Abstract
Pertussis remains a public health concern in South Africa, with an increase in reported cases and outbreaks in recent years. Whole genome sequencing was performed on 32 Bordetella pertussis isolates sourced from three different surveillance programmes in South Africa between 2015 and 2019. Genome sequences were characterized using multilocus sequence typing, vaccine antigen genes (ptxP, ptxA, ptxB, prn and fimH) and overall genome structure. All isolates were sequence type 2 and harboured the pertussis toxin promoter allele ptxP3. The dominant genotype was ptxP3-ptxA1-ptxB2-prn2-fimH2 (31/32, 96.9 %), with no pertactin-deficient or other mutations in vaccine antigen genes identified. Amongst 21 isolates yielding closed genome assemblies, eight distinct genome structures were detected, with 61.9 % (13/21) of the isolates exhibiting three predominant structures. Increases in case numbers are probably not due to evolutionary changes in the genome but possibly due to other factors such as the cyclical nature of B. pertussis disease, waning immunity due to the use of acellular vaccines and/or population immunity gaps.
Keywords: B. pertussis, whole genome sequencing, Illumina sequencing, PacBio sequencing, vaccine antigen genes, genome structural characterization
Full-Text
Data availability
The genome sequences for 32 South Africa B. pertussis isolates are available on NCBI, organized under BioProject accession number PRJNA929342.
Introduction
Pertussis disease caused by Bordetella pertussis is a public health concern in South Africa with peaks observed every 3–5 years [1–3]. Until the start of the coronavirus disease 2019 (COVID-19) pandemic, cases had continued to increase globally over the last 20 years, including in South Africa. The rise in cases in South Africa could be attributed to pathogen adaptation or waning immunity as a result of the switch to the acellular pertussis vaccine in 2009 [4, 5] and/or improved awareness and diagnosis by clinicians and laboratories within the country [6, 7].
Acellular pertussis vaccines may be composed of up to four B. pertussis purified protein antigens (fimbrial antigens, pertactin, filamentous haemagglutinin and pertussis toxin). One of the factors thought to be contributing to the global rise in B. pertussis infections is pathogen adaption, which has increased (predominantly within the genes encoding vaccine antigens) since the introduction of acellular vaccines [8, 9]. This has resulted in the circulation of strains that have diverged from strains used in the vaccine [10–13]. Changes in the pertussis toxin promoter (ptxP) region have been observed, with the majority of isolates now harbouring the novel ptxP3 allele which replaced the ptxP1 allele [9, 14]. Strains harbouring the ptxP3 allele first emerged in the 1980s and have been shown to result in increased production of pertussis toxin in vitro [10]. Mutations within pertactin, filamentous haemagglutinin and pertussis toxin genes have also been described [15–18].
In 2009, South Africa replaced the whole-cell pertussis vaccine with a hexavalent vaccine which protects against diphtheria, tetanus, pertussis, poliomyelitis and Haemophilus influenzae type b, and is given to infants as part of the routine immunization programme [Pentaxim (2009–2014) replaced by Hexaxim (2015 to present); Sanofi Pasteur] [19]. Both Pentaxim and Hexaxim contain pertussis toxin and filamentous haemagglutinin antigens and do not contain pertactin (https://www.sanofi.com). In 2019, according to WHO/UNICEF, the coverage for the first and third dose of the pentavalent vaccine in South Africa was 84 and 77 %, respectively [20]. Between 2013 and 2018, among systematically tested individuals hospitalized with pneumonia, annual B. pertussis incidence was 17 cases per 100 000 population [1]. In addition, in 2018 and 2019, clusters of pertussis disease were observed in several provinces in South Africa [21].
B. pertussis was previously regarded as monomorphic due to lack of observable diversity using available molecular methods. Traditional typing methods such as multilocus sequence typing (MLST) or typing of genes encoding vaccine antigens provide poor discriminatory power [10, 22]. Since the introduction of whole genome sequencing (WGS), typing methods that include parts of and/or the entire genome, such as analysis of the accumulation of SNPs within the genome, whole genome MLST and core genome MLST, have added to the currently available techniques that can be used to classify circulating strains with more resolution compared to the traditional molecular methods [10, 22, 23]. More recently, the combination of long- and short-read WGS technologies have enabled a better understanding of genome arrangement patterns and genomic structural diversity amongst circulating isolates [8, 13, 17]. Using this methodology, unique genomic structures have been described in the USA [13], New Zealand [17] and India [24].
B. pertussis can undergo structural changes caused by alterations in their genome either by insertions, deletions or gene rearrangements, resulting in the circulation of strains that have diverged from vaccine strains [10, 13, 25, 26]. Strains not expressing the vaccine antigens pertactin and filamentous haemagglutinin have been described in the USA following the use of acellular vaccines [16, 25, 27]. Following acellular vaccine introduction, pertactin-deficient strains identified in France were shown to be as virulent as pertactin-expressing strains [28]. These data highlight the potential for the emergence of other vaccine escape mutants following the use of acellular pertussis vaccines and emphasize the importance of genomic surveillance for monitoring B. pertussis strain evolution in response to immune pressure. There are currently no data describing B. pertussis lineages in South Africa. Using WGS, we aimed to describe circulating B. pertussis genotypes and identify new or emerging immune escape mutations in South Africa.
Methods
Study population
B. pertussis isolates (n=32) were sourced from sentinel syndromic surveillance for severe respiratory illness (n=14), paediatric pertussis surveillance (n=11) and diagnostic samples (n=7). Prospective, active, sentinel pneumonia surveillance with B. pertussis testing was implemented in 2012 at sentinel sites in five of the nine provinces and enrols patients of all ages hospitalized with lower respiratory tract illness as well as neonatal sepsis or suspected sepsis in infants ≤3 months of age, irrespective of symptom duration [29]. Respiratory specimens (nasopharyngeal specimens and/or sputum) collected from patients are routinely tested for influenza virus, respiratory syncytial virus and B. pertussis by real-time PCR. Isolates included in this study were collected between 2015 and 2019.
Paediatric pertussis surveillance was conducted at the Chris Hani Baragwanath Academic Hospital (CHBAH) in Gauteng Province from January to December 2015 [30]. All infants aged <12 months who were hospitalized at CHBAH with lower respiratory tract infection or sepsis were approached for enrolment. Nasopharyngeal swabs were collected and tested for B. pertussis by culture and real-time PCR.
In addition, public sector clinicians in South Africa occasionally send respiratory specimens (nasopharyngeal specimens, sputum, tracheal aspirates) from patients of all ages to the National Institute for Communicable Diseases for B. pertussis diagnosis.
B. pertussis culture and phenotypic characterization
All respiratory specimens were tested by real-time PCR for the detection of B. pertussis , Bordetella parapertussis and Bordetella holmesii as previously described [2, 31]. Since B. pertussis culture is not performed routinely at primary diagnostic laboratories, B. pertussis culture was attempted for all respiratory specimens immediately following a positive PCR, where the PCR cycle threshold value was ≤25. Specimens were plated on specialized charcoal agar (Diagnostic Media Products) and plates were incubated under aerobic conditions at 37 °C. Suspected B. pertussis colonies were confirmed using MALDI-TOF MS (Bruker) and real-time PCR [2, 31].
DNA extraction, sequencing and genome assembly
Genomic DNA was extracted from cultures using the Gentra Pure Gene Yeast/Bact. kit (Qiagen) with the Qiagen protocol [25]. DNA was quantified using the Qubit 2.0 fluorometer (Invitrogen) and Qubit dsDNA BR assay kit and sequenced using both the Illumina MiSeq (Illumina) and the PacBio RSII (Pacific Biosciences) sequencing platforms. PacBio sequence data were used to identify genome structural variation. A combination of sequence data from both platforms was used when sequence data were incomplete or when results were not conclusive.
Genomic libraries for Illumina sequencing were prepared (paired-end libraries, 2×300 bp) using the Nextera XT DNA v3 MiSeq sequencing kit. Reads were trimmed at both ends to remove nucleotides with a quality score of <45 bp using CLC Genomics Workbench (v21.0.3; CLC Bio). The passed reads were de novo assembled using CLC Genomics Workbench. Illumina genomes were assessed for quality using QUAST (http://bioinf.spbau.ru/quast) on genome fraction, duplication ratio, total length, N50 value and number of contigs [13]. Acceptable Illumina data included: genome fraction >90 %; total length >3.75 Mb; N50 >5 kb; and contigs between 300 and 350. Libraries for PacBio sequencing were prepared using the SMRTbell template prep kit (Pacific Biosciences) and polymerase binding kit. PacBio sequencing data were de novo assembled, polished and reoriented to the common start position (creating a closed genome) using a CDC in-house Perl script as previously described [13]. The closed, assembled genomes were assessed on genome fraction, total length and N50 value [13]. Acceptable PacBio data included: genome fraction >90 %; total length >3.75 Mb; and N50 >5 kb. The reference genome used was B. pertussis E476 (vaccine strain: Tohama I – accession number: CP010964).
Multilocus sequence typing and vaccine antigen gene typing
Assembled Illumina sequences were used to determine the B. pertussis MLST profile. Sequence type (ST) was assigned using the BIGSdb platform curated by the Institute Pasteur (https://bigsdb.pasteur.fr/bordetella) according to seven housekeeping genes (adk, fumC, glyA, tryB, icd, pepA and pgm).
Detection of the ptxP, ptxA, ptxB, prn and fimH genes, and mutations within the prn gene was performed using a high stringency Basic Local Alignment Search (BLASTn) alignment (https://blast.ncbi.nlm.nih.gov/Blast) on assembled genomes as previously described [13, 25]. Assembled PacBio sequences were analyzed in a similar manner to supplement inconclusive results or incomplete Illumina sequence coverage.
Phylogeny of B. pertussis
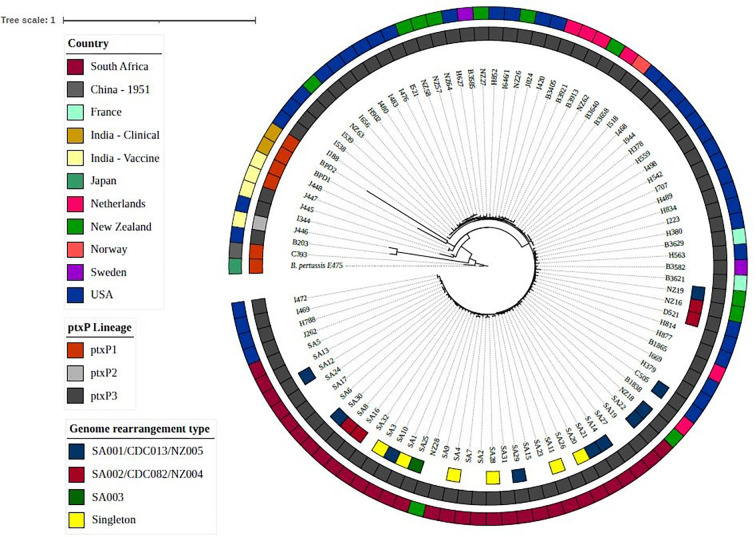
To determine the relationship between South African B. pertussis isolates and publicly available genomes of isolates from other countries, a tree was reconstructed by incorporating genomes from different countries (New Zealand, USA, Netherlands, Sweden, France, Norway, India, Japan and China) available on NCBI (accessed February 2023) that were representative of circulating strains (different ST’s) on each continent between 2012 and 2019, and also represented isolates with varying ptxP types (ptxP1, ptxP2 and ptxP3). These included the reference genome B. pertussis E476 and the additional available vaccine strains from India [24] (Table S1, available in the online version of this article). A maximum-likelihood phylogenetic tree of 99 isolate genomes was reconstructed based on the concatenated alignment of the high-quality SNPs using CSI phylogeny (v1.4) [32] with additional tree annotation performed using iTOL (v6).
Identification of genome structural variation
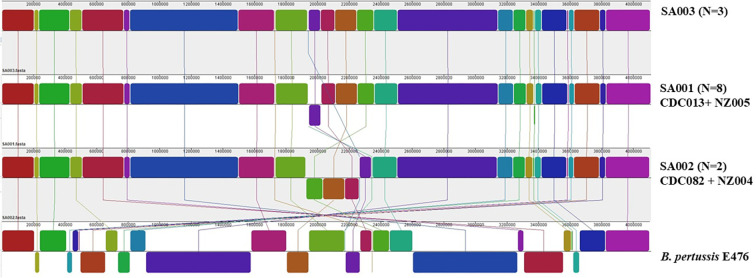
Genome structural variation was determined by aligning complete PacBio assemblies from 21 South African B. pertussis genomes to each other using progressiveMauve (v20150226) with default parameters [32]. The genomes were determined to be collinear if pairwise alignments included no observable inversions or gaps >1500 bp. Alignments were manually inspected and grouped into types based on each unique alignment. One representative sequence from each arrangement type (SA001, SA002 and SA003) as well as the singleton sequences (genome structures not shared with any other isolates) were further aligned, using progressiveMauve, to reported structures from the USA (CDC002, CDC010, CDC013, CDC046, CDC082 and CDC237) and New Zealand (NZ004, NZ005, NZ006, NZ007 and NZ008) [13, 17].
Results
B. pertussis isolates
A total of 32 B. pertussis isolates were available for sequencing. Isolates were obtained from children (n=25) and adults (n=7) from both surveillance as well as clinical diagnostics. All 32 isolates were cultured from sporadic B. pertussis cases. There were no known epidemiological links between the B. pertussis cases.
From 2015 to 2019, a total of 17 097 hospitalized individuals were enrolled into sentinel pneumonia surveillance of whom 228 (1.3 %) tested PCR positive for B. pertussis . We attempted culture on 108 of these PCR-positive samples (Ct≤25). A total of 14 samples (12.9 %, 14/108) yielded a B. pertussis culture. Positive cultures were isolated from individuals ranging from <3 months of age to 64 years of age, and 50.0 % (7/14) of cases were people positive for human immunodeficiency virus (HIV) (Table S2).
On the pertussis paediatric surveillance platform, from 1 January to 31 December 2015 B. pertussis PCR positivity was 2.3 % (42/1839) [30]. A B. pertussis culture was obtained from 26.2 % (11/42) of PCR-positive samples. Cases with positive culture were infants aged <3 months, and the majority were unvaccinated (72.7 %, 8/11) and HIV-unexposed (63.6 %, 7/11).
A total of 770 respiratory samples were received from 2015 to 2019 for B. pertussis diagnostic testing, of which 12.1 % (93/770) tested positive for B. pertussis by PCR. A total of 41 of these positive samples were further cultured and seven samples yielded a B. pertussis culture (7/41, 17.1%), all of which were from infants aged <3 months.
Molecular characterization of B. pertussis
Using WGS data, all 32 B. pertussis isolates were characterized as ST2 (allelic profile: adk1, fumC1, glyA1, tryB3, icd1, pepA1, pgm1). Isolates belonged to the ptxP3 lineage and all genomes harboured an intact pertactin gene with no mutations. The vaccine antigen profile ptxP3-ptxA1-ptxB2-prn2-fimH2 was identified in 31/32 (96.9 %) isolates (Table S2). Overall, based on molecular characterization, all vaccine antigen genes were wild-type. One isolate (SA12) had an SNP within the fimH2 allele (A245G synonymous mutation) which was further classified as fimH2-1. This mutation was confirmed using both the Illumina and PacBio sequence data. The vaccine antigen profile for this isolate was ptxP3-ptxA1-ptxB2-prn2-fimH2-1.
Phylogeny of B. pertussis
All South African isolates clustered within the ptxP3 clade and were phylogenetically distinct from B. pertussis E476 and other global strains (Fig. 1). There were two clusters (Cluster 1: seven South Africa+one New Zealand; and Cluster 2: six South Africa+four USA) where South African isolates clustered with isolates from the USA and New Zealand. Despite having identical ST and vaccine antigen profiles, based on SNPs, the South African isolates were genetically distinct from isolates from other countries (with the exception of the one New Zealand and four USA strains), clustering closely to each other within the tree. In addition, looking at the South African isolates only, there was no geographical clustering of strains and no temporal association of strains was noted.
Fig. 1.
Maximum-likelihood phylogenetic tree for South African B. pertussis isolates and isolates from other geographical locations (2012–2019) based on the concatenated alignment of SNPs of the whole genome. The tree was rooted using the B. pertussis E476 reference (vaccine strain: Tohama I). The inner ring of the figure indicates the genome rearrangement type for South African isolates, the middle ring indicates the ptxP lineage and the outer ring indicates the country of isolation.
B. pertussis genome structural variation
Of the 32 isolates sequenced using the PacBio, 21 (65.6 %) yielded closed genome sequences that could be analyzed for structural variation. The remaining 11 genomes yielded incomplete assemblies with more than one contig and thus could not be used to assess genome structure. Thirteen isolates (13/21, 62 %) could be grouped into three distinct arrangement types [SA001 (n=8), SA002 (n=2) and SA003 (n=3)] based on genome patterns, whilst eight isolates demonstrated unique (‘singleton’) structures distinct from the other arrangement types (Fig. 2 and Table S2). Genomic differences between the arrangement types were due to inversions centred around the replication terminus of the B. pertussis genome and differed from reference strain E476 (Tahoma I). The inversions were flanked by the B. pertussis multi-copy insertion sequence IS481. Comparing the genome structures to isolates from the USA and New Zealand, we found that SA001 was identical in structure to CDC013/NZ005 and SA002 matched structure CDC082/NZ004. In addition, one South African singleton structure (SA20) matched genome structure CDC046/NZ48. There was no concordance between SNP clusters and genome structural clusters amongst South African, USA and New Zealand isolates that shared the same genome structures (Fig. 1). Amongst the SA001/CDC013/NZ005 isolates, we found <18 SNP differences amongst the South African isolates within the structure and <25 SNP differences amongst South African isolates compared with the USA isolates and the New Zealand isolates. In addition, there were no SNP differences amongst the South African isolates within the SA002/CDC082/NZ004 cluster but when the South African isolates were compared to USA and New Zealand isolates we found <18 SNP differences amongst all the isolates.
Fig. 2.
Genome organization and re-arrangement patterns amongst South African B. pertussis isolates collected in 2015–2019 in relation to reference E476 (n=13). Each coloured block represents a region of the genome called a localized co-linear block. A localized co-linear block below the central black line indicates an inversion event.
Discussion
In this B. pertussis genomic characterization study, isolates from South Africa, collected over a 5 year period, were genetically homogeneous using MLST. The dominant vaccine antigen genotype in all isolates except one was ptxP3-ptxA1-ptxB2-prn2-fimH2 and there was no evidence of mutations within the genes encoding vaccine antigens. WGS provided additional discriminatory power and identified three distinct genome structures.
Using MLST, South African B. pertussis isolates were the same sequence type (ST2) that has been dominant globally since the late 1990s [33]. In addition, all harboured the same pertussis toxin promoter allele, ptxP3. Globally, strains harbouring the ptxP3 allele first emerged in the 1980s [10]. Subsequently, over 90 % of globally circulating strains now harbour this allele [34, 35].
In other countries, mutations in vaccine antigen genes of B. pertussis appear to be more common since the introduction of the acellular vaccine, presumably due to vaccine pressure [8–10, 36]. In our setting, only one isolate contained a SNP within the vaccine antigen fimH2 gene. In addition, the South African isolates had an intact pertactin gene and no other mutations were identified within the other vaccine antigen genes. In the USA, pertactin-containing vaccines were introduced in the 1990s. From 2000 to 2013 in the USA, there was a rapid increase in pertactin-deficient B. pertussis isolates, resulting in non-expression of the gene [37]. In Australia the acellular pertactin-containing vaccine was introduced in the 1990s and strains lacking the pertactin gene were first detected between 2008 and 2012 [38]. Additionally, isolates collected between 2012 and 2017 showed spread of pertactin-deficient strains as well as an isolate that was negative for filamentous haemagglutinin. In Spain, between 1986 and 2018 a total of 342 B. pertussis isolates were collected for molecular characterization [39]. Of the 342 isolates, 27 % (93/342) were pertactin-deficient and all of these pertactin-deficient isolates were collected after the introduction of the acellular vaccine. Mutations within pertactin and other vaccine antigen genes were not detected among South African isolates, probably due to the formulation of the acellular pertussis vaccine (Hexaxim; Sanofi Pasteur), which only contains the pertussis toxin and filamentous haemagglutinin antigens, whilst many other countries are using vaccines containing the pertactin antigen as well as two or more other antigens. Maternal vaccination has been recommended for use in South Africa in 2024. The formulation of the vaccine to be used for maternal immunization includes pertactin and fimbriae, so ongoing genomic surveillance following the introduction of this vaccine will be important to monitor potential mutations within vaccine antigen genes amongst circulating B. pertussis strains in South Africa.
Using the traditional typing method of MLST and vaccine antigen profile data, our isolates represented a single clone. However, both methods rely on a very limited set of genes in the genome, limiting their discriminatory power. Determining the complete genomic structures of B. pertussis isolates using long read sequencing technology is relatively new and was first described in 2017 [13]. Using this method, WGS provided additional resolution and identified three distinct genome structures amongst 13 isolates. A combination of both MiSeq and PacBio sequence data proved to be advantageous as we used both data sets to create complete genomes from which we were able to define the genome structure. Studies have shown marked differences amongst B. pertussis genome structures despite having an otherwise monomorphic profile (based on other genome characterization methods). During two state-wide epidemics in the USA in 2010–2012, whole genome data were useful in identifying 16 distinct genome structures from 31 isolates that were otherwise monomorphic based on other typing techniques, and all differed from the vaccine strains [25]. In addition, Weigand et al. provided some evidence of phenotypic diversity amongst genome structures by showing the expansion of ‘cluster 01 (same genome structure)’ within the USA by analysing data from pre-2000 to 2016 [40]. Similarly, in New Zealand, WGS data showed 19 different genome rearrangements amongst 66 isolates [17]. An integrated comparative genomics analysis using isolates collected from the Czech Republic between 2008 and 2012 showed that circulating B. pertussis has substantial variation in genome organization and form separate phylogenetic clusters based on time of collection (historical isolates vs. currently circulating isolates) [41]. When using proteomics, they found that genome rearrangements resulted in changes in gene order, orientation or large deletions, which affected transcriptomic profiles in B. pertussis . Results support the authors’ assumption that genomic rearrangements might affect global expression profiles and phenotypic diversity in B. pertussis . Based on the above-mentioned data, genome structures/rearrangements could be useful signatures of genome evolution. Within our study, two of the South African genomic structures (SA001 and SA002) identified were shared with structures identified in isolates collected in the USA and New Zealand, showing that these structures are not unique to South Africa. In addition, comparing the SNP differences amongst the South African, USA and New Zealand isolates within shared structures, we see that isolates within the same structures are genetically similar (<25 SNP differences). However, based on this analysis we are unable to determine ancestral relationships amongst the South African isolates and isolates from the USA and New Zealand with shared genome structures. Previous studies have demonstrated the low SNP density amongst global B. pertussis isolates, making it difficult to deduce geographical clustering and/or source attribution [10, 42]. Similarly, Weigand et al. have also shown that isolates of B. pertussis with shared genome structures are not automatically similar based on SNPs [40]. There are few data available that describe the importance of structural variation and more studies are required to fully understand the prevalence of arrangement type, and associations with geographical location, phenotype or strain fitness.
This study has some limitations that need to be considered. To our knowledge, there are no earlier B. pertussis genomic data available from South Africa so we are unable to track changes in the genomes over a longer period of time. The absence of earlier data also limits conclusions pertaining to the origin of currently circulating strains. In addition, South Africa changed to the acellular vaccine in 2009, and isolates sequenced in this study were collected between 2015 and 2019. This time period may therefore be too short to observe the effects of the change in vaccine. We had a small sample size of 32 genomes, representing <5 % of B. pertussis PCR-positive samples. Therefore, these data are not representative of all B. pertussis causing disease in the country and we may have missed detecting novel or emerging lineages or vaccine antigen mutations. B. pertussis is a fastidious organism that is difficult to culture in the laboratory, requires specialized culture media that have a short shelf life, and has long incubation periods of 7–10 days, limiting its diagnostic utility. Diagnostic laboratories in South Africa do not routinely perform B. pertussis culture and rely predominantly on PCR testing for B. pertussis diagnosis. In addition, the low culture yield, incomplete geographical representation of sites and small sample numbers do not allow for confidence regarding potential temporal and geographical conclusions. The pneumonia surveillance programme focuses on respiratory viral detection and nasopharyngeal swabs are transported in viral transport medium which is not conducive to B. pertussis culture. Ideally, WGS of B. pertussis directly from clinical specimens needs to be optimized in our setting for future molecular characterization studies [9].
There are currently limited B. pertussis genomic data from Africa and no previously published data from South Africa. These findings now provide baseline genomic data for future studies within our country and in the global context. Although limited and spanning a short time period of 5 years, we show that circulating B. pertussis strains in South Africa do not yet harbour mutations in vaccine antigen genes, as has been observed in other countries. Increases in pertussis cases may be due to other factors such as the cyclical nature of B. pertussis disease, waning immunity due to the use of acellular vaccines that also do not protect against B. pertussis carriage and/or population immunity gaps, and these warrant closer examination within our region. Our findings have implications to help guide recommendations for improved vaccination strategies in low- and middle-income countries.
Supplementary Data
Funding information
This work was supported by the National Institute for Communicable Diseases of the National Health Laboratory Service, the US Centers for Disease Control and Prevention (cooperative agreement number 5U51IP000155) and Sanofi Pasteur (cooperative agreement number PER00059). This work was supported, in part, by a Fogarty International Center Global Infectious Disease research training grant, National Institutes of Health, to the University of Pittsburgh and National Institute for Communicable Diseases (D43TW011255).
Acknowledgements
The authors would like to thank all the individuals who kindly agreed to participate in the study, external collaborators and laboratory staff who contributed to this study.
Conflicts of interest
F.M. reports grants from Sanofi Pasteur. C.C. reports grants from Sanofi Pasteur, US Centers for Disease Control and Prevention, Wellcome, South African Medical Research Council and the Bill and Melinda Gates Foundation. A.vG. and N.W. report grants from Sanofi Pasteur and the Bill and Melinda Gates Foundation; M.C.N. reports grants from the Bill and Melinda Gates Foundation.
Ethical statement
Ethics for pneumonia surveillance (M140824) was approved by the University of Cape Town, Human Research Ethics Committee (HREC) reference 836/2014, University of Witwatersrand HREC reference M140824 and University of Kwa-Zulu Natal Biomedical Research Ethics Committee reference M496/14. Ethics for the paediatric pertussis surveillance and for additional strain typing of B. pertussis was approved by HREC (M131109 and M210676, respectively).
Footnotes
Abbreviations: HIV, human immunodeficiency virus; MLST, multilocus sequence typing; ST, sequence type; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary tables are available with the online version of this article.
References
- 1.Wolter N, Cohen C, Tempia S, Walaza S, Moosa F, et al. Epidemiology of pertussis in individuals of all ages hospitalized with respiratory illness in South Africa, January 2013-December 2018. Clin Infect Dis. 2021;73:e745–e753. doi: 10.1093/cid/ciab089. [DOI] [PubMed] [Google Scholar]
- 2.Moosa F, du Plessis M, Wolter N, Carrim M, Cohen C, et al. Challenges and clinical relevance of molecular detection of Bordetella pertussis in South Africa. BMC Infect Dis. 2019;19:276. doi: 10.1186/s12879-019-3869-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moosa F, Tempia S, Kleynhans J, McMorrow M, Moyes J, et al. Incidence and transmission dynamics of Bordetella pertussis infection in rural and urban communities, South Africa, 2016‒2018. Emerg Infect Dis. 2023;29:294–303. doi: 10.3201/eid2902.221125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witt MA, Katz PH, Witt DJ. Unexpectedly limited durability of immunity following acellular pertussis vaccination in preadolescents in a North American outbreak. Clin Infect Dis. 2012;54:1730–1735. doi: 10.1093/cid/cis287. [DOI] [PubMed] [Google Scholar]
- 5.Baxter R, Bartlett J, Rowhani-Rahbar A, Fireman B, Klein NP. Effectiveness of pertussis vaccines for adolescents and adults: case-control study. BMJ. 2013;347:f4249. doi: 10.1136/bmj.f4249. [DOI] [PubMed] [Google Scholar]
- 6.Dalby T, Harboe ZB, Krogfelt KA. Seroprevalence of pertussis among Danish patients with cough of unknown etiology. Clin Vaccine Immunol. 2010;17:2016–2023. doi: 10.1128/CVI.00270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisman DN, Tang P, Hauck T, Richardson S, Drews SJ, et al. Pertussis resurgence in Toronto, Canada: a population-based study including test-incidence feedback modeling. BMC Public Health. 2011;11:694. doi: 10.1186/1471-2458-11-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fedele G, Ausiello CM. How genomics is changing what we know about the evolution and genome of Bordetella pertussis . Adv Exp Med Biol. 2019;1183:1–17. doi: 10.1007/978-3-030-33249-5. [DOI] [PubMed] [Google Scholar]
- 9.Fedele G, Ausiello CM. Molecular epidemiology of Bordetella pertussis . Adv Exp Med Biol. 2019;1183:19–33. doi: 10.1007/978-3-030-33249-5. [DOI] [PubMed] [Google Scholar]
- 10.Bart MJ, Harris SR, Advani A, Arakawa Y, Bottero D, et al. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. mBio. 2014;5:e01074. doi: 10.1128/mBio.01074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowden KE, Williams MM, Cassiday PK, Milton A, Pawloski L, et al. Molecular epidemiology of the pertussis epidemic in Washington State in 2012. J Clin Microbiol. 2014;52:3549–3557. doi: 10.1128/JCM.01189-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Gent M, Heuvelman CJ, van der Heide HG, Hallander HO, Advani A, et al. Analysis of Bordetella pertussis clinical isolates circulating in European countries during the period 1998–2012. Eur J Clin Microbiol Infect Dis. 2015;34:821–830. doi: 10.1007/s10096-014-2297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weigand MR, Peng Y, Loparev V, Batra D, Bowden KE, et al. The history of Bordetella pertussis genome evolution includes structural rearrangement. J Bacteriol. 2017;199:e00806-16. doi: 10.1128/JB.00806-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Advani A, Hallander HO, Dalby T, Krogfelt KA, Guiso N. Pulsed-Field Gel Electrophoresis Analysis of Bordetella pertussis Isolates Circulating in Europe from 1998 to 2009 from nine countries with distinct vaccination programs. 1998. https://journals.asm.org/journal/jcm [DOI] [PMC free article] [PubMed]
- 15.Williams MM, Sen K, Weigand MR, Skoff TH, Cunningham VA, et al. Bordetella pertussis strain lacking pertactin and pertussis toxin. Emerg Infect Dis. 2016;22:319–322. doi: 10.3201/eid2202.151332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weigand MR, Pawloski LC, Peng Y, Ju H, Burroughs M, Cassiday PK, et al. Screening and Genomic Characterization of Filamentous Hemagglutinin-Deficient Bordetella pertussis. 2018. [DOI] [PMC free article] [PubMed]
- 17.Ring N, Davies H, Morgan J, Sundaresan M, Tiong A, et al. Comparative genomics of Bordetella pertussis isolates from New Zealand, a country with an uncommonly high incidence of whooping cough. Microb Genom. 2022;8:000756. doi: 10.1099/mgen.0.000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hegerle N, Paris A-S, Brun D, Dore G, Njamkepo E, et al. Evolution of French Bordetella pertussis and Bordetella parapertussis isolates: increase of Bordetellae not expressing pertactin. Clin Microbiol Infect. 2012;18:E340–6. doi: 10.1111/j.1469-0691.2012.03925.x. [DOI] [PubMed] [Google Scholar]
- 19.Soofie N, Nunes MC, Kgagudi P, van Niekerk N, Makgobo T, et al. The burden of pertussis hospitalization in HIV-exposed and HIV-unexposed South African infants. Clin Infect Dis. 2016;63:S165–S173. doi: 10.1093/cid/ciw545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO and UNICEF estimates of immunization coverage: 2019 revision. 2020. [ April 24; 2020 ]. https://www.who.int/immunization/monitoring surveillance/data/zaf.pdf accessed.
- 21.Increase in pertussis cases in South Africa. [ April 24; 2021 ]. https://www.nhls.ac.za/increase-in-pertussis-cases-in-south-africa/ n.d. accessed.
- 22.Weigand MR, Peng Y, Pouseele H, Kania D, Bowden KE. Genomic surveillance and improved molecular typing of Bordetella pertussis using wgMLST. n.d. [DOI] [PMC free article] [PubMed]
- 23.Bouchez V, Guglielmini J, Dazas M, Landier A, Toubiana J, et al. Genomic sequencing of Bordetella pertussis for epidemiology and global surveillance of whooping cough. Emerg Infect Dis. 2018;24:988–994. doi: 10.3201/eid2406.171464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alai S, Ghattargi VC, Gautam M, Patel K, Pawar SP, et al. Comparative genomics of whole-cell pertussis vaccine strains from India. BMC Genomics. 2020;21:345. doi: 10.1186/s12864-020-6724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowden KE, Weigand MR, Peng Y, Cassiday PK, Sammons S, et al. Genome structural diversity among 31 Bordetella pertussis isolates from two recent U.S. whooping cough statewide epidemics. mSphere. 2016;1:e00036-16. doi: 10.1128/mSphere.00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Gent M, Bart MJ, van der Heide HGJ, Heuvelman KJ, Mooi FR. Small mutations in Bordetella pertussis are associated with selective sweeps. PLoS One. 2012;7:e46407. doi: 10.1371/journal.pone.0046407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pawloski LC, Queenan AM, Cassiday PK, Lynch AS, Harrison MJ, et al. Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the United States. Clin Vaccine Immunol. 2014;21:119–125. doi: 10.1128/CVI.00717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegerle N, Paris A-S, Brun D, Dore G, Njamkepo E, et al. Evolution of French Bordetella pertussis and Bordetella parapertussis isolates: increase of Bordetellae not expressing pertactin. Clin Microbiol Infect. 2012;18:E340–6. doi: 10.1111/j.1469-0691.2012.03925.x. [DOI] [PubMed] [Google Scholar]
- 29.Cohen C, Moyes J, Tempia S, Groom M, Walaza S, et al. Severe influenza-associated respiratory infection in high HIV prevalence setting, South Africa, 2009-2011. Emerg Infect Dis. 2013;19:1766–1774. doi: 10.3201/eid1911.130546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soofie N, Nunes MC, Kgagudi P, van Niekerk N, Makgobo T, et al. The burden of pertussis hospitalization in HIV-exposed and HIV-unexposed South African infants. Clin Infect Dis. 2016;63:S165–S173. doi: 10.1093/cid/ciw545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatti KM, Sparks KN, Boney KO, Tondella ML, Tatti KM, et al. Novel Multitarget Real-Time PCR Assay for Rapid Detection of Bordetella Species in Clinical Specimens Novel Multitarget Real-Time PCR Assay for Rapid Detection of Bordetella Species in Clinical Specimens. 2011. [DOI] [PMC free article] [PubMed]
- 32.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S-H, Lee J, Sung HY, Yu JY, Kim SH, et al. Recent trends of antigenic variation in Bordetella pertussis isolates in Korea. J Korean Med Sci. 2014;29:328. doi: 10.3346/jkms.2014.29.3.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam C, Octavia S, Bahrame Z, Sintchenko V, Gilbert GL, et al. Selection and emergence of pertussis toxin promoter ptxP3 allele in the evolution of Bordetella pertussis . Infect Genet Evol. 2012;12:492–495. doi: 10.1016/j.meegid.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Ben Fraj I, Kechrid A, Guillot S, Bouchez V, Brisse S, et al. Pertussis epidemiology in Tunisian infants and children and characterization of Bordetella pertussis isolates: results of a 9-year surveillance study, 2007 to 2016. J Med Microbiol. 2019;68:241–247. doi: 10.1099/jmm.0.000892. [DOI] [PubMed] [Google Scholar]
- 36.Carriquiriborde F, Regidor V, Aispuro PM, Magali G, Bartel E, et al. Rare detection of Bordetella pertussis pertactin-deficient strains in argentina. Emerg Infect Dis. 2019;25:2048–2054. doi: 10.3201/eid2511.190329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weigand MR, Williams MM, Peng Y, Kania D, Pawloski LC, et al. Genomic survey of Bordetella pertussis diversity, United States, 2000-2013. Emerg Infect Dis. 2019;25:780–783. doi: 10.3201/eid2504.180812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Z, Octavia S, Luu LDW, Payne M, Timms V, et al. Pertactin-negative and filamentous hemagglutinin-negative Bordetella pertussis, Australia, 2013-2017. Emerg Infect Dis. 2019;25:1196–1199. doi: 10.3201/eid2506.180240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mir-Cros A, Moreno-Mingorance A, Martín-Gómez MT, Abad R, Bloise I, et al. Pertactin-deficient Bordetella pertussis with unusual mechanism of pertactin disruption, Spain, 1986-2018. Emerg Infect Dis. 2022;28:967–976. doi: 10.3201/eid2805.211958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weigand MR, Peng Y, Batra D, Burroughs M, Davis JK, et al. Conserved patterns of symmetric inversion in the genome evolution of Bordetella respiratory pathogens. mSystems. 2019;4:e00702-19. doi: 10.1128/mSystems.00702-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dienstbier A, Pouchnik D, Wildung M, Amman F, Hofacker IL, et al. Comparative genomics of Czech vaccine strains of Bordetella pertussis . Pathog Dis. 2018;76 doi: 10.1093/femspd/fty071. [DOI] [PubMed] [Google Scholar]
- 42.Lefrancq N, Bouchez V, Fernandes N, Barkoff A-M, Bosch T, et al. Global spatial dynamics and vaccine-induced fitness changes of Bordetella pertussis . Sci Transl Med. 2022;14:eabn3253. doi: 10.1126/scitranslmed.abn3253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences for 32 South Africa B. pertussis isolates are available on NCBI, organized under BioProject accession number PRJNA929342.