Abstract
Mutations in the Presenilin (PSEN) genes are the most common cause of early-onset familial Alzheimer's disease (FAD). Studies in cell culture, in vitro biochemical systems, and knockin mice showed that PSEN mutations are loss-of-function mutations, impairing γ-secretase activity. Mouse genetic analysis highlighted the importance of Presenilin (PS) in learning and memory, synaptic plasticity and neurotransmitter release, and neuronal survival, and Drosophila studies further demonstrated an evolutionarily conserved role of PS in neuronal survival during aging. However, molecular pathways that interact with PS in neuronal survival remain unclear. To identify genetic modifiers that modulate PS-dependent neuronal survival, we developed a new Drosophila Psn model that exhibits age-dependent neurodegeneration and increases of apoptosis. Following a bioinformatic analysis, we tested top ranked candidate genes by selective knockdown (KD) of each gene in neurons using two independent RNAi lines in Psn KD models. Interestingly, 4 of the 9 genes enhancing neurodegeneration in Psn KD flies are involved in lipid transport and metabolism. Specifically, neuron-specific KD of lipophorin receptors, lpr1 and lpr2, dramatically worsens neurodegeneration in Psn KD flies, and overexpression of lpr1 or lpr2 does not alleviate Psn KD-induced neurodegeneration. Furthermore, lpr1 or lpr2 KD alone also leads to neurodegeneration, increased apoptosis, climbing defects, and shortened lifespan. Lastly, heterozygotic deletions of lpr1 and lpr2 or homozygotic deletions of lpr1 or lpr2 similarly lead to age-dependent neurodegeneration and further exacerbate neurodegeneration in Psn KD flies. These findings show that LpRs modulate Psn-dependent neuronal survival and are critically important for neuronal integrity in the aging brain.
Keywords: Alzheimer's disease, Presenilin, genetic modifier, lipoproteins, lipid transport and metabolism
Presenilin, the most important genetic factor in Alzheimer's disease, plays an evolutionarily conserved role in protection of neuronal survival during aging. In this study, Kang et al. use Drosophila to identify molecular players that interact with Presenilin in the regulation of neuronal survival. They report that reduction of lipophorin receptors, which are involved in lipid transport, compromises neuronal survival and exacerbates neuronal death caused by reduction of Presenilin expression, highlighting the consequences of disrupted lipid homeostasis in the aging brain
Introduction
Alzheimer's disease (AD) is the most common neurodegenerative disorder, and mutations in the Presenilin (PSEN) genes account for >80% of causative mutations identified in familial AD (FAD) (https://www.alzforum.org/mutations). Genetic studies in mice showed that selective inactivation of the Psen genes in excitatory or inhibitory neurons of the cerebral cortex recapitulates key features of AD, including memory and synaptic plasticity impairment followed by progressive, widespread cortical neurodegeneration, elevated gliosis, and tau hyperphosphorylation, highlighting the importance of Presenilin (PS) in neuronal function and survival (Yu et al. 2001; Saura et al. 2004; Zhang et al. 2009; Wines-Samuelson et al. 2010; Zhang et al. 2010; Wu et al. 2013; Lee et al. 2017; Kang and Shen 2020; Lee et al. 2021, 2023). FAD-linked PSEN1 mutations show impaired essential function in cultured cells, including human iPS cell-derived neurons, knockin (KI) mice, and cell-free systems (Heilig et al. 2010; Heilig et al. 2013; Woodruff et al. 2013; Xia et al. 2015; Xia et al. 2016; Sun et al. 2017; Zhou et al. 2017).
PS is the catalytic subunit of γ-secretase, an intramembrane protease that cleaves type I transmembrane proteins, including Notch and the amyloid precursor protein (APP) (De Strooper et al. 1998; Song et al. 1999; Struhl and Greenwald 1999). Despite Notch being a key mediator of PS function during development, using the same excitatory neuron-specific Camk2a-Cre line to selectively delete Notch1 and Notch2 did not result in neurodegeneration (Shen et al. 1997; Wong et al. 1997; Song et al. 1999; Handler et al. 2000; Saura et al. 2004; Yang et al. 2004; Kim and Shen 2008; Zheng et al. 2012). Furthermore, inactivation of APP and its family members selectively in excitatory neurons of the postnatal forebrain also did not lead to cortical neurodegeneration (Lee et al. 2020). While numerous additional γ-secretase substrates were reported, often using overexpression systems (Haapasalo and Kovacs 2011), the physiological relevance of the γ-secretase-mediated cleavage of the reported putative substrates is less clear.
To identify physiologically relevant molecular pathways that interact with PS in the regulation of neuronal survival in the aging brain, we took advantage of the power of fly genetics. Psn, the PSEN ortholog in Drosophila, shares high sequence homology with human and mouse PSEN, and loss of function in Drosophila Psn induces pupal lethality, maternal neurogenic effects, loss of lateral inhibition within proneural cell clusters, and absence of wing margin formation (Struhl and Greenwald 1999; Ye et al. 1999). We previously developed Psn short hairpin RNA (shRNA) lines and found that conditional knockdown of Psn in adult neurons using the neuron-specific, inducible elav-Gal4 and tubulin-Gal80ts system leads to age-dependent neurodegeneration and increases of apoptosis, demonstrating an evolutionarily conserved role of PS in neuronal protection in the aging brain (Kang et al. 2017). In this study, we developed a simplified version of the neuron-specific Psn KD fly model using elav-Gal4 alone by maintaining flies at 18°C and then shifting to 29°C upon eclosion. Similarly, these Psn KD flies exhibit age-dependent neurodegeneration, which can be reversed by the expression of a wild-type Psn. Through a bioinformatic analysis, using protein motifs from the transmembrane regions of human and Drosophila Notch and the amyloid precursor protein-like (APPL), we ranked Drosophila type I transmembrane proteins. We then tested each of the top 25 ranked genes using two independent RNAi lines, and found that neuron-specific KD of 9 genes enhanced neurodegeneration caused by Psn KD. Four of the 9 genes encode proteins that belong to the low-density lipoprotein receptor (LDLR) family and are involved in lipid transport and metabolism, and 2 encode lipophorin receptor 1 (LpR1) and LpR2. We then performed validation experiments and confirmed that neuron-specific KD of lpr1 or lpr2 results in neurodegeneration and worsens neurodegenerative phenotypes of Psn KD flies. Furthermore, heterozygotic deletions of lpr1 and lpr2 or homozygotic deletions of lpr1 or lpr2 also lead to age-dependent neurodegeneration and further exacerbate neurodegeneration in Psn KD flies. These findings show that lipophorin receptors play essential protective roles in the aging brain and genetically modulate Psn-dependent neuronal survival.
Materials and methods
Fly stocks
Fly stocks were raised on standard cornmeal media, and maintained at 22–25oC and 40–60% relative humidity. UAS-shPsn3 transgenic flies were generated as described (Kang et al. 2017), and elav-Gal4 (elav-Gal4c155; RRID:BDSC_456), P(CarryP)attP2 (RRID:BDSC_36303), UAS-Psn + 14 (RRID:BDSC_63243), lpr1Df (RRID:BDSC_44236), and lpr2Df (RRID:BDSC_44233) flies were obtained from the Bloomington Drosophila Stock Center (BDSC). lpr1/2 deficient flies, UAS-lpr1-J, and UAS-lpr2-E flies were kindly provided by Joaquim Culi (Parra-Peralbo and Culi 2011). UAS-RNAi transgenic flies against top 25 candidate genes were obtained from BDSC, Vienna Drosophila Resource Center, or National Institute of Genetics Fly Stock. Information about the alleles and transgenic flies used in this study can be found in Supplementary Table 1.
Generating neuron-specific Psn and GeneX KD as well as lpr1 and lpr2 transgenic flies
P(CarryP)attP2 flies were crossed with elav-Gal4 or elav-Gal4; UAS-shPsn3 flies to generate the negative controls (elav-Gal4/+) and neuron-specific Psn KD flies as positive controls (elav-Gal4/+; UAS-shPsn3/+), respectively. For the rescue experiments, elav-Gal4; UAS-shPsn3 female flies were crossed with male UAS-Psn + 14 flies to generate elav-Gal4/+; UAS-shPsn3/UAS-Psn + 14 flies. The selected UAS-GeneX-RNAi transgenic flies were crossed with elav-Gal4 or elav-Gal4; UAS-shPsn3 flies to generate neuron-specific GeneX single KD flies (elav-Gal4/+; UAS-GeneX-RNAi/+) or Psn/GeneX double KD flies (elav-Gal4/+; UAS-shPsn3/UAS-GeneX-RNAi or elav-Gal4/+; UAS-shPsn3/+; UAS-GeneX-RNAi/+), respectively. UAS-lpr1-J and UAS-lpr2-E flies were crossed with elav-Gal4/+ to generate neuron-specific lpr1 and lpr2 transgenic flies, respectively. All flies were cultured at 18oC until eclosion and then shifted to 29oC until the end of the experiment.
RT-qPCR
For adult fly head RNA extraction, adult flies were collected and frozen immediately in liquid nitrogen, and heads were removed from bodies by repeated vortexing and sinking the tube into liquid nitrogen to separate heads from bodies. Third instar larval brains and adult brains were dissected in RNase-free PBS and homogenized in lysis buffer using a homogenizing blender (Next Advance). Total RNA was extracted from either 20 adult heads or 15 3rd-instar larval or adult brains, according to the manufacturer's instructions (Quick-RNATM MicroPrep; Zymo Research). The eluted total RNA was reverse transcribed using iScriptTM Select cDNA Synthesis Kit (Bio-Rad) with gene-specific primers for each target gene. Real-time qPCR was performed with ViiATM 7 Real-Time PCR System (Thermo Fisher Scientific) using PowerUP SYBR Green Master Mix (Thermo Fisher Scientific) with two sets of specific primers for each target gene. The sequences of all primers used in the RT and qPCR are listed in Supplementary Table 2. The mRNA levels of target genes were normalized to housekeeping genes of the average value of rp49 and gapdh1 expressions.
Analysis of lifespan
More than 99 flies per genotype were assayed for lifespan as previously described (Kang et al. 2017). Flies were transferred to fresh vials every other day, and each vial housed approximately 30 flies. Lifespans were measured by scoring dead flies remaining in the old vial and were plotted using the Kaplan–Meier method (Kaplan and Meier 1958; Kang et al. 2017). The median lifespan (MedLS) was calculated as the age when half of the flies died. The survival distribution of two genotypic groups was compared using the Log-rank (Mantel–Cox) test.
Bioinformatic analysis
To screen genetic modifiers that modulate PS-dependent neuronal survival, we used a bioinformatics approach to rank order putative γ-secretase substrates, as PS is the catalytic subunit of γ-secretase and numerous γ-secretase substrates have been reported (Haapasalo and Kovacs 2011). While γ-secretase substrates are thought to be type I transmembrane proteins, there are low levels of similarities among those proteins within the transmembrane region. For example, Notch and the amyloid protein precursor (APP) are the well-established physiological substrates of γ-secretase (De Strooper et al. 1998; Song et al. 1999; Struhl and Greenwald 1999), but the similarity among these proteins within the transmembrane region is only 8–27% based on Clastal Omega multiple sequence alignment analysis (https://www.ebi.ac.uk/Tools/msa/clustalo/). To prioritize genes for the genetic modifier testing among the type I transmembrane encoding genes, we first used a transmembrane Hidden Markov Model (TMHMM) prediction (Sonnhammer et al. 1998), a HMM-based (Eddy 1996) method to identify a total of 641 type I transmembrane proteins in Drosophila whole genome. We then used transmembrane sequences of human Notch and APP as well as Drosophila Notch and APPL as inputs to run HHpred (Söding et al. 2005), a HMM-HMM comparison method that uses HMMs to encode protein evolutionary constraint, to look for homologous proteins sharing sequence homology and found 112 proteins. Using the transmembrane sequences of the 112 proteins as a case data set, we used the rest of the type I transmembrane proteins as a control set to train the motif model. To reduce the original amino acid alphabet into the 4-letter alphabet, we used the alphabet reduction strategy (Bacardit et al. 2009) and ran MAFFT (Katoh et al. 2002) on the sequence translated to the reduced alphabet to discover homologous segment. Next, we used the position weight matrices to model the cleavage site of γ-secretase within the transmembrane region and score the type I transmembrane proteins by motif match prediction. Then, we used the DIOPT (DRSC Integrative Ortholog Prediction Tool) database score (Hu et al. 2011) to select 264 genes, based on their mammalian ortholog scores (greater than 3). We further eliminated 44 genes that show less than 3 RPMK values in the central nervous system (CNS) or adult head RNA-seq analysis based on the Flybase database (http://flybase.org/rnaseq/profile_search). Of the final 220 type I transmembrane proteins, we selected the top 25 ranked genes for genetic modifier testing.
Analysis of adult eyes
The analysis of adult fly eyes was performed as previously described (Kang et al. 2017). The adult flies were frozen at the age of 5 days, and the images of eyes were obtained using Leica S9i digital microscope.
Histological analysis
Heads from adult flies at different ages were fixed in 10% formalin overnight, paraffinized, embedded in paraffin, and sectioned from a frontal orientation. Serial sections (8 μm in thickness) spanning the entire brain were collected and placed on glass slides and subjected to further analysis. Brain morphology was evaluated by staining paraffin sections with hematoxylin and eosin (H&E) as previously described (Dias-Santagata et al. 2007; Kang et al. 2017). To evaluate neurodegeneration, the number of vacuoles larger than 10 μm in diameter, which were circular in shape and were distinct from ruptures of paraffin sections, was counted in each of the serial sections throughout the entire brain. Large vacuoles might have been counted more than once in adjacent serial sections, reflecting the more severe neurodegenerative phenotypes in fly brains. The number of brains analyzed in each genotypic, age group is indicated in the figure legend. The number of brain sections used in vacuole quantification is shown in Supplementary Table 4.
TUNEL assay
Cells undergoing apoptosis were detected in the paraffin sections by TUNEL, according to the manufacturer’s instructions (TdT Enzyme DNA Fragmentation Detection Kit; EMD Millipore, Calbiochem, FragEL). Quantification of the TUNEL-positive (TUNEL+) cells was performed by counting cells labeled with visible markers in all the serial sections (8 μm) of the entire fly brain. The number of brains analyzed in each genotypic group is indicated in the figure legend.
Active caspase-3 immunostaining of whole-mount adult brains
Adult male fly brains were dissected in PBS at the age of 25 days, fixed for 30 min in 4% PFA in PBS at room temperature. Fly brains were washed 3 times (10 min for each) in PBS-T (0.1% Triton X-100) and blocked in blocking solution (5% normal goat serum (NGS) in 1× PBS) for 1 h at room temperature followed by incubation in the primary antibody rabbit anti-cleaved caspase-3 (Asp175) (1:200, cat. #9661L, Cell Signaling Technology, RRID: AB_2341188) in blocking solution for 48 h at 4°C. The brains were washed 3 times (10 min for each) in PBS-T followed by incubation of the secondary antibody Goat anti-Rabbit IgG (H + L) Secondary Antibody, Alexa Fluor 488 (1:1,000, cat. #A11034, Invitrogen, RRID: AB_2576217) in blocking solution overnight at 4°C. Brains were washed 3 times in PBS-T and 2 times in PBS (10 min for each) and then mounted in vectashield mounting medium (cat. #H-1000-10, Vector Labs). A total of 15 stacks with 8 μm intervals covering the whole brain were imaged using a FV1000 confocal microscope system (Olympus). Quantification of cleaved caspase-3 cells was performed by counting the number of cleaved caspase-3 positive cells throughout the whole brain using ImageJ 5.0 software (NIH). At least 5 individual brains were analyzed per genotype. The number of brains analyzed in each genotypic group is indicated in the figure legend.
Climbing assay
Climbing ability was assessed as previously described (Rhodenizer et al. 2008; Kang et al. 2017). Briefly, ∼20 flies were gently tapped to the bottom of a plastic vial, and a picture was taken after 20 s. This procedure was repeated 4 times with 1-min intervals between trials to allow the flies to recover from prior tapping. Climbing ability was evaluated by scoring the number of flies that failed to climb over 5 cm from the bottom in each trial.
Statistical analysis
All statistical analysis was performed using Prism (Version 9; GraphPad). All data are presented as the mean ± SEM. The exact sample size (e.g. number of flies) of each experiment is indicated in the figure or figure legends. Statistical analysis was conducted using two-way ANOVA with Bonferroni’s multiple comparisons (Fig. 2b), unpaired Student's t-test (Figs. 2d and 6b), one-way ANOVA with Tukey's multiple comparisons (Figs. 2f, 3b, 4c, 5a, 5c, 5e, 5f, 6d and 7d) and Log-rank (Mantel–Cox) test (Figs. 1c and 5g). All statistical analysis results can be found in figure legends and Supplementary Tables 1, 3, and 4. Significant is shown as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, or NS (not significant).
Fig. 2.
Age-dependent neurodegeneration in neuron-specific Psn KD flies. a) Representative images of H&E stained brain sections of control (elav-Gal4/+) and Psn KD (elav > shPsn3) flies at different ages. Vacuoles indicative of neurodegeneration are marked with yellow arrowheads. Scale bar: 100 μm. Higher power views of the boxed areas show representative vacuoles in the brain sections. Scale bar: 50 μm. b) Quantification of vacuoles in the brain of control and Psn KD flies at the ages of 1, 10, 20, and 30 days. The brains of Psn KD flies show age-dependent, significant increases of vacuoles (F3, 101 = 42.8, P < 0.0001, two-way ANOVA with Bonferroni’s multiple comparisons). At 10 days of age, the number of vacuoles in elav > shPsn3 brains (22.3 ± 2.3) is significantly higher, compared to controls (5.8 ± 1.1, P = 0.0030). At 20 days of age, the number of vacuoles in elav > shPsn3 brains further increases (48.7 ± 3.2), compared to controls (8.3 ± 1.1, P < 0.0001). By 30 days of age, the number of vacuoles in elav > shPsn3 brains (82.2 ± 6.9) is dramatically higher than that in controls (13.7 ± 1.9, P < 0.0001). Vacuoles of circular shapes in intact paraffin brain sections were counted in each of the serial sections throughout the entire brain (10–13 sections per brain), and the total number of vacuoles per brain is shown. For each genotype, age group, 11–16 brains were used for quantification. c) Representative images of TUNEL + cells marked with blue arrowheads in the lateral horn (LH) area of the brain of control and Psn KD flies at the age of 30 days. Scale bar: 50 μm. d) Quantification of TUNEL + cells in all serial sections of the entire brain shows significant increases of TUNEL + apoptotic cells in Psn KD brains (100.6 ± 5.1), compared to controls (22.3 ± 2.4, P < 0.0001, unpaired Student's t-test). Quantification was done using 11 brains per genotype. e) RT-qPCR analysis of Psn mRNA levels in fly heads. Compared to control elav-Gal4/+ flies, Psn mRNA levels are elevated in elav > Psn + 14 transgenic flies (∼40-fold), whereas levels of Psn mRNA are drastically reduced in the presence of shPsn3 in elav > shPsn3, Psn + 14 flies (∼10-fold). The level of Psn mRNA is similar in UAS-shPsn3, UAS-Psn + 14/+, and elav-Gal4/+ flies. The value of Psn mRNA levels in control elav-Gal4/+ flies is set as 1. Psn mRNA levels were normalized to the average value of rp49 and gapdh1 as internal controls. Total RNA was extracted from fly heads at the age of 4 days (20 heads per genotype). Four independent RT-qPCR analyses were done for each genotypic group, using 20 heads per sample for RT-qPCR. f) Quantification analysis shows that the total number of vacuoles in elav > shPsn3, Psn + 14 flies (46.1 ± 3.6) is reduced, compared to elav > shPsn3 flies (109.0 ± 5.8, P < 0.0001, one-way ANOVA with Tukey's multiple comparisons). There is no significant difference between elav > shPsn3, Psn + 14 (46.1 ± 3.6) and control elav-Gal4/+ flies (31.9 ± 2.3, P = 0.2023). The number of vacuoles in UAS-shPsn3, UAS-Psn + 14/+ flies (21.6 ± 2.3) is similar to that in control elav-Gal4/+ flies (P = 0.6588), whereas there are increases of vacuoles in elav > Psn + 14 flies (76.5 ± 7.4, P < 0.0001). For each genotypic group, 12–14 brains were used for quantification. g) Representative images of H&E stained brain sections show that the elav > Psn + 14 transgene reverses the increased vacuoles, marked with yellow arrowheads, in Psn KD flies. Scale bar: 100 μm. Higher power views of the boxed areas show representative vacuoles in the brain sections. Scale bar: 50 μm. All data are expressed as mean ± SEM. Each circle represents data obtained from an individual fly brain in histological analysis. NS, not significant; **P < 0.01, ****P < 0.0001.
Fig. 6.
Overexpression of lpr1 or lpr2 does not rescue neurodegeneration in Psn KD brains. a) Top: Schematic illustrations of gene structures of lpr1 and lpr2. The black boxes represent coding exons, and the gray boxes represent the untranslated region (UTR). Both lpr1 and lpr2 have two transcriptional initiation sites. Primers used in RT-PCR are shown as arrows below common exons, amplifying all isoforms. Middle: mRNA structures of lpr2-E (yellow) and lpr1-J (blue) isoforms. Bottom: Schematic illustrations of LpR2-E and LpR1-J showing conserved domains [red, nonconserved N-terminal domain; green, LA module; blue, EGF module; orange, YWTD β-propeller domain; white, transmembrane region]. b) RT-qPCR analysis of lpr2 and lpr1 mRNA levels in Tg flies. Compared to control flies, lpr2 and lpr1 mRNA levels are elevated in elav > lpr2-E (∼2-fold) and elav > lpr1-J (∼3-fold) Tg flies. The value of lpr2 or lpr1 endogenous mRNA levels in control is set as 1. lpr2 and lpr1 mRNA levels were normalized to the average value of rp49 and gapdh1 as internal controls. Total RNA was extracted from heads of adult flies at the age of 4 days. Four independent RT-qPCR analyses were performed for each indicated group, each sample using 20 adult heads for RT-qPCR. Each circle represents data obtained from an independent experiment. c) Representative images of H&E stained brain sections of indicated genotypes at the age of 30 days. Vacuoles indicative of neurodegeneration are marked with yellow arrowheads. Scale bar: 100 μm. d) Quantification of vacuoles in the brain of indicated genotypes at the age of 30 days. The number of vacuoles in elav > lpr2-E flies (49.4 ± 5.0) is significantly increased, compared to elav-Gal4/+ control (23.6 ± 3.0, P = 0.0168, one-way ANOVA with Dunnett's multiple comparisons). The number of vacuoles in elav > lpr1-J (23.8 ± 4.7, P > 0.9999) is similar to that in elav-Gal4/+ controls. The number of vacuoles in elav > shPsn3; lpr2-E flies (126.3 ± 11.3) is further increased, compared to elav > shPsn3 (87.8 ± 5.0, P = 0.0007), whereas the number of vacuoles in elav > shPsn3; lpr1-J (91.4 ± 7.9) is not significantly altered, compared to elav > shPsn3 (P = 0.9613); 10–19 brains per genotypic group were examined and vacuoles quantified in all serial sections of the entire brain. Each circle represents data obtained from an individual fly brain in histological analysis. All data are expressed as mean ± SEM. NS, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Fig. 3.
Neuron-specific Psn/GeneX KD induces early lethality and developmental phenotypes. a) Representative images of H&E stained brain sections of neuron-specific Psn/GeneX double KD (top) and GeneX KD (bottom) flies at the age of 30 days. elav-Gal4-driven KD of both Notch (N) RNAi lines results in lethality prior to eclosion. Images from one of two independent RNAi lines showing stronger phenotypes are presented. The specific RNAi line of each gene is indicated, and vacuoles are marked with yellow arrowheads. Scale bar: 100 μm. b) Quantification of vacuoles in the brain of indicated genotypic groups. Two individual RNAi lines were tested for each gene. Genes are separated into groups based on lethality phenotypes: lethality during development by both RNAi lines (Notch), a single RNAi line (Delta, Nrx-IV), or early adult lethality by a single RNAi line (tutl, tkv, Lerp). Psn/Dl double KD or Dl KD alone using RNAi line 2 are viable, but Psn/Dl double KD flies show dramatically increased vacuoles (444.7 ± 61.2), compared to Psn KD flies (82.1 ± 3.2, P < 0.0001, one-way ANOVA with Tukey's multiple comparisons). Dl KD using the same RNAi line 2 also displays an elevated number of vacuoles (79.3 ± 7.3), compared to controls (24.3 ± 1.7, P < 0.0001). While elav-Gal4-driven KD of Neurexin IV (Nrx-IV) by RNAi line 1, with or without Psn KD, results in developmental lethality, Nrx-IV KD using RNAi line 2 has no effect on the number of vacuoles in either control or Psn/Nrx-IV KD fly brains. Psn/tutl KD flies by RNAi line 2 exhibit increases of vacuoles (225.9 ± 35.2), compared to Psn KD (P < 0.0001), and tutl KD alone using RNAi lines 1 or 2 does not affect survival or vacuole number. Psn/tkv KD using RNAi line 2 enhances the number of vacuoles (216.3 ± 13.6), compared to Psn KD flies (P < 0.0001), and tkv KD alone also induces more vacuoles (101.4 ± 7.4) than controls (P < 0.0001), whereas tkv KD alone by RNAi line 1 results in early adult lethality. Psn/Lerp KD using RNAi line 1 results in early adult lethality, and Lerp KD alone induces increases in the number of vacuoles (94 ± 7.8), compared to controls (P < 0.0001), whereas Psn/Lerp KD flies using RNAi line 2 are viable and show an elevated number of vacuoles (210.5 ± 12.4), compared to Psn KD alone (P < 0.0001). N/T indicates not tested due to lethality. All data are expressed as mean ± SEM; 10–28 individual brains were analyzed per genotypic group. Each circle represents data obtained from an individual fly brain. The number of vacuoles and full statistical analysis are shown in Supplementary Table 1. ****P < 0.0001.
Fig. 4.
Genetic modifiers of Psn KD-induced neurodegeneration in fly brains. a, b) Representative images of H&E stained brain sections of neuron-specific Psn/GeneX double KD flies a) and neuron-specific GeneX KD flies c) at the age of 30 days (vacuoles are marked by yellow arrowheads). Images from 1 of 2 independent RNAi lines showing stronger phenotypes are presented. The images of control (elav-Gal4/+) and Psn KD (elav > shPsn3) flies are the same as those shown in Fig. 3a. Scale bar: 100 μm. c) Quantification of vacuole numbers in the brain of indicated genotypic groups. Genes are categorized into different groups depending on the effect on neuron-specific Psn KD-induced neurodegeneration (Psn-ND). Left: Enhancing neurodegeneration via both RNAi lines (lpr2, arr, Mul1, and Ptp10D), a single RNAi line (lpr1, robo1, mgl, Bsg, and Ptp99A), no effect (drpr, LRP1, fra, Pvr, crb, and Appl), reducing neurodegeneration via a single RNAi line (GluRIB and robo2), or via both RNAi lines (Miro and mGluR). For example, compared to Psn KD flies (82.1 ± 3.2), KD of lipophorin receptor 2 (lpr2) with Psn together using RNAi line 1 results in an increased number of vacuoles (270.0 ± 53.3, P < 0.0001, one-way ANOVA with Tukey's multiple comparisons), and using RNAi line 2 shows increased number of vacuoles (115.3 ± 11.4, P < 0.0001); KD of metabotropic glutamate receptor (mGluR) with Psn together show reduced number of vacuoles via RNAi line 1 (64.8 ± 4.9, P < 0.0001) or RNAi line 2 (44.3 ± 3.8, P < 0.0001). Right: Compared to controls (24.3 ± 1.7), KD of roundabout 1 (robo1) using RNAi line 1 results in the increased number of vacuoles (207.1 ± 25.5, P < 0.0001), whereas using RNAi line 2 showed a less increased number of vacuoles (86.5 ± 7.5, P < 0.0001). All data are expressed as mean ± SEM; 4–28 brains per genotypic group were examined and vacuoles quantified in all serial sections of the entire brain. Each circle represents data obtained from an individual fly brain. The number of vacuoles and full statistical analysis are shown in Supplementary Table 1. *P < 0.05, **P < 0.01, ***P < 0.001 ****P < 0.0001.
Fig. 5.
Neuron-specific KD of lpr1 or lpr2 enhances neurodegeneration in Psn KD flies. a) RT-qPCR analysis of lpr1 and lpr2 mRNA levels in 4-day-old flies. lpr1 and lpr2 mRNA levels were normalized to the average value of rp49 and gadph1 mRNA levels as internal controls. The value of lpr1 and lpr2 mRNA in control flies is set as 100%. Four independent RT-qPCR analyses were performed for each indicated group with each sample using 20 fly heads, and each circle represents data obtained from an independent experiment. b) Representative images of H&E stained brain sections show increases of vacuoles, marked by yellow arrowheads, in lpr1 KD, lpr2 KD, and Psn/lpr KD flies at the age of 30 days. c) Quantification of vacuoles shows that lpr1 or lpr2 KD worsens neurodegeneration in Psn KD flies. The number of vacuoles in elav > lpr2 line 1 flies (elav > lpr2-IR1, 64.5 ± 4.5) is increased, compared to elav-Gal4/+ control (22.7 ± 1.2, P < 0.0001, one-way ANOVA with Tukey's multiple comparisons). The number of vacuoles in elav > shPsn3; lpr2-IR1 flies (410.2 ± 28.6) is further increased, compared to elav > shPsn3 (75.3 ± 3.4, P < 0.0001). The number of vacuoles in elav > lpr1 line 1 (elav > lpr1-IR1) flies (59.1 ± 2.9) is significantly increased, compared to elav-Gal4/+ controls (P < 0.0001), and the vacuole number in elav > shPsn3; lpr1-IR1 flies (128.0 ± 6.2) is further increased, compared to elav > shPsn3 (P < 0.0001). Vacuoles were counted in serial sections of 11–15 brains per genotypic group. d) Representative images of whole-mount brains at the age of 25 days stained with anti-cleaved active caspase-3 antibody. Compared to elav-Gal4/+ controls, there are more active caspase-3 + cells in the brain of Psn, lpr1, and lpr2 KD flies, whereas Psn/lpr1 and Psn/lpr2 KD further increases active caspase-3 + cells in the brain. Scale bar: 100 µm. Higher power views of the boxed area show representative active caspase-3 immunopositive cells (blue arrowheads). Scale bar: 10 µm. e) Quantification of active caspase-3 + puncta from the entire brain shows significant increases of apoptotic cells in the brain of lpr1 KD (56.4 ± 3.0, P = 0.0073, one-way ANOVA with Tukey's multiple comparisons) or lpr2 KD (64.0 ± 4.3, P = 0.0045) flies, compared to controls (35.0 ± 3.3). Compared to Psn KD flies (65.8 ± 4.3), the number of apoptotic cells is similar in Psn/lpr1 KD flies (79.0 ± 4.1, P = 0.0892) but is markedly increased in Psn/lpr2 KD flies (97.2 ± 6.5, P = 0.0009). The number of active caspase-3 + cells was quantified in 5–6 brains per genotypic group. f) Climbing defects of Psn and lpr KD flies at the age of 30 days. The percentages of the failed climbs are shown. Compared to elav-Gal4/+ control flies (19.0 ± 1.9%), Psn KD flies (72.3 ± 2.4%, P < 0.0001, one-way ANOVA with Tukey's multiple comparisons), lpr2 KD flies (47.9 ± 6.3%, P = 0.0100), and lpr1 KD flies (42.6 ± 7.3%, P = 0.0215) show significantly increased climbing defects. Compared to Psn KD flies, lpr2/Psn KD flies (75.6 ± 6.3%, P = 0.9056) and lpr1/Psn KD flies (60.7 ± 6.6%, P = 0.3299) show similar climbing ability. N = 3–6 independent experiments; ≥ 60 flies per genotype (∼20 flies per experiment) were used in the study. g) Lifespan of the indicated genotypic groups plotted using the Kaplan–Meier method. Compared to elav/+ control flies (55 days, black line), the MedLS of Psn KD (31 days, red line), lpr2 KD (30 days, yellow line), and lpr1 KD (29 days, light green line) is significantly shorter [P < 0.0001, Log-rank (Mantel–Cox) test]. The lifespan is similar between Psn KD flies (31 days) and lpr1/Psn KD flies (30 days, dark green line, P = 0.1696), whereas lpr2/Psn KD flies (28 days, orange line) exhibit a significantly reduced lifespan (P = 0.0011). N = 99–138 flies per genotype. All data are expressed as mean ± SEM. Each circle represents data obtained from an individual fly brain in histological analysis. NS, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Fig. 7.
Germline deletions of lpr1 and lpr 2 worsen neurodegeneration in Psn KD flies. a) Schematic illustration of the gene structures of lpr1 and lpr2. The black boxes represent exons encoding the protein sequences, and the gray boxes represent the UTRs. Both lpr1 and lpr2 have 2 transcription initiation sites. Orange, blue, and pink lines indicate the deleted genomic region in each deficiency line. b, c) Representative images of H&E stained brain sections show age-dependent increases of vacuoles, marked by yellow arrowheads, in flies carrying lpr1, lpr2, or lpr1/2 deficiency mutations alone or together with Psn KD at the age of 1 day b) or 30 days c). Scale bar: 100 μm. d) Quantification of vacuoles shows that lpr deficiency increases vacuoles and worsens neurodegeneration in aged Psn KD flies. At 1 day of age, few vacuoles were detected in the brains of all genotypic groups. At 30 days, compared to elav-Gal4/+ controls (37.4 ± 2.8), there is no difference in vacuole numbers in lpr2 Df/+ flies (39.0 ± 1.9, P > 0.9999, one-way ANOVA with Tukey’s multiple comparisons), or lpr1 Df/+ flies (50.4 ± 3.7, P = 0.3167). lpr1/2 Df/+ flies significantly increase in vacuole number (61.7 ± 9.5, P = 0.001). All 3 homozygotes deficiency flies show increased vacuole numbers, lpr2 Df/Df flies (79.4 ± 6.9, P < 0.0001), lpr1 Df/Df flies (59.9 ± 3, P = 0.001), and lpr1/2 Df/Df flies (74.8 ± 4.6, P < 0.0001). Compared to elav > shPsn3 (94.0 ± 6.4), there is no difference in vacuole numbers in elav > shPsn3; lpr2 Df/+ flies (96.5 ± 10.1, P > 0.9999), or elav > shPsn3; lpr1 Df/+ flies (80.8 ± 3.2, P = 0.8049). However, elav > shPsn3; lpr1/2 Df/+ flies show significantly increased vacuole number (142.0 ± 10.9, P = 0.0008). Compared to elav > shPsn3, elav > shPsn3; lpr2 Df/Df flies (134.5 ± 16, P = 0.0345) show increased vacuole number, but elav > shPsn3; lpr1 Df/Df flies (91.4 ± 6.8, P > 0.9999) show no significant difference. N/T indicates not tested: all elav > shPsn3; lpr1/2 Df/Df flies died between 15 and 23 days of age; 6–24 brains per genotypic group were analyzed. All data are expressed as mean ± SEM. Each circle represents data obtained from an individual fly brain. NS, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Fig. 1.
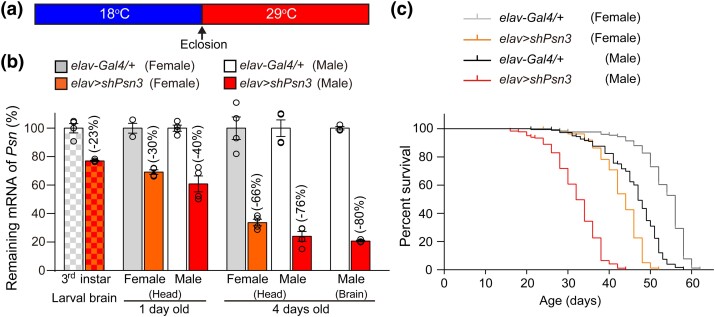
Reduced mRNA levels and shortened lifespan of neuron-specific Psn KD flies. a) Strategy for imposing temporal control of shRNA expression using temperature shift. Flies were crossed and maintained at 18°C, and then offspring following eclosion were collected and shifted to 29°C immediately to increase shRNA expression. b) RT-qPCR analysis shows progressive reductions of Psn mRNA levels in elav > shPsn3 flies. Psn mRNA levels are reduced at 23% in dissected brains of 3rd-instar larvae, indicating basal levels of Psn KD at 18°C, which is sufficient to support Psn function during development. Levels of Psn mRNA are reduced progressively in the head of Psn KD flies from the age of 1 day to 4 days posteclosion at 29°C, and there is a stronger reduction of Psn mRNA in male Psn KD flies than in females. At 4 days of age, in dissected brains of male Psn KD flies, only 20% Psn mRNA remains, compared to controls. Psn mRNA levels are normalized to the average value of rp49 and gapdh1 as internal controls. The value of Psn mRNA levels in control flies was set as 100%. Total RNA was extracted from dissected larval or adult brains or adult heads (15 dissected brains or 20 heads per sample); 3–4 independent RT-qPCR analyses were repeated for each indicated group, each sample using either 15 dissected larval or adult brains or 20 adult heads for RT-qPCR. c) Shortened lifespan of neuron-specific Psn KD flies. Lifespan of female and male control flies (elav-Gal4/+), as well as female and male neuron-specific Psn KD (elav-Gal4/+; UAS-shPsn3/+) flies were plotted using the Kaplan–Meier method. The MedLS of male Psn KD flies (32 days) is significantly shorter, compared to male control flies (47 days; P < 0.0001), and the MedLS of female Psn KD flies (44 days) is also shorter, compared to female control flies [56 days; P < 0.0001, Log-rank (Mantel–Cox) test]; 142–185 flies were tested for each indicated group.
Results
Development of a neuron-specific Psn KD model for genetic modifier screening
Our previously developed adult neuron-specific Psn conditional KD flies using tub-Gal80ts to impose a temporal restriction of Psn shRNA expression (Kang et al. 2017) is cumbersome when testing large numbers of RNAi lines. To develop a simpler Psn KD system that is more conducive for genetic modifier screening, we took advantage of the temperature sensitivity of GAL4 (Duffy 2002) by maintaining flies at 18°C during development and then shifting to 29°C posteclosion. We chose a previously characterized Psn shRNA line, shPsn3, which results in 90% reduction of Psn mRNA while using a ubiquitous Actin-Gal4 driver (Kang et al. 2017). Neuron-specific Psn KD flies (elav-Gal4/+; UAS-shPsn3/+, hereafter, elav > shPsn3) were cultured at 18°C to circumvent pupal lethality associated with Psn loss-of-function mutants (Struhl and Greenwald 1999; Ye and Fortini 1999; Mahoney et al. 2006), and then transferred to 29°C upon eclosion to increase shPsn3 expression (Fig. 1a). RT-qPCR analysis showed that levels of Psn mRNA are progressively reduced, and only 20% of Psn mRNA remains in dissected brains of elav > shPsn3 male flies at 4 days of age (Fig. 1b). The MedLS of elav > shPsn3 male (32 days) and female (44 days) flies is significantly shorter compared to the respective male control flies [47 days; P < 0.0001, Log-rank (Mantel–Cox) test] and female control flies (56 days; P < 0.0001, Fig. 1c). The shorter lifespan of elav > shPsn3 male flies, compared to female flies, is consistent with the stronger reduction of Psn mRNA in elav > shPsn3 male heads (Fig. 1b). We therefore chose male flies for subsequent phenotypic analysis.
We next examined whether elav > shPsn3 flies develop age-dependent neurodegeneration. There were few vacuoles in elav > shPsn3 and control flies posteclosion, but the number of vacuoles increases in an age-dependent manner in elav > shPsn3 brains (22.3 ± 2.3 at 10 days, 48.7 ± 3.2 at 20 days, 82.2 ± 6.9 at 30 days), compared to controls (F3, 101 = 42.8, P < 0.0001, two-way ANOVA with Bonferroni’s multiple comparisons, Fig. 2a and b; Supplementary Table 3). TUNEL assay followed by quantification of TUNEL + apoptotic cells in all serial sections throughout the entire brain also showed a significant increase of TUNEL + apoptotic cells in the brain of elav > shPsn3 flies (100.6 ± 5.1) at the age of 30 days, compared to controls (22.3 ± 2.4, P < 0.0001, unpaired two-tailed Student's t-test, Fig. 2c and d), further confirming neurodegeneration in elav > shPsn3 brains.
To determine whether age-dependent neurodegeneration observed in elav > shPsn3 flies is due to reduction of Psn expression, we crossed elav > shPsn3 flies with UAS-Psn + 14 flies (Ye and Fortini 1999), which express the full-length wild-type Psn cDNA under the control of the UAS promoter. When flies were cultured at 25°C, both male and female elav > Psn + 14 flies exhibited rough eye phenotypes, whereas only male elav > shPsn3 flies showed rough eyes, which were rescued in elav > shPsn3, Psn + 14 flies, indicating that expression of wild-type Psn in elav > shPsn3 flies rescued developmental defects (Supplementary Fig. 1). RT-qPCR analysis showed that compared to control flies, the level of Psn mRNA is elevated 40- and 10-fold in the head of elav > Psn + 14 and elav > shPsn3, Psn + 14 flies, respectively, indicating that Psn shRNA reduced Psn expression by the Psn + 14 transgene (Fig. 2e). Importantly, we found that the number of vacuoles (46.1 ± 3.6) in the brain of elav > shPsn3, Psn + 14 flies is significantly reduced, compared to elav > shPsn3 (109.0 ± 5.8; P < 0.0001, one-way ANOVA with Tukey's multiple comparisons), and there is no significant difference between elav > shPsn3, Psn + 14 flies (46.1 ± 3.6) and controls (31.9 ± 2.3; P = 0.2023; Fig. 2f and g), indicating that fly brains tolerate a 10-fold overexpression of Psn remarkably well. However, 40-fold overproduction of Psn resulted in increases of vacuoles in elav > Psn + 14 flies (76.5 ± 7.4), compared to control flies (31.9 ± 2.3, P < 0.0001), though the neurodegeneration is not as severe as that in elav > shPsn3 flies (Fig. 2f and g). These results demonstrate that elav > shPsn3 flies coupled with a temperature shift paradigm are suitable for screening genetic modifiers of neurodegeneration caused by Psn KD.
Identification of genetic modifiers of Psn KD-dependent neurodegeneration
To look for candidate genes that interact with Psn in modulating neuronal survival, we rank-ordered genes encoding 641 Drosophila type I transmembrane proteins based on the sequence homology with the transmembrane region of human Notch and APP as well as Drosophila ortholog Notch and APPL, the well-characterized γ-secretase substrates, using the HHpred prediction (Söding et al. 2005) and mammalian ortholog scores by DIOPT (DRSC Integrative Ortholog Prediction Tool; Hu et al. 2011) as well as expression levels in the CNS or the adult head (https://flybase.org/rnaseq/profile_search; Supplementary Fig. 2).
We then chose the top 25 candidates and crossed two independent RNAi lines of each gene (UAS-GeneX-RNAi/+) with Psn KD or control flies to generate Psn/GeneX double KD and Psn KD, or neuron-specific GeneX KD and control flies, respectively (Supplementary Fig. 3). Levels of GeneX mRNA remaining in neuron-specific GeneX KD flies (two independent lines for each gene) were determined by RT-qPCR (Supplementary Table 1). Among the 25 genes, we found that neuron-specific KD of Notch (both RNAi lines), Delta (one RNAi line), or Neurexin IV (one RNAi line), either alone or together with Psn KD, results in lethality during development even though the flies were kept at 18°C to minimize GAL4 activity or RNAi expression (Fig. 3). Furthermore, neuron-specific Delta KD using the weaker RNAi line 2 (−51%, Supplementary Table 1) leads to dramatic increases of vacuoles in Dl/Psn KD flies (Fig. 3). We also found that double KD of Psn and turtle (tutl), thickveins (tkv), or lysosomal enzyme receptor protein (Lerp) using one RNAi line results in earlier adult lethality, whereas double KD flies using another RNAi line are viable but display elevated vacuole numbers, compared to single KD (Fig. 3), which may be related to the essential function of tutl, tkv, and Lerp during development (Terracol and Lengyel 1994; Al-Anzi and Wyman 2009; Mummery-Widmer et al. 2009).
Among the remaining 19 candidate genes, we found that elav-Gal4-driven KD of 4 genes, lipophorin receptor 2 (lpr2), arrow (arr), Mitochondrial E3 ubiquitin protein ligase 1 (Mul1), or Protein tyrosine phosphatase 10D (Ptp10D), together with Psn, using both RNAi lines, enhanced neurodegeneration, compared to Psn KD alone (Fig. 4 and Supplementary Table 1 for detailed data and statistical analysis). Specifically, Psn/lpr2 KD flies using RNAi line 1 showed a dramatic increase of vacuoles (270.0 ± 53.3), compared to Psn KD alone (82.1 ± 3.2, P < 0.0001, one-way ANOVA with Tukey's multiple comparisons). Psn/lpr2 KD flies using RNAi line 2 also showed a significant increase in the number of vacuoles (115.3 ± 11.4, P = 0.0357), compared to Psn KD flies (Fig. 4). Moreover, KD of lipophorin receptor 1 (lpr1), roundabout 1 (robo1), Basigin (Bsg), Megalin (mgl), or Protein tyrosine phosphatase 99A (Ptp99A) using RNAi line 1 also elevated neurodegeneration induced by Psn KD (Fig. 4). Psn/lpr1 KD flies showed an increased number of vacuoles (231.2 ± 8.6) compared to Psn KD flies (82.1 ± 3.2, P < 0.0001). Interestingly, among these 9 genes that enhance Psn KD-induced neurodegeneration, lpr2, lpr1, arr, and mgl are orthologs of mammalian LDLR family (Herz 2001; Wehrli et al. 2001; Van Hoof et al. 2005). Furthermore, KD of Mitochondrial Rho (Miro) and metabotropic glutamate receptors (mGluR) using both RNAi lines, KD glutamate receptor IB (GluRIB) and KD roundabout 2 (robo2) using one of the two RNAi lines tested alleviated neurodegeneration induced by Psn KD (Fig. 4). Thus, in this genetic modifier testing, we found that 9 genes enhanced, and 4 genes reduced Psn KD-induced neurodegeneration in fly brains.
We further asked whether neuron-specific KD of these 19 genes alone induces neurodegeneration in adult brains. Interestingly, neuron-specific KD of arr, lpr1, mgl, or lpr2 also results in increases of neurodegeneration (Fig. 4 and Supplementary Table 1 for detailed data and statistical analysis), suggesting that disruption of lipid transport and metabolism affects neuronal survival in aging fly brains. We also found that neuron-specific KD of robo1 using both RNAi lines leads to increases of vacuoles at the age of 30 days, and KD of lpr2, Ptp10D, mgl, GluRIB, or Miro using RNAi line 1 also causes significant increases of vacuoles (Fig. 4).
Neuron-specific lpr1 or lpr2 KD enhances Psn KD-induced neurodegeneration
Volcano plot analysis shows that among all genes/RNAi lines tested, lpr2 RNAi line 1 shows the most prominent effect on Psn KD-induced neurodegeneration (Supplementary Fig. 4). LpR1 and LpR2 share high homology with human very low-density lipoprotein receptor (VLDLR) and LDLR (DIOPT scores: 9–12) with the highest homology between LpR2 and human VLDLR (DIOPT score 12). To verify the initial finding of elevated vacuole numbers in the brain of Psn/lpr KD flies, we performed a validation study by analyzing another batch of newly generated flies. RT-qPCR analysis of the new batch fly heads confirmed substantial decreases of lpr2 and lpr1 mRNA in neuron-specific lpr2 and lpr1 KD flies, compared to controls (Fig. 5a and Supplementary Fig. 5). Indeed, compared to Psn KD flies (75.3 ± 3.4), lpr2 KD greatly enhances neurodegeneration in Psn KD flies, as shown by a much higher number of vacuoles in Psn/lpr2 KD flies using RNAi line 1, IR1 (inverted repeat 1), and a less severe effect using another RNAi line IR2 (410.2 ± 28.6, P < 0.0001; IR2: 113.2 ± 8.9, P = 0.0002, Fig. 5b and c; Supplementary Fig. 5). The increase in vacuole number is not due to more brain sections from these flies, as there is no significant difference in the number of brain sections used in vacuole quantification among the genotypic groups (Supplementary Table 4). We also found that lpr2 KD using both RNAi lines results in increased vacuole numbers (IR1: 64.5 ± 4.5, P < 0.0001; IR2: 42.7 ± 5.5, P = 0.0011), compared to elav-Gal4 controls (22.7 ± 1.2, Fig. 5b and c; Supplementary Fig. 5). The more severe neurodegeneration in Psn/lpr2-IR1 KD flies correlates with a stronger reduction (−63%) of lpr2 mRNA, whereas the less severe neurodegeneration in Psn/lpr2-IR2 KD flies correlates with a smaller reduction (−45%) of lpr2 mRNA (Supplementary Fig. 5).
We also confirmed that lpr1 KD enhances neurodegeneration in Psn KD flies, as shown by increases of vacuoles in Psn/lpr1-IR1 KD flies (128.0 ± 6.2), compared to Psn KD (75.3 ± 3.4, P < 0.0001). lpr1-IR1 KD alone also results in a significant increase of vacuoles (59.1 ± 2.9), compared to elav-Gal4 controls (22.7 ± 1.2, P < 0.0001; Fig. 5b and c; Supplementary Fig. 5). The effects of lpr1 KD on neurodegeneration is less severe relative to lpr2 KD, despite the more dramatic 85% reduction of lpr1 mRNA in lpr1-IR1 KD flies, compared to the 63 and 45% reduction of lpr2 mRNA in lpr2-IR1 and lpr2-IR2 KD flies, respectively (Supplementary Fig. 5). Moreover, lpr1-IR2 KD reduces mRNA levels (−67%) but has no significant effect on the number of vacuoles (23.5 ± 4.3, P = 0.9835), compared to controls. Furthermore, no enhancement of neurodegeneration was detected in Psn/lpr1-IR2 KD flies (73.9 ± 3.2), compared to Psn KD alone (P = 0.9279, Supplementary Fig. 5). These results show that LpR2 plays a more important role than LpR1 in support of PS-dependent neuronal survival.
We further evaluated apoptosis in the whole-mount brain of lpr and Psn KD flies at the age of 25 days using an antibody specific for active caspase-3 to label apoptotic cells, and quantified active caspase-3 + cells throughout the entire brain. Compared to elav-Gal4 control brains (35.0 ± 3.3), there are more apoptotic cells in Psn KD (65.8 ± 4.3, P = 0.0018, one-way ANOVA with Tukey's multiple comparisons), lpr2 KD (64.0 ± 4.3, P = 0.0045), and lpr1 KD (56.4 ± 3.0, P = 0.0073; Fig. 5d and e). While the number of apoptotic cells in Psn/lpr1 KD flies (79.0 ± 4.1, P = 0.0892) is not significantly different from that in Psn KD flies (65.8 ± 4.3), the number of apoptotic cells is significantly elevated in Psn/lpr2 KD flies (97.2 ± 6.5, P = 0.0009), compared to Psn KD flies (Fig. 5d and e), further supporting a more prominent role of LpR2 than LpR1 in support of PS-dependent neuronal survival.
Adult neuron-specific Psn KD flies show age-dependent impairment in locomotor functions measured by climbing ability. We performed a similar climbing assay to evaluate lpr and Psn/lpr KD flies. Consistent with the previous report (Kang et al. 2017), Psn KD flies at the age of 30 days show significantly increased climbing defects, as measured by the percentage of failed climbs (72.3 ± 2.4%, P < 0.0001, one-way ANOVA with Tukey’s multiple comparisons), compared to controls (19.0 ± 1.9%; Fig. 5f). Interestingly, lpr2 KD (47.9 ± 6.3%, P = 0.0100) and lpr1 KD (42.6 ± 7.3%, P = 0.0215) flies also exhibit climbing defects, compared to controls, but the defects are less severe than that in Psn KD flies. However, lpr KD in Psn KD flies does not exacerbate the impairment of climbing ability of Psn KD flies (Psn/lpr2 Psn KD: 75.56 ± 6.26%, P = 0.9056, Psn/lpr1 KD: 60.69 ± 6.56%, P = 0.3299; Fig. 5f).
Furthermore, both lpr2 and lpr1 KD flies display a shortened lifespan, compared to elav-Gal4 controls [MedLS: controls, 55 days; lpr1 KD: 29 days, P < 0.0001; lpr2 KD: 30 days, P < 0.0001, Log-rank (Mantel–Cox) test; Fig. 5g]. The MedLS of Psn/lpr2 KD (28 days, P = 0.0011) is further significantly decreased, compared to Psn KD flies (31 days). In addition, at the age of day 40, the percentage of surviving Psn/lpr2 KD flies (2.9%) is substantially lower than that of Psn KD flies (12.4%), whereas the percentage of surviving flies is similar between Psn/lpr1 KD (10.0%) and Psn KD flies (12.4%, Supplementary Table 5). Moreover, at day 40, the percentage of surviving lpr2 KD flies (13.1%) is lower than that of lpr1 KD flies (21.2%, Supplementary Table 5). These results suggest that lpr2 plays a more important role than lpr1 in mortality.
Neuron-specific overexpression of LpR does not rescue Psn KD-induced neurodegeneration
To investigate whether neuron-specific overexpression of LpR could alleviate neurodegeneration of Psn KD flies, we crossed UAS-lpr flies with Psn KD flies. There are 6 LpR1 and 5 LpR2 isoforms (Fig. 6a) resulting from two transcriptional initiation sites and alternative splicing of two exons (Parra-Peralbo and Culi 2011). Among those isoforms, we selected UAS-lpr1-J and UAS-lpr2-E transgenic flies, which express the longest isoform with nonconserved N-terminal domain, LDL receptor type A module, EGF, and YWTD β-propeller domains (Fig. 6a). Importantly, previous studies showed that overexpression of lpr1-J or lpr2-E rescued deficits in oogenesis, fertility, and lipid uptake caused by the loss of lpr (Parra-Peralbo and Culi 2011). RT-qPCR analysis showed that compared to control flies, the level of lpr1 and lpr2 mRNA in elav > lpr1-J and elav > lpr2-E flies is ∼3- and ∼2-fold, respectively, compared to controls (Fig. 6b). Histological analysis revealed that overexpression of lpr1-J in neurons does not cause neurodegeneration, compared to controls (P > 0.9999, one-way ANOVA with Tukey's multiple comparisons), nor does it impact Psn KD-induced neurodegeneration (P = 0.9613; Fig. 6c and d). However, elav > lpr2-E flies at the age of 30 days exhibit significantly increased vacuoles (49.4 ± 5.0, P = 0.0168), compared to controls (23.6 ± 3.0). Furthermore, more vacuoles were found in elav > shPsn3; lpr2-E flies (126.3 ± 11.3), compared to elav > shPsn3 (87.8 ± 5.0, P = 0.0007; Fig. 6c and d). The increase in vacuole number is not due to more brain sections from these flies, as there is no significant difference in the number of brain sections used in vacuole quantification among the genotypic groups (Supplementary Table 4). These data further support that lpr2 exerts a more potent role than lpr1 in the regulation of neuronal survival.
Germline deletions of lpr1 and lpr2 enhance neurodegeneration induced by Psn KD
To further verify the genetic interaction between lpr1/2 and Psn in the modulation of neuronal survival, we prepared Psn KD flies in the presence of germline deletions of lpr1 and/or lpr2. LpR1 and LpR2 share high sequence homology with each other (DIOPT score 7), and lpr1 and lrp2 are positioned in tandem on the 3rd right chromosome (Rodríguez-Vázquez et al. 2015). It was reported that flies carrying homozygous deletion mutations of lpr1, lpr2, or lpr1/2 are viable, but lpr1/2 deficient female flies are sterile (Parra-Peralbo and Culi 2011). We first tested whether these lpr1, lpr2, and lpr1/2 deficient flies develop age-dependent neurodegeneration by performing neuropathological analysis at the ages of 1 and 30 days (Fig. 7). At the age of 1 day, only a few vacuoles were detected in the brains of all genotypic groups, and there is no significant difference among them (Fig. 7b and d). At the age of 30 days, compared to control flies (elav-Gal4/+, 37.4 ± 2.8), there is no significant increase of vacuoles in heterozygote deficiency flies of lpr2 (39.0 ± 1.9, P > 0.9999, one-way ANOVA with Tukey’s multiple comparisons) and lpr1 (50.4 ± 3.7, P = 0.3167; Fig. 7c and d). However, heterozygosity of double lpr1/2 deficiency resulted in a moderate increase in the number of vacuoles (61.7 ± 9.5, P = 0.001; Fig. 7c and d). Importantly, we found increases of vacuoles in the brains of all homozygotic deficiency flies: lpr2 (79.4 ± 6.9, P < 0.0001), lpr1 (59.9 ± 3.1, P = 0.0003), and lpr1/2 (74.8 ± 4.6, P < 0.0001), compared to the controls (Fig. 7c and d). The increase in vacuole number is not due to more brain sections from these flies, as there is no significant difference in the number of brain sections used in vacuole quantification among the genotypic groups (Supplementary Table 4). These results show the importance of LpR1 and LpR2 in protection of aging brains.
Furthermore, there is no significant increase in the number of vacuoles in Psn KD together with heterozygotic deficiency of lpr2 (96.5 ± 10.1, P > 0.9999, one-way ANOVA with Tukey’s multiple comparisons) and lpr1 (80.8 ± 3.2, P = 0.8049), compared to Psn KD alone (94.0 ± 6.4, Fig. 7c and d). However, Psn KD in the lpr1/2 heterozygotic deficient flies at the age of 30 days exhibited more severe neurodegeneration (142.1 ± 10.9, P = 0.0008) than Psn KD alone (Fig. 7c and d). We also observed stronger neurodegeneration at the age of 30 days in Psn KD together with lpr2 homozygotic deficiency (134.5 ± 16.0, P = 0.0345), but not with lpr1 (91.4 ± 6.8, P > 0.99; Supplementary Table 4). Lastly, Psn KD; lpr1/2-null flies are not viable at the age of 30 days; they start to die at the age of 15 days with a MedLS of 18 days and a maximum lifespan of 23 days. These data further confirm that LpRs modulate Psn-dependent neuronal survival, and LpR2 is more potent than LpR1 in the interaction.
Discussion
More than 400 distinct mutations have been identified in the PSEN1 and PSEN2 genes that are linked or associated with early-onset AD (https://www.alzforum.org/mutations/psen-1). While it has been demonstrated that PSEN orthologs in mice and Drosophila play an evolutionarily conserved role in protection of neurons during aging, molecular pathways that are involved in PS-dependent neuronal survival remain unclear. To investigate the molecular mechanism and identify genetic modifiers that modulate PS-dependent neuronal survival, we developed a new Drosophila Psn model to facilitate modifier screening (Figs. 1 and 2; Supplementary Fig. 1). Following a bioinformatic analysis (Supplementary Fig. 2), we selected and tested the top ranked 25 candidate genes by knocking down its expression selectively in adult neurons, using two independent RNAi lines of each gene, alone or together with Psn KD (Supplementary Fig. 3). Interestingly, 6 genes appear to be important for neuronal survival, as their KD not only enhances neurodegeneration in Psn KD flies but also causes neurodegeneration when KD alone (Fig. 4 and Supplementary Table 1). Specifically, lpr2, lpr1, arr, and mgl encode proteins that belong to the LDLR family. Further validation tests demonstrated that neuron-specific KD or germline deletions of lpr2 or lpr1 greatly exacerbate neurodegeneration in Psn KD flies, whereas loss of LpR function by RNAi or deficiency also results in age-dependent neurodegeneration, increases of apoptosis, climbing defects, and shortened lifespan (Figs. 5–7). Thus, genes involved in lipid homeostasis, such as LpRs, are critically important for neuronal integrity in the aging brain.
Among the 4 genes that showed a protective effect in alleviating neurodegeneration caused by Psn KD, mGluR may be most interesting, as mGluR KD by both RNAi lines markedly reduced the number of vacuoles in Psn KD flies, whereas the reduction of vacuoles in Psn KD flies by KD of Miro, robo2, GluRIB is quite modest (Fig. 4). mGluR is the only metabotropic glutamate receptor in Drosophila (Bogdanik et al. 2004), and it was reported that the heterozygous mGluR null allele or mGluR antagonists prevented short-term memory deficits in two Psn loss-of-function mutants (McBride et al. 2010). Our results further support that neuron-specific KD of mGluR alleviates phenotypes caused by impaired Psn function.
PS and putative γ-secretase substrates in neurodegeneration
PS is the catalytic subunit of γ-secretase, an intramembrane protease that cleaves type I transmembrane proteins, including Notch and APP (De Strooper et al. 1998; Song et al. 1999; Struhl and Greenwald 1999). Among 25 candidate genes we tested, 9 were reported as γ-secretase substrates (N, fra, Dl, Appl, LRP1, mgl, robo1, Nrx-IV, and arr) and 2 as γ-secretase regulators (Bsg and crb) (Zhou et al. 2005; Herranz et al. 2006; Haapasalo and Kovacs 2011). However, despite Notch and APP being the best-established physiological substrates, previous genetic studies in mice showed that selective inactivation of Notch1 and Notch2, which are expressed in adult brains, or all 3 APP family members in excitatory neurons of the postnatal forebrain did not cause neurodegeneration during mouse lifespan (Zheng et al. 2012; Lee et al. 2020), in contrast to age-dependent, progressive, and widespread loss of cortical neurons in Psen or nicastrin conditional knockout mice using the same Camk2a-Cre line (Saura et al. 2004; Tabuchi et al. 2009; Wines-Samuelson et al. 2010). Similar to the lack of neurodegeneration in triple conditional knockout mice lacking all APP families, KD of Appl alone or together with Psn has no effect on the number of vacuoles (Fig. 4). It was also reported that Fra is cleaved by γ-secretase in Drosophila (Neuhaus-Follini and Bashaw 2015), but fra KD does not appear to modulate Psn-dependent neuronal survival (Fig. 4).
Psen1-deficient mice exhibit perinatal lethality and impaired neurogenesis and Notch signaling, but Psen± mice are phenotypically normal (Shen et al. 1997; Handler et al. 2000). Drosophila loss-of-function Psn mutations induced pupal lethality and Notch-like phenotypes (Struhl and Greenwald 1999; Ye and Fortini 1999), whereas adult neuron-specific KD of Psn circumvented developmental phenotypes but resulted in age-dependent neurodegeneration (Kang et al. 2017). While elav-Gal4-driven Psn KD flies maintained at 18°C during development, in which Psn mRNA was reduced ∼20% in 3rd-instar larval brains, are viable and appear normal, though develop age-dependent neurodegeneration, the same elav-Gal4-driven KD of Notch, Dl, or Nrx-IV results in lethality prior to eclosion (Figs. 2–4), consistent with essential functions of these genes in early development (Mohr 1919; Lehmann et al. 1981; Lehmann et al. 1983; Baumgartner et al. 1996). The less severe phenotypes by RNAi line 2 of Dl or Nrx-IV are likely due to the lesser KD efficiency (∼50% mRNA remaining, Supplementary Table 2). While Nrx-IV KD alone or together with Psn does not affect vacuole numbers, Dl/Psn KD flies exhibit striking increases of vacuoles, compared to Psn or Dl KD alone, raising the possibility that this may be at least in part due to developmental requirement (Fig. 3).
Importantly, 10-fold overexpression of Psn mRNA is well tolerated and does not cause neurodegeneration, whereas 80% reduction of Psn mRNA expression leads to age-dependent neurodegeneration. However, 40-fold overproduction of Psn mRNA results in neurodegeneration, albeit at a less severe degree, compared to 80% reduction of Psn expression (Fig. 2). Thus, the fly brain appears to be more sensitive to the partial loss of Psn than to accumulation of high levels of excess Psn in neurons of the adult brain.
PS and lipid homeostasis in neurodegeneration
Among the 19 genes that were not associated with developmental phenotypes under our experimental paradigm, 4 genes, lpr2, lpr1, arr, and mgl, encode proteins that belong to the LDLR family, and modulate neurodegeneration caused by Psn KD (Fig. 4). The mammalian LDLR family proteins are structurally related single transmembrane receptors, and are involved in the binding and internalization of cholesterol-containing lipoprotein particles (Herz and Bock 2002; Holtzman et al. 2012).
Drosophila lpr1 and lpr2 are partially redundant genes with 5–6 isoforms and are required for the uptake and transport of lipoprotein lipophorin and neutral lipids, though lpr1/lpr2 double deficient flies are viable and do not display significant changes in total neutral lipid content in the gut and fat body (Parra-Peralbo and Culi 2011; Palm et al. 2012; Parvy et al. 2012; Rodríguez-Vázquez et al. 2015). Interestingly, lpr1-null or neuron-specific lpr1 KD flies exhibit a significant reduction in the density of lipid droplets in the larval optic neuropil region, suggesting that lpr1 is involved in the maintenance of lipid content and homeostasis in the brain (Yin et al. 2021). Moreover, fluorescently labeled ApoLpp (apolipophorin) revealed the LpR-dependent lipid uptake in a Drosophila neuronal cell line (Matsuo et al. 2019). We found that neuron-specific KD of lpr2 or lpr1, to a lesser extent, dramatically increases neurodegeneration caused by Psn KD (Figs. 4 and 5). Furthermore, apoptotic cells are significantly elevated in lpr2 and lpr1 KD brains, and lpr2 KD further enhanced apoptosis in Psn KD brains (Fig. 5). Interestingly, overexpression of lpr2-E and lpr1-J, previously shown to rescue deficits in oogenesis, fertility, lipid update of lpr deficiency flies (Parra-Peralbo and Culi 2011), failed to rescue neurodegeneration in Psn KD flies (Fig. 6). Rather, overexpression of lpr2-E (2-fold) but not lpr1-J (3-fold) exacerbated neurodegeneration in Psn KD flies, and overexpression of lpr2-E alone but not lpr1-J resulted in neurodegeneration (Fig. 6), further suggesting a more prominent role of LpR2 than LpR1 in supporting neuronal survival during aging.
Furthermore, germline homozygotic lpr2 deletions or heterozygotic deletions of lpr1 and lpr2 also elevated neurodegeneration in aged Psn KD brains (Fig. 7). Consistently, LpR2 appears to play a more important role, compared to LpR1, in the protection of neuronal integrity, based on the more severe neurodegenerative phenotypes in lpr2 KD or deficient flies, despite the greater reduction of lpr1 mRNA driven by both lpr1 RNAi lines (Figs. 5 and 7). Interestingly, neuron-specific KD or germline deletions of lpr2 or lpr1 also caused age-dependent neurodegeneration, though the neurodegenerative phenotypes are not as severe as those in the presence of Psn KD (Figs. 5 and 7). These results suggest that altered lipid transport and metabolism may compromise neuronal survival in the aging fly brain and exacerbate neurodegeneration caused by compromised Psn function, consistent with earlier reports showing that loss of PS function or γ-secretase activity results in disrupted cellular cholesterol levels and homeostasis of lipoproteins as well as intramembrane cleavages of LDLRs and VLDLRs in culture (Herz and Bock 2002; Grimm et al. 2005; Hoe and Rebeck 2005; Landman et al. 2006; Nguyen et al. 2006; Liu et al. 2007; Tamboli et al. 2008; Cho et al. 2019).
Implications to AD and other neurodegenerative diseases
The relevance of altered lipid transport and metabolism in neurodegenerative diseases is highlighted by the strong association between apolipoprotein E4 (apoE4) alleles and increased risks of late-onset AD and other neurodegenerative diseases, such as Parkinson’s and dementia with Lewy bodies (Yamazaki et al. 2019). ApoE is a plasma lipoprotein and a ligand of LDLRs, and its binding with LDLRs regulates the transport and metabolism of cholesterol and other lipids (Mahley 1988). ApoE2, E3, and E4 differ in amino acid residues 112 and 158, and exhibit different affinities in binding to LDLRs, and while ApoE2 is associated with a reduced risk of AD, ApoE4 is associated with an increased risk in a dose-dependent manner (Corder et al. 1993; Strittmatter et al. 1993). Compared to other isoforms, ApoE4 is thought to have a loss of function effect on lipid transport (Yamazaki et al. 2019). Recent studies in Drosophila also showed that compared to ApoE2 and ApoE3, ApoE4 exhibits impaired ability to restore lipid transport (Liu et al. 2017).
Interestingly, genetic association between PSEN1 and apoE has been reported in a single patient carrying the PSEN1 E280A mutation and two copies of the apoE3 Christchurch variant (R136S) (Arboleda-Velasquez et al. 2019). The apoE3 R136S was thought to be protective, as the patient did not develop dementia until age 75, whereas the median age of onset for dementia among a large number of PSEN1 E280A carriers was 49 (Acosta-Baena et al. 2011). Our current study demonstrated that disruption of lpr expression by decreasing or increasing gene dosage results in neuronal death and exacerbates neurodegeneration caused by reduced Psn expression in the aging brain, providing a causal relationship between lipid homeostasis and neurodegeneration in AD. Future studies are needed to elucidate the mechanism by which altered lipid transport and metabolism may modulate PS function and neuronal survival in the aging brain.
Supplementary Material
Acknowledgments
We thank M. Feany for her advice and helpful discussions. Stocks obtained from Bloomington Drosophila Stock Center (NIH P40OD018537), Vienna Drosophila Resource Center, and National Institute of Genetics Fly Stock were used in this study. We thank www.flybase.org for providing databases. We are grateful for the technical support provided by the staff at the Drosophila RNAi Screening Center, R. Binari, C. Hu, and members of the Shen and Perrimon labs for discussion.
Contributor Information
Jongkyun Kang, Department of Neurology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Chen Zhang, Department of Neurology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Yuhao Wang, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Jian Peng, Department of Computer Science, University of Illinois at Urbana-Champaign, Champaign, IL 61801, USA.
Bonnie Berger, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Mathematics, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Norbert Perrimon, Department of Genetics, Harvard Medical School, Boston, MA 02115, USA; Howard Hughes Medical Institute, Boston, MA 02115, USA.
Jie Shen, Department of Neurology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA; Program in Neuroscience, Harvard Medical School, Boston, MA 02115, USA.
Data availability
The authors affirm that all data necessary for confirming the conclusion of the article are present within the article and its online Supplementary material.
Supplementary material is available at GENETICS online.
Funding
This work was supported by grants from the National Institutes of Health (R01NS101745 to J.S. and N.P., 1R35GM141861 to B.B.).
Author contributions
J.K., C.Z., N.P., and J.S. designed the experiments. J.K. and C.Z. performed molecular and histological analysis. Y.W, J.P., and B.B. performed bioinformatic analysis. J.K., C.Z., and J.S. wrote the paper.
Literature cited
- Acosta-Baena N, Sepulveda-Falla D, Lopera-Gómez CM, Jaramillo-Elorza MC, Moreno S, Aguirre-Acevedo DC, Saldarriaga A, Lopera F. 2011. Pre-dementia clinical stages in presenilin 1 e280a familial early-onset Alzheimer’s disease: a retrospective cohort study. Lancet Neurol. 10(3):213–220. doi: 10.1016/S1474-4422(10)70323-9. [DOI] [PubMed] [Google Scholar]
- Al-Anzi B, Wyman RJ. 2009. The Drosophila immunoglobulin gene turtle encodes guidance molecules involved in axon pathfinding. Neural Dev. 4(1):31. doi: 10.1186/1749-8104-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleda-Velasquez JF, Lopera F, O’Hare M, Delgado-Tirado S, Marino C, Chmielewska N, Saez-Torres KL, Amarnani D, Schultz AP, Sperling RA, et al. 2019. Resistance to autosomal dominant Alzheimer’s disease in an apoe3 Christchurch homozygote: a case report. Nat Med. 25(11):1680–1683. doi: 10.1038/s41591-019-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacardit J, Stout M, Hirst JD, Valencia A, Smith RE, Krasnogor N. 2009. Automated alphabet reduction for protein datasets. BMC Bioinformatics. 10(1):6. doi: 10.1186/1471-2105-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner S, Littleton JT, Broadie K, Bhat MA, Harbecke R, Lengyel JA, Chiquet-Ehrismann R, Prokop A, Bellen HJ. 1996. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 87(6):1059–1068. doi: 10.1016/S0092-8674(00)81800-0. [DOI] [PubMed] [Google Scholar]
- Bogdanik L, Mohrmann R, Ramaekers A, Bockaert J, Grau Y, Broadie K, Parmentier ML. 2004. The Drosophila metabotropic glutamate receptor dmglura regulates activity-dependent synaptic facilitation and fine synaptic morphology. J Neurosci. 24(41):9105–9116. doi: 10.1523/JNEUROSCI.2724-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YY, Kwon OH, Park MK, Kim TW, Chung S. 2019. Elevated cellular cholesterol in familial Alzheimer's presenilin 1 mutation is associated with lipid raft localization of β-amyloid precursor protein. PLoS One. 14(1):e0210535. doi: 10.1371/journal.pone.0210535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. 1993. Gene dose of apolipoprotein e type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. 1998. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 391(6665):387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- Dias-Santagata D, Fulga TA, Duttaroy A, Feany MB. 2007. Oxidative stress mediates tau-induced neurodegeneration in Drosophila. J Clin Invest. 117(1):236–245. doi: 10.1172/JCI28769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB. 2002. Gal4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 34(1–2):1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 1996. Hidden Markov models. Curr Opin Struct Biol. 6(3):361–365. doi: 10.1016/S0959-440X(96)80056-X. [DOI] [PubMed] [Google Scholar]
- Grimm MO, Grimm HS, Pätzold AJ, Zinser EG, Halonen R, Duering M, Tschäpe JA, De Strooper B, Müller U, Shen J, et al. 2005. Regulation of cholesterol and sphingomyelin metabolism by amyloid-beta and presenilin. Nat Cell Biol. 7(11):1118–1123. doi: 10.1038/ncb1313. [DOI] [PubMed] [Google Scholar]
- Haapasalo A, Kovacs DM. 2011. The many substrates of presenilin/γ-secretase. J Alzheimers Dis. 25(1):3–28. doi: 10.3233/JAD-2011-101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler M, Yang X, Shent J. 2000. Presenilin-1 regulates neuronal differentiation during neurogenesis. Development. 127(12):2593–2606. doi: 10.1242/dev.127.12.2593. [DOI] [PubMed] [Google Scholar]
- Heilig EA, Gutti U, Tai T, Shen J, Kelleher RJ III. 2013. Trans-dominant negative effects of pathogenic psen1 mutations on γ-secretase activity and aβ production. J Neurosci. 33(28):11606–11617. doi: 10.1523/JNEUROSCI.0954-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig EA, Xia W, Shen J, Kelleher RJ III. 2010. A presenilin-1 mutation identified in familial Alzheimer disease with cotton wool plaques causes a nearly complete loss of gamma-secretase activity. J Biol Chem. 285(29):22350–22359. doi: 10.1074/jbc.M110.116962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H, Stamataki E, Feiguin F, Milán M. 2006. Self-refinement of notch activity through the transmembrane protein crumbs: modulation of gamma-secretase activity. EMBO Rep. 7(3):297–302. doi: 10.1038/sj.embor.7400617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J. 2001. The LDL receptor gene family: (un)expected signal transducers in the brain. Neuron. 29(3):571–581. doi: 10.1016/S0896-6273(01)00234-3. [DOI] [PubMed] [Google Scholar]
- Herz J, Bock HH. 2002. Lipoprotein receptors in the nervous system. Annu Rev Biochem. 71(1):405–434. doi: 10.1146/annurev.biochem.71.110601.135342. [DOI] [PubMed] [Google Scholar]
- Hoe HS, Rebeck GW. 2005. Regulation of apoe receptor proteolysis by ligand binding. Brain Res Mol Brain Res. 137(1–2):31–39. doi: 10.1016/j.molbrainres.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Herz J, Bu G. 2012. Apolipoprotein e and apolipoprotein e receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med. 2(3):a006312. doi: 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. 2011. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 12(1):357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Shen J. 2020. Cell-autonomous role of presenilin in age-dependent survival of cortical interneurons. Mol Neurodegener. 15(1):72. doi: 10.1186/s13024-020-00419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Shin S, Perrimon N, Shen J. 2017. An evolutionarily conserved role of presenilin in neuronal protection in the aging Drosophila brain. Genetics. 206(3):1479–1493. doi: 10.1534/genetics.116.196881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. 1958. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 53(282):457–481. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. Mafft: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Shen J. 2008. Presenilins are required for maintenance of neural stem cells in the developing brain. Mol Neurodegener. 3(1):2. doi: 10.1186/1750-1326-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman N, Jeong SY, Shin SY, Voronov SV, Serban G, Kang MS, Park MK, Di Paolo G, Chung S, Kim TW. 2006. Presenilin mutations linked to familial Alzheimer’s disease cause an imbalance in phosphatidylinositol 4,5-bisphosphate metabolism. Proc Natl Acad Sci U S A. 103(51):19524–19529. doi: 10.1073/pnas.0604954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Bolshakov VY, Shen J. 2021. Inactivation of presenilin in inhibitory neurons results in decreased gabaergic responses and enhanced synaptic plasticity. Mol Brain. 14(1):85. doi: 10.1186/s13041-021-00796-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Bolshakov VY, Shen J. 2023. Presenilins regulate synaptic plasticity in the perforant pathways of the hippocampus. Mol Brain. 16(1):17. doi: 10.1186/s13041-023-01009-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kang J, Ho A, Watanabe H, Bolshakov VY, Shen J. 2020. APP family regulates neuronal excitability and synaptic plasticity but not neuronal survival. Neuron. 108(4):676–690.e8. doi: 10.1016/j.neuron.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Lutz D, Mossalam M, Bolshakov VY, Frotscher M, Shen J. 2017. Presenilins regulate synaptic plasticity and mitochondrial calcium homeostasis in the hippocampal mossy fiber pathway. Mol Neurodegener. 12(1):48. doi: 10.1186/s13024-017-0189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Dietrich U, Jiménez F, Campos-Ortega JA. 1981. Mutations of early neurogenesis indrosophila. Wilehm Roux Arch Dev Biol. 190(4):226–229. doi: 10.1007/BF00848307. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Jiménez F, Dietrich U, Campos-Ortega JA. 1983. On the phenotype and development of mutants of early neurogenesis in Drosophila melanogaster. Wilehm Roux Arch Dev Biol. 192(2):62–74. doi: 10.1007/BF00848482. [DOI] [PubMed] [Google Scholar]
- Liu CX, Ranganathan S, Robinson S, Strickland DK. 2007. Gamma-secretase-mediated release of the low density lipoprotein receptor-related protein 1b intracellular domain suppresses anchorage-independent growth of neuroglioma cells. J Biol Chem. 282(10):7504–7511. doi: 10.1074/jbc.M608088200. [DOI] [PubMed] [Google Scholar]
- Liu L, MacKenzie KR, Putluri N, Maleti-Savati M, Bellen HJ. 2017. The glia-neuron lactate shuttle and elevated ros promote lipid synthesis in neurons and lipid droplet accumulation in glia via apoe/d. Cell Metab. 26(5):719–737.e6. doi: 10.1016/j.cmet.2017.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW. 1988. Apolipoprotein e: cholesterol transport protein with expanding role in cell biology. Science. 240(4852):622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Mahoney MB, Parks AL, Ruddy DA, Tiong SY, Esengil H, Phan AC, Philandrinos P, Winter CG, Chatterjee R, Huppert K, et al. 2006. Presenilin-based genetic screens in Drosophila melanogaster identify novel notch pathway modifiers. Genetics. 172(4):2309–2324. doi: 10.1534/genetics.104.035170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N, Nagao K, Suito T, Juni N, Kato U, Hara Y, Umeda M. 2019. Different mechanisms for selective transport of fatty acids using a single class of lipoprotein in Drosophila. J Lipid Res. 60(7):1199–1211. doi: 10.1194/jlr.M090779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Choi CH, Schoenfeld BP, Bell AJ, Liebelt DA, Ferreiro D, Choi RJ, Hinchey P, Kollaros M, Terlizzi AM, et al. 2010. Pharmacological and genetic reversal of age-dependent cognitive deficits attributable to decreased presenilin function. J Neurosci. 30(28):9510–9522. doi: 10.1523/JNEUROSCI.1017-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr OL. 1919. Character changes caused by mutation of an entire region of a chromosome in Drosophila. Genetics. 4(3):275–282. doi: 10.1093/genetics/4.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA. 2009. Genome-wide analysis of notch signalling in Drosophila by transgenic RNAi. Nature. 458(7241):987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus-Follini A, Bashaw GJ. 2015. The intracellular domain of the frazzled/dcc receptor is a transcription factor required for commissural axon guidance. Neuron. 87(4):751–763. doi: 10.1016/j.neuron.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HN, Son DJ, Lee JW, Hwang DY, Kim YK, Cho JS, Lee US, Yoo HS, Moon DC, Oh KW, et al. 2006. Mutant presenilin 2 causes abnormality in the brain lipid profile in the development of Alzheimer’s disease. Arch Pharm Res. 29(10):884–889. doi: 10.1007/BF02973910. [DOI] [PubMed] [Google Scholar]
- Palm W, Sampaio JL, Brankatschk M, Carvalho M, Mahmoud A, Shevchenko A, Eaton S. 2012. Lipoproteins in Drosophila melanogaster—assembly, function, and influence on tissue lipid composition. PLoS Genet. 8(7):e1002828. doi: 10.1371/journal.pgen.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Peralbo E, Culi J. 2011. Drosophila lipophorin receptors mediate the uptake of neutral lipids in oocytes and imaginal disc cells by an endocytosis-independent mechanism. PLoS Genet. 7(2):e1001297. doi: 10.1371/journal.pgen.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvy JP, Napal L, Rubin T, Poidevin M, Perrin L, Wicker-Thomas C, Montagne J. 2012. Drosophila melanogaster acetyl-coa-carboxylase sustains a fatty acid-dependent remote signal to waterproof the respiratory system. PLoS Genet. 8(8):e1002925. doi: 10.1371/journal.pgen.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodenizer D, Martin I, Bhandari P, Pletcher SD, Grotewiel M. 2008. Genetic and environmental factors impact age-related impairment of negative geotaxis in Drosophila by altering age-dependent climbing speed. Exp Gerontol. 43(8):739–748. doi: 10.1016/j.exger.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Vázquez M, Vaquero D, Parra-Peralbo E, Mejía-Morales JE, Culi J. 2015. Drosophila lipophorin receptors recruit the lipoprotein ltp to the plasma membrane to mediate lipid uptake. PLoS Genet. 11(6):e1005356. doi: 10.1371/journal.pgen.1005356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, Chattarji S, Kelleher RJ III, Kandel ER, Duff K, et al. 2004. Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron. 42(1):23–36. doi: 10.1016/S0896-6273(04)00182-5. [DOI] [PubMed] [Google Scholar]
- Shen J, Bronson RT, Chen DF, Xia W, Selkoe DJ, Tonegawa S. 1997. Skeletal and cns defects in presenilin-1-deficient mice. Cell. 89(4):629–639. doi: 10.1016/S0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. 2005. The hhpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33(Web Server):W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Nadeau P, Yuan M, Yang X, Shen J, Yankner BA. 1999. Proteolytic release and nuclear translocation of notch-1 are induced by presenilin-1 and impaired by pathogenic presenilin-1 mutations. Proc Natl Acad Sci U S A. 96(12):6959–6963. doi: 10.1073/pnas.96.12.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 6:175–182. https://sonnhammer.sbc.su.se/download/papers/1998_ISMB_6:175-182.pdf. [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. 1993. Apolipoprotein e: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Greenwald I. 1999. Presenilin is required for activity and nuclear access of notch in Drosophila. Nature. 398(6727):522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- Sun L, Zhou R, Yang G, Shi Y. 2017. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of aβ42 and aβ40 peptides by γ-secretase. Proc Natl Acad Sci U S A. 114(4):E476–E485. doi: 10.1073/pnas.1618657114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Chen G, Südhof TC, Shen J. 2009. Conditional forebrain inactivation of nicastrin causes progressive memory impairment and age-related neurodegeneration. J Neurosci. 29(22):7290–7301. doi: 10.1523/JNEUROSCI.1320-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamboli IY, Prager K, Thal DR, Thelen KM, Dewachter I, Pietrzik CU, St George-Hyslop P, Sisodia SS, De Strooper B, Heneka MT, et al. 2008. Loss of gamma-secretase function impairs endocytosis of lipoprotein particles and membrane cholesterol homeostasis. J Neurosci. 28(46):12097–12106. doi: 10.1523/JNEUROSCI.2635-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracol R, Lengyel JA. 1994. The thick veins gene of Drosophila is required for dorsoventral polarity of the embryo. Genetics. 138(1):165–178. doi: 10.1093/genetics/138.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D, Rodenburg KW, Van der Horst DJ. 2005. Receptor-mediated endocytosis and intracellular trafficking of lipoproteins and transferrin in insect cells. Insect Biochem Mol Biol. 35(2):117–128. doi: 10.1016/j.ibmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Wehrli M, Dougan ST, Caldwell K, O’Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. 2001. Correction: arrow encodes an LDL-receptor-related protein essential for wingless signalling. Nature. 410(6830):847. doi: 10.1038/35071131. [DOI] [PubMed] [Google Scholar]
- Wines-Samuelson M, Schulte EC, Smith MJ, Aoki C, Liu X, Kelleher RJ III, Shen J. 2010. Characterization of age-dependent and progressive cortical neuronal degeneration in presenilin conditional mutant mice. PLoS One. 5(4):e10195. doi: 10.1371/journal.pone.0010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PC, Zheng H, Chen H, Becher MW, Sirinathsinghji DJ, Trumbauer ME, Chen HY, Price DL, Van der Ploeg LH, Sisodia SS. 1997. Presenilin 1 is required for notch1 and dii1 expression in the paraxial mesoderm. Nature. 387(6630):288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- Woodruff G, Young JE, Martinez FJ, Buen F, Gore A, Kinaga J, Li Z, Yuan SH, Zhang K, Goldstein LS. 2013. The presenilin-1 δe9 mutation results in reduced γ-secretase activity, but not total loss of ps1 function, in isogenic human stem cells. Cell Rep. 5(4):974–985. doi: 10.1016/j.celrep.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Yamaguchi H, Lai FA, Shen J. 2013. Presenilins regulate calcium homeostasis and presynaptic function via ryanodine receptors in hippocampal neurons. Proc Natl Acad Sci U S A. 110(37):15091–15096. doi: 10.1073/pnas.1304171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Kelleher RJ III, Shen J. 2016. Loss of aβ43 production caused by presenilin-1 mutations in the knockin mouse brain. Neuron. 90(2):417–422. doi: 10.1016/j.neuron.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Watanabe H, Wu B, Lee SH, Li Y, Tsvetkov E, Bolshakov VY, Shen J, Kelleher RJ III. 2015. Presenilin-1 knockin mice reveal loss-of-function mechanism for familial Alzheimer’s disease. Neuron. 85(5):967–981. doi: 10.1016/j.neuron.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Zhao N, Caulfield TR, Liu CC, Bu G. 2019. Apolipoprotein e and Alzheimer disease: pathobiology and targeting strategies. Nat Rev Neurol. 15(9):501–518. doi: 10.1038/s41582-019-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Klein R, Tian X, Cheng HT, Kopan R, Shen J. 2004. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev Biol. 269(1):81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Ye Y, Fortini ME. 1999. Apoptotic activities of wild-type and Alzheimer’s disease-related mutant presenilins in Drosophila melanogaster. J Cell Biol. 146(6):1351–1364. doi: 10.1083/jcb.146.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Lukinova N, Fortini ME. 1999. Neurogenic phenotypes and altered notch processing in Drosophila presenilin mutants. Nature. 398(6727):525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- Yin J, Spillman E, Cheng ES, Short J, Chen Y, Lei J, Gibbs M, Rosenthal JS, Sheng C, Chen YX, et al. 2021. Brain-specific lipoprotein receptors interact with astrocyte derived apolipoprotein and mediate neuron-glia lipid shuttling. Nat Commun. 12(1):2408. doi: 10.1038/s41467-021-22751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Saura CA, Choi SY, Sun LD, Yang X, Handler M, Kawarabayashi T, Younkin L, Fedeles B, Wilson MA, et al. 2001. APP processing and synaptic plasticity in presenilin-1 conditional knockout mice. Neuron. 31(5):713–726. doi: 10.1016/S0896-6273(01)00417-2. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wu B, Beglopoulos V, Wines-Samuelson M, Zhang D, Dragatsis I, Südhof TC, Shen J. 2009. Presenilins are essential for regulating neurotransmitter release. Nature. 460(7255):632–636. doi: 10.1038/nature08177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhang C, Ho A, Kirkwood A, Südhof TC, Shen J. 2010. Inactivation of presenilins causes pre-synaptic impairment prior to post-synaptic dysfunction. J Neurochem. 115(5):1215–1221. doi: 10.1111/j.1471-4159.2010.07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Watanabe H, Wines-Samuelson M, Zhao H, Gridley T, Kopan R, Shen J. 2012. Conditional deletion of notch1 and notch2 genes in excitatory neurons of postnatal forebrain does not cause neurodegeneration or reduction of notch mRNAs and proteins. J Biol Chem. 287(24):20356–20368. doi: 10.1074/jbc.M112.349738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yang G, Shi Y. 2017. Dominant negative effect of the loss-of-function γ-secretase mutants on the wild-type enzyme through heterooligomerization. Proc Natl Acad Sci U S A. 114(48):12731–12736. doi: 10.1073/pnas.1713605114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Zhou H, Walian PJ, Jap BK. 2005. CD147 is a regulatory subunit of the gamma-secretase complex in Alzheimer’s disease amyloid beta-peptide production. Proc Natl Acad Sci U S A. 102(21):7499–7504. doi: 10.1073/pnas.0502768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusion of the article are present within the article and its online Supplementary material.
Supplementary material is available at GENETICS online.