Abstract
SNZ1, a member of a highly conserved gene family, was first identified through studies of proteins synthesized in stationary-phase yeast cells. There are three SNZ genes in Saccharomyces cerevisiae, each of which has another highly conserved gene, named SNO (SNZ proximal open reading frame), upstream. The DNA sequences and relative positions of SNZ and SNO genes have been phylogenetically conserved. This report details studies of the expression of the SNZ-SNO gene pairs under various conditions and phenotypic analysis of snz-sno mutants. An analysis of total RNA was used to determine that adjacent SNZ-SNO gene pairs are coregulated. SNZ2/3 and SNO2/3 mRNAs are induced prior to the diauxic shift and decrease in abundance during the postdiauxic phase, when SNZ1 and SNO1 are induced. In snz2 snz3 mutants, SNZ1 mRNA is induced prior to the diauxic shift, when SNZ2/3 mRNAs are normally induced. Under nitrogen-limiting conditions, SNZ1 mRNAs accumulate in tryptophan, adenine, and uracil auxotrophs but not in prototrophic strains, indicating that induction occurs in response to the limitation of specific nutrients. Strains carrying deletions in all SNZ-SNO gene pairs are viable, but snz1 and sno1 mutants are sensitive to 6-azauracil (6-AU), an inhibitor of purine and pyrimidine biosynthetic enzymes, and methylene blue, a producer of singlet oxygen. The conservation of sequence and chromosomal position, the coregulation and pattern of expression of SNZ1 and SNO1 genes, and the sensitivity of snz1 and sno1 mutants to 6-AU support the hypothesis that the associated proteins are part of an ancient response to nutrient limitation.
It is generally thought that the degree of conservation of genes and their respective proteins through species evolution is a measure of their importance for the survival of an organism. In fact, most highly conserved proteins, such as Hsp70, are essential in most organisms (14). We recently identified the Snz1 protein in Saccharomyces cerevisiae, which is the most highly conserved protein present in all three phylogenetic domains, exhibiting 60% identity with Snz proteins in archaea and bacteria (4). Because of the high degree of conservation of Snz1p, it was surprising to us that snz1 deletion mutants exhibited no defects in viability or growth rate under a variety of laboratory conditions.
Although Snz proteins are found in bacteria, archaea, and eucarya including plants and fungi, very little is understood about the specific function of these proteins. Snz proteins contain no distinct functional motifs, although they exhibit very distant relationships with proteins involved in amino acid, vitamin, and nucleic acid biosynthesis, such as bacterial HisF, TrpC, and ThiG, and appear to contain a phosphate-binding region (10). Nevertheless, Snz-related proteins do appear to have a role in stress responses. For example, in the rubber tree Hevea brasiliensis, an SNZ-related gene is induced in response to ethylene and salicylic acid (31). In Bacillus subtilis, a Snz homologue is guanylated during sporulation (23) and is induced in response to oxygen stress (2). In the ascomycete Cercospora nicotianae, Snz1-related protein Sor1 is required for resistance to singlet oxygen-generating photosensitizers (7).
We initially identified Snz1p as a protein whose synthesis increases dramatically as yeast cells enter stationary phase (9). There are three SNZ-related genes in yeast, SNZ1, SNZ2, and SNZ3. Each of these genes is found adjacent to another conserved gene family we have named SNO (Snz-proximal open reading frame [ORF]), recently described as snzB (10). Although Sno proteins, like Snz proteins, have an unknown function, they show some sequence similarity to glutamine amidotransferases (10).
The high degree of conservation of Snz and Sno proteins, the conservation of their proximal chromosome location, and the unique pattern of expression of SNZ1 compelled us to investigate the regulation and function of these gene families and their encoded proteins. We report here that adjacent SNZ and SNO genes are coregulated during growth to stationary phase and during nutrient starvation; that Snz1p and Au Sno1p interact, as determined by a two-hybrid analysis; and that expression of SNZ1 is repressed by expression of SNZ2 and SNZ3 during growth to stationary phase. We have determined that snz1 and sno1 mutants are sensitive to 6-azauracil (6-AU), an inhibitor of purine and pyrimidine biosynthesis (15), and methylene blue, a producer of singlet oxygen. Our results support the hypotheses that SNZ- and SNO-related genes have been linked through evolutionary time and that they are involved in an ancient response to nutrient limitation.
MATERIALS AND METHODS
Strains and media.
The following media were used to cultivate yeast: YPD (1% yeast extract, 2% peptone, 2% glucose), synthetic complete (SC) medium (0.67% Bacto-yeast nitrogen base without amino acids [Difco], 2% glucose; supplemented with auxotrophic requirements but lacking the amino acids for which one is selecting, unless indicated otherwise) (27), and nonsupplemented YNB (2% glucose, 0.17% Bacto-yeast nitrogen base without amino acids and ammonium sulfate [Difco], 16.7% succinate buffer) (18). When indicated (see Results), supplements were added to the YNB media (supplemented YNB) to the following final concentrations: adenine, 0.06 mg/ml; uracil, tryptophan, and histidine, 0.02 mg/ml; and leucine, 0.03 mg/ml. Solid media contained 2% agar. For each liquid-culture experiment, yeast cells were shaken at 250 rpm in 100 ml of medium at 30°C for the time indicated (see Results).
Yeast strains are listed in Table 1. Most of the strains produced for this study were derived from the common laboratory strains W303-1A (MW644) and W303-1B (MW647) (32). All yeast transformations were performed by the lithium acetate protocol or the quick-colony method (11). Transformants were selected on SC media lacking only the auxotrophic requirement used in the selection.
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| MW751 | MATa his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52 | F. Winston |
| MW481 | MATα gal2 mal2 | G. Fink |
| MW644 | MATa leu 2-3,12 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 | R. Rothstein |
| MW647 | MATα leu 2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 | R. Rothstein |
| PJ69-4A | Mata ade2-1 his3-200 leu2-3,112 lys2-801 gal4Δgal80ΔGAL2-ADE2 trp1-901 ura3-52 LYS2:: GAL1-HIS3, MET2::GAL7-LacZ | P. James |
| MW1128 | MATα trp1-1 can1-100 | This study |
| MW1201 | MATα his3-11,15 can1-100 trp1-1 YCplac22(TRP1) | This study |
| MW1203 | MATa ade2-1 can1-100 trp1-1 YCplac22(TRP1) | This study |
| MW1199 | MATa can1-100 trp1-1 YCplac22(TRP1) | This study |
| MW1207 | MATa ura3-1 can1-100 trp1-1 YCplac22(TRP1) | This study |
| MW740 | MATa leu 2-3,112 trp1-1 ade2-1 his3-11,15 can1-100 | This study |
| MW810 | MATa snz2-1 snz3-1 leu 2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 | This study |
| MW926 | MATα snz1Δ2 leu 2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 | E. Braun |
| MW1435 | MATα snz1Δ2 leu 2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 YCplac19(TRP1 URA3) | This study |
| MW980 | MATa snz1Δ3sno1Δ3 snz2Δ3sno2Δ3 snz3Δ3sno3Δ3 leu 2-3,112 trp1-1, ura3-1 ade2-1 his3-11,15 can1-100 | This study |
| MW1071 | MATa his3-11 trp1-1 | This study |
| MW1072 | MATa ade2-1 his3-11 trp1-1 | This study |
| MW983 | MATα, snz1Δ3sno1Δ3 snz2Δ3sno2Δ3 snz3Δ3sno3Δ3 leu 2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 | This study |
| MW908 | MATa snz1Δ3sno1Δ3 leu 2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 | This study |
| MW1286 | MATα snz2Δ3sno2Δ3 snz3Δ3sno3Δ3 leu 2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 YCplac19(TRP1 URA3) | This study |
| MW1283 | MATα trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 YCplac19(TRP1 URA3) | This study |
| MW1359 | MATa/MATα sno1-1/SNO1 cryR/CRY1 HIS3/his3-Δ200 leu2-Δ98/leu2Δ98 can1R/CAN1 CYH2/cyh2R trp1-1/TRP1 | M. Snyder |
| MW1434 | MATa/MATα ssa4::URA3/SSA4 cryR/CRY1 HIS3/his3-Δ200 leu2-Δ98/leu2Δ98 can1R/CAN1 CYH2/cyh2R trp1-1/TRP1 | M. Snyder |
| MW1427 | MATa sno1-1 his3-Δ200 leu2-Δ98 trp1-1 | This study |
To obtain strains with single tryptophan (MW1128), histidine (MW1201), uracil (MW1207), or adenine (MW1203) auxotrophies, PCR fragments containing the appropriate wild-type genes were obtained from common laboratory strain S288C (MW481) (25) and introduced by linear transformation into a W303-1 strain (Table 1). To obtain strains that are prototrophic for tryptophan, W303-1 strains were transformed with the YCplac22 CEN plasmid (12). Escherichia coli XL2-Blue cells were used for propagation of all plasmids and were cultured in Luria broth with ampicillin according to the manufacturer’s recommendations (Stratagene).
Mutant construction.
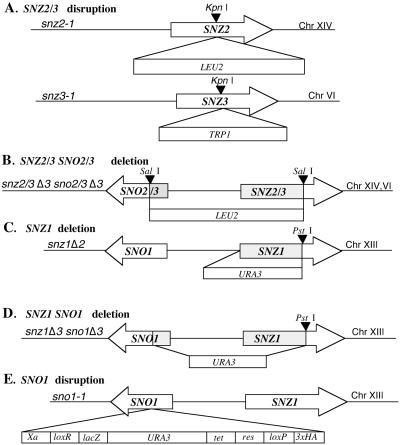
The snz2-1 and snz3-1 disruption alleles were constructed by inserting a 1.8-kb KpnI LEU2 fragment, generated by PCR from YCplac181 (12), into SNZ2 and a 1.4-kb KpnI TRP1 fragment, generated by PCR from YCplac22 (12), into the KpnI site of SNZ3 (Fig. 1A). Chromosome blot analysis and Southern blot analysis were used to confirm the disruption of SNZ2 and SNZ3.
FIG. 1.
SNZ and SNO deletion and disruption mutations. (A) Disruption of SNZ2 and SNZ3. SNZ2 was disrupted with a 1.8-kb LEU2 fragment cloned into the KpnI site, and SNZ3 was disrupted with a 1.1-kb TRP1 fragment cloned into the KpnI site. The resulting alleles are snz2-1 and snz3-1. (B) SNZ2 and -3 and SNO2 and -3 deletion by insertion of a 1.8-kbp LEU2 fragment. The resulting alleles are snz2Δ3sno2Δ3 and snz3Δ3sno3Δ3. (C) SNZ1 deletion by insertion of a 1.1-kbp URA3 fragment. The resulting allele is snz1Δ2. (D) SNZ1 and SNO1 deletion and insertion of a 1.1-kbp URA3 fragment. The resulting allele is snz1Δ3sno1Δ3. (E) SNO1 disrupted with the URA3 fragment carried on transposon mTn-4x. The resulting allele is sno1-1.
The sno2Δ3snz2Δ3 sno3Δ3snz3Δ3 strain was constructed by inserting a 1.8-kb SalI LEU2 fragment generated by PCR from YCplac181 (12) into the SalI sites in SNO3 and SNZ3, respectively. This resulted in the deletion of the SNZ2 and -3 and SNO2 and -3 promoter region as well as 178 bp of the SNO2 and -3 coding region and 575 bp of the SNZ2 and -3 coding region (Fig. 1B). The deletions were confirmed by Southern analysis. The snz1Δ3 sno1Δ3 strain was produced by outward-directed PCR of the snz1Δ1 construct from the pWFY12 plasmid (4). This modification resulted in the deletion of 751 bp of the SNZ1 coding region and 159 bp of the SNO1 coding region as well as the 450 bp between the two genes (Fig. 1D). These strains were mated with sno2Δ3snz2Δ3 sno3Δ3snz3Δ3 strain, and haploid segregants from tetrads were isolated to obtain the snz, sno sextuple mutant (MW980). The construction of the snz1Δ2 strain (Fig. 1C) has been previously described (4).
The heterozygous diploid sno1-1/SNO1 strain, carrying sno1 disrupted by URA3 at amino acid 139 (of 224) (Fig. 1E), and a control ssa4/SSA4 strain (MW1434) were obtained from Mike Snyder (28). Conventional dissection techniques were used to obtain the haploid mutant strains.
Chromosome analysis.
Yeast chromosomes were isolated and separated according to protocols for the Bio-Rad CHEF-DR II gel apparatus. Gels were blotted to GeneScreen membranes (NEN) and probed with either an SNZ1 or an SNZ3 probe, labeled with 32P by using random primers (Pharmacia).
Analysis of mRNA accumulation.
Total RNA was prepared with the Purescript RNA isolation kit (Gentra Systems Inc.), except that glass beads were used to lyse the cells. Briefly, the cells were vortexed with the glass beads for 30 s and put on ice for an additional 30 s, a cycle which was repeated three times. This produced a better yield of total RNA, especially for stationary-phase cells. Electrophoresis, hybridizations, and digitizing of autoradiograms were performed as previously described (4).
Two-hybrid analysis.
Two-hybrid analysis was carried out by using genomic GAL4 activation domain libraries YL2H-C1, -C2, and -C3 and yeast host strain PJ69-4A (17). This experiment was performed once, so it was not an exhaustive search. Amplification of the libraries, resulting in greater than 20 million transformants, was achieved with electrocompetent cells (Gibco BRL) and the Gibco BRL electroporator. The transformants were pooled and inoculated into 3 liters of Terrific Broth medium and grown at 30°C to an optical density (OD) of 1.1. (17). Plasmid DNA was isolated with Maxiprep columns (Qiagen). Plasmids (100 ng) from the three activation domain libraries were separately transformed (17) into PJ69-4A harboring the GAL4-binding domain plasmid, pGBT9 (Clontech), into which the SNZ1 ORF had been subcloned in frame.
Transformants were selected on SC medium lacking leucine and tryptophan (to maintain plasmids) and containing histidine–2 mM 3-aminotriazole (to test the two-hybrid interaction). In PJ69-4A, under these conditions, the auxotrophic requirements can be met only when the Gal4-Snz1 fusion proteins interact and activate transcription of the GAL1-HIS3 reporter gene (17). Two additional reporter genes in PJ69-4A, GAL2-ADE2 and GAL4-lacZ, were used to confirm two-hybrid interacting candidates by screening for growth on medium lacking adenine and by determining β-galactosidase activity. β-Galactosidase assays were performed as described by Miller (22). Plasmids were isolated from cells that showed positive two-hybrid interactions and were retransformed into E. coli DH10B cells. The DNA of isolated plasmids was sequenced (Amersham; Thermosequenase) and further analyzed by using DNASIS 2.0 software (Hitachi) and the National Center for Biotechnology Information Web site (http://www.ncbi.n.lm.nih.gov/BLAST/). Positive controls, i.e., proteins known to interact, p53 and simian virus 40 large T antigen, were obtained from Clontech.
Analysis of the Snz1p/Sno1p complex by nondenaturing gel electrophoresis.
Cell pellets (20 OD at 600 nm [OD600] units) were collected from 8-day stationary-phase cultures and resuspended in radioimmunoprecipitation assay lysis buffer (0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 0.5% deoxycholate, 150 mM NaCl, 50 mM TRIS, pH 8) containing complete protease inhibitors (Boehringer Mannheim). Acid-washed glass beads were added to the cells, and the suspension was vortexed for 10 min at 4°C. After centrifugation in a microcentrifuge for 10 min at 22,000 × g, the supernatant was collected and assayed with bicinchoninic acid (Pierce) to determine protein concentration. Approximately 20 μg of protein was loaded per lane and separated (100 V, 20 h) on a 4-to-20% nondenaturing polyacrylamide gel electrophoresis precast gel (FMC), along with native high-molecular-weight standards (HMW calibration kit proteins; Pharmacia), to determine the size of the complex. After electrophoresis, proteins were blotted onto an Immobilon-P membrane (Millipore) and probed with anti-Snz1,2,3p antibody (rabbit polyclonal antibody; generated to the common N-terminal peptide sequence ALESIPADMRKSGKVC) (QCB). The blot was dried and probed according to Millipore’s rapid chemiluminescence protocol. A peroxidase-conjugated anti-rabbit secondary antibody (Amersham) was used to detect the anti-Snz antibody bound to the blotted protein. The blot was then developed with BLAZE SuperSignal chemiluminescent detection reagent (Pierce). After detection, the blot was stained for total proteins with GelCode Blue (Pierce).
6-AU, methylene blue, and MPA sensitivities.
6-AU sensitivity was evaluated on SC medium lacking uracil and supplemented with 6-AU to a final concentration of 30.0 μg/ml (15). Mycophenolic acid (MPA) sensitivity was scored on SC medium lacking adenine, guanine, and uracil and supplemented with MPA at a final concentration of 30.0 μg/ml (15). When indicated (see Results) uracil was added to the SC medium to a final concentration of 30.0 μg/ml. Strains were incubated at 30°C for 2 days.
To evaluate methylene blue sensitivity, yeast strains were grown overnight in YPD and the growth medium was diluted to a final OD600 of 0.25/ml and then diluted by 1/5 for a series of five dilutions. Then, 6 μl from each of these dilutions was plated onto YPD medium, supplemented with 37.0 μg of methylene blue per ml, and exposed to a light source, when indicated (see Results), for 3 days at 25°C (7).
Sequence analysis.
Sequences were retrieved from databases by using the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov/BLAST/). Identification of SNZ and SNO sequences was carried out with BLAST and BLAST2 software (1a). Genome sequences examined were those for S. cerevisiae Sno1p (Swiss-Prot Q03144), Sno2p (Swiss-Prot P53823), and Sno3p (Swiss-Prot P43544); B. subtilis SnzP (Swiss-Prot P37527) and SnoP (Swiss-Prot P37528); Haemophilus influenzae SnzP (Swiss-Prot P45293) and SnoP (Swiss-Prot P45294); Mycobacterium tuberculosis SnzP (Swiss-Prot O06208) and SnoP (Swiss-Prot e1299817); Methanococcus jannaschii SnzP (Swiss-Prot Q58090) and SnoP (Swiss-Prot Q59055); Pyrococcus horikoshii SnzP (DDBJ d1028463) and SnoP (DDBJ d1028462); Mycobacterium leprae SnzP (Swiss-Prot O07145) and SnoP (Swiss-Prot S72721); Methanobacterium thermoautotrophicum SnzP (Swiss-Prot O26762) and SnoP (GenBank 2621878); and Archaeoglobus fulgidus SnzP (Swiss-Prot O29742) and SnoP (GenBank 2650108). Multiple sequence alignments were made with CLUSTAL W (32a). Percent identity values correspond to the percentages of identical amino acids in multiple sequence alignments, calculated with the ProtST program (1).
RESULTS
The SNZ family in yeast.
SNZ1 was previously identified, based on Southern hybridization and sequence analysis, as a member of a multigene family in yeast (4). The presence of two additional SNZ1 homologues, SNZ2 and SNZ3, was confirmed after publication of the entire yeast genome (13). SNZ1 is situated proximal to the centromere on the right arm of chromosome XIII. SNZ2 and SNZ3 are located in the telomeric regions on the left arms of chromosomes XIV and VI, respectively, within a 7-kb region that is nearly identical between these two chromosomes. This duplicated region is not observed in the telomeric regions of other yeast chromosomes (13). The Snz2p sequence is approximately 99% identical to that of Snz3p and approximately 80% identical to that of Snz1p.
Duplicated genes are often present in variable numbers even in closely related yeast strains (5, 6, 24). To determine whether this was also true for SNZ genes, we assessed the numbers of SNZ genes in several laboratory yeast strains by Southern hybridization of chromosome blots. All of the strains we examined carried a single copy of a gene closely related to SNZ1 but carried variable numbers of SNZ2 and -3 genes. S288C (25), W303 (32), and YPH (30) contain a single SNZ1 homologue and two genes more closely related to SNZ2 and -3 (data not shown). At least one strain, Σ1278, which grows pseudohyphally (19), contains a single copy of SNZ1 and does not contain genes related to SNZ2 or SNZ3. Finally, DS10, which is derived from S288C (34), contains a fourth gene related to SNZ2 and -3 on chromosome II. Although we do not know the chromosomal position of this fourth SNZ gene, its similarity to SNZ2 and SNZ3 suggests that it may have arisen from gene duplication in the telomeric region.
SNZ genes are adjacent to members of a second highly conserved gene family, the SNO genes.
An analysis of the sequences adjacent to the yeast SNZ genes revealed an additional conserved, duplicated gene upstream of each SNZ gene, which we called SNO (for SNZ-proximal ORF). The presence of SNO2 and SNO3 genes adjacent to SNZ2 and SNZ3 was expected because of the chromosomal duplication that included the entire region. The presence of the SNO1 gene adjacent to the more centromeric SNZ1 was surprising and suggested that the original duplication of SNZ1 also included the entire SNO1 gene. Like SNZ2 and SNZ3 genes, SNO2 and SNO3 encode proteins that are almost 100% identical to each other and that are approximately 72% identical to Sno1p. The Sno proteins are predicted to have a molecular mass of 21.5 kDa.
It was recently reported that the Sno proteins are conserved in all three phylogenetic domains (10). Our comparison of the predicted yeast Sno1 protein sequence to the protein sequences of Sno1p homologues extends this observation (Table 2). Yeast Sno1p is 40% identical to the B. subtilis homologue (SnzB) and 37% identical to the M. jannaschii homologue. The M. jannaschii and B. subtilis (SnzB) proteins are 42% identical. Overall, the Sno proteins are less conserved than other highly conserved proteins such as the Hsp70 and Snz proteins (Table 2), suggesting that there are fewer constraints on the structure and sequence of Sno proteins.
TABLE 2.
Comparison of similarities between Sno proteins and other highly conserved proteins
| Protein | % Identity between proteins of:
|
||
|---|---|---|---|
|
S. cerevisiae and
|
M. jannaschii and B. subtilis | ||
| B. subtilis | M. jannaschii | ||
| Sno1p | 40 | 37 | 42 |
| Snz1pa | 58 | 61 | 67 |
| Hsp70a | 48 | 50 | 66 |
| Enolasea | 52 | 59 | 57 |
| EF-2a | 34 | 57 | 36 |
| GAPDHa,b | 27 | 35 | 33 |
| EF-1aa | 52 | 19 | 21 |
a See reference 4.
b GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
On the basis of the microbial genomes that have been entirely sequenced, all species that contain an SNZ gene also contain a SNO gene. In organisms for which the complete genome sequences are available, the proximal location of the SNZ and SNO genes is relatively conserved but the orientations of the two genes differ between eukaryotes and prokaryotes. In yeast, SNZ and SNO genes are adjacent and divergently transcribed (this work and reference 10), whereas in the bacteria B. subtilis, H. influenzae, and M. leprae the SNZ and SNO homologues are in apparent operons (10), i.e., they are adjacent and transcribed in the same direction. In the bacterium M. tuberculosis, SNZ and SNO are separated by a single gene, whose product has some homology to thioesterases. In the archaea P. horikoshii and A. fulgidus SNZ and SNO are also in an apparent operon. These observations suggest that there has been strong selective pressure for the retention of the proximal location of SNZ and SNO genes through evolution. M. jannaschii (10) and M. thermoautotrophicum are the two exceptions; for them the chromosomal location is not conserved between SNZ and SNO genes.
SNZ and SNO gene expression during growth to stationary phase.
We previously reported that SNZ1 was induced at low levels during the postdiauxic phase and induced 8- to 10-fold in stationary phase (4). Similar patterns of SNZ1 mRNA accumulation in other laboratory strains have been observed, indicating that the unique regulation of SNZ1 is not strain dependent (data not shown).
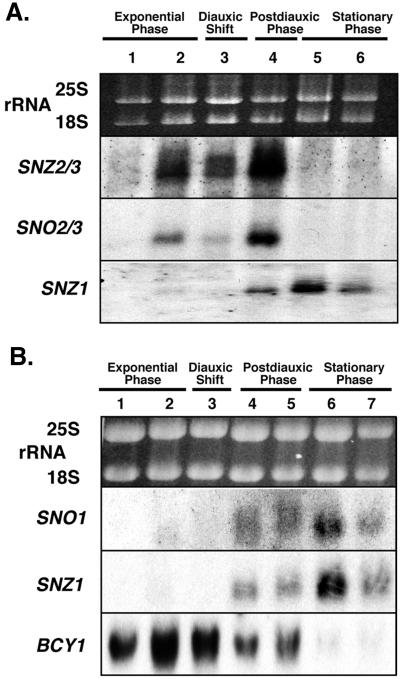
Northern analysis of total RNA was used to assay SNZ2 and SNZ3 mRNA accumulation during growth to stationary phase (Fig. 2A). We refer to these mRNAs as SNZ2/3 because the close identity between SNZ2 and SNZ3 does not allow distinction between their mRNAs. The SNZ2/3 pattern of expression differs noticeably from that of SNZ1 in cells grown to stationary phase (Fig. 2A). SNZ2/3 mRNAs accumulate slightly before the diauxic shift, decrease in abundance at the diauxic shift, and increase for a short period after the diauxic shift. Unlike SNZ1 mRNA, SNZ2/3 mRNAs are not detectable in Northern analysis of total RNA from stationary-phase cells. rRNAs are used as a control for loading because most mRNAs, e.g., that for actin, decrease in abundance in stationary phase and we currently have no RNA that remains constant in both exponential and stationary phases and that therefore could be used as an internal reference. For these analyses, we have used SNZ1 and BCY1 as controls in some blots to allow us to demonstrate the intactness of mRNAs in cells grown to stationary phase (Fig. 2), because the mRNA levels of both SNZ1 and BCY1 in cells grown to stationary phase are known (4).
FIG. 2.
Northern analysis of SNZ and SNO gene expression in yeast cells grown to stationary phase. (A) Northern blot probed with SNZ3, SNO3, and SNZ1 to show the relative timing of SNZ2 and -3 and SNO2 and -3 expression relative to that of SNZ1. Ethidium bromide-stained rRNAs are presented to show relative loading. Lanes: 1, early exponential phase (OD600 = 0.95); 2, late exponential phase (OD600 = 5.0); 3, diauxic shift (OD600 = 7.1; determined by glucose exhaustion); 4, 24 h after the diauxic shift (OD600 = 10.5); 5, stationary phase (5 days after inoculation); 6, stationary phase (8 days after inoculation). (B) Northern blot probed with SNO1, SNZ1, and BCY1 to show the timing of SNO1 expression relative to that of SNZ1. Lanes: 1, early exponential phase (OD600 = 1.4); 2, late exponential phase (OD600 = 6.6); 3, diauxic shift (OD600 = 7.1); 4, 22 h after the diauxic shift (OD600 = 10.5); 5, 48 h after the diauxic shift (OD600 = 19.6); 6, stationary phase (5 days after inoculation); 7, stationary phase (8 days after inoculation). BCY1 was used as a control. Autoradiographs were exposed for 3 days (SNZ2 and -3, SNO2 and -3, BCY1, and SNZ1) or 5 days (SNO1).
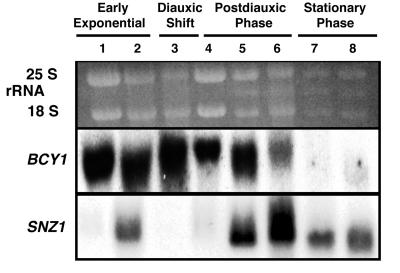
The pattern of SNZ1 expression during growth to stationary phase is altered dramatically in snz2,3 disruption mutants (Fig. 3). In snz2,3 mutants, SNZ1 mRNA levels increase during late exponential phase, decrease at the diauxic shift and shortly thereafter, increase again 24 h after the diauxic shift, and remain high in stationary phase. This novel pattern of SNZ1 mRNA accumulation prior to the diauxic shift in snz2,3 mutants is similar to that of SNZ2/3 mRNA accumulation in control strains. These results suggest that SNZ2 and -3 directly or indirectly regulate SNZ1 expression. SNZ2/3 mRNA accumulation is not altered in an snz1Δ2 mutant grown to stationary phase (data not shown), which indicates that the converse regulation of SNZ2 and -3 by SNZ1 does not occur.
FIG. 3.
Northern analysis of SNZ1 expression in an snz2 snz3 mutant during growth to stationary phase. The Northern blot was probed with SNZ1 to show the relative timing of SNZ1 expression in an snz2-1 snz3-1 strain. Ethidium bromide-stained rRNAs are presented to show relative loading. Lanes: 1, early exponential phase (OD600 = 3.7); 2, late exponential phase (OD600 = 5.7); 3, diauxic shift (determined by glucose exhaustion) (OD600 = 6.2); 4, 2 h after the diauxic shift (OD600 = 7.0); 5, 24 h after the diauxic shift; 6, 48 h after the diauxic shift; 7, stationary phase (5 days after inoculation); 8, stationary phase (8 days after inoculation). BCY1 was used as a control. Autoradiographs were exposed for 4 days (BCY1) or 2 days (SNZ1).
All three SNZ-SNO gene pairs in yeast are divergently transcribed and are separated by 400 to 450 bp. Because the positions of the SNZ and SNO genes are conserved during evolution and because their relative orientations in yeast suggested that they share common promoter elements, we wanted to determine whether they were coregulated. Northern analysis of total RNAs revealed that SNO1 and SNZ1 mRNAs do exhibit the same pattern of expression in cells grown to stationary phase (Fig. 2B). Likewise, SNO2/3 mRNAs, which are also indistinguishable from each other by Northern analysis, accumulate at the same time as SNZ2/3 mRNAs (Fig. 2A). We concluded from this that the SNZ-SNO gene pairs are coregulated and that the ability to coregulate these genes might have been an important factor in maintaining their proximity and orientations during evolution.
Snz and Sno proteins interact.
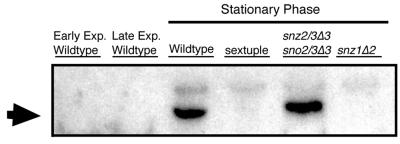
In order to determine if Snz proteins interact as a higher-order complex, we analyzed extracts of wild-type and various snz mutants via nondenaturing gradient polyacrylamide gel electrophoresis. This method involves running samples on a 4-to-20% acrylamide gel for 20 h through a matrix of decreasing pore size until proteins stop at an impassable acrylamide percentage. Because of the long run, proteins that migrate slowly due to low charge still run to the limiting pore size of the gel. A protein’s migration on these gels is based solely on shape and size, allowing specific size comparisons, as opposed to that of proteins analyzed by continuous nondenaturing electrophoresis, where protein migration is based on size, charge, and shape (21). The analysis revealed that Snz proteins are part of a complex, in stationary-phase cells, with an apparent molecular mass of approximately 230 kDa (Fig. 4). The antibody to the common N-terminal peptide that was produced is capable of recognizing all three Snz proteins in yeast (data not shown). However, the complex is only present when Snz1p is present (Fig. 4). Thus, we conclude that Snz1p is part of a protein complex in stationary-phase cells.
FIG. 4.
Western analysis of Snz proteins separated by nondenaturing polyacrylamide gel electrophoresis. Proteins were isolated, seperated, and blotted as described in Materials and Methods. Snz proteins were visualized by using a rabbit polyclonal antibody to the N termini of Snz proteins and a secondary anti-rabbit horseradish peroxidase-conjugated antibody in combination with a chemiluminescence assay (Pierce) as described in Materials and Methods. The arrow indicates the Snz-reactive band. Lanes (from left to right): early exponential phase (OD600 = 2.7; wild-type strain [MW1072]); late exponential phase (OD600 = 7.2; wild-type strain [MW1072]); stationary phase, wild-type strain (MW1072); stationary phase, sextuple mutant (MW983); stationary phase, snz2/3Δ3 sno2/3Δ3 mutant (MW 976); stationary phase, snz1Δ3 sno1Δ3 mutant (MW908).
To learn more about the Snz1p complex, we used two-hybrid analysis to identify proteins that might interact with Snz1p (8). We first examined whether Snz1p could interact with itself by cloning the SNZ1 ORF in frame with the GAL4-binding domain (GAL4-bd) and GAL4 activation domain (GAL4-ad). By themselves, the Gal4-Snz1 activation or binding domain fusion proteins did not activate GAL7-lacZ (Table 3) or allow growth on media lacking histidine or adenine. When cotransformed into yeast cells, the activation and binding domain fusion proteins activated the transcription of a GAL7-lacZ reporter gene (Table 3) and allowed strain PJ69-4A, which is conditionally auxotrophic for histidine and adenine, to grow in the absence of these supplements. These results suggest that Snz1p could function in the cell as a dimer or higher-order oligomer.
TABLE 3.
Two-hybrid analysis of Snz1p: interaction of Snz1p with ORF fused to GAL4 transactivation domain
| ORF | Interaction strength (β-galactosidase units) |
|---|---|
| None | <1 |
| SNZ1 | 21.4 ± 9.2 |
| SNZ2 | 9.2 ± 3.3 |
| SNO1 | 24.1 ± 10.6 |
| TPK1 | <1 |
| SV40a large T antigen ORF | 1.0 ± 0.7 |
a SV40, simian virus 40.
To identify other proteins that interact with Snz1p, yeast cells were transformed with the GAL4-bd–SNZ1 vector and a yeast GAL4-ad library (17). Plasmids from cells that grew on medium lacking histidine were isolated for further study. The results of this screen yielded two plasmids with activating genes SNZ2 and SNO1 (Table 3). The plasmid containing SNO1 includes all of the ORF except for the first 40 nucleotides. The plasmid containing SNZ2 includes more than three-fourths of the coding region. Based on β-galactosidase assays, the interaction between Snz1p and Snz2p is not as strong as the interaction between Snz1p and Snz1p. However, the interactions between Snz1p and Sno1p and between Snz1p and Snz1p are equally strong. These results suggest that Snz1p and Sno1p may function in the cell as an oligomeric complex. The apparent physical interaction between Snz1p and Sno1p, the coregulation of their genes, and their conserved proximity on the chromosome provide strong support for the hypothesis that these two proteins are involved in the same pathway.
SNZ1 is induced in response to starvation for specific nutrients.
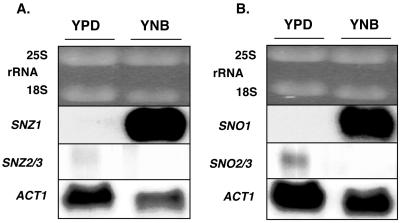
The different patterns of SNZ1 and SNZ2/3 mRNA accumulation during growth to stationary phase are likely to reflect a response to the changing nutritional environment. To determine whether limitation for specific nutrients affects the regulation of SNZ genes, auxotrophic cells (MW644) that have all wild-type SNZ and SNO genes were transferred from rich, glucose-based medium to nonsupplemented, nitrogen-limiting medium (YNB medium). Northern analysis revealed that SNZ1 and SNO1 mRNAs accumulate in cells transferred to nonsupplemented YNB medium whereas SNZ2/3 and SNO2/3 mRNAs did not (Fig. 5). These results indicate that under specific starvation conditions as well as in cells grown to stationary phase, SNZ and SNO genes are coregulated and that SNZ1 and SNZ2 and -3 are controlled by distinct regulatory mechanisms.
FIG. 5.
Northern analysis of SNZ and SNO mRNA accumulation in nitrogen-starved cells. Total RNA was isolated from yeast cells (MW644) grown overnight in YPD medium to an OD600 of 2.06 (YPD lanes). YNB-labeled lanes show RNA from yeast cells transferred from YPD to YNB medium for 90 min. Ethidium bromide-stained rRNAs are shown to indicate relative loading. (A) Northern blot probed with SNZ3 and SNZ1. (B) Northern blot probed with SNO3 and SNO1. Northern blots were probed with ACT1 (actin gene) as a control. Autoradiographs were exposed for 1 day.
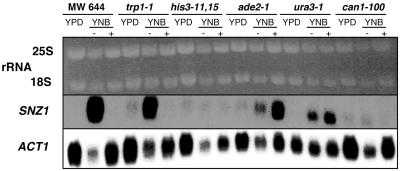
To determine whether the accumulation of SNZ1 mRNA in cells incubated in YNB was due to starvation for specific nutrients or a general nitrogen starvation signal, we examined SNZ1 expression in auxotrophic, W303-derived cells (MW644) transferred from rich, glucose-based medium to YNB medium supplemented with auxotrophic requirements (Fig. 6). Under these conditions, SNZ1 mRNA does not accumulate, suggesting that SNZ1 induction is not due to general nitrogen limitation but to the absence of one or more auxotrophic requirements.
FIG. 6.
Northern analysis of SNZ1 expression in auxotrophic and prototrophic strains in the presence or absence of auxotrophic requirements. A Northern blot of total RNA isolated from cells grown overnight in YPD medium to an OD600 of 2.0 to 3.0 and from cells transferred from YPD to YNB medium for 90 min is shown. The blot was probed with SNZ1. Auxotrophic strains were incubated with (+) or without (−) their specific auxotrophic requirements. Genotypes of the strains are as follows: leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 (MW644); trp1-1 (MW1128); his3-11,15 (MW1201); ade2-1 (MW1203); ura3-1 (MW1207); and can1-100 (MW1199). Ethidium bromide-stained rRNAs are shown to indicate loading. The Northern blot was probed with ACT1 as a control. The autoradiograph was exposed for 1 day.
To identify the nutrient limitation responsible for the dramatic increase in SNZ1 expression, we evaluated SNZ1 mRNA accumulation in W303-derived strains that are auxotrophic for a single nutrient (Table 1 and Fig. 6). SNZ1 mRNA accumulated in the trp1-1 (MW1128), ade2-1 (MW1203), and ura3-1 (MW1207) mutants in YNB medium but did not accumulate in the his3-11 (MW1201) or the prototrophic (MW1199) strains incubated in YNB medium (Fig. 6). SNZ1 mRNA did not accumulate when tryptophan was added to YNB medium in which the trp1-1 mutant (MW1128) was incubated. SNZ1 mRNA accumulation in leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 (MW644) cells incubated in YNB medium with supplements was suppressed with the addition of uracil and adenine. Surprisingly, the SNZ1 mRNA accumulation in ade2-1 (MW1203) and ura3-1 (MW1207) mutants incubated in YNB medium with supplements was not suppressed (Fig. 6). These results indicate that an imbalance between nucleotide levels could have an effect on SNZ1 mRNA accumulation. The difference in SNZ1 mRNA accumulation in the leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 (MW644) cells versus the ura3-1 (MW1207) or ade2-1 (MW1203) cells incubated in YNB medium with supplements cannot be directly determined because of the additional auxotrophies in the MW644 cells which could affect the SNZ1 mRNA levels.
Strains derived from another common laboratory strain, S288C, were examined to determine whether SNZ1 induction in YNB medium was a strain-specific phenomenon. As in W303-derived strains, SNZ1 is induced when S288C strains with multiple auxotrophies, e.g., MW751 (his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52), are incubated in YNB medium but is not induced in a prototrophic strain (MW481) (data not shown). We conclude from these results that SNZ1 induction in response to limitation of specific nutrients is a function of the auxotrophies of a given strain and is not a function of strain background.
snz and sno mutant sensitivity to 6-AU.
To further investigate Snz and Sno protein function, we evaluated the phenotypes of snz and sno mutants. Yeast strains carrying snz1, sno1, snz2,3 sno2,3, and snz1,2,3 sno1,2,3 mutations are viable and grow at rates indistinguishable from those of wild-type cells on rich, glucose-based medium (data not shown). To determine if Snz and Sno proteins were essential under other conditions, we tested these mutants for their ability to survive and grow under a variety of different medium and temperature regimes. None of the snz sno mutants exhibited changes in growth rates or viability during growth to stationary phase on glucose medium, acetate-based medium, or nitrogen-limiting medium (YNB medium) at 30 and 37°C, compared with control cells.
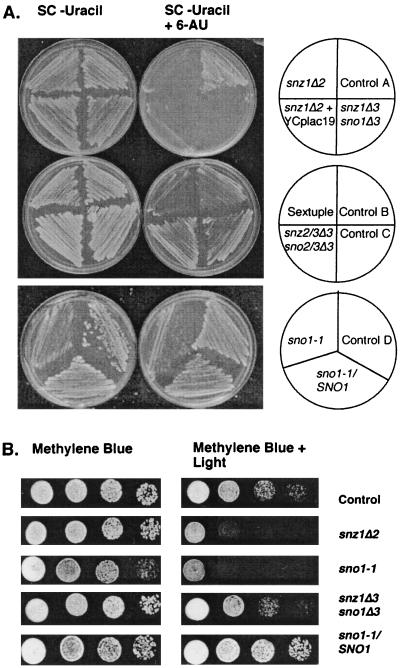
Since SNZ1 mRNA accumulates in ura3 and ade2 mutants starved for uracil and adenine, we tested snz1, sno1, snz2,3 sno2,3, and snz1,2,3 sno1,2,3 mutants for sensitivity to 6-AU, an inhibitor of both pyrimidine and purine biosynthesis. 6-AU inhibits IMP dehydrogenase, encoded by PUR5, and OMP decarboxylase, encoded by URA3 (15). Strains carrying any of the snz1 mutant alleles (Fig. 1), including snz1Δ2 and snz1Δ3 sno1Δ3, and strains carrying sno1 mutations are extremely sensitive to 6-AU (Fig. 7A). The growth inhibition is specific to strains carrying snz1 or sno1 mutations, and is not observed with SNZ1 snz2,3 sno2,3 mutants or wild-type controls (Fig. 7A). The 6-AU sensitivity cosegregates with the snz1 mutation through numerous crosses in both S288C and W303 strain backgrounds.
FIG. 7.
Phenotype analysis of snz and sno mutants. (A) Growth of snz and sno mutants and control strains on minimal medium without uracil, with or without 6-AU. A control is shown for each mutant strain; the control has the same auxotrophic markers as the mutant strain. Strains are as follows: control A, MW740; snz1Δ2 strain, MW926; snz1Δ3sno1Δ3 strain, MW908; snz1Δ2 YCplac19 strain, MW1435; control B MW1071; sextuple snz1,2,3Δ3 sno1,2,3Δ3 mutant strain, MW980; control C, MW1283; snz2,3Δ3 sno2,3Δ3 strain, MW1286; control D, MW1434; heterozygous sno1-1/SNO1 strain, MW1359; sno1-1 strain, MW1427. (B) Growth of snz and sno mutants and control strains on YPD medium supplemented with methylene blue, in the presence or absence of light. Strains are as follows: control strain, MW740; snz1Δ2 strain, MW926; sno1-1 strain, MW1427; snz1Δ3 sno1Δ3 strain, MW908; heterozygous diploid sno1-1/SNO1 strain, MW1359.
The addition of uracil to the medium or the introduction of URA3 on a 2μm plasmid suppresses the 6-AU growth inhibition of snz1 strains (data not shown). However, the introduction of the centromeric plasmid YCplac19 (URA3 TRP1) to the snz1Δ2 mutant does not suppress the 6-AU sensitivity (Fig. 7A). Thus, one copy or a few copies of plasmid-borne URA3 are not sufficient to suppress the 6-AU sensitivity of snz1 mutant strains. However, snz1Δ2 mutants were not complemented by mating to a SNZ1 ura3 strain or by transformation with a CEN plasmid carrying the SNZ1 structural gene and 953 bp of the region upstream of the start codon. This data suggests that the snz1Δ2 mutant allele may cause a dominant-negative effect or an imbalance between Snz1p and Sno1p resulting in a mutant phenotype under these conditions or that the snz1Δ2 mutant allele results in an alteration of SNO1 expression.
We examined complementation by comparison of sno1-1/SNO1 heterozygous diploid and sno1-1 haploid strains which both contain a single gene disrupted by transposon mutagenesis (Fig. 1). To ensure that the 6-AU sensitivity was not due to the URA3 gene used to disrupt SNO1, an ssa4::URA3 mutant was used as a control. The 6-AU sensitivity of the sno1 mutant is complemented in diploids that are heterozygous for SNO1, heterozygous for URA3 at the SNO1 locus, and homozygous at the ura3-52 locus, i.e., they contain only the URA3 gene used to disrupt SNO1. Additionally, sno1 mutants have a slower growth rate on SC-uracil media than the ssa4::URA3 mutant control and the sno1-1 heterozygous diploid (Fig. 7A). We conclude from these results that loss of either Snz1 or Sno1 protein function is responsible for the 6-AU sensitivity observed in these mutants.
To determine whether the 6-AU sensitivity of snz1 mutants was due to inhibition of IMP dehydrogenase or OMP decarboxylase or both, we examined the growth of snz1 mutants in SC medium containing MPA, a specific inhibitor of IMP dehydrogenase (15). The growth of snz1 mutants was not affected by MPA (data not shown), suggesting that the sensitivity occurs through an effect of Snz1p on pyrimidine biosynthesis.
Sensitivity of snz and sno mutants to methylene blue.
A recent report indicated that a mutant of the SNZ1 orthologue in the ascomycete C. nicotianae was sensitive to singlet oxygen generators, including methylene (7). To determine whether S. cerevisiae snz and sno mutants were also sensitive to methylene blue, various mutants and parental controls were grown on methylene blue plates in the light, which stimulates the production of singlet oxygen within the cell. Our results indicated that methylene blue sensitivity is specific to cells carrying snz1 or sno1 mutations (Fig. 7B). Surprisingly, the snz1,2,3Δ3 sno1,2,3Δ3, the snz2,3Δ3 sno2,3Δ3 (data not shown), and the snz1Δ3sno1Δ3 alleles are not sensitive to growth on methylene blue (Fig. 7B). Thus, sensitivity to methylene blue is only exhibited when Snz1p or Sno1p is absent. These results indicate that an imbalance between Snz1p and Sno1p, i.e., the presence of either Snz1p or Sno1p and the absence of the other protein, results in the sensitivity to methylene blue. The sno1-1 mutant sensitivity to methylene blue is complemented in a sno1-1/SNO1 heterozygous mutant (Fig. 7B).
DISCUSSION
We have identified two gene families, named SNZ and SNO, which are highly conserved. Adjacent SNZ and SNO genes are coregulated, and different SNZ-SNO gene pairs are induced at alternate times during growth to stationary phase. The induction of SNZ1 mRNA accumulation in a snz2/3 mutant during late-exponential and postdiauxic phases is reminiscent of the cross-regulation observed in the HSP70 genes in yeast (34). SNZ1 is also induced in auxotrophic mutants in response to the limitation of adenine, uracil, and tryptophan. The observation that Snz1 and Sno1 proteins interact, based on the two-hybrid assay, and the result that both snz1 and sno1 mutants are sensitive to 6-AU and methylene blue, support the hypothesis that these two genes function in the same pathway and that they play a role in nucleic acid metabolism.
Other reports have provided additional clues to Snz and Sno protein function. SNZ genes are induced in response to oxidative stress in B. subtilis (2) and are required for resistance to singlet oxygen in the ascomycete C. nicotianae (7). Results of sequence analysis suggest that Snz proteins are closely related to metabolic enzymes such as ThiG, TrpC, and HisF and that Sno proteins are related to glutamine amidotransferases such as GuaA and HisH (10). Several glutamine amidotransferase reactions are part of both purine and pyrimidine biosynthesis (26, 35); thus, it is possible that Snz1p and Sno1p have an enzymatic function in these pathways.
Mutations in two other yeast genes, PPR1 and PPR2, also result in 6-AU sensitivity (15). PPR1 encodes a transcriptional regulator of pyrimidine pathway genes URA1, URA3, and URA4 (29). ppr1 sensitivity to 6-AU is also suppressed by the addition of uracil to the medium (29). PPR2 encodes TFIIS, a general transcriptional elongation factor, which is sensitive to decreased GTP pools in the cell (3). ppr2 sensitivity to 6-AU is suppressed by the addition of guanine to the medium. The suppression of the snz1 6-AU sensitivity by the addition of uracil suggests that Snz1p, like Ppr1p, may play a role in pyrimidine metabolism. However, the two proteins are not likely to have the same function because, unlike Ppr1p, Snz1p has no motifs that suggest it is a DNA-binding protein.
It is not readily apparent why snz1 and sno1 mutants are sensitive to both methylene blue, a singlet oxygen generator, and 6-AU, a drug which lowers nucleotide pools within the cell. However, there may be some similarities at the subcellular level as a result of these two drugs. Singlet oxygen generators, such as methylene blue, can cause damage to lipids, DNA, and RNA. Singlet oxygen results in modified guanine residues (16, 20), breaks in RNA, cross-linking between strands of RNA, and cross-linking between RNA and proteins (33). It seems possible that methylene blue causes a turnover in RNA or an increase in DNA repair, which results in an alteration of intracellular nucleotide concentrations, in turn resulting in an induction of SNZ1 and SNO1. The sensitivity of either snz1 or sno1 mutants, but not the double mutant, to methylene blue, in addition to the physical interaction of Snz1 and Sno1 proteins, provides evidence that the balance and physical interaction of these proteins may be needed to maintain metabolites that are targets for the drug. Based on our results, we hypothesize that the Snz1p-Sno1p complex, which may have a glutamine amidotransferase activity, plays a role in nucleotide metabolism and is induced in response to stresses that cause an imbalance in, the maintenance of, or a decrease in the concentrations of nucleotides. This hypothesis may seem inconsistent with the fact that SNZ1 and SNO1 are expressed during stationary phase, a time of low concentrations of a carbon source. However, it is conceivable that starvation for a carbon source, in stationary-phase cells, may result in a depletion of other important metabolites such as nucleotides. Thus, it is possible that Snz1p and Sno1p participate in nucleotide metabolism, a process that is likely to be important to starved cells.
Interestingly, although SNZ and SNO genes are found in all three phylogenetic domains, these genes are not present in all organisms. Examples of organisms lacking SNZ and SNO orthologues include E. coli (10), Borrelia burgdorferi, Synechocystis spp., Helicobacter pylori, Mycoplasma pneumoniae, and Mycoplasma genitalium. The majority of the organisms that lack SNZ1 and SNO1 orthologues reside in nutrient-rich environments within the host, e.g., in the stomach or intestine, or adjacent to damaged cells. Thus, one hypothesis that could explain this gene distribution is that SNZ and SNO are important for survival in nutrient-poor conditions and organisms that reside in relatively nutrient-rich conditions may have no selective pressure to maintain these genes. The presence of SNZ and SNO genes in plants seems inconsistent with this hypothesis but in fact may be indicative of the frequency of nutrient limitation experienced by plants due either to poor soils or drought. Snz and Sno proteins have not been identified in animals, which are organisms that are unable to maintain viability under frequent severe starvation conditions. Understanding the specific functions of Snz and Sno proteins allows us to learn more about the life cycles of different organisms as well as the conserved mechanisms used by organisms to deal with strong selective pressures such as starvation.
ACKNOWLEDGMENTS
We thank Mary Anne Nelson and Sepp D. Kohlwein for carefully reading the manuscript and participating in helpful discussions. We thank Fred Winston, Mike Snyder, Phil James, and Rodney Rothstein for generously providing strains and Robert H. White for participating in helpful discussions. We thank Stephanie Atencio, JoAnna Bernacik, and Amy Hahn for excellent technical assistance.
This work was supported by grant MCB9418149, RIMI grant HRD-9550649, and PYI grant MCB-9057514 to M.W.-W. from the National Science Foundation and by MBRS grant 5SO6GM52576-03 from the National Science Foundation and a Patricia Roberts Harris Fellowship to P.A.P.
REFERENCES
- 1.Adachi J, Hasegawa M. ProtST, version 1.1.1. 1994. [Google Scholar]
- 1a.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1995;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann H, Bernhardt J, Schmid R, Mach H, Voelker U, Hecker M. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. doi: 10.1002/elps.1150180820. [DOI] [PubMed] [Google Scholar]
- 3.Archambault J, Lacroute F, Ruet A, Friesen J. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun E L, Fuge E K, Padilla P A, Werner-Washburne M. A stationary-phase gene in Saccharomyces cerevisiae is a member of a novel, highly conserved gene family. J Bacteriol. 1996;178:6865–6872. doi: 10.1128/jb.178.23.6865-6872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson M, Botstein D. Organization of the SUC gene family in Saccharomyces. Mol Cell Biol. 1983;3:351–359. doi: 10.1128/mcb.3.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson M, Celenza J L, Eng F J. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Mol Cell Biol. 1985;5:2894–2902. doi: 10.1128/mcb.5.11.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrenshaft M, Jenns A E, Chung K R, Daub M E. SOR1, a gene required for photosensitizer and singlet oxygen resistance in Cercospora fungi, is highly conserved in divergent organisms. Mol Cell. 1998;1:603–609. doi: 10.1016/s1097-2765(00)80060-x. [DOI] [PubMed] [Google Scholar]
- 8.Fields S, Song O. A novel genetic system to detect protein-protein interaction. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 9.Fuge E K, Braun E L, Werner-Washburne M. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J Bacteriol. 1994;176:5802–5813. doi: 10.1128/jb.176.18.5802-5813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galperin M Y, Koonin E V. Sequence analysis of an exceptionally conserved operon suggests enzymes for a new link between histidine and purine biosynthesis. Mol Microbiol. 1997;24:443–445. doi: 10.1046/j.1365-2958.1997.3671706.x. [DOI] [PubMed] [Google Scholar]
- 11.Gietz R D, Schiestl R H. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 12.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 13.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 14.Gupta R S. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 15.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Ito K, Kawanishi S. Site-specific DNA-damage induced by UVA radiation in the presence of endogenous photosensitizer. Biol Chem. 1997;378:1307–1312. [PubMed] [Google Scholar]
- 17.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston G C. Cell size and budding during starvation of the yeast Saccharomyces cerevisiae. J Bacteriol. 1977;132:738–739. doi: 10.1128/jb.132.2.738-739.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kron S J, Styles C A, Fink G R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kvam E, Berg K, Steen H B. Characterization of singlet oxygen-induced guanine residue damage after photochemical treatment of free nucleosides and DNA. Biochim Biophys Acta. 1994;1217:9–15. [PubMed] [Google Scholar]
- 21.Margolis J, Wrigley C. Improvement of pore gradient electrophoresis by increasing the degree of cross-linking at high acrylamide concentrations. J Chromatogr. 1975;106:204–209. [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 23.Mitchell C, Morris P W, Vary J C. Amino acid sequences of several Bacillus subtilis proteins modified by apparent guanylylation. Mol Microbiol. 1992;6:1579–1581. doi: 10.1111/j.1365-2958.1992.tb00882.x. [DOI] [PubMed] [Google Scholar]
- 24.Mortimer R K, Hawthorne D C. Yeast genetics. In: Rose A H, Harrison J S, editors. The yeasts. New York, N.Y: Academic Press; 1969. pp. 385–460. [Google Scholar]
- 25.Mortimer R K, Johnston J R. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuhard J, Kelln R A. Biosynthesis and conversions of pyrimidines. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 580–599. [Google Scholar]
- 27.Rose M D, Winstron F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 28.Ross-Macdonald P, Sheehan A, Roeder G S, Snyder M. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:190–195. doi: 10.1073/pnas.94.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy S, Katze M G, Parkin N T, Edery I, Hovanessian A G, Sonenberg N. Control of the interferon-induced 68-kilodalton protein kinase by the HIV-1 tat gene product. Science. 1990;247:1216–1219. doi: 10.1126/science.2180064. [DOI] [PubMed] [Google Scholar]
- 30.Sikorski R S, Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivasubramaniam S, Vanniasingham V M, Tan C T, Chua N H. Characterization of HEVER, a novel stress-induced gene from Hevea-brasiliensis. Plant Mol Biol. 1995;29:173–178. doi: 10.1007/BF00019129. [DOI] [PubMed] [Google Scholar]
- 32.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 32a.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuite E M, Kelley J M. Photochemical interactions of methylene-blue and analogs with DNA and other biological substrates. J Photochem Photobiol B Biol. 1993;21:103–124. doi: 10.1016/1011-1344(93)80173-7. [DOI] [PubMed] [Google Scholar]
- 34.Werner-Washburne M, Stone D E, Craig E A. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. [Google Scholar]