Abstract
Between 2010 and 2015 the incidence of vancomycin-resistant Enterococcus faecium (VREfm) in Norway increased dramatically. Hence, we selected (1) a random subset of vancomycin-resistant enterococci (VRE) from the Norwegian Surveillance System for Communicable Diseases (2010–15; n=239) and (2) Norwegian vancomycin-susceptible E. faecium (VSEfm) bacteraemia isolates from the national surveillance system for antimicrobial resistance in microbes (2008 and 2014; n=261) for further analysis. Whole-genome sequences were collected for population structure, van gene cluster, mobile genetic element and virulome analysis, as well as antimicrobial susceptibility testing. Comparative genomic and phylogeographical analyses were performed with complete genomes of global E. faecium strains from the National Center for Biotechnology Information (NCBI) (1946–2022; n=272). All Norwegian VREfm and most of the VSEfm clustered with global hospital-associated sequence types (STs) in the phylogenetic subclade A1. The vanB2 subtype carried by chromosomal Tn1549 integrative conjugative elements was the dominant van type. The major Norwegian VREfm cluster types (CTs) were in accordance with concurrent European CTs. The dominant vanB-type VREfm CTs, ST192-CT3/26 and ST117-CT24, were mostly linked to a single hospital in Norway where the clones spread after independent chromosomal acquisition of Tn1549. The less prevalent vanA VRE were associated with more diverse CTs and vanA carrying Inc18 or RepA_N plasmids with toxin–antitoxin systems. Only 5 % of the Norwegian VRE were Enterococcus faecalis, all of which contained vanB. The Norwegian VREfm and VSEfm isolates harboured CT-specific virulence factor (VF) profiles supporting biofilm formation and colonization. The dominant VREfm CTs in general hosted more virulence determinants than VSEfm. The phylogenetic clade B VSEfm isolates (n=21), recently classified as Enterococcus lactis , harboured fewer VFs than E. faecium in general, and particularly subclade A1 isolates. In conclusion, the population structure of Norwegian E. faecium isolates mirrors the globally prevalent clones and particularly concurrent European VREfm/VSEfm CTs. Novel chromosomal acquisition of vanB2 on Tn1549 from the gut microbiota, however, formed a single major hospital VREfm outbreak. Dominant VREfm CTs contained more VFs than VSEfm.
Keywords: E. faecalis, E. faecium, vanA gene cluster, vanB gene cluster, vancomycin-resistant enterococci, VRE outbreak
Data Summary
Illumina and PacBio reads and/or assemblies are available under the following project numbers: PRJNA858233, PRJNA407052, PRJNA393251 and PRJNA306646. Biosample ID and metadata are provided in File S1, available in the online version of this article. The authors confirm all supporting data, code and protocols are provided within the article or through supplementary data files.
Impact Statement.
This study represents the first comprehensive unveiling of the population structure of Enterococcus faecium in a low-prevalence antimicrobial resistance setting, including both vancomycin-resistant (VREfm) and -sensitive (VSEfm) isolates. Through comparative genomic analysis we have provided new insights into the epidemiology and population structure of and interaction between VREfm and VSEfm, highlighting critical factors for the understanding and prevention of VRE spread. Importantly, our study discloses the virulome profiles of VREfm and VSEfm using an in-house database of 30 experimentally verified virulence factors involved in E. faecium pathogenesis. VREfm exhibited higher virulence factor content than genetically related VSEfm. The overall findings expand our current knowledge of the epidemiology and spread of VREfm and provides new insights into the genomic evolution of clinical strains of VREfm and VSEfm. Finally, we demonstrated the minor role played by Enterococcus faecalis in the spread of VRE in a low-AMR-prevalence setting.
Introduction
Enterococcus faecium and Enterococcus faecalis are opportunistic pathogens residing in the human gut microbiota. They can cause severe infections in immunocompromised hospitalized patients [1]. The remarkable adaptability of enterococcal genomes and their capacity to acquire antimicrobial resistance (AMR) genes have played a pivotal role in transforming them into increasingly important opportunistic pathogens [2–4]. Although E. faecalis causes most infections, the hospital-adapted E. faecium genotype is more prone to develop multidrug resistance (MDR) [3]. The global phylogeny of E. faecium is characterized by the dominance of two distinct phylogenetic clades, A and B. Clade A can be further divided into two subclades: A1 consisting primarily of clinical strains, and A2 consisting of strains mainly found in animals but also some non-hospitalized individuals. Clade B encompasses community isolates [4–6] and was recently reclassified as Enterococcus lactis [7].
E. faecium infections are difficult to treat because of both intrinsic and acquired antimicrobial resistance. Vancomycin is a preferred drug in treating E. faecium infections [1]. The increasing prevalence of enterococcal infections has been associated with a rise of vancomycin resistance [8]. Ten different van gene clusters (vanA, vanB, vanC, vanD, vanE, vanG, vanL, vanM, vanN and vanP) are responsible for vancomycin resistance in enterococci [9]. The vanC gene cluster is intrinsic in Enterococcus casseliflavus and Enterococcus gallinarum [3, 10], while the other van gene clusters have been associated with acquired vancomycin resistance only [9, 11].
VanA- and vanB-type vancomycin-resistant enterococci (VRE) are the most prevalent worldwide and are predominantly found in vancomycin-resistant E. faecium (VREfm) [3]. While the vanA gene cluster is usually part of the Tn1546 transposon and often found on plasmids [12], the widespread vanB2 subtype gene cluster is associated with Tn1549 integrative conjugative elements (ICEs) originally acquired from gut anaerobes [13]. However, the mechanisms driving the dissemination of VREfm are complex and both clonal spread and exchange of mobile genetic elements (MGEs) likely play important roles [14].
Although E. faecium and E. faecalis are not considered highly virulent, both species possess virulence factors (VFs) associated with colonization, host invasion and/or tissue damage [3, 15], or otherwise bypassing the host immune system [16]. In E. faecium most of the VFs are involved in interactions with the extracellular matrix proteins vital in biofilm formation and colonization [17].
Since 1996, clinical infections and carriage of VRE have been notifiable to the Norwegian Surveillance System for Communicable Diseases (MSIS). VRE is defined as E. faecium or E. faecalis harbouring van gene clusters. The annual number of reported VRE cases was <10 before 2010. After 2010, there was a significant increase in the prevalence of VRE, reaching a peak in 2011, followed by a subsequent decrease, although it never returned to the pre-2010 levels. In addition, the Norwegian surveillance system for antibiotic resistance in microbes (NORM/NORM-VET) systematically collects and monitors antimicrobial susceptibility data in human and animal pathogens, including E. faecium and E. faecalis [18]. Vancomycin-susceptible E. faecium and E. faecalis are defined according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints and for E. faecium additionally absence of vanA and vanB. The nationwide programmes ensure standardized collection, antimicrobial susceptibility testing and storage of strains, providing a unique opportunity to obtain both vancomycin-susceptible and -resistant enterococcal isolates for further investigation.
In this study, we aimed (i) to perform a comparative phylogenomic analysis of the Norwegian VRE from 2010–15, invasive Norwegian vancomycin-susceptible E. faecium (VSEfm; 2008 and 2014) and global strain genomes, and (ii) describe the dominant VRE outbreak clones, their MGEs harbouring van gene clusters and the VF profile of E. faecium .
Methods
Samples size, collection descriptions and data collection
A total of 502 E. faecium (n=469), E. faecalis (n=12) and E. lactis (n=21) isolates from two different collections were included. (1) Randomly selected clinical and screening isolates of VRE (2010–throughout June 2015) from MSIS [19–21]. The study period was chosen because of a sudden increase in VRE incidence from 2010 (0.12 in 2009, 1.10 in 2010 and 5.87 cases in 2011 per 100 000 person years), which then gradually decreased to 1.5 in 2015. (2) Blood culture isolates of vancomycin-susceptible enterococci (VSE) from [19] and [20] and inclusion of the VSE collection allow us to compare vancomycin-susceptible E. faecium (VSEfm) and VREfm genomes, before and after the increase of VRE. Ninety per cent of the VSE isolates from [19] and 93 % of the VSE isolates from [20] were available for inclusion (Table 1). The VSE collection included 21 E. lactis isolates previously identified as E. faecium . The VRE collection consisted of 239 isolates of the 783 (31 %) VRE reported to MSIS between 2010 and 2015, of which 87 (11 %) were from clinical infections. The relative proportion of included VRE compared to the total numbers of VRE reported in Norway is illustrated in Fig. S1. The VRE collection included all of the clinical isolates. Weighted across geography and time, up to three faecal carrier isolates per clinical isolate were selected. If there was no clinical isolate in the geography and time category, a random carrier isolate was selected as index [22]. Twenty-two isolates were excluded from this study (5 due to wrong species identity, 14 because they were not available for sequencing and 3 because of repeated low quality of their assemblies). Thus, a total of 227 VREfm and 12 VREfs were included in the study. In addition, two VREfm isolates recovered in 1996 from the first VRE outbreak reported in Norway [23] were included in the phylogenetic analyses (Table 1). All of the isolates in the study are listed in File S1 with anonymized IDs, and the names of hospitals have been changed to IDs comprising a letter (N, C, E and W, referring to the Northern, Central, South-Eastern and Western health regions of Norway, respectively) and a digit. An overview of sequence types (STs) for VRE from 2019 to 20 was obtained from the Norwegian National Advisory Unit on Detection of Antimicrobial Resistance (K-res) to compare the 2010–15 ST distribution to more recent data.
Table 1.
Bacterial isolates included in the study and proportions of phenotypic resistance to ampicillin, linezolid and high-level gentamicin resistance (HLGR)
|
Collection and year |
Isolates, n |
Ampicillin resistant, n (%) |
HLGR, n (%) |
Linezolid resistant, n (%) |
|---|---|---|---|---|
|
VRE |
||||
|
E. faecium 2010–2015 |
227 |
226 (99.5) |
82 (36) |
1 (0.4) |
|
E. faecalis 2010–2015 |
12 |
0 |
8 (67) |
0 |
|
E. faecium 1996 |
2 |
2 (100) |
0 |
0 |
|
VSE |
||||
|
E. faecium 2008 |
93 |
82 (88) |
55 (59) |
0 |
|
E. lactis 2008 |
6 |
1 (17) |
0 |
0 |
|
E. faecium 2014 |
147 |
138 (94) |
61 (41) |
0 |
|
E. lactis 2014 |
15 |
1 (7) |
0 |
1 (7) |
Species identification and antimicrobial susceptibility testing (AST)
A single blood agar culture colony was used for sub-culturing and subsequent AST, genomic DNA extraction for whole-genome sequencing and species identification by matrix-assisted laser desorption/ionization time-of-flight mass spectometry (MALDI-TOF MS) (Bruker Daltonics GmbH, Bremen, Germany). For the VSE, AST data were collected as part of the NORM programme (appendix 5 in the NORM report) [20]. For the VRE, AST was performed at K-res using the same methods as in NORM, performed and interpreted according to the EUCAST disc diffusion method [24], and EUCAST clinical breakpoints [25], respectively. The Clinical and Laboratory Standards Institute (CLSI) agar screening method was used for detection of reduced susceptibility to vancomycin [26].
Whole-genome sequencing
Initially, all samples were subjected to short-read sequencing. First, the DNeasy Blood and tissue kit (Qiagen, Hilden, Germany) was used to extract the genomic DNA. Next, a Qubit fluorometer (Invitrogen) was used to quantify the concentration of total genomic DNA. The Genomics Support Center Tromsø sequenced the samples using the Illumina NextSeq550 system as described previously [27]. A selection of 21 isolates was subsequently chosen for long-read sequencing to use as reference genomes. The selection was based on their position in the phylogenetic tree. The Wizard Genomic DNA Purification kit (Promega, Madison, USA) was used to extract a large quantity of genomic DNA for long-read sequencing. Then, the genomic DNA concentration was quantified with a Qubit fluorometer. Long-read sequencing was performed at the Norwegian Sequencing Centre (University of Oslo). To prepare multiplexed microbial libraries, the SMRTbell Express Template prep kit 2.0 was used according to the Pacific Biosciences (PacBio) protocol. Fragmentation of DNA was carried out using g-tubes (Covaries) resulting in 10–16 kb-sized fragments. To select the final library, BluePippin with an 8 kb cut-off was used. Libraries were sequenced on ~90 % of the 8M SMRT cell on Sequel II using the Sequel II Banding kit 2.0 and sequencing chemistry v2.0. Demultiplex barcodes pipeline was carried out using SMRT Tools (SMRT Link v9.0.0.92188) to demultiplex the reads (minimum barcode score 26). Finally, the circular consensus sequencing (CCS) sequences were produced for demultiplexed data using CCS pipeline (SMRT Link v9.0.0.92188). The resulting PacBio reads length ranged from 10 to 20 kb.
Genomic analyses
For Illumina-sequenced samples Trimmomatic v0.39 was used to perform quality trimming and adaptor removal [28] before output reads files were assessed using FastQC [29].
Next, Unicycler v0.4.7 was used for genome assembly [30] and, finally, quality assessment of the genome assemblies was performed using Quast v5.0.2 [31]. A cut-off maximum of 400 contigs and a minimum of 40× genome coverage were used for Illumina-sequenced samples to consider the assemblies as eligible to be included in the analyses (with the exception of three samples with 30–37× coverage). Moreover, the genome size should not show more than ±10 % fluctuation compared to the smallest and largest complete E. faecium or E. faecalis genome assemblies in the National Center for Biotechnology Information’s (NCBI’s) Refseq database.
For PacBio-sequenced samples, Unicycler was used to assemble the CCS reads. The assemblies that Unicycler was unable to circularize were reassembled using Canu v2.2 [32], corrected with Pilon v1.23 [33] and circularized using circulator v1.5.5 [34]. Finally, we performed quality assessment using QUAST. The prokaryotic genome annotation pipeline of NCBI was used to annotate the assemblies, MGEs and plasmids [35]. Snippy v3.1 was used for variant calling between sequences [36].
Multilocus sequence typing (MLST)
MLST was carried out for all samples using MLST v2.19.0 [37]. To generate minimum spanning trees, core genome MLST was performed using SeqSphere+ software v6.0.2 (Ridom GmbH, Münster, Germany; http://www.ridom.de/seqsphere/). For E. faecium isolates, the scheme included 1423 core genes and a threshold of ≤20 allelic differences for cluster calculation and determination of clonal relatedness [38]. The scheme of 1972 gene targets with ≤7 allelic differences was set up for cluster calculation and clonal relatedness of E. faecalis genomes [39]. Novel STs and cluster types (CTs) were obtained by submission of assemblies for allelic profiling to PubMLST [40] and Ridom SeqSphere+, respectively.
Phylogenetic trees
Phylogenetic trees based on the core genome of the Norwegian E. faecium and E. lactis were constructed using Parsnp v1.2 [41]. The global tree included all Norwegian E. faecium and E. lactis isolates of the study (n=490), as well as all publicly available complete genomes of E. faecium retrieved from the NCBI as of 11 May 2022 (n=272). In addition, a local tree that only included the 490 Norwegian E. faecium and E. lactis was built. Finally, Interactive Tree Of Life (iTOL) was applied to display metadata in the trees [42].
MGEs harbouring the vanB gene cluster
To identify the van type in the VRE assemblies, the NCBI bacterial AMR reference gene database (PRJNA313047) was used in the ABRicate tool v1.0.1 [43]. To locate and extract the sequences of MGEs harbouring vanB gene clusters in individual isolates, the closest PacBio closed VSE genome was used as a reference. The contigs of the Illumina assemblies were sorted according to the references using Mauve [44]. Next, sorted Illumina assemblies were concatenated and blasted against their reference genomes using the basic local alignment search tool (blastn) v2.6.0 [45]. The Artemis Comparison Tool (ACT) [46] was used to visualize the blasts and locate the MGEs harbouring the vanB gene cluster. Finally, one representative from each MGE type was chosen to perform a blast and visualize the results using Easyfig v2.2.2 [47].
Plasmids harbouring the vanA gene cluster
Mob-suite was used to reconstruct plasmids in VanA-type VREfm isolates [48]. Plasmid typing was performed using the PlasmidFinder v2.0.1 online database (https://cge.food.dtu.dk/services/PlasmidFinder/). Then plasmids were blasted against the NCBI bacterial AMR reference gene database (PRJNA313047) using the ABRicate tool v1.0 to find those containing the vanA gene cluster. To compare the plasmids and determine the identity between them, a closed PacBio-sequenced vanA plasmid of each cluster type was utilized as a reference for reads mapping. The mem algorithm in the BWA tool v07.17 [49] was used to map the reads against the reference sequence. Indexing and sorting were performed in SAMtools v1.10 [50] and the resulting BAM file was visualized using Artemis v18.1.0 [46]. Samples whose reads fully covered the reference vanA plasmid were considered to contain plasmids similar to the reference. EasyFig v2.2.2 was used to blast the closed plasmids and generate a comparison figure.
Virulence factor profile
All of the E. faecium and E. lactis genomes were investigated for the presence of the determinants of 30 experimentally confirmed VFs (File S2) [17, 51–60]. The coding sequences of all 30 VFs were used to build a database in ABRicate v1.0.1 [43]. blasting of the E. faecium and E. lactis genomes against the database was performed using the minimum cut-off for identity and coverage at 90 %. Next, the local phylogenetic tree of E. faecium was annotated using iTOL [42]. Since the esp gene contains several repeats [61], only the conserved part of this gene (2190 bp) was used to blast against the assemblies. For scm, a new allele was found in our samples; the new allele is 173 bp longer than the reference allele. These extra nucleotides are in the linker region and between the two conserved domains of the gene. For scm, both alleles were used for blast searches.
Results and discussion
Both Norwegian VREfm and VSEfm are dominated by prevalent global STs
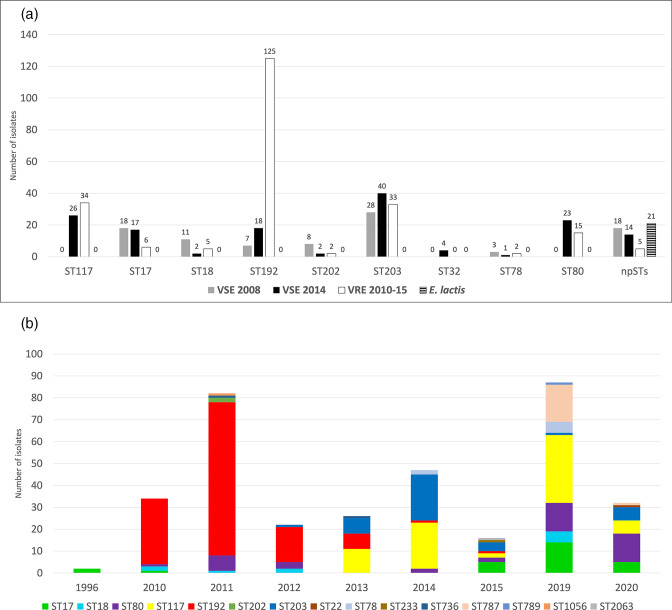
Out of the VREfm 2010–15 isolates, 165 were identified as vanB type, while 62 were identified as vanA type (Fig. 1 and File S1). The majority of the VREfm 2010–15 isolates (n=227) were classified as ST192 (55 %), followed by ST117 (15 %), ST203 (14 %), ST80 (7 %) and ST17 (3 %). Non-prevalent STs (npSTs), including ST18, ST78 and ST202, amounted to 6 % (Figs 1 and 2a). A marked shift in the relative proportions of STs was observed when comparing the VREfm 2010–15 isolates to Norwegian VRE data from 2019 to 2020 [18, 62] (Fig. 2b). The incidence of VRE in 2019 and 2020 was 3.82 and 1.39 cases per 100 000 person years, respectively. While VREfm ST192 was most dominant during 2010–12, it was not observed in 2019–20. In contrast, the prevalence of VREfm ST17 and ST80 increased in the latter years and ST117 started to appear in 2013. All the prevalent STs have been or still are among the dominant STs in European countries. For instance, ST192 was a globally dominant ST mostly related to vanB type VRE in the 2010s [63–65]. ST117 was a dominant ST in Germany over the 1990s and its prevalence increased again after 2010 [63, 66]. ST80 was responsible for the largest VRE outbreak recorded in Germany between 2015 and 2017 with 2900 (vanB-type) cases. ST203, ST17, and ST18 were among the most common STs in Germany from 2000 to 2009, but they began to fade away after a decade (2010–19) [63]. Overall, the major VREfm STs from 2010 were gradually replaced by other STs, showing clonal sweeps of new STs and ST reintroduction (Fig. 2b), consistent with observations from other countries, including Germany and Denmark [63, 67].
Fig. 1.
Number of VRE of different STs by health region in the VRE 2010–2015 collection. The map at the left shows the vanA and the one at the right shows the vanB ST distribution. The four health regions of Norway are coloured in the maps, and pie charts illustrate the frequency of different STs in each region. The STs of the VRE E. faecalis (VREfs) vanB are specified.
Fig. 2.
The frequencies of STs based on collection and year. (a) Frequencies of E. faecium STs shown per sample collection (VSE 2008, VSE 2014 and VRE2010–15) and of VSE E. lactis . The chart illustrates the STs containing at least 1 % of the total number of isolates in this study. STs with <1 % are shown together as non-prevalent STs (npSTs). (b) The prevalence of STs per year shown for VREfm. Data for 2019 and 2020 were added to compare shifts of STs from the period of the study (2010–2015) to more recent data (2019–2020).
The main VSEfm STs are the same as in VREfm 2010–15 but in a different order of prevalence; ST203 (26 %), ST17 (13 %), ST117 (10 %), ST192 (10 %), ST80 (9 %) and ST18 (5 %). The npSTs, including ST32, ST78 and ST202, as well as the 21 E. lactis isolates, covered 27 % of the VSE (Fig. 2a). The presence of each ST varied over time and between VREfm and VSEfm. For instance, ST80 and ST117 were absent among VSEfm 2008 but appeared in VREfm in 2010 and 2013, respectively, and became prevalent STs in VSEfm 2014. ST203, another dominant VREfm ST in 2014, was also present in VSEfm 2014. In contrast, ST17, ST18, ST32 and ST202 were present in VSEfm 2014 but absent in VRE of the same year. Moreover, in VRE 2014, only two isolates out of 47 belonged to the npSTs, while in VSEfm 2014, 34 out of 162 isolates were npSTs, including 15 E. lactis (Fig. 2). Thus, the VSEfm are much more diverse in STs, while the VREfm primarily belong to typical global STs.
Norwegian VREfm are dominated by concurrent major European clusters
In total, 25 vanA-type CTs (19 singletons) and 19 vanB-type CTs (12 singletons) were detected (Fig. S2; Files S1 and S3). The higher diversity and wider geographical dispersion of vanA-type CTs were consistent with smaller outbreaks. We identified four major Norwegian hospital VRE outbreaks during the study period: the vanB-type ST192-CT3/CT26 and ST117-CT24 (Table 2 and File S3), and the vanA-type ST203-CT20 and ST80-CT3097 (Table 3 and File S3). The most prevalent VSEfm CTs are the mixed vanB VRE–VSE clusters, ST117-CT24, ST203-CT3061 and ST80-CT16 (File S3). Three of the predominant VREfm clusters (the vanB-type ST192-CT3/CT26 and ST117-CT24 and vanA-type ST203-CT20) have been reported in other European countries (see Document S1 for details).
Table 2.
Characteristics of Norwegian VREfm clusters and their vanB gene-harbouring MGEs
|
Cluster |
Isolates*, n |
MGE |
MGE insertion location |
Insertion sequence on reference genome (5’–3’) |
|---|---|---|---|---|
|
ST192-CT3/CT26 |
113 |
Tn1549 |
sir gene of tirE operon |
AATATTAAAGGAA |
|
ST117-CT24 |
31 |
Tn1549 |
btuD gene encoding vitamin B12 import ATP-binding protein |
AAAAGTTTTT |
|
ST203-CT3061 |
3 |
Tn1549 |
Between two CDSs encoding hypothetical proteins (HPs) |
TTTTTATAAAAAAA |
|
ST17-CT1709 |
2 |
Tn1549 |
Between CDSs encoding ribonucleoside-diphosphate reductase 2 subunit beta and HP |
TTCAAAAATTTT |
|
ST17-CT6207 |
1 |
Tn1549 |
IS3 family transposase gene |
TTTTTTCTTAAAA |
|
ST80-CT16 |
1 |
Tn1549 |
Between tRNA-Gly and CDS encoding HP |
ATTTTACT |
|
ST6-CT107 |
4 |
Plasmid |
CDS encoding HP |
GATGATGT |
|
ST6-CT1160 |
3 |
Tn1549 |
Between peptidase propeptide and oligopeptide-binding protein (oppA) genes |
TTTTGACA |
|
ST28-CT1162 |
2 |
Tn1549 |
CDS encoding catechol-2,3-dioxygenase |
TTTTAT |
*Singleton VREfs isolates and 15 VREfm isolates with low-quality assembly in the insertion site of Tn1549 are not included in this table.
Table 3.
Characteristics of vanA gene clusters and plasmids in the PacBio-sequenced Norwegian VREfm
|
CT (Reference isolate) |
Isolates, n |
Plasmid size |
CDSs, n |
Plasmid type |
Toxin–antitoxin systems |
Transposon in plasmid |
|---|---|---|---|---|---|---|
|
ST203-CT20 (51271218) |
19 |
55 kb |
73 |
Inc18 |
Epsilon–Zeta |
Tn552 |
|
ST80-CT3097 (51271936) |
10 |
32 kb |
42 |
RepA_N (rep17) |
Axe–Txe |
Tn1546 |
|
ST192-CT188 (51271057) |
4 |
62 kb |
72 |
Inc18 |
Epsilon–Zeta |
Tn1546 |
|
ST18-CT3042 (51276509) |
2 |
43 kb |
51 |
RepA_N (rep17) |
Axe–Txe |
|
|
ST17-CT3037 (51271928) |
2 |
38 kb |
47 |
RepA_N (rep17) |
Axe–Txe and Epsilon–Zeta |
|
|
ST202-CT3079 (51271933) |
1 |
35 kb |
43 |
RepA_N (rep17) |
Axe–Txe |
Tn1546 |
vanB gene clusters in VREfm were carried by de novo-acquired variants of ICE Tn1549
The vanB clusters were carried on ICE Tn1549 variants (Table 2 and Fig. S3) in all vanB-type VREfm from 1996 and from 2010 to 2015. In the ST192-CT3/CT26 isolates that caused the largest outbreak affecting hospitals W1 (n=109) and W2 (n=4) during 2010–13, all but one isolate had an ISL3 element integrated inside the vanB gene cluster in the intergenic region between the vanSB and vanYB genes (variant A in Fig. S3). All Tn1549 in ST17, ST80 and ST203 were also larger than the prototype, mainly due to different IS element insertions (variants B, C and E in Fig. S3).
Acquisitions of Tn1549 have been shown to occur de novo from anaerobic gut microbiota but Tn1549 may also transfer between enterococci [68, 69]. Tn1549 can move between enterococci as part of large chromosomal elements (90–250 kb), in which case the flanking region of Tn1549 should be identical in the donor and recipient isolates [68, 70]. If Tn1549 only transfers between or into enterococci, this should be associated with the transfer of a short coupling sequence from the donor into the recipient genome (5–6 bp) on either the left or right flank of Tn1549 [71]. An identical prototypic Tn1549 was found in one isolate of ST192-CT3/CT26 and one ST117-CT24 isolate that became the dominant clone in the same hospital from 2013. However, the prototypic Tn1549 was integrated into different genomic locations with different flanking sequences in ST192-CT3/CT26 compared to ST117-CT24, suggesting independent ICE Tn1549 acquisitions. While the VREfm isolates of ST117-CT24 are mainly from one hospital in western Norway, the corresponding VSE isolates (n=21) were recovered from nine hospitals covering all four health regions. Thus, this VSE clone has been successful in spreading but likely picked up the vanB ICE Tn1549 in hospital W1, as supported by the finding of a high prevalence of Tn1549 in the non-enterococcal gut flora of admitted patients [72].
vanA gene clusters are carried in unrelated CTs and by different plasmid variants with toxin–antitoxin systems
The vanA gene clusters were carried by different variants of Inc18 or RepA_N family plasmids across different CTs (Table 3 and Fig. S4). Briefly, in ST203-CT20 VREfm a 55 kb vanA Inc18 plasmid with multiple IS integrations was identified. Mapping reads of vanA-type VREfm isolates of this CT against the PacBio-sequenced ST203-CT20 isolate showed that 17 out of 19 vanA plasmids have 100 % coverage to our reference Inc18 plasmid. The vanA gene cluster in this Inc18 plasmid was not part of Tn1546, while other vanA-type clusters like those in ST80-CT3097, ST192-CT188 and ST202-CT3079 were carried by Tn1546. In the second largest cluster, ST80-CT3097, vanA was carried by a 32 kb RepA_N (rep17) plasmid. Other clusters showed vanA Inc18 and RepA_N variants of different sizes (Fig. S4 and Table 3).
Both vanA Inc18 and RepA_N plasmid types may confer increased fitness costs. The persistence of such plasmids has been linked to loss of phenotypic resistance, partial deletions, decreased copy number and toxin–antitoxin systems [73–75]. The partial homology and different sizes of the RepA_N vanA-containing plasmids in our study (Fig. S4) suggest significant rearrangements. Moreover, all the Norwegian VREfm vanA RepA_N plasmids and the two vanA Inc18 plasmids encoded at least one putative toxin–antitoxin system, Axe–Txe and Epsilon–Zeta, respectively, supporting persistence (Table 3) [76, 77].
Norwegian VREfm and successful CTs have enriched virulomes compared to the more diverse VSEfm population
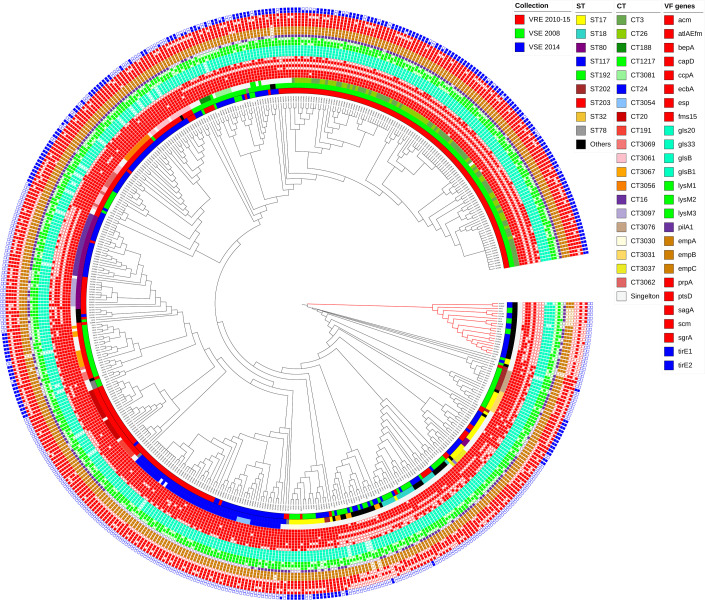
Fig. 3 illustrates the distribution of 26 out of 30 virulence determinants in the Norwegian E. faecium . The VF genes and their function are described in detail in File S2. blast analysis showed that all isolates were negative for boNT/En and epx2 genes encoding exotoxins, which have only been reported in single isolates [53, 55], while positive for fnm and lysM4, which are not shown in the figure. The acm, esp, pilA2, prpA, pstD, scm and srgA genes are involved in colonization and biofilm formation [78–80], tirEs are associated with increased blood survival [60], and gls genes code for general stress proteins [52]. All of these genes are more prevalent in the Norwegian VREfm than in the VSEfm (Table 4 and File S4). All VREfm were positive for all the genes in the empABC operon coding for pilus subunits while for STs containing a mix of VRE and VSE isolates, some VSE lacked empA or empB, and in E. lactis 5/21 (24 %) of the isolates lacked the entire operon (Fig. 3 and File S4).
Fig. 3.
Core genome SNP tree of Norwegian E. faecium and E. lactis annotated with 26 virulence factor genes of E. faecium . Genes of one operon or some genes with similar functional categories are marked with the same colours. However, red is also used for genes that fall into dissimilar functional categories. All of the Norwegian isolates in this study were positive for fnm and lysM4 and negative for bonT/En and epx2, which are not shown in the tree. Annotations shown from the inner layer are sample collection, ST, CT and one layer for each VF gene. The E. lactis clade is highlighted with red branches.
Table 4.
Virulence factor (VF) genes and their distributions (%) in the Norwegian E. faecium and E. lactis
|
VF gene |
Percentage containing VF within major cluster types (CTs) |
|||||||
|---|---|---|---|---|---|---|---|---|
|
ST192-CT3/26 |
ST117-CT24 |
ST203-CT20 |
ST80-CT16 |
ST80-CT3097 |
Percentage with VF in all VREfm |
Percentage with VF in all Efm |
Percentage with VF in all E. lactis |
|
|
(n=113) |
(n=51) |
(n=19) |
(n=23) |
(n=10) |
(n=229) |
(n=469) |
(n=21) |
|
|
VRE |
VRE/VSE |
VRE |
VRE/VSE |
VRE |
VRE |
VRE/VSE |
VSE |
|
|
atlA Efm |
99 |
98 |
100 |
100 |
100 |
99 |
99 |
100 |
|
bepA |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
71 |
|
ccpA |
100 |
98 |
100 |
100 |
100 |
99 |
99 |
100 |
|
empA |
100 |
100 |
100 |
100 |
100 |
100 |
99 |
76 |
|
empB |
100 |
98 |
100 |
100 |
100 |
100 |
98 |
76 |
|
empC |
100 |
98 |
100 |
100 |
100 |
100 |
99 |
76 |
|
sgrA |
100 |
100 |
100 |
100 |
100 |
99 |
94 |
33 |
|
fnm |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
|
ptsD |
99 |
100 |
100 |
100 |
100 |
99 |
97 |
0 |
|
sagA |
100 |
100 |
100 |
100 |
90 |
99 |
99 |
95 |
|
gls20 |
100 |
100 |
94 |
91 |
100 |
98 |
95 |
95 |
|
gls33 |
100 |
100 |
94 |
91 |
100 |
98 |
95 |
95 |
|
glsB |
100 |
100 |
94 |
91 |
100 |
98 |
95 |
95 |
|
glsB1 |
100 |
100 |
94 |
91 |
100 |
98 |
96 |
95 |
|
lysM1 |
71 |
68 |
89 |
69 |
90 |
73 |
72 |
24 |
|
lysM2 |
100 |
100 |
100 |
100 |
100 |
100 |
99 |
95 |
|
lysM3 |
84 |
86 |
42 |
43 |
50 |
47 |
46 |
24 |
|
lysM4 |
100 |
100 |
100 |
100 |
100 |
99 |
100 |
100 |
|
acm |
97 |
100 |
100 |
100 |
100 |
99 |
97 |
28 |
|
ecbA |
0 |
100 |
100 |
0 |
100 |
37 |
42 |
0 |
|
fms15 |
31 |
45 |
15 |
34 |
70 |
33 |
35 |
0 |
|
pilA2 |
99 |
13 |
89 |
100 |
50 |
78 |
67 |
66 |
|
scm |
61 |
72 |
94 |
65 |
50 |
63 |
53 |
0 |
|
esp |
99 |
100 |
68 |
0 |
10 |
90 |
83 |
0 |
|
capD |
0 |
80 |
94 |
0 |
0 |
31 |
50 |
9 |
|
prpA |
100 |
0 |
100 |
0 |
0 |
74 |
68 |
0 |
|
tirE1 |
94 |
0 |
0 |
0 |
0 |
54 |
47 |
0 |
|
tirE2 |
83 |
0 |
0 |
0 |
0 |
48 |
43 |
0 |
|
bonT/En |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
epx2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
The successful VREfm CTs (ST192-CT3/26, ST117-CT24, ST203-CT20, and ST80-CT3097) generally have a high but slightly variable number of virulence determinants (Fig. 3). ST192-CT3/26 (n=113) carries more VFs, in contrast to the ST80-CT16 cluster (containing only 1 VRE out of 23 isolates) lacking 8 VFs (capD, ecbA, esp, prpA, tirE1, tirE2, boNT/En and epx2). Many isolates in the latter cluster also lack fms15, lysM1, lysM3 and scm (Fig. 3 and Table 4).
CT-specific VF profiles were generally observed regardless of the presence or absence of a van gene cluster. Interestingly, clinical VSEfm isolates may have fewer VFs compared to VREfm isolates belonging to the same CT, and npST isolates have fewer VF genes than the predominant STs (see ST203-CT3061 Fig. 3). However, since the virulome of mixed VRE-/VSEfm clusters was highly variable, it was impossible to confirm the significance of the differences statistically.
VREfs incidence is much lower than VREfm
Only 5 % of the Norwegian VRE 2010–15 isolates were VREfs, an observation also found in a previous VRE study [81]. The VREfs isolates (n=12), all vanB-type, clustered in ST6 (n=10) and ST28 (n=2). Nine of those formed three CTs, ST6-CT107 (n=4), ST6-CT1160 (n=3) and ST28-CT1162 (n=2) (Fig. S5). ST6 and ST28 are prevalent clinical STs of E. faecalis [82]. Ampicillin and linezolid resistance were not observed in VREfs, but 8 out of 12 expressed HLGR (Table 1). The VREfs are mainly associated with vanB2-Tn1549 (n=8). However, in ST6-CT107 VREfs (n=4), a vanB1-pTEF1 plasmid remnant was chromosomally integrated (Table 2 and Fig. S3) with 100 % identity and coverage to the typical vanB1-type VRE isolates of V583 (AE016830.1) [83]. The integrated vanB1-pTEF1 plasmid was also found in the genomes of two other V583 derivative isolates [84] and an isolate from the Netherlands (LR961935.1). The VREfm versus VREfs ratio indicates that E. faecium is more prone to acquire and maintain vancomycin resistance. Indeed, transfer of vanB Tn1549 has been shown experimentally to occur from anaerobes to E. faecium [71], while Tn1549 has not been shown to transfer on its own between enterococci and is only occasionally integrated into plasmids. This is one explanation for the low number of VREfs in a low-prevalence setting where vanB is the dominant genotype.
Trends in antimicrobial susceptibility patterns in E. faecium
The ampicillin resistance rate in the Norwegian invasive VSEfm isolates increased from 88 % in 2008 to 94 % in 2014, while it was 99.5 % in VREfm during 2010–15. HLGR on the other hand, showed slightly decreasing prevalence in VSEfm, declining from 59 % in 2008 to 41 % in 2014 and an even lower rate (36 %) in VREfm during 2010–15 (Table 1). Linezolid resistance (chromosomal) was only observed in two single isolates in this study. In comparison, the ampicillin and high-level gentamicin resistance rates in VREfm blood culture isolates from EU/EEA 2012–18 were 99 and 49 %, respectively, in comparison to 89 and 43 %, respectively, for VSEfm [85].
E. lactis is less resistant and has fewer known VFs than E. faecium
Since the E. lactis isolates (n=21) were identified as E. faecium by MALDI-TOF MS, they were also included in the phylogenetic tree of Norwegian E. faecium (n=490) (Fig. S6). In the earlier E. faecium classification, clade A is mainly formed by globally dominant STs, and no clear separation within clade A (A1 and A2 subclades) was observed. Boundaries for subclades in clade A are controversial in E. faecium population structure analysis [4] and may be affected by geographical context. For instance, in VREfm isolates from Latin America, further subclading of A1 was proposed [86]. Thus, we refrain from specifying subclades in our collection (Fig. S6).
Globally, clade A isolates have been shown to be more prone to acquire genes, including resistance genes, while E. lactis (clade B) isolates are usually susceptible [4, 6]. All of the E. lactis (n=21) isolates in this study were npST VSE from 2008 and 2014 (black colour in ST ring of Figs 3 and S6). Our findings highlight significant differences in pheno- and genotype between E. lactis (clade B) and E. faecium (clade A). E. lactis isolates were found to be predominantly susceptible to vancomycin (Fig. S6) and aminoglycosides, whereas resistance to ampicillin and linezolid was limited (Table 1 and File S1). Vancomycin-resistant E. lactis isolates are rarely reported, although E. lactis vanN-type VRE have been observed in Japan (ST669) and the USA (ST240) [87]. Moreover, a lower number of VFs was typical for the E. lactis isolates (n=21), lacking from 13 to 19 of the investigated VF genes. None of the E. lactis isolates were shown to harbour ecbA, esp, fms15, prpA, ptsD, scm, tirE1, tirE2, boNT/En, or epx2, and capD, lysM1, lysM3 and sgrA were found in a minority of isolates (Table 4, Fig. 3 and File S4). Our results reveal differences in E. lactis VF profiles compared to others using a different VF database [88]. Notably, while scm was lacking in Norwegian E. lactis (n=21), it was present in four of nine E. lactis in the study by Roer et al. [88]. Potentially significant differences in prevalence were also observed for srgA and bepA. However, the small number of E. lactis isolates in both studies does not support an overall conclusion.
Study strengths and limitations
The main issues in the global molecular epidemiology of enterococci are the bias caused by (i) the skewed geographical representation and (ii) the dominance of VRE. Most of the examined VRE and VSE genomes are submitted from Europe, followed by Japan, Australia and the USA. Thus, the epidemiology of VRE is less known in other parts of the world (Africa, the Middle East and South Asia). Moreover, most of the studies are biased by an overrepresentation of antibiotic-resistant outbreak isolates. In this study, the sample selection of VRE was performed randomly across time and region, including different types of infection sources and carriers. In addition, VSE isolates were included for genomic comparison. Thus, the current strain collection is more representative of the concomitant VSE and VRE in a defined setting, a low-prevalence AMR European context.
In the global trees and genomic comparisons, we used the complete closed genomes of the E. faecium (n=272 as of 11 May 2022), which included only 2 % of the E. faecium genomes (all assembly levels) available in the NCBI [89]. Excluding 98 % of the genomes, as well as missing data from the rest of the world, may increase the risk of overlooking an association in the global population structure.
Other studies include several putative VFs in the E. faecium virulome [86, 90]. All VF genes included in our study are experimentally confirmed virulence determinants (references listed in File S2), and we believe this provides a more conservative and less speculative approach.
Conclusions
Our study highlights that globally prevalent clones, and particularly concurrent European CTs, influence the population structure of both the vancomycin-resistant and -sensitive Norwegian E. faecium . The prevalent Norwegian VREfm CTs have acquired more virulence determinants than the more diverse nationwide VSEfm population. The majority of the VREfm isolates were vanB type, likely driven by outbreaks in the healthcare setting but also formed by de novo acquisition of vanB from the gut microbiota. VREfs are much rarer than VREfm and are all vanB type.
Supplementary Data
Funding information
This project was supported by a PhD fellowship grant from the Northern Norway Regional Health Authority Medical Research Programme project number HNF1362-17. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
We would like to express our deep gratitude to Kjersti Julin, Ahmed Mekhlif and Theresa Wagner at UiT and Ellen H. Josefsen, Martin O. Christensen and Berit Harbak at K-res for their technical support. We would like to express our appreciation to The Norwegian Surveillance Programme for Antimicrobial Resistance (NORM) for access to information about the VSE strain collections and The Norwegian VRE study group members Silje Bakken Jørgensen (Akershus University Hospital), Annette Onken (Vestre Viken Hospital Bærum), Einar Tollaksen Weme (Vestre Viken Hospital Drammen), Reidar Hjetland (Førde Central Hospital), Ghantous Milad Chedid (Haugesund Hospital), Fabian Åhrberg (Molde Hospital), Hege Elisabeth Larsen (Nordland Hospital), Karianne Wiger Gammelsrud (Oslo University Hospital Ullevål), Iren Löhr (Stavanger University Hospital), Hege Enger (St. Olavs University Hospital), Kari Ødegaard (Innlandet Hospital), Åshild Marvik-Rødland (Vestfold Hospital Trust), Kyriakos Zaragkoulias (Nord-Trøndelag Hospital Trust), Andreas Emmert (Østfold Hospital Trust), Ståle Tofteland (Sørlandet Hospital), Gunnar Skov Simonsen (University Hospital of North Norway) and Einar Nilsen (Ålesund Hospital) for their help in collecting the samples. We would also like to extend our thanks to the Genomics Support Center Tromsø and The Norwegian Sequencing Centre in UiO for genome sequencing, and to Theodor Anton Ross at UiT for language proofreading.
Author contributions
Conceptualization: J.V.B., A.S. and K.H. Data curation: M.A.R., O.K., B.C.H., R.M.N. and K.H. Formal analysis: M.A.R., J.J., J.H. and K.H. Funding acquisition: J.V.B. and K.H. Investigation: M.A.R., J.J., J.V.B., A.S. and K.H. Methodology: M.A.R., J.J., J.V.B., A.S. and K.H. Project administration: J.V.B. and K.H. Supervision: J.J., J.V.B., A.S. and K.H. Validation: M.A.R., J.J., J.V.B., A.S. and K.H. Visualization: M.A.R. Writing – original draft: M.A.R. and K.H. Writing – review and editing: all authors.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The project was approved by the regional ethics committee project under project number 2015/1532/REK sør-øst.
Footnotes
Abbreviations: ACT, Artemis Comparison Tool; AMR, antimicrobial resistance; AST, antimicrobial susceptibility testing; BAM, Binary Alignment and Map; BLAST, Basic Local Alignment Search Tool; CCS, circular consensus sequencing; CDS, coding sequence; CLSI, Clinical Laboratory Standards Institute; CT, cluster type; EUCAST, European Committee on Antimicrobial Susceptibility Testing; HLGR, high-level gentamicin resistance; HP, hypothetical protein; ICE, integrative conjugative element; iTOL, interactive tree of life; K-res, Norwegian National Advisory Unit on Detection of Antimicrobial Resistance; MALDI-TOF, matrix assisted laser desorption ionization - time of flight; MDR, multidrug resistance; MGE, mobile genetic element; MLST, multilocus sequence typing; MSIS, Norwegian Surveillance System for Communicable Diseases; NCBI, National Center for Biotechnology Information; NORM/NORM-VET, Norwegian surveillance system for antibiotic resistance in microbes; npST, non-prevalent ST; SNP, single nucleotide polymorphism; ST, sequence type; VF, virulence factor; VRE, vancomycin resistant enterococci; VREfm, vancomycin resistant Enterococcus faecium; VREfs, vancomycin resistant Enterococcus faecalis; VSE, vancomycin susceptible enterococci; VSEfm, vancomycin susceptible Enterococcus faecium.
References
- 1.Fiore E, Van Tyne D, Gilmore MS. Pathogenicity of enterococci. Microbiol Spectr. 2019;7:4. doi: 10.1128/microbiolspec.GPP3-0053-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramos S, Silva V, Dapkevicius M de LE, Igrejas G, Poeta P. Enterococci, from harmless bacteria to a pathogen. Microorganisms. 2020;8:1118. doi: 10.3390/microorganisms8081118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García-Solache M, Rice LB. The enterococcus: a model of adaptability to its environment. Clin Microbiol Rev. 2019;32:1–28. doi: 10.1128/CMR.00058-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Hal SJ, Willems RJL, Gouliouris T, Ballard SA, Coque TM, et al. The interplay between community and hospital Enterococcus faecium clones within health-care settings: a genomic analysis. Lancet Microbe. 2022;3:E133–E141. doi: 10.1016/S2666-5247(21)00236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arredondo-Alonso S, Top J, McNally A, Puranen S, Pesonen M, et al. Plasmids shaped the recent emergence of the major nosocomial pathogen Enterococcus faecium . mBio. 2020;11:e03284-19. doi: 10.1128/mBio.03284-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio. 2013;4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belloso Daza MV, Cortimiglia C, Bassi D, Cocconcelli PS. Genome-based studies indicate that the Enterococcus faecium Clade B strains belong to Enterococcus lactis species and lack of the hospital infection associated markers. Int J Syst Evol Microbiol. 2021;71:004948. doi: 10.1099/ijsem.0.004948. [DOI] [PubMed] [Google Scholar]
- 8.Sparo M, Delpech G, García Allende N. Impact on public health of the spread of high-level resistance to gentamicin and vancomycin in enterococci. Front Microbiol. 2018;9:3073. doi: 10.3389/fmicb.2018.03073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xavier BB, Coppens J, De Koster S, Rajakani SG, Van Goethem S, et al. Novel vancomycin resistance gene cluster in Enterococcus faecium ST1486, Belgium, June 2021. Euro Surveill. 2021;26:2100767. doi: 10.2807/1560-7917.ES.2021.26.36.2100767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moura TM de, Cassenego APV, Campos FS, Ribeiro AML, Franco AC, et al. Detection of vanC1 gene transcription in vancomycin-susceptible Enterococcus faecalis . Mem Inst Oswaldo Cruz. 2013;108:453–456. doi: 10.1590/S0074-0276108042013009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivertsen A, Pedersen T, Larssen KW, Bergh K, Rønning TG, et al. A silenced vanA gene cluster on a transferable plasmid caused an outbreak of vancomycin-variable enterococci. Antimicrob Agents Chemother. 2016;60:4119–4127. doi: 10.1128/AAC.00286-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arredondo-Alonso S, Top J, Corander J, Willems RJL, Schürch AC. Mode and dynamics of vanA-type vancomycin resistance dissemination in Dutch hospitals. Genome Med. 2021;13:1–18. doi: 10.1186/s13073-020-00825-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium . Clin Microbiol Infect. 2010;16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Chlebowicz MA, Bathoorn E, Rosema S, Couto N, et al. Elucidating vancomycin-resistant Enterococcus faecium outbreaks: the role of clonal spread and movement of mobile genetic elements. J Antimicrob Chemother. 2018;73:3259–3267. doi: 10.1093/jac/dky349. [DOI] [PubMed] [Google Scholar]
- 15.Weiss RA. Virulence and pathogenesis. Trends Microbiol. 2002;10:314–317. doi: 10.1016/s0966-842x(02)02391-0. [DOI] [PubMed] [Google Scholar]
- 16.Mundy LM, Sahm DF, Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clin Microbiol Rev. 2000;13:513–522. doi: 10.1128/CMR.13.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao W, Howden BP, Stinear TP. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr Opin Microbiol. 2018;41:76–82. doi: 10.1016/j.mib.2017.11.030. [DOI] [PubMed] [Google Scholar]
- 18.NORM/NORM-VET 2020 Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. ISSN:1502-2307 (Print) / 1890-9965 (Electronic) Tromsø / Oslo: 2021. [Google Scholar]
- 19.NORM/NORM-VET 2008 Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. ISSN: 1502-2307 (Print) / 1890-9965 (Electronic) Tromsø / Oslo: 2009. [Google Scholar]
- 20.NORM/NORM-VET 2014 Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. ISSN: 1502-2307 (Print) / 1890-9965 (Electronic). ISSN:1502-2307 (Electronic) Tromsø / Oslo: 2015. [Google Scholar]
- 21.Rosvoll TCS, Lindstad BL, Lunde TM, Hegstad K, Aasnaes B, et al. Increased high-level gentamicin resistance in invasive Enterococcus faecium is associated with aac(6´)Ie-aph(2″)Ia-encoding transferable megaplasmids hosted by major hospital-adapted lineages. FEMS Immunol Med Microbiol. 2012;66:166–176. doi: 10.1111/j.1574-695X.2012.00997.x. [DOI] [PubMed] [Google Scholar]
- 22.Grimes DA, Schulz KF. Compared to what? Finding controls for case-control studies. Lancet. 2005;365:1429–1433. doi: 10.1016/S0140-6736(05)66379-9. [DOI] [PubMed] [Google Scholar]
- 23.Dahl KH, Røkenes TP, Lundblad EW, Sundsfjord A. Nonconjugative transposition of the vanB-containing Tn5382-like element in Enterococcus faecium . Antimicrob Agents Chemother. 2003;47:786–789. doi: 10.1128/AAC.47.2.786-789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.European Committee on Antimicrobial Susceptibility Testing – EUCAST Antimicrobial susceptibility testing: EUCAST disk diffusion method, version 7.0. 2019. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2019_manuals/Manual_v_7.0_EUCAST_Disk_Test_2019.pdf
- 25.European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters, version 9.0, 2019. 2019. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf
- 26.Swenson JM, Clark NC, Ferraro MJ, Sahm DF, Doern G, et al. Development of a standardized screening method for detection of vancomycin-resistant enterococci. J Clin Microbiol. 1994;32:1700–1704. doi: 10.1128/jcm.32.7.1700-1704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.AL Rubaye MTS, Janice J, Bjørnholt JV, Jakovljev A, Hultström ME, et al. Novel genomic islands and a new vanD-subtype in the first sporadic VanD-type vancomycin resistant enterococci in Norway. PLoS One. 2021;16:1–15. doi: 10.1371/journal.pone.0255187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews S. Fastqc: a quality control tool for high throughput sequence data. Babraham Bioinform. 2010 [Google Scholar]
- 30.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015;16:1–10. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seemann T. Snippy, Rapid haploid variant calling and core genome alignment. 2015. https://github.com/tseemann/snippy
- 37.Jolley KA, Maiden MCJ. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Been M, Pinholt M, Top J, Bletz S, Mellmann A, et al. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium . J Clin Microbiol. 2015;53:3788–3797. doi: 10.1128/JCM.01946-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumann B, Prior K, Bender JK, Harmsen D, Klare I, et al. A core genome multilocus sequence typing scheme for Enterococcus faecalis . J Clin Microbiol. 2019;57:e01686-18. doi: 10.1128/JCM.01686-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seemann T. Abricate,Github. https://github.com/tseemann/abricate n.d.
- 44.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, et al. ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson J, Nash JHE. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom. 2018;4:e000206. doi: 10.1099/mgen.0.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H. Aligning sequence reads, clone sequences and assembly Contigs with BWA-MEM. arXiv. 2013;1303:3997. [Google Scholar]
- 50.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Somarajan SR, La Rosa SL, Singh KV, Roh JH, Höök M, et al. The fibronectin-binding protein Fnm contributes to adherence to extracellular matrix components and virulence of Enterococcus faecium . Infect Immun. 2015;83:4653–4661. doi: 10.1128/IAI.00885-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choudhury T, Singh KV, Sillanpää J, Nallapareddy SR, Murray BE. Importance of two Enterococcus faecium loci encoding Gls-like proteins for in vitro bile salts stress response and virulence. J Infect Dis. 2011;203:1147–1154. doi: 10.1093/infdis/jiq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong X, Tian S, Yang P, Lebreton F, Bao H, et al. Emerging Enterococcus pore-forming toxins with MHC/HLA-I as receptors. Cell. 2022;185:1157–1171. doi: 10.1016/j.cell.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paganelli FL, Willems RJL, Jansen P, Hendrickx A, Zhang X, et al. Enterococcus faecium biofilm formation: identification of major autolysin AtlAEfm, associated Acm surface localization, and AtlAEfm-independent extracellular DNA Release. mBio. 2013;4:e00154-13. doi: 10.1128/mBio.00154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S, Lebreton F, Mansfield MJ, Miyashita S-I, Zhang J, et al. Identification of a botulinum neurotoxin-like toxin in a commensal strain of Enterococcus faecium . Cell Host Microbe. 2018;23:169–176. doi: 10.1016/j.chom.2017.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Somarajan SR, Roh JH, Singh KV, Weinstock GM, Murray BE. CcpA is important for growth and virulence of Enterococcus faecium . Infect Immun. 2014;82:3580–3587. doi: 10.1128/IAI.01911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cacaci M, Giraud C, Leger L, Torelli R, Martini C, et al. Expression profiling in a mammalian host reveals the strong induction of genes encoding LysM domain-containing proteins in Enterococcus faecium . Sci Rep. 2018;8:12412. doi: 10.1038/s41598-018-30882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ali L, Blum HE, Sakιnç T. Detection and characterization of bacterial polysaccharides in drug-resistant enterococci. Glycoconj J. 2019;36:429–438. doi: 10.1007/s10719-019-09881-3. [DOI] [PubMed] [Google Scholar]
- 59.Zhang X, Top J, de Been M, Bierschenk D, Rogers M, et al. Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J Infect Dis. 2013;207:1780–1786. doi: 10.1093/infdis/jit076. [DOI] [PubMed] [Google Scholar]
- 60.Wagner TM, Janice J, Paganelli FL, Willems RJ, Askarian F, et al. Enterococcus faecium TIR-domain genes are part of a gene cluster which promotes bacterial survival in blood. Int J Microbiol. 2018;2018:1435820. doi: 10.1155/2018/1435820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palmer KL, Schaik W, Willems RJL, Gilmore MS. In: Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Gilmore MS, Clewell DB, Ike Y, editors. Boston: Massachusetts Eye and Ear Infirmary; 2014. Enterococcal genomics. [PubMed] [Google Scholar]
- 62.NORM/NORM-VET 2019 Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. ISSN:1502-2307 (Print) / 1890-9965 (Electronic) Tromsø / Oslo: 2020. [Google Scholar]
- 63.Werner G, Neumann B, Weber RE, Kresken M, Wendt C, et al. Thirty years of VRE in Germany - “expect the unexpected”: The view from the National Reference Centre for Staphylococci and Enterococci. Drug Resist Updat. 2020;53:100732. doi: 10.1016/j.drup.2020.100732. [DOI] [PubMed] [Google Scholar]
- 64.Zhou W, Zhou H, Sun Y, Gao S, Zhang Y, et al. Characterization of clinical enterococci isolates, focusing on the vancomycin-resistant enterococci in a tertiary hospital in China: based on the data from 2013 to 2018. BMC Infect Dis. 2020;20:356. doi: 10.1186/s12879-020-05078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sivertsen A, Billström H, Melefors Ö, Liljequist BO, Wisell KT, et al. A multicentre hospital outbreak in Sweden caused by introduction of a vanB2 transposon into a stably maintained pRUM-plasmid in an Enterococcus faecium ST192 clone. PLoS One. 2014;9:e103274. doi: 10.1371/journal.pone.0103274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weber A, Maechler F, Schwab F, Gastmeier P, Kola A. Increase of vancomycin-resistant Enterococcus faecium strain type ST117 CT71 at Charité - Universitätsmedizin Berlin, 2008 to 2018. Antimicrob Resist Infect Control. 2020;9:109. doi: 10.1186/s13756-020-00754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hammerum AM, Justesen US, Pinholt M, Roer L, Kaya H, et al. Surveillance of vancomycin-resistant enterococci reveals shift in dominating clones and national spread of a vancomycin-variable vanA Enterococcus faecium ST1421-CT1134 clone, Denmark, 2015 to March 2019. Euro Surveill. 2019;24:1900503. doi: 10.2807/1560-7917.ES.2019.24.34.1900503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bender JK, Kalmbach A, Fleige C, Klare I, Fuchs S, et al. Population structure and acquisition of the vanB resistance determinant in German clinical isolates of Enterococcus faecium ST192. Sci Rep. 2016;6:21847. doi: 10.1038/srep21847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Howden BP, Holt KE, Lam MMC, Seemann T, Ballard S, et al. Genomic insights to control the emergence of vancomycin-resistant enterococci. mBio. 2013;4:e00412-13. doi: 10.1128/mBio.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quintiliani R, Courvalin P. Conjugal transfer of the vancomycin resistance determinant vanB between enterococci involves the movement of large genetic elements from chromosome to chromosome. FEMS Microbiol Lett. 1994;119:359–363. doi: 10.1111/j.1574-6968.1994.tb06913.x. [DOI] [PubMed] [Google Scholar]
- 71.Launay A, Ballard SA, Johnson PDR, Grayson ML, Lambert T. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob Agents Chemother. 2006;50:1054–1062. doi: 10.1128/AAC.50.3.1054-1062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nygaard RM, Hegstad K, Kommedal Ø, Lindemann PC. 12th International Meeting on Microbial Epidemiological Markers. Dubrovnik, Croatia: 2019. High prevalence of vanB in non-enterococcal flora including a novel species contributes to persistent outbreaks of vancomycin resistant enterococci in a Norwegian hospital. [Google Scholar]
- 73.Tedim AP, Lanza VF, Rodríguez CM, Freitas AR, Novais C, et al. Fitness cost of vancomycin-resistant Enterococcus faecium plasmids associated with hospital infection outbreaks. J Antimicrob Chemother. 2021;76:2757–2764. doi: 10.1093/jac/dkab249. [DOI] [PubMed] [Google Scholar]
- 74.Wagner TM, Janice J, Sivertsen A, Sjögren I, Sundsfjord A, et al. Alternative vanHAX promoters and increased vanA-plasmid copy number resurrect silenced glycopeptide resistance in Enterococcus faecium . J Antimicrob Chemother. 2021;76:876–882. doi: 10.1093/jac/dkaa541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Freitas AR, Tedim AP, Francia MV, Jensen LB, Novais C, et al. Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986-2012) J Antimicrob Chemother. 2016;71:3351–3366. doi: 10.1093/jac/dkw312. [DOI] [PubMed] [Google Scholar]
- 76.Grady R, Hayes F. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium . Mol Microbiol. 2003;47:1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- 77.Rosvoll TCS, Pedersen T, Sletvold H, Johnsen PJ, Sollid JE, et al. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTbeta-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol Med Microbiol. 2010;58:254–268. doi: 10.1111/j.1574-695X.2009.00633.x. [DOI] [PubMed] [Google Scholar]
- 78.Heikens E, Bonten MJM, Willems RJL. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J Bacteriol. 2007;189:8233–8240. doi: 10.1128/JB.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wagner T, Joshi B, Janice J, Askarian F, Škalko-Basnet N, et al. Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. J Proteomics. 2018;187:28–38. doi: 10.1016/j.jprot.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 80.Revtovich AV, Tjahjono E, Singh KV, Hanson BM, Murray BE, et al. Development and characterization of high-throughput Caenorhabditis elegans - Enterococcus faecium infection model. Front Cell Infect Microbiol. 2021;11:667327. doi: 10.3389/fcimb.2021.667327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Elstrøm P, Astrup E, Hegstad K, Samuelsen Ø, Enger H, et al. The fight to keep resistance at bay, epidemiology of carbapenemase producing organisms (CPOs), vancomycin resistant enterococci (VRE) and methicillin resistant Staphylococcus aureus (MRSA) in Norway, 2006 - 2017. PLoS One. 2019;14:e0211741. doi: 10.1371/journal.pone.0211741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pöntinen AK, Top J, Arredondo-Alonso S, Tonkin-Hill G, Freitas AR, et al. Apparent nosocomial adaptation of Enterococcus faecalis predates the modern hospital era. Nat Commun. 2021;12:1523. doi: 10.1038/s41467-021-21749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis . Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 84.Furlan S, Matos RC, Kennedy SP, Doublet B, Serror P, et al. Fitness restoration of a genetically tractable Enterococcus faecalis V583 derivative to study decoration-related phenotypes of the enterococcal polysaccharide antigen. mSphere. 2022;4:e00310-19. doi: 10.1128/mSphere.00310-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ayobami O, Willrich N, Reuss A, Eckmanns T, Markwart R. The ongoing challenge of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis in Europe: an epidemiological analysis of bloodstream infections. Emerg Microbes Infect. 2020;9:1180–1193. doi: 10.1080/22221751.2020.1769500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rios R, Reyes J, Carvajal LP, Rincon S, Panesso D, et al. Genomic epidemiology of vancomycin-resistant Enterococcus faecium (VREfm) in Latin America: revisiting the global VRE population structure. Sci Rep. 2020;10:5636. doi: 10.1038/s41598-020-62371-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lebreton F, Valentino MD, Schaufler K, Earl AM, Cattoir V, et al. Transferable vancomycin resistance in clade B commensal-type Enterococcus faecium . J Antimicrob Chemother. 2018;73:1479–1486. doi: 10.1093/jac/dky039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roer L, Kaya H, Tedim AP, Novais C, Coque TM, et al. 33rd European Congress of Clinical Microbiology and Infectious Diseases. Copenhagen: 2023. In silico extended virulence profiling of Enterococcus faecium and Enterococcus lactis isolates by use of whole-genome sequencing data; p. 1861. p. [Google Scholar]
- 89.NCBI Enterococcus faecium assemblies. https://www.ncbi.nlm.nih.gov/assembly/?term=enterococcus faecium n.d.
- 90.van Hal SJ, Willems RJL, Gouliouris T, Ballard SA, Coque TM, et al. The global dissemination of hospital clones of Enterococcus faecium . Genome Med. 2021;13:52. doi: 10.1186/s13073-021-00868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.