Abstract
Background
Staphylococcus aureus isolates are the leading cause of diabetic foot infections (DFIs). Identification of specific virulence factors of S. aureus involved in the pathogenesis of DFIs may help control the infection more effectively. Since the most prevalent virulence factor genes are probably related to the DFI pathogenesis, the aim of this study is to evaluate the proportion of virulence factor genes of S. aureus isolates from DFIs.
Materials and methods
We conducted a systematic search of PubMed, Embase, Web of Science, and Scopus to identify all articles reporting the proportion of different types of virulence factors of S. aureus isolates from DFI samples.
Results
Seventeen studies were eligible, in which 1062 S. aureus isolates were obtained from 1948 patients and 2131 DFI samples. Among the toxin virulence factors, hld 100.0% (95% CI: 97.0, 100.0%), hlg 88.0% (95% CI: 58.0, 100.0%), hla 80.0% (95% CI: 31.0, 100.0%), hlgv 79.0% (95% CI: 35.0, 100.0%) and luk-ED 72.0% (95% CI: 42.0, 95.0%) had the highest proportion respectively. Among the genes associated with biofilm formation, both icaA and icaD had the highest proportion 100.0% (95% CI: 95.6, 100.0%).
Conclusion
The results of the present study showed that among the toxin virulence factors, hemolysins (hld, hlg, hla, hlgv) and luk-ED and among the non-toxin virulence factors, icaA and icaD have the greatest proportion in S. aureus isolates from DFIs. These prevalent genes may have the potential to evaluate as virulence factors involved in DFI pathogenesis. Finding these probable virulence factor genes can help control diabetic foot infection more effectively via anti-virulence therapy or preparation of multi-epitope vaccines.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-023-03142-y.
Keywords: Staphylococcus aureus, Virulence factors, Diabetic foot, Foot ulcer, Infections
Introduction
Staphylococcus aureus is a leading cause of serious infections with high morbidity, mortality and health-related costs. Staphylococcus aureus can cause a variety of clinical diseases via various potential virulence factors. These diseases include bacteremia, endocarditis, osteomyelitis, as well as skin and soft tissue, osteoarticular, pulmonary and device-related infections [1]. In a systematic review and meta-analysis, it was reported that the mortality rate from S. aureus bacteremia was 18.1%, 27.0%, and 30.2% at 1 month, 3 months, and 1 year, respectively [2]. S. aureus is also the leading invasive bacterial pathogen in children in many parts of the world [3].
In particular, S. aureus is one of the most common bacteria isolated from diabetic foot infections (DFIs) worldwide. In our recent systematic review and meta-analysis, we reported that the highest pooled proportion of isolated bacteria from DFIs in Iran belongs to S. aureus (24.29%), of which 55% were methicillin resistant strains (MRSA) [4].
Fighting this leading bacterium presents two major challenges. The first problem is that S. aureus expresses many potential toxin and non-toxin virulence factors that intensively target many surfaces and tissues. The second problem is the increasing resistance of S. aureus isolates from DFIs to the most commonly prescribed antibiotics. In fact, MRSA has emerged as one of the major epidemiological and clinical problems [5].
Toxin virulence factors are classified into pore-forming toxins, exfoliative toxins, enterotoxins and epidermal cell differentiation inhibitor toxins. The pore-forming toxins include the single-component α-toxin (α-hemolysin), the phenol-soluble modulins (PSMs), and bi-component leukotoxins, including Panton-Valentine leukocidin (PVL), γ-hemolysin, and leukocidin E/D [6]. Some of the non-toxin virulence factor genes are involved in biofilm formation, such as: icaA, icaD and atl as well as pls. S.aureus produces surface proteins called MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules) and mediates adhesion to the ulcer surface [7]. Typical members of the MSCRAMM family are staphylococcal protein A (SpA), collagen-binding protein, fibronectin-binding proteins A and B (FnbpA and FnbpB), and clumping factor proteins (Clf) A and B [8].
It is the time to focus on new antimicrobial agents for resolving the above-mentioned problems. Among the new therapeutic strategies, anti-virulence therapy has emerged as a new promising strategy [9]. In this method, instead of fighting the bacteria, their pathogenic virulence factors are targeted [9]. Unlike conventional antibiotics, this method may cause lower selective pressure over pathogens and therefore lower emergence and spread of resistance [9].
Given the wide range of different virulence factors mentioned above, an important question arises as to which of these factors of S. aureus can be specifically related to DFI pathogenesis. Several studies measured and characterized the virulence factors of S. aureus isolates from DFIs [10–26]. and some of them introduced potential virulence factors to distinguish colonization from infection [10–12].
Since the identification of the most prevalent virulence factor genes of infecting S. aureus isolates may be related to both their pathogenesis and the differentiation between colonization and infection, the aim of this systematic review and meta-analysis is to evaluate the proportion of virulence factor genes of S. aureus isolates from DFIs.
Materials and methods
Study protocol
This systematic review and meta-analysis was conducted in accordance with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines [27] and a PRISMA checklist was completed. The study protocol has been registered at the Isfahan University of Medical Sciences with the national ethics code of IR.MUI.MED.REC.1399.450.
Data sources and searching strategy
We ran a thorough search in PubMed, Embase, Web of Science, and Scopus following Mesh terms and keywords: ‘virulence’, ‘pathogenicity’, “pathogenicity factor*”, “virulence factor*”, “virulence gene*” And “Staphylococcus aureus”, “S. aureus” And “diabetic foot”, “diabetic feet” And ‘ulcer’, ‘infection’, ‘wound’, ‘osteomyelitis’, ‘cellulitis’, ‘abscess’, ‘gangrene’. There was no publication date and language limit/restriction.
Inclusion and exclusion criteria
This systematic review included original laboratory-based cross-sectional prevalence studies that measured at least one virulence factor gene of S. aureus isolates from human-infected diabetic foot ulcers (grade 2–4). We also excluded all reviews and studies that used animal infections.
Screening and eligibility of studies
The study procedure was carried out by two independent reviewers. Any disagreements were discussed between these reviewers or consulted with a third reviewer. After removing duplicate publications, titles and abstracts of the remaining articles were reviewed for potentially eligible studies. The full text of the remaining studies was then assessed for eligibility. Studies that met the inclusion criteria were considered eligible and were included in the present study. One reviewer extracted the data and a second reviewer verified its accuracy. The following data were extracted: author name, publication date, country, ulcer classification, molecular methods, number of patients, number of DFUs, number of S. aureus isolates, and frequency of each virulence factor.
Critical appraisal of studies
The quality of selected studies was evaluated using standard critical appraisal tools prepared by the Joanna Briggs Institute (JBI) for prevalence studies [28]. The purpose of this appraisal is to assess the methodological quality of a study and to determine the extent to which a study has addressed the possibility of bias in its design, conduct, and analysis. The JBI critical appraisal checklist contains nine questions (Q1-Q9). The scores given by two reviewers were used to make the final decision. A third reviewer was consulted in case of disagreement between their appraisal opinions. Studies with five or more “YES” responses (55% YES response rate) were included in the meta-analysis.
Virulence factor measurements
In the first step, we constructed a list of S. aureus virulence factor genes by precise examining all included studies and studying several reviews and related original articles [6, 8, 29, 30]. For better analysis, we divided the virulence factor genes of S. aureus into two categories: toxin and non-toxin. Toxin and non-toxin virulence factors mentioned in at least three or more studies were included in the meta-analysis. The outcome of interest was the number of isolates possessing each virulence factor gene.
Statistical analysis
The point estimates of the proportion of each virulence factor and its 95% confidence interval (95% CI) were estimated for each study. To estimate the pooled proportions, we used Metaprop, a statistical procedure in STATA (version 14) [31]. A random-effects model including Freeman-Tukey double arcsine transformation of the proportions was used to stabilize variance and reduce the effect of between-study heterogeneity. 95% CIs were computed around study-specific and pooled prevalence of each virulence factor based on the score test statistic and visualized by forest graphs. Between- study heterogeneity was evaluated with Cochran’s Q-test [32] and the percentage of total variation across studies was assessed with the I² measure [33]. Publication bias was tested by Begg’s test, and funnel plot. P values less than 0.05 were considered as statistically significant.
Results
Study selection
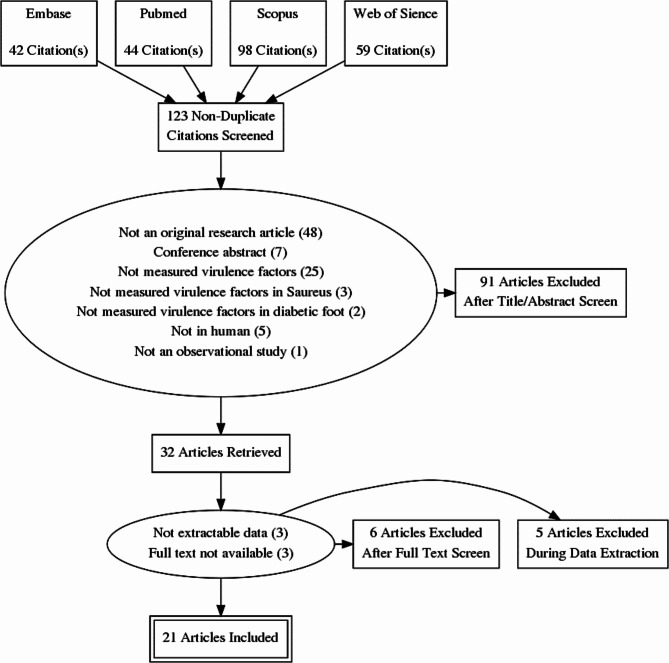
A literature search in electronic databases including PubMed, Embase, Web of Science and Scopus retrieved a total of 243 articles. After removing duplicates (n = 120), 91 studies were excluded in the initial screening of titles and abstracts. Subsequently, 13 additional articles were removed in full-text screening. Twenty-one articles met all eligibility criteria and were included in the systematic review study. Inter-rater agreement between reviewers for study selection was excellent (Kappa statistics = 0.96). The study selection process is detailed in Fig. 1.
Fig. 1.
Preferred reporting items for systematic reviews and meta-analyses (PRISMA) flow diagram depicting the selection process
Characteristics of included studies
A total of 1062 S. aureus isolates from 1948 patients were examined using 2131 DFI samples. The number of S. aureus isolates ranged from 8 [14] to 195 [12]. The number of virulence factor types measured in one study ranged from one [23, 24] to more than thirty [11, 13, 14, 20]. It is interesting that four continents, including Europe (9), Asia (6), Africa (1) and North America (1) had contribution in this topic. Among countries, France contributed the most with five (23.8%) publications [12, 19–22]. All included studies were published within the last 15 years. Six studies [11, 14–18, 24, 25] did not report a clear ulcer classification system. Most articles used PCR methods to measure virulence factor genes (Table 1).
Table 1.
Characteristics of the included studies in this systematic review and meta-analysis
| First author (reference) |
Year | Country | Design | No. of patients | No. of DFI samples | No. of isolates | Ulcer classification system | Methods used for determination of virulence factors |
No. of measured virulence factor types | |
|---|---|---|---|---|---|---|---|---|---|---|
|
Sotto, et al. [10] |
2007 | France | Prospective study | 72 | 72 | 85 | IDSA | Oligonucleotide DNA arrays & PCR | 3 | |
|
Sotto, et al. [11] |
2008 | France |
Prospective longitudinal study |
118 | 118 | 132 | NR | PCR | 33 | |
|
Sotto, et al. [12] |
2012 | France | Prospective study | 195 | 195 | 195 | IDSA/IWGDF | Oligonucleotide DNA arrays | 23 | |
|
Djahmi, et al. [13] |
2013 | Algeria | Prospective study | 128 | 183 | 85 | IDSA-IWGDF | Oligonucleotide DNA arrays | 33 | |
|
Post, et al. [15] |
2014 | Switzerland & France | Retrospective study | 23 | 23 | 23 | NR | PCR | 21 | |
|
Paul, et al. [14] |
2014 | Bangladesh | NR | 8 | 8 | 8 | NR | Multiplex PCR | 36 | |
|
Stappers, et al. [16] |
2015 | Netherlands | RCT | NR | 128 | 113 | NR | Real-time PCR | 2 | |
|
Shettigar, et al. [19] |
2015 | India | Prospective study | 200 | 200 | 86 | IDSA-IWGDF | Multiplex PCR | 3 | |
|
Mottola, et al. [17] |
2016 | Portugal | Transversal observational study | 49 | 49 | 41 | NR | PCR | 9 | |
|
Pobiega, et al. [18] |
2016 | Poland | Laboratory-based study | 68 | 68 | 68 | NR | PCR | 9 | |
|
Dunyach-Remy, et al. [20] |
2017 | France | Prospective study | 276 | 276 | 65 | IDSA–IWGDF | Oligonucleotide DNA arrays | 37 | |
|
Víquez-Molina, et al. [21] |
2018 | Costa Rica | Cross-sectional exploratory study | 379 | 379 | 58 | IDSA | PCR | 4 | |
| Kananizadeh, et al. [22] | 2019 | Iran | Cross-sectional study | 145 | 145 | 30 | Wagner | Multiplex PCR | 2 | |
|
Silva, et al. [25] |
2020 | Portugal | NR | 42 | 42 | 25 | Wagner | PCR | 8 | |
|
Anwar, et al. [23] |
2020 | Iraq | cross-sectional | 46 | 46 | 24 | IWGDF | Multiplex PCR | 1 | |
|
Lin, et al. [24] |
2020 | Taiwan | NR | 112 | 112 | 10 | IDSA | PFGE | 1 | |
|
Al-Bakri, et al. [26] |
2021 | Jordan | cross-sectional | 87 | 87 | 14 | Wagner | Multiplex PCR | 8 | |
Results of quality assessment
Quality of the studies was assessed using JBI tool. Seventeen out of 21 articles received at least five “YES” answers and were included in the meta-analysis (Table S1).
Virulence factor measurements
Among 17 included articles, 15 and 9 articles measure toxin and non-toxin virulence factors respectively. Seven articles reported both toxin and non-toxin virulence factors [11–14, 16, 20, 25]. The following virulence factors were measured in three or more studies, and they were included in the meta-analysis: 24 toxin virulence factors (hla, hlb, hlg, hlgv, hld, luk-SF or PVL, luk-ED, etA, etB, etD, sea, seb, sec, sed, see, seg, seh, sei, sej, sek, seq, tst, edin-A, edin-B) and 19 non-toxin virulence factors (bbp, cna, ebpS, clfA, clfB, fib, fnbA, fnbB, eno, cap5, cap8, agr 1, agr 2, agr 3, agr 4, icaA, icaD, chp, scn).
Toxin virulence factors
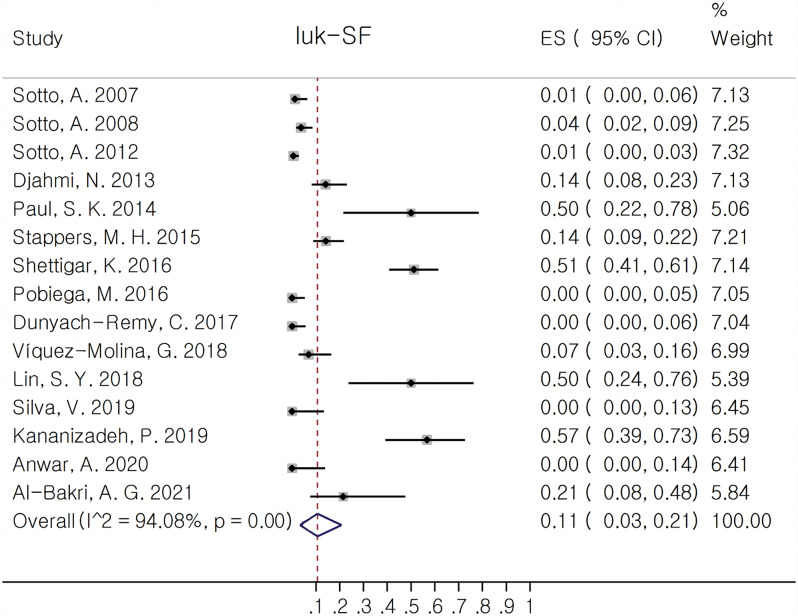
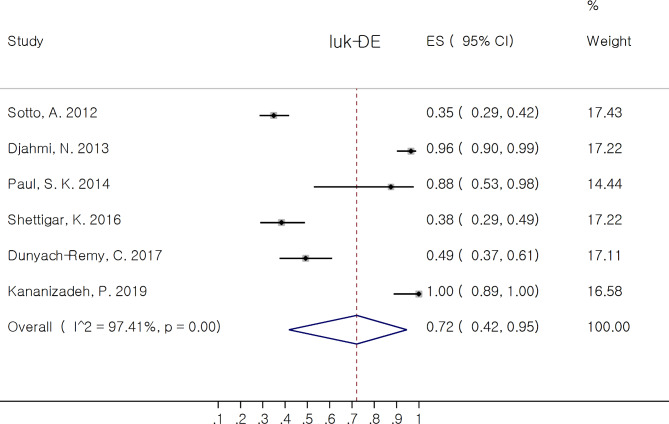
luk-SF (PVL) was the most prevalent reported virulence factor since it was reported in 15 out of 17 included studies. Among pore forming toxins, Bi-Component Leukotoxins had the most contribution. In this group, hld 100.0% (95% CI: 97.0, 100.0%) and hlg 88.0% (95% CI: 58.0, 100.0%) had the most and luk-SF (PVL) 11.0% (95% CI: 3.0, 21.0%) the least pooled estimate of proportion. Among leukocidin family, luk-ED had the most pooled proportion 72.0% (95% CI: 42.0, 95.0%). The corresponded forest plots of luk-SF and luk-ED are depicted in Figs. 2 and 3 respectively.
Fig. 2.
Forest plot showing the pooled proportion of luk-SF (PVL) in Staphylococcus aureus isolates from diabetic foot infection (DFI)
Fig. 3.
Forest plot showing the pooled proportion of luk-ED in Staphylococcus aureus isolates from diabetic foot infection (DFI)
The proportion of toxin virulence factors of S. aureus isolates are reported in Table 2. The prevalence of Exfoliative Toxins (etA, etB and etD), tst, and Epidermal Cell Differentiation Inhibitors Toxins (edinA, edinB) was near zero. The proportion of all Staphylococcal Enterotoxins were below 30%. Among them seg 28.9% (95% CI: 12.9, 47.9%) and sea 28.2% (95% CI: 17.9, 39.7%) had the most and seh 2.5% (95% CI: 0.0, 7.4%) and see 0.0% (95% CI: 0.0, 0.0%) the least pooled estimate of proportion.
Table 2.
Meta-analysis for the proportion of toxin virulence factors of S. aureus isolates from DFIs
| Toxin virulence factor genes | Pooled estimates | Heterogeneity test | ||||||
|---|---|---|---|---|---|---|---|---|
| N | n1 | n2 | Proportion (%) | 95% CI (%) | I2 (%) | |||
| hla | 4 | 183 | 118 | 80.0 | (31.0,100.0) | 97.5 | ||
| hlb | 4 | 360 | 135 | 47.0 | (29.0,66.0) | 87.8 | ||
| hlg | 5 | 304 | 237 | 88.0 | (58.0,100.0) | 96.6 | ||
| hlgv | 5 | 485 | 278 | 79.0 | (35.0,100.0) | 98.9 | ||
| hld | 3 | 158 | 157 | 100.0 | (97.0,100.0) | 51.6 | ||
| luk-SF or PVL | 15 | 998 | 86 | 11.0 | (3.0,21.0) | 94.1 | ||
| luk-ED | 6 | 469 | 222 | 72.0 | (42.0,95.0) | 97.4 | ||
| etA | 7 | 611 | 7 | 0.2 | (0.0,1.3) | 21.3 | ||
| etB | 6 | 543 | 0 | 0.0 | (0.0,0.0) | 0.0 | ||
| etD | 4 | 353 | 19 | 3.8 | (0.0,11.6) | 79.1 | ||
| sea | 8 | 652 | 173 | 28.2 | (17.9,39.7) | 87.5 | ||
| seb | 6 | 372 | 23 | 4.5 | (0.4,11.2) | 75.5 | ||
| sec | 3 | 208 | 24 | 11.3 | (2.0,25.1) | 75.7 | ||
| sed | 3 | 205 | 52 | 13.7 | (0.2,38.5) | 90.7 | ||
| see | 3 | 154 | 0 | 0.0 | (0.0,0.0) | 0.0 | ||
| seg | 4 | 219 | 79 | 28.9 | (12.9,47.9) | 80.5 | ||
| seh | 6 | 372 | 11 | 2.5 | (0.0,7.4) | 66.6 | ||
| sei | 4 | 403 | 120 | 27.4 | (16.5,39.8) | 79.9 | ||
| sej | 3 | 208 | 35 | 8.8 | (0.0,40.8) | 95.1 | ||
| sek | 5 | 375 | 75 | 19.2 | (0.0,55.3) | 98.0 | ||
| seq | 4 | 290 | 72 | 19.3 | (0.0,67.8) | 98.4 | ||
| tst | 8 | 558 | 60 | 9.6 | (4.1,16.9) | 81.6 | ||
| edin-A | 3 | 166 | 0 | 9.6 | (4.1,16.9) | 81.6 | ||
| edin-B | 3 | 158 | 12 | 4.9 | (0.0,23.0) | 87.1 | ||
N: number of studies, n1: total number of isolates in all of the studies that report the respective virulence factor; n2: sum of the number of the isolated bacteria that report the respective virulence factor; CI: confidence interval
Non-toxin virulence factors
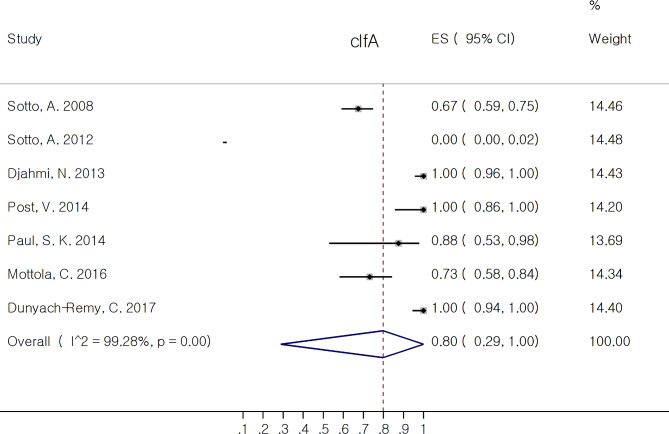
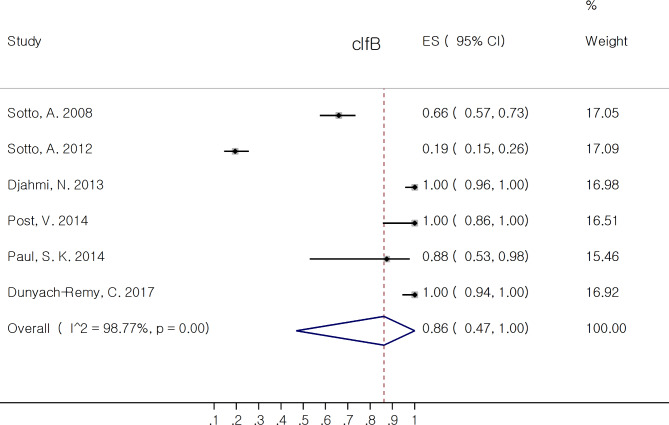
The proportion of non-toxin virulence factors of S. aureus isolates are reported in Table 3. Among MSCRAMM (bbp, cna, ebpS, clfA, clfB, fib, fnbA, fnbB), clfa 79.8% (95% CI: 28.8, 100.0%) and clfb 86.2% (95% CI: 46.9, 100.0%) had the most pooled prevalence. The correspond forest plots of clfA and clfB are depicted in Figs. 4 and 5, respectively. Six out of eight virulence factors (bbp, ebpS, clfA, clfB, fib, and fnbA) had pooled rate of proportion above 50%. Among genes associated with biofilm formation, both icaA and icaD 100.0% (95% CI: 95.6, 100.0%) had the most pooled estimate of proportion. Among agr type, agr1 38.2% (95% CI: 17.7, 60.9%) had the most pooled prevalence.
Table 3.
Meta-analysis for the proportion of non-toxin virulence factors of S. aureus isolates from DFIs
| Non-toxin virulence factor genes | Pooled estimates | Heterogeneity test | ||||||
|---|---|---|---|---|---|---|---|---|
| N | n1 | n2 | Proportion (%) | 95% CI (%) | I2 (%) | |||
| bbp | 6 | 508 | 267 | 54.6 | (20.3,86.8) | 98.2 | ||
| cna | 6 | 508 | 192 | 45.3 | (21.8,69.9) | 96.2 | ||
| ebps | 6 | 508 | 234 | 70.7 | (25.9,99.6) | 98.9 | ||
| clfa | 7 | 549 | 299 | 79.8 | (28.8,100.0) | 99.3 | ||
| clfb | 6 | 508 | 305 | 86.2 | (46.9,100.0) | 98.8 | ||
| fib | 6 | 508 | 246 | 63.1 | (31.8,89.5) | 97.7 | ||
| fnba | 5 | 485 | 219 | 73.3 | (21.5,100.0) | 99.2 | ||
| fnbb | 6 | 508 | 159 | 35.7 | (14.0,60.8) | 96.3 | ||
| cap5 | 4 | 609 | 174 | 35.0 | (13.4,60.3) | 96.7 | ||
| cap8 | 4 | 609 | 179 | 44.6 | (10.6,81.8) | 98.6 | ||
| agr1 | 9 | 687 | 281 | 38.2 | (17.7,60.9) | 96.9 | ||
| agr2 | 7 | 566 | 102 | 19.8 | (13.0,27.5) | 74.6 | ||
| agr3 | 7 | 566 | 69 | 11.7 | (6.6,17.8) | 71.2 | ||
| agr4 | 7 | 566 | 31 | 3.5 | (0.1,9.9) | 86.8 | ||
| icaa | 3 | 333 | 113 | 100.0 | (95.6,100.0) | 47.8 | ||
| icad | 3 | 333 | 113 | 100.0 | (95.6,100.0) | 47.8 | ||
| eno | 3 | 163 | 136 | 91.7 | (70.9,100.0) | 80.7 | ||
| chp | 3 | 158 | 90 | 61.0 | (40.8,79.4) | 78.1 | ||
| scn | 3 | 158 | 143 | 71.5 | (26.6,99.8) | 96.1 | ||
N: number of studies, n1: total number of isolates in all of the studies that report the respective virulence factor; n2: sum of the number of the isolated bacteria that report the respective virulence factor; CI: confidence interval
Fig. 4.
Forest plot showing the pooled proportion of clfA in Staphylococcus aureus isolates from diabetic foot infection (DFI)
Fig. 5.
Forest plot showing the pooled proportion of clfB in Staphylococcus aureus isolates from diabetic foot infection (DFI)
Publication bias
Funnel plots of standard error with the prevalence of luk-SF (Fig. S2) and Begg’s test (p = 0.080) show no evidence of publication bias. We did not draw funnel plot for virulence factors reported in lower than 10 studies. Results of Begg’s test for virulence factors with more than 4 studies including luk-ED (p = 0.260), sea (p = 0.618), tst (p = 0.536), hlb (p = 0.308), hlg (p = 1.0), etA (p = 0.548), seb (p = 0.707), seg (p = 0.734), she (p = 0.707) and sek (p = 0.613) did not imply for publication bias. Among nontoxic virulence factors, Begg’s test results for clfa (p = 1.0), clfb (p = 0.707), fib (p = 0.260), fnbA (p = 0.462), fnbB (p = 1.0), agr1 (p = 0.917), agr2 (p = 0.230), agr3 (p = 0.230) and agr4 (p = 1.0) as well did not show significant evidence of publication bias.
Discussion
Diabetic foot ulcer is one of the most serious complications of diabetes, which significantly affects patients’ quality of life. It can quickly spread into deeper tissue areas and cause critical conditions. Considering that bacteria are always present in the wound environment, making the diagnosis of infection only on the basis of microbial culture may lead to inappropriate prescription of antibiotics, which in turn leads to an increase in the prevalence of resistance to antibiotics, especially methicillin resistant S. aureus (MRSA) [10]. Identification of the most prevalent virulence factors of S. aureus isolates from DFIs may contribute more to the pathogenesis and help distinguish colonization from infection.
There are controversial issues about the role of PVL in skin soft tissue infections caused by S. aureus. In this study, although Luk-SF (PVL) was reported in most of our included articles, the prevalence in DFIs was not high and significant. This observation is consistent with the results of Stapper et al. [16]. Consistent with our findings, Víquez-Molina reported low proportion of several virulence factor genes, including pvl, etA, etB, and tsst in the profile of S. aureus recovered from DFIs [21]. Therefore, our study suggests that PVL toxin may not play a crucial role in the pathogenesis of DFIs, nor may it serve to differentiate colonization from infection. Interestingly, an Iranian study reported an unusual high prevalence of pvl (pvl, 56% and luk-ED 100%) in DFIs [22]. This observation suggests that the prevalence of virulence factors may be region-specific.
Several studies have established the role of luk-ED in the pathogenesis of S. aureus isolates from clinical samples [34–36]. Vasquez et al. identified a domain critical for targeting the Staphylococcus aureus LukED receptor and erythrocyte lysis [37]. Djahmi et al. reported a high prevalence of luk-ED (96.5%) among S. aureus isolates from DFIs. They also found that several virulence factors, including sek, seq, lukED, fnbB, cap8 and agr group 1 genes, were significantly associated with MRSA strains [13]. Another study reported 100% luk-ED positivity in S. aureus isolates from DFIs [22]. Interestingly, Dunyach-Remy et al. found statistical significance in the prevalence of luk-ED from DFU and nares isolates compared to DFU alone. This may imply that luk-ED made a significant contribution to DFI pathogenicity [20]. We also found a high pooled estimate of the proportion of luk-ED (72%).
Although there were a few studies reported the frequency of intercellular adhesions, we found the most pooled proportion for icaA and icaD (100%). This could therefore indicate that these factors may play a role in the formation of the biofilm and the development of the infection.
On the other hand, from a microbiological perspective, distinguishing colonization from infection is one of the key challenges for clinicians in the treatment of DFIs. Misdiagnosis of colonization as an infection can lead to inappropriate antibiotic prescribing, which in turn leads to an increase in the prevalence of antibiotic resistance, particularly methicillin-resistant S. aureus (MRSA) [10]. Sotto et al. (2008) found that the combination of five genes, including sea, sei, lukED, hlgv, and cap8 was useful as predictive markers for distinguishing uninfected diabetic foot ulcers (grade 1) from infected ones (grade 2–4) [11]. This may mean that the genetic profiles of infecting and colonizing S. aureus strains isolated from uninfected and infected diabetic foot ulcers are different. Therefore, by comparing genetic profile of the infecting and colonizing isolates, some virulence factors may be found that have specific role in DFI pathogenesis. In the present study, we found a high pooled estimate of the proportion of luk-ED (72%) and hlgv (79%), but in the case of sea, sei and cap8, we did not obtain the same consistent results. This discrepancy may be due to the fact that in our study each virulence factor was considered individually and not in combination with other virulence factors. Additionally, we did not compare infected and uninfected ulcers.
Limitations and strengths
One of the limitations of this study is that few articles reporting separate results for infected and uninfected ulcers. Therefore, we only considered the results for the infected ulcers. We were also unable to analyze the prevalence of virulence factors associated with MSSA and MRSA because most studies did not separately report the frequency of virulence factors for these isolates. Furthermore, we were unable to analyze the proportion of virulence factors associated with the type of infection (monomicrobial or polymicrobial) because most studies had focused on monomicrobial ulcers. The other limitation is that although numerous virulence factors were examined in all included articles, some of them were mentioned in only one or two articles and therefore were not included in the meta-analysis. The significant heterogeneity among studies could limit the interpretation of the pooled estimates. However, we attempted to address the results of each individual study to compensate for this heterogeneity. Finally, based on reports arising from PCR methods, it is difficult to say that prevalent genes have prevalent expression in a physiological situation and play a specific role in the pathogenicity.
The strengths of this systematic review and meta-analysis are worth noting. It provides a systematic and comprehensive search of all original published studies reporting the proportion of virulence factor genes of S. aureus isolates from DFIs. Furthermore, it is the first meta-analysis to examine the prevalence of virulence factors associated with the specific infection caused by S. aureus.
Conclusion
The results of the present study showed that among the toxin virulence factors, hemolysins (hld (100.0%), hlg (88.0%), hla (80.0%), hlgv (79.0%)) and luk-ED (72, 0%) and among the non-toxin virulence factors, icaA and icaD (100.0%) stand out as having the highest proportion in S. aureus isolates from DFIs. These prevalent genes may have the potential to evaluate as virulence factors involved in DFI pathogenesis. Finding these probable virulence factor genes can help control diabetic foot infection more effectively via anti-virulence therapy or preparation of multi-epitope vaccines.
Moreover, the present study suggests that an effective approach to better distinguish colonization from infection could be to assess the intrinsic virulence potential of infecting strain of isolated bacteria. Therefore, these genes could also be assessed as candidate biomarkers, using an oligonucleotide microarray, to differentiate colonization from infection.
Future studies are recommended to examine the proportion of these prevalent virulence factor genes in the colonizing S. aureus isolates to demonstrate their specificity for DFI pathogenicity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Quality assessment of studies using JBI’s critical appraisal tools designed for prevalence studies
Supplementary Material 2: Funnel plot of positive luk-SF proportions
Acknowledgements
We thank Dr Najmeh Ansari for her kindly help to conducting this study and Tahereh Sadeghi for language editing of the manuscript.
Abbreviations
- DFI
diabetic foot infections
- MRSA
methicillin resistant Staphylococcus aureus
- PSM
phenol-soluble modulins
- PVL
Panton-Valentine leucocidinMSCRAMM:Microbial surface components recognizing adhesive matrix molecules
- SpA
staphylococcal protein A
- Fnbp
fibronectin-binding proteins
- Clf
clumping factor
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- JBI
Joanna Briggs Institute
- IDSA
The Infectious Diseases Society of America
Author Contributions
Conceptualization, S.S. and M.S.; methodology, S.S. and A.T.; software, M.Y.; validation, S.S. and A.T.; formal analysis, M. Y.; writing—original draft preparation, S.S. and A.T.; writing—review and editing, S.S., A.T., M.Y., and M.S.; visualization, M.Y.; supervision, M.S.; project administration, S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Khodaverdian V, et al. Discovery of antivirulence agents against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57(8):3645–52. doi: 10.1128/AAC.00269-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai AD, et al. Staphylococcus aureus bacteraemia mortality: a systematic review and meta-analysis. Clin Microbiol Infect. 2022;28(8):1076–84. doi: 10.1016/j.cmi.2022.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Cassat JE, Thomsen I. Staphylococcus aureus Infections in children. Curr Opin Infect Dis. 2021;34(5):510–8. doi: 10.1097/QCO.0000000000000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shahrokh S et al. Bacterial Profile and Antimicrobial Resistance patterns of infected Diabetic Foot Ulcers in Iran: a systematic review and Meta-analysis of cross-sectional studies. Int J Low Extrem Wounds, 2021: p. 15347346211002715. [DOI] [PubMed]
- 5.Stacey HJ, et al. The prevalence of methicillin-resistant Staphylococcus aureus among diabetic patients: a meta-analysis. Acta Diabetol. 2019;56(8):907–21. doi: 10.1007/s00592-019-01301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunyach-Remy C, et al. Staphylococcus aureus toxins and diabetic foot ulcers: role in pathogenesis and interest in diagnosis. Toxins. 2016;8(7):209. doi: 10.3390/toxins8070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mottola C, et al. Susceptibility patterns of Staphylococcus aureus biofilms in diabetic foot Infections. BMC Microbiol. 2016;16(1):119. doi: 10.1186/s12866-016-0737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghasemian A, et al. The microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) genes among clinical isolates of Staphylococcus aureus from hospitalized children. Iran J Pathol. 2015;10(4):258. [PMC free article] [PubMed] [Google Scholar]
- 9.Fleitas Martínez O, et al. Recent advances in anti-virulence therapeutic strategies with a focus on dismantling bacterial membrane microdomains, toxin neutralization, quorum-sensing interference and biofilm inhibition. Front Cell Infect Microbiol. 2019;9:74. doi: 10.3389/fcimb.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sotto A, et al. Miniaturized oligonucleotide arrays: a new tool for discriminating colonization from Infection due to Staphylococcus aureus in diabetic foot ulcers. Diabetes Care. 2007;30(8):2051–6. doi: 10.2337/dc07-0461. [DOI] [PubMed] [Google Scholar]
- 11.Sotto A, et al. Virulence potential of Staphylococcus aureus strains isolated from diabetic foot ulcers: a new paradigm. Diabetes Care. 2008;31(12):2318–24. doi: 10.2337/dc08-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sotto A, et al. Distinguishing colonization from Infection with Staphylococcus aureus in diabetic foot ulcers with miniaturized oligonucleotide arrays: a French multicenter study. Diabetes Care. 2012;35(3):617–23. doi: 10.2337/dc11-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djahmi N, et al. Molecular epidemiology of Staphylococcus aureus strains isolated from inpatients with infected diabetic foot ulcers in an Algerian University Hospital. Clin Microbiol Infect. 2013;19(9):E398–404. doi: 10.1111/1469-0691.12199. [DOI] [PubMed] [Google Scholar]
- 14.Paul SK, et al. Detection and genetic characterization of PVL-positive ST8-MRSA-IVa and exfoliative toxin d-positive European CA-MRSA-Like ST1931 (CC80) MRSA-IVa strains in Bangladesh. Microb Drug Resist. 2014;20(4):325–36. doi: 10.1089/mdr.2013.0153. [DOI] [PubMed] [Google Scholar]
- 15.Post V, et al. Phenotypic and genotypic characterisation of Staphylococcus aureus causing musculoskeletal Infections. Int J Med Microbiol. 2014;304(5–6):565–76. doi: 10.1016/j.ijmm.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Stappers MHT, et al. Direct molecular versus culture-based assessment of Gram-positive cocci in biopsies of patients with major abscesses and diabetic foot Infections. Eur J Clin Microbiol Infect Dis. 2015;34(9):1885–92. doi: 10.1007/s10096-015-2428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mottola C, et al. Molecular typing, virulence traits and antimicrobial resistance of diabetic foot staphylococci. J Biomed Sci. 2016;23:33. doi: 10.1186/s12929-016-0250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pobiega M, et al. Virulence potential of Staphylococcus aureus strains isolated from diabetic foot ulcers among patients from southern Poland. Curr Vasc Pharmacol. 2016;14(6):547–51. doi: 10.2174/1570161114666160625083742. [DOI] [PubMed] [Google Scholar]
- 19.Shettigar K, et al. Virulence determinants in clinical Staphylococcus aureus from monomicrobial and polymicrobial Infections of diabetic foot ulcers. J Med Microbiol. 2016;65(12):1392–404. doi: 10.1099/jmm.0.000370. [DOI] [PubMed] [Google Scholar]
- 20.Dunyach-Remy C, et al. Link between nasal carriage of Staphylococcus aureus and infected diabetic foot ulcers. Diabetes Metab. 2017;43(2):167–71. doi: 10.1016/j.diabet.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Víquez-Molina G, et al. Virulence factor genes in Staphylococcus aureus isolated from Diabetic Foot Soft tissue and bone Infections. Int J Low Extrem Wounds. 2018;17(1):36–41. doi: 10.1177/1534734618764237. [DOI] [PubMed] [Google Scholar]
- 22.Kananizadeh P, et al. Molecular characteristics of methicillin-resistant staphylococcus aureus (MRSA) isolated from diabetic foot Infection. Iran J Pathol. 2019;14(4):329–37. doi: 10.30699/IJP.2019.101092.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anwar K, Hussein D, Salih J. Antimicrobial susceptibility testing and phenotypic detection of MRSA isolated from diabetic foot Infection. Int J Gen Med. 2020;13:1349–57. doi: 10.2147/IJGM.S278574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin SY, et al. Methicillin-resistant Staphylococcus aureus nasal carriage and Infection among patients with diabetic foot Ulcer. J Microbiol Immunol Infect. 2020;53(2):292–9. doi: 10.1016/j.jmii.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Silva V, et al. Emergence of community-acquired methicillin-resistant Staphylococcus aureus EMRSA-15 clone as the predominant cause of diabetic foot Ulcer Infections in Portugal. Eur J Clin Microbiol Infect Dis. 2020;39(1):179–86. doi: 10.1007/s10096-019-03709-6. [DOI] [PubMed] [Google Scholar]
- 26.Al-Bakri AG et al. Characterization of staphylococci sampled from diabetic foot Ulcer of Jordanian patients. J Appl Microbiol, 2021. [DOI] [PubMed]
- 27.Moher D, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 28.Munn Z, et al. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and cumulative incidence data. Int J Evid Based Healthc. 2015;13(3):147–53. doi: 10.1097/XEB.0000000000000054. [DOI] [PubMed] [Google Scholar]
- 29.Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12(1):547–69. doi: 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes-Robles T, Torres VJ. Staphylococcus aureus pore-forming toxins Staphylococcus aureus: microbiology, pathology, immunology, therapy and prophylaxis, 2017: p. 121–144. [DOI] [PubMed]
- 31.Nyaga VN, Arbyn M, Aerts M. Metaprop: a Stata command to perform meta-analysis of binomial data. Archives of Public Health. 2014;72(1):39. doi: 10.1186/2049-3258-72-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10(1):101–29. doi: 10.2307/3001666. [DOI] [Google Scholar]
- 33.Higgins JP, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Eiff C, et al. Prevalence of genes encoding for members of the staphylococcal leukotoxin family among clinical isolates of Staphylococcus aureus. Diagn Microbiol Infect Dis. 2004;49(3):157–62. doi: 10.1016/j.diagmicrobio.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 35.Rasmussen G, et al. Prevalence of clonal complexes and virulence genes among commensal and invasive Staphylococcus aureus isolates in Sweden. PLoS ONE. 2013;8(10):e77477. doi: 10.1371/journal.pone.0077477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He C, et al. Leukotoxin and pyrogenic toxin Superantigen gene backgrounds in bloodstream and wound Staphylococcus aureus isolates from eastern region of China. BMC Infect Dis. 2018;18(1):1–10. doi: 10.1186/s12879-018-3297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasquez MT, et al. Identification of a domain critical for Staphylococcus aureus LukED receptor targeting and lysis of erythrocytes. J Biol Chem. 2020;295(50):17241–50. doi: 10.1074/jbc.RA120.015757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Quality assessment of studies using JBI’s critical appraisal tools designed for prevalence studies
Supplementary Material 2: Funnel plot of positive luk-SF proportions
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.