Porteous DJ, Wardlaw JM, Deary IJ, 2014a. Alzheimer’s disease susceptibility genes APOE and TOMM40, and brain white matter integrity in the Lothian Birth Cohort 1936. Neurobiology of aging 35(6), 1513 e1525–1533. 10.1016/j.neurobiolaging.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall DM, Harris SE, Bastin ME, Munoz Maniega S, Murray C, Lutz MW, Saunders AM, Roses AD, Valdes Hernandez Mdel C, Royle NA, Starr JM, Porteous DJ, Wardlaw JM, Deary IJ, 2014b. Are APOE varepsilon genotype and TOMM40 poly-T repeat length associations with cognitive ageing mediated by brain white matter tract integrity? Translational psychiatry 4(9), e449. 10.1038/tp.2014.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall DM, Munoz Maniega S, Harris SE, Bastin ME, Murray C, Lutz MW, Saunders AM, Roses AD, Valdes Hernandez Mdel C, Royle NA, Starr JM, Porteous DJ, Deary IJ, Wardlaw JM, 2015. APOE/TOMM40 genetic loci, white matter hyperintensities, and cerebral microbleeds. Int J Stroke 10(8), 1297–1300. 10.1111/ijs.12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall DM, Royle NA, Harris SE, Bastin ME, Maniega SM, Murray C, Lutz MW, Saunders AM, Roses AD, del Valdes Hernandez MC, Starr JM, Porteous DJ, Wardlaw JM, Deary IJ, 2013. Alzheimer’s disease susceptibility genes APOE and TOMM40, and hippocampal volumes in the Lothian birth cohort 1936. PloS one 8(11), e80513. 10.1371/journal.pone.0080513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan CR, Kensinger EA, 2016. Cortical complexity as a measure of age-related brain atrophy. Neuroimage 134, 617–629. 10.1016/j.neuroimage.2016.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Harris SE, Zhang Q, Mcrae AF, Hagenaars SP, Hill WD, Davies G, Ritchie CW, Gale CR, Starr JM, Goate AM, Porteous DJ, Yang J, Evans KL, Deary IJ, Wray NR, Visscher PM, 2018. GWAS on family history of Alzheimer’s disease. Transl Psychiat 8. https://doi.org:ARTN 99 10.1038/s41398-018-0150-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi C, Giannelli M, Tessa C, Mascalchi M, Diciotti S, 2020. Toward a more reliable characterization of fractal properties of the cerebral cortex of healthy subjects during the lifespan. Sci Rep 10(1), 16957. 10.1038/s41598-020-73961-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr., Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R, Mohs RC, Morris JC, Rossor MN, Scheltens P, Carrillo MC, Thies B, Weintraub S, Phelps CH, 2011. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 7(3), 263–269. https://doi.org:S1552-5260(11)00101-4 [pii] 10.1016/j.jalz.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR Jr., Schuff N, Weiner MW, Thompson PM, Alzheimer’s Disease Neuroimaging I, 2009. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp 30(9), 2766–2788. 10.1002/hbm.20708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, 1993. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 43(11), 2412b–2414. [DOI] [PubMed] [Google Scholar]
- Morris JK, Vidoni ED, Johnson DK, Van Sciver A, Mahnken JD, Honea RA, Wilkins HM, Brooks WM, Billinger SA, Swerdlow RH, Burns JM, 2017. Aerobic exercise for Alzheimer’s disease: A randomized controlled pilot trial. PloS one 12(2), e0170547. 10.1371/journal.pone.0170547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JK, Zhang G, Dougherty RJ, Mahnken JD, John CS, Lose SR, Cook DB, Burns JM, Vidoni ED, Okonkwo O, 2020. Collective effects of age, sex, genotype, and cognitive status on fitness outcomes. Alzheimers Dement (Amst) 12(1), e12058. 10.1002/dad2.12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro N, Malpetti M, Cope TE, Bevan-Jones WR, Mak E, Passamonti L, Rowe JB, O’Brien JT, 2020. Cortical Complexity Analyses and Their Cognitive Correlate in Alzheimer’s Disease and Frontotemporal Dementia. J Alzheimers Dis 76(1), 331–340. 10.3233/JAD-200246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Rojas J, Arboleda-Bustos CE, Guerrero E, Neira J, Arboleda H, 2022. Genetic Variants and Haplotypes of TOMM40, APOE, and APOC1 are Related to the Age of Onset of Late-onset Alzheimer Disease in a Colombian Population. Alzheimer disease and associated disorders 36(1), 29–35. 10.1097/WAD.0000000000000477 [DOI] [PubMed] [Google Scholar]
- Payton A, Sindrewicz P, Pessoa V, Platt H, Horan M, Ollier W, Bubb VJ, Pendleton N, Quinn JP, 2016. A TOMM40 poly-T variant modulates gene expression and is associated with vocabulary ability and decline in nonpathologic aging. Neurobiology of aging 39, 217 e211–217. 10.1016/j.neurobiolaging.2015.11.017 [DOI] [PubMed] [Google Scholar]
- Protas HD, Chen K, Langbaum JB, Fleisher AS, Alexander GE, Lee W, Bandy D, de Leon MJ, Mosconi L, Buckley S, Truran-Sacrey D, Schuff N, Weiner MW, Caselli RJ, Reiman EM, 2013. Posterior cingulate glucose metabolism, hippocampal glucose metabolism, and hippocampal volume in cognitively normal, late-middle-aged persons at 3 levels of genetic risk for Alzheimer disease. JAMA neurology 70(3), 320–325. 10.1001/2013.jamaneurol.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, Huentelman MJ, Welsh-Bohmer KA, Reiman EM, 2010. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. The pharmacogenomics journal 10(5), 375–384. 10.1038/tpj.2009.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de Miras J, Costumero V, Belloch V, Escudero J, Avila C, Sepulcre J, 2017. Complexity analysis of cortical surface detects changes in future Alzheimer’s disease converters. Hum Brain Mapp 38(12), 5905–5918. 10.1002/hbm.23773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddarth P, Burggren AC, Merrill DA, Ercoli LM, Mahmood Z, Barrio JR, Small GW, 2018. Longer TOMM40 poly-T variants associated with higher FDDNP-PET medial temporal tau and amyloid binding. PloS one 13(12), e0208358. 10.1371/journal.pone.0208358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH, 2012. Alzheimer’s disease pathologic cascades: who comes first, what drives what. Neurotoxicity research 22(3), 182–194. 10.1007/s12640-011-9272-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH, Khan SM, 2004. A “mitochondrial cascade hypothesis” for sporadic Alzheimer’s disease. Med Hypotheses 63(1), 8–20. 10.1016/j.mehy.2003.12.045S0306987704001252 [pii] [DOI] [PubMed] [Google Scholar]
- Swerdlow RH, Khan SM, 2009. The Alzheimer’s disease mitochondrial cascade hypothesis: an update. Experimental neurology 218(2), 308–315. https://doi.org:S0014-4886(09)00017-X [pii] 10.1016/j.expneurol.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres GG, Dose J, Hasenbein TP, Nygaard M, Krause-Kyora B, Mengel-From J, Christensen K, Andersen-Ranberg K, Kolbe D, Lieb W, Laudes M, Gorg S, Schreiber S, Franke A, Caliebe A, Kuhlenbaumer G, Nebel A, 2022. Long-Lived Individuals Show a Lower Burden of Variants Predisposing to Age-Related Diseases and a Higher Polygenic Longevity Score. International journal of molecular sciences 23(18). 10.3390/ijms231810949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varathan P, Gorijala P, Jacobson T, Chasioti D, Nho K, Risacher SL, Saykin AJ, Yan J, 2022. Integrative analysis of eQTL and GWAS summary statistics reveals transcriptomic alteration in Alzheimer brains. BMC medical genomics 15(Suppl 2), 93. 10.1186/s12920-022-01245-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidoni ED, Johnson DK, Morris JK, Van Sciver A, Greer CS, Billinger SA, Donnelly JE, Burns JM, 2015. Dose-Response of Aerobic Exercise on Cognition: A Community-Based, Pilot Randomized Controlled Trial. PloS one 10(7), e0131647. 10.1371/journal.pone.0131647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidoni ED, Van Sciver A, Johnson DK, He J, Honea R, Haines B, Goodwin J, Laubinger MP, Anderson HS, Kluding PM, Donnelly JE, Billinger SA, Burns JM, 2012. A community-based approach to trials of aerobic exercise in aging and Alzheimer’s disease. Contemp Clin Trials 33(6), 1105–1116. 10.1016/j.cct.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts A, Wilkins HM, Michaelis E, Swerdlow RH, 2019. TOMM40 ‘523 Associations with Baseline and Longitudinal Cognition in APOE varepsilon3 Homozygotes. J Alzheimers Dis 70(4), 1059–1068. 10.3233/JAD-190293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub S, Salmon D, Mercaldo N, Ferris S, Graff-Radford NR, Chui H, Cummings J, DeCarli C, Foster NL, Galasko D, Peskind E, Dietrich W, Beekly DL, Kukull WA, Morris JC, 2009. The Alzheimer’s Disease Centers’ Uniform Data Set (UDS): the neuropsychologic test battery. Alzheimer disease and associated disorders 23(2), 91–101. 10.1097/WAD.0b013e318191c7dd [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins HM, Koppel SJ, Bothwell R, Mahnken J, Burns JM, Swerdlow RH, 2017. Platelet cytochrome oxidase and citrate synthase activities in APOE epsilon4 carrier and non-carrier Alzheimer’s disease patients. Redox Biol 12, 828–832. 10.1016/j.redox.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette AA, Webb JL, Lutz MW, Bendlin BB, Wennberg AM, Oh JM, Roses A, Koscik RL, Hermann BP, Dowling NM, Asthana S, Johnson SC, Alzheimer’s Disease Neuroimaging I, 2017. Family history and TOMM40 ‘523 interactive associations with memory in middle-aged and Alzheimer’s disease cohorts. Alzheimers Dement 13(11), 1217–1225. 10.1016/j.jalz.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C, Roe CM, Buckles V, Fagan A, Holtzman D, Balota D, Duchek J, Storandt M, Mintun M, Grant E, Snyder AZ, Head D, Benzinger TL, Mettenburg J, Csernansky J, Morris JC, 2011. Role of family history for Alzheimer biomarker abnormalities in the adult children study. Arch Neurol 68(10), 1313–1319. 10.1001/archneurol.2011.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Zhao N, Caulfield TR, Liu CC, Bu G, 2019. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nature reviews. Neurology 15(9), 501–518. 10.1038/s41582-019-0228-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotter RA, Nenadic I, Ziegler G, Thompson PM, Gaser C, 2011a. Local cortical surface complexity maps from spherical harmonic reconstructions. Neuroimage 56(3), 961–973. 10.1016/j.neuroimage.2011.02.007 [DOI] [PubMed] [Google Scholar]
- Yotter RA, Thompson PM, Gaser C, 2011b. Algorithms to improve the reparameterization of spherical mappings of brain surface meshes. Journal of neuroimaging : official journal of the American Society of Neuroimaging 21(2), e134–147. 10.1111/j.1552-6569.2010.00484.x [DOI] [PubMed] [Google Scholar]
- Yu L, Lutz MW, Farfel JM, Wilson RS, Burns DK, Saunders AM, De Jager PL, Barnes LL, Schneider JA, Bennett DA, 2017a. Neuropathologic features of TOMM40 ‘523 variant on late-life cognitive decline. Alzheimers Dement 13(12), 1380–1388. 10.1016/j.jalz.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Lutz MW, Wilson RS, Burns DK, Roses AD, Saunders AM, Yang J, Gaiteri C, De Jager PL, Barnes LL, Bennett DA, 2017b. APOE epsilon4-TOMM40 ‘523 haplotypes and the risk of Alzheimer’s disease in older Caucasian and African Americans. PloS one 12(7), e0180356. 10.1371/journal.pone.0180356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun HJ, Im K, Jin-Ju Y, Yoon U, Lee JM, 2013. Automated sulcal depth measurement on cortical surface reflecting geometrical properties of sulci. PloS one 8(2), e55977. 10.1371/journal.pone.0055977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziukelis ET, Mak E, Dounavi ME, Su L, J TOB., 2022. Fractal dimension of the brain in neurodegenerative disease and dementia: A systematic review. Ageing research reviews 79, 101651. 10.1016/j.arr.2022.101651 [DOI] [PubMed] [Google Scholar]


