Abstract
Rhizobium meliloti can occupy at least two distinct ecological niches; it is found in the soil as a free-living saprophyte, and it also lives as a nitrogen-fixing intracellular symbiont in root nodules of alfalfa and related legumes. One approach to understanding how R. meliloti alters its physiology in order to become an integral part of a developing nodule is to identify and characterize genes that are differentially expressed by bacteria living inside nodules. We used a screen to identify genes under the control of the R. meliloti regulatory protein NodD3, SyrM, or SyrA. These regulatory proteins are expressed by bacteria growing inside the root nodule. One gene isolated in this screen was mapped to pSymB and displayed complex regulation. The gene was downregulated by the syrA gene product and also by glucose and succinate. This gene, referred to as agpA, encodes a periplasmic binding protein that is most similar to proteins from the periplasmic oligopeptide binding protein family. It is likely that AgpA binds α-galactosides, because α-galactosides induce the expression of agpA, and agpA mutants cannot utilize or transport these sugars. Activity of an agpA::TnphoA fusion was downregulated by SyrA. Because syrA is known to be expressed at high levels in intracellular symbiotic R. meliloti and at low levels in the free-living bacteria, we propose that AgpA may belong to the class of gene products whose expression decreases when R. meliloti becomes an intracellular symbiont.
Rhizobium meliloti is a gram-negative bacterium which can live as a saprophyte in soil or as a nitrogen-fixing symbiont inside root nodule cells of alfalfa and related legumes. The interactions leading to symbiosis begin when R. meliloti detects flavonoids and other compounds released by host plants and then induces bacterial genes required for the biosynthesis of lipooligosaccharide signaling molecules. These compounds initiate many of the physiological and morphogenic changes seen in the plant early in nodulation. These changes include root hair curling, depolarization of the root hair cell membrane, calcium oscillations in root hairs, and the initiation of cell division in the inner root cortex, which establishes a nodule primordium. An infection thread, which is a plant-derived tubule filled with dividing and growing bacteria, extends down through the root hair and then traverses several cell layers to deliver bacteria to root cells in the developing nodule. Bacteria exit the infection thread, differentiate, and fix atmospheric nitrogen, which is exported from the nodule to the rest of the plant (5, 6, 19, 29, 33, 39, 43, 47).
The R. meliloti nod genes, which are required for the synthesis of the lipooligosaccharide signaling molecules (Nod factors), are carried on a 1,500-kb plasmid, pSymA, and are upregulated by the LysR-type transcriptional activators, NodD1, NodD2, and NodD3. NodD1 and NodD2 become active transcriptional regulators in the presence of specific flavonoid inducers released from host plant roots and seeds (32, 36, 37). nodD3 is part of a cluster of three regulatory genes (nodD3, syrM, and syrA) present on pSymA about 15 kb downstream of nodD1 (32). NodD3 upregulates syrM, and SyrM upregulates nodD3; together they form a self-amplifying regulatory circuit which can activate the induction of the other nod genes (32, 45). The third gene, syrA, encodes a small, hydrophobic protein which upregulates the synthesis of exopolysaccharide (2, 32). Exopolysaccharide is required for R. meliloti to form nitrogen-fixing nodules (13, 23–25).
NodD3, SyrM, and SyrA are synthesized by bacteria inside alfalfa nodules (2, 42, 45). We asked whether these proteins might be responsible for regulating the synthesis of other genes needed for the establishment or maintenance of the nitrogen-fixing symbiosis. To define more fully the roles of NodD3, SyrM, and SyrA in nodulation, we developed a screen which allowed us to identify TnphoA insertions in genes regulated by these proteins. One gene, agpA (α-galactoside permease), isolated in this screen mapped to the symbiotic plasmid, pSymB, and displayed intriguing patterns of expression. agpA is similar to genes which encode periplasmic binding proteins of oligopeptide transport systems in other bacteria. This study describes the isolation and initial characterization of this gene.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and their relevant genotypes are listed in Table 1. R. meliloti was cultured at 30°C, and growth was monitored by measuring the optical density at 595 nm (OD595) of 100 μl of cell culture in a 96-well microtiter dish with a Bio-Rad 3550 plate reader.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| R. meliloti | ||
| Rm1021 | Str | 31 |
| Rm8002 | Str Pho− | 28 |
| SG1001 | agpA::TnphoA; derived from Rm8002 | This laboratory |
| E. coli | ||
| XL1 Blue | Used for cloning and sequencing | Stratagene |
| MM294a | pro28 thi-1 endA1 hsdR17; used to deliver pRK609 to R. meliloti | 28 |
| Plasmids | ||
| pLAFR1 | Tcr; broad-host-range cloning vector | 11 |
| pTE3 | Tcr; pLAFR1 with constitutively active S. typhimurium trp promoter | 7 |
| pRK609 | Nmr Smr; TnphoA | 28 |
| pRmJT5 | Tcr; carries 20-kb region of R. meliloti pSymA including the genes nodD3, syrM, and syrA in pLAFR1 | 44 |
| pMB89 | Derivative of pTE3 with syrA driven by ptrp | 2 |
| pRmE63 | Derivative of pTE3 with nodD3 driven by ptrp | 45 |
| pRmS73 | Derivative of pTE3 with syrM driven by ptrp | 45 |
Growth media.
Bacteria were grown on Luria broth (LB), M9 minimal medium with various carbon sources, or basal medium. Basal medium is M9 minimal medium containing Difco yeast extract at a concentration of 65 μg/ml. XP (5-bromo-4-chloro-3-indolylphosphate-p-toluidine) was used at a final concentration of 60 μg/ml.
Genetic screen for loci under the control of nodD3, syrM, or syrA.
R. meliloti Rm8002/pRmJT5 was randomly mutagenized with TnphoA (30) by introducing plasmid pRK609 via conjugation with Escherichia coli MM294a/pRK609 (28). The mating mix was diluted and plated on an indicator medium, LB plus streptomycin (500 μg/ml) plus neomycin (200 μg/ml) plus XP. The plasmid pRmJT5 overexpresses the regulatory genes nodD3, syrM (44), and syrA (2) and is unstably maintained in the absence of tetracycline selection. Therefore, TnphoA insertions into genes which were under the control of either of these three regulatory genes gave rise to sectored colonies when plated on the indicator medium described above. Colonies which showed sectors were purified on LB plus streptomycin (500 μg/ml) plus neomycin (200 μg/ml) plus tetracycline (10 μg/ml) and further characterized.
DNA sequencing.
DNA from the regions flanking the agpA::TnphoA insertion site was amplified by inverse PCR with primers which hybridized to the left end and center of TnphoA (5′-GCAGAGCGGCAGTCTGATCACCCGTTA and 5′-AAGGTCGCGCGCATTCCCGATGAA, respectively) (35). The PCR fragments were used to screen an R. meliloti lambda library constructed in the vector Lambda Fix II (48). DNA from positive lambda clones was subcloned and sequenced by standard methods.
Alkaline phosphatase assays.
Samples (0.1 to 0.5 ml each) were taken from growing cultures, pelleted, and resuspended in 0.5 ml of 1 M Tris-HCl, pH 8.0, and frozen at −80°C until all samples from an experiment had been taken. The samples were then thawed and permeabilized with a drop of chloroform and 50 μl of 0.1% sodium dodecyl sulfate, and 180 μl of this extract was added to 20 μl of a 4% solution of ortho-nitrophenylphosphate and placed in a 96-well microtiter dish. Controls containing 180 μl of sample extract and 20 μl of water were also placed in the dish. Hydrolysis of ortho-nitrophenylphosphate was monitored at 595 nm with a Bio-Rad 3550 plate reader. The OD595 of the samples was taken every 2 to 10 min, and each reading was corrected by subtracting the OD595 of the respective control at each time point. The alkaline phosphatase-specific activity for each sample was estimated by dividing the slope of the resultant curve (change in OD595 per minute) by the amount of cell material present in the control sample (OD595) at the first time point and multiplying the result by 1,000.
Transport assays.
Stationary-phase cells were harvested after 60 h, washed in M9 salts containing no added sugars, and resuspended to an OD595 of 1.0. A 0.5-ml portion of the cell suspension was added to a microcentrifuge tube containing 10 μl of 10 mM [3H]raffinose (0.1 mCi/ml). Samples (0.1 ml each) were removed every minute and filtered through a Millipore nitrocellulose filter (0.45-μm pore size). The filter was immediately rinsed three times with 5 ml of M9 salts containing no sugar and counted with a scintillation counter.
Isolation of periplasmic proteins.
Periplasmic proteins were isolated from R. meliloti by the chloroform shock method (1a). A 1.5-ml portion of an overnight culture of R. meliloti was centrifuged for 30 s in a microcentrifuge, and the supernatant was removed. Twenty microliters of chloroform was added to the cell pellet and vortexed briefly. Following 20 min of incubation at room temperature, the mixture was resuspended in 200 μl of 0.01 M Tris-HCl and immediately spun for 30 s in a microcentrifuge. The supernatant, which contained the periplasmic proteins, was removed, and 20 μl was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
RESULTS
A screen for genes under the control of nodD3, syrM, or syrA.
The regulatory genes nodD3, syrM, and syrA are closely linked on the symbiotic plasmid pSymA and are expressed by bacteria growing inside nodules. nodD3 is expressed by bacteria growing in the meristematic and infection zones of the nodule (45), and syrM and syrA are highly expressed in the older tissue, where bacteroids are differentiating and fixing nitrogen (2, 45). These expression patterns suggested that these loci may control genes involved either in establishing and maintaining the symbiosis or in fixing nitrogen. In order to isolate insertions in genes encoding membrane or extracellular proteins whose expression is regulated by nodD3, syrM, or syrA, an R. meliloti strain carrying the plasmid pRmJT5 (Tcr), which overproduces NodD3, SyrM, and SyrA (32, 44), was mutagenized with TnphoA. The TnphoA-mutagenized cells were plated on LB medium which contained XP and lacked tetracycline. Plasmid pRmJT5(Tcr) is unstable in the absence of tetracycline selection. Therefore, if the TnphoA present in a strain was upregulated or downregulated by the proteins overproduced on pRmJT5, then colonies plated on the nonselective indicator medium should generate sectors. These sectors would arise from a population of cells which lost pRmJT5 during colony development and therefore expressed the TnphoA fusion differently than neighboring cells in the colony which retained the plasmid. By this method, 62 independent strains were isolated. These strains gave rise to sectored colonies on nonselective plates and therefore contained insertions whose expression was dependent on the plasmid bearing nodD3, syrM, or syrA.
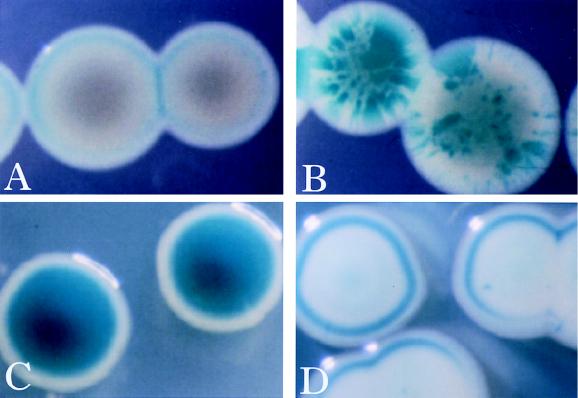
Strain SG1001/pRmJT5 displayed dramatic sectoring patterns when it was plated on nonselective indicator medium. The colonies were white without sectors when they were grown on selective medium but white with blue sectors when they were grown on nonselective indicator medium (Fig. 1A and B). These results indicated that the gene fused to TnphoA was downregulated by genes on plasmid pRmJT5 when it was present.
FIG. 1.
Colony phenotypes of strain SG1001 (agpA::TnphoA). Strain SG1001 (agpA::TnphoA)/pRmJT5 on LB XP indicator plates with (A) and without (B) tetracycline. Plasmid pRmJT5 is not stably maintained without tetracycline, and its loss allows the generation of sectors containing bacteria which express the agpA::TnphoA fusion. (C and D) Strain SG1001 (agpA::TnphoA) and strain SG1001 (agpA::TnphoA)/pMB89 on LB XP indicator plates, respectively. syrA is overexpressed from pMB89, resulting in downregulation of the agpA::TnphoA fusion.
The TnphoA insertion in strain SG1001 is downregulated by syrA.
pRmJT5 overproduces nodD3, syrM, and syrA because there are multiple copies of the plasmid per cell (2, 45). We determined which of these three genes affected the expression of the TnphoA insertion in strain SG1001 by introducing plasmids which overexpressed each of the three proteins. The overexpression of nodD3, syrM, or syrA was achieved by placing each gene downstream of a constitutively active trp promoter from Salmonella typhimurium (7). The plasmids which overproduced NodD3 and SyrM had minor effects on the expression of the agpA::TnphoA fusion (data not shown). However, the plasmid which overproduced SyrA strongly repressed expression of the fusion (Fig. 1C and D). The minor effects of the overexpression of nodD3 and syrM were probably due to the effects of the proteins on the expression of syrA (2, 32, 45).
Cells which overproduced syrA, either from pRmJT5 or from pMB89, formed white colonies which did not sector. However, such colonies were often ringed with blue near the periphery (Fig. 1A and D). Often, the rings formed when colonies were close to one another or close to the wall of the petri dish. This suggested that the rings arose in response to a local accumulation of a metabolic byproduct or to a local depletion of a substance from the growth medium. We were unable to differentiate between these two possibilities with preliminary studies (data not shown).
Strain SG1001 contains an insertion in a gene encoding a periplasmic binding protein.
Inverse PCR (35) was used to amplify DNA flanking the TnphoA insertion in strain SG1001. This DNA was then used to make a probe and map the insertion sites. Southern analysis indicated that the TnphoA insertion site in strain SG1001 was on the symbiotic megaplasmid pSymB (8).
Chromosomal fragments which flanked the TnphoA insertion in strain SG1001 were subcloned from a lambda library and sequenced. This revealed that the transposon had inserted into a gene encoding a 77-kDa protein similar to the periplasmic binding protein component of the oligopeptide family of permeases (46, 49) (Fig. 2A). Bacterial permease systems typically consist of a ligand binding protein which is in the periplasm of gram-negative bacteria or tethered to the outer face of the cytoplasmic membrane of gram-positive bacteria by a covalently attached lipid (12) together with two different transmembrane proteins and at least one protein capable of binding and hydrolyzing ATP. While the overall identity between AgpA and other members of the oligopeptide binding protein family from S. typhimurium is only about 15%, the identity in subregions of the protein is much higher. These highly similar subregions are diagnostic for periplasmic binding protein components of the oligopeptide permease family of transporters (46, 49).
FIG. 2.
(A) AgpA is similar to oligopeptide binding proteins of periplasmic transport systems. The deduced amino acid sequence of AgpA was compared to the sequences of other members of the oligopeptide binding protein family with the Pileup program of the Wisconsin Genetics Computer Group sequence analysis package, Clustal W and SeqVu. The peptides used in the lineup are R. meliloti AgpA (Rm AgpA), Agrobacterium tumefaciens agrocinopine transport protein AccA (At AccT) (17), B. subtilis oligopeptide transport protein Spo0K (Bs Spo0K) (41), E. coli dipeptide transport protein DppA (Ec DppA), E. coli nickel transport protein NikA (Ec NikA) (34), Streptococcus pneumoniae oligopeptide pheromone binding protein AmiA (Sp AmiA) (1), and S. typhimurium oligopeptide binding protein OppA (St OppA) (18). The small inverted triangle indicates the location of the PhoA fusion site in R. meliloti SG1001. (B) MelA from R. meliloti is similar to MelA, an α-galactosidase, of E. coli. The deduced amino acid sequence of R. meliloti MelA was compared to that of E. coli MelA with the Pileup program of the Wisconsin Genetics Computer Group sequence analysis package.
The agpA gene was preceded by an open reading frame which encodes a 55-kDa protein similar (31% identity) to the MelA α-galactosidase of E. coli (Fig. 2B) (27).
Regulation of agpA.
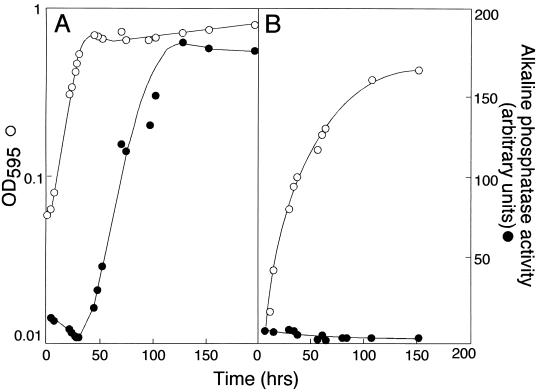
The agpA::TnphoA fusion is induced in the central part of colonies when cells are plated on LB medium (Fig. 1C). Because the growth of the bacteria in this part of the colony has slowed or stopped, we tested the possibility that the TnphoA fusion was induced as strain SG1001 entered stationary phase. We found that the agpA::TnphoA fusion was induced as the growth rate slowed at the end of the exponential phase when cells were in LB medium (Fig. 3A). However, the fusion was not induced when cells grown in M9 glucose minimal medium entered stationary phase (Fig. 3B). By growing cells in M9 salts containing various components of LB medium, we found that the agpA::TnphoA fusion was induced as cells entered stationary phase when yeast extract was included in the medium (data not shown).
FIG. 3.
The agpA::TnphoA fusion is induced as strain SG1001 enters stationary phase when cells are grown in LB medium but not when they are grown in M9 glucose medium. Strain SG1001 was grown in LB medium (A) and M9 glucose medium (B), and the OD of the culture and the specific activity of the AgpA′::′PhoA were monitored.
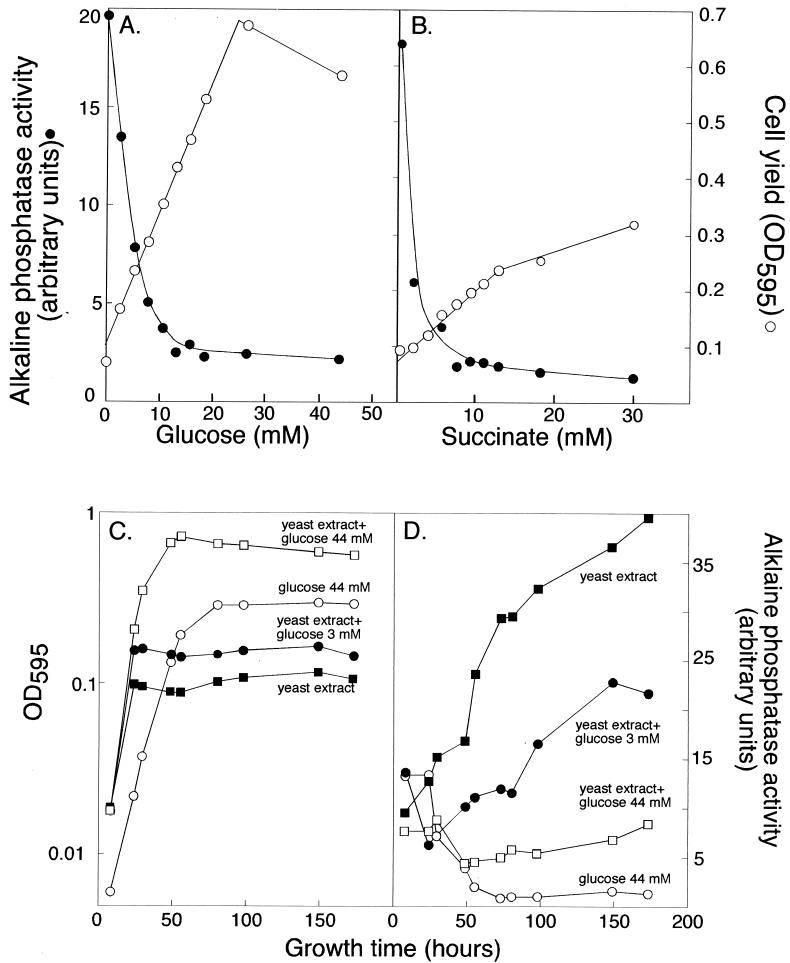
While conducting experiments on the yeast extract-dependent induction of agpA, we noticed that the expression of agpA was influenced by carbon sources in the medium. When cultures were grown in basal medium (which includes a small amount of yeast extract) containing various amounts of glucose or succinate, cell yield increased with increased concentrations of the carbon source, but AgpA′::′PhoA activity decreased (Fig. 4A and B). The activity of the AgpA′::′PhoA fusion was also monitored as cultures grew in basal medium or basal medium plus glucose, and results indicated that increasing the amount of glucose present in the culture medium decreased the rate at which AgpA::PhoA activity protein accumulated in the cells (Fig. 4C and D). Previous work has indicated that succinate and glucose are preferred carbon sources for Rhizobium species, as they have been shown to repress the utilization of other carbon sources, such as lactose and myo-inositol (20, 38). Our results show that succinate and glucose also downregulate the expression of the agpA gene.
FIG. 4.
Expression of the agpA::TnphoA fusion in strain SG1001 is repressed by glucose and succinate. Cell yield and the specific activity of the AgpA′::′PhoA fusion were determined after strain SG1001 had been cultured for 96 h in basal medium containing various concentrations of glucose (A) and succinate (B). (C) Growth of strain SG1001 in M9 salts supplemented with various combinations of glucose and yeast extract. The concentrations of glucose and yeast extract of the different media are indicated on the graph. (D) Specific activity of the AgpA′::′PhoA fusion in strain SG1001 as cells grew in basal medium and basal medium supplemented with various concentrations of glucose. The specific activity data are from the cultures whose growth is shown in panel C.
The agpA gene product is involved in the transport of α-galactosides.
The agpA gene encodes a protein which is similar to those in the periplasmic oligopeptide binding protein family. Despite the name of this family, its members are involved in the transport of a variety of substrates (46, 49). For example, besides oligopeptides and dipeptides, family members which transport nickel (34), heme (16), and substituted sugars (17, 21) have been characterized.
Our initial experiments showed no evidence that the agpA gene product was involved in oligopeptide transport (data not shown). Because the agpA gene is preceded by an open reading frame which encodes a protein similar to α-galactosidases, we sought to determine whether strain SG1001 is able to grow on α-galactosides. For this experiment, we grew strain SG001 and its parental strain, Rm8002, overnight in a noninducing M9 glucose medium, and the next day we diluted cells into basal media with and without the sugars to be tested. The yeast extract in the basal medium was necessary because it allowed the cells to grow to a measurable density, even if they were unable to catabolize a particular sugar. We found that strain SG1001 was unable to use melibiose and raffinose, both α-galactosides, as carbon sources. This strain was, however, able to catabolize glucose and galactose under the same experimental conditions. The parental strain, Rm8002, was able to utilize all four compounds.
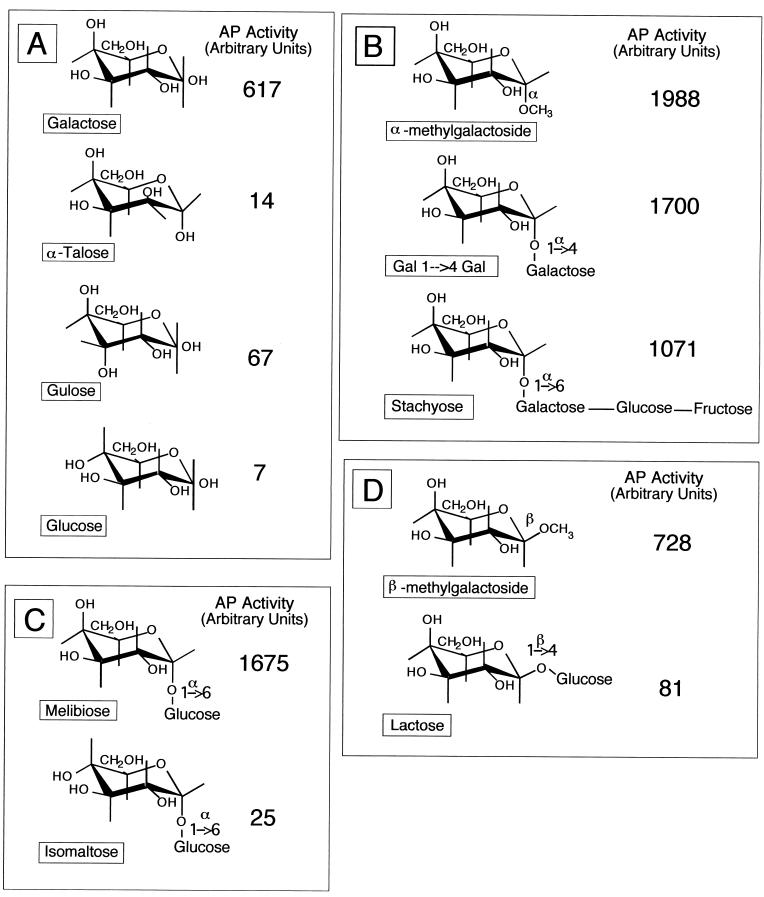
We tested different sugars for their ability to induce expression of the agpA::TnphoA fusion. Cells were grown in basal medium containing one of a variety of sugars at a concentration of 0.4% (wt/vol). After 96 h, the cells were harvested and the alkaline phosphatase activity arising from the AgpA′::′PhoA fusion was measured. Yeast extract was included in all the media tested to ensure that we had enough cells to assay for alkaline phosphatase activity and to ensure a small but measurable amount of AgpA′::′PhoA activity. The results from one such experiment are summarized in Fig. 5. We found that galactose was a good inducer of the agpA gene; isomers of galactose, such as glucose, talose, and gulose, were poor inducers of the gene (Fig. 5A). α-Galactosides were also good inducers of the gene, even if the terminal galactose is linked to a trisaccharide, as in stachyose (Fig. 5B). α-Glucosides, such as isomaltose, were poor inducers of agpA (Fig. 5C). We found that β-galactosides can act as inducers if the moiety linked to the terminal galactose is small. For example, β-methyl-galactoside is a good inducer of agpA, whereas lactose is not (Fig. 5D).
FIG. 5.
The agpA::TnphoA fusion in strain SG1001 is induced by α-galactosides. Strain SG1001 was grown in basal medium supplemented with the indicated sugars. Alkaline phosphatase specific activity was determined 96 h after inoculation, when cells were in stationary phase. The supplemental sugars were present at a concentration of 0.4% (wt/vol). Specific activity of the AgpA′::′PhoA fusion when strain SG1001 was grown in the presence of galactose or isomers of galactose (A); α-galactosides (B); the α-galactoside melibiose or an isomer of melibiose, the α-glucoside isomaltose (C); and β-galactosides (D).
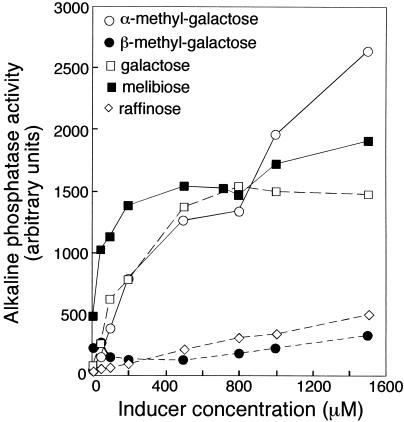
α-Galactosides induced the agpA gene when they were present at 0.4% (wt/vol), a concentration of about 20 mM. We sought to determine which sugars are the most-effective inducers by calculating the concentrations at which they were able to induce the agpA::TnphoA fusion. We inoculated strain SG1001 into basal medium, and sugars were added in concentrations ranging from 10 μM to 1.6 mM. Cells were harvested after 96 h, and the specific activity of the AgpA′::′PhoA fusion was measured. The resulting dose-response curves, shown in Fig. 6, indicated that melibiose was the most-effective inducer of the fusion and that it was active at a concentration of 10 μM.
FIG. 6.
Dose-response curve of agpA::TnphoA induction to various inducing galactosides. Strain SG1001 was inoculated into basal medium containing various galactosides which induce the AgpA′::′PhoA fusion and harvested for alkaline phosphatase activity assays after 96 h of growth. Sugars were present at concentrations ranging from 10 μM to 1.6 mM.
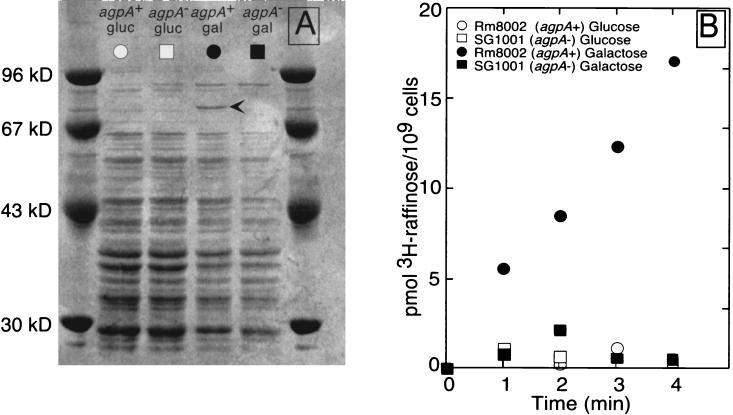
We hypothesized that the agpA gene was part of a transport system involved in α-galactoside uptake, because the sequence of the agpA gene suggested that it encoded a periplasmic binding protein for a transport system and the experiments described above suggested that it was involved in the utilization of α-galactosides. Localization of the agpA gene product and studies of raffinose transport supported this hypothesis. Strain SG1001 and the parental strain, Rm8002, were grown in M9 glucose medium, which represses agpA expression, or in M9 galactose medium, which induces agpA expression, and periplasmic proteins were isolated and separated by electrophoresis (Fig. 7A). When strain Rm8002 (agpA+) was grown in M9 galactose medium, the inducing medium, it expressed a periplasmic protein of about 80 kDa which is close to 75 kDa, the predicted size of the mature AgpA protein. This protein was absent from strain SG1001 (agpA::TnphoA) when it was grown under inducing conditions and from both strains when they were grown in M9 glucose. Only those cells which expressed the 80-kDa protein (i.e., strain Rm8002 grown in inducing medium) were able to transport the α-galactoside raffinose as assayed by uptake of [3H]raffinose (Fig. 7B). For these reasons, we believe that the 80-kDa protein is mature AgpA and the periplasmic binding protein of an α-galactoside transport system.
FIG. 7.
AgpA is a periplasmic protein needed for transport of the α-galactoside raffinose. (A) Periplasmic proteins from strains Rm8002 (agpA+) and SG1001 (agpA) grown in M9 glucose, which represses agpA expression, or in M9 galactose, which induces agpA expression. (B) Transport of raffinose by strains Rm8002 and SG1001. Raffinose uptake assays were performed with cells from the cultures used for isolating the periplasmic proteins shown in panel A.
DISCUSSION
By screening for R. meliloti genes which are regulated by nodD3, syrM, or syrA, we have isolated and characterized a TnphoA insertion in a gene, agpA, which encodes the binding protein component of a periplasmic transport system. This gene is downregulated by syrA.
The periplasmic transport systems, of which there are more than 50, cluster into seven families (46). The sequence of agpA indicates that it encodes a member of the oligopeptide permease transport system family. The oligopeptide permease family includes transport systems from gram-negative and gram-positive bacteria which transport a variety of substrates—small peptides (14, 18), peptide pheromones (22, 26, 41), nickel (34), heme (16), and modified sugar molecules such as aminoglycoside antibiotics (21), agrocinopine, and agrocin 84 (17).
Strain SG1001, which contains an agpA::TnphoA fusion, fails to utilize or transport α-galactosides. In addition, the agpA::TnphoA fusion is induced by α-galactosides, and the agpA gene is downstream of a gene which is similar to an α-galactosidase gene from E. coli. For these reasons, it is plausible that the agpA gene product binds α-galactosides such as melibiose, raffinose, and stachyose in the periplasm and is involved in their transport across the bacterial cell membrane.
The fact that the agpA::TnphoA fusion in strain SG1001 is induced by α-galactosides despite the inability of this strain to utilize or transport α-galactosides raises the question of how the cells are able to sense and induce agpA in response to such sugars. It may be that the inducing molecules cross the cell membrane, via a transport system for which they have low affinity, at a rate which is too low to support growth or to be easily detected by uptake assays but which is sufficient to allow the sugar to interact with a regulatory protein and to induce agpA::TnphoA fusion. Alternatively, it may be that α-galactosides are detected extracellularly and that the information is transduced to the internal compartment, leading to agpA induction. The agpA gene is induced to high levels by either α-galactosides or galactose, but not by a β-galactoside such as lactose. This finding indicates that the molecules responsible for detecting and initiating agpA induction in response to galactose do not have access to the galactose released by the hydrolysis of lactose, perhaps because α-galactosides and galactose are sensed in the extracellular compartment by molecules that do not recognize lactose. Alternatively, the sensing molecules may be cytoplasmic but may fail to respond to the galactose released during hydrolysis of lactose because the galactose pool is small or because the galactose is rapidly channeled into central metabolic pathways.
We were able to show that the agpA gene is downregulated by syrA. syrA encodes an 88-amino-acid protein (2) which is similar to two other small proteins found in Rhizobium species: ExoX (15, 40) and Psi (4). All three of these proteins are hydrophobic in their amino-terminal halves and hydrophilic in their carboxy-terminal halves. It has been suggested that ExoX and Psi are anchored in the membrane by their N termini and have their hydrophilic C termini in the cell cytoplasm (4). In addition to their similar structures, SyrA, ExoX, and Psi regulate the production of exopolysaccharides. The regulatory effects of ExoX and Psi on exopolysaccharide production are posttranslational. It has been suggested that the proteins interact with other membrane proteins required for the synthesis of exopolysaccharide and inhibit their activity in some manner (15, 40, 50). If SyrA also exerts its effects posttranslationally, then the downregulation of agpA by syrA is likely to be indirect because SyrA overproduction results in lowered expression of the agpA::TnphoA fusion, and fusion expression is likely to be controlled at the transcriptional level. One possible scenario is that SyrA downregulates the activity of a transport system which brings in the α-galactosides which induce the agpA gene, or perhaps SyrA interferes with proteins involved in sensing or responding to α-galactosides.
Recent work has shown that the syrA gene is highly expressed by R. meliloti inside nodule cells (2). Therefore, it may be that the α-galactoside transport system, of which agpA is a part, is downregulated in these intercellular bacteria. We have shown that succinate and glucose also downregulate the expression of the agpA gene. Succinate and other dicarboxylic acids are present in nodule cells and are used by bacteroids to fuel nitrogen fixation (3, 9, 10). These acids may also act to downregulate agpA inside nodule cells. Whether or not repression of agpA by succinate and glucose is dependent on syrA is currently unknown.
The α-galactoside transport system may be expressed by bacteria growing in the rhizosphere of host plants, because α-galactosides such as raffinose and stachyose, both of which induce agpA, are present at high levels in many legume seeds and thus may also be present in the rhizosphere of germinating seeds or young plants. Such sugars are catabolized by R. meliloti, and their utilization may have a role in growth and survival of these bacteria in the rhizosphere of their host plants. Whether subsequent downregulation of the α-galactoside transport system occurs when R. meliloti is growing inside nodule cells and whether this regulation relates to metabolism or to other aspects of symbiosis may be revealed in further studies.
ACKNOWLEDGMENTS
S.R.L. is an investigator at the Howard Hughes Medical Institute. This research was additionally supported by NIH research service award GM16211 to D.J.G. and by DOE grant DE-FG-03-90ER20010 to S.R.L.
We thank the members of our laboratory for useful suggestions and discussion. In particular, we thank Mike Willits for the R. meliloti lambda library and Melanie Barnett for providing information on syrA prior to publication.
REFERENCES
- 1.Alloing G, Trombe M C, Claverys J P. The ami locus of the gram-positive bacterium Streptococcus pneumoniae is similar to binding protein-dependent transport operons of gram-negative bacteria. Mol Microbiol. 1990;4:633–644. doi: 10.1111/j.1365-2958.1990.tb00632.x. [DOI] [PubMed] [Google Scholar]
- 1a.Ames G F, Prody C, Kustu S. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J Bacteriol. 1983;160:1181–1183. doi: 10.1128/jb.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett M J, Swanson J A, Long S R. Multiple genetic controls on Rhizobium meliloti syrA, a regulator of exopolysaccharide abundance. Genetics. 1998;148:19–32. doi: 10.1093/genetics/148.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolton E, Higgisson B, Harrington A, O’Gara F. Dicarboxylic acid transport in Rhizobium meliloti: isolation of mutants and cloning of dicarboxylic acid transport gene. Arch Microbiol. 1986;144:142–146. [Google Scholar]
- 4.Borthakur D, Downie J A, Johnston A W B, Lamb J W. psi, a plasmid-linked Rhizobium-phaseoli gene that inhibits exopolysaccharide production and which is required for symbiotic nitrogen fixation. Mol Gen Genet. 1985;200:278–282. [Google Scholar]
- 5.Brewin N J. Development of the legume root nodule. Annu Rev Cell Biol. 1991;7:191–226. doi: 10.1146/annurev.cb.07.110191.001203. [DOI] [PubMed] [Google Scholar]
- 6.Denarie J, Debelle F, Prome J C. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 7.Egelhoff T T, Long S R. Rhizobium meliloti nodulation genes: identification of nodDABC gene products, purification of nodA protein, and expression of nodA in Rhizobium meliloti. J Bacteriol. 1985;164:591–599. doi: 10.1128/jb.164.2.591-599.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eiken P. Stanford University honors thesis, Stanford, Calif. 1996. [Google Scholar]
- 9.Finan T M, Oresnik I, Bottacin A. Mutants of Rhizobium meliloti that are defective in succinate metabolism. J Bacteriol. 1988;170:3396–3403. doi: 10.1128/jb.170.8.3396-3403.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finan T M, Wood J M, Jordan D C. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J Bacteriol. 1983;154:1403–1413. doi: 10.1128/jb.154.3.1403-1413.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Use of a cos derivative of pRK290 in constructing a clone bank of Rhizobium meliloti DNA. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 12.Gilson E, Alloing G, Schmidt T, Claverys J P, Dudler R, Hofnung M. Evidence for high affinity binding-protein dependent transport systems in Gram positive bacteria and in Mycoplasma. EMBO J. 1988;7:3971–3974. doi: 10.1002/j.1460-2075.1988.tb03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez J E, York G M, Walker G C. Rhizobium meliloti polysaccharides: synthesis and symbiotic function. Gene. 1996;179:141–146. doi: 10.1016/s0378-1119(96)00322-8. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin E W, Higgins C F. Uptake of cell wall peptides by Salmonella typhimurium and Escherichia coli. J Bacteriol. 1987;169:3861–3865. doi: 10.1128/jb.169.8.3861-3865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray J X, Djordjevic M A, Rolfe B G. Two genes that regulate exopolysaccharide production in Rhizobium sp. strain NGR234: DNA sequences and resultant phenotypes. J Bacteriol. 1990;172:193–203. doi: 10.1128/jb.172.1.193-203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson M S, Slaughter C, Hansen E J. The hbpA gene of Haemophilus influenzae type b encodes a heme-binding protein conserved among heme-dependent Haemophilus species. Infect Immun. 1992;60:2257–2266. doi: 10.1128/iai.60.6.2257-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayman G T, von Bodman S B, Kim H, Jiang P, Farrand S K. Genetic analysis of the agrocinopine catabolic region of Agrobacterium tumefaciens Ti plasmid pTiC58, which encodes genes required for opine and agrocin 84 transport. J Bacteriol. 1993;175:5575–5584. doi: 10.1128/jb.175.17.5575-5584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiles I D, Higgins C F. Peptide uptake by Salmonella typhimurium. Eur J Biochem. 1986;158:561–567. doi: 10.1111/j.1432-1033.1986.tb09791.x. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch A M. Developmental biology of legume nodulation. New Phytol. 1992;122:211–237. doi: 10.1111/j.1469-8137.1992.tb04227.x. [DOI] [PubMed] [Google Scholar]
- 20.Jelesko J G, Leigh J A. Genetic characterization of a Rhizobium meliloti lactose utilization locus. Mol Microbiol. 1994;11:165–173. doi: 10.1111/j.1365-2958.1994.tb00298.x. [DOI] [PubMed] [Google Scholar]
- 21.Kashiwagi K, Miyaji A, Ikeda S, Tobe T, Sasakawa C, Igarashi K. Increase of sensitivity to aminoglycoside antibiotics by polyamine-induced protein (oligopeptide-binding protein) in Escherichia coli. J Bacteriol. 1992;174:4331–4337. doi: 10.1128/jb.174.13.4331-4337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koide A, Hoch J A. Identification of a second oligopeptide transport system in Bacillus subtilis and determination of its role in sporulation. Mol Microbiol. 1994;13:417–426. doi: 10.1111/j.1365-2958.1994.tb00436.x. [DOI] [PubMed] [Google Scholar]
- 23.Leigh J A, Reed J W, Hanks J F, Hirsch A M, Walker G C. Rhizobium meliloti mutants that fail to succinylate their calcofluor-binding exopolysaccharide are defective in nodule invasion. Cell. 1987;51:579–587. doi: 10.1016/0092-8674(87)90127-9. [DOI] [PubMed] [Google Scholar]
- 24.Leigh J A, Signer E R, Walker G C. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leigh J A, Walker G C. Exopolysaccharides of Rhizobium: synthesis, regulation and symbiotic function. Trends Genet. 1994;10:63–67. doi: 10.1016/0168-9525(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 26.Leonard B A B, Podbielski A, Hedberg P J, Dunny G M. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci USA. 1996;93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liljestrom P L, Liljestrom P. Nucleotide sequence of the melA gene, coding for alpha-galactosidase in Escherichia coli K-12. Nucleic Acids Res. 1987;15:2213–2220. doi: 10.1093/nar/15.5.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long S, McCune S, Walker G C. Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J Bacteriol. 1988;170:4257–4265. doi: 10.1128/jb.170.9.4257-4265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long S R. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulligan J T, Long S R. A family of activator genes regulates expression of Rhizobium meliloti nodulation genes. Genetics. 1989;122:7–18. doi: 10.1093/genetics/122.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mylona P, Pawlowski K, Bisseling T. Symbiotic nitrogen fixation. Plant Cell. 1995;7:869–885. doi: 10.1105/tpc.7.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro C, Wu L-F, Mandrand-Berthelot M A. The nik operon of Escherichia coli encodes a periplasmic binding protein-dependent transport system for nickel homologous to the peptide permease family. Mol Microbiol. 1993;9:1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- 35.Ochman H, Gerber A S, Hartl D L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philips D A, Wery J, Joseph C M, Jones A D, Teuber L R. Release of flavonoids and betaines from seeds of seven Medicago species. Crop Sci. 1995;35:805–808. [Google Scholar]
- 37.Phillips D A. Synthesis, release and transmission of alfalfa signals to rhizobial symbionts. Plant Soil. 1994;161:69–80. [Google Scholar]
- 38.Poole P S, Blyth A, Reid C J, Walters K. myo-Inositol catabolism and catabolite repression in Rhizobium leguminosarum bv. viciae. Microbiology. 1994;140:2787–2795. [Google Scholar]
- 39.Pueppke S G. The genetic and biochemical basis for nodulation of legumes by rhizobia. Crit Rev Biotechnol. 1996;16:1–51. doi: 10.3109/07388559609146599. [DOI] [PubMed] [Google Scholar]
- 40.Reed J W, Capage M, Walker G C. Rhizobium meliloti exoG and exoJ mutations affect the ExoX-ExoY system for modulation of exopolysaccharide production. J Bacteriol. 1991;173:3776–3788. doi: 10.1128/jb.173.12.3776-3788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudner D Z, LeDeaux J R, Ireton K, Grossman A D. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J Bacteriol. 1991;173:1388–1398. doi: 10.1128/jb.173.4.1388-1398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma S B, Signer E R. Temporal and spatial regulation of the symbiotic genes of Rhizobium meliloti in planta revealed by transposon Tn5-gusA. Genes Dev. 1990;4:344–356. doi: 10.1101/gad.4.3.344. [DOI] [PubMed] [Google Scholar]
- 43.Spaink H P. The molecular basis of infection and nodulation by rhizobia: the ins and outs of sympathogenesis. Annu Rev Phytopathol. 1995;33:345–368. doi: 10.1146/annurev.py.33.090195.002021. [DOI] [PubMed] [Google Scholar]
- 44.Swanson J, Tu J K, Ogawa J M, Sanga R, Fisher R, Long S R. Extended region of nodulation genes in Rhizobium meliloti 1021. I. Phenotypes of Tn5 insertion mutants. Genetics. 1987;117:181–189. doi: 10.1093/genetics/117.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swanson J A, Mulligan J T, Long S R. Regulation of syrM and nodD3 in Rhizobium meliloti. Genetics. 1993;134:435–444. doi: 10.1093/genetics/134.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tam R, Saier M H., Jr Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Brussel A A N, Bakhuizen R, van Spronsen P C, Spaink H P, Tak T, Lugtenberg B J J, Kijne J W. Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science. 1992;257:70–72. doi: 10.1126/science.257.5066.70. [DOI] [PubMed] [Google Scholar]
- 48.Willits, M. Personal communication.
- 49.Wu L F, Mandrand-Berthelot M A. A family of homologous substrate-binding proteins with a broad range of substrate specificity and dissimilar biological functions. Biochimie. 1995;77:744–750. doi: 10.1016/0300-9084(96)88192-2. [DOI] [PubMed] [Google Scholar]
- 50.Zhan H, Leigh J A. Two genes that regulate exopolysaccharide production in Rhizobium meliloti. J Bacteriol. 1990;172:5254–5259. doi: 10.1128/jb.172.9.5254-5259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]