Abstract
Loss of heterozygosity (LOH) can occur when a heterozygous mutant cell loses the remaining wild-type allele to become a homozygous mutant. LOH can have physiological consequences if, for example, the affected gene encodes a tumor suppressor. We used fluorescent reporters to study the mechanisms of LOH induction by X-rays, a type of ionizing radiation (IR), in Drosophila melanogaster larval wing discs. IR is used to treat more than half of patients with cancer, so understanding its effects is of biomedical relevance. Quantitative analysis of IR-induced LOH at different positions between the telomere and the centromere on the X chromosome showed a strong sex dependence and the need for a recombination-proficient homologous chromosome, whereas, paradoxically, position along the chromosome made little difference in LOH incidence. We propose that published data documenting high recombination frequency within centromeric heterochromatin on the X chromosome can explain these data. Using a focused screen, we identified E2F1 as a key promotor of LOH and further testing suggests a mechanism involving its role in cell-cycle regulation. We leveraged the loss of a transcriptional repressor through LOH to express transgenes specifically in cells that have already acquired LOH. This approach identified JNK signaling and apoptosis as key determinants of LOH maintenance. These studies reveal previously unknown mechanisms for the generation and elimination of cells with chromosome aberrations after exposure to IR.
Keywords: Drosophila, radiation, loss of heterozygosity
Introduction
Loss of heterozygosity (LOH) occurs when a heterozygous cell loses an allele to become homozygous. LOH can have physiological consequences if, for example, the lost allele encodes a tumor suppressor. Possible mechanisms for LOH include mutations in the gene, aneuploidy of whole or segments of chromosomes, and mitotic recombination between the locus and the centromere (producing −/− and +/+ twin spots from a −/+ cell). We are using Drosophila melanogaster as a model to study LOH that follows exposure to X-rays, a type of ionizing radiation (IR). IR causes DNA strand breaks and its ability to induce apoptosis is the reason IR is used to treat half of patients with cancer. Understanding IR's effects on genome integrity and underlying mechanisms may help us mitigate this side effect of radiation therapy to make it safer.
Classical and recent studies of IR-induced LOH in Drosophila employed visible adult markers such as multiple wing hair (mwh) and forked (f) that alter the number or morphology of actin-based hairs on the cell surface (Baker et al. 1978). Starting with heterozygotes of recessive alleles, induced homozygosity is detectable on a cell-by-cell basis. In these studies, exposure to IR occurred in the larval stages while the resulting LOH was scored many days later in the adult. To be able to monitor steps in between, we have been developing fluorescent LOH reporters that can be used at all developmental stages.
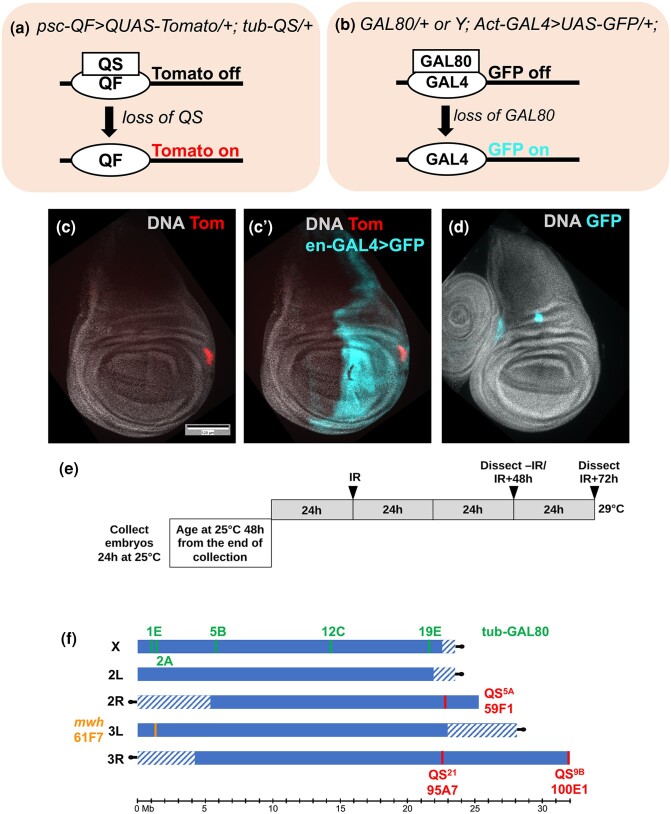
In our published work (Brown et al. 2020), we used the QF/QS module wherein QF binds its recognition sequence QUAS to promote transcription while QS represses QF (Riabinina et al. 2015). psc-QF drives the expression of QUAS-tdTomato (Tom) throughout the larval wing disc and in other tissues (Fig. 1a) unless repressed by ubiquitously expressed QS. Loss of QS by LOH results in de-repression of QF and cell-autonomous Tom expression (Fig. 1c and c’). Our published study produced a surprising result; IR-induced mwh LOH was lower by more than 10-fold compared with IR-induced QS9B LOH detected under identical experimental conditions in adult wings (Brown et al. 2020). Classical studies suggest that segmental aneuploidy (loss of a chromosome segment) is the primary mechanism for IR-induced LOH (Baker et al. 1978). If breakage and loss of the corresponding chromosome tip are indeed the mechanisms, mwh LOH would accompany the loss of more genes compared with QS9B LOH because mwh is further from the telomere than QS9B (Fig. 1f). Resulting mwh LOH cells may be less fit than QS9B LOH cells, explaining the difference in IR-induced LOH incidence we saw. Alternatively, homozygous mutants for mwh, which encodes an actin-bundling protein, may be less fit while the loss of QS, which has no known role in Drosophila, may be neutral and it is this difference in gene function rather than the amount of chromosome lost that explains the differences in IR-induced LOH incidence. To address these possibilities and to identify regulators of IR-induced LOH, we studied LOH induction at 7 additional loci (Fig. 1f) and performed a focused genetic screen.
Fig. 1.
Fluorescent reporters for loss of heterozygosity. a) Loss of QS results in de-repression of psc-QF-driven Tom expression, reproduced from Brown et al. 2020. b) Loss of GAL80 results in de-repression of act-GAL4-driven GFP expression. c and c′) An example of a wing disc with a Tom clone. This disc also expresses en-GAL4 > GFP that was used to express RNAi and transgenes in the posterior compartment of the disc. The genotype was en-GAL4 > UAS-GFP, psc-QF > Tom, QS5A/+ from the cross of en-GAL4 > UAS-GFP, psc-QF > Tom, QS5A/CyO-GFP with w1118. Larvae were treated as in panel E but without the temperature shift. d) An example of a wing disc with GFP clones. The genotype was GAL805B/+; Act-GAL4, UAS-GFP/+ from the cross of GAL805B/GAL805B with Act-GAL4, UAS-GFP/CyO RFP Tb. Larvae were treated as in panel E but without the temperature shift. e) The experimental protocol used to express RNAi and transgenes conditionally on the focused screen. Placing the larvae in a 29°C incubator inactivated GAL80ts and initiated UAS-transgene expression. For experiments that do not involve transgene expression, the same protocol was followed but larvae were maintained at 25°C without a temperature shift. f) Chromosomal location of QS5A, QS9B, QS21, mwh, and GAL80 studied here. Chromosome lengths and the boundaries of centromeric heterochromatin (hatched regions) are adapted from Fig. 2 of Hoskins et al. (2015). The locations of mwh and insertion loci were computed based on the Gene Map Table on Flybase (FB2023_05, released 2023 September 26, gene_map_table_fb_2023_05.tsv.gz). Scale bar = 120 microns in c and d.
Materials and methods
Drosophila stocks and methods
Drosophila stocks used are in Supplementary Table 1 and include those with UAS-puc (Martín-Blanco et al. 1998), UAS-Wg (Lawrence et al. 1996), and recombinant chromosome 3 encoding tub-QS9B and tub-GAL80ts (Brown et al. 2020). QUAS lines are a gift from Perez-Garijo et al. (2013). Virgin females and males were crossed and cultured for 3 d on Nutri-Fly German Formula food (Genesee Scientific) at 25°C before egg collections. Eggs were collected and larvae were raised on Nutri-Fly Bloomington Formula food (Genesee Scientific) at 25°C unless otherwise noted. The cultures were monitored daily for signs of crowding, typically seen as “dimples” on the food surface, and were split as needed. Larvae in food were placed in petri dishes and irradiated in a Faxitron Cabinet X-ray System Model RX-650 (Lincolnshire, IL, USA) at 115 kVp and 5.33 rad/s. Larval sex was determined using gonad size observed under a dissecting microscope; male gonads are large and free of the fat body while female gonads are hard to find (Kerkis 1931).
To induce neutral clones, embryos were collected at 25°C for 24 h and aged for 3 d from the end of collection. Larvae were heat shocked in a 37°C bath with Lab Armor beads (Thermo Fisher) for 30 min, returned to 25°C, and dissected 3 d after heat shock.
Tissue preparation and imaging
Larval wing discs were dissected in PBS, fixed in 4% paraformaldehyde in PBS for 30 min, and washed 3 times with PBTx (0.1% Triton X-100). The discs were stained with 10 μg/ml Hoechst33342 in PBTx for 5 min, washed 3 times, and mounted on glass slides in Fluoromount G (Southern Biotech).
For Ci staining, fixed wing discs were incubated with 1:300 Rat anti-Ci (2A1 concentrate, Developmental Studies Hybridoma Bank) overnight at 4°C in block (PBTx + 5% normal goat serum), rinsed 3 times in PBTx and, and stained with 1:400 anti-rat TRITC secondary antibody (Jackson) in block for 2 h at room temperature. For Dcp1 staining, the same procedure was followed but with 1:100 rabbit anti-cleaved Dcp1 (Cell Signaling #9578) and 1:500 anti-rabbit secondary antibody (Jackson). The discs were rinsed 3 times in PBTx and mounted as described above.
Wing discs were imaged on a Leica DMR compound microscope using a Q-Imaging R6 CCD camera and Ocular or Micro-Manager software. Images were quantified in FiJi/Image J, focusing on the columnar layer of the wing disc proper; clones in the peripodial layer were excluded from analysis.
Statistical analysis
For sample size justifications, we used a simplified resource equation from Charan and Kantharia (2013): E = total number of animals − total number of groups, where E-value of 10–20 is considered adequate. When we compare 2 groups (−IR and +IR, for example, where the former is the control group and the latter is the treated group), n = 6 per group, or E = 11, would be adequate. All samples subjected to statistical analysis meet or exceed this criterion. To compute P-values, 2-tailed Student t-tests were used.
Results
IR-induced LOH shows sex dependence but locus independence
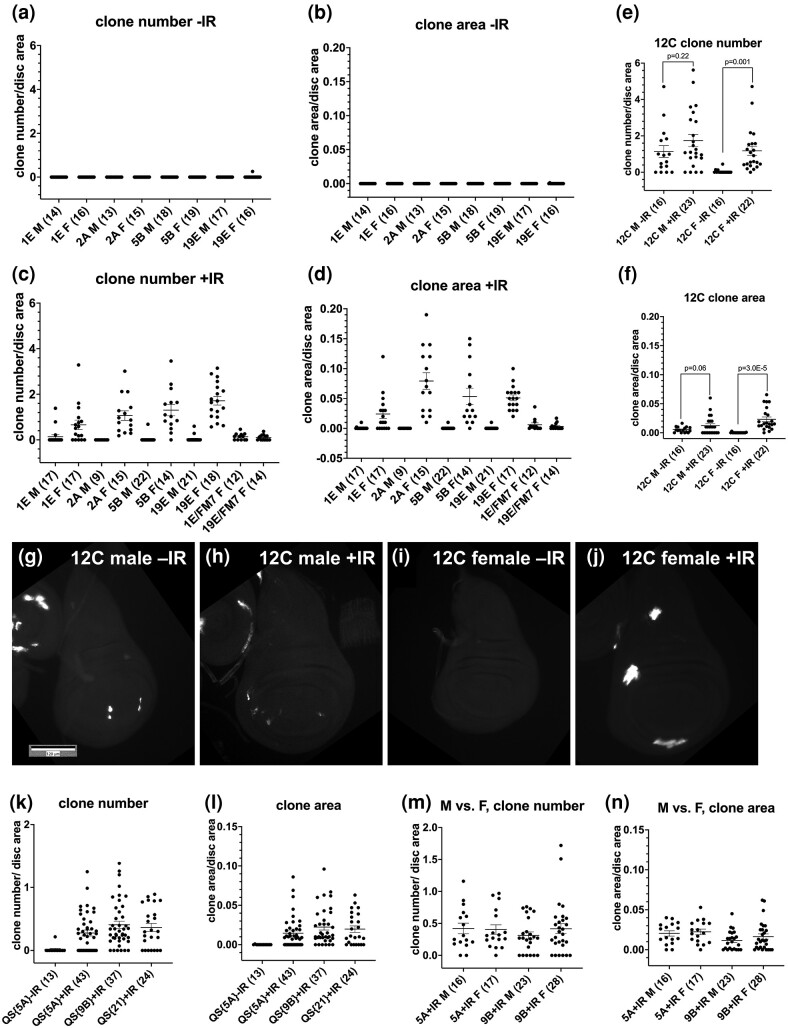
We used 4,000 Rads (R) of X-rays, a typical dose in Drosophila (for example, Brodsky et al. 2004; Wells and Johnston 2012) because it kills more than half of the cells in wing discs, yet larvae can still develop into viable adults. Thus, 4,000R causes significant damage but is still compatible with recovery and regeneration. To ask if the distance from the chromosome tip matters, we used 5 GAL80 insertions along the X chromosome which were used before to detect spontaneous (without IR) LOH (Fig. 1, b, d, and f; Siudeja et al. 2015). For 4 of the 5 GAL80 insertions, wing discs from un-irradiated larvae show very few cells with GFP signal that is as bright as what is seen after IR (Fig. 2a and b, Supplementary Fig. 1). In feeding 3rd instar larval stage, cells of the wing disc that will ultimately form the adult structures are in a single layer columnar epithelium with uniform cell size. Therefore, clone area serves as a proxy for cell number as in previous studies (Brown et al. 2020). We normalize clone number and area to the total disc area so that we can control for variations in disc size and total cell number. Occasional cells with faint GFP seen in unirradiated discs (arrow in Supplementary Fig. 1a–a″) are consistent with a recent report of spontaneous GAL80 “loss” in larval brains through poorly understood mechanisms (Goupil et al. 2022). Importantly, we noticed in early experiments with GAL80 that only about half of the irradiated discs had GFP expressing clones. GAL80 insertions are on the X chromosome, which led us to hypothesize a sex dependence in GAL80 LOH, like what was reported for spontaneous LOH of X-linked loci (Siudeja et al. 2015). Analysis of wing discs from males and female larvae (see Materials and Methods) shows that IR-induced increases in GFP+ cells were confined almost exclusively to females (Fig. 2c and d).
Fig. 2.
IR-induced LOH shows sex dependence for the X chromosome but locus independence. Larvae were treated as in Fig. 1e but without the temperature shift and wing discs were fixed and stained for DNA. In panels C–N, male and female larvae were separated before dissection and processed separately. The graphs show data from at least 2 biological replicates per sample, with the total number of wing discs examined for each sample shown in parentheses. Error bars represent mean ± 1 SEM. Scale bar = 120 microns. P-values were calculated using a 2-tailed t-test. a–d) Quantification of GFP+ clone number (a, c) and clone area (b, d) with and without irradiation for 4 GAL80 insertions on the X chromosome. c and d) Quantification of GFP+ clone number (e) and clone area (f) with and without irradiation for 12C6 GAL80 insertion on the X chromosome. g–j) Representative examples of wing discs for the 12C insertion. k–n) QS insertions are not significantly different from each other in IR-induced LOH clone number (k) or clone area (l) and show no sex dependence (m and n; P > 0.2 for all M/F pairs). The genotypes were: a–j) For 1E/FM7 and 19E/FM7, homozygous virgin GAL80/GAL80 females were crossed to yw FM7a; Act-GAL4, UAS-GFP/CyO-GFP males, and female larvae without CyO-GFP were dissected. All others were GAL80/+; Act-GAL4, UAS-GFP/+ that result from GAL80/GAL80 X Act-GAL4, UAS-GFP/CyO RFP Tb. 9B = psc-QF > Tom/+; QS9B/+ that results from psc-QF > Tom; QS9B X to w1118. 5A = psc-QF > Tom/QS5A that results from psc-QF > Tom/CyO-GFP X QS5A/CyO-GFP. 21 = psc-QF > Tom/+; QS21/+ that results from psc-QF > Tom/CyO-GFP X QS21/QS21.
The 12C insertion is an anomaly in that it shows the greatest incidence of GFP+ cells without IR via unknown mechanisms, and a further increase after IR (Fig. 2e and f). Even in this case, IR induced increase was significant only in the females (Fig. 2g–j, quantified in e and f). Thus, IR-induced GAL80 LOH shows sex dependence for all 5 insertions on the X chromosome, similar to spontaneous LOH (Siudeja et al. 2015). To our surprise, however, IR-induced LOH did not decrease with proximity to the centromere. If anything, clone number increased with increasing proximity to the centromere, but this trend was not observed for the clone area (Fig. 2c and d). This is unlike spontaneous LOH (Siudeja et al. 2015); GAL805B showed spontaneous LOH in 75% of adult Drosophila intestines while the corresponding number for GAL8019E was 40%. In contrast, IR-induced LOH incidence was nearly identical for GAL805B and GAL8019E (Fig. 2c and d, P = 0.193 for clone number and 0.861 for clone area). We conclude that IR-induced LOH on the X chromosome shows sex dependence (like spontaneous LOH) but chromosomal position independence (unlike spontaneous LOH).
Data for QS insertions that are further from the telomere than QS9B or mwh (Fig. 1f) support the idea that IR-induced LOH is not affected by distance from the chromosome end. We saw no significant difference in IR-induced LOH among the 3 QS insertions (Fig. 2k and l), although additional insertions closer to the centromere remain to be studied.
In females that showed robust X-ray-induced LOH induction, the chromosome bearing the GAL80 transgene was in trans to a wild-type chromosome, allowing for recombination to occur. To address the possible role of recombination in LOH induction, we studied the effect of placing an FM7 balancer chromosome in trans to the GAL80-inserted chromosome. We found that this reduced the LOH incidence for both GAL80 insertions tested when compared with no-FM7 samples of the same insertions (1E and 19E; the last 2 samples in Fig. 2c and d). The reduction was significant for 19E in terms of both clone number (P = 2.6E − 8) and area (P = 1.3E − 8). The reduction was significant for 1E in terms of clone number (P = 0.044) but not for area (P = 0.068), presumably because of the lower clone area in 1E to begin with. We conclude that the presence of a recombination-proficient homologous chromosome is important for optimal LOH induction, which could explain the low LOH incidence for X-linked loci in males (Fig. 2c and d). Lack of sex dependence for QS insertions on autosomes (Fig. 2m and n) further supports this idea because males and females would have a recombination-proficient homologous chromosome in this case, although the effect of a balancer chromosome remains to be studied. Possible explanations for why recombination-based LOH induction on the X would be insensitive to chromosome position are given in Discussion.
A focused screen identified E2F1 as a regulator of LOH
Results so far address cis-acting determinants, namely, chromosome position of the LOH loci and the presence of a homologous X chromosome. To identify trans-acting regulators, we performed a focused screen through known regulators of DNA damage and stress responses, cell growth, cell survival, and cell competition (Supplementary Table 1), using published constructs that have been shown to produce a phenotype, when possible. We used QS in this screen so that we can use the GAL4/UAS system in parallel to knockdown/overexpress genes. We used QS9B because it shows more robust IR-induced LOH incidence than QS5A, although the differences were not statistically significant.
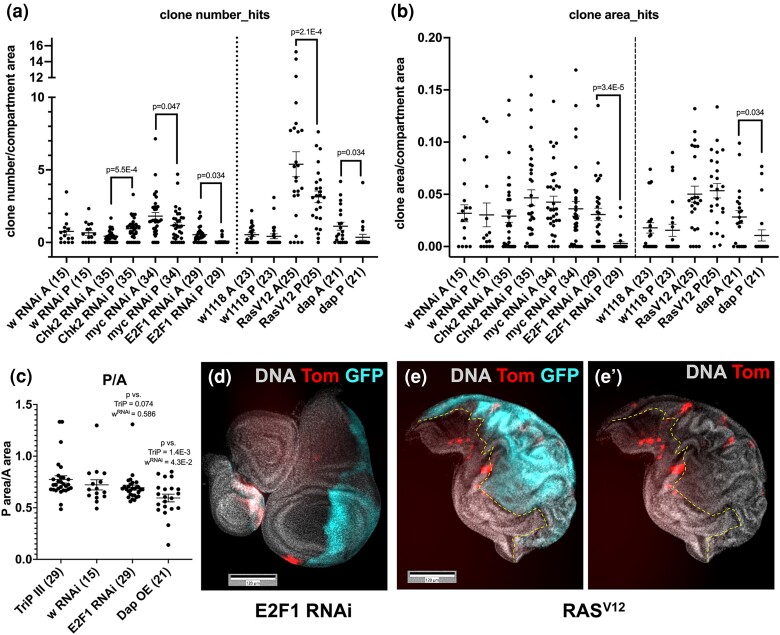
Transgenes were expressed in the posterior (P) half of wing discs using the protocol in Fig. 1e. As we saw no Tom clones without IR, only +IR data are shown (Fig. 3 and Supplementary Fig. 2). Three controls were used: the TRiP III line that was the recipient of many RNAi constructs and RNAi for the w gene (both as controls for RNAi) and w1118 (control for overexpression or OE). After normalizing compartment area to control for differences in compartment size, IR-induced LOH clone number or area is not significantly different between A and P compartments in all 3 controls (Fig. 3a and b and Supplementary Fig. 2). Most genes tested did not affect IR-induced LOH, including those with known roles in cell competition such as duox and Xrp1 (Supplementary Fig. 2). This group is of interest because a prior publication suggested that cells with segmental aneuploidy, a possible mechanism for LOH generation, may be eliminated by cell competition (McNamee and Brodsky 2009). We interpret negative results with caution, however, because our protocol for conditional and transient expression (Fig. 1e) may not deplete gene function sufficiently even if the same construct produced a phenotype under different expression conditions before.
Fig. 3.
A focused screen identified E2F1 as a regulator of LOH. Wing discs from 3rd instar larvae were dissected 3 d after irradiation, fixed and stained for DNA. Larvae were treated as in Fig. 1e. The graphs show data from at least 2 biological replicates per sample, with the total number of wing discs examined per sample shown in parentheses. The mean ± 1 SEM is shown for each sample. P-values were calculated using a paired 2-tailed t-test. a and b) Results of the focused screen showing clone number (a) and clone area (b). RNAi against w is shown as the control for RNAi constructs and w1118 is shown as the control for overexpression constructs. c) The area of the P compartment where transgenes were expressed was normalized to the area of the A compartment (contralateral control). d–e′) Representative examples of wing discs expressing E2F1RNAi or RASV12 in the P compartment (GFP+). The genotypes were en-GAL4 > UAS-GFP, psc-QF > Tom/+; tub-QS9B, tub-GAL80ts/+ with a UAS transgene on the second or the third chromosome, depending on the transgene.
The screen found mild effects with RNAi against Chk2 or Myc and with overexpression of oncogenic RasV12 (Fig. 3a and b). Specifically, Chk2 RNAi increased LOH clone number, while Myc RNAi and RasV12 decreased LOH clone number. The effect sizes, however, were small (1.5- to 2-fold) and clone area was not significantly affected, suggesting compensating changes in the size of individual clones. In addition, expression of RasV12 in the P compartment produced epithelial folds in both compartments in irradiated discs (Fig. 3e and e’), making the quantification of clone area challenging. Because of this and small effect sizes, we have not followed up on Chk2, Myc, and Ras.
In contrast, RNAi against E2F1 profoundly affected both LOH clone number and clone area with a reduction of the former by 6-fold and the latter by >10-fold (Fig. 3d, quantified in a and b). Drosophila E2F1, like its mammalian homologs, promotes cell proliferation by promoting G1/S transition via activation of cyclin E expression and G2/M transition via activation of Cdc25String (Reis and Edgar 2004). E2F1 also promotes IR-induced apoptosis in the wing discs when p53 has been depleted; that is, its role in apoptosis is negligible in a wild-type background (Wichmann et al. 2006). Therefore, we addressed the more likely possibility that the requirement for E2F1 in LOH clone formation/maintenance is through its role in the cell cycle. To this end, we expressed the Cdk inhibitor Dap to inhibit the G1/S transition, using the same temperature shift protocol as in Fig. 1e. Dap phenocopied E2F1 RNAi (Fig. 3a and b), supporting the idea that E2F1 is important for the proliferation of cells with LOH. In contrast, depletion of E2F1 had little effect on neutral (non-LOH) clones induced by heat-shock-induced recombination (Supplementary Fig. 3).
We normalize LOH clone number and area to the compartment area. If E2F1 depletion or Dap overexpression in the P compartment affected all cells within this compartment equally regardless of their LOH status, we should see little change in normalized LOH clone number or area. Yet, we see multiple-fold reductions by both measures. We infer that manipulations of cell-cycle regulators have a more severe effect on cells with LOH than on cells in the rest of the P compartment. This effect was seen even when conditional E2F1 RNAi or Dap overexpression was mild enough to have little or no effect on compartment size (Fig. 3c).
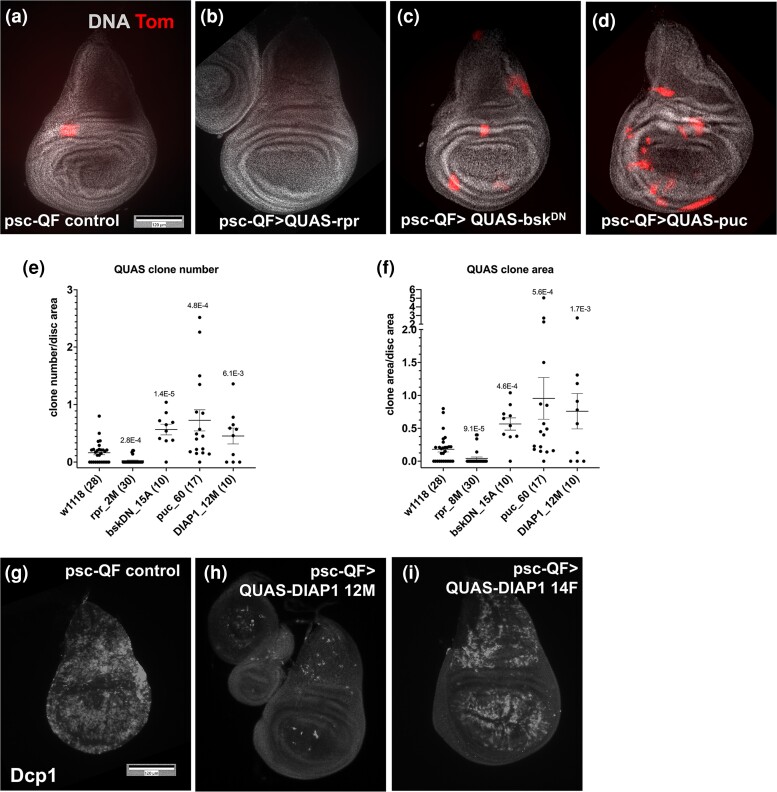
The manipulations thus far alter gene expression in the whole compartment and identified differential effects on LOH vs non-LOH cells. To address cell-autonomous needs in LOH cells more directly, we took advantage of QS loss and consequent QF de-repression to express QUAS transgenes and assayed their effect. This protocol has the added advantage of identifying needs AFTER the clones have formed and have initiated QF-mediated gene expression. As proof of concept, we used a published QUAS-rpr transgene (Perez-Garijo et al. 2013) to express this pro-apoptotic gene specifically in LOH clones, with the expectation that such clones will be lost to cell death. The results show the system works as expected; QUAS-rpr reduced both clone number and clone area to near completion (Fig. 4a and b, quantified in e and f).
Fig. 4.
Cell-autonomous requirements for LOH. a–d) Larvae were treated as in Fig. 1e but without the temperature shift and wing discs were fixed and stained to visualize DNA. The discs shown are from irradiated larvae. e and f) Quantification of Tom+ clone number (e) and clone area (f) in irradiated discs. The graphs show data from at least 2 biological replicates per sample, with the total number of wing discs examined per sample shown in parentheses. The mean ± 1 SEM is shown for each sample. P-values relative to w1118 controls were calculated using a 2-tailed t-test. g–i) Larvae were cultured and irradiated as in (a–f) but wing discs were dissected 4 h after IR, fixed and stained for DNA and with an antibody against cleaved active Dcp1 as a surrogate marker for apoptosis. Scale bars = 120 microns. The genotypes are as shown below. Stock information is in Supplementary Table 1. a, g) Control = psc-QF > Tom/+; QS9B/+ that results from psc-QF > Tom; QS9B X to w1118. h–i) psc-QF > Tom/QUAS transgene; QS9B/+. b–d) psc-QF > Tom/+; QS9B/QUAS transgene.
Using QUAS transgenes, we tested the role of JNK and apoptotic signaling in maintaining cells with LOH. We first assessed transgene functionality by ubiquitous expression with psc-QF (without QS) and assaying for expected phenotypes. For rpr, dominant negative bsk (JNK) and puc (phosphatase and JNK inhibitor), we found lines that produced significant lethality (Supplementary Table 2). For Drosophila inhibitor of apoptosis protein 1 (DIAP1), even the strongest effect on lethality was modest, so we assayed for inhibition of apoptosis instead. Of 3 lines tested, 12M reduced IR-induced cleaved Dcp1 signal to near completion (Fig. 4h), 14F showed a modest effect (Fig. 4i), and 3M showed variable effect from disc to disc. Therefore, QUAS-DIAP1 12M line was used in subsequent experiments. The results show that cell-autonomous inhibition of JNK or apoptosis in cells with LOH led to a significant increase in both Tom clone number and area. JNK activity, assayed by target reporter puc-lacZ, was induced by 3,000–4,000R of X-rays throughout the wing disc for up to 24 h after irradiation, but decayed subsequently and was absent by 96 h after irradiation (Pinal et al. 2018). Our data suggest that in cells that survived LOH long enough to activate QUAS transgenes, JNK, or a downstream effect of JNK signaling continues to operate to eliminate them.
Discussion
We report here studies on the role of chromosomal position and trans-acting regulators on IR-induced LOH in Drosophila larval wing discs. We found that position along the chromosome makes little difference, while the presence of a homologous chromosome is required. We identified E2F1 as a positive regulator of LOH clone incidence and propose that cells with LOH are more sensitive to inhibition of cell proliferation than their neighbors. And we identified JNK signaling and apoptosis as cell-autonomous mechanisms that eliminate cells that survived LOH induction. This contrasts with the previous finding that JNK plays only a backup role in reducing IR-induced Minute (M/+) bristles, a role that becomes apparent only in p53 mutant background (McNamee and Brodsky 2009).
Spontaneous LOH in the adult Drosophila intestine is attributed to mitotic recombination, based on several lines of evidence. First, sex dependence is interpreted as arising from recombination-based mechanisms that require the presence of a homologous chromosome such as crossing over, gene conversion, or break-induced replication (Siudeja et al. 2015). Second, adult guts with spontaneous GAL805B LOH were about twice as frequent as those with spontaneous GAL8019E LOH. An increase in LOH with distance from the centromere is predicted for recombination-based mechanisms, because a distal locus would experience more frequent exchanges between homologous chromosomes. This study noted, however, that the frequency of spontaneous LOH at 19E was higher than predicted from meiotic maps and higher than observed for a centromere-proximal locus on chromosome 3R, suggesting additional factors at play. Third, whole-genome sequencing confirmed that spontaneous LOH of chromosome 2 loci in the adult intestine occurs through mitotic recombination (Zouabi et al. 2022).
In contrast, IR-induced LOH may rely on more than one mechanism. Classical studies by Baker et al. (1978) monitored several adult visible markers such as yellow (y) and forked (f) in heterozygous adult females that were irradiated as larvae or pupae with 1,000R from a Co-60 source. In adult female abdomens monitored for y or f, both X-linked genes, 40% of LOH clones induced by IR (i.e. the difference between −/+IR samples) consisted of twin clones (likely products of mitotic recombination), while the remaining 60% were single clones. Examination of single clones that arose spontaneously (without radiation) in the same study found that 20% of cells in such clones showed Minute bristles, which result from haploinsufficiency of loci encoding ribosomal proteins. The authors suggest that single LOH clones arose by aneuploidy that turned wild-type (+/+) cells into heterozygotes for ribosomal protein genes (M/+). In another set of studies, McNamee and Brodsky (2009) used the Minute bristle phenotype and assayed gain of heterozygosity, from +/+ to M/+, rather than loss of heterozygosity, in bristles from adults that result from irradiated larvae. Recombination-based mechanisms induced by IR would not be detectable in this assay as they would change +/+ to +/+. Regardless, taken together, these results suggest that both aneuploidy and recombination-based mechanisms operate to produce IR-induced LOH in tissues that produce the adult body wall.
Our data suggest a greater contribution by recombination-based mechanisms for IR-induced LOH because of sex dependence for X-lined insertions and because a balancer chromosome reduced the LOH incidence. How then do we explain the lack of dependence on distance from the centromere? Seminal studies by Becker (1969) found that X-ray-induced recombination as measured by twin spots was at least 10-folds higher for the X chromosome than chromosome III, which he attributed to the higher heterochromatin content of the former. Using inversion chromosomes with altered amount of heterochromatin between the centromere and the locus of the phenotypic marker, Becker confirmed that most X-ray-induced recombination events occurred in centromeric heterochromatin. This region (shown as a hatched region in Fig. 1f) is outside the euchromatic region with all GAL80 insertions we studied, which can explain why we see little position effect for IR-induced LOH.
The presence of a balancer chromosome reduced but did not eliminate IR-induced LOH (Fig. 2c and d), suggesting that non-recombination-based mechanisms such as aneuploidy also operate. Studies by Baker, Brodsky, and colleagues did not distinguish among various types of aneuploidies, which could include loss of whole or segments of chromosomes and terminal deletions. Overexpression of the telomere capping protein HipHop has been shown to increase the survival of cells with terminal deletions that result from dividing experimentally induced dicentric chromosomes (Kurzhals et al. 2017). Overexpression of HipHop, however, did not increase the incidence of IR-induced LOH (Supplementary Fig. 2), suggesting that terminal deletions may not be the mechanism. The finding that distance from the telomere did not affect the QS LOH incidence supports this idea. Instead, relevant types of aneuploidy may include X-ray-induced segmental losses that are well-documented in numerous studies in Drosophila and mammals (for example, Pastink et al. 1987, 1988; Eeken et al. 1994; Chick et al. 2005). Such losses can explain our finding that the LOH of X-linked loci requires a homologous chromosome. We suggest that without part or whole of the X chromosome in XY males, a cell may not survive to express GFP, reducing the level of LOH observed in males. In other words, the presence of 1 X chromosome allows cells to survive with partial or complete loss of the other X chromosome.
Supplementary Material
Acknowledgments
The authors thank the Perrimon, Bardin, and Steller laboratories for the fly stocks. Additional stocks from the Bloomington Drosophila Stock Center (NIH P40OD018537) and Vienna Drosophila Resource Center were used in this study. A monoclonal antibody against Ci deposited by R. Holmgren was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA, USA.
Contributor Information
Jeremy Brown, Department of Molecular, Cellular and Developmental Biology, 347 UCB, University of Colorado, Boulder, CO 80309-0347, USA; Department of Cell and Developmental Biology, University of Colorado, Anschutz Medical Campus, 13001 E. 17th Pl., Aurora, CO 80045, USA.
Tin Tin Su, Department of Molecular, Cellular and Developmental Biology, 347 UCB, University of Colorado, Boulder, CO 80309-0347, USA; University of Colorado Cancer Center, Anschutz Medical Campus, 13001 E. 17th Pl., Aurora, CO 80045, USA.
Data availability
Drosophila stocks are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at GENETICS online.
Funding
This work was supported by the National Institutes of Health grant R35 GM130374 to T.T.S.
Literature cited
- Baker BS, Carpenter AT, Ripoll P. 1978. The utilization during mitotic cell division of loci controlling meiotic recombination and disjunction in Drosophila melanogaster. Genetics. 90(3):531–578. doi: 10.1093/genetics/90.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HJ. 1969. The influence of heterochromatin, inversion-heterozygosity and somatic pairing on x-ray induced mitotic recombination in Drosophila melanogaster. Mol Gen Genet. 105(3):203–218. doi: 10.1007/BF00337472. [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM. 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol. 24(3):1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Bush I, Bozon J, Su TT. 2020. Cells with loss-of-heterozygosity after exposure to ionizing radiation in Drosophila are culled by p53-dependent and p53-independent mechanisms. PLoS Genet. 16(10):e1009056. doi: 10.1371/journal.pgen.1009056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charan J, Kantharia ND. 2013. How to calculate sample size in animal studies? J Pharmacol Pharmacother. 4(4):303–306. doi: 10.4103/0976-500X.119726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick WS, Mentzer SE, Carpenter DA, Rinchik EM, Johnson D, You Y. 2005. X-ray-induced deletion complexes in embryonic stem cells on mouse chromosome 15. Mamm Genome. 16(9):661–671. doi: 10.1007/s00335-005-0011-5. [DOI] [PubMed] [Google Scholar]
- Eeken JC, de Jong AW, Loos M, Vreeken C, Romeyn R, Pastink A, Lohman PH. 1994. The nature of X-ray-induced mutations in mature sperm and spermatogonial cells of Drosophila melanogaster. Mutat Res. 307(1):201–212. doi: 10.1016/0027-5107(94)90293-3. [DOI] [PubMed] [Google Scholar]
- Goupil A, Heinen JP, Salame R, Rossi F, Reina J, Pennetier C, Simon A, Skorski P, Louzao A, Bardin AJ, et al. 2022. Illuminati: a form of gene expression plasticity in Drosophila neural stem cells. Development. 149(22):dev200808. doi: 10.1242/dev.200808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE, Booth BW, Pfeiffer BD, George RA, Svirskas R, et al. 2015. The release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25(3):445–458. doi: 10.1101/gr.185579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkis J. 1931. The growth of the gonads in Drosophila melanogaster. Genetics. 16(3):212–224. doi: 10.1093/genetics/16.3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzhals RL, Fanti L, Ebsen ACG, Rong YS, Pimpinelli S, Golic KG. 2017. Chromosome healing is promoted by the telomere cap component hiphop in Drosophila. Genetics. 207(3):949–959. doi: 10.1534/genetics.117.300317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Sanson B, Vincent JP. 1996. Compartments, wingless and engrailed: patterning the ventral epidermis of Drosophila embryos. Development. 122(12):4095–4103. doi: 10.1242/dev.122.12.4095. [DOI] [PubMed] [Google Scholar]
- Martín-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A. 1998. Puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12(4):557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee LM, Brodsky MH. 2009. p53-independent apoptosis limits DNA damage-induced aneuploidy. Genetics. 182(2):423–435. doi: 10.1534/genetics.109.102327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastink A, Schalet AP, Vreeken C, Paradi E, Eeken JC. 1987. The nature of radiation-induced mutations at the white locus of Drosophila melanogaster. Mutat Res. 177(1):101–115. doi: 10.1016/0027-5107(87)90026-1. [DOI] [PubMed] [Google Scholar]
- Pastink A, Vreeken C, Schalet AP, Eeken JC. 1988. DNA sequence analysis of X-ray-induced deletions at the white locus of Drosophila melanogaster. Mutat Res. 207(1):23–28. doi: 10.1016/0165-7992(88)90006-1. [DOI] [PubMed] [Google Scholar]
- Perez-Garijo A, Fuchs Y, Steller H. 2013. Apoptotic cells can induce non-autonomous apoptosis through the TNF pathway. Elife. 2:e01004. doi: 10.7554/eLife.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinal N, Martin M, Medina I, Morata G. 2018. Short-term activation of the Jun N-terminal kinase pathway in apoptosis-deficient cells of Drosophila induces tumorigenesis. Nat Commun. 9(1):1541. doi: 10.1038/s41467-018-04000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis T, Edgar BA. 2004. Negative regulation of dE2F1 by cyclin-dependent kinases controls cell cycle timing. Cell. 117(2):253–264. doi: 10.1016/S0092-8674(04)00247-8. [DOI] [PubMed] [Google Scholar]
- Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, Potter CJ. 2015. Improved and expanded Q-system reagents for genetic manipulations. Nat Methods. 12(3):219–22, 5 p following 222. doi: 10.1038/nmeth.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siudeja K, Nassari S, Gervais L, Skorski P, Lameiras S, Stolfa D, Zande M, Bernard V, Frio TR, Bardin AJ. 2015. Frequent somatic mutation in adult intestinal stem cells drives neoplasia and genetic mosaicism during aging. Cell Stem Cell. 17(6):663–674. doi: 10.1016/j.stem.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells BS, Johnston LA. 2012. Maintenance of imaginal disc plasticity and regenerative potential in Drosophila by p53. Dev Biol. 361(2):263–276. doi: 10.1016/j.ydbio.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann A, Jaklevic B, Su TT. 2006. Ionizing radiation induces caspase-dependent but Chk2- and p53-independent cell death in Drosophila melanogaster. Proc Natl Acad Sci U S A. 103(26):9952–9957. doi: 10.1073/pnas.0510528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouabi LA, Stefanutti M, Riddiford N, Rubanova N, Bohec M, Servant N, Bardin A. Molecular underpinnings and environmental drivers of spontaneous loss of heterozygosity in Drosophila intestinal stem cells. bioRxiv 500951. 10.1101/2022.07.21.500951, preprint: not peer reviewed. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Drosophila stocks are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at GENETICS online.