Abstract
Rearrangements within the AUTS2 region are associated with a rare syndromic disorder with intellectual disability, developmental delay, and behavioral abnormalities as core features. In addition, smaller regional variants are linked to wide range of neuropsychiatric disorders, underscoring the gene's essential role in brain development. Like many essential neurodevelopmental genes, AUTS2 is large and complex, generating distinct long (AUTS2-l) and short (AUTS2-s) protein isoforms from alternative promoters. Although evidence suggests unique isoform functions, the contributions of each isoform to specific AUTS2-linked phenotypes have not been clearly resolved. Furthermore, Auts2 is widely expressed across the developing brain, but cell populations most central to disease presentation have not been determined. In this study, we focused on the specific roles of AUTS2-l in brain development, behavior, and postnatal brain gene expression, showing that brain-wide AUTS2-l ablation leads to specific subsets of the recessive pathologies associated with mutations in 3′ exons (exons 8–19) that disrupt both major isoforms. We identify downstream genes that could explain expressed phenotypes including hundreds of putative direct AUTS2-l target genes. Furthermore, in contrast to 3′ Auts2 mutations which lead to dominant hypoactivity, AUTS2-l loss-of-function is associated with dominant hyperactivity and repetitive behaviors, phenotypes exhibited by many human patients. Finally, we show that AUTS2-l ablation in Calbindin 1-expressing cell lineages is sufficient to yield learning/memory deficits and hyperactivity with abnormal dentate gyrus granule cell maturation, but not other phenotypic effects. These data provide new clues to in vivo AUTS2-l functions and novel information relevant to genotype–phenotype correlations in the human AUTS2 region.
Keywords: AUTS2 syndrome, behavior, brain development, gene expression, isoform function
Introduction
AUTS2 was first discovered as the gene disrupted by translocation in a pair of Autistic twins (Sultana et al. 2002), and since that initial finding, mutations in this gene have been associated with a remarkably wide variety of neurological and developmental disorders. Individuals carrying genomic rearrangements in the AUTS2 region typically express “AUTS2 syndrome” phenotypes, typified by short stature, distinctive facial features, developmental delay, and intellectual disability (ID), with some combination of autism spectrum disorders (ASD), feeding difficulties, seizures, attention deficit hyperactivity disorder (ADHD), and other features (Beunders et al. 2015, 2016; Sanchez-Jimeno et al. 2021). Mutations within AUTS2 exon 9 that delete in-frame or otherwise disrupt the essential HX domain of the protein have been associated with Rubinstein–Taybi syndrome and related syndromic phenotypes (Liu et al. 2021; Palumbo et al. 2021); one of these variants, a missense mutation within the HX domain (T534P) was also reported in a second individual with profound ID, epilepsy, and brain pathology including microcephaly (Fair et al. 2023). In addition to these mutations, mutations throughout the AUTS2 region have been associated with speech and language disorders (Amarillo et al. 2014; Palumbo et al. 2021), and susceptibility to substance abuse (Schumann et al. 2011; Chen et al. 2013; Dang et al. 2014). The association of this single genetic region with such a wide variety of neurodevelopmental disorders suggests a central and widespread role in brain development.
Like many essential neurodevelopmental genes, AUTS2 is large and complex, spanning more than 1.5 Mb and encoding long (AUTS2-l) and short (AUTS2-s) protein isoforms from conserved alternative promoters (Weisner et al. 2019; Monderer-Rothkoff et al. 2021; Geng et al. 2022). Analysis of mouse mutations and neurons in vitro have suggested a wide variety of molecular functions including regulation of the actin cytoskeleton (Hori et al. 2014) and ubiquitin-mediated protein degradation (Geng et al. 2022), as well as nuclear functions including the regulation of neurodevelopmental gene expression through chromatin remodeling (Liu et al. 2021; Li et al. 2022; Gao et al. 2014) and RNA stabilization (Castanza et al. 2021). Studies with embryonic stem cell (ESC)-derived neurons (Liu et al. 2021; Monderer-Rothkoff et al. 2021) and brain organoids (Fair et al. 2023) have shown that Auts2 is involved in basic steps of neurogenesis and neuron maturation, and combined in vivo and in vitro studies have pointed to a special function in synapse formation the regulation of dendritic spines (Hori et al. 2020). These studies have provided intriguing hints that the functions of AUTS2-l and AUTS2-s proteins are distinct, but likely intertwined. For example, AUTS2-s is expressed at early stages of neuron commitment in vitro, whereas AUTS2-l rises later as the cultured neurons begin to mature (Monderer-Rothkoff et al. 2021). At the molecular level, AUTS2-l is the only isoform that associates with PRC1 complexes (Gao et al. 2014; Liu et al. 2021; Monderer-Rothkoff et al. 2021), while AUTS2-s, which can also function as a transcriptional activator (Monderer-Rothkoff et al. 2021), must operate via alternative mechanisms. Some evidence suggests that the 2 isoforms might act additively or cooperatively; for example, human mutations in exons 1–7, which are uniquely coding in AUTS2-l, are associated with many of the AUTS2-syndrome phenotypes with mild expression, while mutations in 3′ exons encoding C-terminal amino acids that are shared by AUTS2-l and AUTS2-s (exons 8–19) generate more severe forms of syndromic disease (Beunders et al. 2013, 2015). Consistently, animals homozygous for brain-wide conditional knockout (cKO) of Auts2 exon 7 are born live with subtle developmental phenotypes (Gao et al. 2014), whereas mice homozygous for germline or brain-wide 3′ mutations die perinatally with significant brain pathologies (Hori et al. 2014; Castanza et al. 2021).
Therefore, Auts2 is clearly required for normal brain development and its malfunction or dysregulation can lead to neurodevelopmental disease. However, the contributions of AUTS2 protein isoforms to specific aspects of disease presentation have not been clearly elucidated. The distinct and intertwined functions of AUTS2 isoforms must be clarified before mechanisms of AUTS2-linked neurological disorders can be understood and before genotype:phenotype correlations in this essential genomic region can be resolved. In this study, we used the published Auts2 exon 7 cKO mutant (Gao et al. 2014) to address several open questions about the developmental functions of AUTS2-l. For example, although 3′ mutations are associated with cerebellar pathologies (Weisner et al. 2019; Yamashiro et al. 2020) the impact of isolated loss of AUTS2-l function on cerebellar development has not been investigated. Additionally, although recessive phenotypes have been described for animals inheriting cKO of Auts2 exon 6 in forebrain excitatory neurons (Li et al. 2022), AUTS2 is expressed more broadly across the developing brain (Bedogni et al. 2010; Weisner et al. 2019; Yamashiro et al. 2020), and potential dominant phenotypes associated with the isolated loss of AUTS2-l function have not been investigated. We designed experiments to address these questions with the goal of clarifying functions of AUTS2-l in the developing brain, and to leverage these data to suggest in vivo functions for AUTS2-s as well.
Materials and methods
Animals and tissue collection
The Auts2-ex7 cKO allele (Auts2tm1.1Dare; Gao et al. 2014) was provided as a kind gift from the laboratory of Dr. Danny Reinberg (NYU). Nes-cre (B6.Cg-Tg(Nes-cre)1Kln/J), Calb1-cre (B6;129S-Calb1tm2.1(cre)Hze/J), and Cmv-cre (B6.C-Tg(CMV-cre)1Cgn/J) were purchased from the Jackson Laboratory. Nes-cre mice were crossed to Auts2tm1.1Dare homozygotes, and these mice were used to generate homozygotes, heterozygotes, and wild-type littermates for behavioral testing and tissue collection. We also generated a mouse line stably inheriting a germline deletion of Auts2 exon 7 by crossing Cmv-cre females to Auts2tm1.1Dare males. The Cmv-cre, ex7−/+ F1 offspring were then bred to generate the stable deletion/ex7 KO line. Because the cre allele is no longer required after F1 generation, this stable KO line is labeled here as ex7 KO, with ex7−/+ or ex7−/− being used to describe heterozygotes or homozygotes, respectively. Wild type (WT), Nes-cre, ex7−/− and heterozygote germline KO ex7−/+ mice were tested for developmental milestones including body weight, righting reflex, eye opening, open field test, novel object recognition (NOR), and sociability and social memory tests as previously described (Weisner et al. 2019; Chen et al. 2022). This study strictly followed the Guide for the Care and Use of Laboratory Animals of the Use Committee of the University of Illinois (Animal Assurance Number: A3118-01; approved IACUC protocol number 18240) and the Pacific Northwest Research Institute (A3357-01; IACUC protocol 201-20). Animals were maintained under standard conditions (12-hr light/dark cycle, group housed); all tests were done between 1 and 5 hr prior to lights-off or 1–5 hr prior to lights-on. Supplementary Table 1 summarizes the genotypes and ages of mice used in this study for all behavioral tests, pathology, and/or RNA-seq gene expression analysis.
Behavioral tests
Righting reflex
Righting reflex test was modified from previous studies (Castelhano-Carlos et al. 2010; Chen et al. 2022) with the following modifications. On postnatal day 5, pups were placed belly up on flat surface and scored based on time lapsed before they get back to their feet on the ground: score 0 = failed to turn back within 15 sec; score 1 = pups turned between 10 and 15 sec; and score 2 = pups turned within 10 sec.
Eye opening
Eye opening was measured on postnatal day 13 following previous study (Chen et al. 2022). In brief, scores from 0–2 are assigned. Score 0 = both eyes are closed; score 1 = one eye is open or both eyes partially open; and score 2 = both eyes are fully open.
Open field test
Mouse was placed in a novel open arena, 35-cm × 30-cm paper box for 10 min and videotaped for analysis using DeepLabCut 2.2.3 (DLC) (Mathis et al. 2018). Tracking framework was trained to recognize tail base, middle back, nose tip, left ear and right ear of the mouse. A skeleton, lines connecting between points, was included to connect the tail base to middle back, middle back to nose tip, and both ears each to the nose tip. We used default neural network and iteration settings. Videos were cropped using GUI to include only the arena. Middle back point coordinates were used to calculate total travel distance in pixels from 18,000 frames (10 min).
Stereotypic behavior (grooming)
The average grooming time per episode during the open field test was measured following previously described protocols (Chen et al. 2022).
NOR
Published protocols were followed (Botton et al. 2010; Chen et al. 2022). In brief, tasks include 3 sessions: habituation, training, and retention. During habituation, the mouse completed open field test for 10 min. After a 24-hr interval, the same mouse was introduced to the same arena with 2 novel objects for training session and was allowed to explore the objects for 10 min. The total time a mouse actively explored the objects was quantified as total exploration time for NOR-exploratory activity. During the retention session 24 hr later, one of the familiar objects that appeared in training session was exchanged for a new object in the same arena, and the mouse was allowed to explore for 5 min. The exploratory index was calculated as percentage of time the mouse spent in exploring novel object/total time spent exploring both objects in the retention session. Exploratory behavior was defined as sniffing or touching the object.
Three-chamber test
We followed protocols described in published studies (Hori et al. 2015; Chen et al. 2022) with minor modifications. A Plexiglass box was divided into 3 chambers: object chamber, middle chamber, and social chamber. The tested mouse was introduced to the empty box and allowed to explore for 10 min. Then a novel object (Lego toy or items of similar size/complexity) was introduced to the object chamber, and a sexually immature juvenile female mouse (P21–P29) was placed in the social chamber as stranger mouse 1. Both the object and the stranger mouse were confined under a small stainless steel wire cup. The tested mouse was again allowed to explore the box for another 10 min. Time of the mouse spent in each chamber was quantified, and the percentage of time spent in the social chamber/time spent in both object chamber and social chamber was quantified for sociability behavior. Lastly, a novel sexually immature juvenile female mouse (P21–P29) was introduced to replace the object as stranger mouse 2, and the tested mouse was allowed to explore the box now with a familiar mouse (stranger mouse 1) and a novel mouse (stranger mouse 2) freely for 10 min. Social memory was quantified by percentage of time the tested subject spent in novel mouse chamber/total time spent in either familiar mouse chamber or novel mouse chamber during the final 10-min session.
RNA collection, RNA-seq, qPCR, and RiboTagging
RNA and qRT-PCR
RNA and cDNA were prepared as previously described (Weisner et al. 2019). qRT-PCR was done using custom-designed primers for specific Auts2 isoforms (Supplementary Table 2), and expression values were normalized relative to the Pgk1 control in each sample and compared using standard methods (Livak and Schmittgen 2001).
RNA-seq
Forebrain of 3 P0 ex7−/− and hippocampus of 3 P14 Nes-cre, ex7−/− animals were collected along with WT littermates of each age, and RNA generated as previously described (Weisner et al. 2019). RNA-seq libraries were generated and analyzed as previously described (Chen et al. 2022). Briefly, libraries were constructed with Illumina TruSeq RNA library Prep Kits v2; and sequenced on Illumina Hi-Seq 4000 sequencer for 150-bp paired end reads (17–30-M reads/sample) through Genewiz sequencing service (South Plainfield, NJ). STAR v2.7.5a was used to map reads to mm10/GRCm38 genome and ensemble v102 annotation. Differential expression analysis was done using EdgeR v3.42.4 (Robinson et al. 2009) as previously described (Saul et al. 2017) using Benjamini–Hochberg correction to assign false discovery rate (FDR) values to particular genes. Genes with FDR< 0.05 were considered as significantly differentially expressed. For functional analysis, we used ToppCluster annotation tool (Kaimal et al. 2010) with default statistical parameters, focusing on genes identified as having at least 1.5-fold change (FC) in mutants compared to WT controls with FDR less than 0.05. Visualization of RNA-seq data was performed with “ggplot2” R package (Wickham, 2016).
RiboTag immunoprecipitation
Calb1-cre (B6;129S-Calb1tm2.1(cre)Hze/J) mice were crossed to Rpl22-HA strain (B6N.129-Rpl22tm1.1Psam/J) (Sanz et al. 2009) from Jackson Laboratories. Immunoprecipitation (IP) enrichment was performed on RNA isolated from dissected brain tissue as previously described (Sanz et al. 2009). In brief, samples were suspended in homogenization buffer [50-mM Tris, pH 7.5, 100-mM KCl, 12-mM MgCl2, 1% NP40 substitute, 1-mM DTT, 200-U/mL Promega RNasin, 1-mg/mL heparin, 100 μg/mL cycloheximide, 2× PIC (Roche)]. HA-antibody (ab9110, Abcam) was added to the supernatant after 10,000 g × 10-min centrifugation at 4°C, and incubated at 4°C with rotation for 4 hr. Then protein G beads pre-equilibrated with homogenization buffer were added to the supernatant-antibody mix to bind overnight at 4°C. Beads were washed with high salt buffer (50-mM Tris, pH 7/5, 300-mM KCl, 12-mM MgCl2, 1% NP40 substitute, 1-mM DTT, 100 μg/mL cycloheximide) 3 times, and 900 μL of trizol was added to the beads to release RNA from the HA-labeled polysomes.
Analysis of published AUTS2 chromatin immunoprecipitation (ChIP) data
Data from 2 previously published AUTS2 ChIP datasets (E16.5 forebrain, Oksenberg et al. 2014; P1 whole brain, Gao et al. 2014) were downloaded from the SRA/Gene Expression Omnibus (GEO) data repositories (accession numbers SRR1292304/SRR1292309 and GSE60411, respectively). The raw data were mapped to the mm10 genome build using STAR v2.7.5a aligner (Dobin et al. 2013) and the mapped reads were analyzed using HOMER 4.11 software (Heinz et al. 2010). Peaks were assigned to transcription start sites of 2 nearest genes in the telomeric and centromeric directions, respectively, with bedops v2.4.40 (Neph et al. 2012), and these nearest genes were compared to P0 FB or P14 HC differentially expressed genes (DEGs) (Supplementary Table 2). The mapped peaks with nearest genes and associated DEGs are listed in Supplementary Tables 3 (P1 ChIP dataset) and 4 (E16.5 ChIP dataset). Hypergeometric overlap P-values were calculated using the function hypergeo.overlap.test() in version 1.16.1 of the R package msaul (https://github.com/msaul/msaul), as previously reported (Seward et al. 2022).
Immunohistochemistry and histopathology
Isolated tissues were fixed in fresh 4% paraformaldehyde, embedded in paraffin, and cut into 5-μ sections using a Leica RM2155 microtome and Super Plus charged slides (Leica). Cerebellum (CB) sections were sectioned sagittally; hippocampus (HC) was sectioned coronally. For histology experiments, deparaffinization, rehydration were followed and paraffin-embedded slides were stained with hematoxylin and eosin (H&E) as previously described (Weisner et al. 2019). For immunohistochemistry (IHC) experiments, antigen retrieval was also performed followed by blocking in Antibody Diluent Reagent Solution (Life Technologies). Primary antibodies were tested and used at the optimal ratio in the same antibody diluent and incubated overnight at 4°C. Secondary antibodies (1:200) were also diluted in the same antibody diluent and incubated for 1 hr at room temperature. Hoechst 3342 was used for nuclear stain prior to scanning with a Leica Sp8 confocal microscope and LASX imaging software. Primary antibodies were as follows: AUTS2 (Sigma, HPA000390), Calbindin D-28K (Sigma C9848, 1:300), Calbindin (Santa Cruz sc-365360, 1:300), Calretinin (Santa Cruz sc-365989, 1:300), Calretinin (ZYM MD, 1:300), Tbr2 (Invitrogen 14-4875-52, 1:100), Parvalbumin (R&D system AF5058, 1:50–1:100), cFOS (Santa Cruz s-166940, 1:200), and GAD (R&D system AF2086-SP, 1:100); and secondary antibodies (Thermo-Fisher Scientific): Goat anti-mouse IgG (Alexa Fluor 488, A11001), Goat anti-Rabbit IgG (Alexa Fluor 594, A11012), Donkey anti-Rat IgG (Alexa Fluor 594, A21209), Donkey anti-Sheep IgG (Alexa Fluor 594, Abcam, ab150180), and Donkey anti-Sheep IgG (Novus Biologicals, DyLight488, NBP173002). All measurements and counts were done on multiple sections from at least N = 3 animals for each genotype.
Cell number counts, structure thickness, and analysis
The number of cells stacked in vertical columns of the dentate gyrus (DG) granule cell layer (GCL) structure was counted. The thickness of both CalB+ and CalR+ layers was measured and the number of cells per column of CalB+ and CalR+ cells were counted. The ratio of CalB+ to CalR+ layer or cell counts was reported. For cell density counts, the thickness of each structure (DG GCL or cornu Ammonis (CA) pyramidal cell layer was measured, and the number of cells inside a square, with side-length equal to the thickness of structure, was counted. Cell density was calculated by dividing the total cell number counted by the squared area. Analysis was performed with NanoZoomer NDP view Digital Pathology software. All measurements were taken from at least N = 3 animals of each genotype. Specific numbers of measurements taken are described in each figure legends, respectively. Statistical significance was determined through 2-tailed t-test, and Welch's correction was applied if variances within testing and control groups were greatly different. P-value ranges are reported for each figure, respectively.
Western blot
Proteins were extracted with Radioimmunoprecipitation assay buffer buffer and quantified with the Bicinchoninic acid test. NuPage 4–12% Bis-Tris gel was used for electrophoresis separation of proteins. PVDF membranes were used for immunoblots and blotted with 5% milk in PBS with 0.2% Tween 20 (PBST), then primary antibody in 1% milk in 0.2% PBST. Primary antibodies used: AUTS2 (Sigma, HPA000390), Actin (Santa Cruz, sc-47778). Quantification was performed with FIJI ImageJ.
Statistical analysis
All statistical analysis and graph presentation are performed with GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA, www.graphpad.com) unless specially noted. Two-tailed unpaired t-tests were used to determine statistical significance for behavioral tests, qRT-PCR, and cell counts and outliers were excluded based on specified outlier tests reported in figure legends. P-values less than 0.05 were considered significant; ranges and other details are indicated in figure and figure legends. Number used for each test are reported in Methods, with further information in the text, figure legends, or in the Supplementary Tables in context with discussion of the data. Graphs are presented as mean ± standard error of the mean (SEM). Statistical analysis for RNA-seq data is detailed above.
Results
Conditional deletion of Auts2 exon 7 leads to AUTS2-l ablation, but transient up-regulation of intact AUTS2-s transcripts and protein in neonatal brain
Animals carrying a LoxP-flanked, conditional knockout allele of Auts2 exon 7 (ex7 cKO) have been described previously (Gao et al. 2014). In that study, the ex7 cKO mice were crossed to animals carrying the Nes-cre allele, which expresses cre recombinase in neuronal and glial precursors beginning around E11 (Tronche et al. 1999), corresponding to the time when Auts2 transcripts are first detected in the mouse brain (Bedogni et al. 2010), although there are also reports that cre activity may be most active at a later stage (Liang et al. 2012). We generated animals of that same genotype and also created a stable exon 7 KO line from F1 animals carrying both the ex7 cKO and Cmv-cre alleles (Schwenk et al. 1995; see Methods).
The first report of Nes-cre, ex7−/− animals confirmed that AUTS2-l protein was efficiently ablated across the neonatal brain, while AUTS2-s remained intact (Gao et al. 2014). Before proceeding with phenotypic analysis, we confirmed and quantitated AUTS2 isoform transcript and protein expression, collecting RNA and protein from mutants and WT littermates at P0 and P14 to capture the early postnatal developmental trajectory. We chose P0 because expression of AUTS2-l is still highly expressed at that postnatal stage (Liu et al. 2021) although obvious brain pathologies have not yet developed. On the other hand, hippocampal and cerebellar pathologies have been consistently documented for Auts2 3′ mutants during the second and third postnatal weeks (Weisner et al. 2019; Hori et al. 2020; Yamashiro et al 2020; Castanza et al. 2021). During this postnatal period, hippocampal dentate gyrus granule cell (DG gc) and cerebellar PC have migrated into position and are beginning to mature and establish adult synaptic connectivity (Brandt et al. 2003; Sudarov and Joyner 2007; Nicola et al. 2015; Berg et al. 2019), functions in which AUTS2-l has been particularly implicated (Hori et al. 2014; Monderer-Rothkoff et al. 2021; Li et al. 2022). At P0 we collected forebrain (FB) of ex7−/− and WT littermates and combined the midbrain and hindbrain (MH) regions for testing from the same animals. At P14, we collected hippocampus (HC) and cerebellum (CB) of Nes-cre, ex7−/− mice for both molecular testing and pathology.
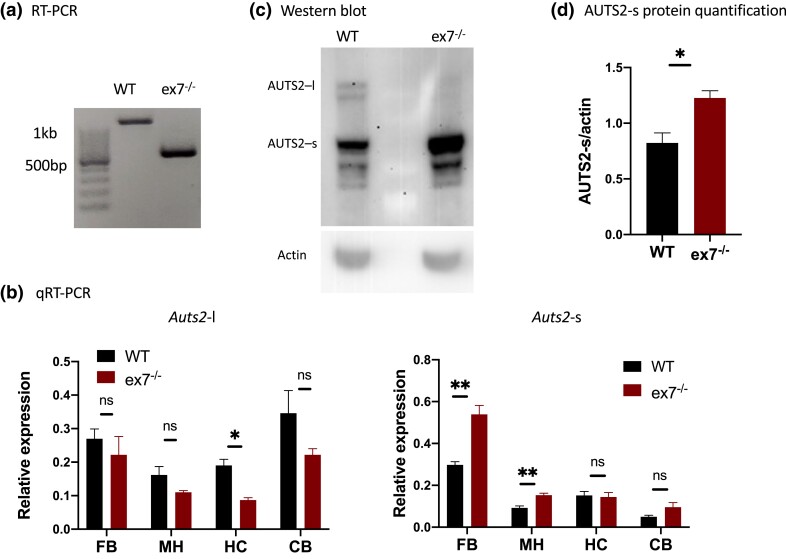
We confirmed exon 7 deletion in mutant Auts2 transcripts using reverse transcript PCR (RT-PCR) to generate cDNA fragments from the P0 samples and examining PCR product size (Fig. 1a) followed by Sanger sequencing (not shown). We then examined the level of each Auts2 transcript with quantitative RT-PCR (qRT-PCR) (Supplementary Table 1). Deletion of exon 7, at 472 bp, puts AUTS2-l transcripts out of frame (as detailed by Gao et al. 2014); however, we found that mutant transcript levels were not significantly different from WT in P0 FB or MH samples (Fig. 1b, left panel) suggesting inefficient clearance by nonsense mediated decay. In contrast, AUTS2-l transcripts were significantly reduced in P14 HC RNA (Fig. 1b, right panel). We also examined AUTS2 isoform expression in western blots with protein P0 ex7−/− mutant and WT littermate brains with a well-tested AUTS2 antibody, specificity of which was recently validated in 3′ KO mutants (Yamashiro et al. 2020; Castanza et al. 2021). Consistent with published reports (Gao et al. 2014; Liu et al. 2021), the western blots also showed selective removal of AUTS2-l protein from P0 ex7−/− brains (Fig. 1c). Antibody specificity was further confirmed by the significant but selective diminution of AUTS2 IHC signal in in Nes-cre, ex7−/− mutant brains (Supplementary Fig. 1a, discussed further below). In contrast, we were surprised to find that AUTS2-s encoding transcripts were significantly up-regulated in both P0 FB and MH mutant fractions (Fig. 1b, right panel), and that in western blots, the AUTS2-s protein was significantly more abundant in the P0 mutant than WT brain (Fig. 1, c and d). In contrast to P0, at P14 AUTS2-s was expressed at WT levels in mutant hippocampus; AUTS2-s transcript levels trended higher in the P14 mutant cerebellum, but the differences did not reach statistical significance (Fig. 1b, right panel). The data suggested that AUTS2-l regulates expression of AUTS2-s, either directly or indirectly but transiently, in the neonatal brain.
Fig. 1.
Expression of Auts2 isoforms in mutant and wild type brain. a) Confirmation of exon 7 deletion in ex7−/− brains. Reverse transcript (RT)-PCR products from WT mouse brain are ∼500 bp longer than those from ex7−/− mouse, confirming the deletion of the 470-bp exon 7 as expected. b) AUTS2-l (left) and AUTS2-s (right) transcript expression characterization by qRT-PCR in RNA from WT, ex7−/− and Nes-cre, ex7−/− animals. RNA from P0 ex7−/− forebrain (FB) and combined midbrain-hindbrain (MH), or from P14 Nes-cre, ex7−/− hippocampus (HC) and cerebellum (CB) was analyzed. N = 3 for each genotype; 1 outlier was excluded from P14 Nes-cre, ex7−/− HC samples based on Grubbs test, G = 1.155, alpha = 0.05. c) Representative western blot analysis of AUTS2-l and major AUTS2-s protein isoform levels in P0 FB of WT and ex7−/− animals with actin as loading control. AUTS2-l was ablated in ex7−/− (as shown in Gao et al. 2014) while AUTS2-s was expressed at higher level in ex7−/− compared to WT. d) Quantification of AUTS2-s bands intensity from western blot in panel (c) using actin as loading control. N = 3 for each genotype. Two-tailed unpaired t-test was used to test statistical significance, *P < 0.05, **P < 0.01, ****P < 0.0001, ns = not significant.
AUTS2-l LOF is associated with a subset of the pathological and behavioral phenotypes associated with 3′ Auts2 alleles
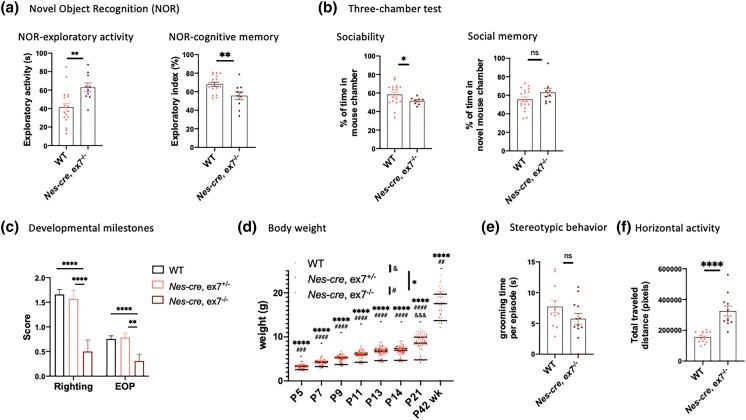
In the first published reports, Nes-cre, ex7−/− mice were found to be smaller than littermates with delayed development of the righting response, deficits in negative geotaxis, and abnormal ultrasonic vocalizations (Gao et al. 2014). Adult behaviors and brain pathology were not investigated. However, behavioral and brain pathology phenotypes have since been associated with mutations that dysregulate, ablate or alter the structure of both Auts2 isoforms (Tables 1 and 2). These mutants have included cKO alleles and 16Gso, a translocation that down-regulates expression of both AUTS2 isoforms as well as neighboring gene, Galnt17 (Weisner et al. 2019). The phenotypes of Galnt17 KO mutants have been described (Chen et al. 2022), and with the caveat that the mis-expression of Galnt17 could possibly contribute to some mutant phenotypes, 16Gso will be compared to ex7 mutants in the following discussion as an allele that affects the expression of both AUTS2 isoforms.
Table 1.
Developmental and behavioral phenotypes in Auts2 ex7 cKO mutants compared to other published Auts2 alleles. Significant differences are highlighted in bold letters.
| Phenotype |
Nes-cre
ex7−/− |
ex7+/− a |
Calb1-cre
ex7−/− |
Emx1-cre
ex6−/− b |
Cag-cre, Auts2del8/+ c |
Auts2
neo/+ d |
16Gso/16Gsoe |
|---|---|---|---|---|---|---|---|
| Cre expression timing | ∼E11 | Germline | Cell-type specificf | ∼E10 | Germline | Germline | n/a |
| Cre expression location | Brain-wide | All cells | Lineage-specific | FB excitatory neurons and glia | All cells | All cells | n/a |
| Postnatal growth | Reduced | Reduced | Normal | Normal | Reduced | Reduced | Reduced |
| Developmental milestonesg | Delayed | Delayed | Delayed | .. | .. | .. | Delayed |
| Stereotypic behaviors | Normal | Increased | .. | Increased | .. | .. | Increased |
| Travel distance, open field | Increased | Increased | Increased | Normal | Reduced | Reduced | Normale |
| Exploratory activity (NOR) | Increased | Normal | Normal | .. | .. | Normal | Reduced |
| Cognitive memory (NOR) | Deficient | Normal | Deficient | Normal | Deficient | Deficient | Deficient |
| Sociability | Deficient | Normal | Normal | Normal | Deficient | Normal | Deficient |
| Social memory | Normal | Normal | .. | Deficient | Deficient | Normal | .. |
.., not reported/not tested; n/a, not applicable (no cre involved). Significant differences are highlighted in bold letters.
a Mouse line generated from Cmv-cre, ex7−/+ F1 animals to carry a permanent, germline KO of Auts2 exon 7.
d Hori et al. (2015); homozygotes for this genotype do not survive.
e Weisner et al. (2019); activity levels were normal in homozygotes, but significantly increased in heterozygotes measured as home cage activity.
f The Calb-cre is expressed at different developmental time points in different cell types, as discussed in text.
g Developmental milestones included the time of righting reflex development and/or eye opening as discussed in the text.
Table 2.
Pathology observed in hippocampus (HC) or cerebellum (CB) in Auts2-ex7 mutations compared with those identified in published Auts2 mouse mutants.
| Brain region and phenotypes | Nes-cre, ex7−/− | Calb1-cre, ex7−/− | Emx1-cre, ex6−/− a | Emx1-cre, ex8−/− b | Emx1-cre, ex15−/− c | 16Gso/16Gsod |
|---|---|---|---|---|---|---|
| Hippocampus | ||||||
| cre allele timing | ∼E11e | Cell-type specific | ∼E10e | ∼E10e | ∼E10e | n/a |
| cre allele location | Brain-wide | Lineage-specific | FB excitatory neurons | FB excitatory neurons | FB excitatory neurons | n/a |
| DG—overall size | Reduced | Normal | Reduced | .. | Reduced | Reduced |
| DG—mature/immature gc ratio | Reduced | Reduced | Normal | .. | .. | Reduced |
| DG—TBR2+ INP numbers | Normal | Normal | Reduced | .. | Reduced | Increased |
| DG—ectopic TBR2+ INP | Increased | Normal | Increased | .. | .. | .. |
| DG—GCL overall cell numbers | Reduced | Normal | Reduced | .. | Reduced | Normal |
| Suprapyramidal bundle thickness | Reduced | .. | Reduced | .. | .. | .. |
| CA1− c-Fos expression | Increased | Normal | .. | Increased | .. | Increased |
| PV+ interneuron cell number | Reduced | Normal | .. | .. | .. | .. |
| HMN collateral axon track | Normal | Normal | .. | .. | Reduced | Normalf |
|
Nes-cre, ex7−/− |
Calb1-cre, ex7−/− | .. | En1-cre, ex8−/−b | .. | 16Gso/16Gsod | |
| Cerebellum | ||||||
| Cre allele timing | ∼E11 | .. | .. | ∼E9 | .. | n/a |
| Cre allele location | Brain-wide | .. | .. | Rhombomere 1-derived | .. | n/a |
| Lobule X | Present g | Absent | .. | |||
| PC numbers/survival | Normalg | .. | .. | Reduced | .. | Reduced |
| P14 Molecular Layer thickness | Reduced g | .. | .. | Reduced | .. | .. |
| P14 EGL thickness | Increased | .. | .. | .. | .. | .. |
| Adult Molecular Layer thickness | Normal | .. | .. | .. | .. | .. |
| P14, adult PC dendritic branching | Normalg | .. | .. | Reduced/abnormal | .. | Reduced |
n/a, not applicable (no cre involved or cre not expressed in this region); .., not tested or not reported; FB, forebrain; DG, dentate gyrus; gc, DG granule cells; GCL, DG granule cell layer; pyc, pyramidal cells; in, interneurons; INP, intermediate progenitors; CA1, cornu Ammonis 1; HMN, hilar mossy neurons; PC, Purkinje cells; EGL, external granule layer. Significant differences are highlighted in bold letters.
b HC pathology examined in Emx1-cre, ex8−/−, Hori et al. (2020); cerebellum pathology examined in En1-cre, ex8−/− mice, Yamashiro et al. (2020).
d Weisner et al. (2019); this mutation also affects expression of Galnt17, which has specific effects on PCs in the vermis, Chen et al. 2022.
e Embryonic day 11 (E11) or 8 (E8).
f HMN were normal in number but showed significantly reduced expression of dopamine receptor, DRD2, Weisner et al. (2019).
g Also observed in P16 and adult ex7−/− germline KO mice.
Auts2 3′ mutations including 16Gso have been associated with specific recessive brain pathologies, primarily centered on the development and survival of cerebellar PC (Weisner et al. 2019; Yamashiro et al. 2020), as well as hippocampal DG gc and their intermediate progenitor precursors (Weisner et al. 2019; Hori et al. 2020; Castanza et al. 2021); pathologies of both cell types have been observed consistently in mutant animals during the first 2 postanal weeks and persist into adulthood. Some of these same hippocampal phenotypes were also recently reported for Emx1-cre, ex6−/−mice (Li et al. 2022) (Table 2). With the goal of identifying the specific contribution of AUTS2-l to those documented phenotypes and to include phenotypes not limited to forebrain excitatory cells, we generated Nes-cre, ex7−/− mice for phenotypic testing. We also collected collect heterozygous ex7−/+ and homozygous ex7−/− offspring for phenotypic testing. Like Nes-cre, ex7−/− mice, the ex7−/− homozygotes were viable. Unlike Nes-cre, ex7−/− mice, which were long-lived and otherwise healthy although sterile, ex7−/− mice were less robust, with some but not all animals surviving into adulthood (Table 1). Heterozygotes of both genotypes were healthy and long-lived.
With the explicit goal of understanding the role of AUTS2-l in previously described phenotypes, we used homozygotes and heterozygotes of both mutant strains to investigate brain pathology and behavior (as summarized in Supplementary Table 1).
A subtle signature of cerebellar developmental delay
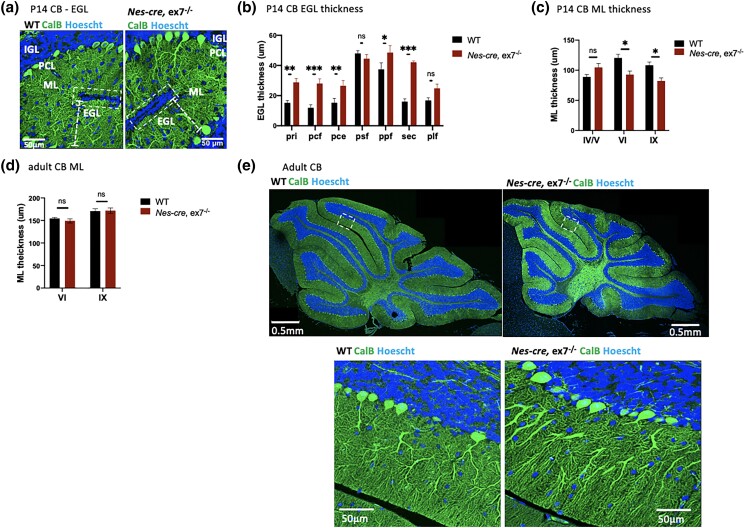
Focusing first on cerebellar pathology, we observed subtle abnormalities in the Nes-cre, ex7−/− mice, including a significantly thicker external granule layer (EGL) at the base of the fissures in mutants than in WT littermates at P14 that suggested a delay in granule neuron migration (Sudarov and Joyner 2007) (Fig. 2, a and b). In addition, the P14 mutant CB molecular layer was decreased in thickness compared to WT littermates (Fig. 2, a and c), suggesting insufficient PC dendritic branching at this stage. Certain cerebellar lobules were more severely affected than others: for example, in the vermis, lobules VI and IX showed significantly decreased molecular layer thickness while lobules IV/V did not (Fig. 2c). However, in the molecular layer thickness in those same lobules was normal in mutant adults (Fig. 2d). This finding could suggest a delay in PC dendritic growth, although we did not detect any obvious abnormalities in overall dendritic branching or structure of PCs in Nes-cre, ex7−/− adults (Fig. 2e). Previously reported phenotypes including PC loss and significantly stunted PC dendrites in both neonates and adults (Weisner et al. 2019; Yamashiro et al. 2020) were not observed at any stage. Furthermore, severe reduction or absence of specific cerebellar lobules seen in En1-cre, ex8−/− mice (Yamashiro et al. 2020), was not observed in the Nes-cre, ex7−/− mice (Table 2). For example, lobule X, which was absent in neonatal and adult En1-cre, ex8−/− mice, was present in P14 and adult Nes-cre, ex7−/− animals (Fig. 2e).
Fig. 2.
Cerebellar pathology in Nes-cre, ex7−/− mutant mice. a) Representative image of the base of the fissures in P14 WT and Nes-cre, ex7−/− cerebellum; white boxes enclose the external granule layer (EGL) at the base of the fissures; dashed lines show the thickness of the molecular layer (ML); PCL, Purkinje cell layer; IGL, inner granule layer. b) Quantification of EGL thickness at the base of different fissures in P14 WT and Nes-cre, ex7−/− cerebellum vermis. Three measurements were averaged for each section examined; N = 3–6 matching sections from 3 mice for each genotype. pri, primary fissure; pcf, preculminate fissure; pce, precentral fissure; psf, posterior superior fissure; ppf, prepyramidal fissure; sec, secondary fissure; plf, posterolateral fissure; c) quantification of ML thickness in P14 WT and Nes-cre, ex7−/− vermis. Three measurements were averaged for each location examined on per section; N = 5–6 matching sections from 3 mice for each genotype. IV/V, VI, and IX indicate lobules examined. d) Quantification of ML thickness in adult WT and Nes-cre, ex7−/− vermis. Three measurements were averaged for each section examined; N = 6–8 matching sections from 3 mice for each genotype. VI and IX indicate lobules examined. e) Representative image of CB in adult WT and Nes-cre, ex7−/− mice (upper panels); dashed boxes circle zoomed in view of lobule VI (lower panels); 2-tailed t-test was used to test statistical significance, *P < 0.05, **P < 0.01, ****P < 0.0001, ns = not significant.
However, En1-cre is first active around E9 (Kimmel et al. 2000) while Nes-cre begins around E11 (Liang et al. 2012), and AUTS2-l LOF during the E9-E11 window could still possibly be associated with these cerebellar defects despite low Auts2 expression at this stage (Bedogni et al. 2010). Therefore, we examined CB of the germline ex7−/− mutants and observed the presence of all vermis lobules (Supplementary Fig. 1b, top panels), grossly normal PC structure (Supplementary Fig. 1b, bottom panels) and subtle defects in molecular layer (ML) thickness (Supplementary Fig. 1b), confirming that isolated AUTS2-l LOF is not sufficient to yield the dramatic defects in cerebellar development associated with 3′ alleles.
A dramatic impact on hippocampal dentate gyrus development
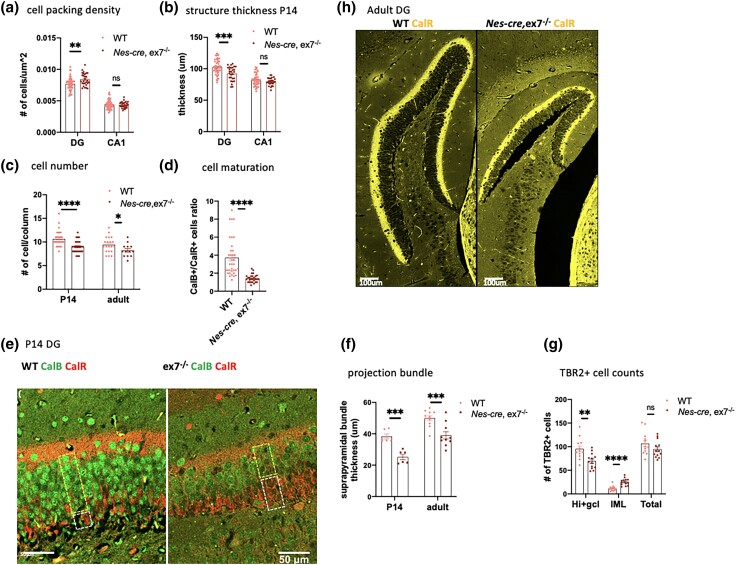
AUTS2-l LOF leads to a delay or block in the maturation of DG gc
In contrast to CB, the mutant HC did display many of the dramatic cellular pathologies described for 3′ mutations, including some also observed in in Emx1-cre, ex6−/− animals (Li et al. 2022). We observed significantly increased cell-packing density in the DG granule cell (gc) layer in mutant hippocampus (Fig. 3a), although we did not see increased packing density in Nes-cre, ex7−/− CA1 pyramidal cell (pc) cells as reported for the 16Gso mutant (Weisner et al. 2019) (Fig. 3a, Table 1). Concordantly, we also observed decreased structural thickness in the DG gc layer in both P14 and adult animals and but not in CA1. (Fig. 3b, Supplementary Fig. 1c).
Fig. 3.
Hippocampal (HC) pathology in Nes-cre, ex7−/− mutants. a) Dentate gyrus (DG) granule cells were more densely packed in P14 Nes-cre, ex7−/− HC as compared to WT, but CA1 pyramidal cells were normally packed. N = 30–53 counts on matching sections from 3–6 mice for each genotype. b) DG structure thickness was significantly decreased in P14 Nes-cre, ex7−/− mice compared to WT animals, although CA1 structure thickness was not significantly different in WT and mutant mice. N = 28–50 measurements on matching sections from 3–6 mice for each genotype. c) The total number of cells per column in the DG was significantly reduced in both P14 and adult Nes-cre, ex7−/− HC compared to WT littermates. N = 28–36 counts for P14 and 13–20 counts for adult on matching sections from 3 mice for each genotype. d) The ratio of CalB+/CalR+ cells was significantly decreased in Nes-cre, ex7−/− mutants DG compared to WT littermates. N = 28–36 counts on matching sections from 3 mice for each genotype. e) Representative picture of the CalB+ and CalR+ layers in DG of P14 WT and Nes-cre, ex7−/− HC. The upper box highlights the CalB+ cell layer and the lower box shows CalR+ cell layer in each sample. f) The thickness of the suprapyramidal bundle of the DG gc-CA3 projection was significantly reduced in P14 and adult Nes-cre, ex7−/− animals compared to WT HC. Three measurements were averaged for each section examined. N = 6 sections for P14 and 10–11 sections for adult from 3 mice for each genotype. g) The number of TBR2+ cells was significantly reduced in the hilus and DG gc layer (Hi + gcl) and increased in molecular layer (ML) of the DG in P14 Nes-cre, ex7−/− HC compared to WT controls (Supplementary Fig. 2a), although the total number of TBR2+ cells was not significantly different between WT and mutant animals. N = 12–13 matching sections from 3 mice for each genotype. h) HMN collateral axon tracks stained with CalR show a bright signal and normal thickness in adult Nes-cre, ex7−/− HC. Two-tailed t-test was used to test statistical significance: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = not significant.
Counting the number of cells stacked in layered columns in the DG GCL, we found that the numbers of DG gc were indeed significantly lower compared to WT littermates in both P14 and adult animals (Fig. 3c). In addition, the GCL layered column structure was less well organized in P14 mutant animals than in WT, a phenotype that persisted into adulthood (data not shown). The reduced DG size thus appeared to be driven by both increased density in and a smaller total number of cells stacked in the GCL (Fig. 3a–c). The increased density of the DG gc could reflect deficient dendritic growth and branching, as reported in human ASD patient postmortem studies (Amaral et al. 2008) and as also noted for Emx1-cre, ex8−/− mice (Hori et al. 2020). On the other hand, the decreased total number of cells could reflect either a deficiency in the progenitor pool or a reduced survival of cells at later stages. Of note, in the 16Gso mutant HC we found increased packing density and reduced DG GCL thickness, but not an overall reduction in gc numbers (Weisner et al. 2019), suggesting reduced dendritic branching without a loss in gc precursors or committed immature DG gc (Table 2).
Likely related to both the abnormal cell packing and reduced cell numbers, we documented a clear decrease in the numbers of maturing/mature CalB+ DG gc relative to immature postmitotic calretinin-positive (CalR+) cells in the mutant DG GCL at P14 (Fig. 3, d and e), suggesting that the transition from the CalR+ to CalB+ stage was delayed or blocked in the mutant mice. A similar finding was also documented for 16Gso mutants (Weisner et al. 2019), but in contrast, P14 Emx1-cre, ex6−/− mice were reported to display normal relative numbers of immature and mature DG cells (Li et al. 2022) (Table 2). Additionally, in the Nes-cre, ex7−/− P14 HC, CalB+ neurons expressed the CalB protein at substantially reduced levels (Fig. 3e). As observed for Emx1-cre, ex6−/− mutants, the main suprapyramidal axon bundle emanating from the DG gc neurons was significantly reduced in thickness in both P14 animals and adults (Fig. 3f; Supplementary Fig. 1d) and the numbers of TBR2+ intermediate progenitors (INP) located ectopically outside the DG GCL in the ML was increased at P14 (Fig. 3g; Supplementary Fig. 1e) suggesting abnormal migration patterns for the INP pool (Table 2). Emx1-cre, ex15−/− mice, which ablate both AUTS2 isoforms, also showed significantly reduced overall numbers of TBR2+ INP (Castanza et al. 2021), but we did not observe that phenotype (Fig. 3g), suggesting that INP proliferation or survival, or the transition from proliferation to the postmitotic state, are functions that require the participation of AUTS2-s.
Effects on hippocampal cell types beyond the DG gc
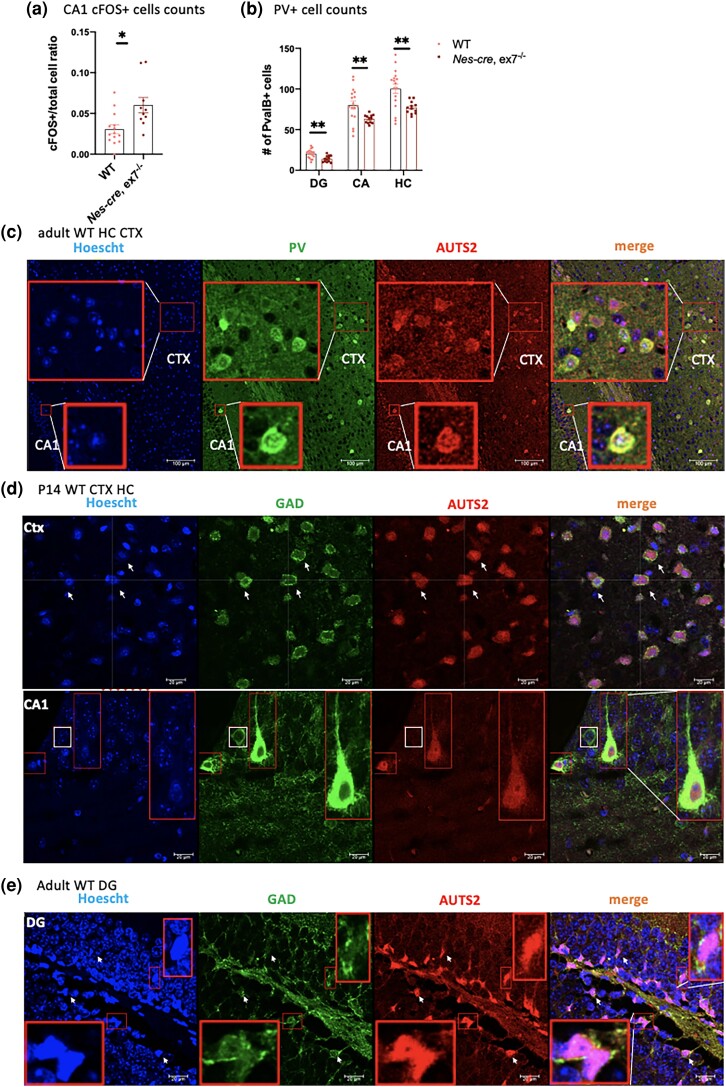
Looking beyond the DG gc abnormalities in hippocampus, we first examined hilar mossy neurons (HMNs), which were decreased in number with substantially reduced calretinin-positive (CalR+) axon tracts in Emx1-cre, ex15−/− animals (Castanza et al. 2021). Possibly related to the loss of HMN function (which is critical to hippocampal excitatory:inhibitory (E/I) balance (Scharfman and Myers 2012)), increased expression of the immediate early gene (IEG) cFOS was documented in cornu Ammonis 1 (CA1) neurons and in other 3′ Auts2 mutants, suggesting abnormal levels of excitation (Weisner et al. 2019; Hori et al. 2020; Castanza et al. 2021) (Table 2). However, in the Nes-cre, ex7−/− HC, we saw normal numbers of HMN and robust CalR+ axon tracts that did not appear different from those in WT littermates (Fig. 3h). Consistent with this finding, AUTS2 protein expression was significantly diminished in the Nes-cre, ex7−/−DG gc compared to WT mice, but remained robust in the HMN suggesting that AUTS2-s is highly expressed in those cells (Supplementary Fig. 1a). Given these observations, we were surprised to observe a significantly increased percentage of cFOS+ cells in the Nes-cre, ex7−/− CA1 region (Fig. 4a; Supplementary Fig. 1f, right panels) and looked for other potential causes of this signal of CA1 hyperactivation. Indeed, looking more closely at the hippocampal inhibitory circuit that includes HMNs, we found that adult mutant Nes-cre, ex7−/− DG and CA1 regions contained significantly fewer parvalbumin-expressing (PV+) interneurons than WT littermates (Fig. 4b; Supplementary Fig. 1f, left panels). AUTS2 is most widely expressed in excitatory cells, but several studies have shown that the gene/protein is also expressed in inhibitory interneurons across the brain, including both forebrain and cerebellum (Bedogni et al. 2010; Weisner et al. 2019; Yamashiro et al. 2020). Co-staining with AUTS2 and PV antibodies, we found that PV+ cells in the adult hippocampus co-expressed AUTS2 (Fig. 4c). PV first appears in cortical and hippocampal interneurons postnatally, and not all interneurons destined for PV+ fate will have made the transition by P14 (Alcántara et al. 1996). However, we also found some but not all GABAergic marker GAD67 expressing cells also co-expressed AUTS2 in P14 hippocampus and cortex (Fig. 4d). Furthermore, although AUTS2 expression drops to low levels or disappears in most neurons as animals mature (Bedogni et al. 2010), AUTS2 protein was strongly co-expressed with PV and GAD67 in some cells in P60 adults (Fig. 4, c and e). This finding is consistent with recent single nucleus sequencing data showing that AUTS2 transcripts are most highly expressed in interneurons within the adult motor cortex (Bakken et al. 2021, summarized in the human protein atlas https://www.proteinatlas.org/ENSG00000158321-AUTS2/single+cell+type). These results suggested that AUTS2-l LOF could directly impact the development, survival, and/or mature function of inhibitory cells.
Fig. 4.
Hippocampal PV+ interneurons express AUTS2 and are reduced in numbers in Nes-cre, ex7−/− mice. a) Quantification of cFOS+/total cell in CA1 of WT and Nes-cre, ex7−/− brain showed significantly increased cFOS+/total cell ratio in Nes-cre, ex7−/− HC compared to WT animals. N = 10–16 matching sections were examined from 3 mice for each genotype. b) Hippocampal PV+ cells were significantly reduced in number in DG and CA regions of Nes-cre, ex7−/− mutants compared to WT littermates, as were overall counts. N = 12–17 matching sections were examined from 3 mice for each genotype. c) PV+ cells showed abundant AUTS2 co-expression in adult animal HC and cortex (image showing somatosensory area). Red boxes show zoomed in view of indicated area in cortex (somatosensory) and CA1; scale bar = 100 μm. d) In P14 cortex (image showing parietal association area) and HC, some (arrows and zoomed images) but not all (box) GAD+ interneurons co-expressed AUTS2 expression at high levels. The crosshairs and arrows show examples of overlap of GAD+ AUTS2+ cells. Boxes show zoomed in view of indicated area; scale bar = 20μm. e) In adult DG, GAD+ cells showed abundant expression of AUTS2. Boxes show zoomed in view of indicated area; scale bar = 20μm. Two-tailed t-test was used to test statistical significance, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Brain-wide AUTS2-l LOF is associated with specific developmental and behavioral phenotypes
Along with brain pathologies, several dominant and recessive behavioral phenotypes have been associated with 3′ Auts2 alleles (Table 1); a subset of these phenotypes was found in Emx1-cre, ex6−/− mice. We tested ex7−/+ animals to identify dominant phenotypes together with the more robust Nes-cre, ex7 heterozygotes and homozygotes to identify both dominant and recessive phenotypes. As with pathology studies, we focused particularly on behavioral and developmental tests that have been applied to other Auts2 mutant mice to ask if these traits were recapitulated after isolated AUTS2-l LOF. This analysis identified a specific subset of AUTS2-l associated behaviors in the Nes-cre, ex7−/− animals. For example, we found NOR deficits, in the Nes-cre, ex7−/− mice, but only as a recessive trait (Fig. 5a, right panel; Fig. 6a), whereas NOR deficits are dominantly expressed in animals carrying 3′ alleles (Hori et al. 2015, 2020) (Table 1). Here we should also note that Emx1-cre, ex6−/− mice did not display NOR deficits (Li et al. 2022); in further contrast to Emx1-cre, ex6−/− mice, we found reduced sociability in Nes-cre, ex7−/− animals (Fig. 5b, left panel; Table 1) but not deficits in social memory (recognizing a familiar mouse) (Fig. 5b, right panel; Table 1). Furthermore, ex7−/+ animals did not display either sociability or social memory deficits, suggesting reduced sociability was presented as recessive phenotype only (Fig. 6b; Table 1).
Fig. 5.
Developmental and behavioral characterization of Nes-cre, ex7−/− mutants. a) Novel object recognition test showed that Nes-cre, ex7−/− animals displayed increased exploratory activity (left panel) and impaired cognitive memory (right panel) compared to WT animals. N = 10–18 animals for each genotype. b) Three-chamber test (see Methods) showed that Nes-cre, ex7−/− animals displayed decreased sociability (left panel) but normal social memory (right panel) compared to WT animals. N = 9–18 animals for each genotype. One outlier was excluded from Nes-cre, ex7−/− group through Grubb's test for both sociability and social memory measurements, with G = 2.543 and 2.372, respectively, at alpha = 0.05. c) Developmental delay tests of righting reflex and eye-opening (EOP) scores (see Methods). Nes-cre, ex7−/− mice showed significantly reduced scores in both righting reflex and EOP tests compared to WT mice and Nes-cre, ex7−/+ animals, but not Nes-cre, ex7−/+ to WT mice. WT N = 44–45; Nes-cre, ex7−/+ N = 21–24; Nes-cre, ex7−/− N = 14. d) Body weight measurements at different developmental time points. P5-adult (P42–P49) animals were measured for body weight. WT N = 14–47; Nes-cre, ex7−/+ N = 6–25; Nes-cre, ex7−/− N = 10–14; & = statistical significance between WT and Nes-cre, ex7−/+ comparisons, #statistical significance between Nes-cre, ex7−/+ and Nes-cre, ex7−/− comparisons; *statistical significance between WT and Nes-cre, ex7−/− comparisons. e) Stereotypic behavior was measured with grooming time per episode and not significantly different in WT and Nes-cre, ex7−/− animals. N = 12 for each genotype. f) Horizontal activity was significantly increased in Nes-cre, ex7−/− animals compared to WT controls. N = 11–15 for each genotype. Two-tailed unpaired t-test was used to test statistical significance, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns= not significant.
Fig. 6.
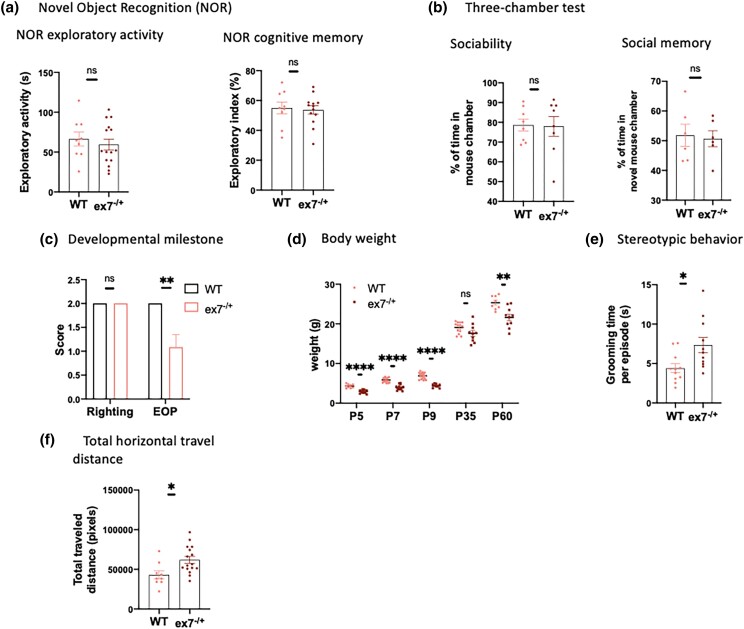
Behavioral characterization of Cmv-cre, ex7−/+ animals, abbreviated as ex7−/+ in figures. a) Novel object recognition test showed that ex7−/+ animals displayed normal exploratory activity (left panel) and cognitive memory (right panel) compared to WT animals. N = 9–16 for exploratory activity and 9–13 for cognitive memory for each genotype. One outlier was excluded from ex7−/+ group in exploratory activity test with Grubb's test, G = 2.903 at alpha = 0.05. b) Three-chamber test (see Methods) showed that ex7−/+ animals displayed normal sociability (left panel) and social memory (right panel) compared to WT animals. N = 8 for sociability and 6 for social memories for each genotype. c) Developmental delay tests of righting reflex and eye-opening (EOP) scores (see Methods). Ex7−/+ mice showed normal establishment of righting reflex but reduced scores in EOP test compared to WT mice N = 10–14 for righting reflex measurement and 12–13 for EOP test for each genotype. d) Body weight measurements at different developmental time points. P5–P60 animals were measured for body weight. WT N = 8–17; ex7−/+ N = 9–15. e) Stereotypic behavior was measured with grooming time per episode and significantly increased in ex7−/+ animals. N = 11 for each genotype. f) Horizontal activity was significantly increased in ex7−/+ animals compared to WT controls. N = 9–16 for each genotype. Two-tailed t-test was used to determine statistical significance; *P < 0.05, **P < 0.01, ****P < 0.0001, ns = not significant (P > 0.05).
We found that ex7−/+ (Fig. 6c) but not Nes-cre, ex7−/+ (Fig. 5c) mice showed a dominant delay in eye opening time but not in establishing righting reflex, the 2 major developmental milestones examined (Table 1). Other dominant phenotypes included reduced postnatal growth (both ex7−/+ (Fig. 6d) and Nes-cre, ex7−/+ heterozygotes) (Figs. 5d and 6d), and excessive stereotypic grooming behaviors [in ex7−/+ mice (Fig. 6e), but not in Nes-cre, ex7−/− (Fig. 5e) animals]. Noting a phenotype that has not been associated with Auts2 cKO of any type in the past, we quantified total traveling distance in the open field test and confirmed that both heterozygous ex7−/+ (Fig. 6f) and homozygous Nes-cre, ex7−/− animals (Fig. 5f) were significantly more active than WT littermates (Figs. 5f and 6f). This hyperactive phenotype stood in stark contrast to the reduced activity levels reported for 3′ Auts2 mutant mice (Hori et al. 2015, 2020) (Table 1). The results suggested that AUTS2-l LOF contributes centrally to a hyperactive phenotype, but also suggested that AUTS2-s might modulate the expression of this trait.
Targeting AUTS2-l LOF to the Calb1-lineage yields specific subsets of AUTS2-linked phenotypes
Published data suggest a role for AUTS2-l in neuron migration and maturation, functions that could be executed in both excitatory and inhibitory neurons. Both maturing/mature DG gc and several types of hippocampal interneurons, including PV+ cells, express CalB a least transiently during their development, (Alcántara et al. 1996); we therefore reasoned that these hippocampal cell populations could be targeted using the Calb1-cre allele (Daigle et al. 2018). AUTS2-s is expressed as the dominant isoform in throughout pre- and early postnatal brain development, but AUTS2-l is also detected in the embryonic brain, persisting at lower levels into adulthood (Yamashiro et al. 2020; Castanza et al. 2021; Liu et al. 2021; Monderer-Rothkoff et al. 2021). We were particularly interested in AUTS2-l expression in DG gc and PV+ interneurons at P14 because developmental abnormalities were detected in Nes-cre, ex7−/− mutants in those cells at that stage.
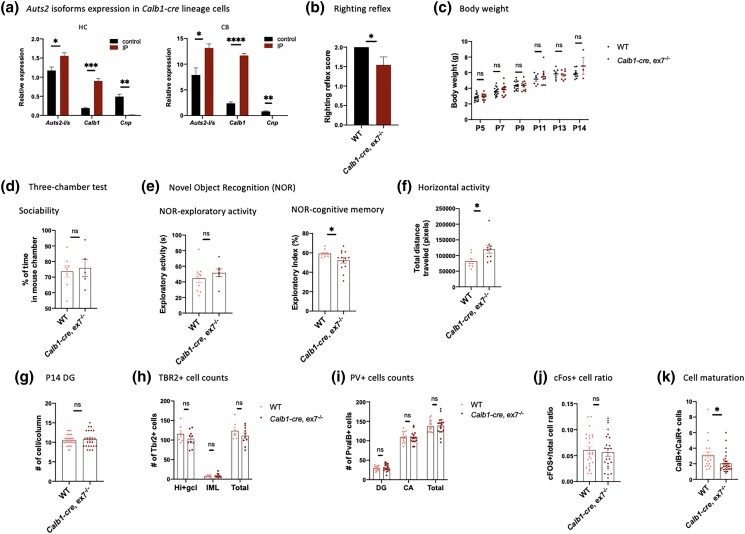
We confirmed isoform expression in the P14 CalB+ cell lineages by crossing Calb1-cre mice with mice carrying the RiboTag allele (Sanz et al. 2009) and isolating ribosome-associated RNA from CalB-lineage cells by immunoprecipitation (IP). Since cerebellar PCs are also of the CalB-lineage, we used this opportunity to test isoform expression in P14 PC by collecting RNA from both HC and CB of the P14 mice. We isolated ribosome-associated (RiboTagged) RNA from both brain regions by IP and used qRT-PCR to confirm transcript expression. First, we confirmed the robust enrichment of Calb1 transcripts in the RiboTagged RNA, as well as the depletion of transcripts for Cnp, a marker of oligodendrocytes (Fig. 7a). Next, we tested for relative levels of AUTS2-l and AUTS2-s transcripts. In unselected P14 HC RNA, AUTS2-s was in highest abundance overall, but the AUTS2-l/AUTS2-s ratio was significantly higher in the IP-enriched compared to the unselected RNA (Fig. 7a, left panel). In contrast, in P14 CB RNA, AUTS2-l transcripts were highly expressed in the unselected samples and were further enriched in the IP-enriched RNA (Fig. 7a, right panel). Therefore, in P14 HC and CB, AUTS2-l transcripts are expressed and relatively enriched in cells of the CalB+ lineage. The data indicated that AUTS2-l could play an important role in phenotypes expressed in those cells, at least in those associated with postnatal developmental events.
Fig. 7.
Auts2 expression in Calb1-cre lineage cells and characterization of Calb1-cre, ex7−/− mutant. a) RT-qPCR showing Auts2 isoforms expression in Calb1-cre lineage cells. Auts2 long/short isoform ratio was significantly higher in RiboTag immunoprecipitated (IP) samples compared to unselected samples from both Hippocampus (HC, left panel) and Cerebellum (CB, right panel), and enrichment of Calb1-cre lineage cells was verified with enrichment of Calb1 transcripts and ablation of Cnp transcripts in IP samples compared to unselected samples. N = 3–6 animals for each group and 2 animals were pooled for each IP experiments. b) Developmental delay test of righting reflex showed significantly reduced test scores in Calb1-cre, ex7−/− animals compared to WT controls. N = 11–13 animals for each genotype. c) Body weight measurements at different developmental time points. P5–P14 animals were measured for body weight. WT N = 7–15, Calb1-cre, ex7−/− N = 3–13. d) Three-chamber test showed normal sociability in Calb1-cre, ex7−/− animals compared to WT control animals. N = 5–8 animals for each genotype. e) Novel object recognition test showed normal exploratory activity (left panel) but impaired cognitive memory (right panel) in Calb1-cre, ex7−/− animals compared to WT controls. N = 7–10 for exploratory activity and 10–13 for cognitive memory for each genotype. One outlier was excluded from WT in cognitive memory test with Grubbs’ test, G = 2.434 at alpha = 0.05. f) Horizontal activity was significantly increased in Calb1-cre, ex7−/− animals compared to WT controls. N = 9–16 for each genotype. Calb1-cre, ex7−/− mice showed significantly increased in total horizontal travel distance compared to WT mice. N = 8–10 animals for each genotype. g) The total number of cells per column in the DG was not significantly different in P14 Calb1-cre, ex7−/− animals compared to WT controls. N = 24–28 counts on matching sections from 3 mice for each genotype. h) The number of TBR2+ cells if not significantly differentially distributed between the hilus together with DG gc layer (Hi + gcl) and the molecular layer (ML) of the DG in P14 Calb1-cre, ex7−/− HC compared to WT controls (Supplementary Fig. 2), and the total number of TBR2+ cells was not significantly different between WT and mutant animals. N = 7–10 counts on matching sections from 3 mice for each genotype. i) Hippocampal PV+ cells were not significantly different in numbers in different regions of HC in Calb1-cre, ex7−/− and WT animals. N = 16–17 counts of matching sections from 3 mice for each genotype. j) Quantification of cFOS+/total cell in CA1 of WT and Calb1-cre, ex7−/− brain showed significantly increased cFOS+/total cell ratio in Calb1-cre, ex7−/− HC compared to WT. N = 24–26 areas on matching sections were examined from 3 mice for each genotype. k) The ratio of CalB+/CalR+ cells was significantly decreased in Calb1-cre, ex7−/− mutants HC DG compared to WT. N = 21–33 counts of matching sections from 3 mice for each genotype. Two-tailed t-test was used to test statistical significance, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = not significant.
To test this hypothesis, we generated Calb1-cre, ex7−/− animals and examined morphology, behavior, and brain pathology in comparison to WT littermates. Young Calb1-cre, ex7−/− mice showed delayed establishment of the righting reflex (Fig. 7b), but they were not smaller than WT littermates (Fig. 7c) in contrast to animals with germline or brain-wide ex7 cKO, or to Emx1-cre, ex6−/− mice (Li et al. 2022). Furthermore, Calb1-cre, ex7−/− mice showed no difference from WT littermates in terms of social preference (Fig. 7d), or levels of exploratory activity (Fig. 7e, left panel). On the other hand, they displayed clear deficits in NOR (Fig. 7e, right panel) and hyperactivity in the open field test compared to WT littermates (Fig. 7f). Therefore, the ablation of AUTS2-l in CalB+ cells alone was sufficient to recapitulate the cognitive deficits and hyperactivity seen in brain-wide AUTS2-l KO animals, but not reduced social interaction or other key mutant phenotypes. Because we did not observe dramatic cerebellar pathology in whole brain Nes-cre, ex7−/− animals, we focused on examining HC pathology in Calb1-cre, ex7−/− animals. Looking at brain pathology, we found that in contrast to other Auts2 mutants including 3′ mutants, Nes-cre, ex7−/− and Emx1-cre, ex6−/− animals, the hippocampus of Calb1-cre, ex7−/− mice was not smaller than that of normal littermates, and the total numbers of cells in the DG GCL was not reduced (Fig. 7g). This finding may be related to the fact that TBR2+ cells in Calb1-cre, ex7−/− hippocampus were of normal numbers and were distributed normally rather than being ectopically positioned (Fig. 7h; Supplementary Fig. 2a), as they were in Nes-cre, ex7−/− mice. Furthermore, we found that PV+ cell counts were not decreased in the Calb1-cre mutants compared to littermate controls (Fig. 7i; Supplementary Fig. 2b, left panels). To ask whether there might still be a malfunction in hippocampal inhibitory circuits signaled by hyperexcitation, we tested c-Fos expression in Calb-cre, ex7−/− mice but found no difference between the mutants and WT littermate controls (Fig. 7j; Supplementary Fig. 2b, right panels). On the other hand, the ratio of CalB+/CalR+ neurons in the DG GCL was significantly lower in the Calb1-cre, ex7−/− mutants relative to WT littermates (Fig. 7k; Supplementary Fig. 2c). This finding indicated that the delay or failure of DG gc to fully mature was driven by AUTS2-l LOF within the DG gc themselves, and that this block was centrally related to expression of NOR deficits in these animals.
AUTS2-l LOF leads to disturbed expression of neurodevelopmental genes
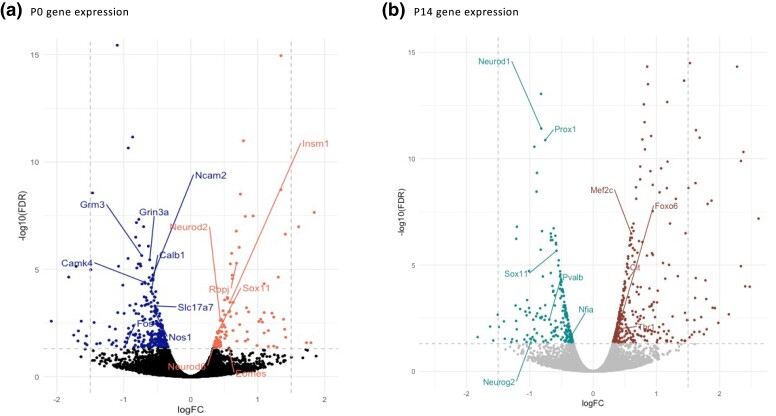
AUTS2 has been implicated in chromatin remodeling, regulation of transcription and RNA binding/stability, all of which can be predicted to impact RNA transcription. Indeed, Auts2 chromatin-binding assays (Gao et al. 2014; Oksenberg et al. 2014) and transcriptomic analyses involving Auts2 mutants of various types (Weisner et al. 2019; Hori et al. 2020; Castanza et al. 2021; Liu et al. 2021; Li et al. 2022) have confirmed a role in regulating the expression of neurodevelopmental genes. However, transcriptomic changes have not been examined after brain-wide AUTS2-l LOF in vivo. With the goal of linking transcriptomic changes due to AUTS2-l LOF to behavioral and brain pathologies, we carried out RNA-seq comparing gene expression in P14 HC (Nes-cre, ex7−/−) and WT littermates to correlate with the observed pathology. To gain clues to the trajectory of this pathology we also examined gene expression in FB samples (from ex7−/− and WT littermates), at P0, a time when DG gc precursors are actively migrating into the hippocampal formation (Mathews et al. 2010) and AUTS2-l is relatively highly expressed in the forebrain (Liu et al. 2021). Hundreds of DEGs were identified in both sample sets, although P0 and P14 DEGs showed relatively little overlap: only 56 genes were commonly detected among the 397 P0 and 546 P14 DEGs. Furthermore, more than half of these overlapping DEGs (31 genes) were oppositely expressed at P0 and P14 (Supplementary Table 2, Supplementary Fig. 3; some examples discussed in the text are highlighted in Fig. 8). DEGs at both time points were strongly enriched in shared functional categories/pathways related to neuron differentiation and function. However, we also observed enrichment of several categories that were unique to each time point and some shared categories that were oppositely up- or down-regulated at the 2 stages (Table 3).
Fig. 8.
Gene expression volcano plot of RNA-seq data from P0 FB and P14 HC ex7−/− compared to WT controls. a) P0 gene expression, left = down-regulated genes, right= up-regulated genes, bottom of each figure= not differentially expressed; b) P14 gene expression, left = down-regulated genes, right = up-regulated genes, bottom = not differentially expressed. For both P0 and P14 plots, view is zoomed to regions with −log10 (FDR) between 0 and 15, and log FC between −2.5 and 2.5. Dashed lines represent log FC = −1.5, 1.5, and FDR = 0.05; plots with all genes are in Supplementary Fig. 3 and gene expression data are fully presented in Supplementary Table 1. Selected genes discussed in the main text were highlighted in the volcano plot.
Table 3.
Functional category enrichments for DEGs from Auts2-ex7−/− P0 forebrain and P14 hippocampus.
| Enrichment −logP | |||||
|---|---|---|---|---|---|
| ex7−/− | Nes-cre, ex7−/− | ||||
| P0 FB | P14HC | ||||
| Functional category | up | down | up | down | |
| GOBP | Positive regulation of transcription by RNA polymerase II | 10 | - | - | - |
| GOBP | Pallium development | 10 | - | - | - |
| MGI | Decreased brain size | 5.25 | - | - | - |
| GOBP | Forebrain development | 10 | - | 10 | 4.97 |
| GOBP | Axonogenesis | 10 | - | 10 | - |
| GOBP | Synapse organization | 10 | - | 10 | - |
| GOBP | Synapse organization | 10 | - | 10 | - |
| MGI | Abnormal motor learning | 10 | - | 10 | - |
| GOBP | Calcium-ion regulated exocytosis | 5.82 | - | 10 | - |
| GOBP | chemotaxis | 5.85 | - | - | 5.32 |
| GOMF | Glutamate receptor activity | - | 10 | - | - |
| GOBP | Positive regulation of synaptic transmission | - | 10 | - | - |
| GOBP | Actin filament-based process | - | 10 | - | - |
| GOCC | Postsynaptic density | - | 10 | - | - |
| MGI | Abnormal emotion/affect behavior | - | 10 | - | - |
| MGI | Abnormal excitatory postsynaptic currents | - | 10 | - | - |
| PATH | KEGG_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | - | 10 | - | - |
| MGI | Abnormal brain interneuron morphology | - | 5.09 | - | - |
| MGI | Abnormal cognition | - | 10 | 10 | - |
| GOMF | Ion channel activity | - | 10 | 5.2 | - |
| PATH | KEGG_CALCIUM_SIGNALING_PATHWAY | - | 10 | 5.2 | |
| GOCC | Postsynaptic membrane | - | 10 | 4.56 | - |
| GOBP | Secretion by cell | - | 10 | 5.78 | - |
| GOMF | Gated channel activity | - | 4.66 | 4.52 | - |
| GOCC | Dendritic shaft | - | 4.63 | 4.22 | - |
| GOBP | Regulation of neurotransmitter transport | - | - | 10 | - |
| GOBP | Dendrite development | - | - | 10 | - |
| GOBP | Adenylate cyclase-activating G protein-coupled receptor signaling pathway | - | - | 10 | - |
| GOBP | Positive regulation of cell migration | - | - | - | 10 |
| GOBP | glial cell differentiation | - | - | - | 10 |
| GOBP | Regulation of neurogenesis | - | - | - | 10 |
| GOBP | Regulation of neuron differentiation | - | - | - | 10 |
| MGI | Abnormal dentate gyrus morphology | - | - | - | 10 |
| GOCC | Hippocampal mossy fiber to CA3 synapse | - | - | - | 10 |
| GOBP | Regulation of neurogenesis | - | - | - | 5.90 |
Functional enrichments were determined for up- and down-regulated FDR< 0.05 DEGs from comparisons of ex7−/− mutant P0 FB (forebrain) and Nes-cre, ex7−/− P14 HC (hippocampus) to WT littermates using the ToppCluster Suite (Kaimal et al. 2010). Representative and most descriptive terms for the most highly enriched categories are shown for each comparison, highlighting categories that are shared by, or differentially regulated in the P0 and P14 samples. In the ToppCluster output, all categories with −logP ≥ 10 are listed with a value of 10 (in columns 3–6). GOBP, GO biological process; GOMF, GO molecular function; GOCC, GO cellular component; PATH, pathways; MGI, mouse genome informatics Mouse Phenotypes.
Interested in understanding how this functional shift might be regulated, we focused on differentially expressed transcription factors (TF) and chromatin regulators and found genes with well-established roles in forebrain and hippocampal development. For example, “positive regulation of transcription by RNA polymerase II” and “pallium development” were 2 of the most highly enriched functional categories for P0 up-regulated DEGs (Table 3) including well known regulators such as Eomes (encoding TBR2), Insm1, Neurod2, Neurod6, and Notch-related TF, Rbpj (Fig. 8); up-regulation of categories regulated by these factors including forebrain development, axon formation and synaptogenesis was also observed. However, among the P0 up-/P14 down-regulated group was Sox11, which promotes proliferation and inhibits differentiation of hippocampal excitatory neuronal precursors at early stages (Wang et al 2013; Hoshiba et al. 2016; Liu et al. 2019), but later acts to promote hippocampal DG gc dendrite formation (Hoshiba et al. 2016; Abulaiti et al. 2022). Concordant with up-regulation of Sox11 expression in P0 FB, genes associated with mature neuron functions, especially neurotransmission and synaptic function (Nos1, Grm3, Grin3a, Slc17a7, Calb1, Camk4, Fos, Ncam2), were down-regulated in P0 FB (Fig. 8a).
In contrast, genes involved in mature neuron functions including ion channel activity, calcium signaling, neurotransmitter transport, and dendrite development were robustly up-regulated in the P14 hippocampus. A closer look revealed that the up-regulated genes in the latter category specialize in the development of synapses and dendritic spines (i.e. Foxo6, Mef2c, Srcin1, and Cit). P14 HC down-regulated genes included genes involved in DG gc differentiation and maturation (Lef1, Neurod1, Neurog2, Prox1, Zbtb20, Reln, Sox11), neurite differentiation and outgrowth (Bdnf, Ntf3, and Ngf) axon pathfinding (Sema3c, Sema5a, and Slit2) cell migration (Cxcl12, Lef1, Prox1), and glial cell differentiation (Cadm2, Fgf10, Nfia, Nfib, Plp1, Sox11) (Table 3; Supplementary Table 2). The Pvalb gene was also significantly down-regulated in P14 HC, consistent with the finding of reduced PV+ cell numbers. The data were also consistent with pathology including the reduced numbers of mature DG gc we observed in Nes-cre, ex7−/− hippocampus (Fig. 4e) and pointed to some possible molecular drivers of these pathological phenotypes.
Association of P0 and P14 DEGs with AUTS2 chromatin-binding peaks reveals hundreds of novel candidate AUTS2 target genes
AUTS2-l binds to chromatin and has been implicated in the transcriptional regulation of neurodevelopmental genes (Gao et al. 2014; Liu et al. 2021). Two AUTS2 chromatin immunoprecipitation (ChIP) datasets are publicly available, both of which were generated with antibodies directed at C-terminal amino acids shared by AUTS2-l and AUTS2-s, and targeting chromatin from embryonic day 16.5 forebrain (E16.5 FB) (Oksenberg et al. 2014) or P1 whole brain (Gao et al. 2014), respectively. However, the relationship between the 2 ChIP datasets has not been systematically examined, and the correlation between AUTS2 chromatin binding and mutant brain gene expression remains unclear. To understand how the isolated loss of AUTS2-1 might impact gene expression in mutant brains, we looked for overlaps between DEGs and AUTS2 binding peaks after reanalyzing both datasets (Supplementary Tables 3 and 4). We found a significant overlap between the 2 ChIP datasets (775 directly overlapping peaks of approximately 5000 peaks total for each set, log P = 1E−1611, hypergeometric test). However, most peaks in each dataset mapped to unique positions, suggesting that AUTS2 DNA binding patterns could depend on developmental stage and/or brain region. Examining associations between DEGs from P0 FB and P14 hippocampus of Nes-cre, ex7−/− mice and peaks from each dataset, we found a significant enrichment of ChIP peaks of both types located in regions that were either directly flanking or within P0 or P14 DEGs (Table 4). The highest correlation between peaks and DEGs at P0 involved down-regulated DEGs, consistent with the idea that AUTS2-l serves as a component of an activating complex. On the other hand, in P14 hippocampus, up-regulated DEGs were more likely to be associated with AUTS2 peaks (Table 4). In these and other respects, the 2 AUTS2 ChIP peak sets gave qualitatively similar results; because of the stronger association with DEGs, we will focus the following discussion on overlaps with the E16.5 dataset.
Table 4.
Hypergeometric P-values for overlaps between published AUTS2 ChIP peaks and DEGs.
| DEG set | E16.5 FBa | # DEGs | P1 whole brainb | # DEGs | E16.5/P1 overlap | # DEGs |
|---|---|---|---|---|---|---|
| P0 FB—all | 2.22E−27 | 100 | 5.66E−27 | 138 | 1.64E−05 | 26 |
| P0 FB—down | 9.92E−25 | 78 | 7.17E−20 | 98 | 1.09E−02 | 14 |
| P0 FB—up | 2.12E−05 | 22 | 3.66E−09 | 40 | 1.79E−05 | 12 |
| P14 HC—all | 5.97E−49 | 156 | 7.64E−38 | 193 | 2.90E−02 | 22 |
| P14 HC—down | 1.11E−14 | 51 | 2.47E−09 | 60 | 5.71E−02 | 9 |
| P14 HC—up | 8.08E−37 | 105 | 1.14E−31 | 133 | 8.79E−02 | 13 |
a From Oksenberg et al. (2014).
b From Gao et al. (2014).
AUTS2 peaks nearest DEGs included promoter-proximal and more distally located intronic and intergenic peaks; most DEG-associated peaks were distally located. Examples included peaks within or flanking neurogenesis regulators Nfia and Sox11 that overlapped with ENCODE-validated long-range enhancers for those genes (Gorkin et al. 2020) (Table 5). Notably given this evidence that they are direct regulatory targets of AUTS2, Nfia and Sox11 were among the 31 DEGs that were oppositely regulated at P0 and P14 (see above). In addition to these genes, other DEGs at both time points contained distal E16.5 AUTS2 peaks within introns or in intergenic locations overlapping with validated DEG-linked enhancers (Supplementary Table 4; Table 5). For example, E16.5 AUTS2 peaks included a peak within the neighboring Alpk1 gene that is an ENCODE-validated distal enhancer for P14 down-regulated DEG Neurog2, 2 intergenic enhancers validated for up-regulated DEG Sox5, and intronic enhancers within both up- and down-regulated DEGs (Table 5).
Table 5.
Candidate AUTS2 regulatory targets, defined as P0 or P14 DEGs with E16.5 FB AUTS2 binding peaks located either within the genes, or flanking the DEGs and validated as DEG gene-linked enhancers.
| P0 FB | P14 HC, CONTINUED | ||||
|---|---|---|---|---|---|
| Gene | Up/down | Peak typea | Gene | Up/down | Peak typea |
| 2610507B11Rik | Down | pr | Sema5ab | in+ (v) | |
| Ablim1 | Down | in | Sgpp2 | in | |
| Arap2 | Down | in+ | Slc2a13 | in+ | |
| Arrdc3 | Down | pr | Slc7a11 | Down | in |
| Atp2c1 | Down | pr | Slit2 | Down | in |
| Brinp3b | Down | in+ | Sox11 | Down | ig (v) |
| Btaf1b | Down | pr | Stxbp6 | Down | in |
| Cap2 | Down | in | Tanc1 | Down | in |
| Car10 | Down | in | Vgll3 | Down | in+ |
| Ccdc85a | Down | in | Zbtb20b | Down | in+ |
| Cntn1 | Down | in | Adora2ab | Up | ig+ (v) |
| Cntnap5ab | Down | in | Agap2 | Up | in+ |
| Cpne2 | Down | pr | Arnt2b | Up | in+ |
| Edardd | Down | ig (v) | Arpp21 | Up | In |
| Fgf12 | Down | in+ | Asic1 | Up | ig (v) |
| Glra3 | Down | in | Atp2b4 | Up | in |
| Grm1 | Down | in+ | Brinp3b | Up | in+ |
| Hspa8 | Down | pr | Cacna2d2 | Up | in |
| Ncam2 | Down | in | Camk2d | Up | in+ (v) |
| Nos1 | Down | in+ | Car10 | Up | in |
| Nsf | Down | pr | Ccdc187 | Up | in |
| Ntng1b | Down | in | Cdc42bpb | Up | pr |
| Pomc | Down | ex | Cdh18 | Up | in+ |
| Ppm1h | Down | in | Chn2 | Up | in (v) |
| Rasgrf1 | Down | in | Cicb | Up | pr,in |
| Robo1 | Down | in+ (v) | Cobl | Up | in (v) |
| Scoc | Down | pr | Dok5 | Up | in |
| Smap1 | Down | utr | Dpp10 | Up | in+ |
| Stxbp6 | Down | in | Dpysl5 | Up | in |
| Syt1b | Down | in | Dzip1 | Up | pr |
| Syt17 | Down | in | Fign | Up | in |
| Xkr4 | Down | pr | Gap43 | Up | in |
| Cdc42ep4 | Up | pr | Garnl3 | Up | in |
| Clk4 | Up | in | Grik3b | Up | in+ (v) |
| Hmga1 | Up | pr | Gsgl1 | Up | in+ |
| Lars2 | Up | in | Ipcef1 | Up | in |
| Lpin1 | Up | pr,in | Kcnk9 | Up | ig (v) |
| Mtcl1 | Up | pr | Kif21b | Up | in |
| Polr1d | Up | in | Lasp1 | Up | pr |
| Slc7a11 | Up | in | Ldb2 | Up | in+ |
| Sox11 | Up | ig+ (v) | Lingo1 | Up | in+ (v) |
| Tmem242 | Up | pr | Nav3b | Up | in |
| Ttc28 | Up | in | Nrg1 | Up | in+ |
| P14 HC | Pbx3 | Up | in | ||
| Bdnf | Down | in (v) | Pitpnm3 | Up | in |
| Bhlhe22 | Down | pr,ig (v) | Ptgfrn | Up | in |
| Cadm2b | Down | in+ | Satb2b | Up | in |
| Ccdc85a | Down | in | Scn5a | Up | in |
| Dgkb | Down | in+ | Scoc | Up | in |
| Dgkh | Down | pr | Sema3a | Up | in |
| Laptm4b | Down | in | Sez6 | Up | in (v) |
| Lars | Down | in | Sgcz | Up | in+ |
| Nfiab | Down | in+ (v) | Sox5b | Up | ig+ (v) |
| Pdgfrl | Down | in+ | Sulf2 | Up | in |
| Ptpro | Down | in (v) | Tiam2 | Up | in+ (v) |
| Relnb | Down | in+ | Tmem132d | Up | in |
| Rnf128 | Down | in | Tox2 | Up | in+ |
| Tshz2 | Up | in | |||
| Xylt1 | Up | in |
a Peak location; in, single intron; in+, multiple introns; ig, intergenic; pr, promoter region; (v), 1 or more peaks associated with the gene overlap with ENCODE-validated enhancers (Gorkin et al. 2020).
b These candidate target genes are confirmed ASD-related genes according to the SFARI gene database (https://gene.sfari.org).
Discussion
Data presented here associate the isolated loss of AUTS2-l with specific subsets of Auts2-linked behavioral, pathological, and transcriptomic phenotypes for the first time; together they reveal new information about the in vivo functions of the long isoform, and several new hints regarding short AUTS2 isoform functions as well. At the level of behavior, we found deficits in both cognitive memory and social interaction as recessive traits in Nes-cre, ex7−/− mice; these same deficits are expressed as dominant traits in animals carrying mutations in 3′ exons that are coding in both AUTS2 protein isoforms, indicating an important role for AUTS2-s in full expression of cognitive and social phenotypes. These findings are consistent with the relatively mild expression of AUTS2-syndrome phenotypes in human patients carrying 5′ variants including mutations within exons 6 and 7 (Beunders et al. 2013, 2015; Sengun et al. 2016), compared to the severe ID and ASD phenotypes associated with heterozygosity for 3′ alleles (Beunders et al. 2013, 2016; Liu et al. 2021). Also consistent with human patients, the mouse Auts2 exon 7 cKO mutants and 3′ mutant heterozygotes display some common dominant phenotypes highlighting an important role for AUTS2-l, including growth retardation and developmental delay. Additionally, we report 1 striking contrast between 5′ and 3′ Auts2 LOF alleles: exon 7 deletion—whether generated with germline or brain-wide cre alleles—is associated with a marked, dominant hyperactivity, whereas 3′ heterozygotes have been reported to display reduced activity levels (Hori et al. 2015).
ADHD has been proposed as a central AUTS2-syndrome phenotype (Sanchez-Jimeno et al. 2021; Biel et al. 2022), and the mechanisms that drive this distinction between Auts2 mutant alleles are thus of special interest. Although additional work will be required to solve this puzzle, we hypothesize that this striking difference could relate to the transient overexpression of AUTS2-s in the ex7−/− mutant neonatal brain. In this scenario, overexpression of AUTS2-s during late gestation or early postnatal life could drive the development of hyperactivity, with the reduced expression of AUTS2-s in 3′ mutants leading to the opposite phenotype. Considering how this pathological overexpression of AUTS2-s might be actuated, we note that the group reporting AUTS2 ChIP in E16.5 forebrain chromatin found multiple AUTS2 chromatin-binding sites within the Auts2 gene (Oksenberg et al. 2014); our remapped data in the mm10 genome build confirmed significant peaks in introns 2, 3, 4, 5, and 13 (Supplementary Table 4). These internal AUTS2 binding peaks were not detected in P1 whole brain ChIP (Gao et al. 2014), suggesting the possibility of a brain region-specific suppression of AUTS2-s encoding transcripts by AUTS2-l binding.
Published data have shown that the AUTS2 isoforms act sequentially during neurogenesis, with AUTS2-l arising after the initial expression of AUTS2-s in neuron progenitors both in vitro (Monderer-Rothkoff et al. 2021) and in vivo (Castanza et al. 2021; Liu et al. 2021) to instigate neuron maturation, neurite outgrowth and synaptogenesis (Hori et al. 2014, 2020). Based on our data and these previous observations, we hypothesize that AUTS2-l binding at 1 or more of the Auts2 intronic enhancers represses AUTS2-s expression in postmitotic neurons, initiating the transcriptomic cascade required to extend axons and dendrites and develop synaptic connections. Additional data will be required to support this hypothesis, but the finding that AUTS2-l LOF leads to AUTS2-s overexpression is an important clue to the timing, interactions, and the implementation of isoform functions across the developing brain. This finding may also have implications for human patients with 5′ mutations that ablate or impact function of AUTS2-l, some of which could possibly also drive aberrant AUTS2-s overexpression in the developing brain.
The neurodevelopmental pathologies we have documented here are also consistent with the notion that AUTS2 isoforms have distinct and possibly developmental stage-specific functions. For example, although AUTS2-l transcripts are highly expressed in postnatal cerebellum and enriched in postnatal Purkinje cells, the ablation of AUTS2-l, either in neuronal progenitors at E11 with the Nes-cre allele or in germline KO mice, resulted in subtle PC pathologies that were mostly corrected in adults, and not the much more dramatic effects on cerebellar and PC differentiation observed in 3′ Auts2 mutants (Yamashiro et al. 2020). These observations do not rule out PC functional abnormalities—for example, electrophysiological deficiencies—but indicate that the dramatic defects in cerebellar and PC development that have been documented for 3′ mutants (Weisner et al. 2019; Yamashiro et al. 2020) must depend on the activities of AUTS2-s.
In contrast, we found that AUTS2-l is essential to hippocampal development, with Nes-cre, ex7−/− mice displaying several key hippocampal pathologies documented for 3′ mutants (Hori et al. 2020; Castanza et al. 2021) and 16Gso mice (Weisner et al. 2019). These shared phenotypes included an overall reduction in hippocampal size and DG gc numbers (Weisner et al. 2019; Castanza et al. 2021; Li et al. 2022), ectopically positioned TBR2+ intermediate progenitors suggesting abnormal migration (Li et al. 2022), and the failed or stalled progression of DG gc from the immature CalR+ to the maturing CalB+ stage (Weisner et al. 2019) (Table 2). In addition to the lower numbers, we found that CalB levels were noticeably reduced in the mutant CalB+ cells; CalB is important for memory consolidation, intracellular calcium level moderation and neuronal protection (Karádi et al. 2012), and the reduced CalB expression levels suggest a functional deficiency in neurons that did advance to the CalB+ stage. Consistent with this collection of pathological findings, differential gene expression indicated delayed neuron maturation in mutant P0 forebrain and reduced expression of genes involved in axon, dendrite and synapse development in mutant hippocampus at P14.
Despite the evidence of delayed DG gc maturation, gene expression data from P14 hippocampus also indicated increased levels of neurotransmitter activity, supported by increased hippocampal c-Fos protein expression in P14 mutants and in adults. This suggested a failure of hippocampal inhibitory circuits although HMNs, which fail to differentiate or survive in Emx1-cre, ex15−/−animals (Castanza et al. 2021), appeared normal in Nes-cre, ex7−/−mice. These data indicate that AUTS2-s is likely required for the differentiation and survival of the HMNs. However, hippocampal inhibitory circuits were still defective in Nes-cre, ex7−/− mice despite HMN survival, correlated with reduced numbers of AUTS2-expressing PV+ hippocampal interneurons.
Loss or malfunction of hippocampal PV+ interneurons, which control not only hippocampal excitation but also DG gc differentiation during critical postnatal stages (Song et al. 2013), might have been considered as a major driver of AUTS2− associated hippocampal pathology and cognitive memory deficits. However, Calb1-cre, ex7−/− animals displayed full NOR deficiencies without the loss of PV+ interneurons or the correlated increase in hippocampal c-Fos expression. Furthermore, in Calb1-cre, ex7−/− mice, overall DG size and migration of TBR2+ intermediate progenitors were both normal, ruling out a required role for those pathologies in the cognitive defects. Rather, our data point to the delay or block in DG gc maturation—observed in both Nes-cre and Calb1-cre, ex7−/− animals—as being central to cognitive deficits in these mice. This delay is likely to reflect the loss of a critical, cell-intrinsic AUTS2-l function within the postmitotic progenitors, as they shift from the immature CalR+ stage to express AUTS2-l and CalB, begin to extend axons and dendrites, and establish synaptic connections that will persist into adulthood. The robust association between the Auts2 exon 7 mutant DEGs and published AUTS2 ChIP peaks suggests hundreds of putative direct AUTS2-l target genes, most of which are presented here with both differential expression and AUTS2 binding-site association for the first time. Of note, a number of the P0 or P14 DEGs that either contain AUTS2 ChIP peaks or are flanked by distal peaks that are validated DEG enhancers (Gorkin et al. 2020) are established ASD candidate genes (https://gene.sfari.org; (marked by asterisk in Table 5). Others, including Sox11 (Tsurusaki et al. 2014) are associated with rare syndromic disorders that include ASD as a core symptom, and additional ASD-associated genes are flanked by AUTS2 peaks that have not yet been validated as DEG-associated enhancers (i.e. Cntnap5, Eomes, Fezf2, Foxp1, Foxp2, Rfx3, Tle3, Tbr1, Tshz3, and Zfp462; Supplementary Tables 3 and 4). These and other putative direct AUTS2-l target genes have known functions that could together explain a host of the key behavioral and histopathological mutant phenotypes we have described in this report. Interestingly, although most AUTS2 peaks are positioned near gene promoters (Gao et al. 2014; Oksenberg et al. 2014; Liu et al. 2021), the majority of DEG-associated AUTS2 ChIP peaks were found at distal intronic or intergenic sites including the validated distal DEG-linked enhancers. These distal AUTS2 binding peaks were associated with a mixture of up- and down-regulated DEGs, including several that were oppositely regulated in P0 FB and P14 HC, suggesting that the effects of AUTS2-l binding could vary with brain region, cell type, and developmental time.
These data thus provide novel clues about the functions of both major AUTS2 isoforms, although many important questions remain to be answered. For example, although Auts2 mutant brain pathology is most pronounced in hippocampal dentate gyrus and cerebellar Purkinje cells, other brain regions and cell types express Auts2 at some time in their development, and likely contribute to mutant phenotypes in humans and mice. Therefore, a full view of behavioral and developmental pathologies will require analysis of isoform function in other brain regions and cell types across the developing brain. Furthermore, evidence suggests that the functions of AUTS2 isoforms are tightly coordinated and probably intertwined, but the biological function of AUTS2-s—the most widely and highly expressed of the 2 major Auts2 isoforms—is still mostly a matter of conjecture. Understanding the broad impact of AUTS2 on neurodevelopment and susceptibility to disease will require a deeper understanding the actions of both AUTS2 isoforms and their interactions within a range of neuronal cell types across the developing brain.
Supplementary Material
Acknowledgments
The authors are grateful to Jennifer Yoo, Derek Sargent, Andrew Look, Manasi Inamdar, and Kian Patton for expert assistance and Kian Patton, Xue Geng, and Stephanie Ceman for comments on the manuscript. We also thank Dr. Danny Reinberg and Dr. James Stafford for providing the Auts2tm1.1Dare floxed mice and for very helpful discussions and advice.
Contributor Information
Yunshu Song, Pacific Northwest Research Institute, Seattle WA 98122, USA; Department of Cell and Developmental Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Christopher H Seward, Pacific Northwest Research Institute, Seattle WA 98122, USA.
Chih-Ying Chen, Pacific Northwest Research Institute, Seattle WA 98122, USA.
Amber LeBlanc, Pacific Northwest Research Institute, Seattle WA 98122, USA.
Analise M Leddy, Pacific Northwest Research Institute, Seattle WA 98122, USA.
Lisa Stubbs, Pacific Northwest Research Institute, Seattle WA 98122, USA; Department of Cell and Developmental Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Data availability
Sequencing data described in this paper have been submitted to the GEO database under accession number GSE231599.
Supplemental material available at GENETICS online.
Funding
This study was funded by the National Institutes of Mental Health, grant MH114600 (awarded to L.S.) and by the generous support of the Pacific Northwest Research Institute.
Literature cited
- Abulaiti X, Wang A, Zhang H, Su H, Gao R, Chen J, Gao S, Li L. 2022. Disrupted mossy fiber connections from defective embryonic neurogenesis contribute to SOX11-associated schizophrenia. Cell Mol Life Sci. 79(3):180. doi: 10.1007/s00018-022-04206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcántara S, De Lecea L, Del Río JA, Ferrer I, Soriano E. 1996. Transient colocalization of parvalbumin and calbindin D28k in the postnatal cerebral cortex: evidence for a phenotypic shift in developing nonpyramidal neurons. Eur J Neurosci. 8(7):1329–1339. doi: 10.1111/j.1460-9568.1996.tb01595.x. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. 2008. Neuroanatomy of autism. Trends Neurosci. 31(3):137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Amarillo IE, Li WL, Li X, Vilain E, Kantarci S. 2014. De novo single exon deletion of AUTS2 in a patient with speech and language disorder: a review of disrupted AUTS2 and further evidence for its role in neurodevelopmental disorders. Am J Med Genet A. 164(4):958–965. doi: 10.1002/ajmg.a.36393. [DOI] [PubMed] [Google Scholar]
- Bakken TE, Jorstad NL, Hu Q, Lake BB, Tian W, Kalmbach BE, Crow M, Hodge RD, Krienen FM, Sorensen SA, et al. 2021. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature. 598(7879):111–119. doi: 10.1038/s41586-021-03465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Nelson BR, Frederick EA, Shiba N, Daza RA, Hevner RF. 2010. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr Patterns. 10(1):9–15. doi: 10.1016/j.gep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg DA, Su Y, Jimenez-Cyrus D, Patel A, Huang N, Morizet D, Lee S, Shah R, Ringeling FR, Jain R, et al. 2019. A common embryonic origin of stem cells drives developmental and adult neurogenesis. Cell. 177(3):654–668.e15. doi: 10.1016/j.cell.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beunders G, De Munnik SA, Van Der Aa N, Ceulemans B, Voorhoeve E, Groffen AJ, Nillesen WM, Meijers-Heijboer EJ, Frank Kooy R, Yntema HG, et al. 2015. Two male adults with pathogenic AUTS2 variants, including a two-base pair deletion, further delineate the AUTS2 syndrome. Eur J Hum Genet. 23(6):803–807. doi: 10.1038/ejhg.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beunders G, van de Kamp J, Vasudevan P, Morton J, Smets K, Kleefstra T, de Munnik SA, Schuurs-Hoeijmakers J, Ceulemans B, Zollino M, et al. 2016. A detailed clinical analysis of 13 patients with AUTS2 syndrome further delineates the phenotypic spectrum and underscores the behavioural phenotype. J Med Genet. 53(8):523–532. doi: 10.1136/jmedgenet-2015-103601. [DOI] [PubMed] [Google Scholar]
- Beunders G, Voorhoeve E, Golzio C, Pardo LM, Rosenfeld JA, Talkowski ME, Simonic I, Lionel AC, Vergult S, Pyatt RE, et al. 2013. Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am J Hum Genet. 92(2):210–220. doi: 10.1016/j.ajhg.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel A, Castanza AS, Rutherford R, Fair SR, Chifamba L, Wester JC, Hester ME, Hevner RF. 2022. AUTS2 syndrome: molecular mechanisms and model systems. Front Mol Neurosci. 15:858582. doi: 10.3389/fnmol.2022.858582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botton PH, Costa MS, Ardais AP, Mioranzza S, Souza DO, da Rocha JBT, Porciúncula LO. 2010. Caffeine prevents disruption of memory consolidation in the inhibitory avoidance and novel object recognition tasks by scopolamine in adult mice. Behav Brain Res. 214(2):254–259. doi: 10.1016/j.bbr.2010.05.034. [DOI] [PubMed] [Google Scholar]
- Brandt MD, Jessberger S, Steiner B, Kronenberg G, Reuter K, Bick-Sander A, Von Der Behrens W, Kempermann G. 2003. Transient calretinin expression defines early postmitotic step of neuronal differentiation in adult hippocampal neurogenesis of mice. Mol Cell Neurosci. 24(3):603–613. doi: 10.1016/S1044-7431(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Castanza AS, Ramirez S, Tripathi PP, Daza RAM, Kalume FK, Ramirez JM, Hevner RF. 2021. AUTS2 regulates RNA metabolism and dentate gyrus development in mice. Cerebral Cortex. 31(10):4808–4824. doi: 10.1093/cercor/bhab124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelhano-Carlos MJ, Sousa N, Ohl F, Baumans V. 2010. Identification methods in newborn C57BL/6 mice: a developmental and behavioural evaluation. Lab Anim. 44(2):88–103. doi: 10.1258/la.2009.009044. [DOI] [PubMed] [Google Scholar]
- Chen CY, Seward CH, Song Y, Inamdar M, Leddy AM, Zhang H, Yoo J, Kao WC, Pawlowski H, Stubbs LJ. 2022. Galnt17 loss-of-function leads to developmental delay and abnormal coordination, activity, and social interactions with cerebellar vermis pathology. Dev Biol. 490:155–171. doi: 10.1016/j.ydbio.2022.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Liao DL, Lai CH, Chen CH. 2013. Genetic analysis of AUTS2 as a susceptibility gene of heroin dependence. Drug Alcohol Depend. 128(3):238–242. doi: 10.1016/j.drugalcdep.2012.08.029. [DOI] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R, et al. 2018. A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell. 174(2):465–480.e22. doi: 10.1016/j.cell.2018.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Zhang Q, Zhu Y-S, Lu X-Y. 2014. The evidence for the contribution of the autism susceptibility candidate 2 (AUTS2) gene in heroin dependence susceptibility. J Mol Neurosci. 54(4):811–819. doi: 10.1007/s12031-014-0421-5. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics. 29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair SR, Schwind W, Julian DL, Biel A, Guo G, Rutherford R, Ramadesikan S, Westfall J, Miller KE, Kararoudi MN, et al. 2023. Cerebral organoids containing an AUTS2 missense variant model microcephaly. Brain. 146(1):387–404. doi: 10.1093/brain/awac244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Lee P, Stafford JM, Von Schimmelmann M, Schaefer A, Reinberg D. 2014. An AUTS2-polycomb complex activates gene expression in the CNS. Nature. 516 (7531):349–354. doi: 10.1038/nature13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Z, Wang Q, Miao W, Wolf T, Chavez J, Giddings E, Hobbs R, DeGraff DJ, Wang Y, Stafford J, et al. 2022. AUTS2 controls neuronal lineage choice through a novel PRC1-independent complex and BMP inhibition. Stem Cell Rev Rep. 19:531–549 doi: 10.1007/s12015-022-10459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkin DU, Barozzi I, Zhao Y, Zhang Y, Huang H, Lee AY, Li B, Chiou J, Wildberg A, Ding B, et al. 2020. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature. 583(7818):744–751. doi: 10.1038/s41586-020-2093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Nagai T, Shan W, Sakamoto A, Abe M, Yamazaki M, Sakimura K, Yamada K, Hoshino M. 2015. Heterozygous disruption of autism susceptibility candidate 2 causes impaired emotional control and cognitive memory. PLoS One. 10(12):1–13. doi: 10.1371/journal.pone.0145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Nagai T, Shan W, Sakamoto A, Taya S, Hashimoto R, Hayashi T, Abe M, Yamazaki M, Nakao K, et al. 2014. Cytoskeletal regulation by AUTS2 in neuronal migration and neuritogenesis. Cell Rep. 9(6):2166–2179. doi: 10.1016/j.celrep.2014.11.045. [DOI] [PubMed] [Google Scholar]
- Hori K, Yamashiro K, Nagai T, Shan W, Egusa SF, Shimaoka K, Kuniishi H, Sekiguchi M, Go Y, Tatsumoto S, et al. 2020. AUTS2 regulation of synapses for proper synaptic inputs and social communication. IScience. 23(6):101183. doi: 10.1016/j.isci.2020.101183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshiba Y, Toda T, Ebisu H, Wakimoto M, Yanagi S, Kawasaki H. 2016. Sox11 balances dendritic morphogenesis with neuronal migration in the developing cerebral cortex. J Neurosci. 36(21):5775–5784. doi: 10.1523/JNEUROSCI.3250-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. 2010. Toppcluster: a multiple gene list feature analyzer for comparative enrichment clustering and networkbased dissection of biological systems. Nucleic Acids Res. 38(Web Server):96–102. doi: 10.1093/nar/gkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karádi K, Janszky J, Gyimesi C, Horváth Z, Lucza T, Dóczi T, Kállai J, Ábrahám H. 2012. Correlation between calbindin expression in granule cells of the resected hippocampal dentate gyrus and verbal memory in temporal lobe epilepsy. Epilepsy Behav. 25(1):110–119. doi: 10.1016/j.yebeh.2012.06.007. [DOI] [PubMed] [Google Scholar]
- Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. 2000. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 14(11):1377–1389. doi: 10.1101/gad.14.11.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sun X, You Y, Li Q, Wei C, Zhao L, Sun M, Meng H, Zhang T, Yue W, et al. 2022. Auts2 deletion involves in DG hypoplasia and social recognition deficit: the developmental and neural circuit mechanisms. Sci Adv. 8(9):eabk1238. https://www.science.org. doi: 10.1126/sciadv.abk1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Hippenmeyer S, Troy Ghashghaei H. 2012. A Nestin-Cre transgenic mouse is insufficient for recombination in early embryonic neural progenitors. Biol Open. 1(12):1200–1203. doi: 10.1242/bio.20122287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P-P, Xu Y-J, Dai S-K, Du H-Z, Wang Y-Y, Li X-G, Teng Z-Q, Liu C-M. 2019. Polycomb protein EED regulates neuronal differentiation through targeting SOX11 in hippocampal dentate gyrus. Stem Cell Rep. 13(1):115–131. doi: 10.1016/j.stemcr.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Aldinger KA, Cheng CV, Kiyama T, Dave M, McNamara HK, Zhao W, Stafford JM, Descostes N, Lee P, et al. 2021. NRF1 association with AUTS2-polycomb mediates specific gene activation in the brain. Mol Cell. 81(22):4663–4676.e8. doi: 10.1016/j.molcel.2021.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mathews EA, Morgenstern NA, Piatti VC, Zhao C, Jessberger S, Schinder AF, Gage FH. 2010. A distinctive layering pattern of mouse dentate granule cells is generated by developmental and adult neurogenesis. J Comp Neurol. 518(22):4479–4490. doi: 10.1002/cne.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, Bethge M. 2018. Deeplabcut: markerless pose estimation of user-defined body parts with deep learning. Nat Neurosci. 21(9):1281–1289. doi: 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- Monderer-Rothkoff G, Tal N, Risman M, Shani O, Nissim-Rafinia M, Malki-Feldman L, Medvedeva V, Groszer M, Meshorer E, Shifman S. 2021. AUTS2 isoforms control neuronal differentiation. Mol Psychiatry. 26:666–681. doi: 10.1038/s41380-019-0409-1. [DOI] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, et al. 2012. BEDOPS: high-performance genomic feature operations. Bioinformatics. 28(14):1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola Z, Fabel K, Kempermann G. 2015. Development of the adult neurogenic niche in the hippocampus of mice. Front Neuroanat. 9:1–13. doi: 10.3389/fnana.2015.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg N, Haliburton GDE, Eckalbar WL, Oren I, Nishizaki S, Murphy K, Pollard KS, Birnbaum RY, Ahituv N. 2014. Genome-wide distribution of Auts2 binding localizes with active neurodevelopmental genes. Transl Psychiatry. 4(9):e431. doi: 10.1038/tp.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo P, Di Muro E, Accadia M, Benvenuto M, Di Giacomo MC, Castellana S, Mazza T, Castori M, Palumbo O, Carella M. 2021. Whole exome sequencing reveals a novel AUTS2 in-frame deletion in a boy with global developmental delay, absent speech, dysmorphic features, and cerebral anomalies. Genes (Basel). 12(2):1–10. doi: 10.3390/genes12020229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2009. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Jimeno C, Blanco-Kelly F, López-Grondona F, Pozo RL-D, Moreno B, Rodrigo-Moreno M, Martinez-Cayuelas E, Riveiro-Alvarez R, Fenollar-Cortés M, Ayuso C, et al. 2021. Attention deficit hyperactivity and autism spectrum disorders as the core symptoms of AUTS2 syndrome: description of five new patients and update of the frequency of manifestations and genotype–phenotype correlation. Genes (Basel). 12(9):9. doi: 10.3390/GENES12091360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, Stanley McKnight G, Amieux PS. 2009. Cell-type-specific isolation of ribosome-associated MRNA from complex tissues. Proc Natl Acad Sci U S A. 106(33):13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul MC, Seward CH, Troy JM, Zhang H, Sloofman LG, Lu X, Weisner PA, Caetano-Anolles D, Sun H, Zhao SD, et al. 2017. Transcriptional regulatory dynamics drive coordinated metabolic and neural response to social challenge in mice. Genome Res. 27(6):959–972. doi: 10.1101/gr.214221.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfman HE, Myers CE. 2012. Hilar mossy cells of the dentate gyrus: a historical perspective. Front Neural Circuits. 6:1–17. doi: 10.3389/fncir.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivières S, Aliev FA, Khan AA, Amin N, et al. 2011. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S Am. 108(17):7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron1 U, Rajewsky K. 1995. A cremtransgenic mouse strain for te ubiquitous deletion of ioxflanked gene segments including deletion in germ cells. Nucleic Acids Res. 23(24):, 5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengun E, Yararbas K, Kasakyan S, Alanay Y. 2016. AUTS2 syndrome in a 68-year-old female: natural history and further delineation of the phenotype. Am J Med Genet A. 170(12):3231–3236. doi: 10.1002/ajmg.a.37882. [DOI] [PubMed] [Google Scholar]
- Seward CH, Saul MC, Troy JM, Dibaeinia P, Zhang H, Sinha S, Stubbs LJ. 2022. An epigenomic shift in amygdala marks the transition to maternal behaviors in alloparenting virgin female mice. PLoS One. 17(2):e0263632. doi: 10.1371/journal.pone.0263632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Sun J, Moss J, Wen Z, Sun GJ, Hsu D, Zhong C, Davoudi H, Christian KM, Toni N, et al. 2013. Parvalbumin interneurons mediate neuronal circuitry-neurogenesis coupling in the adult hippocampus. Nat Neurosci. 16(12):1728–1730. doi: 10.1038/NN.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarov A, Joyner AL. 2007. Cerebellum morphogenesis: the foliation pattern is orchestrated by multi-cellular anchoring centers. Neural Dev. 2(1):1. doi: 10.1186/1749-8104-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R, Yu CE, Yu J, Munson J, Chen D, Hua W, Estes A, Cortes F, de la Barra F, Yu D, et al. 2002. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics. 80(2):129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schütz G, et al. 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 23(1):99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y, Koshimizu E, Ohashi H, Phadke S, Kou I, Shiina M, Suzuki T, Okamoto N, Imamura S, Yamashita M, et al. 2014. De novo SOX11 mutations cause Coffin–Siris syndrome. Nat Commun. 5:4011. doi: 10.1038/ncomms5011. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lin L, Lai H, Parada LF, Lei L. 2013. Transcription factor Sox11 is essential for both embryonic and adult neurogenesis. Dev Dyn. 242(6):638–653. doi: 10.1002/dvdy.23962. [DOI] [PubMed] [Google Scholar]
- Weisner PA, Chen C-Y, Sun Y, Yoo J, Kao W-C, Zhang H, Baltz ET, Troy JM, Stubbs L. 2019. A mouse mutation that dysregulates neighboring Galnt17 and Auts2 genes is associated with phenotypes related to the human AUTS2 syndrome. G3 (Bethesda). 9(11):3891–3906. doi: 10.1534/g3.119.400723. September, g3.400723.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. Ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag. [Google Scholar]
- Yamashiro K, Hori K, Lai ESK, Aoki R, Shimaoka K, Arimura N, Egusa SF, Sakamoto A, Abe M, Sakimura K, et al. 2020. AUTS2 governs cerebellar development, purkinje cell maturation, motor function and social communication. IScience. 23(12):101820. doi: 10.1016/j.isci.2020.101820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data described in this paper have been submitted to the GEO database under accession number GSE231599.
Supplemental material available at GENETICS online.