Abstract
Nigrospora (Xylariales, Apiosporaceae) consists of species of terrestrial plant endophytes and pathogens. Nigrospora has also been reported in marine environments such as mangroves, sea fans, and macroalgae. However, limited research has been conducted on Nigrospora associated with macroalgae. Here, we isolated Nigrospora species from three types of algae (brown, green, and red algae) from Korean islands (Chuja, Jeju, and Ulleung) based on phylogenetic analyses of multigenetic markers: the internal transcribed spacers (ITS), beta-tubulin (BenA), and translation elongation factor 1 (TEF1-α). A total of 17 Nigrospora strains were isolated from macroalgae and identified as nine distinct species. The majority of Nigrospora species (seven) were found on brown algae, followed by red algae (three), and then green algae (two). To our understanding, this study represents the first account of N. cooperae, N. covidalis, N. guilinensis, N. lacticolonia, N. osmanthi, N. pyriformis, and N. rubi occurring in marine environments. Additionally, this study provides the first report of the occurrence of N. cooperae, N. covidalis, N. guilinensis, N. lacticolonia, and N. osmanthi in South Korea. This study will provide valuable insights for future research exploring the functions of fungi in macroalgal communities.
Keywords: Marine fungi, taxonomy, seaweed, algicolous fungi, Ascomycota
1. Introduction
Nigrospora Zimm. is characterized by large dark conidiospores [1]. Since the first report of the type species, N. panici, from leaves in Indonesia [1], Nigrospora species have been reported globally [2–6]. The phylogenetic analysis based on the internal transcribed spacer (ITS), beta-tubulin (BenA), and translation elongation factor 1-a (TEF1-α) affirmed the placement of Nigrospora in Apiosporaceae of Xylariales [7]. Up to date, 44 Nigrospora species have been recorded in the MycoBank database (https://www.mycobank.org/; accessed on 2023.06.14). Among them, 33 species have DNA sequence data in GenBank.
Nigrospora is usually reported to occur in terrestrial environments such as indoors [8,9], lichens [5,10], and plants [4,11], but it has also been reported to occur in marine environments. Specifically, Nigrospora oryzae and Nigrospora sphaerica have predominantly been isolated from marine organisms such as corals [12], mangroves [13–15], macroalgae [16–19], sea fans [19,20], and sponges [21,22]. Furthermore, Nigrospora camelliae-sinensis has been isolated from a mangrove [23], and Nigrospora aurantiaca has been found in sponges [24]. Most of these studies aimed at discovering bioactive compounds derived from Nigrospora rather than exploring its diversity or ecological interactions.
Macroalgae are integral components of marine ecosystems, providing habitats for diverse organisms and contributing to carbon sequestration [25–27]. Microbial associations with macroalgae have been extensively studied [28,29]. Bacterial communities associated with macroalgae have been found to play roles in nutrient supply to macroalgae, defense against unwanted colonization, and even morphogenesis of macroalgae [28]. However, fungal contributions to macroalgae remain poorly understood. Given the significant value of macroalgae and their microorganisms, it is crucial to investigate the relationship between macroalgae and fungi to gain a comprehensive understanding of their ecological significance and potential application.
In this study, we investigated i) Nigrospora species inhabiting macroalgae collected from three Korean islands (Chuja, Jeju, and Ulleung) and ii) whether they have a specific correlation with algal types. We isolated 17 Nigrospora strains from brown, green, and red algae. Nine Nigrospora species were identified at the species level based on both multigenetic markers (ITS, BenA, and TEF1-α) and morphological analysis. Seven of these species are, for the first time, reported to be associated with macroalgae and marine environments. Brown algae exhibited the highest level of Nigrospora species diversity of the three algal types.
2. Materials and methods
2.1. Sampling and fungal isolation
Fourteen macroalgae samples were collected from three islands (Chuja, Jeju, and Ulleung) in South Korea in August of 2018 and 2021 (Table 1). Macroalgae were morphologically identified according to [30] and Marine Bio-Resource Information System (https://www.mbris.kr/pub/info/encyclopedia/algae.do; accessed on 2023.05.17).
Table 1.
List of strains, collection information, and GenBank accession numbers of sequences used in the phylogenetic analysis. Newly reported strains are indicated in bold and holotypes are indicated by “*”.
| Species | Strain number | Habitat/host | Country | GenBank accession numbers |
||
|---|---|---|---|---|---|---|
| ITS | BenA | TEF1-α | ||||
| Apiospora sargassi | KUC21287 | Sargassum fulvellum | Jeju, South Korea | MF615227 | MF615232 | MN868934 |
| Nigrospora aurantiaca | CGMCC 3.18130* = LC 7302 | Nelumbo sp. | China | KX986064 | KY019465 | KY019295 |
| SFC20230324-M01 | Polyopes sp. (Rhodophyta) | Chuja, South Korea | OQ726356 | OQ735177 | OQ735194 | |
| SFC20230324-M02 | Hypnea sp. (Rhodophyta) | Chuja, South Korea | OQ726355 | OQ735178 | OQ735195 | |
| N. bambusae | CGMCC 3.18327* = LC 7114 | Bamboo (leaf) | China | KY385307 | KY385319 | KY385313 |
| N. cooperae | BRIP 72440a* | Heteropogon contortus | Australia | OP035048 | OP039540 | OP039539 |
| SFC20230324-M03 | Ishige sp. (Phaeophyceae) | Chuja, South Korea | OQ726361 | OQ735179 | OQ735196 | |
| N. camelliae-sinensis | CGMCC 3.18125* = LC 3500 | Camellia sinensis | China | KX985986 | KY019460 | KY019293 |
| N. covidalis | CGMCC 3.20538* | Lithocarpus sp. | China | OK335209 | OK431479 | OK431485 |
| SFC20230324-M04 | Ishige sp. (Phaeophyceae) | Chuja, South Korea | OQ726371 | OQ735180 | OQ735197 | |
| N. chinensis | CGMCC 3.18127* = LC 4575 | Machilus breviflora | China | KX986023 | KY019462 | KY019422 |
| N. falsivesicularis | CGMCC 3.19678* | Saccharum officinarum | China | MN215778 | MN329942 | MN264017 |
| N. globosa | CGMCC 3.19633* | Soil of cave | China | MK329121 | MK336134 | – |
| N. globospora | CGMCC 3.20539* | Petasites hybridus | China | OK335211 | OK431481 | OK431487 |
| N. gorlenkoana | CBS 480.73* | Vitis vinifera | Kazakhstan | KX986048 | KY019456 | KY019420 |
| N. guangdongensis | CFCC 53917 | Cunninghamia lanceolata (needle) | China | MT017509 | MT024495 | MT024493 |
| N. guilinensis | CGMCC 3.18124* = LC 3481 | Camellia sinensis | China | KX985983 | KY019459 | KY019292 |
| SFC20230324-M05 | Sargassum sp. (Phaeophyceae) | Ulleung, South Korea | OQ726362 | OQ735181 | OQ735198 | |
| N. hainanensis | CGMCC 3.18129* = LC 7030 | Musa paradisiaca (leaf) | China | KX986091 | KY019464 | KY019415 |
| N. lacticolonia | CGMCC 3.18123* = LC 3324 | Camellia sinensis | China | KX985978 | KY019458 | KY019291 |
| SFC20230324-M06 | Sargassum sp. (Phaeophyceae) | Chuja, South Korea | OQ726357 | OQ735182 | OQ735199 | |
| N. macarangae | MFLUCC 19-0141* | Macaranga tanarius | Taiwan | MW114318 | – | – |
| NCYUCC 19-0177 | Macaranga tanarius | Taiwan | MW114319 | – | – | |
| N. magnoliae | MFLUCC 19-0112* | Magnolia liliifera | China | MW285092 | MW438334 | – |
| N. musae | CBS 319.34* | Musa paradisiaca (fruit) | Australia | KX986076 | KY019455 | KY019419 |
| N. oryzae | LC 7306 | Nelumbo sp. (leaf) | China | KX986068 | KY019612 | KY019408 |
| LC 2689 | Rhododendron sp. | China | KX985942 | KY019469 | KY019423 | |
| LC 4265 | Rhododendron sp. | China | KX985994 | KY019518 | KY019335 | |
| LC 4338 | Camellia sp. | China | KX986008 | KY019532 | KY019349 | |
| SFC20230324-M07 | Ulva sp. (Chlorophyta) | Jeju, South Korea | OQ726369 | OQ735183 | OQ735200 | |
| SFC20230324-M08 | Myagropsis sp. (Phaeophyceae) | Ulleung, South Korea | OQ726367 | OQ735184 | OQ735201 | |
| SFC20230324-M09 | Chondria sp. (Rhodophyta) | Ulleung, South Korea | OQ726368 | OQ735185 | OQ735202 | |
| SFC20230324-M10 | Grateloupia sp. (Rhodophyta) | Chuja, South Korea | OQ726366 | OQ735186 | OQ735203 | |
| N. osmanthi | CGMCC 3.18126* = LC 4350 | Osmanthus sp. | China | KX986010 | KY019461 | KY019421 |
| SFC20230324-M11 | Codium sp. (Chlorophyta) | Jeju, South Korea | OQ726360 | OQ735187 | OQ735204 | |
| SFC20230324-M12 | Ulva sp. (Chlorophyta) | Jeju, South Korea | OQ726358 | OQ735188 | OQ735205 | |
| SFC20230324-M13 | Codium sp. (Chlorophyta) | Jeju, South Korea | OQ726359 | OQ735189 | OQ735206 | |
| N. philosophiae-doctoris | CGMCC 3.20540* | Disporum sessile | China | OK335213 | OK431483 | OK431489 |
| N. pyriformis | CGMCC 3.18122* = LC 2045 | Citrus sinensis | China | KX985940 | KY019457 | KY019290 |
| SFC20230324-M14 | Sargassum sp. (Phaeophyceae) | Chuja, South Korea | OQ726363 | OQ735190 | OQ735207 | |
| SFC20230324-M15 | Polyopes sp. (Rhodophyta) | Chuja, South Korea | OQ726364 | OQ735191 | OQ735208 | |
| SFC20230324-M16 | Laurencia sp. (Rhodophyta) | Chuja, South Korea | OQ726365 | OQ735192 | OQ735209 | |
| N. rubi | CGMCC 3.18326* = LC 2698 | Rubus sp. | China | KX985948 | KY019475 | KY019302 |
| SFC20230324-M17 | Sargassum sp. (Phaeophyceae) | Chuja, South Korea | OQ726370 | OQ735193 | OQ735210 | |
| N. saccharicola | CGMCC 3.19362* | Saccharum officinarum | China | MN215788 | MN329951 | MN264027 |
| N. sacchari-officinarum | CGMCC 3.19335* | Saccharum officinarum | China | MN215791 | MN329954 | MN264030 |
| N. singularis | CGMCC 3.19334* | Saccharum officinarum | China | MN215793 | MN329956 | MN264032 |
| N. sphaerica | LC 2840 | Harpullia longipetala | China | KX985965 | KY019492 | KY019318 |
| LC 2958 | Cleyera japonica | China | KX985966 | KY019493 | KY019319 | |
| LC 4372 | Rhododendron arboreum | China | KX986012 | KY019535 | KY019351 | |
| LC 6969 | Musa paradisiaca (leaf) | China | KX986077 | KY019584 | KY019386 | |
| N. vesicularifera | CGMCC 3.19333* | Saccharum officinarum | China | MN215812 | MN329975 | MN264051 |
| N. vesicularis | CGMCC 3.18128* = LC 7010 | Musa paradisiaca (leaf) | China | KX986088 | KY019463 | KY019294 |
| N. zimmermanii | CBS 290.62* | Saccharum officinarum (leaf) | Ecuador | KY385309 | KY385317 | KY385311 |
Macroalgae samples were cut into 0.5 × 0.5 cm2 pieces and placed on dichloran rose bengal chloramphenicol (DRBC) agar (Difco, Sparks, MD, USA) media supplemented with sterilized seawater (SSW). Fungal colonies grown from the samples were isolated and transferred to potato dextrose agar (PDA; Difco, Sparks, MD, USA) media supplemented with SSW. The living cultures of each isolate were stocked in 20% (v/v) glycerol at −80 °C and deposited into the Seoul National University Fungus Collection (SFC).
2.2. Molecular analyses (DNA extraction, PCR amplification, sequencing, and phylogenetic analysis)
The mycelium of each fungal isolate grown on PDA was ground by a Bead Ruptor Elite Homogenizer (OMNI International, Kennesaw, GA, USA). DNA extraction was conducted using an AccuPrep® Genomic DNA Extraction Kit (Bioneer, Daejeon, South Korea) following the manufacturer’s protocol with a small modification where cetyltrimethylammonium bromide (CTAB) extraction solution (Biosesang, Incheon, South Korea) was used instead of the TL buffer included in the kit.
The ITS region was amplified by PCR using a C1000 thermal cycler (Bio-Rad, Richmond, CA, USA) with the primer sets ITS1F/ITS4 [31,32]. BenA and TEF1-α were subsequently amplified with Bt2a/Bt2b [33] and EF1-728F/EF2 [34,35] primers, respectively. The PCR conditions were as follows: initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 95 °C for 40 s, annealing at 55 °C for 40 s, and extension at 72 °C for 60 s, followed by a final extension at 72 °C for 5 min. Purification was done using an Expin™ PCR SV kit (GeneAll Biotechnology, Seoul, South Korea), following the manufacturer’s protocol. Sanger sequencing was performed in both forward and reverse directions using the PCR primers in an ABI prism 3730xl Genetic Analyzer (Life Technologies, Gaithersburg, MD, USA) at Macrogen (Seoul, South Korea). Obtained sequences were merged using the De novo assemble function in the Geneious Prime software ver. 2023. 1. 1. (Biomatters Ltd., San Diego, CA, USA) and were then proofread and edited manually. The proofread sequences were deposited in GenBank (Table 1).
The generated sequences, reference GenBank Nigrospora sequences, and an outgroup sequence of Apiospora sargassi (KUC 21287) were aligned by each genetic marker (ITS, BenA, and TEF1-α) using MAFFT v7.490 [36] in the Geneious Prime software ver. 2023.1.1. (Biomatters Ltd., San Diego, CA, USA). The alignments were then concatenated. The best model test was investigated in MEGA 7.0.26 [37] to conduct the maximum likelihood analysis. The phylogenetic tree was inferred through RAxML analysis [38] with 1,000 replications using the GTR GAMMA model in the Geneious Prime software.
2.3. Morphological observation
For an effective observation and measurement of microscopic features, Nigrospora strains were initially subcultured on PDA and subsequently transferred to both PDA and synthetic nutrient-poor agar media (SNA; KH2PO4 1 g, KNO3 1 g, MgSO4·7H2O 5 g, KCl 0.5 g, Glucose 0.2 g, Saccharose 0.2 g, and Bacto agar 20 g per 1 L). The colonies on PDA were incubated for 7 d at 25 °C in the dark to observe the culture morphology. The color of the colonies was determined using the Methuen Handbook of Color [39]. Representative strains of each species were cultivated on SNA for conidial structure observation. All observations were done using a Nikon 80i light microscope (Tokyo, Japan). At least 30 measurements were obtained per strain to calculate the mean size of the microscopic structures. The colonies and the conidial structures were measured using ImageJ software [40].
3. Results
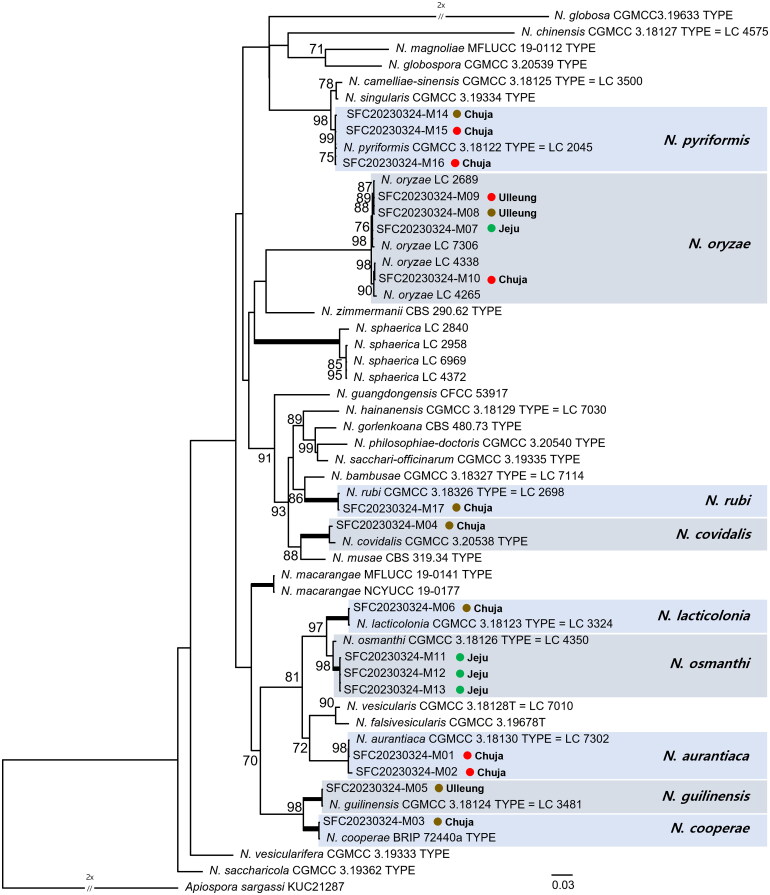
Seventeen Nigrospora strains were isolated from macroalgae, and ITS and two protein-coding genes (BenA and TEF1-α) were used to infer a maximum likelihood tree to identify the strains at the species level. The total number of molecular characters was 1,853 (586 in ITS, 402 in BenA, and 865 in TEF1-α). Nine Nigrospora species (N. aurantiaca, N. cooperae, N. covidalis, N. guilinensis, N. lacticolonia, N. oryzae, N. osmanthi, N. pyriformis, and N. rubi) were identified through the phylogenetic analysis (Figure 1). Each strain matched its corresponding species with at least 98% bootstrap support.
Figure 1.
The maximum likelihood tree of Nigrospora species with outgroup Apiospora sargassi (KUC21287). ITS, BenA, and TEF1-α genetic markers were used in phylogenetic analyses. The newly collected strains are enclosed in colored boxes. Bootstrap values of more than 70% are shown at the nodes. Branches that lead to nodes of bootstrap values of 100 are indicated by a bold line. Colored circles indicate the color of algae (brown, green, and red) where each strain was isolated, followed by the island on which the strain was collected.
Nigrospora species were categorized based on the algal types to which they were associated (Figure 1). The macroalgae were morphologically identified as brown (Sargassum spp., Ishige okamurae, and Myagropsis myagroides), green (Ulva sp. and Codium fragile), and red algae (Hypnea sp., Polyopes sp., Laurencia sp., Grateloupia sp., and Chondia sp.) (Table 1). The largest number of Nigrospora species (seven spp.)—N. cooperae, N. covidalis, N. guilinensis, N. lacticolonia, N. oryzae, N. pyriformis, and N. rubi—were isolated from brown algae. Three Nigrospora species (N. aurantiaca, N. oryzae, and N. pyriformis) were isolated from red algae. Nigrospora oryzae and N. osmanthi were isolated from green algae. Nigrospora oryzae appeared on all types of algae, and N. pyriformis was detected in both brown and red algae (Figure 1).
4. Taxonomy
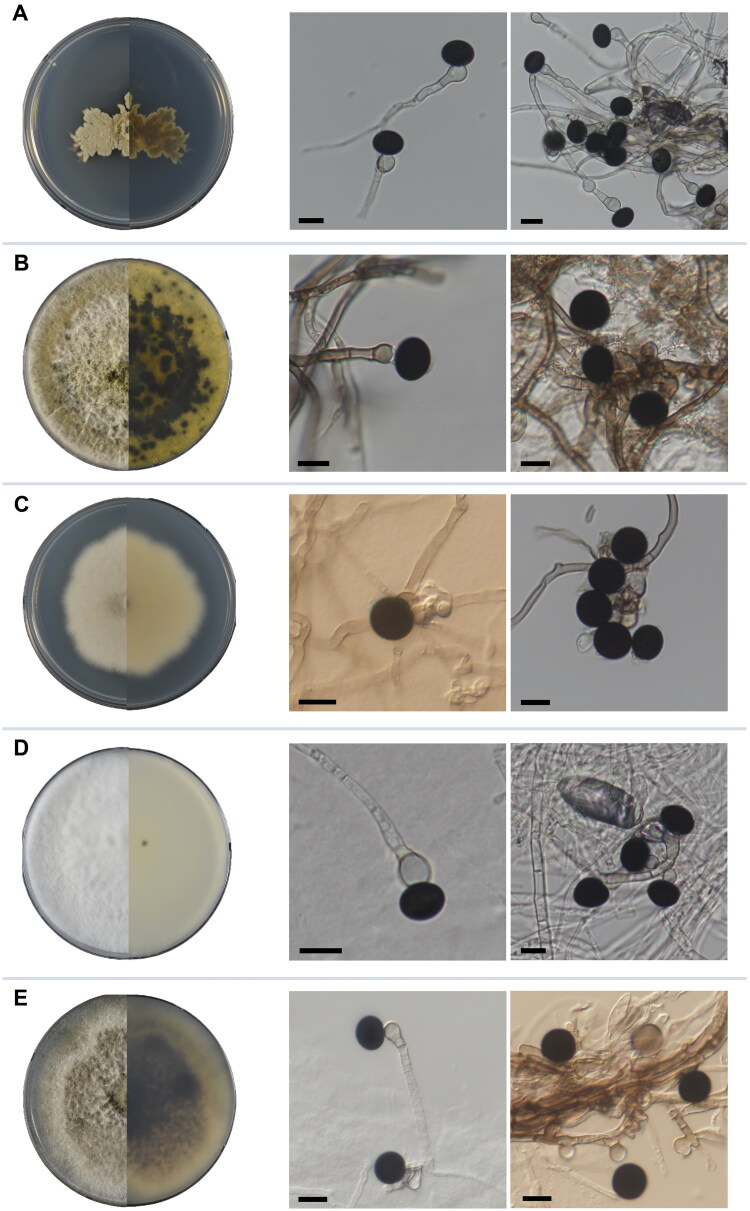
Nigrospora cooperae Y.P. Tan, Bishop-Hurley, Bransgr. & R.G. Shivas (2022) (Figure 2A).
Figure 2.
Cultures of five Nigrospora species from this study. On the left, surface (left) and reverse (right) sides of strains on PDA are shown in halves. On the right, conidial structures on SNA media are shown. (A) N. cooperae, (B) N. covidalis, (C) N. guilinensis, (D) N. lacticolonia, and (E) N. osmanthi. Scale bar: 10 μm.
Sexual morph: Undetermined. Asexual morph on SNA: Hyphae branched, guttulate, septate, pale brown, 1.6–5.4 μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells monoblastic, discrete, pale brown, doliiform to ampulliform to subglobose, 5.6–13.9 × 4–7 μm (av. = 8.6 ± 2.07 × 5.47 ± 0.83). Conidia solitary, spherical or ellipsoidal, aseptate, black, shiny, smooth-walled, spherical 10.3–14 μm (av. = 12.2 ± 0.84), ellipsoidal 11.4–14.6 × 8.2–11.4 μm (av. = 12.67 ± 0.84 × 9.63 ± 0.79).
Culture characters on PDA: Colonies sparse, velvety, fimbriate, irregular at the margin, surface gray-green (1C4) to bile yellow (30C5), reverse concolorous, not producing pigments in PDA, with prominent exudates, reaching 18–45 mm diameter in 7 d at 25 °C.
Materials examined: South Korea. South Sea, Chuja island, 33°57′11″N, 126°18′07″E, from Ishige sp. (Phaeophyceae), 31 August 2021, M. S. Park & Y. W. Lim (SFC20230324-M03, stored in a metabolically inactive state).
Notes: SFC20230324-M03 produces prominent exudates, and the conidiogenous cells of this isolate displayed a range of length variations. However, the original description of N. cooperae does not produce any prominent exudates, and the length variation (7–10 μm) of conidiogenous cells in the holotype is lower than our isolate [41].
Nigrospora covidalis M. Raza, Qian Chen & L. Cai. (2017) (Figure 2B).
Sexual morph: Undetermined. Asexual morph on SNA: Hyphae branched, septate, guttulate, hyaline to pale brown, 1.8–4.6 μm diam. Conidiophores monoblastic, flexuous or straight, pale brown, and some conidiophores reduced to conidiogenous cells. Conidiogenous cells monoblastic, discrete, pale brown, doliiform to ampulliform, 5.4–12.7 × 3.7–8.8 μm (av. = 8.24 ± 1.81 × 6.32 ± 1.32). Hyaline vesicles delimited the conidia from their conidiogenous cells. Conidia sparse, solitary, spherical or ellipsoidal, aseptate, mostly black, discrete on aerial hyphae, spherical 10.9–15.2 μm diam. (av. = 12.9 ± 1.21), ellipsoidal 11.8–15.8 × 8.8–12.3 μm (av. = 13.86 ± 1.12 × 11.03 ± 0.81).
Culture characters on PDA: Colonies floccose, surface white (1A1) to light grey (1D1), sometimes deep green (1C8), reverse pale grey (1B1) to grayish yellow (1B4) with black patches, mostly producing yellow pigments in PDA, reaching 90 mm diameter in 4–5 d at 25 °C.
Materials examined: South Korea. South Sea, Chuja islands, 33°57′11″N, 126°18′07″E, from Ishige sp. (Phaeophyceae), 31 August 2021, M. S. Park & Y. W. Lim (SFC20230324-M04, stored in a metabolically inactive state).
Notes: Nigrospora covidalis can be morphologically distinguished from N. musae, which is phylogenetically a sister species, by the smaller size of its conidia [7,41]. Even though the absence of vesicles is a taxonomic key to delimiting N. covidalis from N. musae [7], hyaline vesicles are observed in this isolate. Furthermore, SFC20230324-M04 produces yellow pigment and grew faster on PDA media compared to what is reported in the original description [42].
Nigrospora guilinensis Mei Wang & L. Cai (2017) (Figure 2C).
Sexual morph: Undetermined. Asexual morph on SNA: Hyphae branched, smooth, hyaline to pale brown, septate, 1.9–6.1 μm diam. Conidiophores usually reduced to conidiogenous cells, aggregated in clusters on hyphae. Conidiogenous cells monoblastic, determinate, hyaline to pale brown, smooth-walled, doliiform to ampulliform, in clusters on aerial mycelia, 5.0–13.6 × 3.7–12.9 μm (av. = 8.02 ± 2.12 × 6.53 ± 1.93). Conidia solitary, spherical or ellipsoidal, aseptate, black, shiny, smooth-walled, spherical, 10.8–13.4 μm diam. (av. = 12 ± 0.7), ellipsoidal, 11.4–14.2 × 8.7–11.2 μm (av. = 12.77 ± 0.66 × 9.97 ± 0.63).
Culture characters on PDA: Colonies wooly, cottony, margin irregular, undulate, surface and reverse white (1A1) with a few black patches, sometimes producing red pigment, reaching 54–68 mm diameter after 7 d at 25 °C.
Materials examined: South Korea. East Sea, Ulleung island, 37°30′52″N, 130°47′41″E, from Sargassum sp. (Phaeophyceae), 29 August 2018, M. S. Park & Y. W. Lim (SFC20230324-M05, stored in a metabolically inactive state).
Notes: Nigrospora guilinensis can be distinguished from closely related species by morphological characteristics such as the ability to produce diffusible pigment on PDA and the arrangement of conidiogenous cells [7]. Nevertheless, pigment production is not consistently observed in SFC20230324-M05, and the isolate forms wider conidiogenous cells (6–11 × 4–7.5 μm) compared to that reported in the original description [7].
Nigrospora lacticolonia Mei Wang & L. Cai. (2017) (Figure 2D).
Sexual morph: Undetermined. Asexual morph on SNA: Hyphae branched, smooth, hyaline, septate, 1.5–4.6 μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells sometimes aggregated in clusters on hyphae, pale brown, smooth-walled, mostly spherical, sometimes doliiform, 5–13.2 × 4–8 μm (av. = 8.63 ± 1.89 × 6.09 ± 1.02). Conidia sparse, solitary, spherical or ellipsoidal, aseptate, black, shiny, smooth-walled, spherical 9.7–15.1 μm diam. (av. = 12.28 ± 1.45), ellipsoidal 10.2–15.2 × 7.9–12.1 μm (av. = 12.52 ± 1.13 × 9.73 ± 1.03).
Culture characters on PDA: Colonies floccose, entire edge, surface, and reverse white (1A1), without any patches, reaching 90 mm diameter in 3–4 d at 25 °C.
Materials examined: South Korea. South Sea, Chuja island, 33°57′11″N, 126°18′07″E, from Sargassum sp. (Phaeophyceae), 31 August 2021, M. S. Park & Y. W. Lim (SFC20230324-M06, stored in a metabolically inactive state).
Notes: Nigrospora lacticolonia derived its name from the creamy white colonies on PDA [7]. Similarly, the isolate SFC20230324-M06, from Sargassum sp., colonized PDA in white. SFC20230324-M06 sparsely produces conidia and does not show a prominent tendency to aggregate conidiogenous cells in clusters. Moreover, narrower ellipsoidal conidia are observed than those in the original description (13.5–17.5 × 10.5–13.5 μm) [7].
Nigrospora osmanthi Mei Wang & L. Cai. (2017) (Figure 2E).
Sexual morph: Undetermined. Asexual morph on SNA: Hyphae branched, guttulate, septate, hyaline to pale brown, 1.8–6.4 μm diam. Conidiophores mostly reduced to conidiogenous cells. Conidiogenous cells monoblastic, discrete, determinate, brown, subspherical, ampulliform to cylindrical, 4.8–15.3 × 4.1–11.9 μm (av. = 8.58 ± 3.28 × 6.34 ± 1.76). Conidia solitary, globose or subglobose, aseptate, initially pale brown, becoming black with age, shiny, smooth-walled, sometimes formed directly from the mycelia, 9.9–16.7 μm diam. (av. = 13.07 ± 1.29).
Culture characters on PDA: Colonies flat, floccose, undulate, surface initially white (1A1), becoming grayish green (1D3), abundant aerial mycelium, reverse concolorous with dark patches, reaching 90 mm diameter in 5 d at 25 °C.
Materials examined: South Korea. South Sea, Jeju island, 33°23′53″N, 126°14′24″E, from Codium sp. (Chlorophyta), 15 August 2021, M. S. Park & Y. W. Lim (SFC20230324-M11, stored in a metabolically inactive state); ibid., from Ulva sp. (Chlorophyta), 15 August 2021, M. S. Park & Y. W. Lim (SFC20230324-M12, stored in a metabolically inactive state) ibid., from Codium sp. (Chlorophyta), 15 August 2021, M. S. Park & Y. W. Lim (SFC20230324-M13, stored in a metabolically inactive state).
Notes: Three Nigrospora osmanthi strains (SFC20230324-M11, SFC20230324-M12, and SFC20230324-M13) have bigger conidiogenous cells (5.5–12 μm) and smaller conidia (13.5–16.5 μm) than those of the strains in the original description [7]. Regarding growth on PDA, our isolates achieve a 90 mm diameter on plates within a span of 5 d at 25 °C, whereas the strains in the original description take 10 days to reach the same diameter [7].
5. Discussion
Nigrospora can be identified at the genus level by large dark conidiospores. However, morphological variation sometimes appears in strains despite them belonging to the same species, and different species can share similar morphological characteristics. Therefore, multigenetic marker analysis is imperative for the detection and identification of Nigrospora, given that the phylogeny of Nigrospora has been well-established using multigenetic markers, including ITS, BenA, and TEF1-α [7]. A total of nine Nigrospora species were identified in this study using multigenetic markers analysis and morphological data. This study provided the first report of five of these species in South Korea (N. cooperae, N. covidalis, N. guilinensis, N. lacticolonia, and N. osmanthi), and their morphological characteristics are provided in the taxonomy section.
Two Nigrospora species, N. oryzae and N. sphaerica, are frequently encountered. Nigrospora oryzae has been consistently reported from macroalgae [16,43] and was also commonly detected in this study on three islands and on three types of algae. However, Nigrospora sphaerica was not detected on marine macroalgae in this study, although it has been commonly reported in various marine environments [44,45]. Taritla et al. [18] isolated a strain L18/35 from the macroalga Sargassum muticum and identified it as N. sphaerica, but we confirmed that the ITS sequence (MF457920) of the strain L18/35 did not match that of N. sphaerica provided on NCBI. This creates doubt regarding the viability of its association with macroalga. Nigrospora aurantiaca has been reported on sponges [24] but was only isolated from red algae in this study. Until this study, there were no records of N. cooperae, N. covidalis, N. guilinensis, N. lacticolonia, N. osmanthi, N. pyriformis, and N. rubi in marine environments. These findings indicate that some Nigrospora species can adapt and inhabit both terrestrial and marine habitats, but further research is required to elucidate the underlying mechanisms of such environmental adaptation.
Although secondary metabolites produced by Nigrospora isolated from macroalgae have received limited attention in previous studies, numerous valuable secondary metabolites have been isolated from Nigrospora species [19]. One such example is nigrosporone B, which has shown anti-cancer, anti-bacterial, cytotoxic, and anti-malarial activities [46]. Nigrospora aurantiaca produces a red pigment known as bostrycin that can be used as a natural dye [47], and the same color pigment was observed in our N. aurantiaca strains SFC20230324-M01 and SFC20230324-M02 as well. Notably, Nigrospora species isolated from marine environments also produce a diverse range of secondary metabolites, many of which exhibit beneficial properties such as antimicrobial, antitumor, and cytotoxic activities [48–50].
The diversity of Nigrospora species was found to be highest in brown algae, followed by red algae and green algae. With the exception of N. oryzae and N. pyriformis, all species were exclusively isolated from a specific algal type (Figure 1). It is too early to conclude that Nigrospora species have a symbiotic relationship with algae due to the limited number of studied samples. However, considering that Arthrinium spp., a sister genus of Nigrospora, improves the survival of brown algae by providing antioxidants in response to decreased photosynthetic activity [51], it is possible that Nigrospora may also interact with algae. Moreover, pyrenocines isolated from Phaeosphaeria sp. can protect macroalgae against protistan pathogens, such as Olpidiopsis pyropia (Oomycota), through the collapse of the zoosporangia of O. pyropia [52]. Therefore, further investigations are required to elucidate the ecological role of Nigrospora associated with macroalgae and whether it acts as an endophyte or a pathogen. This study provides insights and discusses the possibility of biologically meaningful interactions between Nigrospora and macroalgae. Further studies aimed at comprehending the role of algicolous Nigrospora will greatly contribute to the effective management of macroalgal aquaculture and pathogenicity.
Acknowledgments
We thank Editage (www.editage.co.kr) for English language editing.
Funding Statement
This work was supported by the management of Marine Fishery Bio-resources Center (2023), funded by the National Marine Biodiversity Institute of Korea (MABIK).
Disclosure statement
The authors have declared that no competing interests exist.
Data availability
All the data that support the findings of this study are available within the article.
References
- 1.Zimmerman A. Ueber einige an tropischen kulturpflanzen beobachtete pilze III. Zentralblatt für bakteriologie. Parasitenkunde. 1902;8:216–221. [Google Scholar]
- 2.Fan Y-M, Huang W-M, Li W, et al. Onychomycosis caused by Nigrospora sphaerica in an immunocompetent man. Arch Dermatol. 2009;145(5):611–612. doi: 10.1001/archdermatol.2009.80. [DOI] [PubMed] [Google Scholar]
- 3.Dutta J, Gupta S, Thakur D, et al. First report of nigrospora leaf blight on tea caused by Nigrospora sphaerica in India. Plant Disease. 2015;99(3):417–417. doi: 10.1094/PDIS-05-14-0545-PDN. [DOI] [PubMed] [Google Scholar]
- 4.Uzor PF, Ebrahim W, Osadebe PO, et al. Metabolites from Combretum dolichopetalum and its associated endophytic fungus Nigrospora oryzae—evidence for a metabolic partnership. Fitoterapia. 2015;105:147–150. doi: 10.1016/j.fitote.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 5.Oh SY, Yang JH, Woo JJ, et al. Diversity and distribution patterns of endolichenic fungi in jeju island, South Korea. Sustainability. 2020;12(9):3769. doi: 10.3390/su12093769. [DOI] [Google Scholar]
- 6.de Queiroz Brito AC, de Mello JF, de Almeida Souza AE, et al. Richness of Nigrospora spp. (Apiosporaceae) in manihot esculenta in Brazil and the description of three new species. Mycol Progress. 2023;22(6):37. doi: 10.1007/s11557-023-01887-4. [DOI] [Google Scholar]
- 7.Wang M, Liu F, Crous PW, et al. Phylogenetic reassessment of Nigrospora: ubiquitous endophytes, plant and human pathogens. Persoonia. 2017;39(1):118–142. doi: 10.3767/persoonia.2017.39.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jothish PS, Nayar TS.. Airborne fungal spores in a sawmill environment in Palakkad district, Kerala, India. Aerobiologia. 2004;20(1):75–81. doi: 10.1023/B:AERO.0000022981.70984.b7. [DOI] [Google Scholar]
- 9.Nayar TS, Jothish PS.. An assessment of the air quality in indoor and outdoor air with reference to fungal spores and pollen grains in four working environments in Kerala, India. Aerobiologia. 2013;29(1):131–152. doi: 10.1007/s10453-012-9269-8. [DOI] [Google Scholar]
- 10.Tripathi M, Joshi Y. Endolichenic fungi: A case study from Uttarakhand. Endolichenic fungi: present and future trends. Singapore: Springer; 2019. pp. 119–145. doi: 10.1007/978-981-13-7268-1_6. [DOI]
- 11.Lee DJ, Lee JS, Lee HB, et al. Four endophytic ascomycetes new to Korea: Cladosporium anthropophilum, C. pseudocladosporioides, Daldinia eschscholtzii, and Nigrospora chinensis. The Korean J Mycol. 2019;47(3):187–197. doi: 10.4489/KJM.20190023. [DOI] [Google Scholar]
- 12.Sun XP, Xu Y, Cao F, et al. Isoechinulin-type alkaloids from a soft coral-derived fungus Nigrospora oryzae. Chem Nat Compd. 2014;50:1153–1155. doi: 10.1007/s10600-014-1189-0. [DOI] [Google Scholar]
- 13.Zhang QH, Tian L, Sun ZL, et al. Two new secondary metabolites from the marine-derived fungus Nigrospora sphaerica. J Asian Nat Prod Res. 2015;17(5):497–503. doi: 10.1080/10286020.2015.1009899. [DOI] [PubMed] [Google Scholar]
- 14.Ukwatta KM, Lawrence JL, Wijayarathna CD.. The study of antimicrobial, anti-cancer, anti-inflammatory and α-glucosidase inhibitory activities of nigronapthaphenyl, isolated from an extract of Nigrospora sphaerica. Mycology. 2019;10(4):222–228. doi: 10.1080/21501203.2019.1620892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ola ARB, Lapailaka T, Wogo HE, et al. Bioactive secondary metabolites from the mangrove endophytic fungi Nigrospora oryzae. Indones J Chem. 2021;21(4):1016–1022. doi: 10.22146/ijc.63129. [DOI] [Google Scholar]
- 16.de Felício R, Pavão GB, de Oliveira ALL, et al. Antibacterial, antifungal and cytotoxic activities exhibited by endophytic fungi from the Brazilian marine red alga Bostrychia tenella (Ceramiales). Revista Brasileira de Farmacognosia. 2015;25(6):641–650. doi: 10.1016/j.bjp.2015.08.003. [DOI] [Google Scholar]
- 17.Rajulu MG, Rajamani T, Murali TS, et al. The fungal endobiome of seaweeds of the Andaman islands, India. Curr Sci. 2022;123(12):1508–1514. doi: 10.18520/cs/v123/i12/1508-1514. [DOI] [Google Scholar]
- 18.Taritla S, Kumari M, Kamat S, et al. Optimization of physicochemical parameters for production of cytotoxic secondary metabolites and apoptosis induction activities in the culture extract of a marine algal-derived endophytic fungus Aspergillus sp. Front Pharmacol. 2021;12:542891. doi: 10.3389/fphar.2021.542891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu T, Song Z, Hou Y, et al. Secondary metabolites of the genus Nigrospora from terrestrial and marine habitats: chemical diversity and biological activity. Fitoterapia. 2022;161:105254. doi: 10.1016/j.fitote.2022.105254. [DOI] [PubMed] [Google Scholar]
- 20.Dong JJ, Bao J, Zhang XY, et al. Alkaloids and citrinins from marine-derived fungus Nigrospora oryzae SCSGAF 0111. Tetrahedron Lett. 2014;55(16):2749–2753. doi: 10.1016/j.tetlet.2014.03.060. [DOI] [Google Scholar]
- 21.Ding B, Yin Y, Zhang F, et al. Recovery and phylogenetic diversity of culturable fungi associated with marine sponges Clathrina luteoculcitella and Holoxea sp. in the South China Sea. Mar Biotechnol (NY). 2011;13(4):713–721. doi: 10.1007/s10126-010-9333-8. [DOI] [PubMed] [Google Scholar]
- 22.Yao J, Shi Y, Liu Y, et al. Highly oxidized ergosterol derivatives from the fungus Nigrospora oryzae. Chem Nat Compd. 2019;55(2):390–392. doi: 10.1007/s10600-019-02700-z. [DOI] [Google Scholar]
- 23.Huang DY, Nong XH, Zhang YQ, et al. Two new 2, 5-diketopiperazine derivatives from mangrove-derived endophytic fungus Nigrospora camelliae-sinensis S30. Nat Prod Res. 2022;36(14):3651–3656. doi: 10.1080/14786419.2021.1878168. [DOI] [PubMed] [Google Scholar]
- 24.Said Hassane C, Fouillaud M, Le Goff G, et al. Microorganisms associated with the marine sponge Scopalina hapalia: a reservoir of bioactive molecules to slow down the aging process. Microorganisms. 2020;8(9):1262. doi: 10.3390/microorganisms8091262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hay ME. Marine chemical ecology: chemical signals and cues structure marine populations, communities, and ecosystems. Ann Rev Mar Sci. 2009;1(1):193–212. doi: 10.1146/annurev.marine.010908.163708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause-Jensen D, Lavery P, Serrano O, et al. Sequestration of macroalgal carbon: the elephant in the blue carbon room. Biol Lett. 2018;14(6):20180236. doi: 10.1098/rsbl.2018.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Froehlich HE, Afflerbach JC, Frazier M, et al. Blue growth potential to mitigate climate change through seaweed offsetting. Curr Biol. 2019;29(18):3087–3093.e3. doi: 10.1016/j.cub.2019.07.041. [DOI] [PubMed] [Google Scholar]
- 28.Egan S, Harder T, Burke C, et al. The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiol Rev. 2013;37(3):462–476. doi: 10.1111/1574-6976.12011. [DOI] [PubMed] [Google Scholar]
- 29.Ren CG, Liu ZY, Wang XL, et al. The seaweed holobiont: from microecology to biotechnological applications. Microb Biotechnol. 2022;15(3):738–754. doi: 10.1111/1751-7915.14014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim CM, et al. The compilation inventory of national biological resources. National Institute of Biological Resources. 2011. https://www.nibr.go.kr/aiibook/ecatalog5.jsp?Dir=108&catimage=&callmode=admin [Google Scholar]
- 31.Gardes M, Bruns TD.. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 32.White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- 33.Glass NL, Donaldson GC.. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carbone I, Kohn LM.. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91(3):553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 35.O'Donnell K, Kistler HC, Cigelnik E, et al. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci USA. 1998;95(5):2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K, Rozewicki J, Yamada KD.. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar S, Stecher G, Tamura K.. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kornerup A, Wanscher J. Methuen handbook of colour. London: Eyre Methuen; 1978. ISBN-13: 978-0413334008 [Google Scholar]
- 40.Schneider CA, Rasband WS, Eliceiri KW.. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan YP, Bishop-Hurley SL, Shivas RG, et al. Fungal planet description sheets: 1436–1477. Persoonia Mol Phylogeny Evol Fungi. 2022;49:261–350. doi: 10.3767/persoonia.2022.49.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Q, Bakhshi M, Balci Y, et al. Genera of phytopathogenic fungi: GOPHY 4. Stud Mycol. 2022;101(1):417–564. doi: 10.3114/sim.2022.101.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calado MDL, Silva J, Alves C, et al. Marine endophytic fungi associated with Halopteris scoparia (Linnaeus) Sauvageau as producers of bioactive secondary metabolites with potential dermocosmetic application. PLoS One. 2021;16(5):e0250954. doi: 10.1371/journal.pone.0250954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang QH, Tian L, Zhou LD, et al. Two new compounds from the marine Nigrospora sphaerica. J Asian Nat Prod Res. 2009;11(11):962–966. doi: 10.1080/10286020903339614. [DOI] [PubMed] [Google Scholar]
- 45.Passarini MR, Santos C, Lima N, et al. Filamentous fungi from the atlantic marine sponge Dragmacidon reticulatum. Arch Microbiol. 2013;195(2):99–111. doi: 10.1007/s00203-012-0854-6. [DOI] [PubMed] [Google Scholar]
- 46.Kornsakulkarn J, Choowong W, Rachtawee P, et al. Bioactive hydroanthraquinones from endophytic fungus Nigrospora sp. BCC 47789. Phytochem Lett. 2018;24:46–50. doi: 10.1016/j.phytol.2018.01.015. [DOI] [Google Scholar]
- 47.Suwannarach N, Kumla J, Nishizaki Y, et al. Optimization and characterization of red pigment production from an endophytic fungus, Nigrospora aurantiaca CMU-ZY2045, and its potential source of natural dye for use in textile dyeing. Appl Microbiol Biotechnol. 2019;103(17):6973–6987. doi: 10.1007/s00253-019-09926-5. [DOI] [PubMed] [Google Scholar]
- 48.Trisuwan K, Rukachaisirikul V, Sukpondma Y, et al. Pyrone derivatives from the marine-derived fungus Nigrospora sp. PSU-F18. Phytochemistry. 2009;70(4):554–557. doi: 10.1016/j.phytochem.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Shang Z, Li XM, Li CS, et al. Diverse secondary metabolites produced by marine-derived fungus Nigrospora sp. MA75 on various culture media. Chem Biodivers. 2012;9(7):1338–1348. doi: 10.1002/cbdv.201100216. [DOI] [PubMed] [Google Scholar]
- 50.Ding L, Yuan W, Peng Q, et al. Secondary metabolites isolated from the sponge-associated fungus Nigrospora oryzae. Chem Nat Compd. 2016;52(5):969–970. doi: 10.1007/s10600-016-1837-7. [DOI] [Google Scholar]
- 51.Heo YM, Oh SY, Kim K, et al. Comparative genomics and transcriptomics depict marine algicolous Arthrinium species as endosymbionts that help regulate oxidative stress in brown algae. Front Mar Sci. 2021;8:753222. doi: 10.3389/fmars.2021.753222. [DOI] [Google Scholar]
- 52.Vallet M, Strittmatter M, Murúa P, et al. Chemically-mediated interactions between macroalgae, their fungal endophytes, and protistan pathogens. Front Microbiol. 2018;9:3161. doi: 10.3389/fmicb.2018.03161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data that support the findings of this study are available within the article.