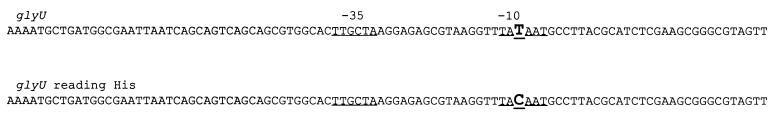Abstract
We previously described Escherichia coli mutator tRNAs that insert glycine in place of aspartic acid and postulated that the elevated mutation rate results from generating a mutator polymerase. We suggested that the proofreading subunit of polymerase III, ɛ, is a likely target for the aspartic acid-to-glycine change that leads to a lowered fidelity of replication, since the altered ɛ subunits resulting from this substitution (approximately 1% of the time) are sufficient to create a mutator effect, based on several observations of mutD alleles. In the present work, we extended the study of specific mutD alleles and constructed 16 altered mutD genes by replacing each aspartic acid codon, in series, with a glycine codon in the dnaQ gene that encodes ɛ. We show that three of these genes confer a strong mutator effect. We have also looked for new mutator tRNAs and have found one: a glycine tRNA that inserts glycine at histidine codons. We then replaced each of the seven histidine codons in the mutD gene with glycine codons and found that in two cases, a strong mutator phenotype results. These findings are consistent with the ɛ subunit playing a major role in the mutator effect of misreading tRNAs.
Mutator genes confer elevated rates of spontaneous mutations. Mutators have been used to help define DNA repair systems and to identify pathways of mutagenesis in prokaryotic as well as eukaryotic cells (24). The findings that certain human inherited cancer susceptibilities result from mutator phenotypes caused by defects in mismatch repair systems (5, 12, 18, 30) have sparked renewed interest in mutators. Defects in the mismatch repair genes lead to a strong mutator phenotype (25), as do combined defects in the GO system that repairs or prevents the incorporation of 7,8-dihydro-8-oxoguanidine in DNA (19, 20, 22). One of the strongest bacterial mutators in Escherichia coli results from a defect in the ɛ subunit of DNA polymerase III (Pol III), which is responsible for the editing function of Pol III (10, 31). The mutD/dnaQ gene encodes the ɛ subunit, and mutations in this gene result in the observed mutator effect. Some of these mutations give very strong effects; an example is the mutD5 allele, which increases the mutation frequency as much as 105-fold in rich medium (6, 9). There are surprisingly few published sequence changes resulting from mutD mutations, and some of the published assignments have been disputed (15, 33).
Although mutators have been actively investigated for several decades, new pathways of mutagenesis are still being discovered. We recently described a novel pathway of mutagenesis mediated by mutator tRNAs that are encoded by the E. coli genes mutA and mutC (21, 32). We postulated that the mutator tRNA effect is exerted by mistranslation that generates a mutator polymerase, through an alteration in the ɛ subunit of DNA Pol III (32). The mutator tRNAs are glycine tRNAs with anticodons altered to read the aspartic acid codon and which insert glycine at approximately 1% efficiency. There are 16 aspartic acid codons in the ɛ subunit. In this study, we investigated their importance by replacing each of the 16 codons, in series, with a glycine codon. Three cases resulted in a strong mutator. We also report the construction of an additional mutator tRNA that inserts glycine at histidine codons. We describe the replacement of seven histidine codons in the mutD gene with glycine codons and show that two of the constructed mutants are strong mutators. We discuss these results with regard to the mechanism of the mutator tRNAs and the primary structure of the ɛ subunit.
MATERIALS AND METHODS
Bacterial strains and methods.
E. coli CC105 has been described by Michaels et al. (21) and Cupples and Miller (7). Strain XL2-Blue MRF′ (Stratagene, La Jolla, Calif.) was used for cloning. All genetic methods were as described in reference 23.
DNA synthesis and sequencing.
Oligonucleotides were synthesized on a Beckman oligo1000 DNA synthesizer using solid-phase cyanoethyl phosphoramidite chemistry. All oligonucleotides were deprotected in ammonium hydroxide and used without further purification. DNA sequencing was carried out by using [α-32P]dATP and a SequiTherm cycle sequencing kit (Epicentre Technologies, Madison, Wis.) with reagents supplied by the manufacturer. For sequencing cDNA clones we used PCR conditions as described by the manufacturer.
Cloning of the mutD gene.
The DNA encoding dnaQ/mutD was PCR amplified from chromosomal DNA from CC105, isolated by standard procedures. Primers for amplification were based on sequences from GenBank (accession no. X04027, K00985, and M30201), and the gene, along with an extra 202 bp from the 5′ end and 28 bp from the 3′ end, was amplified. The 202-bp-long fragment from the 5′ end of the PCR-amplified DNA segment contained the promoter of the mutD gene and 137 bp of the 5′ end of the rnh gene. The conditions for PCR were as described previously (32), and we used PCR SuperMix (Gibco/BRL, Life Technologies, Gaithersburg, Md.). The PCR-amplified DNA segment was end polished by using chemicals and procedures supplied by Stratagene and blunt-end cloned into the PvuII site of pBR329 (GenBank accession no. L08859 and J01753). After cloning, the entire insert was sequenced to ensure that there were no PCR-introduced point mutations.
Cloning of tRNA genes.
The tRNA genes glyV, glyW, glyU, glyT, tyrT, serU, and pheV were cloned into the PvuII site of pBR329 by the same strategy as used for cloning the mutD gene.
Site-directed mutagenesis of the mutD gene.
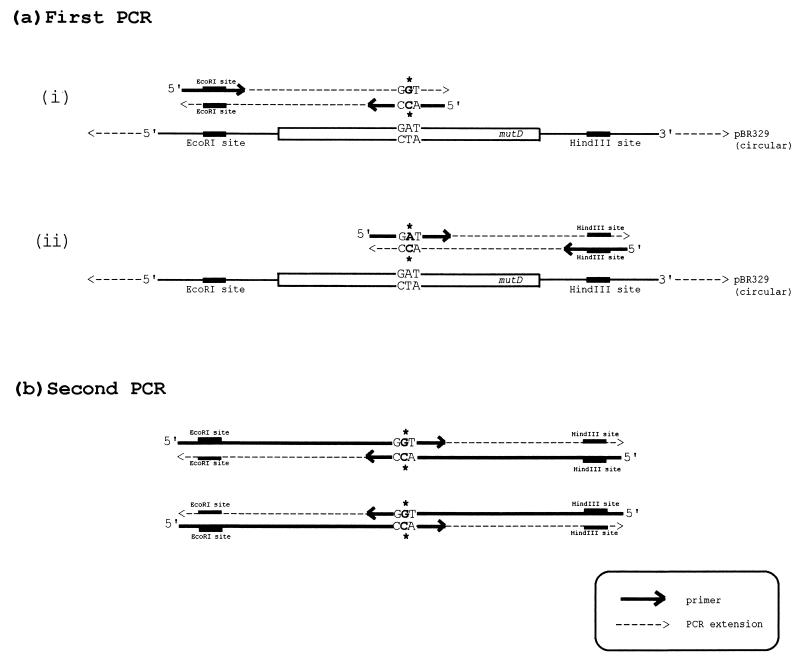
We performed site-directed mutagenesis by overlap extension as described by Ho et al. (14). For each mutant, two primers complementary to both strands of the mutD gene were synthesized. Primers were complementary to the mutD sequence except for the desired nucleotide change: GA(T/C)→GG(T/C) for the Asp→Gly replacement and CA(T/C)→GG(T/C) for the His→Gly replacement. Primers covered about 10 to 15 bases from the targeted nucleotide codon on each side and always ended before an A in the sequence. In addition, we made two primers complementary to the opposite strands of pBR329 and covering the restriction sites EcoRI and HindIII that flank the mutD insert in the recombinant plasmid. These four primers were used in specific combinations of a two-step PCR amplification which yielded a DNA segment containing the mutated mutD gene flanked by the fragments of pBR329 with the EcoRI and HindIII sites (Fig. 1). The fragment was digested with EcoRI and HindIII and cloned into pBR329; each mutated clone was sequenced to ensure that the desired mutation was introduced and that there were no extra point mutations. Conditions for PCR amplification were as in reference 32. Plasmids containing mutated mutD genes were next transformed into CC105 to measure mutation frequencies.
FIG. 1.
Strategy for site-directed mutagenesis. For each mutant, two primers complementary (except for a desired mutation) to the sequence of mutD were synthesized. We also made primers complementary to the sequence of pBR329 in the region of EcoRI and HindIII sites. In the first PCR, we obtained two fragments of the mutD gene, each containing the desired mutation. These fragments were agarose purified and used in the second PCR, in which the entire mutD gene (together with flanking pBR fragments) was amplified. The DNA obtained in the second PCR was digested with EcoRI and HindIII and cloned into pBR329.
Site-directed mutagenesis of tRNA genes.
For site-directed mutagenesis of tRNA genes, we used the same strategy as for site-directed mutagenesis of the mutD gene. Since we had problems with recovery of all the site-directed tRNA genes we attempted to synthesize, we did three independent ligations for each site-directed mutagenized tRNA, for each ligation performed two transformations, and sequenced on average five clones for the tRNA.
Mutation frequency measurements.
Mutation frequency measurements were determined as described in reference 23.
Nucleotide sequence accession numbers.
The GenBank accessions numbers for the sequences of the tRNA genes cloned in this study are as follows: glyV, X53236; glyW, J01624; glyU, M54893 and M38664; glyT, J01717; serU, M10746 and M10748; tyrT, X90989, K01197, J01720, K01198, K01217, K01300, and M10704; and pheV, X52799.
RESULTS
mutD mutants constructed by replacement of aspartic acid with glycine.
In a previous report (32), we described a mutator effect of E. coli tRNAs that insert glycine in place of aspartic acid. We suggested that the effect is exerted by generating a mutator ɛ subunit of E. coli DNA Pol III. In this study, we investigated the number of crucial aspartic acids in the ɛ protein that are sensitive to substitution by glycine. Using site-directed mutagenesis (see Materials and Methods), we replaced each aspartic acid codon in the mutD gene with a glycine codon, constructing 16 mutD genes, each with a single amino acid substitution. The newly constructed mutD genes were cloned onto pBR329 plasmids and transformed into strain CC105, which carries a lacZ mutation that reverts via the AT→TA transversion (7). We compared the mutator phenotype of the constructed mutD genes by measuring the frequency of Rifr and Lac+ mutants in strains harboring the plasmid with the mutD genes (Table 1). Of 16 mutD genes encoding different glycine-for-aspartic acid substitutions, three (D12G, D103G, and D167G) resulted in strong mutator effects, with increases in Rifr frequency over the wild-type value on the order of 1,000- to 5,000-fold. D129G displayed a weaker effect. The same constructs also showed an increase in Lac+ reversion, where the frequency of the AT→TA transversion at one specific site was measured (Table 1). Note that these effects are seen in the presence of the chromosomal wild-type mutD/dnaQ gene. Many mutD mutations are dominant in partial heterodiploids (6).
TABLE 1.
Mutation frequencies in strain CC105 harboring pBR329-derived plasmids with cloned mutated mutD genesa
| Strain | Mutation frequency (108)
|
|
|---|---|---|
| Rifr | Lac+ | |
| Wild type | 3.8 | 0.9 |
| D12G | 17,664.0 | 713.0 |
| D55G | 8.1 | 0.7 |
| D59G | 6.1 | 0.6 |
| D70G | 10.6 | 4.4 |
| D75G | 5.8 | 3.5 |
| D84G | 3.6 | 1.1 |
| D88G | 4.7 | 0.8 |
| D103G | 4,133.3 | 326.0 |
| D108G | 2.7 | 0.7 |
| D117G | 6.1 | 0.6 |
| D129G | 254.4 | 82.0 |
| D146G | 17.3 | 1.6 |
| D155G | 5.3 | 1.0 |
| D167G | 5,173.3 | 290.4 |
| D219G | 5.1 | 1.0 |
| D230G | 4.1 | 0.9 |
Each mutant carries an altered mutD gene, which results in an aspartic acid-to-glycine change at the indicated amino acid position in the ɛ subunit. Between 4 and 10 single colonies of each strain were inoculated into LB medium with tetracycline or ampicillin and grown overnight. Samples were plated on LB medium with rifampin and on lactose minimal plates to measure mutants and on LB medium without rifampin to measure total viable cells. The Lac+ revertants of CC105 strain result from AT→TA transversions (7).
Construction of additional synthetic tRNA genes.
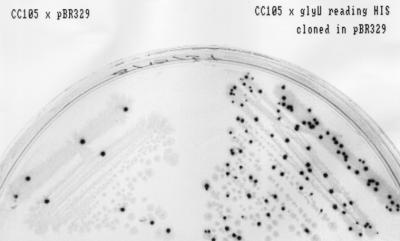
We attempted to synthesize 20 different misreading tRNAs by altering the tRNA-encoding genes glyW, glyV, gluU, glyT, tyrT, serU, and pheV. However, we recovered only 13 of the desired constructs. In nine of these cases, the plasmids recovered after ligation contained different extra point mutations in the tRNA-encoding genes. The same extra point mutations were not recovered repeatedly. In seven other cases, we simply failed to recover any clones. It is possible that synthesizing fully expressed misreading tRNAs on a multicopy plasmid is lethal in many cases. Table 2 summarizes these results. We tested the mutator phenotype of each of the misreading tRNAs that we successfully recovered. In one case we found a phenotype similar to that previously described for the mutA and mutC genes. Here, the glyU gene was altered so that a glycine tRNA now recognizes histidine codons, inserting glycine in place of histidine. Although we found an extra point mutation in the promoter region (Fig. 2), it still displays a mutator effect in both the papillation assay (Fig. 3) and the Lac+ reversion test in strain CC105 (49.9 ± 24.7 Lac+ revertants/108 cells, compared with 0.81 ± 0.4 for CC105 cells with pBR). The mutator effect of the altered glyU tRNA was smaller (3.0 ± 0.5 Lac+ revertants/108 cells) than in the case of the mutC mutator but still significant.
TABLE 2.
Miscoding tRNAs
| Type of tRNA | Description |
|---|---|
| Constructed | Phe (pheV) reading Leu (UUG) |
| Tyr (tyrT) reading Phe (UUC) | |
| Ser (serU) reading Leu (UUG) | |
| Phe (pheV) reading Thr (ACU) | |
| Failed to construct | Ser (serU) reading Phe (UUC) |
| Gly (glyW) reading Leu (CUC) | |
| Gly (glyW) reading His (CAC) | |
| Gly (glyV) reading Phe (UUC) | |
| Gly (glyV) reading Glu (GAA) | |
| Gly (glyW) reading Glu (GAA) | |
| Gly (glyT) reading Lys (AAG) | |
| Constructed but carrying extra point mutations | Gly (glyU) reading His (CAC) |
| Gly (glyU) reading Phe (UUC) | |
| Tyr (tyrT) reading His (CAC) | |
| Gly (glyV) reading His (CAC) | |
| Gly (glyW) reading Phe (UUC) | |
| Gly (glyT) reading Arg (CGA) | |
| Gly (glyV) reading Ser (AGC) | |
| Phe (phe V) reading Ala (GCC) | |
| Gly (glyT) reading Cys (UGU) |
FIG. 2.
The promoter region of the glyU gene and the change found in the glyU gene that reads histidine codons. The diagram of the sequence is adopted from reference 17.
FIG. 3.
Mutator phenotype of glyU that reads histidine codons. CC105 with plasmid pBR329 (control) and with plasmid pBR329 with the cloned altered glyU was plated on minimal glucose, P-Gal (phenyl β-d-galactoside), X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactoside), and tetracycline plates. Single colonies of the strain with the altered glyU gene contain more blue papillae (28), since they form more Lac+ revertants than the strain with pBR329 alone.
mutD mutants constructed by replacement of histidine with glycine.
We changed each histidine codon in the mutD gene to a glycine codon, generating seven additional mutD genes. After plasmids containing these genes were transformed into CC105, we compared Rifr and Lac+ mutation frequencies and found that two mutants (H98G and H162G) showed a strong mutator phenotype and one (H66G) displayed a very weak effect (Table 3).
TABLE 3.
Mutation frequencies in strain CC105 harboring pBR329 plasmids with cloned mutated mutD genesa
| Strain | Mutation frequency (108)
|
|
|---|---|---|
| Rifr | Lac+ | |
| Wild type | 3.8 | 0.9 |
| H24G | 2.5 | 0.4 |
| H28G | 7.2 | 5.2 |
| H49G | 4.4 | 2.7 |
| H66G | 49.0 | 27.7 |
| H98G | 1,282.2 | 142.4 |
| H162G | 3,680.4 | 66.6 |
| H225G | 4.0 | 0.5 |
Each mutant carries an altered mutD gene, which results in a histidine-to-glycine change at the indicated amino acid position in the ɛ subunit. For details, see the footnote to Table 1.
DISCUSSION
In this report, we have described 23 mutants with an engineered change in the ɛ subunit of E. coli DNA Pol III. In each of 16 cases, one of the aspartic acid residues in the ɛ subunit was ultimately changed to glycine. This allowed us to determine whether any sites in this protein are sensitive to the exact substitution caused by miscoding tRNAs present in mutA and mutC strains. The altered tRNAs in mutA and mutC strains insert glycine at aspartic acid codons approximately 1% of the time and result in mutation rates about 1% of that found in mutD strains defective in the ɛ subunit. We postulated that the mutator effect seen in mutA and mutC strains was in fact due to generating 1% altered ɛ subunits. Murphy and Humayun (27) have shown that mutA and mutC strains also display a UVM (UV modulator of mutagenesis) phenotype, which results in an increase of mutagenesis at the site of a 3,N4-ethenocytosine lesion in M13 single-stranded DNA transfected into E. coli and that the mutA and mutC mutator effect is recA dependent. They argued that the UVM phenotype could not result from a minor fraction (1%) of the ɛ subunits being altered to create a mutD effect. However, it is possible that the mutator effect of mutA and mutC does not occur by the same mechanism as the UVM effect. Moreover, Grollman and coworkers (26) have shown that a mutD strain indeed displays the UVM phenotype.
The dominance of many strong mutD mutator alleles (6) indicates that altered ɛ subunits can cause mutations even in the presence of normal subunits. Are there targets in the ɛ protein for the Asp-to-Gly change produced by mutA and mutC that would yield a mutator subunit? As Table 1 shows, there are three aspartic acid residues which yield a mutator ɛ subunit when exchanged for glycine.
We searched for other tRNAs which would give mutator effects, but despite the construction and examination of numerous tRNA genes with altered anticodons (Table 2), we detected a mutator effect in only one case, that of a glyU gene altered so that the resulting tRNA now inserts glycine in place of histidine. The resulting mutator effect is somewhat weaker than that seen for mutA and mutC but is nonetheless significant (Results and Fig. 3). We therefore altered each of the codons specifying histidine in the mutD gene so that in each of seven cases the ɛ subunit would carry a glycine in place of one of the histidines. Table 3 shows that replacement of either of two histidines with glycine results in a strain with a strong mutator effect.
There are two explanations why the effect of a tRNA that inserts glycine in place of histidine is smaller than the effect of a tRNA which replaces aspartic acids with glycine. First, there are three strong mutD mutants made by replacement of aspartic acids with glycine and only two mutD mutants made by replacement of histidines with glycine, which means there are more targets resulting in the mutator phenotype for the tRNA inserting glycine in place of aspartic acid than for the tRNA inserting glycine in place of histidine. In addition, the aspartic acid replacement gives a stronger mutator, especially in the case of the D12G replacement (Table 1). Another explanation for the smaller mutator effect of the glyU reading histidine tRNA would be the point mutation in the promoter region of glyU gene that reads histidine, resulting in lower levels of this tRNA. The continued failure to clone this gene without secondary point mutations raises the possibility that the cell does not tolerate the elevated expression of this missense tRNA and that the only way to maintain the plasmid in the cell is through lowering of expression by a change in the promoter.
The results presented above shed some light on the conflicting interpretation of the primary structure of the ɛ subunit. The existence of three conserved motifs in a large number of polymerase-associated exonucleases (Exo motifs) was proposed by Bernad et al. (3) on the basis of the protein alignment of different proofreading polymerases with the Klenow fragment of E. coli Pol I, whose three-dimensional structure was elucidated by Steitz and coworkers (2, 13, 29) and Derbyshire et al. (8). Each Exo motif contains highly conserved amino acids involved in the two-metal-ion catalysis reaction defined by Steitz and coworkers for the Klenow fragment of Pol I. For many proofreading polymerases, the role of critical amino acids in the three Exo motifs was confirmed by site-directed mutagenesis (for a review, see reference 16). For the ɛ subunit, the two main Exo I residues were predicted to be D12 and E14 (reference 3 and Fig. 4). Replacement of D12 and E14 with alanine results in an ɛ protein with a strong, dominant mutator phenotype, and cells containing the sole copy of the mutated gene (dnaQ926) are inviable unless they also carry a dnaE antimutator or a plasmid with the mismatch repair gene mutL (11). The mutD5 allele, which produces a strong mutator effect, generates the T15I substitution and mutD14 generates the T16I substitution (11, 15). In the work reported here, the D12G substitution resulted in a very strong mutator phenotype. For the Exo II motif, the crucial amino acids were predicted to be N99 and D103 (reference 3 and Fig. 4). As expected, both H98G and D103G turned out to be strong mutators (Table 1).
FIG. 4.
Nucleotide and amino acid sequences of the mutD/dnaQ gene and the ɛ protein. For the mutD nucleotide sequence, numbering is given at the right relative to the ATG translation start site (+1); the amino acid sequence of ɛ, given below in single-letter code, is also numbered at the right. Putative Exo I, Exo II (3), and two Exo III motifs (3, 4) are underlined; Exo IIIɛ (1) is underlined with a dotted line (the C-terminal homology of the ɛ subunit with gram-positive bacteria ends somewhere between E192 and V206 in the alignment of Barnes et al. [1]), and crucial amino acids in all putative Exo motifs are doubly underlined.
There are at least three different protein alignments of the Exo III region within the ɛ subunit. Two different regions of the ɛ protein have been proposed (3, 4), pointing to Y152 and D155 or V215 and D219, respectively (Fig. 4), as the residues that parallel Y497 and D501 of Exo III from the Klenow fragment. On the other hand, Barnes et al. (1) examined the structure of the Exo site of Pol III from Bacillus subtilis by site-directed mutagenesis and proposed the existence of an alternative Exo IIIɛ motif. They showed that three residues of this domain, His565, Asp533, and Asp574, are crucial for proofreading activity. These authors also point out that these residues are highly conserved among the Pol III Exo domains of three gram-positive bacteria, B. subtilis, Staphylococcus aureus, and Mycoplasma pulmonis, and that the same motif can be found in the ɛ subunit of E. coli Pol III (1).
For the E. coli ɛ protein, the critical amino acids of the Exo IIIɛ domain according to Bernad et al. (3) would be H162, D129, and D167 (Fig. 4). Our results support this hypothesis since all three changes (H162G, D129G, and D167G) diminish the proofreading activity of the ɛ subunit, whereas replacements of the amino acids (D155 and D219) as predicted by Bernad et al. (3) and Blanco et al. (4) do not affect the proofreading activity of ɛ. The results presented above are also in agreement with the work of Horner and Cox (15), who found that D167N and H162T replacements had a mutator effect. On the other hand, Horner and Cox have found a mutator mutD gene with the H225T replacement, whereas we found that H225G had no mutagenic effect (Table 3). All of the strong mutator effects described above are explained by changes in the Exo motifs of proofreading polymerases.
ACKNOWLEDGMENT
This work was supported by grant GM 32184 to J.H.M. from the National Institutes of Health.
REFERENCES
- 1.Barnes M H, Spacciapoli P, Li D H, Brown N C. The 3′-5′ exonuclease site of DNA polymerase III from Gram-positive bacteria: definition of a novel motif structure. Gene. 1995;165:45–50. doi: 10.1016/0378-1119(95)00530-j. [DOI] [PubMed] [Google Scholar]
- 2.Beese L S, Steitz T A. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernad A, Blanco L, Lazaro J M, Martin G, Salas M. A conserved 3′-5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 4.Blanco L, Bernad A, Salas M. Evidence favouring the hypothesis of a conserved 3′-5′ exonuclease active site in DNA-dependent DNA polymerases. Gene. 1992;112:139–144. doi: 10.1016/0378-1119(92)90316-h. [DOI] [PubMed] [Google Scholar]
- 5.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earibino C, Lipford J, Linblom A, Tannergard P, Bollag R J, Godwin A J, Ward D C, Nordenskjold M, Fishel R, Kolodner R D, Liskay R M. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary nonpolyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 6.Cox E C, Horner D L. Dominant mutators in Escherichia coli. Genetics. 1982;100:7–18. doi: 10.1093/genetics/100.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cupples C G, Miller J H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derbyshire V, Grindley N D F, Joyce C M. The 3′-5′ exonuclease of DNA polymerase I of Escherichia coli: contribution of amino acid at the active site to the reaction. EMBO J. 1991;10:17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degnen G E, Cox E C. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J Bacteriol. 1974;117:477–487. doi: 10.1128/jb.117.2.477-487.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echols H, Lu C, Burges P M. Mutator strains of Escherichia coli, mutD and dnaQ, with defective exonucleolytic editing by DNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1983;80:2189–2192. doi: 10.1073/pnas.80.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fijalkowska I J, Schaaper R M. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fishel R, Lescoe M K, Rao M R S, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 13.Freemont P S, Friedman J M, Beese L S, Sanderson M R, Steitz T A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci USA. 1988;85:8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 15.Horner, D. L., and E. C. Cox. Personal communication.
- 16.Joyce C M, Steitz T A. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 17.Komine Y, Adachi T, Inokuchi H, Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990;212:579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- 18.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M. Mutations of a mutS homolog in hereditary nonpolyposis colectoral cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 19.Maki H, Sekiguchi M. MutT protein specifically hydrolyzes a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 20.Michaels M L, Cruz C, Grollman A P, Miller J H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaels M L, Cruz C, Miller J H. mutA and mutC: two mutator loci in Escherichia coli that stimulate transversions. Proc Natl Acad Sci USA. 1990;87:9211–9215. doi: 10.1073/pnas.87.23.9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaels M L, Miller J H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 24.Miller J H. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol. 1996;50:625–643. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- 25.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 26.Moriya M, Zhang W, Johnson F, Grollman A P. Mutagenic potency of exocyclic DNA adducts: marked differences between Escherichia coli and simian kidney cells. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy H S, Humayun M Z. Escherichia coli cells expressing a mutant glyV (glycine tRNA) gene have a UVM-constitutive phenotype: implications for mechanisms underlying the mutA or mutC mutator effect. J Bacteriol. 1997;179:7507–7514. doi: 10.1128/jb.179.23.7507-7514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nghiem Y, Cabrera M, Cupples C G, Miller J H. The mutY gene: a mutator locus in Escherichia coli that generates GC→TA transversions. Proc Natl Acad Sci USA. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ollis D L, Brick P, Hamlin R, Xuong N G, Steitz T A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 30.Papadopoulos N, Nicolaides N C, Wei Y F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter J C, Hamilton S R, Petersen G M, Watson P, Lynch H T, Peltomaki P, Mecklin J-P, de la Chapelle A, Kinzler K W, Vogelstein B. Mutation of a mutL homolog in hereditary nonpolyposis colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 31.Scheuermann R, Tam S, Burgers P M, Lu C, Echols H. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci USA. 1983;80:7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slupska M M, Baikalov C, Lloyd R, Miller J H. Mutator tRNAs are encoded by the Escherichia coli mutator genes mutA and mutC: a novel pathway for mutagenesis. Proc Natl Acad Sci USA. 1996;93:4380–4385. doi: 10.1073/pnas.93.9.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takano K, Nakabeppu Y, Maki H, Horiuchi T, Sekiguchi M. Structure and function of dnaQ and mutD mutators of Escherichia coli. Mol Gen Genet. 1986;205:9–13. doi: 10.1007/BF02428026. [DOI] [PubMed] [Google Scholar]