Abstract
5-methylcytosine (5mC) is the most important DNA modification in mammalian genomes. The ideal method for 5mC localization would be both non-destructive of DNA and direct, without requiring inference based on detection of unmodified cytosines. Here, we present Direct Methylation Sequencing (DM-Seq), a bisulfite-free method for profiling 5mC at single-base resolution using nanogram quantities of DNA. DM-Seq employs two key DNA modifying enzymes: a neomorphic DNA methyltransferase and a DNA deaminase capable of precise discrimination between cytosine modification states. Coupling these activities with novel deaminase-resistant adapters enables accurate detection of only 5mC via a C-to-T transition in sequencing. By comparison, we uncover a PCR-related underdetection bias with the hybrid enzymatic-chemical TAPS approach. Importantly, we show that DM-Seq, unlike bisulfite-sequencing, unmasks prognostically important CpGs in a clinical tumor sample by not confounding 5mC with 5-hydroxymethylcytosine. DM-Seq thus offers an all-enzymatic, non-destructive, faithful, and direct method for the reading of 5-methylcytosine alone.
The methylation of cytosine bases in cytosine-guanine (CpG) dinucleotides is critical for diverse biological processes including gene expression, imprinting, and the suppression of mobile genetic elements 1. Given its role in shaping transcriptional programs, the landscape of 5-methylcytosine (5mC) can define cell lineages2, and dysregulation of cytosine methylation is a hallmark of diseases such as cancer3,4.
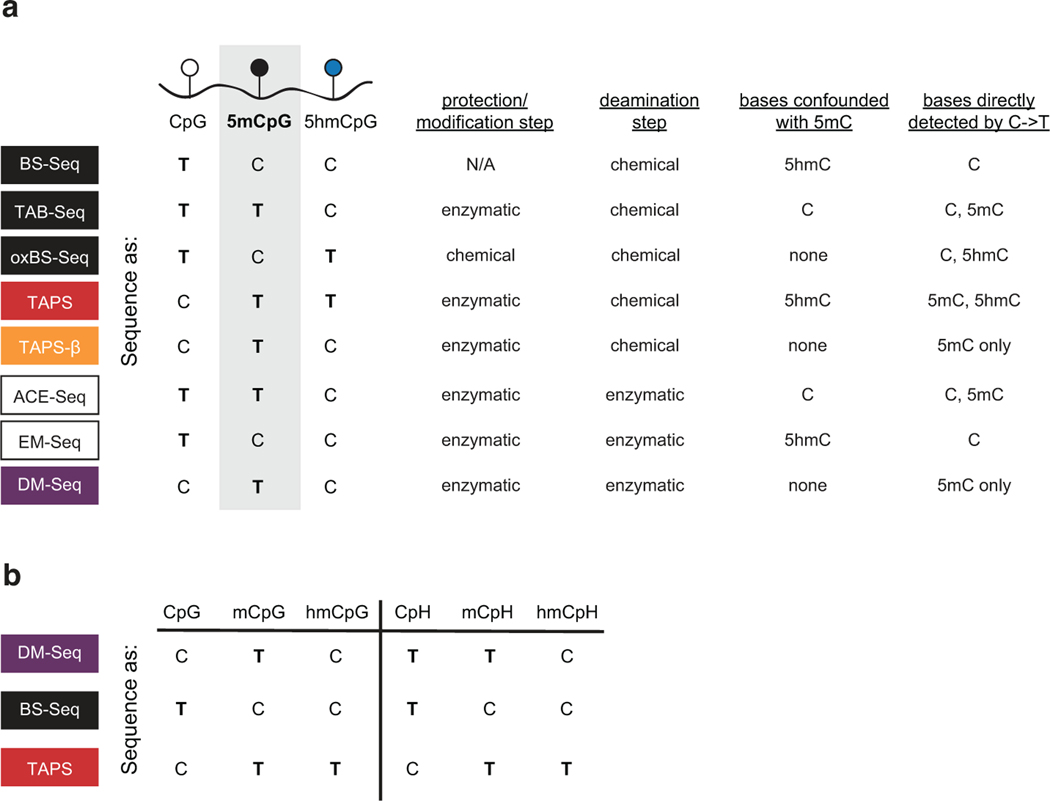
Mapping 5mC has most commonly been accomplished using chemical deamination methods. In bisulfite-based sequencing (BS-Seq), the reaction of sodium bisulfite with unmodified cytosines results in a C-to-T transition. As 5mC reacts slowly with bisulfite, its presence can be indirectly inferred by bases that remain as a C5. BS-Seq, however, poses at least two limitations that impact our ability to resolve 5mC. First, bisulfite significantly damages DNA6. Second, the indirect inference of 5mC has created major challenges. As one striking example, 5-hydroxymethylcytosine (5hmC), the product of TET-mediated oxidation of 5mC, also remains as a C in BS-Seq7. Reliance on BS-Seq was a major reason why 5hmC had escaped detection for decades although it accounts for more than 20% of modified cytosines in some cell types8. Parsing 5mC and 5hmC is functionally important. For example, in human glioblastomas (GBM) these modifications can have antagonistic functions on gene expression and impact prognoses9.
Several recently developed epigenetic sequencing technologies partially address the limitations associated with BS-Seq. Important advances have come from both bisulfite-dependent and bisulfite-independent techniques (Extended Data Fig. 1)10. For example, resolving 5mC and 5hmC is possible with oxidative bisulfite sequencing (oxBS-Seq), although low microgram DNA input remains an impractical limitation for many samples11,12. The chemical deamination method TET-assisted pyridine borane sequencing (TAPS) can also map modified cytosines13,14. TAPS requires efficient oxidation of 5mC by TET enzymes as well as chemical conversion to the non-aromatic nucleobase analog dihydrouracil (DHU). In parallel, methods employing enzymatic rather than chemical deamination have been developed. Enzymes are ideal tools for epigenetic sequencing as they are accurate and non-destructive10. APOBEC-Coupled Epigenetic Sequencing (ACE-Seq) was the first technology employing a DNA deaminase, APOBEC3A (A3A), to selectively deaminate unmodified Cs and 5mCs, while leaving protected 5hmCs unconverted15. Similarly, Enzymatic-Methylation Sequencing (EM-Seq) utilizes TET and β-glucosyltransferase (βGT) enzymes prior to enzymatic deamination to achieve a readout akin to bisulfite merging 5mC and 5hmC16.
Despite the promise of enzymatic deamination approaches, to date, these methods have been limited by the fact that both C and 5mC are deaminated by A3A. Motivated to develop an accurate and non-destructive pipeline for studying 5mC alone, we envisioned that direct 5mC detection could be achieved if unmodified CpGs could be protected, leaving only 5mC subject to deamination in the CpG context (Fig. 1a). Towards this goal, we considered the possibility of pairing an engineered methyltransferase (MTase*) with an S-adenosyl-L-methionine (SAM) analog (Fig. 1b) to create a modified cytosine base resistant to A3A deamination, creating a direct strategy for localizing 5mCs within genomes.
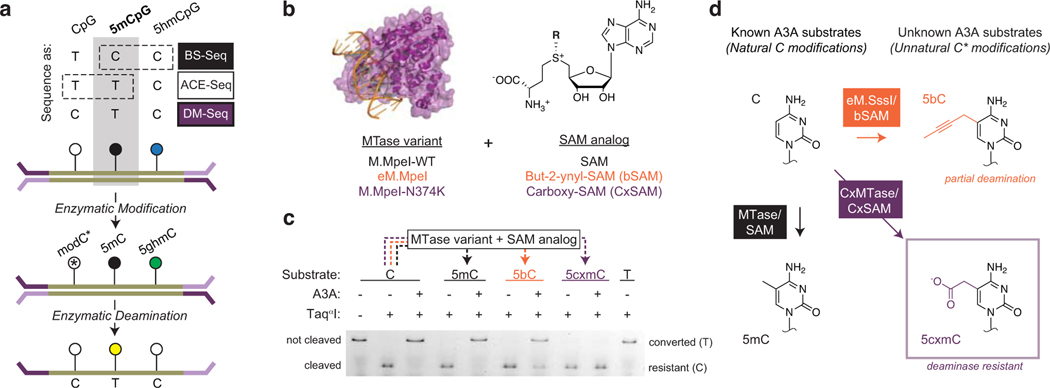
Figure 1. Direct Methylation Sequencing (DM-Seq) is enabled by 5cxmC generation.
a) Top: Sequencing methods for localizing C, 5mC, and 5hmC differ in their use of chemical (e.g. BS-Seq) or enzymatic (e.g. ACE-Seq) deamination, with 5mC signal confounded by either 5hmC or C (boxes with dashed lines). Bottom: Proposed workflow for DM-Seq. DM-Seq was envisioned as an all-enzymatic workflow for the direct detection of only 5mC. This goal could be realized by coupling an engineered DNA MTase (MTase*) with a SAM analog to create a sterically bulky cytosine base that resists deamination by APOBEC3A (A3A). C*, modified C generated from MTase and SAM analog; 5ghmC, Glucosylated 5hmC. In the proposed workflow, only 5mC alone is converted T at CpG sites. b) MTase variants and SAM analogs including two candidates for DM-Seq. c) Restriction enzyme coupled assay for assessing A3A deamination of unnatural cytosine analogs. An oligonucleotide with a single TaqαI restriction site (TCGA) is modified by the appropriate MTase variant to create 5mC, 5bC, or 5cxmC (dashed lines). Modified DNA is then deaminated by A3A. TaqαI only cleaves DNA if C is protected from A3A deamination. Experiment was performed twice with similar results. See Extended Data Fig. 2 for assay schematic and ESI-MS validation of 5bC and 5cxmC substrates. d) Summary of reactivity of various cytosine derivatives towards A3A. Left: Cytosine (C) is modified to 5-methylcytosine (5mC) by WT MTases and SAM. Both C and 5mC are favorable substrates for enzymatic deamination by A3A. Right: structures of 5-(but-2-ynyl)cytosine (5bC) and 5-carboxymethylcytosine (5cxmC), with previously uncharacterized reactivity towards A3A. Box: 5cxmC satisfies the criteria required for the DM-Seq strategy: efficient MTase* transfer and complete protection from A3A deamination.
Here, we describe how the full Direct Methylation Sequencing (DM-Seq) approach was realized in order to directly sequence 5mC alone. Additional comparisons to TAPS uncovered unexpected biases in 5mC detection, likely a result of poor polymerase amplification of the DHU base. Ultimately, using nanogram quantities of input DNA, we show that DM-Seq outperforms BS-Seq in terms of both sequencing coverage and accurate quantification of 5mC, including at prognostically important CpG sites within a human glioblastoma tumor.
RESULTS
5cxmC is a candidate modified base for DM-Seq
Having previously exploited two classes of cytosine-modifying enzymes (glucosyltransferases and AID/APOBEC deaminases) to build ACE-Seq15, we envisioned that the addition of an MTase* could enable DM-Seq. In evaluating the feasibility of this idea, we were encouraged for two reasons. First, WT CpG-specific MTases have been applied in sequencing to detect modifications that are significantly more sparse than 5mC17. Second, multiple MTases have been engineered to transfer extended alkyl chains that could feasibly render previously unmodified CpGs resistant to enzymatic deamination. One example is a Q142A/N370A mutant of the CpG MTase M.SssI (eM.SssI) that has been shown to utilize multiple SAM-analogs, including but-2-ynyl-SAM (bSAM)18,19. We also recently discovered a CpG-specific carboxymethyltransferase (CxMTase), M.MpeI N374K, which uses the naturally occurring E. coli metabolite, carboxy-S-adenosyl-L-methionine (CxSAM), to form 5-carboxymethylcytosine (5cxmC)20.
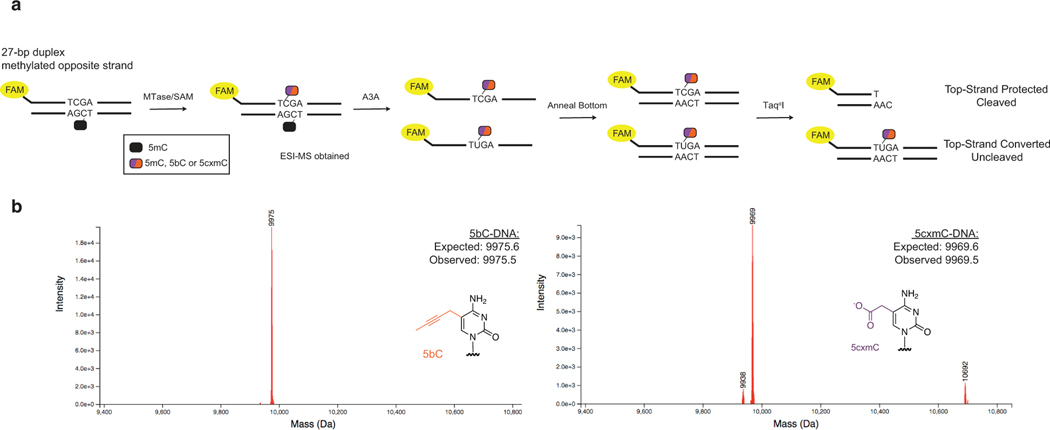
There would be at least two requirements for DM-Seq to succeed: efficient transfer of the protecting group to unmodified CpGs and complete protection of the newly generated modified base from A3A-mediated deamination. We reasoned that the second requirement was critical to prioritize because transfer could be improved while deamination is difficult to prevent. We therefore focused analysis on two candidate MTase* and SAM-analog pairs, a M.MpeI Q136A/N374A variant (eM.MpeI, analogous to eM.SssI) and bSAM, as well as our CxMTase and CxSAM. Using an oligonucleotide containing a single CpG site embedded within a TaqαI (TCGA) restriction site, we demonstrated that each MTase* and SAM-analog pair resulted in efficient conversion of the CpG to a modified CpG (Extended Data Fig. 2), yielding DNA with 5-(but-2-ynyl)-cytosine or 5cxmC, respectively. We then subjected the oligonucleotides to enzymatic deamination with A3A and analyzed for deamination by restriction cleavage with TaqαI (Fig. 1c). While C and 5mC are readily deaminated by A3A, we newly show that 5bC can be partially deaminated by A3A, and 5cxmC, which has features of both size and negative charge that can be disfavored by A3A22,23, appears to resist enzymatic deamination (Fig. 1d). The promising properties of 5cxmC led us to focus on the CxMTase:CxSAM enzyme:substrate pair for further development.
DNA carboxymethylation is limited by opposite-strand biases
Having identified our candidate MTase* and SAM-analog pair, we next aimed to assess the efficiency of DNA carboxymethylation using sequencing. We sheared unmodified 48.5-kb lambda phage genomic DNA (gDNA) and ligated Illumina Y-shaped adapters containing 5mC bases. These samples were subjected to WT M.MpeI or M.Mpel N374K and different SAM substrate conditions. 5mC and 5cxmC generation was assessed by BS-Seq across the 3113 CpG dyads. For the WT M.MpeI, in the absence of SAM, 0.2% of Cs in the CpG context were detected as modified, while in the presence of SAM, 97.2% of CpGs were modified (Fig. 2a). When M.MpeI N374K was used with SAM, the majority of CpG sites were detected as modified. However, only 48.9% of the CpGs were detected as modified, a disappointing result inconsistent with our prior restriction digestion assay that suggested complete DNA carboxymethylation20.
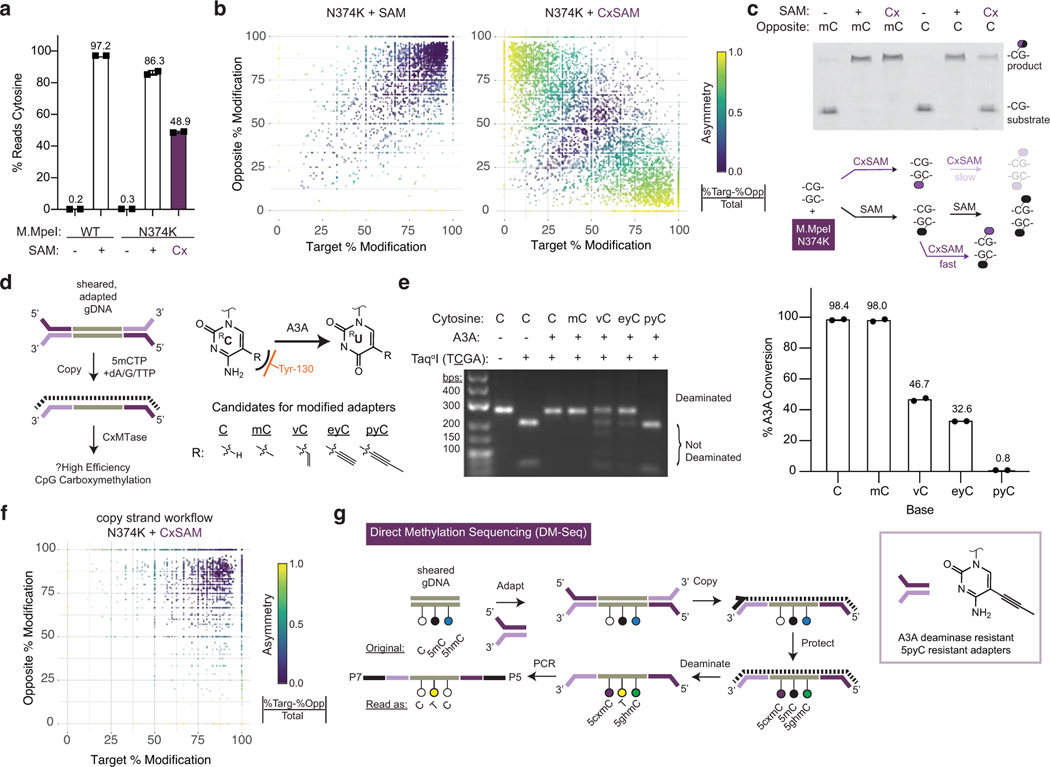
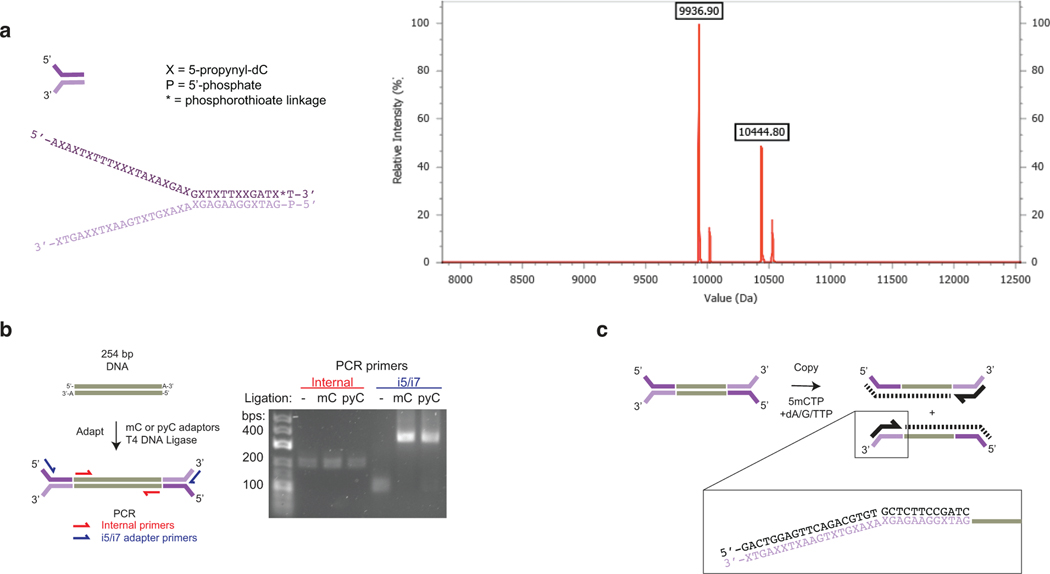
Figure 2. The challenge of asymmetric DNA carboxymethylation is overcome with copy-strand synthesis enabled by 5pyC adapters.
a) Lambda gDNA sequencing experiment. Lambda gDNA was incubated with WT M.MpeI or M.MpeI N374K in the presence of no SAM, SAM, or CxSAM. DNA was treated with bisulfite (BS) before PCR and Illumina sequencing (n=2 independent experiments). Data are presented as mean values +/− SD. b) Scatter plot showing relationship between CpGs on opposite strands within the same dyad. Data is filtered for CpGs with at least 5 sequencing reads.c) Top: Oligonucleotide modification assay (see Extended Data Fig. 3). An oligonucleotide with a labelled top-strand containing an unmodified CpG across from either a 5mCpG or unmodified CpG was reacted with M.MpeI N374K and no SAM, SAM, or CxSAM. Shown is the denaturing gel after digestion with a modification-sensitive restriction enzyme that reports on the modification status of the top strand. Experiment was performed twice with similar results. Bottom: Model for incomplete transfer. Symmetrical modification proceeds readily with SAM but is slow with CxSAM due to inefficient transfer across from a 5cxmC. Carboxymethylation is efficient when the opposite strand contains a 5mC. d) Left: Envisioned scheme where A3A-resistant adapters (purple) initiate universal copy strand synthesis. A copy strand (dotted line) containing 5mCs serves as a favorable substrate for DNA carboxymethylation. Right: Structures of unnatural cytosine analogs explored. The 5-position modifications are anticipated to interact with the steric gate Tyr-130 residue in A3A (orange) (see Extended Data Fig. 4). e) Left: PCR product is generated using modified-dCTPs in place of dCTP. dsDNA is deaminated, PCR amplified, and restriction digested to analyze deamination status at a single TCGA TaqαI site. Right: Deep sequencing of same PCR products as in gel (n=2). f) Scatter plot as in b) showing improvement of carboxymethylation with copy strand synthesis. Data corresponds to Extended Data Fig. 6. g) Full DM-Seq workflow. Sheared gDNA is end-prepped and adapted to A3A resistant 5pyC adapters. A copy strand made with 5mCTPs is synthesized before glucosylation and carboxymethylation. A3A deaminates 5mCpGs to Ts which can be detected upon PCR amplification. Box: 5pyC adapters are synthetically accessible and permissive for A3A-dependent sequencing.
To understand the mechanistic basis for inefficient transfer, we focused on understanding CpGs within the same dyad but on opposite-strands, as some MTases, such as DNMT1, are impacted by opposite-strand modified cytosines24. We found that M.MpeI N374K transfer of SAM was mostly symmetrically clustered (purple), suggesting no strong influence of the target-strand on the opposite-strand within the same CpG dyad (Fig. 2b). In contrast, with CxSAM, many CpGs were asymmetrically modified (yellow). These data are consistent with a model where the first carboxymethylation event at a CpG dyad is efficient, but a second carboxymethylation event on the opposite-strand is slow (Fig. 2c, bottom).
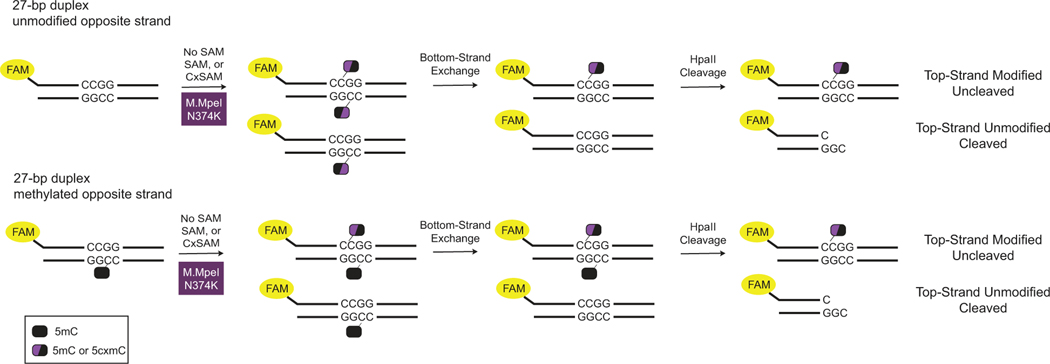
To understand the impact of the opposite-strand modifications that appeared to be limiting to our initial DM-Seq strategy, we devised a new oligonucleotide assay (Extended Data Fig. 3). In this assay, we perform the MTase* reaction using a fluorophore-labelled top-strand containing a single CpG embedded within a methylation-sensitive HpaII (CCGG) restriction site and duplexed to a chemically synthesized bottom-strand containing either an unmodified CpG or 5mCpG. After strand exchange with an excess of an unmodified bottom-strand, digestion with HpaII can detect top-strand modification. When the duplex was reacted with M.MpeI N374K and SAM, the top-strand CpG could be readily modified opposite either an unmodified CpG or 5mCpG (Fig. 2c). Critically, M.MpeI N374K could fully transfer CxSAM across from a 5mCpG, but only partially with an opposite-strand unmodified CpG, recapitulating our sequencing results.
5pyC adapters enable efficient carboxymethylation
Our biochemistry unveiled that the presence of a hemimethylated CpG is particularly favorable for DNA carboxymethylation. This finding offered us a potential solution to realize our initial objective. In our newly envisioned workflow, after adapter ligation, a primer complementary to the adapter could initiate synthesis of a copy-strand containing all 5mCs in lieu of unmodified Cs with the resulting hemimethylated duplex favorable for CxMTase activity (Fig. 2d). In the proposed workflow, the copy-strand is not deaminase-resistant and thus would not be amplified upon library preparation.
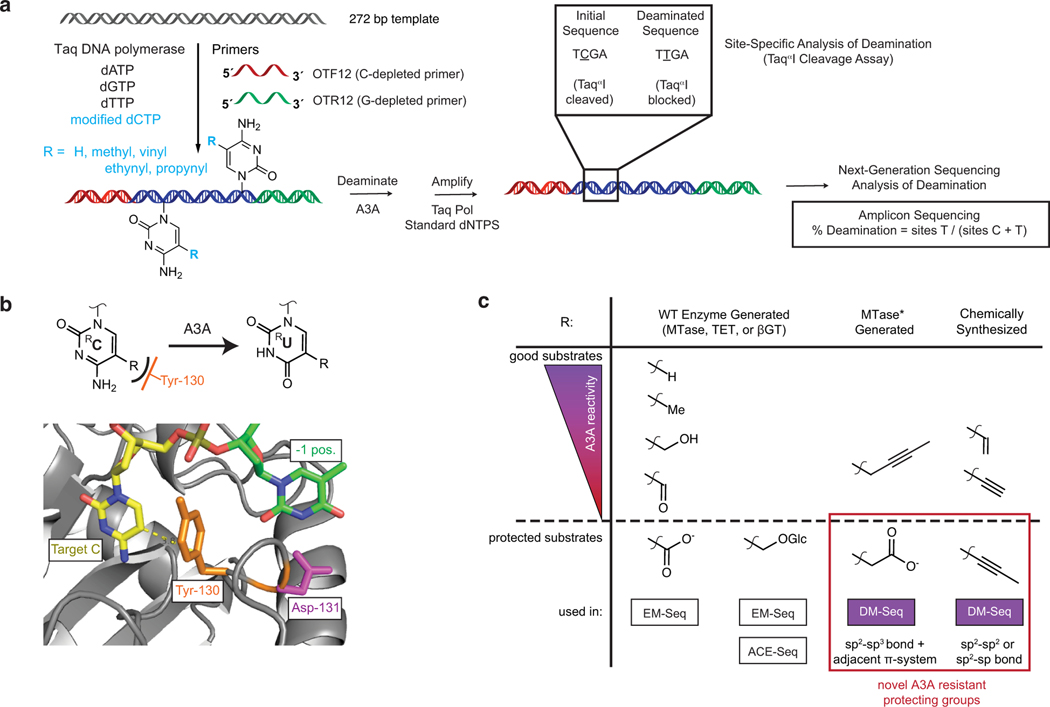
This alternative workflow, however, posed new potential challenges. Traditional adapters contain 5mC, which readily converts to T with enzymatic deamination. Our new workflow with pre-conversion adapter ligation would require custom adapters resistant to deamination by A3A, leading us to reflect on A3A selectivity further. Biochemical studies suggest that the enzyme discriminates against bulky modifications at the 5-position of cytosine,23 a feature which is borne out by structural work highlighting a “steric gate” residue Y130 (orange) abutting the C5-C6 face of the target cytosine (Fig. 2d) 26. To identify candidate analogs suitable for A3A-resistant adapters, we explored a series of dCTP analogs with increasing steric bulk at C5, including C, 5mC, 5-vinylcytosine (5vC), 5-ethynylcytosine (5eyC), and 5-propynylcytosine (5pyC)27, generating duplex DNA with exclusively modified cytosines by PCR. After reacting with A3A, the DNA was reamplified and interrogated for cleavage at a specific TaqαI restriction site within the amplicon or by deep sequencing (Extended Data Fig. 4a). Using this assay approach, amplicons with either C or 5mC show resistance to TaqαI, indicating deamination (Fig. 2e, left). The larger 5vC and 5eyC both showed intermediate resistance, while the 5pyC-containing template appeared resistant to A3A. In agreement with the qualitative assay, the C and 5mC templates were fully deaminated, 5eyC and 5vC incompletely deaminated, and the 5pyC substrate showed <1% deamination by sequencing. Integrating across the series of chemically-modified cytosines, along with the enzymatically-generated 5bC and 5cxmC, our results offer a more complete and structurally informed model for how the hybridization of C5-bond linkages, steric bulk, and charge all collaborate to shape selective enzymatic deamination by A3A (Extended Data Fig. 4b–c).
Our results support the candidacy of 5pyC as a suitable modification for use in DNA deaminase-resistant adapters, especially given its synthetic accessibility28. We therefore synthesized Illumina TruSeq Y-shaped adapters, with all Cs replaced with 5pyC bases, and validated that ligation was not impacted by the presence of 5pyC bases (Extended Data Fig. 5). We next evaluated the copy-strand workflow using sheared, unmodified lambda phage gDNA. With SAM transfer, the inclusion of the copy step had minimal impact on the efficiency of CpG protection (Extended Data Fig. 6). By contrast, CxSAM transfer improved significantly, and the asymmetric protection evident in the absence of the copy-strand was no longer present (Fig. 2f). The analogous experiment with 5pyC adapter ligation and A3A deamination also showed efficient transfer (Extended Data Fig. 6), providing a roadmap for a new DM-Seq workflow using 5pyC adapters.
DM-Seq detection of heterogeneous samples
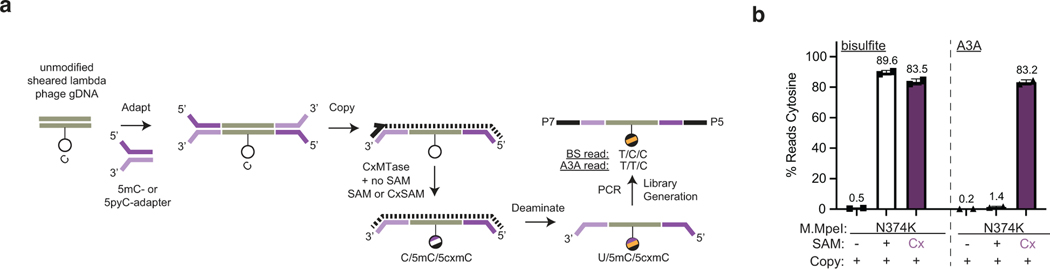
Building on this new design, we optimized our pipeline to further improve DNA carboxymethylation efficiency. In the final DM-Seq workflow (Fig. 2g), 5pyC adapters are ligated to sheared gDNA and copied to create a strand exclusively containing 5mCs in place of C. The gDNA is then protected by the CxMTase (acting on unmodified CpGs) and glucosylation by βGT (for 5hmCs). Subsequent deamination by A3A is performed before PCR amplification and sequencing.
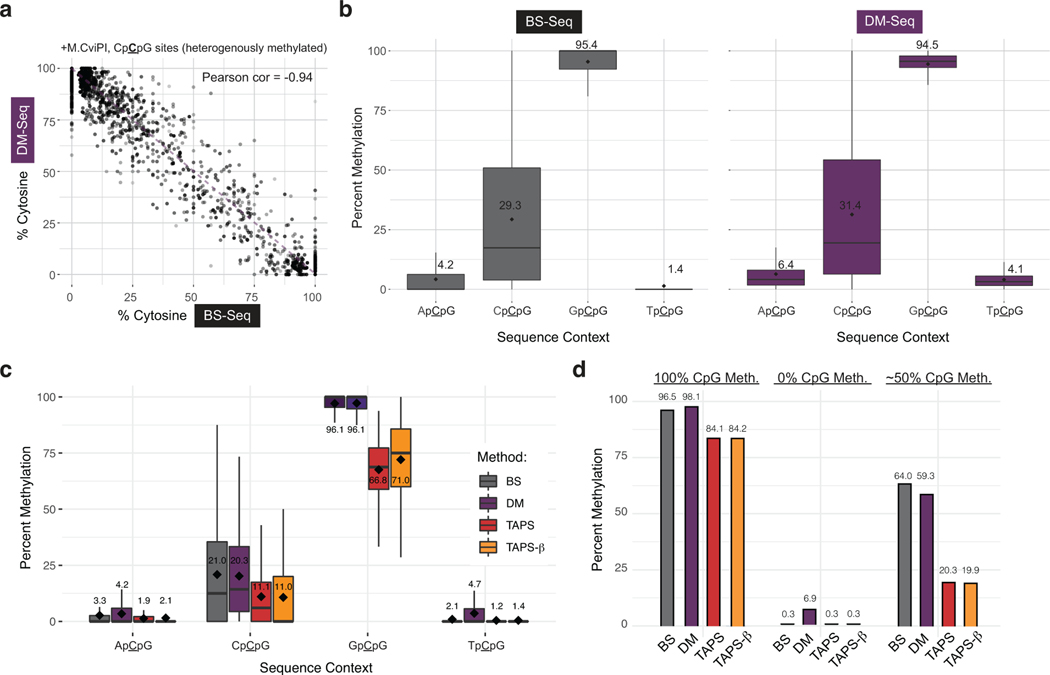
We sought to quantify the fidelity of this workflow using three lambda phage gDNA samples: native gDNA as a standard with unmodified CpGs, gDNA methylated at CpG sites with M.SssI, and gDNA methylated at GpC sites with the MTase M.CviPI. Sheared gDNA samples were split and then analyzed with either 5mC-containing adapters and BS-Seq or 5pyC-containing adapters and DM-Seq (Fig. 3a). After deamination, amplifiable DNA content was 22-fold more across DM-Seq samples as compared to BS-Seq by qPCR (avg Ct = 17.0 vs 12.5, Fig. 3a). Focusing next on the genome-wide comparison of methods (Fig. 3b), for the unmodified lambda phage gDNA, we found a low rate of CpG non-conversion by BS-Seq (0.23%), and a high rate of protection from deamination with DM-Seq (96.7%), validating the efficiency of the copy-strand protocol for CpG conversion to 5cxmCpG. For the gDNA sample treated with M.SssI, 91.3% of CpGs were protected from deamination with BS-Seq, with a comparable level (93.1%) deaminated by A3A in DM-Seq. In the M.CviPI MTase condition, detection of 5mCpG at the genome wide level was similar for BS-Seq and DM-Seq (Fig. 3b). M.CviPI-treated gDNA provided an added opportunity to compare heterogeneous levels of methylation, as this enzyme is known to have detectable but variable off-target activity at CpCpG sites29. At highly methylated sites, we detected 95.4% of GpCpGs as methylated by BS-Seq and 94.5% as methylated by DM-Seq, while off-target CpCpG showed strong correlation at the level of individual CpG sites and globally (Extended Data Fig. 7a–b).
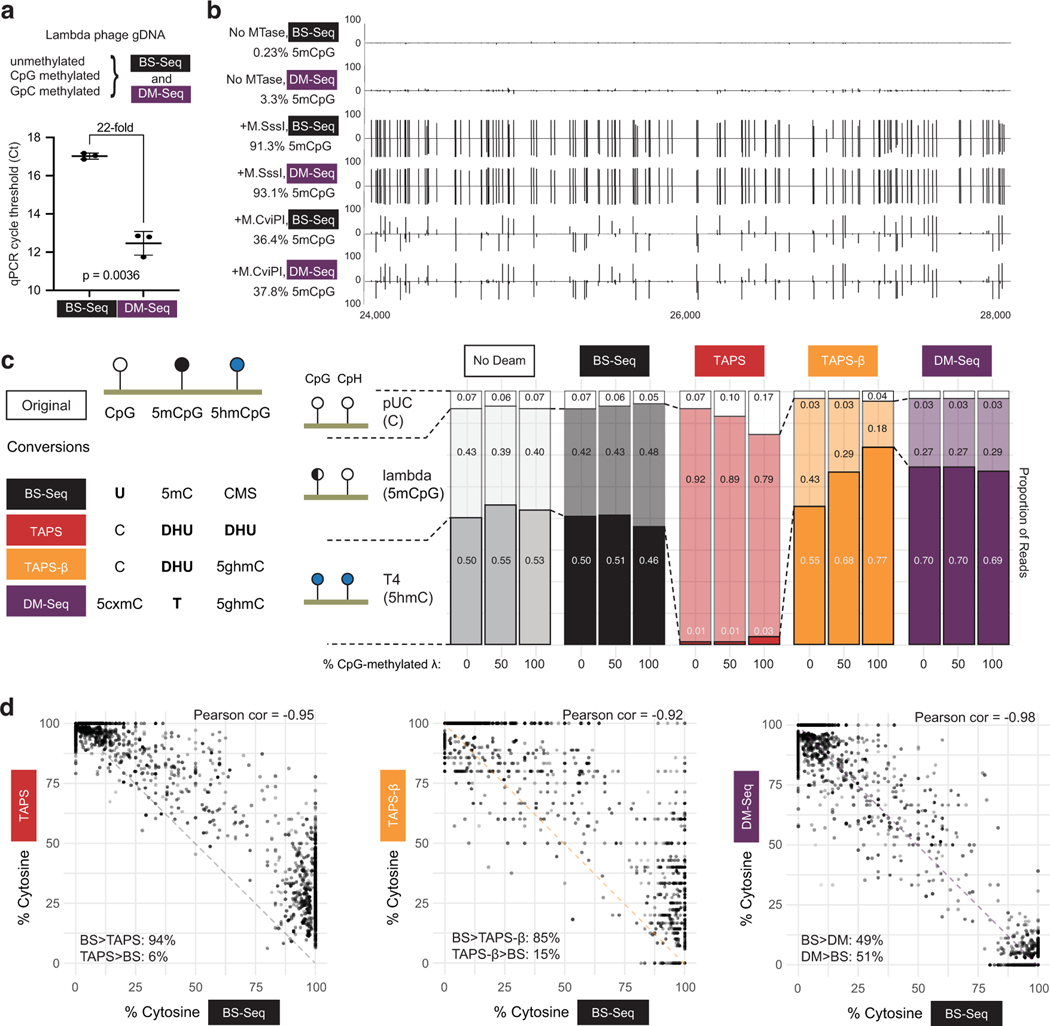
Figure 3. DM-Seq accurately detects 5mCpGs at single-base resolution and is more accurate than TAPS.
a) Difference in Ct between DM-Seq and BS-Seq determined by qPCR. p-value represents paired two-tailed t-test (n = 3 MTase conditions). Data are presented as mean values +/− SD. b) Shown is the genome browser view for coordinates 24,000–28,000 in the lambda phage genome for all CpGs. Lambda gDNA was modified with SAM and no MTase, M.SssI (CpG), or M.CviPI (GpC). Numbers on left represent total efficiency across the entire 48.5 kB genome. c) Comparison of multiple deamination-dependent sequencing workflows. At left is a schematic showing the state of specific DNA modifications after conversion and prior to library generation (cytosine methylene sulfonate, CMS; dihydrouracil, DHU; glucosylated 5hmC, 5ghmC). A mixture of 3 sheared DNA samples: unmodified pUC19 DNA, variably methylated lambda phage (0%, ~50%, or 100% CpG methylated), and T4-hmC phage (with all C bases replaced by 5hmC) was subjected to either no deamination, BS-Seq, DM-Seq, TAPS, or TAPS-β workflows. Plotted is the distribution of reads mapping to each genome under each condition, with the read fraction listed. d) Correlation of BS-Seq to DM-Seq, TAPS, TAPS-β on a M.CviPI GpC-methylated modified substrate. The dashed line shows the readout if BS-Seq signal inversely correlates with DM-Seq, TAPS, or TAPS- β as anticipated, with skew between methods suggested by asymmetrical distribution around this line. In the bottom corner of each plot, for sites where the two methods are not in agreement, the percent of sites where one method detects a higher level of modification than the other method are given.
Comparison of DM-Seq to TAPS uncovers DHU bias
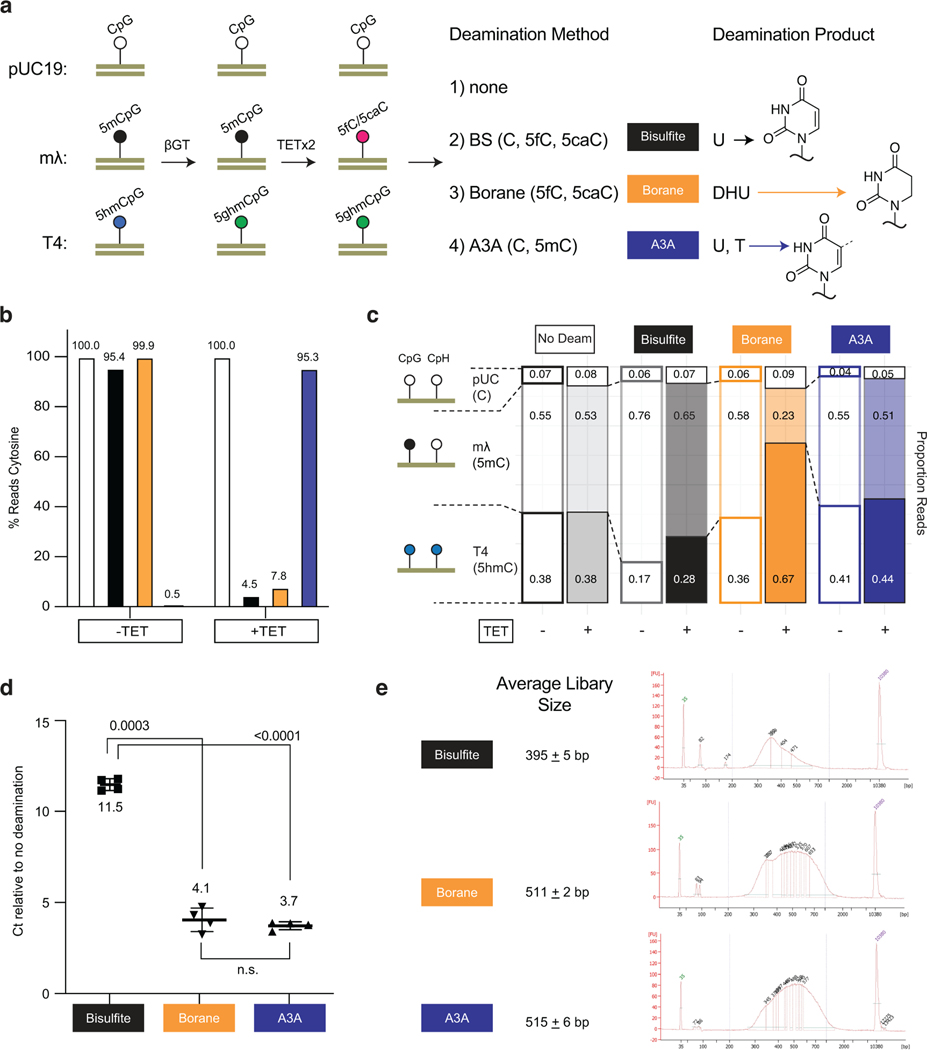
Given the burgeoning interest in bisulfite-free epigenetic sequencing technologies, we saw an opportunity to directly compare DM-Seq to other methods. In particular, we focused on TAPS-β, a variation of TAPS where 5hmCs are protected from TET oxidation, resulting in the chemical reductive deamination of only 5mCs to DHU14. To this end, we combined unmethylated pUC19 plasmid, CpG-methylated lambda gDNA, and T4-hmC phage gDNA to quantify the behavior of C, 5mC, and 5hmC in a single mixture. To assess correlations between methods, we included 0% methylated, ~50% CpG-methylated, 100% CpG-methylated, or 100% GpC-methylated lambda substrate. The ~50% CpG-methylated substrate was made by combining 0% methylated and 100% CpG-methylated lambda DNA. We then subjected these substrates to five possible conditions: no deamination, BS-Seq, DM-Seq, TAPS, or TAPS-β.
We first examined the relative proportion of reads mapping to each of the three DNA genomes (Fig. 3c), and noted two unexpected trends unique to the borane-based technologies. First, and most strikingly, TAPS shows nearly complete obliteration of mapping of the T4-hmC phage (dark red, 1–3% of reads compared to ~50% in the no deamination control). Notably, in the T4-hmC phage, every C including those in non-CpG (CpH) contexts are modified 5hmC bases, thus resulting in a high density of DHU. These findings contrast with TAPS-β, where the 5hmCs are protected from conversion to DHU and T4-hmC reads are detectible. Second, unlike BS-Seq and DM-Seq, both TAPS and TAPS-β both show depletion of the lambda gDNA reads as a function of the level of CpG methylation, with 43% of the total TAPS-β reads mapping to the lambda genome when unmethylated, but only 18% when full-methylated.
We next focused on the GpC-methylated gDNA (Fig. 3d). While, DM-Seq and BS-Seq strongly correlate (Pearson coefficient = −0.98), TAPS and TAPS-β showed weaker correlations with BS-Seq, and additionally showed consistent skew in the data. While DM-Seq is equally likely to detect that an individual CpG is more or less methylated relative to BS-Seq, TAPS and TAPS-β 5mC levels are lower than those detected by BS-Seq at the vast majority of sites (94% or 85% of individual CpGs sites, respectively) (Fig. 3d, Extended Data Fig. 7c). Importantly, we note that while underestimation of 5mC could be explained partially by incomplete conversion efficiencies (discussed below), decreased coverage of the T4-hmC and lambda genomes with more highly modified DNA cannot be.
We sought to further investigate the mechanisms responsible for 5mC detection bias in TAPS. We considered that DHU-containing DNA might be less efficiently amplified, as this non-planar, non-aromatic analog of uracil has been shown to stall numerous polymerases including the SMRT polymerase13,30 and to disrupt base stacking in nucleic acids31,32. With CpG-methylated lambda gDNA, while BS-Seq and DM-Seq detected 96.5% and 98.1% modification, respectively, TAPS and TAPS-β reported ~84% methylation (Extended Data Fig. 7d). However, with the mixed unmethylated and methylated lambda gDNA sample, while BS-Seq and DM-Seq correlated with one another (64.0% vs 59.3%), both TAPS and TAPS-β significantly underestimated the methylation levels (20.3% and 19.9%). The fact that 5mC was underestimated much more significantly with the mixed lambda gDNA sample than with the fully methylated lambda sample is consistent with preferential amplification of the unmethylated lambda gDNA relative to the modified, DHU-containing lambda gDNA. To further understand both read depletion and 5mC detection accuracy, we additionally used our samples with fully-methylated lambda gDNA and performed TET-oxidation followed by BS-Seq or A3A treatment (to measure 5fC/5caC levels) or borane-mediated deamination (Extended Data Fig. 8). This analysis supports the conclusion that the observed bias in amplification and 5mC detection requires both TET-mediated oxidation of 5mC and borane-mediated deamination, consistent with DHU as the source of bias.
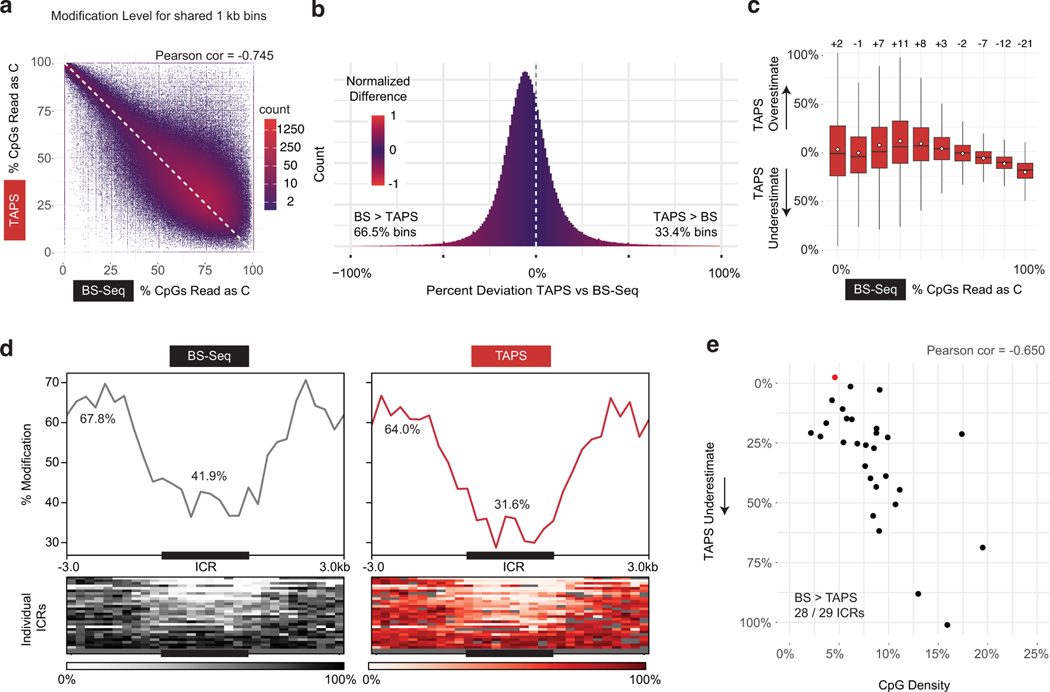
Although our conversion efficiency values are better than those published by other groups who have attempted to replicate TAPS33, our values (range 84–92% across experiments) remained lower than the those reported by the group that initially advanced TAPS (>97%)13. We reasoned that if bias was related to DHU generation and not incomplete conversion efficiency, there should also be evidence of bias in published data sets. We elected to further investigate an established matched mouse embryonic stem cell (mESC) dataset where TAPS was optimized and BS-Seq was also performed13. When we reanalyzed non-overlapping 1 kB bins across the genome, a modest correlation between TAPS and BS-Seq can be observed (Pearson correlation −0.745, Extended Data Fig. 9a). Consistent with our data, TAPS underestimates modification levels at 66.5% of these bins relative to BS-Seq (Extended Data Fig. 9b). We further considered whether bias was a function of modification density, as we observed with lambda gDNA samples. Indeed, lowly modified 1kB bins have an equal probability of being underestimated or overestimated by TAPS, while TAPS detects ~21% lower levels on average than BS-Seq with bins where >90% of CpGs are modified (Extended Data Fig. 9c). Finally, we considered whether methylated regions in cis would be especially prone to TAPS bias in ESCs. Indeed, when examining an established set of imprinting control regions (ICRs)15, we find that TAPS detected lower levels of modification at 28 of 29 ICRs (Extended Data Fig. 9d), with an average of 41.9% CpG modification detected by BS-Seq and 31.6% by TAPS (Extended Data Fig. 9e). Interestingly, the level of deviation between BS-Seq and TAPS increased as a function of CpG density at the ICRs (Pearson coefficient = −0.65). These data show that TAPS modification bias is reproduced in an existing mammalian dataset generated by an independent research group, with multiple trends consistent with DHU being responsible for the observed bias.
DM-Seq is superior to BS-Seq in characterizing a human tumor
We chose to apply DM-Seq on a human GBM sample because this cancer has been extensively characterized for its heterogeneous cytosine methylation patterns34,35. Although mammalian brain tissue is typically enriched with 5hmC, two independent studies utilizing oxBS-Seq concluded that 5hmC is highly depleted in GBMs9,36. GBMs thus offer a complex mammalian genome where DM-Seq and BS-Seq could be directly compared with limited interference from 5hmC, which BS-Seq cannot parse. At the same time, despite relatively low overall abundance of 5hmC, 5hmC at a limited set of CpG sites have been implicated as an important disease biomarker9, offering the possibility that direct detection of 5mC with DM-Seq could provide a prognostically-relevant signal through the accurate sequencing of 5mC alone.
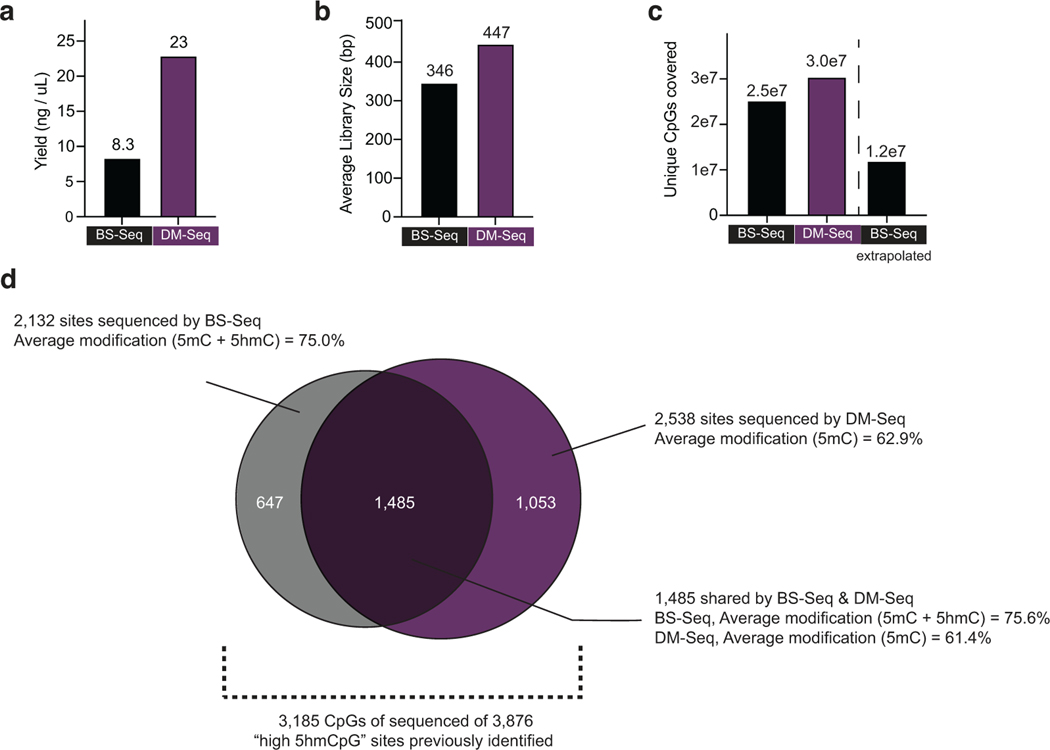
We obtained gDNA from a surgically resected human GBM and added our three spike-in controls to validate DM-Seq efficiency (Fig. 4a). We used >35–100-fold less DNA input than previously used to characterize GBM by oxBS-Seq9,36. The sample was sheared, evenly split, and processed by either DM-Seq or BS-seq pipelines. Overall, the non-destructive nature of DM-Seq was evident in the generated libraries, with a 2.8x greater library yield and a greater average library size (447 vs 346 bp) (Extended Data Fig. 10). Despite the higher yield with DM-Seq, for rigorous comparison of the two methods, we normalized the libraries and aimed to sequence equally, targeting ~1x coverage on a single Illumina NextSeq run (223,862,027 reads for DM-Seq and 223,430,253 reads for BS-Seq).
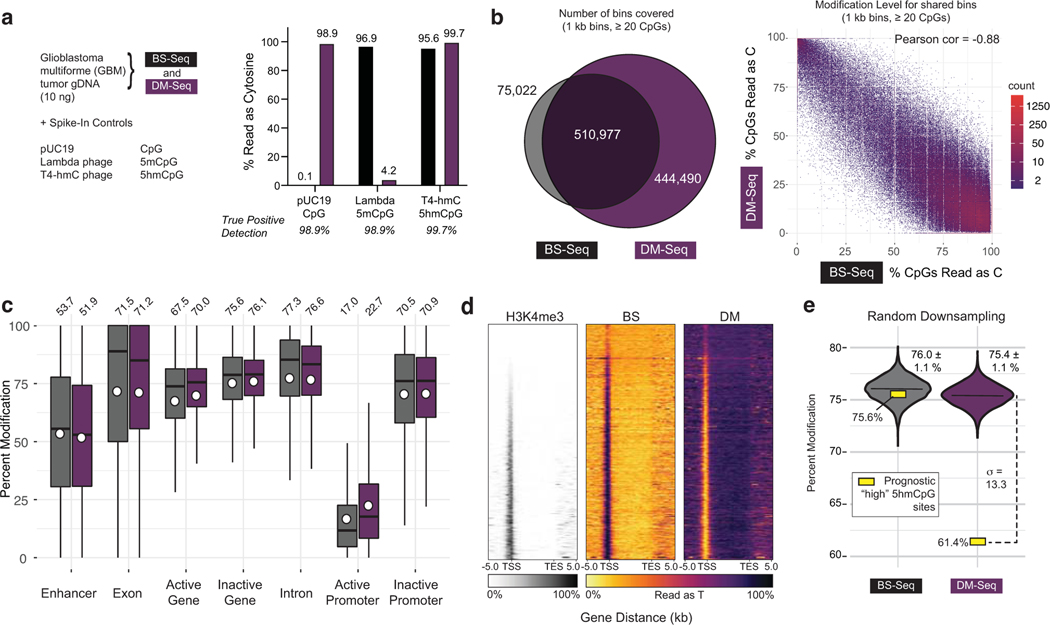
Figure 4. DM-Seq directly detects 5mCpGs in human glioblastoma.
a) Experimental design with spike-in controls showing accuracy of C, 5mC, and 5hmC detection. b) Binned CpG analysis using non-overlapping 1 kB bins with at least 20 CpGs covered. Left: Venn diagram showing bins covered by BS-Seq and DM-Seq. Right: Correlation between DM-Seq and BS-Seq in the 510,977 shared bins. c) Percent cytosine modification at various genomic features. The box shows the lower quartile, median, and upper quartile. Minimum and maximum values are shown by the whiskers. Circles are the mean values displayed above each boxplot. d) Heatmap representation of all annotated genes for H3K4me3 ChIP-Seq, BS-Seq, and DM-Seq. Genes are ranked by their average H3K4me3 signal. e) Observed DM-Seq and BS-seq signal at 3,876 previously defined “high 5hmCpG sites” (yellow square, DM-Seq: 61.4%, BS-Seq: 75.6%). The violin plot shows data from the shared BS and DM-Seq CpGs randomly downsampled 10,000 times to the same coverage as BS and DM-Seq at these sites. Data represents mean ± 1 standard deviation (BS-Seq = 76.0 ± 1.1%; DM-Seq = 75.4 ± 1.1%). The dotted line shows the number of standard deviations (13.3) between the downsampled (violin) and observed (yellow box) data at these prognostically significant CpGs. Extended data from these downsamplings are shown in Supplementary Table 4 and Extended Data Fig. 10.
Analysis of the BS-Seq spike-ins showed accurate conversion of unmodified Cs as Ts while 5mC and 5hmC were detected as Cs (Fig. 4a). With DM-Seq, CpGs in the unmodified pUC DNA sample were 98.9% sequenced as cytosine, validating efficient generation of 5cxmC and its protection from deamination by A3A. Efficient deamination of 5mC was confirmed with 95.8% of the CpGs in the in vitro methylated lambda gDNA sample reading as T relative to 96.9% expected based on BS-Seq, and the T4-hmC sample was protected from deamination with 99.7% bases reading as C. Thus, the true positive detection rates for each of the three modified bases was 98.9% or higher. This accurate detection of the pUC19 and T4-hmC spike in controls specifically provides strong evidence that copy-strands containing all 5mCs are not being measurably amplified in DM-Seq.
Despite compensating for the 2.8-fold lower library yields with BS-Seq, DM-Seq still provided more information content than BS-Seq in the normalized sequencing libraries (Extended Data Fig. 10). DM-Seq captured 5.9-fold more 1-kB non-overlapping, unique bins with at least 20 CpGs (444,490 vs 75,022) as compared to BS-Seq (Fig. 4b). Given that the majority of BS-Seq signal comes from 5mC and not 5hmC in GBM, we next explored correlations between the two data sets. Focusing on the 510,977 shared bins showed a strong inverse correlation between signals for BS-Seq (5mC + 5hmC) and DM-Seq (5mC alone) (Fig. 4b, Pearson = −0.88). Given the predominance of 5mC in GBM, we found that profiles generated by the two methods track with one another across various genomic elements (Fig. 4c). Notably, DM-Seq signals were distinct for active and inactive promoters (22.7 ± 19.1% vs 70.9 ± 20.7%), as defined by H3K4me3 ChIP Seq, with patterns that are mirrored relative to BS-Seq37, a relationship that extends when rank-ordering the enrichment level of H3K4me3 signal (Fig. 4d).
Having established that DM-Seq and BS-Seq are strongly correlated, we next sought to investigate the utility of direct 5mC mapping with DM-Seq by focusing on a limited subset of CpGs that have been shown to harbor DNA modifications of prognostic value. In prior work using oxBS-Seq microarrays, a candidate list of 3,876 CpGs were identified as the top 1% 5hmC sites across 30 GBMs, with an average 5hmC level of 10.1%9. These “high 5hmC sites” were disproportionally enriched in certain genomic elements, correlated with gene expression, and could be used to predict a 3.3-fold difference in patient survival. In our GBM tumor, 2,538 and 2,132 of these sites were sequenced by DM-Seq and BS-Seq, respectively, with 1,485 CpGs sequenced across both datasets (Extended Data Fig. 10). At these common sites, DM-Seq reported 61.4% 5mC modification, 14.3% lower than the 75.6% modification level observed by BS-Seq, quantifying the ‘blind spot’ that BS-Seq harbors to 5hmC at functionally important CpGs within this cancer (Fig. 4e). To determine if the ‘low 5mC’ level at these CpG sites could serve as a distinct signal, akin to the ‘high 5hmC’ detected by oxBS-Seq, we performed multiple downsamplings of 1,485 random CpG sites from either BS-Seq or DM-Seq (Fig. 4e). While the measured value for the ‘high 5hmC sites’ fell within the expected range with BS-Seq (76.0 ± 1.1%), these CpGs were major outliers in DM-Seq (75.4 ± 1.1%). These results highlight how direct detection of 5mC from DM-Seq, rather than a pooled 5mC/5hmC signal from BS-Seq, could advance efforts to sequence prognostically significant CpGs.
DISCUSSION
Here, we describe DM-Seq—the first non-destructive and enzyme-only workflow for directly detecting 5mCpGs at single-base resolution. To create DM-Seq, we originally envisioned employing an engineered MTase*:SAM-analog enzyme:substrate pair which could create an unnatural DNA base capable of resisting enzymatic deamination by A3A (Fig. 1a). We identified the CxMTase M.MpeI N374K and CxSAM pair as a favorable pair, but unexpectedly uncovered opposite-strand biases to DNA carboxymethylation that led us to employ several additional innovations. Specifically, we found that ligation of DNA deaminase-resistant adapters containing unnatural 5pyC, followed by copying of gDNA with 5mC in lieu of unmodified C, created an ideal substrate for CxMTase activity. The application of our CxMTase offers an important precedent, as engineering or evolution principles can be applied to invent enzymes that expand our sequencing toolbox beyond native activities. Opportunities abound to further improve the enzyme:substrate pair in DM-Seq as there are many SAM-analogs besides CxSAM38, and new DNA CxMTases could even be, in principle, evolved in vivo given the natural availability of CxSAM as a secondary metabolite in E. coli39.
Our work highlights how structure-activity studies on DNA-modifying enzymes can be harnessed to devise new sequencing pipelines. In existing DNA deaminase-based pipelines, the ability of A3A to discriminate against the natural DNA modifications, glucosylated 5hmC and 5-carboxylcytosine (5caC), have been exploited. We now reveal two new unnatural cytosine analogs that can be effectively used in sequencing applications: 5cxmC and 5pyC (Extended Data Fig. 4). Our findings that 5cxmC is protected from enzymatic deamination are corroborated by an independent study40, although their assay designs mask issues with opposite-strand modification that would have prevented whole-genome sequencing applications, as we have demonstrated here. We also anticipate added utility for 5pyC, as our novel 5pyC adapters can more generally be exploited to improve other non-destructive, DNA deaminase-dependent sequencing workflows.
While both borane and enzymatic deamination methods have been touted as non-destructive, we provide the first quantitative data, to our knowledge, that has shown on the same matched substrate that their relative effects on total DNA preservation are essentially equivalent (Extended Data Fig. 8d–e). This quantitative trend is likely explained by their contrasting mechanisms, where only bisulfite-catalyzed deamination required cytosine to undergo electrophilic activation creating unstable sulfonated intermediates prone to depyrimidation. We unexpectedly found that TAPS underestimates modified bases, a feature that may be attributed to DHU generation. Regions with a PPhigh density of DNA modifications in cis may be especially prone to underamplification. Although we anticipate that TAPS will remain a useful technology, as demonstrated by recent applications41, this behavior is critical to further understand, as caution should likely be taken when comparing new TAPS results to existing reference datasets based on BS-Seq or when deconvoluting mixtures of DNA. It remains to be seen whether quantitative correction for bias or the use of alternative polymerases, such as those evolved to copy across from sulfonated DHU42, could improve TAPS.
We believe that the development of DM-Seq highlights the limitations of current methods for 5mC localization, a topic of critical importance. Our GBM analysis revealed how directly sequencing 5mC with DM-Seq at specific CpGs can provide distinctive prognostic information. Extension of the technology to profiling sparse DNA samples, such as cell-free DNA for early cancer detection, can be readily imagined. Given its non-destructive and DHU-free workflow, DM-Seq could also be coupled to rapidly evolving third-generation sequencing platforms including nanopore for PCR-free or long-read sequencing. Ultimately, by directly mapping 5mC, rather than modified cytosines in aggregate, DM-Seq can allow for the biological function of 5mC alone to be better ascertained.
METHODS
Protein expression and purification.
The E. coli strain ER1821 (NEB) was used for all cloning and expression to overcome methylation-associated toxicity. The CxMTase M.MpeI N374K was expressed with an N-terminal fusion of maltose binding protein (MBP). Cloning of pMG81-MBP-M.MpeI-N374K-His was performed by Golden Gate Assembly43. Purification of MBP-A3A-His, M.MpeI-WT-His, M.MpeI-N374K-His were performed as previously described20,25. MBP-M.MpeI-N374K-His and M.MpeI-Q136A/N374A-His (eM.MpeI) were purified using the same single Cobalt column and high salt wash strategy as previously published without further purification.
Synthesis of SAM and cytosine analogs.
The synthesis and characterization of both bSAM and CxSAM have been previously described20,44. The synthesis and characterization of 5-ethynylcytosine and 5-vinylcytosine triphosphates have been previously reported27. The triphosphosphates of cytosine (Promega), 5-methylcytosine (NEB), and 5-propynylcytosine (TriLink) were purchased.
Oligonucleotide assay for A3A deamination.
A fluorescein (FAM)-labelled 27 bp top-strand oligonucleotide with a single unmethylated TCGA and unlabeled complementary bottom-strand oligonucleotide with a single methylated TCGA were used (Supplementary Table 1). 1 μM of the duplexed oligonucleotide was reacted in a final volume of 10 μL with no enzyme and no SAM, 1 μM M.MpeI WT with 40 μM SAM, 1 μM M.MpeI Q136A/N374A and 40 μM bSAM, or 1 μM M.MpeI N374K and 40 μM CxSAM at 37°C for 1 hr, before heat inactivation at 95°C for 5 min. The sample was treated with 1 μL of Proteinase K (NEB) and incubated at 37°C for 15 min before purification with an oligonucleotide spin column (Zymo) and elution in 10 μL 0.1x low EDTA TE. ESI-MS was obtained (Novatia) to validate the efficiency of 5bC and 5cxmC generation. 1 μL of the resulting DNA was snap cooled and then incubated with 6 μM MBP-A3A-His under ramping conditions for 2 hrs in a final volume of 50 μL, as previously described25. DNA was purified using an oligonucleotide spin column (Zymo) and DNA input was normalized. DNA was annealed to 10 μM (excess) unmethylated and mismatched opposite-strand in 1x CutSmart Buffer (NEB) in a total volume of 9.5 μL. The mismatched opposite strand was a safeguard ensuring that a deaminated TUGA top strand would not be cut. DNA was then digested with TaqαI (NEB) following recommended conditions in a total of 10 μL for 1 hour at 65°C. The samples were diluted 2-fold in 95% formamide and subjected to 20% denaturing polyacrylamide gel electrophoresis (PAGE) in Tris-Borate-EDTA (TBE) buffer, followed by imaging of the FAM signal using a Typhoon imager.
Lambda gDNA.
Dam–/Dcm– lambda phage gDNA was obtained (Thermo Fisher). The CpG- and GpC-modified lambda genomic DNA were modified with two rounds of DNA methylation as previously described for CpG-modified DNA25. The CpG-methylated DNA was modified using M.SssI (NEB) while GpC-methylated DNA was modified using M.CviPI (NEB) in recommended buffers.
General gDNA Processing.
Purified gDNA was quantified by Qubit (Thermo Fisher). All gDNA was sheared to ~350 bp using a Covaris sonicator before SPRI purification (Beckman, 1.2x). DNA was then end repaired with NEBNext Ultra End Prep Kit. The four DNA libraries were separately ligated with IDT xGen Y-shaped adapters containing either all 5mC (used in BS-Seq) or all 5pyC (used in A3A workflows including DM-Seq, custom synthesis from IDT) modifications using an NEBNext Ultra II Prep Kit, purified by SPRI beads (Beckman, 1.2X), and then re-quantified by Qubit. DNA was stored in the −20°C freezer after this step. All libraries, regardless of deamination method, are quantified (Qubit) and characterized by BioAnalyzer (High Sensitivity Kit, Agilent) before sequencing. Sequencing was performed using a MiSeq Reagent Kit v2 Nano (Illumina) except for human glioblastoma tumor libraries.
Initial DM-Seq Assay without copy-strand synthesis.
Dam–/Dcm– lambda phage gDNA was obtained (Thermo Fisher) and quantified by Qubit (Thermo Fisher). All gDNA was sheared to ~350 bp using a Covaris sonicator before SPRI purification (Beckman, 1.2x). DNA was then end-repaired with NEBNext Ultra End Prep Kit and ligated to 5mC-containing xGEN Y-shaped adapters (IDT). 10 ng 5mC-adapter ligated gDNA was reacted with untagged 0.5 μM M.MpeI-N374K and 160 μM CxSAM in M.MpeI reaction buffer (50 mM NaCl, 10 mM Tris-HCl pH 7.9, 10 mM EDTA) and incubated for 1 hour at 37°C followed by denaturation for 5 min at 95°C. 1 μL of Proteinase K was added (NEB) and incubated at 37°C for 15 min. The samples were SPRI purified (1.2x) and subjected to standard BS-Seq (Diagenode, see below). The library was amplified using indexing primers (IDT) and HiFi HotStart Uracil+ Ready Mix (KAPA) before purification over SPRI beads (0.8X) to yield final libraries.
PCR-based analysis for identifying cytosine analogs resistant to deamination.
A template DNA (see Supplementary Table 1) was synthesized (IDT). Modified dCTPs were used along with dATP, dTTP, and dGTP in a PCR reaction using the C-depleted primer OTF12 and G-depleted primer OTR12 (see Supplementary Table 1) and Taq Polymerase (NEB) in a final volume of 50 μL (see Supplementary Table 2 for all PCR methods). These 254 bp PCR products were purified over an oligonucleotide spin column (Qiagen) and quantified by Qubit (Thermo Fisher). In a volume of 6 μL, 2 ng of DNA was snap cooled and then incubated with 6 μM MBP-A3A-His under ramping conditions for 2 hrs in a final volume of 50 μL, as previously described25. Reaction products were purified using oligonucleotide spin columns (Zymo) and eluted into 10 μL. 1 μL of reaction product was PCR-amplified using Taq Polymerase using OTF2 and OTR2 primers containing Illumina TruSeq partial adapters in a total volume of 50 μL. For initial assessment of deamination, 5 μL of the crude PCR product was digested with TaqαI (NEB) following recommended conditions and visualized on a 2% Tris-Borate-EDTA (TBE) agarose gel pre-stained with SYBR Safe. For Next-Generation Sequencing analysis, the amplicons were indexed using another round of PCR in a total volume of 10 μL (IDT TruSeq primers, KAPA HiFI polymerase) before SPRI purification (0.8x) to yield final libraries.
Oligonucleotide assay for opposite-strand impacts on CxMTase.
A fluorescein (FAM)-labelled 27 bp top-strand oligonucleotide with a single unmethylated CCGG and unlabeled complementary bottom-strand oligonucleotides with either an unmethylated or methylated CCGG were used (Supplementary Table 1), as previously described20. 200 nM of the duplexed oligonucleotide was reacted in a final volume of 5 μL with 1 μM M.MpeI N374K and either 40 μM SAM or CxSAM substrate at 37°C for 30 min, before heat inactivation at 95°C for 5 min. 25x excess of unmethylated bottom-strand was added, and the duplex was reannealed before restriction digestion with HpaII (NEB) in a final volume of 50 μL. The samples analyzed by PAGE as described for oligonucleotide assay above.
Synthesis of modified adapters and evaluation of ligation efficiency.
5pyC adapters were synthesized by standard phosphoramidite chemistry (IDT). The purified oligonucleotides were further characterized by mass spectrometry (Extended Data Fig. 5a). For comparison to standard 5mC-containing adapters (IDT xGen Y-shaped adapters), a 254 bp DNA product with unmodified cytosine (described above, Supplementary Table 1) was ligated to adapters according to manufacturer instructions (NEB Ultra II). DNA was then purified by SPRI beads (Beckman) before amplification was attempted with either internal primers OTF2/R2 and Taq Polymerase (NEB) as above or Illumina TruSeq primers (IDT) and HiFi HotStart Polymerase (KAPA). Samples were visualized on a 2% TAE agarose gel.
Initial unoptimized copy-strand workflow.
10 ng of gDNA ligated to 5mC- or 5pyC-containing adapters was used as input. A methylated copy-strand was created. First, 1 μM of copy primer was annealed (Supplementary Table 1, v1), in a total volume of 10 μL in CutSmart Buffer and 1 mM final concentration individually of dATP/dGTP/dTTP (Promega) and dmCTP (NEB). 5 units of Klenow (exo-) polymerase (NEB) was then added and incubated for 30 min at 37°C. After purification (Zymo Oligo Clean & Concentrator), libraries were mixed with 1 μM untagged M.MpeI-N374K and 160 μM CxSAM in carboxymethylation buffer (50 mM NaCl, 10 mM Tris-HCl pH 7.9, 10 mM EDTA) and incubated for 1 hour at 37°C followed by denaturation for 5 min at 95°C. 1 μL of Proteinase K was added (NEB) and samples were incubated at 37°C for 15 min. The samples were purified using SPRI beads (1.2x) and eluted in 1 mM Tris-Cl, pH 8.0. DNA was then subjected to BS-Seq (Diagenode, see below) or to snap-cooling and A3A deamination in a final volume of 50 μL as previously described 25. Purified DNA was then amplified using indexing primers (IDT) and HiFi HotStart Uracil+ Ready Mix (KAPA) before purification over SPRI beads (0.8X) to yield final libraries. The non-optimized workflow was used in Figure 2.
Tumor DNA.
Patient glioblastoma tissue was collected at the Hospital of the University of Pennsylvania after informed patient consent under a protocol approved by the University of Pennsylvania’s Institutional Review Board. The patient had a surgically resected left temporal tumor. Fresh surgically resected glioblastoma tissue was placed in sterile phosphate buffered saline and taken immediately to the University of Pennsylvania Department of Pathology to confirm a preliminary diagnosis of grade IV glioma by the attending neuropathologist (M.N.). Tumor gDNA was extracted using the Agencourt FormaPure Kit with some protocol alterations as follows. Tissue was lysed at 70°C for 1 hour, proteinase K digestion was performed at 55°C for 1 hour, and tubes were briefly spun and incubated at 80°C for an additional hour. Tissue was lysed in tissue lysis buffer (Qiagen) at 56°C overnight, and nucleic acid was extracted by using the FormaPure protocol beginning with the bind 1 step.
Spike-In Controls:
The spike-in control contains a 1:1:1 (m:m:m) mixture of unmethylated pUC19 plasmid DNA (NEB), CpG-methylated lambda phage gDNA (see above), and T4-hmC phage gDNA extracted as previously described 15,25. Unsheared tumor gDNA was pooled 1:100 (m:m) with the spike-in mixture before gDNA processing (see above). To calculate true positive detection rates, it was assumed that pUC19 and T4-hmC were 100% pure of CpGs and 5hmCpGs, respectively, so DM-Seq detection was determined by the percentage of bases sequencing as cytosine. For 5mCpGs, incomplete modification by M.SssI was taken into account and true positives = % T by DM-Seq / % C by BS-Seq.
Optimized DM-Seq workflow.
See extended Supplementary Note for detailed discussion of specific steps in the optimized and final DM-Seq workflow. Briefly, 10 ng of gDNA ligated to 5pyC-containing adapters was used as input for DM-Seq. A methylated copy-strand was created. 1 μM fully-methylated copy primer was annealed (Supplementary Table 1, v2) in a total volume of 10 μL in CutSmart Buffer and 1 mM final concentration (individually) of dATP/dGTP/dTTP (Promega) and dmCTP (NEB). 1 μl or 8 units Bst polymerase, large fragment (NEB) was added and incubated for 30 min at 65°C. The 5hmCs were then glucosylated with 40 μM UDP-Glucose and 1 μL or 10 units of T4 Phage β-glucosyltransferase (NEB) for 1 hour at 37°C in a final volume of 20 μL. Incompletely copied or uncopied fragments were degraded with 1 μL or 10 units Mung Bean Nuclease (NEB) for 30 min at 30°C. After SPRI bead purification (1.2x), libraries were mixed with 0.5 μM MBP-M.MpeI-N374K and 160 μM CxSAM in carboxymethylation buffer (50 mM NaCl, 10 mM Tris-HCl pH 7.9, 10 mM EDTA) and incubated overnight at 37°C followed by denaturation for 5 min at 95°C. 1 μL or 0.8 units of Proteinase K (NEB) was subsequently added and incubated at 37°C for 15 min. The samples were purified using SPRI beads (1.2x) and eluted in 1 mM Tris-Cl, pH 8.0. DNA was then subjected to snap-cooling and A3A deamination in a final volume of 50 μL as previously described 25 before SPRI beads purification (1.2x). DM-Seq libraries were amplified using indexing primers (IDT) and HiFi HotStart Uracil+ Ready Mix (KAPA Biosystems) before purification over SPRI beads (0.8X) to yield final libraries. This optimized workflow was used in Figures 3 and 4. Alternative deamination conditions using commercially-available APOBEC (NEB) and formamide denaturation were used without other alterations for the data generated in Extended Data Fig. 7 and 8.
Bisulfite Sequencing.
BS-Seq was performed on 10 ng gDNA ligated to 5mC-containing adapters (xGen, IDT), with no added copy or DM-Seq specific steps, using manufacturer instructions (Diagenode). Purified BS-Seq libraries were amplified using indexing primers (IDT) and HiFi HotStart Uracil+ Ready Mix (KAPA Biosystems) before purification over SPRI beads (0.8X) to yield final libraries.
TET-assisted Pyridine Borane Sequencing (TAPS) and TET-assisted Pyridine Borane Sequencing with βGT blocking (TAPS-β).
TAPS and TAPS-β were performed as previously described except for the source of TET enzyme (NEB, EM-Seq Conversion Module). 1 ng of sheared DNA input (consisting of fully unmodified C pUC19 DNA, 5mCpG-modified lambda phage gDNA, and fully 5hmC-modified T4 phage gDNA) was ligated to C-containing Y-shaped adaptors using the same protocol as described above (IDT). For TAPS-β, the ligated DNA was added to a 20 μL reaction containing 1x NEB CutSmart buffer (50 mM Potassium Acetate 20 mM Tris-acetate, 10 mM Magnesium Acetate, and 100 μg/ml BSA), 0.04 nM UDP-glucose, and 10 U T4-βGT. The glucosylated DNA was then purified with SPRI beads (1.2x). This glucosylation step was omitted for standard TAPS. The purified DNA was then incubated in a 50 μL reaction containing 1x NEB EM-Seq TET buffer (50 mM Tris pH 8.0, 1 mM DTT, 5 mM sodium-L-ascorbate, 20 mM αKG, 2 mM ATP, and 50 mM ammonium iron (II) sulfate hexahydrate) and 16 μg TET2 (NEB). The reaction was then incubated at 37°C for 80 min. Following oxidation, 0.8 U of Proteinase K (NEB) was added, and the mixture was incubated for 30 min at 37°C. The oxidized DNA was then purified with SPRI beads (1.2x) and input into a second round of TET oxidation. The oxidized DNA was added to a 50-μL reaction containing 600 mM sodium acetate (pH 4.3) and 1 M pyridine borane (Alfa Aesar). The reaction was incubated at 37 °C and 850 r.p.m. in a ThermoMixer (Eppendorf) placed in a chemical fume hood for 16 hrs and purified by Zymo-IC column (Zymo Research) with Oligo Binding Buffer (Zymo Research). The libraries were amplified using indexing primers (NEB) and HiFi Hotstart Uracil+ Ready Mix (KAPA Biosystems) before purification with SPRI beads (1.2x) to yield final libraries.
Comparison of sequencing technologies.
1 ng of sheared DNA input (consisting of fully unmodified C pUC19 DNA, 5mCpG modified Lambda control gDNA, and fully 5hmC-modified T4 phage gDNA) was used for each condition. The 5mCpG modified lambda control gDNA was one of 4 possibilities: fully unmethylated, fully CpG-methylated, fully GpC-methylated, or ~50% CpG-methylated. The ~50% CpG-methylated DNA was made by mixing equal amounts of unmethylated and fully CpG-methylated lambda DNA. DNA was end-prepped and ligated with the same protocol as above except for the adapters used, which was different for each method (no deamination, TAPS, TAPS-β: C adapters, BS-Seq: 5mC adapters, DM-Seq: 5pyC adapters). Each method was then performed according to the protocols described above before sequencing. Bioinformatics were performed as below using a standard Bismark based alignment for all pipelines. Bioinformatic statistics including number of reads and conversion efficiencies are provided in the Supplementary Data File.
Comparison of deamination methods.
1 ng of sheared DNA input (consisting of fully unmodified C pUC19 DNA, fully 5mCpG modified Lambda control gDNA, and fully 5hmC-modified T4 phage gDNA) was used for each condition. DNA was added to a 20 μL reaction containing 1x NEB CutSmart buffer (50 mM Potassium Acetate 20 mM Tris-acetate, 10 mM Magnesium Acetate, and 100 μg/ml BSA), 0.04 nM UDP-glucose, and 10 U T4-βGT. The glucosylated DNA was then purified with SPRI beads (1.2x). The purified DNA was then in a 50 μL reaction containing 1x NEB EM-Seq TET buffer (50 mM Tris pH 8.0, 1 mM DTT, 5 mM sodium-L-ascorbate, 20 mM αKG, 2 mM ATP, and 50 mM ammonium iron (II) sulfate hexahydrate) and 16 μg TET2 (NEB). The reaction was then incubated at 37°C for 80 min. Following oxidation, 0.8 U of Proteinase K (NEB) was added and the mixture was incubated for 30 min at 37°C. The oxidized DNA was then purified with SPRI beads. The DNA was then subjected to a second round of TET oxidation, proteinase K treatment, and SPRI purification. The no TET control was carried through mock oxidation reactions without TET enzyme. The purified oxidized sample was then end-prepped and ligated with the same protocol as above except for the adapters used, which was different for each method (no deamination, TAPS, TAPS-β: C adapters, BS-Seq: 5mC adapters, DM-Seq: 5pyC adapters). BS deamination, A3A deamination, pyridine borane deamination, or a no deamination control were then performed as above. Bioinformatics were performed as below using a standard Bismark alignment for all pipelines. Bioinformatic statistics including number of reads and conversion efficiencies are provided in the Supplementary Data File. The deamination methods were also compared by qPCR and BioAnalyzer using this same workflow, by using unamplified libraries as input to determine Ct values (KAPA SYBR FAST ROX, Applied Biosystems).
General bioinformatics.
Reads were quality and length trimmed with Trim Galore! Reads were aligned with Bismark and deduplicated with Picard 45. All data was analyzed single-end. Reads were filtered if 3 consecutive CpHs were non-converted using Bismark’s existing filter_non_conversion command. Locus-specific amplicons (cytosine analog experiment, see above) were not deduplicated or filtered. Filtering served two purposes (in different experiments). For BS-Seq with copy-strand synthesis, the consecutive CpH conversion eliminated reads from copy-strand amplification which contained all mCpHs, unlike the lambda gDNA template. BS-Seq without copy-strand synthesis was not filtered. For DM-Seq, the copy-strand does not amplify because the copy primer 5mCs are deaminated to Ts by A3A. DM-Seq filtering additionally eliminates dsDNA hairpins which can cause A3A non-deamination, similar to previously described enzymatic deamination protocols15,25. Only reads with MAPQ ≥ 30 were analyzed. GraphPad Prism 7 was used to report percent CpG modification unless otherwise specified. See Supplementary Table 3 for library statistics. A custom R script was employed to visualize target and opposite-strand modifications as well as GpC specific analysis. Pearson correlations at the individual CpG level for BS-Seq vs DM-Seq was obtained using the cor function in R.
TAPS mammalian bioinformatics.
Data was obtained as mm9 bedfiles from GSE112520 where TAPS and BS-Seq were performed on mESCs. For correlation analyses, the raw TAPS and BS-seq signals were calculated for 1 kB non-overlapping genomic bins. Pearson correlation was obtained using the cor function in R. ICR locations were previously provided described as mm10 but required liftover (https://liftover.broadinstitute.org/) to convert to mm9. Heatmaps were produced using deepTools (v3.5.1). In the heatmaps, genes were scaled to 3 kb and binned at 300 bases.
Glioblastoma bioinformatics.
BS-Seq and DM-Seq GBM libraries were sequenced 75-bp single end on an Illumina NextSeq using a High Output kit v2.5. Reads were processed as above. For correlation analyses, the raw DM-Seq and BS-seq signals were calculated for 1 kB non-overlapping genomic bins. Only bins containing at least 20 CpGs were analyzed. Venn diagrams were generated using the VennDiagram package in R. Pearson correlation was obtained using the cor function in R, using the same bins as described for generating the Venn diagram. Heatmaps were produced using deepTools (v3.5.1). In the heatmaps, genes were scaled to 10 kb and binned at 500 bases. Only genes with all bins visualized between DM-Seq and BS-Seq are shown. Genomic elements were defined as follows: exons and introns: UCSC RefGene for genome build hg19. Promoters: +/− 1kb from the transcription start site. Active promoters: H3K4me3 ChIP-Seq performed on glioblastoma stem cells37. Enhancers: Non-promoter regions with a H3K27Ac ChIP-Seq signal34. Signals in genomic elements were determined using bedtools with each genomic element as defined as an individual bin (v.2.25.0). The 3,876 “high 5hmC” CpG sites were previously defined identified by OxBS-Seq and used to calculate percent modification by both BS-Seq and DM-Seq9. Downsampling was performed to obtain 1,485 CpGs from the 16,438,445 total CpGs covered by both BS-Seq and DM-Seq in R. Random downsampling was performed to randomly extract 10,000 CpGs from the obtained BS-Seq and DM-Seq data and extended results are reported in Extended Data Fig. 10 and Supplementary Table 4.
Research Use Only Statement.
IDT products are for research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser or user is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.
Data availability.
Sequencing data supporting the findings of this study are available in the NCBI Gene Expression Omnibus (GEO, GSE225975). The plasmid encoding MBP-t-M.MpeI-N374K-His has be made available from Addgene (197985). Relevant DNA sequences are provided in Supplementary Information.
Code availability.
Software utilized for each analysis is detailed in the relevant Methods section. Scripts have been deposited on Github (https://github.com/twang518/DM-Seq).
Extended Data
Extended Date Figure 1. Chemical and enzymatic sequencing methods for resolving DNA modifications.
a) Methods differ in their use of protection or modification steps to alter C, 5mC or 5hmC. They differ in deamination steps with chemical or enzymatic reagents. In each method, C, 5mC, or 5hmC are detected based on the pattern of C-to-T changes in sequencing, resulting in different possible bases that can be confounded with 5mC. b) Shown are the anticipated sequencing results for C, 5mC and 5hmC in CpG versus CpH contexts.
Extended Date Figure 2. TaqαI assay for assessment of modified cytosine deamination by A3A.
a) A fluorophore-labelled top-strand is duplexed to a complementary bottom-strand containing a methylated cytosine. The methylated cytosine is represented with a black oval. The substrate is reacted with either WT M.MpeI + SAM, eM.MpeI + bSAM, or M.MpeI N374K + CxSAM. The half-purple/half-orange oval represents a modified cytosine that can either be 5mC, 5bC, or 5cxmC after the action of the MTase variant and SAM analog. The substrate is then deaminated with A3A before duplexing a complement strand. The restriction enzyme TaqαI only cleaves DNA if C is protected from A3A deamination. b) ESI-MS validating generation of 5bC and 5cxmC substrates before A3A reaction. No unmodified C substrate was detected.
Extended Date Figure 3. HpaII assay for assessment of opposite strand effects in carboxymethylation.
A fluorophore-labelled top-strand is duplexed to a complementary strand containing either an unmodified or methylated cytosine (represented with a black oval). The duplex is incubated with M.MpeI N374K and either no SAM, SAM, or CxSAM. The half-black/half-purple oval represents the modified cytosine on the labelled top-strand resulting after the action of the M.MpeI N374K and the SAM analog. Excess of unmodified bottom strand exchanges away the modification on the bottom strand. HpaII cleavage interrogates the modification status of the top strand.
Extended Date Figure 4. Structurally-informed identification of both 5cxmC and 5pyC as new protected cytosines useful for A3A dependent sequencing.
a) Generation of homogenously-modified PCR substrates containing unnatural cytosines. A DNA template is amplified with a C-depleted forward primer (red) and G-depleted reverse primer (green) as well as dA/G/TTP and a modified dCTP (blue). DNA is then A3A deaminated before amplification. Amplicons are interrogated with the TaqαI restriction enzyme or by Next-Generation Sequencing quantifying all C sites. b) Active site of human A3A (PDB: 5SWW) showing gating tyrosine (orange) which abuts the C5-C6 face of the target cytosine (yellow) and is anticipated to limit the size of the 5-position substituent (dashed yellow line). A cartoon representation is also shown above. c) Summary of cytosine analogs and deamination by A3A. Left: WT MTases, TETs, and βGT make naturally-occurring modified cytosines which have different reactivities towards A3A. 5caC and 5ghmC are used in the existing methods, EM-Seq and ACE-Seq, to protect from A3A deamination. Right: 5cxmC and 5pyC are identified as novel, protected A3A substrates, both employed in DM-Seq. Despite their shared utility, 5cxmC and 5pyC also contrast in their bond types at the 5-position of cytosine, which are determined by their contrasting modes of biochemical and chemical synthesis, respectively.
Extended Date Figure 5. 5pyC adapters improve DNA carboxymethylation efficiency through the synthesis of a 5mC copy strand.
a) Structure of 5pyC adapters. ESI-MS characterizing 5pyC adapters. Expected m/z of the two strands: 10,444.2 and 9,936.6. The phosphorothioate linkage (*) substitutes a sulfur in place of a phosphate in the backbone of the oligonucleotide to minimize nuclease degradation. b) 5pyC adapter ligation experiment. Template DNA was ligated to 5mC- or 5pyC-containing Y-shaped adapters. The template DNA was detected by amplification with internal primers (red) or successful ligation was detected by amplification with Illumina indexing primers (blue). Experiment was performed once. c) Schematic of copy strand synthesis. A copy strand is made by incubation of a copy primer, polymerase, and dA/G/TTPs with 5mdCTP.
Extended Date Figure 6. Copy strand synthesis improves DNA carboxymethylation efficiency.
a) Experimental scheme. Sheared lambda gDNA is ligated to 5mC- or 5pyC-containing adapters. A copy strand with 5mCs is synthesized before reaction with the CxMTase and either no SAM, SAM, or CxSAM, with the product of this reaction represented by the oval with mixed colors. Subsequent BS or A3A deamination shows efficiency of DNA modification. Data are presented as mean values +/− SD (n=2 independent experiments). b) Next-Generation Sequencing quantifying efficiency of CxMTase with methylation or carboxymethylation after copy strand synthesis.
Extended Date Figure 7. Comparison of different sequencing methods on M.CviPI-modified gDNA.
The M.CviPI-methylated lambda phage gDNA shows near complete modification at GpCpG sequencing contexts given the known GpC preference for M.CviPI. The enzyme has known off-target and heterogenous activity at CpCpG sites. The dashed line shows the readout if BS-Seq signal inversely correlates with DMSeq as anticipated. a) Correlation of BS-Seq to DM-Seq in an M.CviPI-modified substrate. b) Comparison of BS-Seq and DM-Seq modification status by 5’ sequence context. The box shows the lower quartile, median, and upper quartile. Minimum and maximum values are shown by the whiskers. Data in a-b) corresponds to experiment shown in Fig. 3a–b. c) Comparison of BS-Seq, TAPS, TAPS-β, and DM-Seq modification status by 5’ sequence context. The box shows the lower quartile, median, and upper quartile. Minimum and maximum values are shown by the whiskers. d) Percent modification in CpG contexts of BSSeq, TAPS, TAPS-β, and DM-Seq of 3 different methylated lambda phages. Data in c-d) corresponds to experiment shown in Fig. 3c–d.
Extended Date Figure 8. Comparison of deamination methods show that TAPS bias is dependent on both TET and borane-mediated deamination.
a) Workflow for comparing deamination methods. A mixture of unmodified pUC19 DNA, 100% CpG methylated lambda phage, and T4-hmC phage (where all C bases are 5hmC) was glucosylated. Samples were then subjected to either two rounds of TET treatment or no TET treatment. DNA was ligated to the appropriate adapter and subjected to one of four conditions: no deamination, BS, pyridine borane or A3A. The pyridine borane workflow is equivalent to TAPS-β. The bases deaminated by each method (detected as T by sequencing) are noted, with structures of the deamination products at right, including the non-aromatic DHU. b) Percent reads C as determined by the methylated lambda phage spike-in. The sample with TET and bisulfite indicates efficient conversion of 5mC to 5fC/5caC by TET. c) Proportion of reads mapping to each spike-in are shown. Only borane deamination shows decreased reads mapping to the methylated lambda phage, with depletion dependent upon TET oxidation. d) qPCR detection of amplifiable DNA library after each deamination method. Shown are the p-values from paired two-tailed t-test (n = 4 for each deamination condition, 3e-4 between BS and borane, 2e-5 between BS and A3A). Data are presented as mean values +/− SD. e) Mean library size ± standard deviation for each deamination method. A representative BioAnalyzer trace is shown for each deamination method.
Extended Date Figure 9. Existing mammalian TAPS vs BS-Seq data suggest bias.
a) Binned CpG analysis using non-overlapping 1 kB bins. Correlation between TAPS and BS-Seq in mESCs in existing datasets (GSE112520). b) Histogram showing ~2-fold as many 1 kB bins with greater modification detected by BS-Seq than TAPS. Percent Deviation = (TAPS % reads T – BS-Seq % reads C) / (BS-Seq % reads C). c) Percent deviation of TAPS vs BS-Seq as a function of % modification of CpGs in a given 1 kB bin. The box shows the lower quartile, median, and upper quartile. Minimum and maximum values are shown by the whiskers. d) ICRs show underdetection of 5mC by TAPS relative to BS-Seq. At bottom is the heatmap representation of individual ICRs. The percent modification outside of ICRs (64.2% vs 60.1%) represents the genome-wide average for each method using 1 kB bins vs just at the ICR (41.9% vs 31.6%). e) Plot of the CpG density in individual ICRs versus the percent TAPS underestimates the level of 5mC relative to BS-Seq. 28 of 29 ICRs show lower modification density by TAPS than by BS-Seq. The one exception is shown in red. The associated correlation coefficient tracks the % underestimate as a function of CpG density.
Extended Date Figure 10. Mammalian genome DM-Seq metrics.
DM-seq and BS-seq data from gDNA derived from a patient glioblastoma. a) Final library yield. b) Average size of library fragments (adapters included) determined using a Bioanalyzer. c) Unique CpGs covered by BS-Seq and DM-Seq. The extrapolated BS-Seq bar takes into account if the sequencer was loaded with the same volume of each library rather than by normalizing the amount of DNA loaded. d) High 5hmCpG sites, previously identified by oxBS-Seq of 30 tumors. The Venn diagram shows the portion of these CpG sites that were covered by BS-seq or DM-Seq with this glioblastoma sample. The metrics for the sites sequenced by either BS-Seq or DM-Seq alone are similar to those at the sites that were sequenced by both methods. The analysis in Fig. 4e focuses on the 1,485 shared CpG sites sequenced by both methods.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Yemin Lan, Wanding Zhou, Tim Christopher, and the Penn Center for Personalized Diagnostics for useful discussions and reagents. We also thank Kabirul Islam for generously providing but-2-ynyl-SAM. This work was supported by the National Institutes of Health through R01-HG10646 (to R.M.K. and H.W.). E.K.S., K.N.B., and J.E.D. were NSF Graduate Research Fellows.
Footnotes
COMPETING INTERESTS
The University of Pennsylvania has patents pending for CxMTase enzymes, DNA deaminase-resistant adapters and the DM-Seq pipeline. R.M.K. has served as a scientific advisory board member for Cambridge Epigenetix (CEGX). W.G. is an employee of CEGX and T.W. was supported by a fellowship from CEGX. A.D. and N.D. are employees of Integrated DNA Technologies, Inc. The remaining authors declare no competing interests.
REFERENCES
- 1.Schubeler D. Function and information content of DNA methylation. Nature 517, 321–326 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Luo C, Hajkova P. & Ecker JR Dynamic DNA methylation: In the right place at the right time. Science 361, 1336–1340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, Zuzarte PC, Borgida A, Wang TT, Li T, Kis O, Zhao Z, Spreafico A, Medina TDS, Wang Y, Roulois D, Ettayebi I, Chen Z, Chow S, Murphy T, Arruda A, O’Kane GM, Liu J, Mansour M, McPherson JD, O’Brien C, Leighl N, Bedard PL, Fleshner N, Liu G, Minden MD, Gallinger S, Goldenberg A, Pugh TJ, Hoffman MM, Bratman SV, Hung RJ & De Carvalho DD Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MDM, Niu B, McLellan MD, Uzunangelov V, Zhang J, Kandoth C, Akbani R, Shen H, Omberg L, Chu A, Margolin AA, Van’t Veer LJ, Lopez-Bigas N, Laird PW, Raphael BJ, Ding L, Robertson AG, Byers LA, Mills GB, Weinstein JN, Van Waes C, Chen Z, Collisson EA, Cancer Genome Atlas Research Network, Benz CC, Perou CM & Stuart JM Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158, 929–944 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL & Paul CL A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U. S. A. 89, 1827–1831 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka K. & Okamoto A. Degradation of DNA by bisulfite treatment. Bioorg. Med. Chem. Lett. 17, 1912–1915 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR & Rao A. The Behaviour of 5-Hydroxymethylcytosine in Bisulfite Sequencing. PLoS ONE 5, e8888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kriaucionis S. & Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson KC, Houseman EA, King JE, von Herrmann KM, Fadul CE & Christensen BC 5-Hydroxymethylcytosine localizes to enhancer elements and is associated with survival in glioblastoma patients. Nat. Commun. 7, 13177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T, Loo CE & Kohli RM Enzymatic approaches for profiling cytosine methylation and hydroxymethylation. Mol. Metab, 101314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W. & Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B. & He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Siejka-Zielinska P, Velikova G, Bi Y, Yuan F, Tomkova M, Bai C, Chen L, Schuster-Bockler B. & Song CX Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat. Biotechnol. 37, 424–429 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Hu Z, Cheng J, Siejka-Zielińska P, Chen J, Inoue M, Ahmed AA & Song CX Subtraction-free and bisulfite-free specific sequencing of 5-methylcytosine and its oxidized derivatives at base resolution. Nat. Commun. 12, 618–2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schutsky EK, DeNizio JE, Hu P, Liu MY, Nabel CS, Fabyanic EB, Hwang Y, Bushman FD, Wu H. & Kohli RM Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat. Biotech. 36, 1083–1090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaisvila R, Ponnaluri VKC, Sun Z, Langhorst BW, Saleh L, Guan S, Dai N, Campbell MA, Sexton BS, Marks K, Samaranayake M, Samuelson JC, Church HE, Tamanaha E, Corrêa IR, Pradhan S, Dimalanta ET, Evans TC, Williams L. & Davis TB EM-seq: Detection of DNA Methylation at Single Base Resolution from Picograms of DNA. bioRxiv, 2019.12.20.884692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, Wu X, Shen L. & Zhang Y. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol. 32, 1231–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stasevskij Z, Gibas P, Gordevicius J, Kriukiene E. & Klimasauskas S. Tethered Oligonucleotide-Primed Sequencing, TOP-Seq: A High-Resolution Economical Approach for DNA Epigenome Profiling. Mol. Cell 65, 554–564.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kriukienė E, Labrie V, Khare T, Urbanavičiūtė G, Lapinaitė A, Koncevičius K, Li D, Wang T, Pai S, Ptak C, Gordevičius J, Wang SC, Petronis A. & Klimašauskas S. DNA unmethylome profiling by covalent capture of CpG sites. Nat. Commun. 4, 2190 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Wang T. & Kohli RM Discovery of an Unnatural DNA Modification Derived from a Natural Secondary Metabolite. Cell. Chem. Biol. 28, 97–104.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C. & Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y. & Kohli RM AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 8, 751–758 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schutsky EK, Nabel CS, Davis AKF, DeNizio JE & Kohli RM APOBEC3A efficiently deaminates methylated, but not TET-oxidized, cytosine bases in DNA. Nucleic Acids Res. 45, 7655–7665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seiler CL, Fernandez J, Koerperich Z, Andersen MP, Kotandeniya D, Nguyen ME, Sham YY & Tretyakova NY Maintenance DNA Methyltransferase Activity in the Presence of Oxidized Forms of 5-Methylcytosine: Structural Basis for Ten Eleven Translocation-Mediated DNA Demethylation. Biochemistry 57, 6061–6069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, Luo M, Berrios KN, Schutsky EK, Wu H. & Kohli RM Bisulfite-Free Sequencing of 5-Hydroxymethylcytosine with APOBEC-Coupled Epigenetic Sequencing (ACE-Seq). Methods Mol. Biol. 2198, 349–367 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Shi Carpenter, Banerjee Shaban, Kurahashi Salamango, Mccann Starrett, Duffy Demir, Amaro Harki, Harris & Aihara. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat Struct Mol Biol 24, 131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghanty U, DeNizio JE, Liu MY & Kohli RM Exploiting Substrate Promiscuity to Develop Activity-Based Probes for TET Family Enzymes. J. Am. Chem. Soc. 140, 17329–17332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chinchilla R. & Najera C. The Sonogashira reaction: a booming methodology in synthetic organic chemistry. Chem. Rev. 107, 874–922 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Kelly TK, Liu Y, Lay FD, Liang G, Berman BP & Jones PA Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 22, 2497–2506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Cheng J, Siejka-Zielińska P, Weldon C, Roberts H, Lopopolo M, Magri A, D’Arienzo V, Harris JM, McKeating JA & Song CX Accurate targeted long-read DNA methylation and hydroxymethylation sequencing with TAPS. Genome Biol. 21, 54–6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sipa K, Sochacka E, Kazmierczak-Baranska J, Maszewska M, Janicka M, Nowak G. & Nawrot B. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA 13, 1301–1316 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalluge JJ, Hashizume T, Sopchik AE, McCloskey JA & Davis DR Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res. 24, 1073–1079 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onodera A, González-Avalos E, Lio CJ, Georges RO, Bellacosa A, Nakayama T. & Rao A. Roles of TET and TDG in DNA demethylation in proliferating and non-proliferating immune cells. Genome Biol. 22, 186–1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, Curry WT, Martuza RL, Rivera MN, Rossetti N, Kasif S, Beik S, Kadri S, Tirosh I, Wortman I, Shalek AK, Rozenblatt-Rosen O, Regev A, Louis DN & Bernstein BE Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157, 580–594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klughammer J, Kiesel B, Roetzer T, Fortelny N, Nemc A, Nenning KH, Furtner J, Sheffield NC, Datlinger P, Peter N, Nowosielski M, Augustin M, Mischkulnig M, Ströbel T, Alpar D, Ergüner B, Senekowitsch M, Moser P, Freyschlag CF, Kerschbaumer J, Thomé C, Grams AE, Stockhammer G, Kitzwoegerer M, Oberndorfer S, Marhold F, Weis S, Trenkler J, Buchroithner J, Pichler J, Haybaeck J, Krassnig S, Mahdy Ali K, von Campe G, Payer F, Sherif C, Preiser J, Hauser T, Winkler PA, Kleindienst W, Würtz F, Brandner-Kokalj T, Stultschnig M, Schweiger S, Dieckmann K, Preusser M, Langs G, Baumann B, Knosp E, Widhalm G, Marosi C, Hainfellner JA, Woehrer A. & Bock C. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat. Med. 24, 1611–1624 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raiber EA, Beraldi D, Martínez Cuesta S, McInroy GR, Kingsbury Z, Becq J, James T, Lopes M, Allinson K, Field S, Humphray S, Santarius T, Watts C, Bentley D. & Balasubramanian S. Base resolution maps reveal the importance of 5-hydroxymethylcytosine in a human glioblastoma. NPJ Genom. Med. 2, 6–6. eCollection 2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Q, Wu TP, Gimple RC, Li Z, Prager BC, Wu Q, Yu Y, Wang P, Wang Y, Gorkin DU, Zhang C, Dowiak AV, Lin K, Zeng C, Sui Y, Kim LJY, Miller TE, Jiang L, Lee CH, Huang Z, Fang X, Zhai K, Mack SC, Sander M, Bao S, Kerstetter-Fogle AE, Sloan AE, Xiao AZ & Rich JNN (6)-methyladenine DNA Modification in Glioblastoma. Cell 175, 1228–1243.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J. & Zheng YG SAM/SAH Analogs as Versatile Tools for SAM-Dependent Methyltransferases. ACS Chemical Biology 11, 583–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Xiao H, Bonanno JB, Kalyanaraman C, Brown S, Tang X, Al-Obaidi NF, Patskovsky Y, Babbitt PC, Jacobson MP, Lee Y. & Almo SC Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature 498, 123–126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong J, Chen KK, Xie NB, Ji TT, Yu SY, Tang F, Xie C, Feng YQ & Yuan BF Bisulfite-Free and Single-Base Resolution Detection of Epigenetic DNA Modification of 5-Methylcytosine by Methyltransferase-Directed Labeling with APOBEC3A Deamination Sequencing. Anal. Chem. (2022). [DOI] [PubMed] [Google Scholar]
- 41.Siejka-Zielińska P, Cheng J, Jackson F, Liu Y, Soonawalla Z, Reddy S, Silva M, Puta L, McCain MV, Culver EL, Bekkali N, Schuster-Böckler B, Palamara PF, Mann D, Reeves H, Barnes E, Sivakumar S. & Song CX Cell-free DNA TAPS provides multimodal information for early cancer detection. Sci. Adv. 7, eabh0534 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Millar D, Christova Y. & Holliger P. A polymerase engineered for bisulfite sequencing. Nucleic Acids Res. 43, e155 (2015).stylefix [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS ONLY REFERENCES
- 43.Engler C, Kandzia R. & Marillonnet S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PloS one 3, e3647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arora S, Horne WS & Islam K. Engineering Methyllysine Writers and Readers for Allele-Specific Regulation of Protein-Protein Interactions. J. Am. Chem. Soc. 141, 15466–15470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krueger F. & Andrews SR Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data supporting the findings of this study are available in the NCBI Gene Expression Omnibus (GEO, GSE225975). The plasmid encoding MBP-t-M.MpeI-N374K-His has be made available from Addgene (197985). Relevant DNA sequences are provided in Supplementary Information.