Small-molecule stabilization of weak ubiquitin-enzyme interactions to selectively target the ubiquitin-proteasome system.
Abstract
Pharmacological control of the ubiquitin-proteasome system (UPS) is of intense interest in drug discovery. Here, we report the development of chemical inhibitors of the ubiquitin-conjugating (E2) enzyme CDC34A (also known as UBE2R1), which donates activated ubiquitin to the cullin-RING ligase (CRL) family of ubiquitin ligase (E3) enzymes. A FRET-based interaction assay was used to screen for novel compounds that stabilize the noncovalent complex between CDC34A and ubiquitin, and thereby inhibit the CDC34A catalytic cycle. An isonipecotamide hit compound was elaborated into analogs with ~1000-fold increased potency in stabilizing the CDC34A-ubiquitin complex. These analogs specifically inhibited CDC34A-dependent ubiquitination in vitro and stabilized an E2~ubiquitin thioester reaction intermediate in cells. The x-ray crystal structure of a CDC34A-ubiquitin-inhibitor complex uncovered the basis for analog structure-activity relationships. The development of chemical stabilizers of the CDC34A-ubiquitin complex illustrates a general strategy for de novo discovery of molecular glue compounds that stabilize weak protein interactions.
INTRODUCTION
The ubiquitin-proteasome system (UPS) controls the stability, localization, and interactions of much of the proteome and thereby governs cell behavior (1). Covalent attachment of the small highly conserved protein ubiquitin to a substrate protein generates a variety of specific signals that dictate the fate of the substrate (2). The addition of ubiquitin to a substrate is catalyzed by a conserved enzymatic cascade. Ubiquitin is first activated as a high-energy thioester linkage to the catalytic cysteine of a ubiquitin-activating (E1) enzyme in an adenosine 5′-triphosphate (ATP)–dependent fashion. The activated ubiquitin is then transferred as a subsequent thioester to the catalytic cysteine of a ubiquitin-conjugating (E2) enzyme. Last, ubiquitin is covalently conjugated as an isopeptide bond to a substrate lysine residue by a ubiquitin ligase (E3) enzyme. E3 enzymes fall into two broad classes: those that ligate ubiquitin to substrate via a catalytic thioester intermediate, termed homology to E6AP C terminus (HECT) and RING-between-RING (RBR) E3 enzymes, and those that bridge the E2 enzyme to the substrate for direct ubiquitin ligation, termed RING domain enzymes (3). Different E3 enzymes recognize particular binding site motifs on their cognate substrates, termed degrons, often in a manner that depends on phosphorylation or other posttranslational modifications, thereby coupling the UPS to myriad signaling events (4, 5).
The UPS enzyme cascade has been diversified through evolution to allow the specific targeting of thousands of substrate proteins (6). In humans, two E1 enzymes serve to charge 38 different E2 enzymes, which, in turn, are paired with more than 800 known and predicted E3 enzymes in a partially redundant fashion. Ubiquitin conjugation can be dynamically reversed by more than 100 deubiquitinating enzymes (DUBs) that also act in a substrate-specific manner. Different E2-E3 enzyme combinations within the UPS are able to generate distinct types of ubiquitin chains through repeated conjugation to one of the seven different lysine residues on the surface of ubiquitin or to the ubiquitin N terminus. Linkage specificity is determined by the orientation of donor and acceptor ubiquitin moieties, which are poised appropriately by low-affinity interactions with E2 or E3 enzymes during the conjugation reaction (7–9). Different types of poly-ubiquitin chains direct the substrate to different fates through the action of some 150 different ubiquitin binding domain (UBD) proteins that recognize the ubiquitin tag in different contexts. For instance, Lys48-linked chains enable substrates to be efficiently recognized and rapidly degraded by the 26S proteasome, Lys63-linked chains modulate the assembly of protein complexes, and substrate monoubiquitination is used to control vesicle trafficking events. Collectively, this vast system of ubiquitin writers, erasers, and readers embodies a complex and as yet still largely uncharted ubiquitin code that dynamically shapes the proteome (6).
The cullin-RING ligases (CRLs) represent the largest family of E3 enzymes by virtue of their organization as modular multi-subunit complexes (10, 11). The cullin subunit acts as a scaffold that connects a diverse set of substrate-binding adaptor subunits to an E2-binding RING domain subunit (Fig. 1A). The archetypal SKP1/cullin/F-box protein (SCF) complexes were the first CRLs discovered and are built from a CUL1 scaffold, a SKP1 subunit that binds a large set of substrate recruitment factors called F-box proteins, and the RING domain subunits, RBX1 or RBX2. All CRLs operate in conjunction with two dedicated E2 enzymes called CDC34A (UBE2R1) and CDC34B (UBE2R2), which are able to efficiently build K48-linked chains that target substrates to the 26S proteasome. A dynamic electrostatic interaction between CDC34A and CUL1 enables highly processive chain assembly (12), while other SCF-associated E2 and E3 enzymes prime substrates with an initial ubiquitin moiety before chain elongation (13–15). Yet other factors are required for CRL activation, including a ubiquitin-like protein modifier called NEDD8 that must be covalently attached to the cullin subunit in order for the RBX1-E2 complex to access substrates (11). The F-box proteins that dictate substrate specificity of SCF complexes are now implicated in many diseases. For example, FBXW7 targets oncogenes such as cyclin E, MYC, and NOTCH and is one of the most commonly mutated tumor suppressor genes in cancer (16). Conversely, another F-box protein called SKP2 targets tumor suppressor proteins and is often overexpressed in cancer (17).
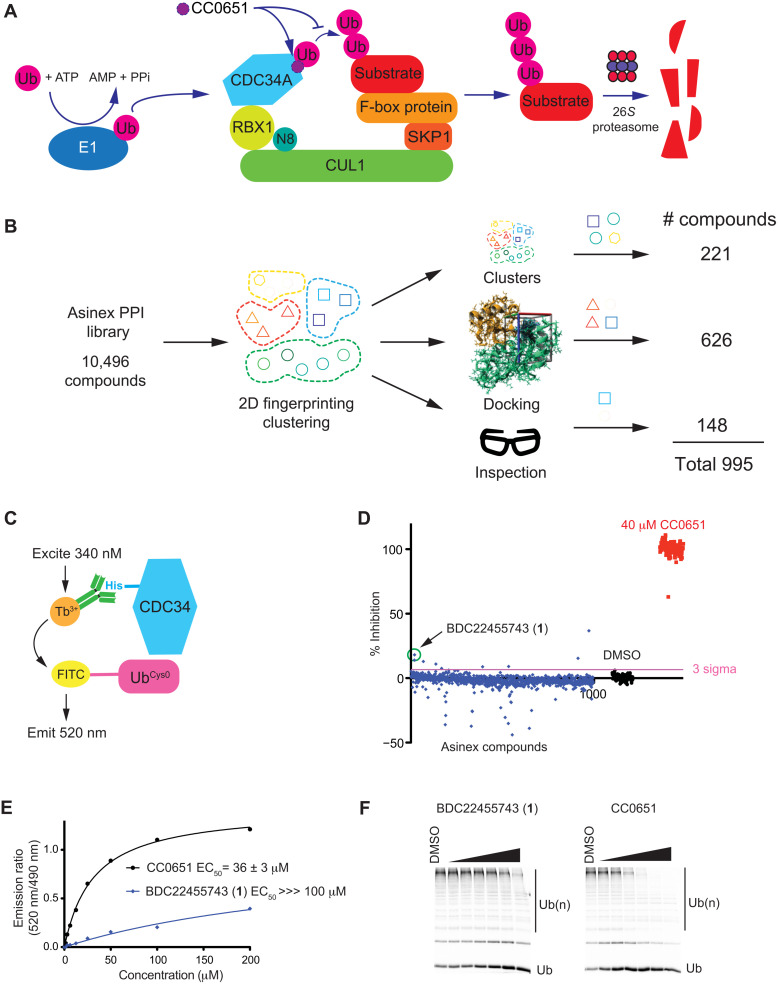
Fig. 1. Library design, TR-FRET assay, screen data, and hit validation.
(A) Schematic of CDC34A/SCF-mediated substrate ubiquitination. CC0651 inhibits ubiquitin (Ub) transfer by stabilizing the noncovalent CDC34A-donor ubiquitin complex. (B) Selection of compounds for custom CDC34A screening library. (C) Schematic of TR-FRET assay for detection of the CDC34A-ubiquitin interaction. (D) Bioactivity results from screen of CDC34A custom library (blue) with CC0651-positive controls (red) and DMSO-negative controls (black). Inhibition values from emission ratio measurements (520 nm/490 nm) were normalized to CC0651 (100%) and DMSO controls (0%). (E) Dose-response inhibition by CC0651 and hit 1 in TR-FRET assay. Median effective concentration (EC50) values represent the mean ± SD (n = 3). (F) Dose-response inhibition by CC0651 and hit 1 in ubiquitination assay. Concentrations tested were 1.1, 3.3, 11.1, 33.3, 100, and 300 μM for each compound.
Given the pervasive influence of the UPS in cellular regulation, the discovery of small molecules that modulate UPS activity is an area of intense focus for therapeutic development (18). General inhibitors of the 26S proteasome such as bortezomib are approved for treatment of multiple myeloma and other cancers, despite the relatively narrow therapeutic window. An inhibitor of the E1 enzyme that activates NEDD8 for conjugation, MLN4924 (pevonedistat), has recently been granted breakthrough therapy status by the U.S. Food and Drug Administration for the treatment of high-risk myelodysplastic syndrome (19). However, the UPS presents an inherent challenge to drug development. While the deep ATP-binding cleft of E1 enzymes is amenable to small-molecule inhibition, E2s, E3s, DUBs, and UBDs tend to lack obvious druggable pockets. Unlike classical enzymes that act on small-molecule metabolites or chemical group donors, UPS enzyme mechanism must accommodate the large ubiquitin moiety, and hence is characterized by flat, expansive active sites. Furthermore, UPS enzyme activity and substrate selection are controlled largely by protein interactions, which are inherently difficult to interdict by small molecules (20, 21). Nevertheless, inhibitors of different UPS enzymes have been identified, many of which directly or indirectly interfere with protein interactions. A number of small molecules that engage substrate recognition sites on E3 enzymes have been identified, for example, the nutlin family of MDM2 inhibitors that stabilize p53 (22). More rarely, small molecules may also block substrate interactions through allosteric effects (23). With respect to direct inhibition of the enzymatic cascade, chemical inhibition of E2-E3 complex formation is a promising strategy, as exemplified by inhibitors of the interaction between UBE2M and the DCN1 subunit of the NEDD8 E3 complex (24, 25). A small-molecule inhibitor that forms a covalent adduct with an E2 enzyme catalytic cysteine has also been described (26). Last, fragment-based screening approaches have identified small chemical moieties that bind to allosteric sites remote from the E2 catalytic cysteine as new routes for inhibitor development (27, 28).
A novel strategy for modulation of the UPS seeks to control the protein interactions of the UPS with nonconventional small-molecule ligands, referred to as protein degraders (29). In particular, heterobifunctional ligands called PROTACs have been used to direct protein neosubstrates to a noncognate E3 enzyme, thereby catalyzing ubiquitination and degradation of the protein of interest (30). Despite the promise of PROTACs in cell-based models, the structural complexity of PROTACs presents additional pharmacokinetic and pharmacodynamic challenges. A related approach exploits the unique ability of particular compounds, sometimes termed molecular glues, to stabilize either weak native protein interactions or even completely noncognate protein interactions (31, 32). The plant hormone auxin was the first compound discovered to stabilize an E3-substrate interaction (33). The exemplar molecular glues for ectopic substrate degradation, termed immunomodulatory imides (IMiDs), are based on thalidomide and stabilize an interaction between the cereblon (CRBN) subunit of the CUL4A-DDB1-CRBN E3 complex and the Ikaros family transcription factors IKZF1 and IKZF3, leading to their proteasome-dependent degradation (34). Drugs based on the IMiD scaffold, such as lenalidomide (revlimid), are now approved for multiple myeloma, myelodysplastic syndromes, and various lymphomas (35). While a few other molecular glues that target neo-substrates for degradation have been found, to date, their discovery has mainly relied on serendipity (31).
As opposed to activation of neo-substrate degradation, it is also possible to inhibit UPS enzymes by stabilization of weak noncovalent interactions with ubiquitin itself. This effect was first demonstrated through characterization of a compound called CC0651 that was discovered as an inhibitor of a complex in vitro reaction for ubiquitination of the cyclin-dependent kinase (CDK) inhibitor p27 by the SCFSKP2 ubiquitin ligase (36). Unexpectedly, CC0651 was found to stabilize a low-affinity noncovalent CDC34A-ubiquitin interaction and thereby freeze the catalytic cycle. The structure of a CDC34A-ubiquitin-CC0651 complex revealed that CC0651 engages an extended composite pocket formed by the donor ubiquitin and CDC34A (37). This composite pocket is not present in the CDC34A apo structure and is induced by CC0651, with possible allosteric effects on the CDC34A-ubiquitin catalytic intermediate (36, 37). Despite its novel mode of action, the potency of CC0651 in vitro is only modest (~5 μM) and its efficacy in cells is poor. Structure-activity relationship (SAR) analysis through the synthesis of more than 150 analogs failed to markedly improve the potency of CC0651 (37). Furthermore, the three chiral centers present in CC0651 unduly complicate analog synthesis for SAR. These drawbacks raised the question of whether we could discover an alternate inhibitor scaffold with greater potential for optimization through a de novo screen for molecular glues that stabilize the CDC34A-ubiquitin interaction. Here, we report the results of a focused screening campaign for these compounds and the optimization of a screen hit.
RESULTS
Design of a CDC34A screening collection
We constructed a chemically and structurally diverse library using the commercial provider Asinex as a source enriched for compounds that target protein-protein interactions (PPIs). To reduce the library size to a tractable number for a focused in vitro screening campaign, we selected ~1000 compounds from the full 10,496-member Asinex PPI library based on a three-pronged approach to maintain diversity. Compounds were first segregated by two-dimensional (2D) fingerprinting and Tanimoto distance into 100 structurally diverse clusters. Virtual screening using AutoDock Vina (38) and a previously published CDC34A-ubiquitin-CC0651 complex (37) as a template was used to enrich for compounds hypothetically able to engage the CC0651 binding pocket. Compounds with a docking score higher than that of CC0651 itself were chosen as a main component of the library (626 compounds). To retain maximal chemical and structural diversity from the parent library, representative members of underrepresented clusters in the docked set were then added to the library (221 compounds). An additional set of compounds was then selected by chemists based on interesting chemical and structural features (148 compounds). This total collection of 995 compounds formed the custom CDC34A screening collection (Fig. 1B; see table S1 for compound identifiers and structures). This custom CDC34A library was assessed for pan-assay interference compounds (PAINS) using a previously described method (39) as implemented by the SwissADME web tool (40). Only 11 of the 995 compounds were flagged as containing substructures with a potential propensity for assay interference (table S1).
Assay for chemical stabilizers of a noncovalent CDC34A-ubiquitin complex
To screen for compounds that stabilize the low-affinity interaction between the donor ubiquitin binding site of CDC34A and ubiquitin, we exploited an assay based on time-resolved Förster resonance energy transfer (TR-FRET). This assay used an N-terminal His-tagged version of CDC34A recognized by an anti-His6 antibody coupled to Tb3+ and an N-terminal cysteine mutant of ubiquitin (denoted UbCys0) stoichiometrically labeled with 5′-iodoacetamide-fluorescein. In response to titration with CC0651, excitation of Tb3+ at 340 nm resulted in fluorescence energy transfer to the fluorescein moiety and emission at 520 nm (Fig. 1C). A Z-factor score of 0.78 based on the CC0651-positive control and dimethyl sulfoxide (DMSO)–negative control indicated that the assay was suitable for screening purposes.
We carried out a single-point TR-FRET screen against the 995-compound library (Fig. 1D; see table S1 for primary data values). A 3-sigma threshold identified seven hits, one of which, BDC22455743 (1), was validated by multipoint dose-response analysis in the TR-FRET assay (Fig. 1E) and a gel-based in vitro ubiquitination assay that monitored substrate-independent poly-ubiquitin chain formation by gel electrophoresis (Fig. 1F). The other six hits showed no inhibitory activity in the in vitro ubiquitination reactions and thus were not studied further (fig. S1). To verify hit compound identity, fresh BDC22455743 was obtained by in-house synthesis, its structure was ascertained by analytical methods, and its activity was recapitulated in the TR-FRET and ubiquitination assays (see below).
Hit compound structure and shared features with CC0651
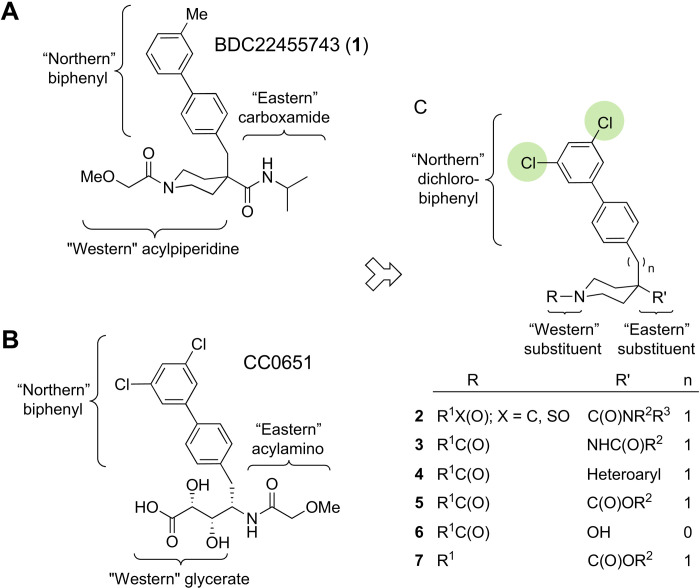
BDC22455743 (1) harbors a central isonipecotamide scaffold that can be subdivided into three distinct portions: a “northern” lipophilic biphenyl moiety, an “eastern” carboxamide group, and a “western” acylpiperidine unit (Fig. 2A). Similarly, CC0651 lends itself to segmentation into a northern biphenyl moiety, an eastern acylamino group, and a western glycerate unit (Fig. 2B). Although the inhibitors 1 and CC0651 were identified from completely different chemical libraries and screening tactics, the two compounds bear structural resemblances. The similarity between the northern meta-substituted biphenyls in 1 and CC0651 suggested that the binding modes might be similar. Moreover, the eastern amide pharmacophores are analogous, albeit with opposite amide bond orientations. Structural divergence between 1 and CC0651 was greatest at the western acylpiperidine and glycerate units, respectively. We exploited the shared features of 1 and CC0651 in the design of analog series 2 to 7 (Fig. 2C; see table S2 for all analogs). Neither 1 nor any of its analogs synthesized in this study had any PAINS-like characteristics (table S2).
Fig. 2. Structure comparison of hit 1 and CC0651.
(A) Structure of isonipecotamide hit 1 with definition of northern, eastern, and western regions. (B) Structure of CC0651 with corresponding regions indicated. (C) Summary of isonipecotamide analog series with indicated replacements in eastern and western regions.
Integration of the analogous northern dichlorobiphenyl and central isonipecotamide units into a single scaffold allowed elaboration of more potent CDC34A inhibitors. The implicit substitution of glycerate for acylpiperidine fortuitously eliminated chirality in the E2 inhibitor and thereby allowed short and robust synthetic routes (see below). In contrast, synthesis of CC0651 requires eight steps because of the glycerate subunit chemical reactivity and the need for synthetic discrimination between eight diastereomers (37). The isonipecotamide scaffold displays high synthetic modularity owing to increased opportunity for orthogonal transformations and flexible order of assembly for optimal analog access at the diversification points. These considerations motivated our pursuit of the isonipecotamide theme despite the weak initial activity of 1.
Synthetic strategy for elaboration of the BDC22455743 hit scaffold
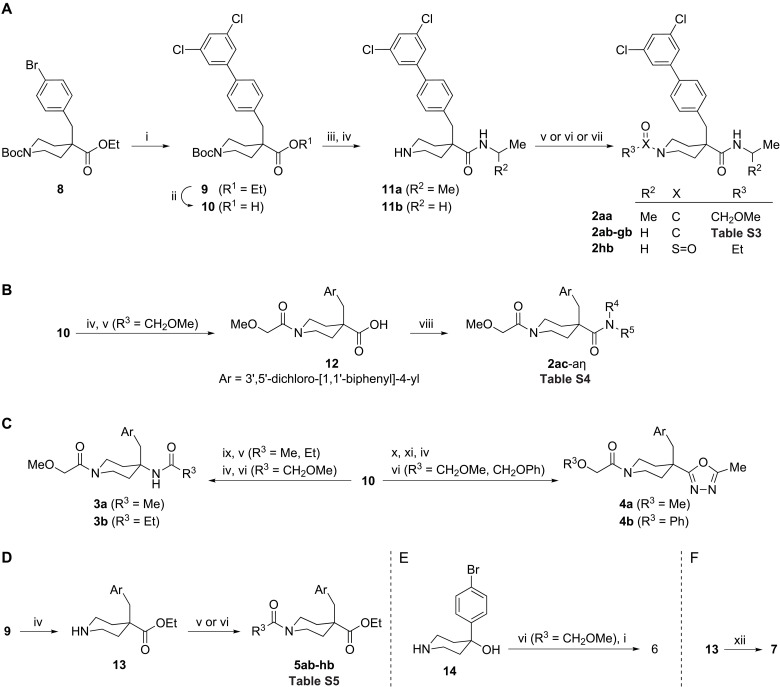
To enable the SAR exploration of isonipecotamide 1, we developed synthetic routes that exploited the commercial multifunctional precursor 8 (Fig. 3). The resulting set of two to six step syntheses enabled structural exploration about the northern biphenyl, eastern carboxamide, and western acylpiperidine moieties of 1.
Fig. 3. Synthesis of isonipecotamide analogs 2 to 7 from precursors 8 and 14.
(A) Synthesis of 2aa to 2hb from 8. Biphenyl 9 and carboxylic acid 10 were obtained by cross-coupling followed by alkaline hydrolysis. Subsequent amidation and Boc-group removal yielded piperidine-4-carboxamides 11a to 11b. Piperidine acylation using suitable acid chlorides, carboxylic acids, as mediated by [Dimethylamino(triazolo[4,5-b]pyridin-3-yloxy)methylidene]-dimethylazanium hexafluorophosphate (HATU)], or ethanesulfonyl chloride yielded 2aa to 2hb. (B) Synthesis of 2ac to 2aη from 10. Piperidine 10 Boc removal followed by reaction with methoxyacetyl chloride gave isonipecotic acid 12, which was activated using HATU and coupled to a series of amines to yield isonipecotamides 2ac to 2aη. (C) Synthesis of 3 to 4 from 10. Conversion of acid 10 into a piperidin-4-amine followed by reaction with acid chlorides yielded amides that were subjected to Boc-group removal and acylation by methoxyacetic acid, as mediated by HATU, to yield 3a to 3b. The active ester of 10 was reacted with acethydrazide followed by in situ generated PPh3Cl2 resulting in an oxadiazole, which, after Boc removal and acylation by methoxy and phenoxyacetic acid, as mediated by HATU, gave 4a to 4b. (D) Synthesis of 5ab to 5hb from 9. Piperidine 9 Boc removal followed by piperidine acylation using acid chlorides or carboxylic acids, as mediated by HATU, yielded esters 5ab to 5hb. (E) Synthesis of 6 from 14. Piperidin-4-ol 14 was acylated by methoxyacetic acid, as mediated by HATU, and subjected to cross-coupling to yield biaryl 6. (F) Synthesis of 7 from 13. Reaction of piperidine 13, pentanal, and sodium cyanoborohydride gave amine 7. Reagents and conditions: (i) 3,5-Cl2C6H3B(OH)2, PdCl2(dppf), NaHCO3; (ii) LiOH, dioxane, H2O, 80°C, 12 to 24 hours; (iii) H2NCH(R2)Me, HATU, DIEA; (iv) TFA, DCM; (v) R3C(O)Cl, TEA (or DIEA), DCM; (vi) R3CO2H, HATU, DIEA, DMF (or MeCN); (vii) R3SO2Cl, TEA, DCM; (viii) HNR4R5, HATU, DIEA, DMF; (ix) DPPA, DIEA, PhMe, 100°C; (x) AcNHNH2, HATU, DIEA, MeCN; (xi) PPh3, C2Cl6, DIEA, MeCN; (xii) BuC(O)H, NaBH3CN, MeOH.
The synthesis of isonipecotamide analogs in the 2 series was achieved using two variations of a five-step synthesis starting from the commercial isonipecotate 8, which has aryl bromide, ester, and protected amine functional groups as orthogonal synthetic handles to facilitate structural variation about a 4-benzylisonipecotate core (Fig. 3, A and B). Intermediate 9, which harbors a 3,5-dichlophenyl unit, was obtained by Suzuki reaction using 3,5-dichlorophenylboronic acid as a coupling partner. Subsequent hydrolysis to the acid 10, isopropyl or ethylamine coupling, and acidolysis yielded isonipecotamides 11a to 11b (Fig. 3A and table S3). These N-H-piperidines were subjected to a series of acylation reactions to furnish isonipecotamides 2aa to 2hb that vary on the western flank.
Access to a set of isonipecotamide analogs bearing variation at the eastern carboxamide nitrogen was facilitated by a variant route in which piperidine-4-carboxylate amidation was conducted last, by way of N-acylisonipecotic acid 12 (Fig. 3B and table S4). Attempts to access carboxylic acid 12 directly from its cognate ethyl ester failed because of preferential hydrolytic cleavage at the methoxyacetamide moiety, presumably due to steric hindrance. Instead, a fully deprotected γ-amino acid intermediate from Boc-piperidine 10 was subjected to selective reaction at nitrogen using methoxyacetyl chloride. With acid 12 in hand, amide bond formation with ethylamine was used to recapitulate the synthesis of isonipecotamide 2ab and validate the route. Deploying carboxylic acid 12 in conjunction with an array of amines yielded analogs 2ac to 2aη (table S4).
To access scaffold-modified analogs 3 to 7, a broader array of synthetic transformations was deployed. Reorientation of the eastern carboxamide in 2 to mimic CC0651 required an alternative 4-aminopiperidine scaffold, which was generated by a Curtius reaction on isonipecotic acid 10 (Fig. 3C). The resulting N-1-Boc-piperidin-4-yl amine intermediate was elaborated into reversed-amide analogs 3a to 3b by sequential acylation reactions. Oxadiazole bioisosteric substitution for the piperidin-4-yl carboxamide was achieved by preparation of 1,2-diacylhydrazine from acid 10 followed by cyclodehydration (Fig. 3C). Subsequent piperidine nitrogen liberation followed by acylation reactions gave piperidin-4-yl-1,3,4-oxadiazole analogs 4a to 4b. Ethyl esters 5 were varied in the western acylpiperidine unit by isonipecotate 9 Boc removal to give piperidine-4-carboxylate 13, which was subjected to a series of acylation reactions yielding isonipecotates 5ab to 5hb (Fig. 3D and table S5). A more radical departure from the isonipecotamide scaffold required an alternative commercial starting material, 4-(4-bromophenyl)piperidin-4-ol 14 (Fig. 3E), the acylation of which proceeded selectively at nitrogen. Subsequent generation of the biaryl unit by Suzuki coupling resulted in piperidin-4-ol 6. Variation in the western carbonyl on the piperidine nitrogen was achieved using reductive amination chemistry. By using N-H-piperidine ethyl ester 13 as a secondary amine input, reaction with an alkyl aldehyde in the presence of reductant furnished isonipecotate 7 (Fig. 3F).
In summary, a network of synthetic routes allowed the creation of a custom analog library around the isonipecotamide theme. Compounds were chosen to explore the diversity in structures 2 to 7 (Fig. 2C) in biochemical and cellular assays.
Overview of structural exploration and activity relationships
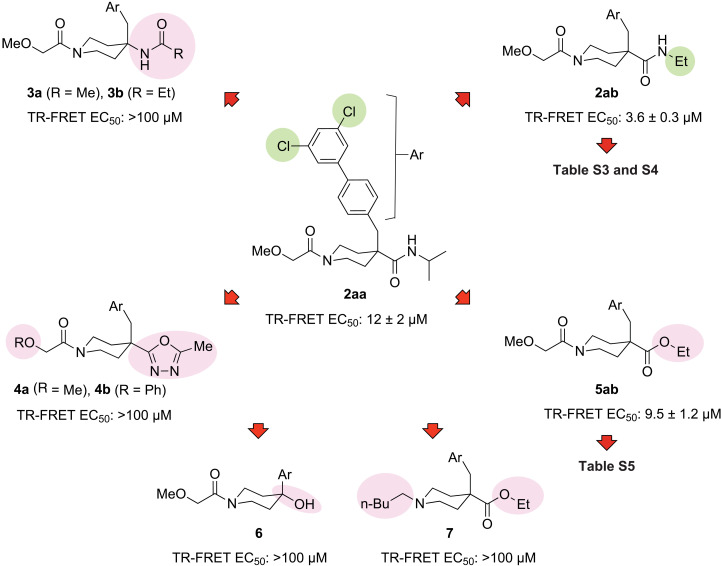
Engagement of CC0651 in a ternary complex with ubiquitin and CDC34A depends on an optimal biphenyl moiety, namely, 4-(3,5-dichlorophenyl)phenyl (37). Our initial exploration of hit 1 bearing a northern 3,5-dichlorophenyl (Fig. 2C) was conducted with isonipecotamide 2aa (Fig. 4), which displayed a marked 27-fold improvement [median effective concentration (EC50) = 327 μM for 1 versus 12 μM for 2aa] in potency in the TR-FRET assay (see fig. S2A for raw data traces). The observed potency increase supported the notion that hit 1 was engaging the same composite binding pocket as CC0651 and motivated further pursuit of the isonipecotamide theme. Our custom CDC34A library also contained a few close yet inactive hit 1 analogs (fig. S3), the inactivity of which was readily rationalized by a lack of the required linear [1,1′-biphenyl]-4-yl geometry or suitable biphenyl unit 3,5-substitution, as previously established by CC0651 biphenyl SAR (37).
Fig. 4. Overview of SAR of isonipecotamide analog series.
Representative analogs are shown. Analog 2ab was the point of departure for further optimization. Ar = 3′,5′-dichloro-[1,1′-biphenyl]-4-yl. EC50 values represent the mean ± SD (n = 3).
We next probed the eastern carboxamide of 2aa in the form of ethylamine analog 2ab (Fig. 4). Size reduction in the amine group relative to the bulky 2aa isopropyl counterpart afforded a three-fold potency increase to 4 μM for 2ab. Salient features of isonipecotamide series 2 were explored by comparison to the activity of scaffold-modified analogs 3 to 7. The design of isonipecotamides series 2 adopts the northern dichlorobiphenyl unit of CC0651 but leaves an eastern amide directionality difference (i.e., a reverse amide), which was tested in piperidine-4-amines 3a and 3b. This change abolished TR-FRET activity, revealing that the eastern amide bond directionality in 1 is critical for isonipecotamide activity. The potency of CC0651 is known to depend on the hydrogen-bond accepting capacity of its secondary amide functional group (37). Despite the modified amide bond directionality, we suspected that isonipecotamides in the 2 series would engage in similar H-bonding. As predicted, the electron-poor heterocyclic bioisosteres deployed in 4a and 4b reduced H-bonding capacity and abolished activity. The ethyl ester analog 5ab exhibited similar potency to the parent compound 2aa in the TR-FRET assay, while the more apt comparison between the ethyl amide 2ab and the ethyl ester 5ab revealed a two- to threefold loss in potency upon substitution of the nitrogen atom for oxygen. The piperidin-4-ol 6 lacks both the methylene spacer between the piperidine and biaryl subunits, and the secondary carboxamide functional group (see Fig. 2C, entry 6: R′ = OH, n = 0) and was completely inactive. Removal of the piperidin-1-yl acyl carbonyl in 7 also resulted in complete abrogation of activity compared to the moderately active analog 5.
As the isonipecotate 5 appeared to be the second most active scaffold, additional analogs were generated. Modification of ethyl ester 5ab while avoiding loss of the western acyl carbonyl, as in 7, was explored using a set of acyl analogs (table S5 and fig. S2). Substitution of the methoxyacetyl group with related alkyl acyl groups was reasonably well tolerated, as measured in the TR-FRET assay (table S5). In contrast, related carboxylic acid synthetic intermediates such as 12 lacked activity (table S5).
In summary, exploration of hybrid molecules derived with features of both hit 1 and CC0651 (Fig. 2C) led to compound 2ab with >80-fold improved activity compared to hit 1 and eightfold improved activity relative to CC0651 (4 μM versus 31 μM) in the TR-FRET assay. This overall result validated our hypothesis that the glycerate portion of CC0651 can be replaced by an isonipecotamide counterpart. Departure from first-generation hybrids 2 by cumulative scaffold modification in analogs 3 to 7 illustrated that the only tolerable change within the isonipecotamide core is subtle modification of the NH group. As modifications in the vicinity of the central piperidine proved detrimental, subsequent SAR investigation was focused on preserving the 1-acylpiperidine-4-carboxamide scaffold in 2.
Refinement of SAR
After a first round of structure-activity investigation, compound 2ab emerged as the optimal analog. To further refine the SAR, we explored an additional set of 38 analogs of 2ab. Modifications were focused at the western acylpiperidine [Fig. 2C, entry 2: R = R1X(O)] and the eastern carboxamide groups [Fig. 2C, entry 2: R′ = C(O)NR2R3]. As above, potency was assessed using the TR-FRET assay (see fig. S2 for raw TR-FRET data traces).
From a set of western acylpiperidine analogs 2bb to 2hb (table S3), the compounds 2bb, 2cb, 2gb, and 2hb were essentially equipotent (2 to 4 μM) to the parent 2ab (4 μM). Analogs 2eb and 2fb had heterocyclic appendages that resulted in a four- to sixfold loss in activity (17 to 23 μM). Complete removal of the acyl group, in the form of the N-H-piperidine synthetic intermediate 11b (Fig. 3A), also led to activity loss (13 μM). Most notably, the isobutyl carbamate 2db gave a 10-fold increase in potency (0.4 μM). Overall, the scope of suitable western acyl units on the piperidine is broad, ranging from the lipophilic alkyls 2ab to 2db, to the aryl 2gb, and to the polar sulfonamide 2hb.
Structural exploration of the eastern isonipecotamide carboxamide unit uncovered a unified set of structural preferences and constraints (table S4). Introduction of various alkyl alcohol moieties to the 2ab lipophilic ethylamine group abrogated activity (2ai, 2at, and 2au). Although the primary amide 2ac lacked activity, the methyl 2ad (20 μM), ethyl 2ab (4 μM), propyl 2ae (4 μM), cyclopropyl 2al (5 μM), and cyclopropylmethyl 2ah (7 μM) in a secondary N-alkyl series were well tolerated. Further elongation in the pentyl analog 2ak (26 μM) and branching at the N-alkyl α-position in 2aa (12 μM) and tert-butyl 2as (15 μM) were detrimental, as were four-, five-, and six-membered saturated ring substitutions in 2am to 2ar and the aniline-derived 2aw.
Further insight into the eastern region was revealed by good tolerance of dimethylamine and diethylamine derivatives 2ax (10 μM) and 2ay (6 μM). Ring variants that incorporated azetidine 2az (4 μM), pyrrolidine 2aα (3 μM), piperidine 2aη (1 μM), or morpholine 2aζ (3 μM) were equal to or improved upon the parent 2ab analog. Ether and organofluorine substitutions had either modest or detrimental effects on activity compared to their cognate hydrocarbons, as shown for 2aj, 2af, 2ag, 2aβ, and 2aγ. As with the acyclic alcohol analogs, pyrrolidin-3-yl carbinol variants 2aδ and 2aε also reduced activity. In summary, systematic exploration of the western acyl and eastern carboxamide based on 2ab not only allowed up to 10-fold improvement in activity but also uncovered a diverse series of analogs of comparable or slightly better activity.
Isonipecotamide analog potency and specificity for inhibition of in vitro ubiquitination
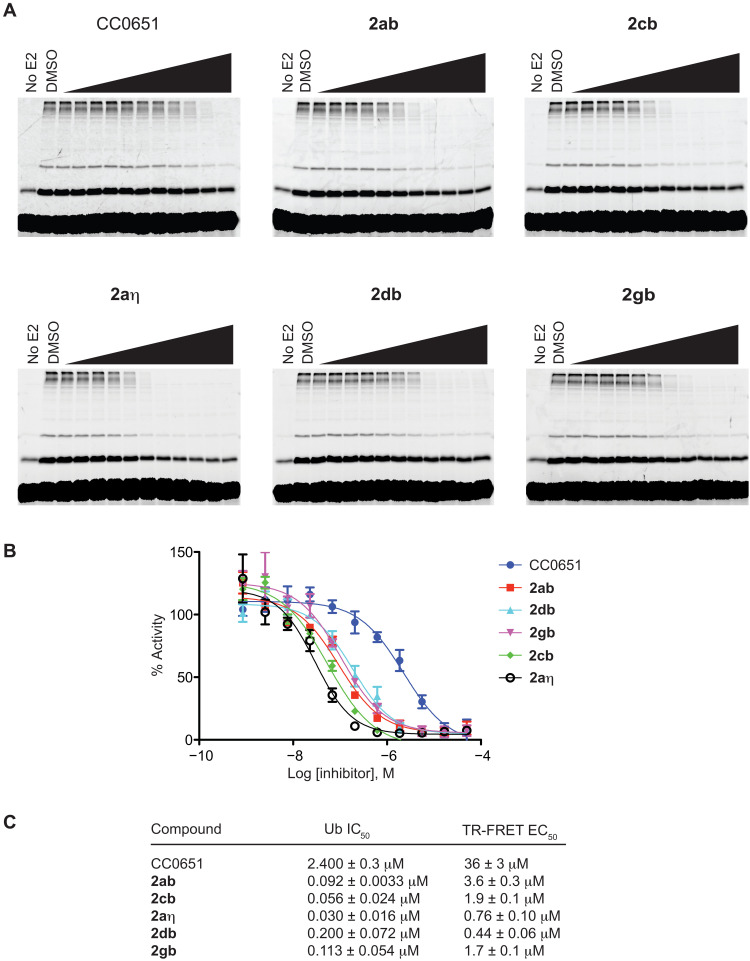
We next investigated the activity of various inhibitors in an in vitro ubiquitination reaction using purified recombinant proteins for E1 enzyme, CDC34A, and CUL1-RBX1 complex in the presence of fluorescein-labeled ubiquitin and ATP (37). This CDC34A-CUL1-RBX1 reaction mixture efficiently catalyzes the assembly of free ubiquitin chains without the need for SKP1, an F-box protein, or substrate, as shown previously for the corresponding yeast proteins (41). Chain assembly was assessed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and quantified by fluorescence imaging of ubiquitin conjugates. We used the established inhibitor CC0651 as a benchmark and evaluated five of the most potent analogs in the TR-FRET assay: the parent isonipecotamide 2ab (4 μM) and the 2cb (2 μM), 2aη (1 μM), 2db (0.4 μM), and 2gb (2 μM) analogs. Each inhibitor was evaluated in 10-point dose-response curves, and median inhibitory concentration (IC50) values estimated by graphical analysis (Fig. 5, A to C, and tables S3 and S4). The selected isonipecotamide compounds showed between 12- and 80-fold improvement over CC0651, with IC50 values ranging from 0.03 to 0.2 μM versus 2.4 μM for CC0651. We observed a strong correlation between potency in the TR-FRET assay and the in vitro ubiquitination assay that monitored assembly of free ubiquitin chains.
Fig. 5. Inhibition of CDC34A-mediated in vitro ubiquitination by most potent isonipecotamide 2ab analogs.
(A) Gel-based assay for poly-ubiquitin chain formation. Concentrations tested ranged from 0.00084 to 50 μM in threefold increments. Experiment was performed twice, and representative gels from one experiment are shown. (B) Quantitation of poly-ubiquitin chain formation. Percent inhibition of poly-ubiquitin chain formation for each inhibitor concentration represents the mean from two experiments. (C) Comparison of TR-FRET EC50 and ubiquitination IC50 values. EC50 values represent the mean ± SD (n = 3). IC50 values represent the mean ± variance (n = 2).
To ascertain that the inhibitors also prevented substrate-level ubiquitination, we tested for activity in an SCFSKP2 ubiquitination assay with full-length p27 as the substrate. To allow substrate recognition by SCFSKP2, fluorescein-labeled p27 was phosphorylated by preincubation with CDK2–cyclin A. We note that an advantage of the free ubiquitin chain formation assay was that the strong ubiquitination signal allowed us to use a low E2 concentration (100 nM), which, in turn, allowed better discrimination between inhibitors of different potencies. However, at this low E2 concentration, full-length substrate ubiquitination was inefficient. We therefore used 250 nM CDC34A in the p27 ubiquitination assay, which still allowed us to discern differences in inhibitor potency. We observed clear inhibition of p27 ubiquitination with the same approximate rank order of inhibitor potency in the TR-FRET and free chain ubiquitination assays (fig. S4, A to C). We note that the apparent inhibition values were somewhat higher in this assay because of the higher E2 concentrations used. These results demonstrated that stabilization of the CDC34A-donor ubiquitin complex inhibits both CDC34A-dependent ubiquitin chain formation and substrate-level polyubiquitination.
Previous studies have shown that CC0651 displays near-absolute specificity for CDC34A over its functionally redundant isoform UBE2R2, also known as CDC34B (36, 37). To assess the specificity of our isonipecotamide analogs, we examined their effectiveness in in vitro ubiquitination reactions carried out with UBE2R2 instead of CDC34A. Like CC0651, all of the isonipecotamide inhibitors were completely ineffective at inhibiting UBE2R2-mediated ubiquitin chain formation in the SCF reactions (fig. S5). We then extended this specificity comparison to a panel of 20 additional recombinant human E2 enzymes in a TR-FRET assay. As a control to demonstrate E2 functionality, we showed that E2 enzymes formed a detectable interaction with ubiquitin in ATP-dependent charging reactions (fig. S6A). Titration of recombinant CDC34A, UBE2R2, and UBE2D1 (also known as UBCH5) in the presence of the parent isonipecotamide 2ab (2 μM) demonstrated that saturable binding measured by TR-FRET occurred only for CDC34A (fig. S6B). We then tested each of the 21 E2s in a single-point TR-FRET assay in the presence or absence of 2ab at 20 μM (fig. S6C). Only CDC34A yielded an inhibitor-dependent interaction with ubiquitin, thereby demonstrating specificity of the isonipecotamide for the CDC34A-ubiquitin complex.
Isonipecotamide analog potency and specificity in cells
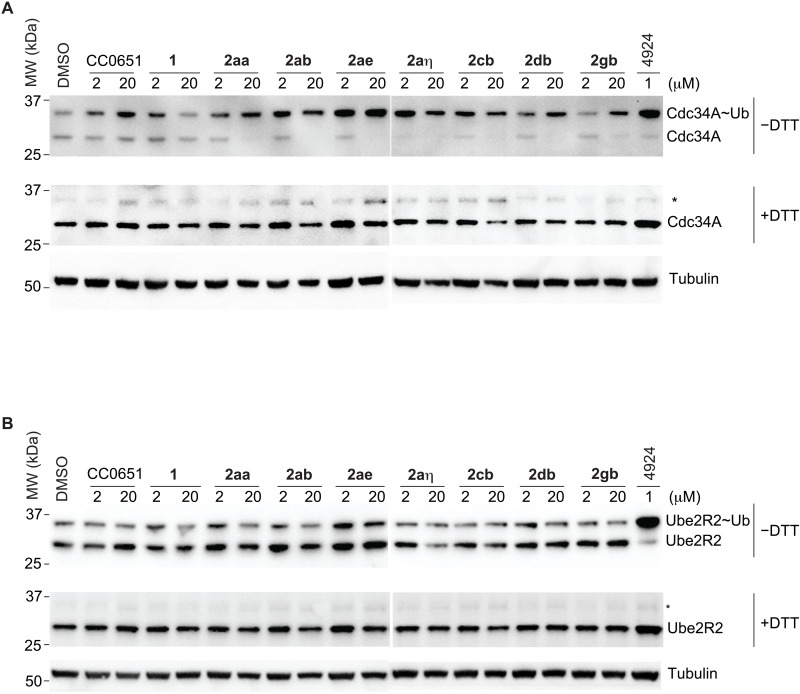
A key feature of the mechanism of action of CC0651 is that it does not prevent formation of a high-energy E2~ubiquitin thioester intermediate but instead impairs discharge of the ubiquitin thioester to a substrate lysine residue (36). To investigate the ability of isonipecotamide analogs to inhibit CDC34A discharge in live cells, we assayed for stabilization of the CDC34A~ubiquitin thioester intermediate. PC3 prostate cancer cells were synchronized in G1 phase, incubated with each inhibitor for 24 hours, and then lysed under either nonreducing conditions to preserve the CDC34~ubiquitin thioester or reducing conditions to hydrolyze the thioester. We analyzed the above five potent analogs in comparison to CC0651, the initial hit 1, and its more active 2aa derivative. All of the isonipecotamides were far more potent at stabilizing the CDC34A~ubiquitin thioester intermediate than CC0651 or the original hit compound (Fig. 6A). As expected, in a control experiment, none of the inhibitors stabilized the UBE2R2~ubiquitin thioester intermediate, consistent with in vitro specificity for CDC34A (Fig. 6B). These results show not only the improved potency of the isonipecotamide but also that these inhibitors are able to cross the cell membrane and effectively inhibit physiological levels of CDC34A.
Fig. 6. Inhibition of thioester discharge in PC3 cells by most potent isonipecotamide 2ab analogs.
(A) CDC34A~ubiquitin discharge. (B) UBE2R2 thioester discharge. Duplicate blots were probed with either anti-CDC34A, anti-UBE2R2, or, as a loading control, anti-tubulin. Asterisks indicate residual E2-thioester species in reducing (+DTT) conditions. 4924 indicates MLN4924.
Structural basis for isonipecotamide interaction with CDC34A and ubiquitin
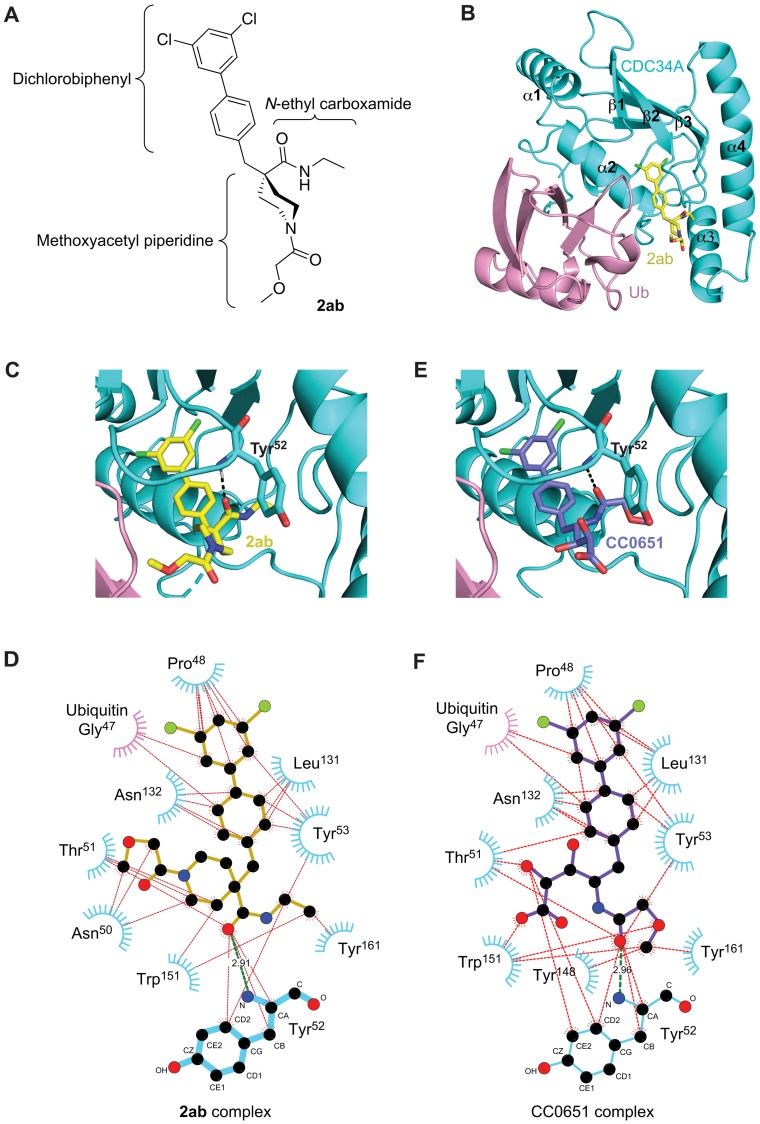
To determine the mode of isonipecotamide binding, the parent analog 2ab was selected for structural investigation by x-ray crystallography in complex with CDC34A and ubiquitin. Suitable 2ab-containing crystals that diffracted to 2.47-Å resolution were identified, which allowed a structure determination by molecular replacement using the previously determined individual CDC34A [Protein Data Bank (PDB) code 2OB4] and ubiquitin (PDB code 1UBQ) atomic structures as search models (see Materials and Methods). The crystal asymmetric unit contained four CDC34A-ubiquitin-2ab ternary complexes, which were essentially identical [root mean square deviation (RMSD) = 0.37 to 0.44 Å2 over 158 CDC34A residues]. In addition, well-defined unbiased electron density maps allowed unambiguous modeling of 2ab inhibitor coordinates in a highly similar binding configuration (fig. S7, A to C). Thus, our discussion is focused on a single complex composed of chain A (CDC34A with isonipecotamide 2ab) and chain B (ubiquitin). The arrangement of CDC34A and ubiquitin was essentially unchanged from the previous determined ternary CDC34A-ubiquitin-CC0651 complex (RMSD = 0.46 Å2), with 2ab occupying the same composite binding pocket as CC0651, i.e., nestled between CDC34A and ubiquitin (Fig. 7, A to C). The binding surface of 2ab on CDC34A was composed of the α1-β1 linker, the C-terminal end of helix α2, the β2-β3 linker, the C-terminal end of helix α3, and the α3-α4 linker. The remainder of the 2ab binding pocket was composed of the β3 strand of ubiquitin.
Fig. 7. Structure of 2ab in complex with CDC34A and ubiquitin.
(A) Chemical structure of 2ab drawn to match the orientation depicted in (B). (B) Ribbon representation of CDC34A (cyan) and ubiquitin (pink) with 2ab (carbon atoms in yellow, oxygen in red, nitrogen in blue, and chlorine in green). (C) Zoomed-in view of 2ab in the CDC34A-ubiquitin binding pocket (carbon atoms shown in yellow). See fig. S8A for a stereo view with side-chain details. (D) Ligplot representation of 2ab interactions in the CDC34A-ubiquitin binding pocket. The amide backbone of Tyr52 from CDC34A makes a hydrogen bond with a carbonyl of 2ab (green line) in addition to a network of hydrophobic interactions (red lines). Phe28, Ile45, Phe58, Phe77, Met81, and Ile128 of CDC34A make minor contributions but are not shown for clarity. (E) Zoomed-in view of CC0651 in the CDC34A-ubiquitin binding pocket (carbon atoms shown in purple). See fig. S8B for a stereo view with side-chain details. (F) Ligplot representation of CC0651 interactions in the CDC34A-ubiquitin binding pocket.
The northern dichlorobiphenyl moiety of 2ab is embedded in the deepest portion of the binding pocket in a hydrophobic cleft lined by Gly47 of ubiquitin and Pro48, Tyr53, Leu131, and Asn132 of CDC34A (Fig. 7, C and D; see fig. S8A for a detailed stereo view). In addition, Phe28, Ile45, Phe58, Phe77, Met81, and Ile128 of CDC34A also line the pocket, but these residues contribute less than 10 Å2 per residue. As anticipated, dichlorobiphenyl binding geometry was identical in CC0651 (Fig. 7, E and F; see fig. S8B for a detailed stereo view), explaining the ease with which hit 1 potency was improved by introduction of the dichlorobiphenyl moiety.
The eastern N-ethyl carboxamide carbonyl moiety of 2ab engages in a single hydrogen bond with the backbone amide N-H of Tyr52, while the N-ethyl group orients into a spacious pocket lined by the CDC34A residues Tyr53, Tyr148, Trp151, Tyr161, Thr162, and Ile165. The western methoxyacetyl piperidine substituent of 2ab fits into a solvent-accessible cleft lined by Asn50, Thr51, Tyr52, and Trp151 of CDC34A and Lys48 of ubiquitin. The lysine side chain of ubiquitin is oriented 3.8 Å from the methoxy group of 2ab but is disordered in two of the four copies of the complex observed in the crystal environment such that it is unlikely to contribute substantially to inhibitor binding. We note that among the structural subunits of 2ab, the methoxyacetyl ether group displayed the greatest conformational flexibility across the four complexes in the crystal asymmetric unit (fig. S7C). This observation is consistent with the paucity of specific interactions that constrain the solvent-facing N-acyl moiety.
Structure-based interpretation of isonipecotamide SAR
The crystallographic model explained the observed SAR trends in the isonipecotamide 2 analog series. For the western acyl moiety, the flat SAR among 2bb to 2hb was consistent with the solvent-exposed nature of this region. For example, although 2gb and 2hb respectively incorporated a large hydrophobic benzoyl group and a compact polar ethanesulfonyl group, the two compounds were equipotent. This structural tolerance affords the opportunity for further derivatization of the scaffold without a risk of reduced activity, for example, in future optimization of pharmacokinetics - absorption, distribution, metabolism, and excretion (PK-ADME) properties or additional chemical handles for chemical probe design.
At the outset, SAR trends for the eastern alkyl carboxamide region suggested that a well-defined pocket was engaged, which was subsequently confirmed by crystallography. The complete loss of activity that resulted from the introduction of polar hydroxyl groups in 2ai, 2at, and 2au is explained by incompatibility with the hydrophobic pocket formed from Tyr53, Tyr161, and Ile165 residues of CDC34A. Conversely, the introduction of alkyl moieties optimized engagement of the pocket and improved activity, such as for the methyl, ethyl, and propyl series of 2ad, 2ab, and 2ae as well as the azetidine, pyrrolidine, and piperidine series of 2az, 2aα, and 2aη. Moreover, the potency of the secondary amide analogs suggested that availability of a hydrogen bond donor in this region was not important for binding.
The internal carbonyl groups of both CC0651 and 2ab play critical roles as hydrogen bond acceptors and were essential for activity. In contrast, the reoriented-amide analogs 3 were inactive, revealing the importance of locating the carbonyl group at a 4-atom distance (i.e., on Cδ) from the anchoring biphenyl for CC0651 and at a 3-atom distance (i.e., on Cγ) for 2ab.
Although the binding mode of the biaryl moiety is nearly identical for CC0651 and 2ab, contact between the piperidine flank and the solvent-accessible cleft in the CC0651 binding pocket cantilevers the γ/δ amide atoms by 0.7 Å toward Tyr52 of CDC34A. Analogs in the 3 amide series cannot accommodate the resulting clash and thus lose potency. In contrast, the isonipecotamides in the 1 to 2 series capitalize on an optimal carbonyl distance through Cβ-Cγ bond rotation.
Overall, we observed a strong concordance between the SAR across the isonipecotamide analog series and the crystallographic structure. It is likely that all of the analogs engage the CDC34A-donor ubiquitin pocket in a highly similar manner.
DISCUSSION
The UPS has emerged as an area of intense focus for therapeutic small-molecule development (18). However, the druggability of the UPS presents a challenge because ubiquitin is a large ligand that must be manipulated through flat enzyme surfaces that are not amenable to conventional inhibition strategies (21). Furthermore, UPS enzyme and substrate interactions are regulated through PPIs that are also difficult to inhibit with small molecules (20). Inhibition of enzyme function by stabilization of protein interactions is an underexploited strategy in drug discovery (42). This concept has recently been extended with the identification of compounds that stabilize interactions between E3 targeting subunits and substrates to enable therapeutically programmed substrate degradation (35, 43). The discovery of CC0651 as an inhibitor that acts by stabilization of the noncovalent CDC34A-donor ubiquitin complex showed that this approach is also feasible for UPS enzyme-ubiquitin interactions (37). CC0651 emerged fortuitously from a screen with a complex in vitro enzyme assay based on a 12-component mixture of E1, E2, E3, ubiquitin, substrate, and substrate kinase (36). While target-rich assays increase the likelihood of obtaining novel mechanism-of-action hits, substantial effort is then needed to decipher inhibitor mechanism. In this study, we sought to directly identify novel small-molecule stabilizers of the CDC34A-donor ubiquitin interaction.
To screen for stabilizers of the CDC34A-ubiquitin interaction, we used a sensitive TR-FRET PPI assay that monitored the proximity of CDC34A and ubiquitin. A focused screen against a small library enriched for protein interaction modulators yielded the isonipecotamide 1 as a weak hit that was 10-fold worse than CC0651 but which nevertheless had a promising biphenyl moiety analogous to the critical dichlorobiphenyl group of CC0651. An SAR campaign ultimately yielded analogs that were nearly 1000-fold more potent than the original hit and 100-fold more potent than CC0651 in the TR-FRET assay. This potency increase was mirrored in CDC34A-dependent in vitro ubiquitination assays for free ubiquitin chain formation and substrate polyubiquitination, as well as in a cell-based CDC34A thioester stabilization assay. In all assays, all inhibitors tested were exquisitely specific for CDC34A compared to its closely related UBE2R2 isoform and an extensive panel of 20 other human E2 enzymes. Structure determination of the representative isonipecotamide 2ab in complex with CDC34A and ubiquitin fully rationalized the critical chemical features identified through SAR and explained the specificity for CDC34A compared to other human E2 enzymes. The isonipecotamides represent a structurally novel E2 inhibitor class with strong potential for further development. Compared to CC0651, synthesis is simplified by the absence of chirality and is amenable to modular assembly through short, simple, and flexible synthetic routes. Moreover, the sugar-like glycerate scaffold of CC0651 is replaced with a more drug-like heterocycle in the isonipecotamides. The similarity of the joint pocket formed by CDC34A and ubiquitin in both the CC0651 and 2ab ternary structures demonstrates that, despite the weak noncovalent nature of the CDC34A-donor ubiquitin interaction, the pocket is virtually identical and thus amenable to further structure-guided inhibitor optimization. In total, these results provide proof of concept that molecular glue compounds that stabilize the E2-donor ubiquitin interaction can be directly identified in screens with a suitable PPI assay.
This screening strategy developed here can be readily applied to other noncovalent UPS enzyme-ubiquitin interactions. Genetic and biochemical evidence suggests that all UPS enzymes have evolved weak noncovalent interactions with ubiquitin that guide the various reaction paths for the ubiquitin thioester (7–9). These weak interactions can be mutationally stabilized in ubiquitin variants that bind more tightly and thereby inhibit reactions catalyzed by E2, E3, and DUB enzymes (44). If these myriad weak UPS enzyme-ubiquitin interactions create spacious joint pockets analogous to the CDC34A-donor ubiquitin pocket, then it should be possible to identify small molecules that specifically stabilize these interactions. The combinatorial redundancy of the UPS presents a further challenge for therapeutic intervention, as demonstrated by the nonessential nature of CDC34A and other E2 enzymes (14, 36, 37). The availability of inhibitors against each UPS enzyme will be critical for the development of combinatorial small-molecule strategies to modulate UPS activity in different disease contexts. In addition to UPS enzymes, related enzyme systems that catalyze conjugation of ubiquitin-like modifiers (45), such as for NEDD8, SUMO, ISG15, and UFM1, may also be amenable to the same screening strategy described here. Notably, because only one E2 enzyme exists for each ubiquitin-like modifier, this lack of redundancy may make these enzymes ideal targets for intervention using the interaction stabilization screening approach. Other recent examples suggest that the elaboration of weak PPI stabilizers into highly selective inhibitors holds promise as a general strategy for drug discovery (46).
MATERIALS AND METHODS
Design of a custom CDC34A screening library
2D extended connectivity fingerprints (ECFP4) were computed for each ligand in the Asinex PPI library (www.asinex.com/ppi), and a distance matrix was calculated using the Tanimoto distance function. Ligands were then clustered using k-medoids into 100 clusters. Fingerprints, distance function calculation, and clustering were performed as implemented in RDkit (www.rdkit.org) and Knime (www.knime.com) (47). 2D structure-data files of the ligand structures were converted to 3D structure files suitable as input for virtual screening by docking. Atoms and bonds were autotyped using the autosmiles algorithm (48), adding hydrogens, and assigning charges. An initial 3D structure was generated using semi-empirical quantum mechanics (QM) implemented in MOPAC (49), followed by simulated annealing energy minimization and another stage of semi-empirical QM. Flexible ligand docking was performed using AutoDock VINA (38) with default parameters and the CC0651-CDC34A-ubiquitin crystal structure (37) as a template. The docking site was limited to the CC0651 binding pocket. The entire workflow including ligand and receptor structure preparation and docking setup was implemented in the YASARA Structure (www.yasara.com) molecular modeling package (50). From all docked conformers, the best docking score was retained for each compound, and those compounds with better scores than CC0651 were selected for the CDC34A custom library. Representative ligands from underrepresented clusters were added manually to increase structural and chemical diversity.
Protein expression and purification
Full-length CDC34A (CDC34AFL, residues 1 to 236) and the core catalytic domain (CDC34ACAT, residues 7 to 184) were fused to a tobacco etch virus (TEV)–cleavable N-terminal His6 tag and expressed from the pProEx-HTb plasmid in Escherichia coli BL21(DE3) codon+ cells (Agilent Technologies) by induction with 0.25 mM isopropyl-β-d-thiogalactopyranoside for 14 to 18 hours at 18°C. Cell pellets were resuspended in 20 mM HEPES (pH 7.5), 400 mM NaCl, and 5 mM β-mercaptoethanol and lysed by passage through a cell homogenizer (Avestin Inc.). Following centrifugation at 30,000g, the supernatant was applied to a HiTrap nickel chelating HP column (GE Healthcare) equilibrated in lysis buffer with 5 mM imidazole. Proteins were eluted with buffer containing 300 mM imidazole and incubated overnight with TEV protease and 2 mM dithiothreitol (DTT). Cleaved protein was dialyzed in HiTrap loading buffer, flowed over a subtractive HiTrap nickel chelating column, and then concentrated for injection onto a 120-ml Superdex S75 column (GE Healthcare) equilibrated in 20 mM HEPES (pH 7.5), 100 mM NaCl, and 5 mM β-mercaptoethanol. Fractions at >95% purity were concentrated to 20 to 35 mg/ml. All other human E2s were expressed using a pET28a-LIC plasmid (51) that contained an N-terminal His6 tag with a thrombin cleavage site and purified as above. As the His6 tag was required for in vitro assays, fractions from the after the chelating column were concentrated and loaded onto a 120-ml Superdex S75 column equilibrated in 20 mM HEPES (pH 7.5), 100 mM NaCl, and 5 mM β-mercaptoethanol followed by concentration to 20 to 30 mg/ml.
TR-FRET assay
A modified version of a previously described TR-FRET assay was used (37). A master mix containing 0.5 μM His-CDC34AFL, 10 μM fluorescein-labeled ubiquitin, and 2 nM anti–His6-Tb3+ antibody (Invitrogen) in buffer [25 mM HEPES (pH 7.5), 100 mM NaCl, 0.01% Brij 35 (Thermo Fisher Scientific), and 2.5 mM β-mercaptoethanol] was dispensed into a 384-well plate (Corning 3573) in 24.6-μl aliquots. Library compounds dissolved in DMSO (40 μM final concentration), DMSO-only negative control, and CC0651-positive control (40 μM) were added as 0.4-μl volumes, mixed, and incubated at room temperature for 45 min. TR-FRET signal was measured on BMG PHERAstar (BMG Labtech) using an excitation filter for Tb3+ at 340 nm and emission filters for Tb3+ at 490 nm and fluorescein at 520 nm. Results were normalized to DMSO (negative) and CC0651 (positive) controls. Dose-response curves were acquired as for the screening assay except that the concentrations of His6-CDC34AFL and fluorescein-labeled ubiquitin were 0.05 and 2 μM, respectively, and bovine serum albumin (BSA; 0.1 mg/ml; Sigma-Aldrich) was added to the buffer. Compounds were serially diluted to a range of 0.2 to 200 μM and incubated for 30 min at room temperature, followed by TR-FRET measurement on Synergy Neo (BioTek Instruments) using the same filter set. Results were analyzed using GraphPad Prism 9 software.
For E2 titration assays, 2 nM anti–His6-Tb3+ antibody conjugate (Invitrogen) and 2 μM fluorescein-labeled ubiquitin in a buffer of 25 mM HEPES (pH 7.5), 100 mM NaCl, 0.01% Brij 35 (Thermo Fisher Scientific), BSA (0.1 mg/ml) (Sigma-Aldrich), and 1 mM DTT (BioShop) were dispensed into a 384-well plate (Corning). His6-tagged E2 enzymes were added over a range of 0 to 500 nM to give a final volume of 24.5 μl to which 0.5 μl of a 1 mM solution of compound 2ab in DMSO (20 μM final) was added and incubated for 30 min at room temperature before measurement as above. Single-point TR-FRET assays were carried out in the same manner except without DTT. Charging reactions contained 100 nM E1 and 20 μM compound 2ab or DMSO and were initiated by addition of 10 mM MgCl2 and 5 mM ATP in a final volume of 25 μl, followed by incubation for 60 min and measurement as above.
In vitro ubiquitination assays
Substrate-independent ubiquitination assays were performed by titrating compounds or DMSO solvent into 1 μM UBE1, 0.1 μM CDC34A or UBE2R2, and 0.25 μM CUL1-RBX1 complex in buffer [25 mM HEPES (pH 7.5), 100 mM NaCl, and 1 mM DTT] for 15 min at room temperature (37). Reactions were initiated by addition of 10 μM fluorescein-labeled ubiquitin, 3 mM ATP, and 10 mM MgCl2, incubated at room temperature for 60 min, and resolved on a 15% SDS-PAGE gel. Products were visualized on ChemiDoc MP (Bio-Rad) and quantified using Image Lab 6.0 software (Bio-Rad), and the amount of poly-ubiquitin chain product was normalized to DMSO controls. Results were analyzed by GraphPad Prism 9 software. Substrate-dependent ubiquitination assays were performed by titrating compounds or DMSO solvent into 1 μM UBE1, 0.25 μM CDC34A, 0.25 μM CUL1-RBX1-SKP1-SKP2-CKS1, 1 μM phosphorylated fluorescein-labeled p27, and 1.5 μM CDK2–cyclin A in buffer containing 25 mM HEPES (pH 7.5), 100 mM NaCl, and 0.5 mM DTT for 10 min at room temperature. Fluorescein-labeled p27 was pre-phosphorylated by incubation with CDK2–cyclin A for 1 hour at room temperature in the reaction buffer containing 3 mM ATP and 10 mM MgCl2. Ubiquitination reactions were initiated by addition of 20 μM ubiquitin, 3 mM ATP, and 10 mM MgCl2 and incubated at room temperature for 4 hours. Products were resolved, quantified, and analyzed as above. Signals for polyubiquitinated p27 species were normalized to DMSO control.
Ubiquitin~CDC34A thioester assay
Human PC3 prostate adenocarcinoma cells (American Type Culture Collection, CRL-1435) were grown in F-12K medium supplemented with 10% fetal bovine serum and penicillin/streptomycin (100 U/ml). Cells were synchronized in G0-G1 by serum starvation, released by addition of 10% serum, treated with CDC34A inhibitors for 24 hours before lysis in buffer [50 mM MES (pH 4.5), 150 mM NaCl, 0.2% NP-40, and protease inhibitor cocktail (Roche)], and clarified by centrifugation at 18,000g. Each sample was divided into two aliquots to which nonreducing (no DTT) or reducing (with 100 mM DTT) SDS-PAGE sample buffer was added. Reduced samples were heated at 90°C for 5 min. Proteins were resolved on NuPAGE bis-tris 4 to 12% gradient gels (Thermo Fisher Scientific), transferred onto nitrocellulose membrane, and immunoblotted with either anti-CDC34A (G11, sc-166738) or anti-UBE2R2 (H51, sc-376097) antibodies (Santa Cruz Biotechnology) followed by anti-mouse-HRP (horseradish peroxidase) (ab97041, Abcam) or anti-rabbit-HRP (AP182PMI, MilliporeSigma), respectively. Blots were visualized with ECL substrate (Froggabio UltraScence) on a Bio-Rad ChemiDoc MP imaging system.
Chemical synthesis
The critical 1-tert-butyl 4-ethyl 4-(4-bromobenzyl)piperidine-1,4-dicarboxylate (isonipecotate 8) and 4-(4-bromophenyl)piperidin-4-ol (piperidin-4-ol 14) starting materials (Sigma-Aldrich) were transformed into analog series using multistep syntheses over several batches at 0.3 to 3 mmol per batch. Standard organic synthesis and workup techniques were used. High-purity compounds were obtained through a combination of normal-phase medium-performance liquid chromatography (MPLC) and reversed-phase preparatory high-performance liquid chromatography (HPLC). Compounds were characterized and validated by liquid chromatography–mass spectrometry, nuclear magnetic resonance, high-resolution mass spectrometry, and thin-layer chromatography. Final target compounds were obtained as solids in quantities ranging from 5 to 102 mg at ≥95% purity as ascertained by HPLC-ELS (evaporative light scattering). Aliquots of the synthetic analogs were dissolved in DMSO as 10 to 100 mM solutions. Details of each synthetic route and data for compound validation are provided in the Supplementary Materials.
X-ray crystallography
All procedures were essentially as described (37). Crystals of a CDC34ACAT-ubiquitin-2ab complex were grown in hanging drops by mixing 1 μl of protein solution containing 500 μM CDC34ACAT, 500 μM ubiquitin, and 600 μM 2ab with 1 μl of well solution [29% PEG3350 (polyethylene glycol, molecular 3350), 40 mM dl-malic acid, 0.1 M HEPES (pH 7.0), and 5 mM DTT] at 20°C. For cryoprotection, a single crystal was soaked in well solution supplemented with 25% ethylene glycol. Diffraction data were collected at 100 K at 0.97919-Å wavelength on beamline NE-CAT 24-ID-E (APS, Chicago, IL) and processed with HKL2000 (52). Molecular replacement was performed with Phaser using the crystal structures of CDC34A (PDB code 3RZ3) and ubiquitin (PDB code 1UBQ) as search models (53). Model building and refinement were performed using Coot and Refmac5 (54). Data collection and refinement statistics are provided in table S6. Protein structure figures were generated using PyMOL (www.pymol.org/).
Statistical analysis
EC50 values for inhibitor-induced binding of CDC34A to ubiquitin assessed by TR-FRET assay represent mean values ± SD (n = 3) (Figs. 1E and 4). IC50 values for small-molecule inhibition of in vitro ubiquitination reactions represent mean values ± SD (n = 2) (Fig. 5B and fig. S6B).
Acknowledgments
We thank S. Beha and Z. Seghiri for technical assistance.
Funding: This publication was supported by grants from the Canadian Institutes of Health Research (FDN-167277 to M.T. and FDN-143277 to F.S.), a Canadian Cancer Society Impact Award (704116 to F.S.), a Genome Canada/Genome Quebec Genomics Technology Platform Award (to M.T. and P. Thibault), and Canada Research Chairs in Structural Biology (to F.S.) and in Systems and Synthetic Biology (to M.T.). Research conducted at the Northeastern Collaborative Access Team beamlines are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
Author contributions: A.M., F.S., and M.T. conceived the study; D.S.-C., S.O., A.M.v.d.S., S.M., C.J., and J.C.-H. designed the custom CDC34A library, performed chemical screens, and carried out screen data analysis; D.S.-C. and C.J. designed synthetic routes, performed analog synthesis, and characterized all compounds; D.F.C., S.O., and S.K. developed and performed biochemical assays; X.T. and T.B. carried out cell-based assays; D.F.C. and S.O. performed x-ray structure determinations; D.S.-C., D.F.C., S.O., A.M.v.d.S., X.T., S.K., S.M., G.P., J.C.-H., A.M., F.S., and M.T. wrote the manuscript; A.M., F.S., and M.T. obtained funding and provided supervision.
Competing interests: The authors declare that they have no competing interests. F.S. is a founder and consultant of Repare Therapeutics, a company that is focused on DNA damage–based therapeutics and is completely unrelated to the work presented in this paper. Repare Therapeutics did not provide any funding or support for this work.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Atomic coordinates and structure files have been submitted to the PDB for release on publication (PDB: 7M2K).
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Legends for tables S1 and S2
Tables S3 to S6
Supplementary Materials and Methods
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.Rape M., Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 19, 59–70 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Baek K., Scott D. C., Schulman B. A., NEDD8 and ubiquitin ligation by cullin-RING E3 ligases. Curr. Opin. Struct. Biol. 67, 101–109 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng N., Shabek N., Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Lucas X., Ciulli A., Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr. Opin. Struct. Biol. 44, 101–110 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Varshavsky A., N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. U.S.A. 116, 358–366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yau R., Rape M., The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Saha A., Lewis S., Kleiger G., Kuhlman B., Deshaies R. J., Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickliffe K. E., Lorenz S., Wemmer D. E., Kuriyan J., Rape M., The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart M. D., Ritterhoff T., Klevit R. E., Brzovic P. S., E2 enzymes: More than just middle men. Cell Res. 26, 423–440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshaies R. J., Joazeiro C. A., RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Lydeard J. R., Schulman B. A., Harper J. W., Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 14, 1050–1061 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R. J., Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957–968 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott D. C., Rhee D. Y., Duda D. M., Kelsall I. R., Olszewski J. L., Paulo J. A., de Jong A., Ovaa H., Alpi A. F., Harper J. W., Schulman B. A., Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell 166, 1198–1214.e24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill S., Reichermeier K., Scott D. C., Samentar L., Coulombe-Huntington J., Izzi L., Tang X., Ibarra R., Bertomeu T., Moradian A., Sweredoski M. J., Caberoy N., Schulman B. A., Sicheri F., Tyers M., Kleiger G., Robust cullin-RING ligase function is established by a multiplicity of poly-ubiquitylation pathways. eLife 8, e51163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dove K. K., Kemp H. A., di Bona K. R., Reiter K. H., Milburn L. J., Camacho D., Fay D. S., Miller D. L., Klevit R. E., Two functionally distinct E2/E3 pairs coordinate sequential ubiquitination of a common substrate in Caenorhabditis elegans development. Proc. Natl. Acad. Sci. U.S.A. 114, E6576–E6584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis R. J., Welcker M., Clurman B. E., Tumor suppression by the Fbw7 ubiquitin ligase: Mechanisms and opportunities. Cancer Cell 26, 455–464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skaar J. R., Pagan J. K., Pagano M., Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skaar J. R., Pagan J. K., Pagano M., SCF ubiquitin ligase-targeted therapies. Nat. Rev. Drug Discov. 13, 889–903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barghout S. H., Schimmer A. D., E1 enzymes as therapeutic targets in cancer. Pharmacol. Rev. 73, 1–56 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Scott D. E., Bayly A. R., Abell C., Skidmore J., Small molecules, big targets: Drug discovery faces the protein-protein interaction challenge. Nat. Rev. Drug Discov. 15, 533–550 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Rennie M. L., Chaugule V. K., Walden H., Modes of allosteric regulation of the ubiquitination machinery. Curr. Opin. Struct. Biol. 62, 189–196 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Burgess A., Chia K. M., Haupt S., Thomas D., Haupt Y., Lim E., Clinical overview of MDM2/X-targeted therapies. Front. Oncol. 6, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orlicky S., Tang X., Neduva V., Elowe N., Brown E. D., Sicheri F., Tyers M., An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat. Biotechnol. 28, 733–737 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott D. C., Hammill J. T., Min J., Rhee D. Y., Connelly M., Sviderskiy V. O., Bhasin D., Chen Y., Ong S. S., Chai S. C., Goktug A. N., Huang G., Monda J. K., Low J., Kim H. S., Paulo J. A., Cannon J. R., Shelat A. A., Chen T., Kelsall I. R., Alpi A. F., Pagala V., Wang X., Peng J., Singh B., Harper J. W., Schulman B. A., Guy R. K., Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase. Nat. Chem. Biol. 13, 850–857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou W., Ma L., Ding L., Guo Q., He Z., Yang J., Qiao H., Li L., Yang J., Yu S., Zhao L., Wang S., Liu H. M., Suo Z., Zhao W., Potent 5-Cyano-6-phenyl-pyrimidin-based derivatives targeting DCN1-UBE2M interaction. J. Med. Chem. 62, 5382–5403 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Chen H., Wu G., Gao S., Guo R., Zhao Z., Yuan H., Liu S., Wu J., Lu X., Yuan X., Yu Z., Zu X., Xie N., Yang N., Hu Z., Sun Q., Zhang W., Discovery of potent small-molecule inhibitors of ubiquitin-conjugating enzyme UbcH5c from α-Santonin derivatives. J. Med. Chem. 60, 6828–6852 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Hewitt W. M., Lountos G. T., Zlotkowski K., Dahlhauser S. D., Saunders L. B., Needle D., Tropea J. E., Zhan C., Wei G., Ma B., Nussinov R., Waugh D. S., Schneekloth J. S. Jr., Insights into the allosteric inhibition of the SUMO E2 enzyme Ubc9. Angew. Chem. Int. Ed. Engl. 55, 5703–5707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morreale F. E., Bortoluzzi A., Chaugule V. K., Arkinson C., Walden H., Ciulli A., Allosteric targeting of the fanconi anemia ubiquitin-conjugating enzyme Ube2T by fragment screening. J. Med. Chem. 60, 4093–4098 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu T., Yoon H., Xiong Y., Dixon-Clarke S. E., Nowak R. P., Fischer E. S., Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat. Struct. Mol. Biol. 27, 605–614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai A. C., Crews C. M., Induced protein degradation: An emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16, 101–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber S. L., The rise of molecular glues. Cell 184, 3–9 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Che Y., Gilbert A. M., Shanmugasundaram V., Noe M. C., Inducing protein-protein interactions with molecular glues. Bioorg. Med. Chem. Lett. 28, 2585–2592 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Dharmasiri N., Dharmasiri S., Estelle M., The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Lu G., Middleton R. E., Sun H., Naniong M., Ott C. J., Mitsiades C. S., Wong K. K., Bradner J. E., Kaelin W. G., The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chamberlain P. P., Cathers B. E., Cereblon modulators: Low molecular weight inducers of protein degradation. Drug Discov. Today Technol. 31, 29–34 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Ceccarelli D. F., Tang X., Pelletier B., Orlicky S., Xie W., Plantevin V., Neculai D., Chou Y. C., Ogunjimi A., al-Hakim A., Varelas X., Koszela J., Wasney G. A., Vedadi M., Dhe-Paganon S., Cox S., Xu S., Lopez-Girona A., Mercurio F., Wrana J., Durocher D., Meloche S., Webb D. R., Tyers M., Sicheri F., An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell 145, 1075–1087 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Huang H., Ceccarelli D. F., Orlicky S., St-Cyr D. J., Ziemba A., Garg P., Plamondon S., Auer M., Sidhu S., Marinier A., Kleiger G., Tyers M., Sicheri F., E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin. Nat. Chem. Biol. 10, 156–163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trott O., Olson A. J., AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baell J. B., Holloway G. A., New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Daina A., Michielin O., Zoete V., SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seol J. H., Feldman R. M., Zachariae W., Shevchenko A., Correll C. C., Lyapina S., Chi Y., Galova M., Claypool J., Sandmeyer S., Nasmyth K., Deshaies R. J., Shevchenko A., Deshaies R. J., Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13, 1614–1626 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrei S. A., Sijbesma E., Hann M., Davis J., O’Mahony G., Perry M. W. D., Karawajczyk A., Eickhoff J., Brunsveld L., Doveston R. G., Milroy L. G., Ottmann C., Stabilization of protein-protein interactions in drug discovery. Expert. Opin Drug Discov. 12, 925–940 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Simonetta K. R., Taygerly J., Boyle K., Basham S. E., Padovani C., Lou Y., Cummins T. J., Yung S. L., von Soly S. K., Kayser F., Kuriyan J., Rape M., Cardozo M., Gallop M. A., Bence N. F., Barsanti P. A., Saha A., Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction. Nat. Commun. 10, 1402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ernst A., Avvakumov G., Tong J., Fan Y., Zhao Y., Alberts P., Persaud A., Walker J. R., Neculai A. M., Neculai D., Vorobyov A., Garg P., Beatty L., Chan P. K., Juang Y. C., Landry M. C., Yeh C., Zeqiraj E., Karamboulas K., Allali-Hassani A., Vedadi M., Tyers M., Moffat J., Sicheri F., Pelletier L., Durocher D., Raught B., Rotin D., Yang J., Moran M. F., Dhe-Paganon S., Sidhu S. S., A strategy for modulation of enzymes in the ubiquitin system. Science 339, 590–595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cappadocia L., Lima C. D., Ubiquitin-like protein conjugation: Structures, chemistry, and mechanism. Chem. Rev. 118, 889–918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sijbesma E., Visser E., Plitzko K., Thiel P., Milroy L. G., Kaiser M., Brunsveld L., Ottmann C., Structure-based evolution of a promiscuous inhibitor to a selective stabilizer of protein-protein interactions. Nat. Commun. 11, 3954 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazanetz M. P., Marmon R. J., Reisser C. B., Morao I., Drug discovery applications for KNIME: An open source data mining platform. Curr. Top. Med. Chem. 12, 1965–1979 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Jakalian A., Jack D. B., Bayly C. I., Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 23, 1623–1641 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Stewart J. J., MOPAC: A semiempirical molecular orbital program. J. Comput. Aided Mol. Des. 4, 1–105 (1990). [DOI] [PubMed] [Google Scholar]
- 50.Krieger E., Koraimann G., Vriend G., Increasing the precision of comparative models with YASARA NOVA-a self-parameterizing force field. Proteins 47, 393–402 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Sheng J., Yu H., Li J., Sheng G., Zhou L., Lu Y., Cloning and expression of the human augmenter of liver regeneration at low temperature in Escherichia coli. J. Biochem. Biophys. Methods 70, 465–470 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Otwinowski Z., Minor W., Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 53.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Legends for tables S1 and S2
Tables S3 to S6
Supplementary Materials and Methods
Tables S1 and S2