ABSTRACT
Household contacts (HHCs) of patients with active tuberculosis (ATB) are at higher risk of Mycobacterium tuberculosis (M. tuberculosis) infection. However, the immune factors responsible for different defense responses in HHCs are unknown. Hence, we aimed to evaluate transcriptome signatures in human peripheral blood mononuclear cells (PBMCs) of HHCs to aid risk stratification. We recruited 112 HHCs of ATB patients and followed them for 6 years. Among the HHCs, only 2 developed ATB, while the remaining HHCs were classified into three groups: (1) HHC-1 group (n = 23): HHCs with consistently positive T-SPOT.TB test, negative chest radiograph, and no clinical symptoms or evidence of ATB during the 6-year follow-up period; (2) HHC-2 group (n = 15): HHCs with an initial positive T-SPOT result that later became negative without evidence of ATB; (3) HHC-3 group (n = 14): HHCs with a consistently negative T-SPOT.TB test and no clinical or radiological evidence of ATB. HHC-2 and HHC-3 were combined as HHC-23 group for analysis. RNA sequencing (RNA-seq) in PBMCs, with and without purified protein derivative (PPD) stimulation, identified significant differences in gene signatures between HHC-1 and HHC-23. Gene ontology analysis revealed functions related to bacterial pathogens, leukocyte chemotaxis, and inflammatory and cytokine responses. Modules associated with clinical features in the HHC-23 group were linked to the IL-17 signaling pathway, ferroptosis, complement and coagulation cascades, and the TNF signaling pathway. Validation using real-time PCR confirmed key genes like ATG-7, CXCL-3, and TNFRSF1B associated with infection outcomes in HHCs. Our research enhances understanding of disease mechanisms in HHCs. HHCs with persistent latent tuberculosis infection (HHC-1) showed significantly different gene expression compared to HHCs with no M. tuberculosis infection (HHC-23). These findings can help identify HHCs at risk of developing ATB and guide targeted public health interventions.
KEYWORDS: Household contacts, peripheral blood mononuclear cell, RNA sequence, tuberculosis, latent tuberculosis infection
Introduction
Tuberculosis remains the primary global cause of death as an infectious disease, particularly prevalent in less developed regions, where the number of newly infected individuals exceeds 10 million annually. In 2022, an estimated 10.6 million people fell ill with tuberculosis (TB) worldwide, and a total of 1.3 million people died from TB. Worldwide, TB is the second leading infectious killer after COVID-19 [1]. Latent tuberculosis infection (LTBI) refers to a condition in which the immune system mounts a continuous response to M. tuberculosis antigens, yet no signs of active TB (ATB) are clinically evident. LTBI is a complex and diverse state resulting from the intricate interplay between the host's immune response and the infecting organism. It is estimated that the lifetime risk of reactivation of LTBI ranges from 5% to 10%, and a significant proportion of ATB cases are traced back to the reactivation of LTBI [2].
Infection by M. tuberculosis is primarily contracted through close contact with individuals suffering from respiratory or pulmonary TB (PTB), who generate droplet aerosols. Household contacts (HHCs) of patients with pulmonary TB are at significantly higher risk of developing ATB or LTBI compared to other community members. Factors such as cohabitation with a pulmonary TB patient for 8–12 weeks, spousal relationship, and sharing a confined living space with the patient are potential risk factors for LTBI among HHCs [3]. The prevalence of LTBI among HHCs of ATB patients was found to be 45.5% in low-middle income countries and 30% in high-income countries, with corresponding ATB prevalence rates of 3.1% and 3.0% respectively [4]. Another systematic review reported that ATB (both bacteriologically confirmed and clinically diagnosed) occurred in 4.5% of HHCs, while LTBI was present in 51.4% of HHCs of pulmonary TB patients [5]. Despite extensive exposure to infectious pulmonary TB cases, some HHCs remain uninfected, demonstrating resistance to infection [6]. Studying these individuals, who exhibit diverse host responses ranging from ATB and LTBI to non-TB infection, can yield valuable insights into the pathogenic mechanisms of M. tuberculosis.
The diagnosis of LTBI is made with either the tuberculin skin test (TST) or the T-cell-based gamma interferon (IFN-γ) release assay (IGRA). Long-time-used TST has encountered considerable difficulties, mainly due to the disability of its mixed antigens tuberculin purified protein derivative (PPD) to distinguish the true ATB patients from those vaccinated with BCG or sensitized with Nontuberculous Mycobacteria (NTM) [3]. The IGRAs have shown their superior diagnostic performance over TST by using at least two specific antigens (ESAT-6 & CFP 10) present exclusively in M. tuberculosis but absent in BCG strains and most NTM [4,5]. The T-SPOT test, classified as an IGRA, evaluates the immune response against M. tuberculosis by quantifying the number of T-cells activated by M. tuberculosis-specific antigens (ESAT-6 and CFP-10). This test is capable of detecting LTBI as it targets effector T-cells that persist following a previous M. tuberculosis infection, rather than directly detecting the presence of M. tuberculosis. Additionally, T-SPOT can identify the loss of M. tuberculosis-specific effector memory T-cells once the pathogen has been successfully eradicated [6]. Therefore, during the long-term follow-up of household contacts (HHCs) after exposure with M. tuberculosis, the host could develop different outcomes including active disease, latent infection, or no infection, which depends on both bacterial and host immune factors that could be indicated by different follow-up results of T-SPOT [7]. The mechanisms accounting for these diverse outcomes following M. tuberculosis infection remain unclear, necessitating further research to shed light on the immunological basis of active disease, latent infection, or pathogen clearance [6,8,9].
In recent years, several genome-wide transcriptomic studies have been conducted to identify variations in host gene expression profiles in relation to different states of M. tuberculosis infection [10–12]. However, while most studies have primarily focused on discerning differences between patients with active TB and individuals with LTBI [10,13], little is known about the diverse host responses among household contacts (HHCs). Thus, the objective of this study was to conduct a prospective investigation in mainland China, aiming to evaluate transcriptome signatures in human peripheral blood mononuclear cells (PBMCs) of HHCs with distinct clinical outcomes. These outcomes were determined through long-term follow-up utilizing T-SPOT and other diagnostic tests. The findings of this study will assist in pinpointing pivotal molecules and signaling pathways influencing the divergent outcomes observed in HHCs, ultimately enhancing our understanding of the pathogenesis of tuberculosis infection. Furthermore, this research will contribute to the identification of novel markers that can be utilized for risk stratification and prediction among HHCs.
Materials and methods
Study design and participants
A prospective cohort of HHCs was recruited to identify RNA-transcript signatures associated with different stages of HHCs of PTB patients. Active TB during longitudinal assessment was diagnosed based on microbiological confirmation of MTB by culture or positive Xpert MTB/RIF (Cepheid, Sunnyvale, CA, USA), and all the PTB patients in this study had positive sputum smears for acid-fast bacilli. The median time for these PTB patients from the onset of symptoms to the sputum smear turning negative is 35 days (interquartile range: 21–63 days). The inclusion criteria comprised HHCs (≥14 years old) who had been in contact with ATB patients within 3 months of the ATB diagnosis. Trained interviewers administered standardized questionnaires to gather sociodemographic information and health status data, encompassing demographics, biological relationship with patients, clinical symptoms, medical history, and Bacillus Calmette-Guérin (BCG) vaccination status (determined by the presence of BCG scars). Exclusion criteria encompassed missing key data, Human Immunodeficiency Virus (HIV) infection, and a history of TB. Baseline data included chest radiographs, T-SPOT.TB, and tuberculin skin tests (TST) to assess the M. tuberculosis infection status of all participants. Follow-up evaluations were conducted biennially over a 6-year period, which entailed medical history reviews, systemic assessments, chest radiographs, T-SPOT.TB and TST.
The study protocol was approved by the ethics committees of Huashan Hospital of Fudan University, Shanghai, China. All participants in the trial agreed to participate in this study and provided written informed consent.
PBMC preparation, stimulation, and RNA extraction
Peripheral blood (8 mL) was collected from each participant from the cubital vein into a heparinized vacuum blood collection vessel (BD Vacutainer). The peripheral blood mononuclear cells (PBMCs) were isolated within 4 h of collection using lymph cell separation medium (Lymphoprep). The Countess automatic cell counter (Life Technologies, USA) was employed to determine the number of viable cells via trypan blue staining. Subsequently, the PBMCs of each subject were adjusted to 2.5 × 106 cells in 1 mL of AIM-V medium (Life Technologies, USA) and incubated in 24-well plates. The PBMCs were then stimulated with 10 µg/mL of M. tuberculosis purified protein derivative (PPD, MycosResearch LLC, USA) or left unstimulated, incubated at 37°C with 5% CO2 for 4 h. After the incubation period, the PBMCs were collected and suspended in Trizol reagent (Invitrogen, California, USA). Total RNA was extracted immediately following the manufacturer's instructions, and any genomic DNA contamination was removed using ribonuclease-free DNase I (American Life Technology Corporation). The concentration of RNA was quantified using a nanodrop instrument (Thermo Fisher Scientific Corporation), and its integrity and quality were evaluated using an Agilent2100 biological analyzer (Agilent Technology, USA). RNA with a RNA Integrity Number (RIN) ≥ 7.0 and a 28S/18S ratio > 0.7 were selected for library preparation and RNA sequence analysis.
RNA library preparation, sequencing, and data analysis
We used the Illumina TruSeq RNA sample preparation kit following the manufacturer's recommended procedures to generate a cDNA library from RNA samples extracted from each group. Sequencing was performed using the Illumina Hiseq 2000 instrument. Detailed information about library preparation, sorting, and data processing is available in the supplementary method.
To analyze the differentially expressed genes between the four clinical groups, the folding change (FC) of each individual gene was calculated by comparing the PPD-stimulated and non-stimulated samples. Subsequently, the FC values were analyzed using Student's t-test through pairwise comparison. Additionally, the ratio of the average FC values between the different groups was calculated. The differentially expressed genes between the two clinical groups were determined and selected for further analysis based on the criteria of Student's t-test (P < 0.05) and a ratio of >2.0 (14,22). Subsequently, the selected genes were subjected to functional analysis. The detailed procedure for biological information analysis is provided in the supplementary method.
Cibersort [14] analysis
CIBERSORT is a machine learning approach for characterizing the cell composition of a tumour biopsy from gene expression data (http://cibersort.stanford.edu) and is a useful method for the high-throughput characterization of various cell types. Usually, a feature matrix containing 22 functionally defined human immune subgroups (LM22) is used for verification. Here, we used the CIBERSORT method to calculate the proportions of 22 immune cells in different group of HHCs.
qRT-PCR analysis
RT–PCR was used to verify the expression level of differentially expressed genes. Total RNA was extracted from PPD-stimulated and unstimulated PBMC using TRIzol reagent (Life Technologies, USA) following the manufacturer's instructions. The purified RNA was reverse transcribed into cDNA using the Prime ScriptH RT kit (TaKaRa) following the manufacturer's instructions. Subsequently, qRT-PCR was performed using SYBRTM Green PCR Master Mix (TaKaRa) under the standard conditions of the ABI 7500 real-time PCR system (Applied Biosystems). The 2−11Ct method was used to compare with the expression of the housekeeping gene GAPDH. The qRT-PCR primers for the target gene can be found in the supplementary method.
T-SPOT®.TB test (T-SPOT)
All recruited candidates were screened by T-SPOT®.TB kit (Oxford Immunotec Ltd, Oxford, UK). The T-SPOT test was performed according to the manufacturer’s instructions. Spot-forming cells (SFCs) in T-SPOT Panel A (T-SPOT A) and T-SPOT Panel B (T-SPOT B), representing antigen-specific T-cells secreting IFN-γ, were counted with an automated ELISPOT reader (AID-GmbH, Germany). The results were double-checked by other laboratory workers and, if necessary, corrected by manual counting. The laboratory technicians were blinded to the subject identifiers.
RNA-seq data pre-processing, differential expression analysis, and functional enrichment analysis
The quality of the RNA-seq data obtained was assessed using FastQC. The raw sequencing reads were pre-processed by trimming the adapter sequences and by removing the >20 base pair (bp) long low-quality reads (Phred quality score <20). The filtered clean reads were aligned to the human reference genome (GRCh38) using Tophat2. Then, uniquely mapped reads were assigned to each annotated gene using featureCounts. Differential expression and statistical analyzes were performed using DESeq2 from the R package. The fold change of gene expression level altered by PPD stimulation was expressed as log2 transformed Cy3/Cy5 intensities for each probe. Differentially expressed genes (DEGs) were identified based on the P-values <0.05 by Student’s t-test and the fold change >2.0. Principal component analysis was performed on all normalized expression data using prcomp package with R. The PCA plot demonstrated top 2 PCs for each sample. To determine the Gene Ontology (GO) terms and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways associated with the DEGs, functional enrichment analysis (PCA) was conducted using the clusterProfiler package, with the threshold set to P-value <0.05. GO terms were used to describe gene functions and to classify the DEGs into three functional categories, namely biological process (BP), cellular component (CC), and molecular function (MF).
Weighted correlation network analysis (WGCNA)
A co-expression network was constructed (β-value = 18), in which genes were clustered into branches of highly expressed genes and modules were identified by the tree cut algorithm with the additional PAM stage. The minimum modules size is 30. This modules number of module size was selected based on the soft thresholding (β value), obtained by the pickSoftThreshold function in the WGCNA package. Two binary variables were generated and used to calculate the module trait relationships in both PPD-stimulated and unstimulated samples. Additionally, a between-group Kruskal–Wallis test was applied to the Module Eigengene (ME) values to detect overall differential gene expression. The modules were then functionally annotated using cluster Profiler package.
Protein–protein interaction (PPI) network construction
The PPI data were obtained from the Search Tool for the Retrieval of Interacting Genes (STRING) database. The PPI network was constructed using the TB-specific DEGs, with a confidence score of >0.9. The gene networks were subsequently generated using STRING: functional protein association networks v11[15] to further assess the complex associations among the TB-specific DEGs.
Results
Characteristics of the recruited subjects
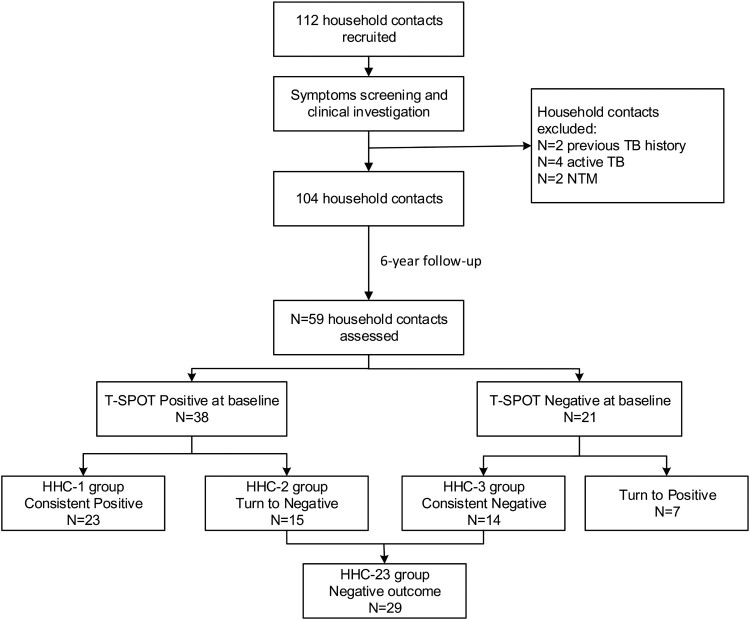
This prospective cohort study followed 112 household contacts (HHCs) of sputum smear-positive ATB patients for 6 years to observe the development of ATB. Among the HHCs, only 2 developed ATB, while the remaining HHCs were classified into three groups based on several examination results, including clinical symptom assessment, blood routine, erythrocyte sedimentation rate, chest X-ray, sputum smear, and T-SPOT (Figure 1). These three groups were defined as follows: (1) HHC-1 group (n = 23): HHCs with consistently positive T-SPOT.TB test, negative chest radiograph, and no clinical symptoms or evidence of ATB during the 6-year follow-up period; (2) HHC-2 group (n = 15): HHCs with an initial positive T-SPOT result that later became negative without evidence of ATB; (3) HHC-3 group (n = 14): HHCs with a consistently negative T-SPOT.TB test and no clinical or radiological evidence of ATB.
Figure 1.
Flow chart of the study process and T-SPOT results of household contacts during follow-up.
In our further analysis, we opted to merge HHC-2 and HHC-3 into a singular group, denoted as HHC-23. This amalgamation aimed to facilitate a comparison of the transcriptional profiles between HHC-1 and the combined HHC-23 cohort. The rationale for this grouping was twofold: (1) Following prolonged and close contact with ATB patients, individuals in the HHC-1 cohort eventually developed LTBI, whereas those in the HHC-23 group remained uninfected by TB. (2) Our pairwise comparison analysis among the HHC-1, HHC-2, and HHC-3 groups (not displayed) revealed highly similar expression profiles for HHC-2 and HHC-3, both of which significantly differed from the HHC-1 group.
Different gene expression patterns and functional characterization between persistent LTBI (HHC-1) and those initial or finally eliminating M. tuberculosis (HHC-23).
In order to explore the difference of host immune response between the two groups, we selected 4 and 8 (4 from HHC-2 and 4 from HHC-3) PBMC samples from HHC-1 and HHC-23 group, respectively. Subsequently, we employed RNA-seq technology to determine the transcriptional profiles of the entire genome from PBMCs of individuals in different groups, both with and without PPD stimulation. Analysis of gene expression differences between the two groups was performed to investigate the underlying reasons for the varying reactions. PBMCs from the remaining subjects in these two groups were designated as a validation set for quantitative real-time PCR (qRT-PCR) verification.
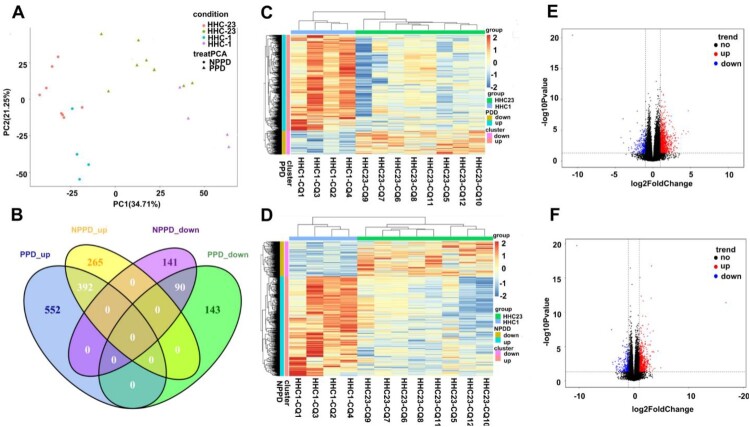
The principal component analysis (PCA) plot, based on gene expression profiles, distinctly distinguished between the HHC-1 and HHC-23 groups in both PPD-stimulated and unstimulated samples (Figure 2A). The hierarchical clustering heatmap analysis also illustrated the gene expression profiles of HHC-1 was highly distinct from the HHC-23 group, in both PPD-stimulated and unstimulated samples (Figure 2C,D). The distribution of all DEGs was then plotted on a volcano map, based on the -log10 false discovery rate (-log10FDR) and log2FC values (Figure 2E,F).
Figure 2.
Transcriptional patterns of peripheral blood mononuclear cells (PBMC) defining PPD-stimulated and un-stimulated of HHC-1 and HHC-23 group. RNA-seq were performed using 4 and 8 (4 from HHC-2 and 4 from HHC-3) PBMC samples from HHC-1 and HHC-23 group, respectively. A. Principal component analysis diagram. B. Venn diagrams showing overlaps of TB-specific genes changes between HHC-1 and HHC-23 group. Unsupervised hierarchical clustering of transcribed genes and differentially expressed over 1.5-fold of PPD-stimulated (C) and unstimulated samples (D). Volcano plot shows the upregulated and downregulated transcribed genes between HHC-1 and HHC-23 group of PPD-stimulated (E) and unstimulated samples (F).
The Venn diagram in Figure 2B reveals a total of 1,177 DEGs were identified in the PPD-stimulated samples, with 944 upregulated and 233 downregulated DEGs in the HHC-1 group in comparison to the HHC-23 group; in the unstimulated samples, 888 DEGs were significantly differentially expressed in the HHC-1 group in comparison to the HHC-23 group, with 657 upregulated and 231 downregulated genes. Notably, 482 DEGs (41.0%) overlapped between PPD-stimulated and unstimulated samples. Among the up-regulated and down-regulated genes, 695 (59.0%) DEGs were specific to PPD-stimulated, while 406 (34.5%) DEGs were specific to unstimulated samples.
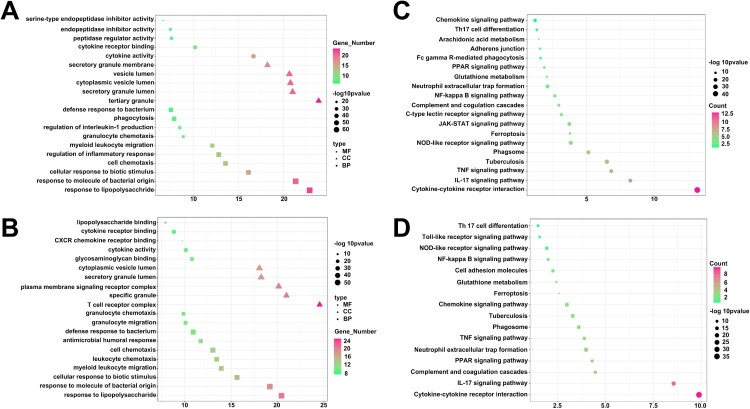
GO analysis was conducted to demonstrate the biological pathways, networks, and functional categories of differentially expressed genes (DEGs) between the HHC-1 and HHC-23 groups. As depicted in Figure 3, the top 5 enriched Gene Ontology (GO) terms in biological processes (BP), cellular components (CC), and molecular functions (MF). The GO-enrichment analysis revealed that various DEGs were implicated in cellular responses to bacteria, leukocyte chemotaxis, regulation of inflammatory responses, and cytokine responses (Figure 3A,B). These processes are typically activated during the early stages of infection and are involved in the host's response to bacterial invasion.
Figure 3.
Different gene expression and functional enrichment analysis of HHC-1 and HHC-23 group. DEGs was applied to GO analysis in BP, CC and MF using PPD-stimulated (A) and unstimulated samples (B). DEGs was applied to KEGG using PPD-stimulated (C) and unstimulated samples (D).
KEGG analysis was conducted to identify pathways in PPD-stimulated and unstimulated samples between the HHC-1 and HHC-23 groups, with a false discovery rate (FDR) lower than 0.01. The results of this analysis revealed the top 10 pathways that showed a significant correlation in the HHC-1 group compared to the HHC-23 group, as shown in Figure 3C,D. In the PPD-stimulated samples, the KEGG analysis for all differentially expressed genes in the HHC-1 vs HHC-23 comparison identified several highly correlated pathways, including cytokine-cytokine receptor interaction (FDR = 3.72 × 10−6, ratio = 33/70), IL-17 signaling pathway (FDR = 1.29 × 10−5, ratio = 32/54), TNF signaling pathway (FDR = 7.59 × 10−3, ratio = 34/62), and tuberculosis (FDR = 7.05 × 10−4, ratio = 16/30). These results suggest that the difference in the mentioned pathways contributes to the different close-contact endings observed between the HHC-1 and HHC-23 groups. These pathways are associated with host immune responses to bacterial invasion, suggesting their importance in eliminating M. tuberculosis. In summary, the findings from the KEGG analysis and the corresponding GO term results indicate that the crucial role of host immune responses to bacterial invasion in the control of M. tuberculosis infection, as reflected in these pathways.
Finally, the CIBERSORT algorithm illustrated that in PPD-stimulated samples, activated Mast cells were higher in the HHC-1 group compared to the HHC-23 group (p = 0.02), while CD8+ T-cells were higher in the HHC-23 group (p = 0.03) (Supplementary Figure 1).
3. Construction of WGCNA Network
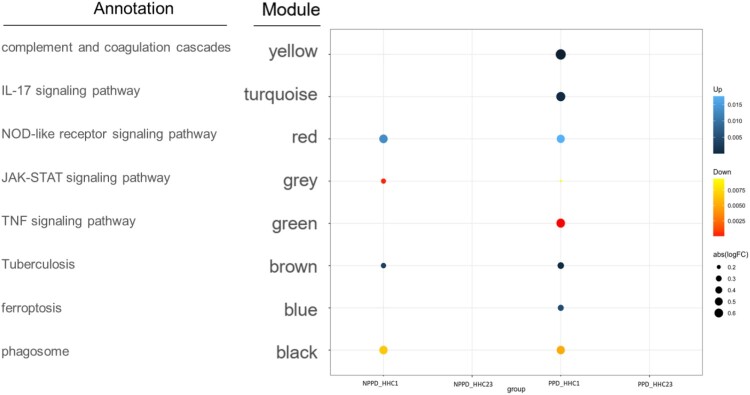
WGCNA analysis was performed to construct co-expressed networks and identify co-expression modules associated with host defense against M. tuberculosis. The genes were clustered based on their co-expression patterns, resulting in a network consisting of 8 co-expression modules. To summarize the gene co-expression within each module, the ME value was calculated as the first principal component, serving as a representative measure. The correlation between ME values and variables representing the transcriptional signatures of the HHC-1 and HHC-23 groups (Supplementary Figure 2) was then examined.
Upon analysis, we observed that four key modules were strongly associated with enhanced host defense against M. tuberculosis (HHC-1 vs HHC-23) in both PPD-stimulated and unstimulated samples. Specifically, the turquoise, blue, and yellow modules showed positive correlations with clinical outcomes in the HHC-23 group, indicative of stronger host defense against M. tuberculosis. Conversely, the green module exhibited a negative correlation with the HHC-23 group in PPD-stimulated samples. These correlations were visually depicted in Figure 4. Furthermore, the turquoise, blue, and yellow modules were found to be enriched with specific Gene Ontology (GO) terms. Notably, the turquoise module showed enrichment for the “IL-17 signaling pathway,” the blue module for “ferroptosis,” and the yellow module for “complement and coagulation cascades.” On the other hand, the green module was associated with the “TNF signaling pathway.” These results suggest that innate immune responses following TB infection play a crucial role in determining the infection outcome after M. tuberculosis infection. Specifically, the “IL-17 signaling pathway,” “ferroptosis,” “complement and coagulation cascades,” and “TNF signaling pathway” are identified as key immune response processes influencing these outcomes.
Figure 4.
Modular transcriptional signatures of HHC-23 compared to HHC-1 group. Fold enrichment scores derived using QuSAGE are depicted, with red and blue indicating modules over- or under-expressed. Colour intensity and size represent the degree of enrichment.
4. Validation of the signature gene set for discriminating HHC-1 and HHC-23 group
For further analysis, we constructed a protein–protein interactions (PPI) network of the expressed genes in the four selected modules (Supplementary Figure 3). We identified the top 5 hub genes out of a total of 20 genes. To verify the expression of the differentially transcribed genes identified by RNA-seq, we conducted a quantitative real-time PCR (qRT-PCR) using a test set comprising 15 subjects from the HHC-1 group and 19 subjects from the HHC-23 group (9 in HHC-2 and 10 in HHC-3).
Out of the 20 differentially expressed genes, 16 showed consistent results with the RNA-seq data and demonstrated a significant difference between the two groups (80.0% agreement, 16/20). The qRT-PCR results are presented in Table 1. The most significant differences in expression were observed for ATG-7, CXCL-3, and TNFRSF1B, suggesting their key roles in the distinct host responses.
Table 1.
Validation results of significantly regulated genes in PPD-stimulated PBMCs between HHC-1 and HHC-23 group in a test set by qRT-PCR.
| GO terms | Gene symbol | Median fold change | Ratio | P-value | |
|---|---|---|---|---|---|
| HHC-23 | HHC-1 | ||||
| IL-17 signaling pathway | CXCL3 | 10.88 | 3.16 | 3.44 | <0.0001 |
| NFKBIA | 8.46 | 7.89 | 1.07 | 0.135 | |
| CXCL2 | 11.11 | 10.30 | 1.08 | 0.035 | |
| IL8 | 12.85 | 10.59 | 1.21 | 0.009 | |
| CSF2 | 5.89 | 4.38 | 1.34 | 0.006 | |
| Complement and coagulation cascades | SERPINA1 | 7.11 | 6.27 | 1.13 | 0.011 |
| PLAUR | 7.85 | 7.23 | 1.09 | 0.043 | |
| SERPINE1 | 0.80 | 1.51 | 0.53 | 0.161 | |
| PROCR | 3.59 | 2.60 | 1.38 | 0.022 | |
| SERPINB2 | 10.47 | 9.58 | 1.09 | 0.014 | |
| Ferroptosis | ACSL5 | 6.73 | 6.14 | 1.10 | 0.035 |
| ATG7 | 4.42 | 2.54 | 1.74 | <0.0001 | |
| MAP1LC3A | 2.79 | 2.98 | 0.94 | 0.160 | |
| SLC3A2 | 5.89 | 5.97 | 0.99 | 0.257 | |
| FTL | 12.37 | 11.48 | 1.08 | 0.002 | |
| TNF signaling pathway | TNFRSF1B | 3.81 | 8.24 | 0.46 | <0.0001 |
| CCl20 | 9.41 | 10.95 | 0.86 | 0.004 | |
| MMP14 | 7.42 | 8.08 | 0.92 | 0.001 | |
| ICAM1 | 6.39 | 7.11 | 0.90 | 0.016 | |
| SOCS3 | 5.74 | 5.31 | 1.08 | 0.012 | |
Fold change: the fold change was calculated by dividing gene expression level of PPD-stimulated sample by gene expression level of unstimulated sample by qRT-PCR.
Ratio: the ratio was calculated by dividing mean fold change of one group by mean fold change of another group.
Discussion
The primary focus of this study was to investigate the immune responses in human PBMCs of HHCs of patients with ATB. RNA-seq was used to identify differentially expressed genes between HHCs with persistent LTBI and those who either cleared the M. tuberculosis infection initially or finally. Several key pathways and genes associated with these different infection outcomes were identified.
Infection with M. tuberculosis can result in various outcomes including active disease, latent infection, or clearance, which profoundly depend on both bacterial and host factors. Infections with M. tuberculosis elicit complex immune responses, and the reason behind the immune system’s failure to eradicate the pathogen from the body, as well as the reactivation of M. tuberculosis in some individuals but not others. This study recruited HHCs of patients with ATB as the study subjects. The overall incidence rate of active disease in household contacts was estimated to be 11.0 per 1000 person-years [16], with the majority remaining asymptomatic without developing the active disease. These subjects’ immune systems may be able to control or eradicate the bacteria, despite repeated and prolonged exposure. Therefore, analyzing gene expression in these subjects may help identify profiles that correlate with resistance or susceptibility to M. tuberculosis.
In this study, we employed a novel approach by using PPD-stimulated PBMCs to minimize unrelated background noise and maximize TB-specific host responses. T-SPOT tests were utilized to screen individuals with latent infection who were HHCs. Although the cross-reactivity to PPD skin testing caused by BCG vaccination is relatively small in adults, our previous study still showed that in areas of high TB endemicity and BCG coverage in China, the IGRA is a most promising test for LTBI diagnosis due to the improvement of its specificity and convenience [17, 18]. Furthermore, individuals who tested positive for T-SPOT in the HHC group and had known exposure to smear-positive pulmonary ATB patients were considered to have acquired recent infection, while those T-SPOT positives without established TB exposure were deemed to have experienced remote infection [19,20]. Moreover, among the HHCs in our cohort, those with positive T-SPOT results at baseline exhibited different outcomes of M. tuberculosis infection based on the dynamic change of their T-SPOT results. Individuals with persistent positive T-SPOT results were found to have latent infection but could not clear the pathogen. On the other hand, individuals with converted T-SPOT results, although the M. tuberculosis pathogen in the HHC-23 group load could not be directly determined, may have had their host immune response not only controlled the replication of M. tuberculosis but also potentially eradicate it. This speculation is supported by the quantification of T-cell response measured by IFN-gamma to M. tuberculosis-specific antigens, which may reflect the bacterial burden of the subjects [21]. Following successful treatment of active disease and subsequent reduction in the number of M. tuberculosis, the frequency of effector T-cells also decreased [21–23]. For instance, chemoprophylaxis with isoniazid and rifampicin is known to decrease IFN-γ levels in individuals with LTBI, and some individuals even achieved reversion of the IGRA results [24–27]. Therefore, the different outcomes of IGRA results in HHCs may indicate varying abilities of the host innate and adaptive immune systems to eradicate or control the bacteria during M. tuberculosis infection.
IL-17, a cytokine, plays a critical role in the induction of the adaptive immune response against M. tuberculosis. M. tuberculosis microorganisms have been found to manipulate Th17-related cytokine signaling pathways to promote their survival in primary Tb [28]. Furthermore, active TB patients have exhibited a significantly lower frequency of Th17 cells compared to healthy subjects, indicating a potential protective role of Th17 cells [29]. Published studies have suggested that M. tuberculosis microorganisms can manipulate Th17-related cytokine pathways to facilitate their survival in primary TB [30–32]. Furthermore, there is evidence indicating that immune responses involving the signal transducer and activator of transcription 3 (STAT3) signaling pathway and Th17-like T-cell subsets are dysregulated or impaired in patients with TB [32]. Consequently, these mechanisms could be utilized by the bacteria in individuals with LTBI to ensure their survival and counteract the immune system's efforts to eradicate them. Ferroptosis, a regulated form of necrosis, is characterized by the build-up of free iron and toxic lipid peroxides. Ferroptosis has been identified as a significant mechanism of necrosis in M. tuberculosis infection, with its excessive activation contributing to M. tuberculosis dissemination [33]. M. tuberculosis infection of macrophages could inducs ferroptosis in mice, and infection by M. tuberculosis is restricted when ferroptosis is inhibited [33]. The emerging critical roles of ferroptosis in infectious diseases emphasize its potential as a therapeutic target and diagnostic biomarker in this field [34–36]. The complement and coagulation cascades contribute to a multi-faceted immune response against M. tuberculosis invasion in macrophages. While the activated complement system contributes to the immune response in clearing M. tuberculosis, the bacteria can evade clearance by the immune system and persist within macrophages using various mechanisms [37]. These pathways were found inhibited in patients with persist latent infection and enhanced in patients initial or finally eliminating M. tuberculosis, suggesting that these pathways are related to the host defense of M. tuberculosis, especially under the condition of granuloma formation. Further mechanisms need to be explored, and not a single pathway can determine the outcome.
In our study, we observed up-regulation of three genes, namely ATG-7, TNFRSF1B, and CXCL-3, in HHCs who eliminated M. tuberculosis infection, as opposed to HHCs with persistent latent tuberculosis infection (HHC-1). This suggests an association between these three genes and host immune defense against M. tuberculosis. Notably, ATG-7 is a crucial autophagy protein essential for mycobacteria elimination in macrophages [38,39], Previous studies demonstrated that ATG-7 deficient mice were susceptible to mycobacterial infection. The TNFRSF1B gene influences the biological activity of TNF-α by encoding TNFR-2 [40,41], and research on knockout mice has revealed the involvement of TNFR-2 in Mycobacterium bovis bacillus Calmette-Guérin infection [42]. Additionally, previous studies have indicated dowmregulation of TNFRSF1B gene expression in patients with ATB compared to those with LTBI [43,44]. The CXCL-3 gene primarily encodes the ligand for CXCR-2 and contributes to the innate immune stress response against tuberculosis. It has been shown that CXCL-3 gene expression is significantly increased in patients with ATB compared to healthy individuals [4546].
Limitation
This study possesses a few limitations. First, the sample size is relatively small, and it is recommended for future studies to involve larger samples and increase the frequency of follow-up. However, the subjects from the three groups were well matched after adjustment for age, sex, HIV coinfection, and previous TB disease. Further, the PCA plot, hierarchical clustering, and Venn diagrams showed that the transcriptomic signatures were significantly different in different groups. Second, a more refined study population should be selected to enhance the homogeneity of the study population and minimize the impact of individual factors on the outcomes. In our study, to avoid the occurrence of errors to a certain extent, we confirmed the expression of differential genes in an independent validation cohort. Lastly, although investigation of the spectrum of LTBI using transcriptome analysis can accurately discover the underlying host immune mechanisms involved and assess the risk of TB development among individuals with LTBI under clinical settings, further in-depth functional investigations are still necessary to ascertain the roles and mechanisms of the aforementioned pathways and molecules in relation to immunity against TB infection.
Conclusion
Our research has enhanced the understanding of the disease mechanism associated with different clinical outcomes of household contacts (HHCs) infected with tuberculosis. Significantly divergent transcriptional profiles were observed in HHCs with persistent LTBI (HHC-1) compared to HHCs who successfully eliminated M. tuberculosis infection (HHC-23). The distinct infection outcomes among HHCs of active TB patients are primarily linked to the expression of genes involved in the IL-17 signaling pathway, ferroptosis, complement and coagulation cascades, and TNF signaling pathway. Furthermore, ATG-7, CXCL-3, and TNFRSF1B of these pathways have been identified as key genes.
These findings contribute to a better comprehension of the pathogenesis of tuberculosis and have the potential to identify HHCs at risk of developing active TB. Ultimately, this could allow for the targeting of public health interventions.
Supplementary Material
Funding Statement
This work is supported by National Key Research and Development Program of China (2022YFC2009801), Autonomous deployment project of Shanghai Sci-Tech InnoCenter for Infection and Immunity (SSIII-202305), Shanghai Science and Technology Committee (21NL2600100), and Major projects of Wuxi Municipal Health Commission (Z202111), Wuxi Taihu Lake Talent Plan.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Global tuberculosis report . https://www.who.int/publications-detail-redirect/9789240083851; 2023.
- 2.Horsburgh CR, Rubin EJ.. Clinical practice. Latent tuberculosis infection in the United States. N Engl J Med. 2011;364(15):1441–1448. doi: 10.1056/NEJMcp1005750 [DOI] [PubMed] [Google Scholar]
- 3.Richeldi L. An update on the diagnosis of tuberculosis infection. Am J Respir Crit Care Med. 2006;174(7):736–742. doi: 10.1164/rccm.200509-1516PP [DOI] [PubMed] [Google Scholar]
- 4.Lalvani A, Pareek M.. Interferon gamma release assays: principles and practice. Enferm Infecc Microbiol Clin. 2010;28(4):245–252. doi: 10.1016/j.eimc.2009.05.012 [DOI] [PubMed] [Google Scholar]
- 5.Diel R, Loddenkemper R, Nienhaus A.. Evidence-based comparison of commercial interferon-gamma release assays for detecting active TB: a metaanalysis. Chest. 2010;137(4):952–968. doi: 10.1378/chest.09-2350 [DOI] [PubMed] [Google Scholar]
- 6.Walzl G, Ronacher K, Hanekom W, et al. . Immunological biomarkers of tuberculosis. Nat Rev Immunol. 2011;11(5):343–354. doi: 10.1038/nri2960 [DOI] [PubMed] [Google Scholar]
- 7.Dheda K, Schwander SK, Zhu B, et al. . The immunology of tuberculosis: from bench to bedside. Respirology. 2010;15(3):433–450. doi: 10.1111/j.1440-1843.2010.01739.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marais BJ, Raviglione MC, Donald PR, et al. . Scale-up of services and research priorities for diagnosis, management, and control of tuberculosis: a call to action. Lancet. 2010;375(9732):2179–2191. doi: 10.1016/S0140-6736(10)60554-5 [DOI] [PubMed] [Google Scholar]
- 9.Barry CE, 3rd, Boshoff HI, Dartois V, et al. . The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7(12):845–855. doi: 10.1038/nrmicro2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo Y, Xue Y, Mao L, et al. . Activation phenotype of mycobacterium tuberculosis-specific CD4+ T cells promoting the discrimination between active tuberculosis and latent tuberculosis infection. Front Immunol. 2021;12:721013), doi: 10.3389/fimmu.2021.721013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thuong NTT, Dunstan SJ, Chau TTH, et al. . Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. PLoS Pathog. 2008;4(12):e1000229), doi: 10.1371/journal.ppat.1000229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobsen M, Repsilber D, Gutschmidt A, et al. . Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J Mol Med (Berl). 2007;85(6):613–621. doi: 10.1007/s00109-007-0157-6 [DOI] [PubMed] [Google Scholar]
- 13.Albayrak N, Dirix V, Aerts L, et al. . Differential expression of maturation and activation markers on NK cells in patients with active and latent tuberculosis. J Leukoc Biol. 2022;111(5):1031–1042. [DOI] [PubMed] [Google Scholar]
- 14.Zhao M, Shen S, Xue C.. A Novel m1A-Score Model Correlated With the Immune Microenvironment Predicts Prognosis in Hepatocellular Carcinoma. Front Immunol. 2022;13:805967), doi: 10.3389/fimmu.2022.805967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D, Gable AL, Lyon D, et al. . STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross JM, Xie Y, Wang Y, et al. . Estimating the population at high risk for tuberculosis through household exposure in high-incidence countries: a model-based analysis. EClinicalMed. 2021;42:101206), doi: 10.1016/j.eclinm.2021.101206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soysal A, Bakir M.. T-SPOT.TB assay usage in adults and children. Expert Rev Mol Diagn. 2011;11(6):643–660. doi: 10.1586/erm.11.46 [DOI] [PubMed] [Google Scholar]
- 18.Shao L, Zhang W, Zhang S, et al. . Potent immune responses of Ag-specific Vgamma2Vdelta2+ T cells and CD8+ T cells associated with latent stage of Mycobacterium tuberculosis coinfection in HIV-1-infected humans. AIDS. 2008;22(17):2241–2250. doi: 10.1097/QAD.0b013e3283117f18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Diao N, Lu C, et al. . Evaluation of the diagnostic potential of IP-10 and IL-2 as biomarkers for the diagnosis of active and latent tuberculosis in a BCG-vaccinated population. PLoS One. 2012;7(12):e51338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Shao L, Mo L, Chen J, Wang F, Meng C, Zhong M, Qiu L, Wu M, Weng Xet al. : Evaluation of gamma interferon release assays using Mycobacterium tuberculosis antigens for diagnosis of latent and active tuberculosis in Mycobacterium bovis BCG-vaccinated populations. Clinl Vaccine Immunol CVI 2010, 17(12):1985-1990. doi: 10.1128/CVI.00294-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lalvani A. Counting antigen-specific T cells: a new approach for monitoring response to tuberculosis treatment? Clin Infect Dis. 2004;38(5):757–759. doi: 10.1086/381763 [DOI] [PubMed] [Google Scholar]
- 22.Carrara S, Vincenti D, Petrosillo N, et al. . Use of a T cell-based assay for monitoring efficacy of antituberculosis therapy. Clin Infect Dis. 2004;38(5):754–756. doi: 10.1086/381754 [DOI] [PubMed] [Google Scholar]
- 23.Pathan AA, Wilkinson KA, Klenerman P, et al. . Direct ex vivo analysis of antigen-specific IFN-gamma-secreting CD4 T cells in Mycobacterium tuberculosis-infected individuals: associations with clinical disease state and effect of treatment. J Immunol. 2001;167(9):5217–5225. doi: 10.4049/jimmunol.167.9.5217 [DOI] [PubMed] [Google Scholar]
- 24.Lee SW, Lee SH, Yim JJ.. Serial interferon-gamma release assays after chemoprophylaxis in a tuberculosis outbreak cohort. Infection. 2012;40(4):431–435. [DOI] [PubMed] [Google Scholar]
- 25.Chee CB, KhinMar KW, Gan SH, et al. . Latent tuberculosis infection treatment and T-cell responses to Mycobacterium tuberculosis-specific antigens. Am J Respir Crit Care Med. 2007;175(3):282–287. doi: 10.1164/rccm.200608-1109OC [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson KA, Kon OM, Newton SM, et al. . Effect of treatment of latent tuberculosis infection on the T cell response to Mycobacterium tuberculosis antigens. J Infect Dis. 2006;193(3):354–359. doi: 10.1086/499311 [DOI] [PubMed] [Google Scholar]
- 27.Theron G, Peter J, Lenders L, et al. . Correlation of Mycobacterium tuberculosis specific and non-specific quantitative Th1 T-cell responses with bacillary load in a high burden setting. PLoS One. 2012;7(5):e37436), doi: 10.1371/journal.pone.0037436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong Z, Wen Z, Darnell JE.. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264(5155):95–98. doi: 10.1126/science.8140422 [DOI] [PubMed] [Google Scholar]
- 29.Zeng G, Zhang G, Chen X.. Th1 cytokines, true functional signatures for protective immunity against TB? Cell Mol Immunol. 2018;15(3):206–215. doi: 10.1038/cmi.2017.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyadova IV, Panteleev AV.. Th1 and Th17 cells in tuberculosis: protection, pathology, and biomarkers. Mediators Inflamm. 2015: 854507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torrado E, Cooper AM.. IL-17 and Th17 cells in tuberculosis. Cytokine Growth Factor Rev. 2010;21(6):455–462. doi: 10.1016/j.cytogfr.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen H, Chen ZW.. The crucial roles of Th17-related cytokines/signal pathways in M. tuberculosis infection. Cell Mol Immunol. 2018;15(3):216–225. doi: 10.1038/cmi.2017.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amaral EP, Costa DL, Namasivayam S, et al. . A major role for ferroptosis in Mycobacterium tuberculosis-induced cell death and tissue necrosis. J Exp Med. 2019;216(3):556–570. doi: 10.1084/jem.20181776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nisa A, Kipper FC, Panigrahy D, et al. . Different modalities of host cell death and their impact on Mycobacterium tuberculosis infection. Am J Physiol Cell Physiol. 2022;323(5):C1444–C1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wufuer D, Li Y, Aierken H, et al. . Bioinformatics-led discovery of ferroptosis-associated diagnostic biomarkers and molecule subtypes for tuberculosis patients. Eur J Med Res. 2023;28(1):445), doi: 10.1186/s40001-023-01371-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geng S, Hao P, Wang D, et al. . Zinc oxide nanoparticles have biphasic roles on Mycobacterium-induced inflammation by activating autophagy and ferroptosis mechanisms in infected macrophages. Microb Pathog. 2023;180:106132), doi: 10.1016/j.micpath.2023.106132 [DOI] [PubMed] [Google Scholar]
- 37.Jagatia H, Tsolaki AG.. The role of complement system and the immune response to tuberculosis infection. Medicina (Kaunas). 2021;57(2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castillo EF, Dekonenko A, Arko-Mensah J, et al. . Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci USA. 2012;109(46):E3168–E3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson RO, Manzanillo PS, Cox JS.. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150(4):803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214(2):149–160. doi: 10.1002/path.2287 [DOI] [PubMed] [Google Scholar]
- 41.D'Haens G, Daperno M.. Advances in biologic therapy for ulcerative colitis and Crohn's disease. Curr Gastroenterol Rep. 2006;8(6):506–512. doi: 10.1007/s11894-006-0041-5 [DOI] [PubMed] [Google Scholar]
- 42.Jacobs M, Brown N, Allie N, et al. . Tumor necrosis factor receptor 2 plays a minor role for mycobacterial immunity. Pathobiology. 2000;68(2):68–75. doi: 10.1159/000028116 [DOI] [PubMed] [Google Scholar]
- 43.Ai J-W, Zhang H, Zhou Z, et al. . Gene expression pattern analysis using dual-color RT-MLPA and integrative genome-wide association studies of eQTL for tuberculosis suscepitibility. Respir Res. 2021;22(1):23), doi: 10.1186/s12931-020-01612-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenum S, Dhanasekaran S, Lodha R, et al. . Approaching a diagnostic point-of-care test for pediatric tuberculosis through evaluation of immune biomarkers across the clinical disease spectrum. Sci Rep. 2016;6:18520), doi: 10.1038/srep18520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang N, Luo X, Huang J, et al. . The landscape of different molecular modules in an immune microenvironment during tuberculosis infection. Brief Bioinform. 2021;22(5. [DOI] [PubMed] [Google Scholar]
- 46.Yu EA, John SH, Tablante EC, et al. . Host transcriptional responses following ex vivo re-challenge with Mycobacterium tuberculosis vary with disease status. PLoS One. 2017;12(10):e0185640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.