Abstract
In many epidemiological and environmental health studies, developing an accurate exposure assessment of multiple exposures on a health outcome is often of interest. However, the problem is challenging in the presence of multicollinearity, which can lead to biased estimates of regression coefficients and inflated variance estimators. Selecting one exposure variable as a surrogate of multiple highly correlated exposure variables is often suggested in the literature as a solution to handle the multicollinearity problem. However, this may lead to loss of information, since the exposure variables that are highly correlated tend to have not only common but also additional effects on the outcome variable. In this study, a two-stage latent factor regression method is proposed. The key idea is to regress the dependent variable not only on the common latent factor(s) of the explanatory variables, but also on the residuals terms from the factor analysis as the explanatory variables. The proposed method is compared to the traditional latent factor regression and principal component regression for their performance of handling multicollinearity. Two case studies are presented. Simulation studies are performed to assess their performances in terms of the epidemiological interpretation and stability of parameter estimates.
KEYWORDS: Multicollinearity, variance inflation factor, factor analysis, principal component analysis
1. Introduction
In many environmental and epidemiological studies, the observed exposures are often highly correlated. Estimating the exposure-specific effects of the multiple correlated exposures on the health outcome in a statistical regression model can be challenging, since multicollinearity among the exposures can lead to biased and unstable estimated regression coefficients for the correlated covariates because of the overlapping information they share [10,12–14,17,25,39].
Selecting one exposure variable as a surrogate of multiple highly correlated exposure variables is often suggested in the literature as a solution to handle the multicollinearity problem. However, selecting one exposure variable to represent all may result in loss of information [12], since exposure variables that are highly correlated tend to have not only common but also additional effects on the outcome variable. Another strategy is to conduct residual or sequential regression to disentangle unique from shared contributions of the exposure variables [11]. This method assumes that one variable is functionally more important than the other and regress the less important variables against the prioritized variable, and then replaces the less important variables with the residuals from the regression [20]. Subsequent multiple regression analyses will be unbiased since the explanatory variables are no longer statistically collinear. However, prioritized variable based on a researcher's own instincts and intuition can be subjective and also may not be relevant to the true functional importance of the variables [12].
Principal component regression (PCR) has been suggested in literature to overcome the multicollinearity problem (for example, [1,7,16,27,36,38]). The idea of PCR is that instead of regressing the dependent variable on the explanatory variables directly, the principal components of the explanatory variables based on principal component analysis (PCA) [19,23,32], are used as regressors. The PCA method aims at explaining as much of the variation in the data by finding linear combinations that are independent of each other without losing too much information in the process. Principal components are obtained according to the size of their variance; therefore, the first principal component explains the largest amount of variation among the variables and the last component explains the least. The PCA achieves the dimensional reduction by dropping the principal components of low variance (i.e. eigenvalues less than one). However, this could result in a loss of information, since it is possible that principal components associated with small eigenvalues carry important information. Also, PCA looks for linear combinations of the original features to explain the highest variance, so the interpretation of the principal components can be challenging [12]. In fact, the very serious potential pitfalls in the application of PCR method for dealing with multicollinearity have been demonstrated in Hadi and Ling [15]. However, the pitfalls of PCR have been continuously overlooked in applied quantitative research.
An alternative method to handle the multicollinearity problem is to use latent factor regression (LFR) by regressing the outcome variable on the latent variable(s) underlying the multiple exposure variables, extracted based on the factor analysis. The conventional LFR method assumes that the dependence of the outcome variable on the exposure variables are only through the latent variable(s), but not the error terms [36]. However, exposure variables that are highly correlated tend to not only have common influence on the outcome but also may have an additional and unique effect on the outcome. For example, in environmental health studies, environmental pollutants, such as PM2.5 (ambient particulate matter with diameters that are generally 2.5 micrometers and smaller), PM10 (ambient particulate matter with a diameter of 10 micrometers or less) and NO2 (nitrogen dioxide, a gaseous air pollutant composed of nitrogen and oxygen), have been shown be all related to numerous health outcomes including mortality, emergency department visits and hospital admissions (for example [9,26,29]). Many epidemiological studies have quantified the health impact resulting from exposure to a single pollutant (for example [9,26,29]). However, the air that we are exposed to is a complex mixture of multiple pollutants [37]. Quantifying the health effect of the multipollutants that we breathe is an important public health goal and has been an active research area in environmental epidemiology, but challenging due to the potential collinearity among some of the pollutants. One way to tackle the problem is to disentangle unique from shared contributions of the exposure effects on the outcome variable. As another example, body fat can be predicted by various physical measurements, such as weight, abdomen or neck circumferences. Those physical measurements are often highly correlated, so they may have a joint and common effect on body fat. Each physical measurement may also have an additional and unique influence on body fat. Hence, the question arises as to the additional influence of an exposure variable when all common influences are accounted for.
To this end, we propose a new LFR method, which is implemented in two-stages. In the first stage, factor analysis is performed to extract the latent factor(s) and the residuals of multiple correlated exposure variables. The second stage is a multiple regression model, which regresses the disease outcome on the latent factor(s) and residual components extracted from the first stage. The conventional LFR method only regresses the outcome variable on the latent factor(s) extracted based on the factor analysis, which can not capture the unique contribution of the exposure variables. Therefore, the novelty of the proposed method lies in including the exposure-specific residuals from factor analysis in addition to the latent factor as the explanatory variables n the multiple regression. We compare the new LRF method with the conventional LRF method and also the PCR method through simulation studies and two case studies in terms of their epidemiological interpretation and stability of parameter estimates.
The rest of the paper is organized as follows. Section 2 reviews the impact of multicollinearity on the parameter estimates and the commonly used diagnostic methods. Section 3 introduces the proposed LFR method and provides a review of the PCR. Section 4 presents the results of real data analysis for two case studies to demonstrate the performance of LFR in comparison to other methods for handling the multicollinearity issue. Section 5 presents a simulation study to investigate the properties of LFR in comparison to PCR. Finally, concluding remarks are provided in Section 6.
2. Multicollinearity
2.1. Impact of multicollinearity
Previous research showed that the high degree of correlation between explanatory variables may inflate the standard errors of regression coefficients, thus leading to insignificant coefficients [10,12,14,39], and sometimes it may change the coefficients to opposite sign of their true values [24]. The unreliable and biased regression coefficients caused by multicollinearity make the interpretation of the covariate effects unrealistic and unconvinced [39].
More specifically, in the context of a multiple linear regression model, let denotes the response variable, , where n denotes the sample size and denotes the vector of the covariates for the ith subject, and is the unobserved random error following a normal distribution with mean zero and variance . The linear regression model is expressed as,
| (1) |
where denotes the regression coefficients for the explanatory variables. The least-squares estimators of the regression coefficients are , where is nearly singular when the columns of the design matrix are nearly linearly dependent. Therefore, can be unstable when the covariates are highly correlated, since small changes in the data may result in large change of . In addition, some of the elements of the can be large yielding large variances or covariances of the elements of . Mathematically, when no collinearity exists between the explanatory variables, all the eigenvalues ( ) of the correlation matrix will be one. In contrast, when there is multicollinearity among the explanatory variables, at least one eigenvalue is near zero. The estimated variance of the regression coefficient is written as,
| (2) |
From Equation (2), we can see that when any of the eigenvalues are small that nearly equal to 0, will be large.
The effect of multicollinearity on the coefficient estimates can also be inferred from Equation (2):
so
| (3) |
Equation (3) shows that if is small due to multicollinearity, the vector of estimated coefficients will be heavily biased from the true values [35].
2.2. Diagnosis of multicollinearity
Variance inflation factor (VIF) is commonly used to measure the severity of multicollinearity, which indicates how much the variance of the explanatory variable is inflated due to multicollinearity in explaining the variance of the response variable [31]. VIF can be computed as, where denotes the squared correlation between the kth covariate and other covariates, namely, the proportion of variance that the kth covariate shared with other covariates in the regression model [28].
The rule of thumb for deciding severe multicollinearity is ad hoc. Generally, VIF greater than 10 indicates that the multicollinearity can not be ignored, but some other researchers suggest that a small VIF can also raise concerns for the validity of the analysis, such as inflated standard errors of the estimated regression coefficients [12]. The threshold of VIF equal to 5 or 8 is also adopted in some studies [39]. Other studies showed that VIFs as low as 2 can have a negative impact on the reliability and interpretation of multiple regression [12].
3. Methodology
In this section, we first introduce the new LFR in Section 3.1 proposed to analyze the effect of multiple highly correlated exposure variables on the outcome variable. In Section 3.2, a brief review of PCR is provided.
3.1. A two-stage latent factor regression
The proposed new LFR method comprises two-stages. First, factor analysis is adopted to extract the latent factors and also the residuals for multiple exposures. Second, a multiple regression model is implemented to regress the outcome variable on the latent factors and residuals of multiple exposures extracted based on the factor analysis.
3.1.1. First stage
In the first stage of the proposed latent factor regression (LFR), factor analysis is used for extracting the common structure (i.e. factors) underlying the explanatory variables and the additional sources of variation for each of the explanatory variable [21]. More specifically, suppose there are p highly correlated explanatory variables of interest, denoted as , so is a matrix, where n denotes the sample size. The explanatory variables are all standardized to avoid the problem of having one variable with a large variance influence the determinations of factor loadings. The factor analysis model can also be expressed in matrix notation as,
| (4) |
where is a matrix of factor scores following a multivariate normal with p denoting the number of latent common factors. and , where is the identity variance-covariance matrix. is a matrix of factor loadings, which can be regarded as weights of the effect of the factors on the observed variables. The higher the value of the loading, the greater the effect of the factor on a particular variable. is a error matrix with and , where , is the specific variance associated with variable . Each column of specific the latent variable unique to each variable of . Also, the errors must be uncorrelated and independent from the factor scores: . The covariance of can be expressed as
| (5) |
The parameters and can be estimated by the principal component method or maximum likelihood method [21].
For example, in the scenario with three highly correlated explanatory variables, denoted as . The factor analysis method with a single latent factor ( ) can be expressed as follows:
| (6) |
where the coefficients are the loadings of the kth variable on the latent factor . The additional sources of variation for the kth response is denoted as , which is associated only with the kth response. Note that all the common factor and residual components are unobservable.
3.1.2. Second stage
In the second stage of the LFR model, the extracted components in the first stage including the estimated common factor and the residual components =( , and ) are included as the explanatory variables for modeling the dependent variable. In literature, only the latent factor is included in a regression model as an explanatory variable, which omits the possible contribution of the residual components ( , and ) for predicting the outcome variable. Therefore, in our proposed model, the residuals ( , and ) are also included as the covariates in addition to the common factor , for modeling the response variable.
For a response variable with mean and link function , the disease model can be expressed as , where refers to the vector of regression coefficients corresponding to for the estimated latent common latent factor , and the estimated residual effects extracted based on the first stage of the LFR method.
3.2. Principal component regression
Principal component regression (PCR) is also a two-stage approach similarly as the procedure described in Section 3.1, but the first stage uses PCA. The main idea of PCA is to reduce the dimensionality of variables, meanwhile retaining the variation present in the data set as much as possible [21,23,32]. A new set of uncorrelated variables, namely the principal components, are transformed from the data set and ordered according to their contribution to the variation. Therefore, the first few of the ordered PCs can preserve most of the variation to represent the original variables [23].
To perform PCA for reducing the dimensionality of the correlated exposure variables, the first step is to center the data on the means of each variable, by subtracting the mean of a variable from all values of that variable. The distributions of each variable should be checked for normality, and data transformations should be used where necessary to correct for skewness. Outliers should be removed from the data set as they can dominate the results of a PCA.
Each of the PCs, denoted as is given by a linear combination of the variables , i.e.
| (7) |
where . The PCs are uncorrelated with (i.e. perpendicular to) each other. In matrix notation, the PCs are written as
| (8) |
where the rows of matrix A are called the eigenvectors, which specify the orientation of the PCs relative to the original variables. The elements of an eigenvector, that is, the values within a particular row of the matrix , are the weights . These values are called the loadings, and they describe how much each variable contributes to a particular PC. Large loadings indicate that a particular variable has a strong relationship to a particular PC. The sign of loading indicates whether a variable and a principal component are positively or negatively correlated.
The variance-covariance matrix of the PCs can be calculated:
| (9) |
where the elements in the diagonal of matrix are the eigenvalues, that is, the variance explained by each PC. The off-diagonal elements of matrix are zero, since the principal components are independent and therefore have zero covariance. The components associated with small eigenvalues have very small variances, so they can be omitted. In the second stage of the PCR, the outcome variable is regressed on the first τ PCs, , where .
4. Real data applications
4.1. Case study : air pollution study
4.1.1. Data description
This study is motivated by an environmental health study by Lee et al. to examine the impact of air pollutants measured in the year 2010 on the risk of respiratory diseases in Greater Glasgow, Scotland in the year 2011 [29]. The air pollutants include ambient particulate matter that is less than in diameter (PM2.5), ambient particulate matter that is in the air with a diameter of 10 micrometers or less (PM10) and nitrogen dioxide, or NO2, a gaseous air pollutant composed of nitrogen and oxygen. The study population was nearly 1.2 million people who lived in the city of Glasgow and the River Clyde estuary, which comprised 271 administrative units with about 4000 people in each region on average.
The response variable is the counts of hospital admissions with diagnosed respiratory diseases (codes J00-J99 and R09.1 of the International Classification of Disease Tenth revision) from each of the 271 administrative units. In order to adjust for the different sizes and demographic structures of populations in each region, the expected counts of hospital admissions were calculated using external standardization based on age and sex. The pollution concentrations were yearly averaged using dispersion models with the source from the Department for the Environment, Food and Rural Affairs. The median value of the modeled average concentrations at a resolution of 1km grid squares in 2010 in each region was used as the measure of explanatory variables. The pollution concentrations in the year prior to the observed hospital admissions were used to ensure the causal relationship that air pollution exposures should occur before the disease diagnosis [29]. Socio-economic deprivation is the key confounder, since previous research showed that socio-economically deprived areas have worse health on average than more affluent areas. The proportion of the working age population in receipt of Job Seekers Allowance (denoted JSA) from each region was included as a confounder as a proxy of the unemployment rate.
As shown in Figure 1, PM2.5, PM10 and NO2 are highly and nearly linearly correlated with VIFs equal to 119.56, 81.16 and 30.84, respectively. In particular, PM2.5 and PM10 are almost perfectly linearly related, with the Pearson correlation coefficient equal to 0.994. NO2 is also highly related to PM2.5 and PM10 with the Pearson correlation coefficients equal to 0.983 and 0.974, respectively. The original aim of the study by Lee et al. [29] was to propose a localized conditional autoregressive model for modeling the local spatial dependence of the data, and the air pollutants entered the model separately to circumvent the issue of multicollinearity. The study found PM2.5, PM10and NO2 were related to the increased risks of respiratory diseases individually in the studied area [29]. Our current study is not to investigate the methods of modeling spatial correlation of the disease incidence, but to demonstrate the advantage of LFR models for uncovering the common and residual effects of highly correlated exposure variables on the disease outcome.
Figure 1.
The density curves, pairwise scatter plots and the estimated Pearson correlation coefficients between PM , PM and NO in case study #1.
4.1.2. Results of the analysis
To model the impact of three pollutants on the respiratory disease outcome in the study region, we considered negative binomial (NB) regression for modeling the number of respiratory disease cases. Let and denote the observed and expected numbers of respiratory disease cases in the ith region, respectively. The disease model can be formulated as with where represents the relative risk of the disease in the ith region, where n = 271. The regression coefficient for JSA is denoted as γ. The explanatory variables relating to the air pollutants are denoted as with representing the corresponding regression coefficients.
A number of competing models were considered depending on how was defined, which are listed as follows. For all these models, the linearity assumptions of the effects of explanatory variables were examined by comparing the model fits with generalized additive models by flexibly modeling the effects of explanatory variables as smoothing spline functions. The results suggested that the model fit was not significantly different. As a result, all the covariates are modeled as having linear effects.
Naive: The Naive model includes PM2.5 and NO2 as the explanatory variables. The effect of PM10 can not be estimated when it was included in the model, so it was not included in the Naive model as an explanatory variable.
Single Exposure Analysis: include each pollutant as an explanatory variable, separately.
- PCR: In the first stage of the PCR method, PCA was performed to extract PCs for the three exposure variables. The PCs explained and of the variation of all three pollutants, respectively. We consider either the first or the first two PCs as candidate explanatory variables in modeling the disease outcome. Two PCR models were considered including
- PCR1: including the first PC score as an explanatory variable.
- PCR2: including the first two PC scores as explanatory variables.
-
LFR: Under the LFR method, the choice of the number of common factors is determined by examining the number of eigenvalues of the correlation matrix greater than the unity. The estimated eigenvalues are for the three pollutants, respectively. Moreover, the common factor accounts for a proportion of the total sample variance. As a result, only one common factor is identified in this application. The factors loadings are estimated as , so the commonalities are estimated as , which indicates the common factor accounts for a large percentage of sample variance of each pollutant variable.
Figure 2 displays the distributions of the common factor ( ) and residual components ( ) extracted based on the factor analysis. The plots indicate all the latent common and residual components were roughly symmetric without substantial skewness. Nevertheless, the residual components had almost negligible variance ( ) and was not statistically significant. Therefore, and were not included as explanatory variables in the second stage of the LFR method. Two LFR models were therefore considered including- LFR1: including the extracted latent common factor ( ) based on the factor analysis as an explanatory variable.
- LFR2: including the extracted latent common factor ( ) and residual component ( ) based on the factor analysis as the explanatory variables.
Figure 2.
The histograms and density distributions of the extracted common factor ( ) and residual components ( ) based on the factor analysis of the three pollutants (PM2.5, PM10 and NO2) in Case Study .
Table 1 presents the estimated parameters and the overall model fit for the competing models. Under the Naive model, the effect of PM2.5 was estimated as with p-value=0.046, indicating PM2.5 decreased the respiratory disease risk, which is against intuition and existing literature. In contrast, the results of the single exposure analysis indicate that each of the three pollutants significantly increased the respiratory disease risk. However, the model fits were slightly worse than the Naive model, according to the deviance and AIC comparisons.
Table 1.
Estimated parameters and the overall model fit of the naive, single exposure analysis, PCR and LFR methods for Case Study .
| Model | Estimate | Std. Error | P-value | deviance | AIC |
|---|---|---|---|---|---|
| Naive Model | |||||
| Intercept | 0.154 | 0.335 | 0.646 | 277.66 | 2286.57 |
| PM2.5 | 0.114 | 0.057 | 0.046 | ||
| NO2 | 0.028 | 0.011 | 0.010 | ||
| Single Exposure Analysis | |||||
| Only include PM2.5 | |||||
| Intercept | 0.976 | 0.097 | 0.001 | 278.22 | 2291.17 |
| PM2.5 | 0.030 | 0.011 | 0.006 | ||
| Only include PM10 | |||||
| Intercept | 0.992 | 0.101 | 0.001 | 278.28 | 2291.03 |
| PM10 | 0.022 | 0.008 | 0.005 | ||
| Only include NO2 | |||||
| Intercept | 0.816 | 0.040 | 0.001 | 278.21 | 2288.57 |
| NO2 | 0.007 | 0.002 | 0.001 | ||
| Principal Component Regression | |||||
| PCR1 | |||||
| Intercept | 0.703 | 0.028 | 0.001 | 278.25 | 2290.20 |
| PC1 | −0.023 | 0.008 | 0.003 | ||
| PCR2 | |||||
| Intercept | 0.703 | 0.028 | 0.001 | 277.70 | 2288.07 |
| PC1 | 0.023 | 0.008 | 0.003 | ||
| PC2 | 0.158 | 0.078 | 0.043 | ||
| Latent Factor Regression | |||||
| LFR1 | |||||
| Intercept | 0.705 | 0.028 | 0.001 | 278.22 | 2291.17 |
| 0.037 | 0.013 | 0.006 | |||
| LFR2 | |||||
| Intercept | 0.700 | 0.028 | 0.001 | 277.66 | 2286.58 |
| 0.037 | 0.013 | 0.005 | |||
| 0.178 | 0.070 | 0.010 | |||
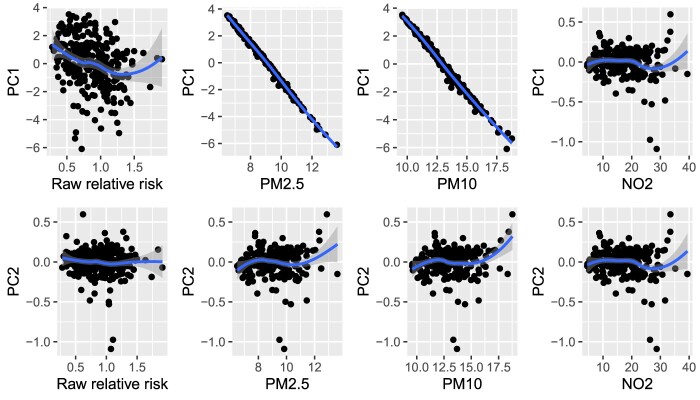
The results of the PCR1 and PCR2 indicate that the first two PCs significantly decreased the risk of respiratory disease. This phenomenon is also reflected in Figure 3, which displays the scatter plots of the first two principal components against the raw relative risk and each of the three pollutants, respectively. The figure indicates that the PC1 had a moderate negative relationship with raw relative risk of respiratory hospitalization and a strong and negative relationship with PM2.5 and PM10, but no clear relationship with NO2. PC2 had no apparent relationship with raw relative risk or any of the pollutants. Therefore, although PCA could help reduce the dimensionality of data into lower space, the interpretation of the components based on PCR method in a meaningful way could be challenging and misleading, since the procedure extracting PCs have nothing to do with how these variables affect the response variable [4].
Figure 3.
Scatter plots of the first two PCs based on the PCA method against the raw relative risk of hospital admission for respiratory condition and each of the three pollutants (PM2.5, PM10 and NO2), respectively, for case study . The blue lines and 95% gray shaded confidence band are generated based on loess nonparametric smoothing method.
Comparison of LFR1 and LFR2 indicates that LFR2 (AIC=2286.58) provided a better model fit than LFR1 (AIC=2291.17), which suggests the residual component of NO2 had additional contribution for predicting the respiratory disease risk. The results based on the LFR2 method indicate the latent common factor of the three pollutants ( ) significantly increased the risk of disease outcome ( , p-value ), and NO2 significantly increased the relative risk of respiratory diseases even after accounting for the effect of the common factor ( , p-value ). In other words, NO2 had a greater effect on increasing the risk of respiratory disease compared to the other two pollutants.
4.2. Case study : body fat study
4.2.1. Data description
Accurate measurement of body fat is inconvenient and costly. A study was conducted to show that physical measurements, i.e. weight, abdomen or neck circumferences, using only a scale and a measuring tape provides a convenient way of estimating body fat [22]. The dataset contains the estimates of the percentage of body fat and various body circumference measurements for 252 men [22]. The response variable is the percent body fat, which was accurately estimated by an underwater weighing technique. The predictors include weight (lbs), neck circumference (cm) and abdomen circumference (cm) ‘at the umbilicus and level with the iliac crest’. This data can be downloaded from the mfp R package [2].
Figure 4 shows that the body fat, weight, abdomen and neck circumferences are approximately symmetrically distributed and they are all significantly and positively correlated in a linear fashion, with VIFs equal to 6.61, 4.75, and 3.24, respectively. The Pearson correlation coefficients for weight and abdomen circumference, weight and neck circumference, abdomen and neck circumferences are equal to 0.89, 0.83 and 0.75, respectively.
Figure 4.
The density curves, pairwise scatter plots and the estimated Pearson correlation coefficients between body fat, weight, abdomen and neck circumferences in the case study .
4.2.2. Results of the analysis
Table 2 presents that under the naive model, only abdomen circumference had a positive linear relationship with body fat. By contrast, weight and neck circumference had significant negative relationships with body fat, demonstrating the impact of multicollinearity on the estimation of the relationship between predictors and the dependent variable, in the presence of multicollinearity. Our study also demonstrated that even VIF in the multiple linear regression model could result in unrealistic and untenable interpretations of the effect of predictors.
Table 2.
Estimated parameters and the overall fit of the naive, single exposure analysis, PCR and LFR methods for Case Study .
| Model | Estimate | Std. Error | P-value | AIC | |
|---|---|---|---|---|---|
| Naive Model | |||||
| Intercept | 35.015 | 5.800 | 0.001 | 0.72 | 1470.71 |
| weight | 0.121 | 0.024 | 0.001 | ||
| abdomen | 0.997 | 0.056 | 0.001 | ||
| neck | 0.436 | 0.207 | 0.036 | ||
| Single Exposure Analysis | |||||
| Only include weight | |||||
| Intercept | 12.052 | 2.581 | 0.001 | 0.38 | 1672.43 |
| weight | 0.174 | 0.014 | 0.001 | ||
| Only include abdomen | |||||
| Intercept | 39.280 | 2.660 | 0.001 | 0.66 | 1517.79 |
| abdomen | 0.631 | 0.029 | 0.001 | ||
| Only include neck | |||||
| Intercept | 45.015 | 7.223 | 0.001 | 0.24 | 1721.51 |
| neck | 1.689 | 0.190 | 0.001 | ||
| Principal Component Regression | |||||
| PCR1 | |||||
| Intercept | 19.151 | 0.387 | 0.001 | 0.46 | 1634.13 |
| PC1 | 3.499 | 0.238 | 0.001 | ||
| PCR2 | |||||
| Intercept | 19.151 | 0.325 | 0.001 | 0.62 | 1547.58 |
| PC1 | 3.499 | 0.200 | 0.001 | ||
| PC2 | 6.637 | 0.648 | 0.001 | ||
| Latent Factor Regression | |||||
| LFR1 | |||||
| Intercept | 19.151 | 0.408 | 0.001 | 0.40 | 1661.04 |
| 5.362 | 0.413 | 0.001 | |||
| LFR2 | |||||
| Intercept | 19.151 | 0.280 | 0.001 | 0.72 | 1471.25 |
| 5.167 | 0.283 | 0.001 | |||
| 11.182 | 0.664 | 0.001 | |||
For the single exposure analysis, all physical measures had significantly positive relationships with body fat. However, the model fits for the single exposure analysis yield much lower and higher AIC compared to the naive model. This demonstrates that selecting one exposure as a predictor to enter the regression model could lead to the loss of information for predicting the response variable.
For the PCR method, PCA was firstly performed to extract PCs for the three physical measurements. The three PCs explained , and of the variation of three physical measurements, respectively. Figure 5 displays scatter plots of the first two PCs based on the PCA method against the body fat and each of the three physical measurements, i.e. weight, abdomen and neck circumferences, respectively. The figure indicates that the PC1 had a positive and linear relationship with body fat and all physical measurements. However, PC2 had a somewhat negative linear relationship with bodyfat and no discernible relationship with weight and abdomen circumferences, but a positive relationship with neck circumference. Table 2 shows PC1 had a significant positive relationship with bodyfat, but the , which is much lower than the for the naive model, which is 0.72, The AIC for the PC1 (1634.13) is also much higher than the AIC for the naive model (AIC = 1470.71). For the model including both PC1 and PC2 as predictors in the multiple linear regression, PC2 was significantly and negatively associated with the body fat with an improved model fit ( , AIC = 1547.58), but the model fit was still not comparable with the naive model.
Figure 5.
Scatter plots of the first two principal components based on the PCA method against the body fat and each of the three physical measurements, i.e. weight, abdomen and neck circumferences, respectively, in case study . The blue lines and 95% gray shaded confidence band are generated based on the loess nonparametric smoothing method.
Under the LFR method, the choice of the number of common factors is determined based on examining the number of eigenvalues of the correlation matrix greater than the unity. The estimated eigenvalues are for the three physical measurement variables, respectively. Moreover, the common factor accounts for a proportion of the total sample variance. As a result, only one common factor is identified in this application. The factors loadings are estimated as , so the commonalities are estimated as , which indicates the common factor accounts for a large percentage of sample variance of physical measurement variables.
Figure 6 displays the distributions of the common factor ( ) and residual components ( ) based on the factor analysis, which indicate all the latent and residual components were roughly symmetric without substantial skewness. When including all the common factor and residual components in the second stage of the LFR method, the effect of can not be estimated due to singularities and effect of was not statistically significant (p-value=0.114), so only the common factor ( ) or both and were included in the multiple linear regression model as explanatory variables.
Figure 6.
The histograms and density distributions of the extracted common factor ( ) and residual components ( ) based on the factor analysis method applied to the three physical measures (weight, abdomen circumference, and neck circumference) in case study .
Table 2 shows that LFR2 provided a comparable model fit to the naive model with and AIC=1471.25. LFR2 also substantially outperformed LFR1 in terms of overall model fit, which highlights the importance of residual components from factor analysis for predicting the outcome variable. The results based on the LFR2 method indicate the common latent factor of the three physical measurements ( ) had a significant positive association with body fat ( , p-value ), and abdomen circumference had an additional contribution for predicting the body fat with the estimated regression coefficient equal to 11.182 (p-value ), even after accounting for the effect of the common factor. In other words, abdomen had a greater predictive power of body fat compared to the other two physical measurements.
5. Simulation study
This section investigates the properties and performance of the LFR in comparison to PCR. The primary purpose is to evaluate the methods in terms of their interpretability of the effects of multiple highly correlated exposures on the outcome variable.
5.1. Data generation
Data are generated from LRF2 model with the simulation setting mimicking the real data application presented in Section 4. The simulation process is detailed as follows.
-
Step 1: Simulate three highly correlated explanatory variables , as follows.
- Generate the variance covariance matrix for , i.e. , where is the vector of factor loadings and is the vector of variances for . We assume follows standardized multivariate normal distribution with , k = 1, 2, 3. Hence, , so . To vary the degree of the correlation among the covariates, we set and , where with and then from 0.01 to 0.07 at an increment of 0.01, so that the VIF of ranges between about 35 to 10.
- Generate from multivariate normal distribution with mean and variance covariance matrix .
Step 2: Extract the latent common factor ( ) and exposure-specific residual components ( , and ) by conducting factor analysis of the explanatory variables .
Step 3: To mimic case study , we assume , have no effect on the outcome variable. Simulate outcome variable from a negative binomial regression model with logarithm of the expected mean count , where and are the regression coefficients . The shape parameter of the negative binomial distribution is set as 34 to mimic the real data application.
Step 4: Fit the Naive, PCR and LFR models to the simulated data.
Step 5: Repeat Steps 1-4 for R = 500 times to evaluate the performances of the methods.
Additional simulation studies were conducted to examine if the performance of LFR compared to PCR depends on the distributional assumption of the response variable . To this end, in Step 3 of the above-mentioned data generation process, binary regression and linear regression in the second stage of the LFR model are also considered, as follows.
For the binary regression model, we consider generating the response variable from a logistic regression model , where and .
For the linear regression model, the response variable is generated from normal distribution with mean where and and variance .
5.2. Method comparison metrics
The primary goal of the study is to evaluate the methods for their practicality and interpretability of the effects of multiple correlated exposure variables, as well as their overall model fit. The performances of the methods are therefore evaluated in terms of the directions of the estimated regression coefficients, which are measured as the percentage of the estimated regression coefficients greater than zero based on the 500 simulated samples.
The overall model fit is evaluated based on both in-sample and out-of-sample predictions. The in-sample model fit is evaluated based on the average AIC over the 500 simulated samples. The out-of-sample predictions are evaluated based on M-fold cross-validation (CV), by splitting the data set into M folds, then using M−1 folds as the training set, and the remaining fold as the test set. Here, we consider M = 10. The root mean square error (RMSE) of the predicted response variable based on the rth simulated dataset is then calculated. , where denotes the mth testing dataset and denotes the training dataset, which includes the observations except for the data from the mth fold. denotes the predicted expected mean of the outcome variable. The average RMSE over all the R simulated datasets can be then defined as .
5.3. Results
5.3.1. True model: LFR2 for modeling the impact of highly correlated exposure variables on a count outcome variable
Table 3 presents the average estimated regression coefficients under the Naive, LFR and PCR methods over 500 datasets simulated from the two-stage LFR2 model with increasing multicollinearity. The outcome variable in the second stage is simulated from a NB model as described in Section 5.1. The data were simulated to mimic the case study presented in Section 4, such that the three exposure variables have a joint and positive effect and the last exposure has an additional positive effect on the outcome variable.
Table 3.
The average estimated regression coefficients, the percentage of the estimated regression coefficients greater than zero, and the percentage of the estimated regression coefficients are significantly different from zero at level of significance, over 500 datasets simulated from the LFR2 method, with the second stage model being a negative binomial regression model.
| VIF | 34 | 29 | 26 | 21 | 17 | 15 | 13 | 12 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| Naive Model | |||||||||
| 0.031 | 0.044 | 0.050 | 0.056 | 0.064 | 0.060 | 0.064 | 0.069 | 0.065 | |
| 0.036 | 0.026 | 0.023 | 0.019 | 0.015 | 0.015 | 0.012 | 0.011 | 0.010 | |
| 0.168 | 0.173 | 0.177 | 0.179 | 0.185 | 0.183 | 0.186 | 0.193 | 0.190 | |
| 0.322 | 0.258 | 0.200 | 0.144 | 0.084 | 0.094 | 0.068 | 0.026 | 0.054 | |
| 0.312 | 0.306 | 0.320 | 0.328 | 0.340 | 0.328 | 0.376 | 0.364 | 0.350 | |
| 0.996 | 0.998 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.074 | 0.146 | 0.144 | 0.192 | 0.276 | 0.280 | 0.354 | 0.410 | 0.424 | |
| 0.092 | 0.064 | 0.064 | 0.064 | 0.052 | 0.078 | 0.072 | 0.072 | 0.070 | |
| 0.692 | 0.854 | 0.916 | 0.978 | 0.996 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Principal Component Regression | |||||||||
| PCR1 | |||||||||
| 0.002 | 0.000 | 0.001 | 0.005 | 0.002 | 0.003 | 0.003 | 0.000 | 0.004 | |
| 0.518 | 0.496 | 0.510 | 0.454 | 0.476 | 0.518 | 0.526 | 0.502 | 0.474 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| PCR2 | |||||||||
| 0.002 | 0.000 | 0.001 | 0.005 | 0.002 | 0.003 | 0.003 | 0.000 | 0.004 | |
| 0.000 | 0.002 | 0.006 | 0.007 | 0.003 | 0.009 | 0.003 | 0.004 | 0.001 | |
| 0.518 | 0.496 | 0.510 | 0.454 | 0.476 | 0.518 | 0.526 | 0.502 | 0.474 | |
| 0.496 | 0.518 | 0.476 | 0.516 | 0.516 | 0.518 | 0.478 | 0.488 | 0.508 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.326 | 0.516 | 0.658 | 0.826 | 0.908 | 0.954 | 0.978 | 0.988 | 0.992 | |
| Latent Factor Regression | |||||||||
| LFR1 | |||||||||
| 0.100 | 0.102 | 0.102 | 0.101 | 0.102 | 0.102 | 0.103 | 0.103 | 0.104 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| LFR2 | |||||||||
| 0.099 | 0.101 | 0.100 | 0.099 | 0.100 | 0.100 | 0.100 | 0.100 | 0.101 | |
| 0.200 | 0.201 | 0.201 | 0.198 | 0.201 | 0.196 | 0.197 | 0.202 | 0.199 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.978 | 0.988 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.522 | 0.700 | 0.828 | 0.960 | 0.996 | 1.000 | 0.998 | 1.000 | 1.000 | |
The results indicate that under the naive method, the first two exposures are negatively associated with the outcome, but the last exposure has a positive effect on the outcome, which clearly shows the multicollinearity among the covariates can severely impact the estimation of the effects of exposures. The significance of the exposure variables decreases rapidly, as the degree of multicollinearity increases. The weak exposure variables ( and ) are more vulnerable to erroneous exclusion than the strong one, i.e. .
Under the PCR methods, regardless PCR1 or PCR2 methods, the effects of PCs are almost negligible but statistically significant, and about half of the estimated regression coefficients over 500 simulated datasets are negative. As a result, although the PCR method can mitigate the impact of multicollinearity on estimating the covariate effects, the regression coefficients for PCs could result in unrealistic and untenable interpretations. By contrast, under the LFR1 method, the latent factor is positively associated with the outcome variable with the estimated effect close to the true regression coefficient for the common latent factor. Compared to the LFR1 method, the LFR2 method accurately estimates the regression coefficients for both the common and residual components. The latent factor remains statistically significant as the degree of multicollinearity increases, while the significance for the residual component decreases gradually. This is not surprising, since as the multicollinearity among exposures increases, the latent common factor could capture the majority variability of the exposure variables leaving little unexplained for the residual or error terms. In contrast, with decreased multicollinearity, the effect of exposure-specific residual components could potentially capture more unique contributions of the exposure variables in explaining the variability of the outcome variable.
Figure 7 presents the average AIC scores (left panel) and average 10 fold cross-validation RMSE (right panel) for the considered methods over 500 datasets simulated from the two-stage LFR2 model. The results indicate that LFR2 consistently outperforms the other methods at varying levels of VIF in terms of the overall model fit measured by AIC and RMSE. The naive model performs slightly worse than LFR2 followed by PCR2. LFR1 and PCR1 perform substantially worse than the LFR2 and naive methods in terms of overall model fit when the exposures are not extremely highly correlated. Not surprisingly, as the VIF increases, all the methods perform similarly in terms of overall model fit, since the first PC for the PCR method and common factor for LFR method can capture the majority of the variability of the exposure variables.
Figure 7.
The average AIC scores (left panel) and average 10 fold cross validation RMSE (right panel) over the 500 datasets simulated from the LFR2 method, with the second stage model being a NB regression.
5.3.2. True model: LFR2 for modeling the impact of highly correlated exposure variables on a binary outcome variable
In the scenario when the outcome variable is a binary indicator simulated from the LFR2 method, the results (Table 4) indicate that the directions of the associations between predictors and outcome variable under the naive method are unstable, with about half of the estimated regression coefficients ( and ) being positive and half negative. Under the PCR method, although the first PC is significantly associated with the outcome variable, the magnitude is very small and almost half of its estimated regression coefficients are negative. In comparison to the naive and PCR methods, the LFR methods give the regression coefficients very close to the true values. Most of the estimated regression coefficients are estimated as positive. The common factor is mostly significantly and positively associated with the outcome, and the residual component becomes more significantly associated with the outcome variable, as the multicollinearity among exposures decreases. These results are consistent with findings when the true model is LFR2 for count outcome. This demonstrates that the LFR2 approach can enhance the reliability and interpretation of multiple regression in the presence of multicollinearity.
Table 4.
The average estimated regression coefficients, the percentage of the estimated regression coefficients greater than zero, and the percentage of the estimated regression coefficients are significantly different from zero at level of significance, over 500 datasets simulated from the LFR2 method, with the second stage model being a logistic regression model.
| VIF | 34 | 29 | 25 | 21 | 17 | 15 | 13 | 12 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| Naive Model | |||||||||
| 0.171 | 0.206 | 0.295 | 0.275 | 0.332 | 0.338 | 0.318 | 0.365 | 0.350 | |
| 0.082 | 0.141 | 0.108 | 0.135 | 0.072 | 0.076 | 0.075 | 0.031 | 0.050 | |
| 0.763 | 0.861 | 0.930 | 0.945 | 0.951 | 0.965 | 0.954 | 0.963 | 0.974 | |
| 0.412 | 0.374 | 0.320 | 0.330 | 0.252 | 0.240 | 0.236 | 0.194 | 0.206 | |
| 0.422 | 0.400 | 0.428 | 0.366 | 0.450 | 0.452 | 0.442 | 0.452 | 0.420 | |
| 0.868 | 0.920 | 0.950 | 0.968 | 0.990 | 0.996 | 0.998 | 1.000 | 1.000 | |
| 0.070 | 0.044 | 0.092 | 0.066 | 0.096 | 0.104 | 0.088 | 0.126 | 0.162 | |
| 0.046 | 0.046 | 0.050 | 0.068 | 0.034 | 0.054 | 0.046 | 0.040 | 0.068 | |
| 0.196 | 0.324 | 0.408 | 0.552 | 0.648 | 0.734 | 0.784 | 0.820 | 0.864 | |
| Principal Component Regression | |||||||||
| PCR1 | |||||||||
| −0.006 | 0.010 | 0.009 | 0.021 | 0.004 | 0.008 | 0.003 | 0.015 | 0.010 | |
| 0.486 | 0.510 | 0.510 | 0.532 | 0.502 | 0.480 | 0.508 | 0.486 | 0.480 | |
| 0.984 | 0.976 | 0.988 | 0.986 | 0.980 | 0.986 | 0.990 | 0.992 | 0.994 | |
| PCR2 | |||||||||
| 0.006 | 0.010 | 0.009 | 0.021 | 0.004 | 0.008 | 0.003 | 0.015 | 0.010 | |
| 0.022 | 0.002 | 0.033 | 0.030 | 0.013 | 0.049 | 0.050 | 0.028 | 0.000 | |
| 0.486 | 0.510 | 0.510 | 0.532 | 0.502 | 0.480 | 0.508 | 0.486 | 0.480 | |
| 0.522 | 0.498 | 0.508 | 0.494 | 0.502 | 0.486 | 0.516 | 0.492 | 0.488 | |
| 0.984 | 0.976 | 0.990 | 0.986 | 0.982 | 0.988 | 0.990 | 0.992 | 0.994 | |
| 0.100 | 0.158 | 0.222 | 0.328 | 0.344 | 0.444 | 0.510 | 0.514 | 0.560 | |
| Latent Factor Regression | |||||||||
| LFR1 | |||||||||
| 0.505 | 0.503 | 0.511 | 0.509 | 0.514 | 0.506 | 0.507 | 0.498 | 0.495 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.984 | 0.974 | 0.986 | 0.980 | 0.976 | 0.982 | 0.980 | 0.982 | 0.980 | |
| LFR2 | |||||||||
| 0.502 | 0.502 | 0.507 | 0.506 | 0.509 | 0.503 | 0.506 | 0.500 | 0.502 | |
| 0.886 | 0.994 | 1.061 | 1.043 | 1.029 | 1.030 | 1.005 | 1.009 | 1.016 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.806 | 0.874 | 0.918 | 0.960 | 0.982 | 0.986 | 0.996 | 0.996 | 0.998 | |
| 0.970 | 0.968 | 0.976 | 0.980 | 0.976 | 0.982 | 0.980 | 0.980 | 0.988 | |
| 0.132 | 0.240 | 0.324 | 0.454 | 0.548 | 0.656 | 0.710 | 0.776 | 0.834 | |
In terms of the overall model fit, the average AIC and 10-fold RMSE for the LFR2 are consistently lower than the other methods, except when the exposure variables are extremely highly correlated, as shown in Figure 8. Interestingly, the naive method performs the worst in terms of the overall model fit, when the exposure variables are extremely highly correlated. The PCR1 and LFR1 methods perform worse than the other methods, especially when the exposure variables are not extremely highly correlated.
Figure 8.
The average AIC scores (left panel) and average 10-fold cross-validation RMSE (right panel) over the 500 datasets simulated from the LFR2 method, with the second stage model being a logistic regression.
5.3.3. True model: LFR2 for modeling the impact of highly correlated exposure variables on a continuous outcome variable
In the scenario when the outcome variable is simulated from a multiple linear regression model with the common factor and residuals extracted based on LFR method as the predictors, the results (Table 5 and Figure 9) are very similar to the previous simulation scenarios with count or binary outcomes, especially the first scenario when the outcome variable is simulated from a NB regression.
Table 5.
The average estimated regression coefficients, the percentage of the estimated regression coefficients greater than zero, and the percentage of the estimated regression coefficients are significantly different from zero at level of significance, over 500 datasets simulated from the LFR2 method, with the second stage model being a normal linear regression model.
| VIF | 33 | 29 | 25 | 21 | 17 | 15 | 13 | 12 | 11 |
|---|---|---|---|---|---|---|---|---|---|
| Naive Model | |||||||||
| 0.172 | 0.215 | 0.240 | 0.270 | 0.313 | 0.315 | 0.332 | 0.312 | 0.328 | |
| 0.180 | 0.135 | 0.119 | 0.094 | 0.081 | 0.063 | 0.058 | 0.061 | 0.053 | |
| 0.859 | 0.858 | 0.875 | 0.891 | 0.930 | 0.922 | 0.947 | 0.941 | 0.952 | |
| 0.324 | 0.242 | 0.190 | 0.150 | 0.098 | 0.084 | 0.052 | 0.054 | 0.042 | |
| 0.294 | 0.310 | 0.326 | 0.330 | 0.346 | 0.392 | 0.352 | 0.324 | 0.346 | |
| 0.992 | 1.000 | 0.998 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.078 | 0.104 | 0.136 | 0.178 | 0.246 | 0.274 | 0.358 | 0.336 | 0.424 | |
| 0.100 | 0.074 | 0.070 | 0.064 | 0.068 | 0.082 | 0.052 | 0.074 | 0.060 | |
| 0.736 | 0.864 | 0.904 | 0.986 | 0.996 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Principal Component Regression | |||||||||
| PCR1 | |||||||||
| 0.018 | 0.011 | 0.014 | 0.002 | 0.008 | 0.034 | 0.013 | 0.024 | 0.019 | |
| 0.472 | 0.482 | 0.480 | 0.496 | 0.488 | 0.446 | 0.520 | 0.464 | 0.470 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| PCR2 | |||||||||
| 0.018 | 0.011 | 0.014 | 0.002 | 0.008 | 0.034 | 0.013 | 0.024 | 0.019 | |
| 0.040 | 0.032 | 0.022 | 0.022 | 0.066 | 0.003 | 0.022 | 0.017 | 0.001 | |
| 0.472 | 0.482 | 0.480 | 0.496 | 0.488 | 0.446 | 0.520 | 0.464 | 0.470 | |
| 0.482 | 0.468 | 0.482 | 0.510 | 0.448 | 0.500 | 0.486 | 0.492 | 0.508 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.342 | 0.520 | 0.634 | 0.804 | 0.916 | 0.940 | 0.968 | 0.978 | 0.994 | |
| Latent Factor Regression | |||||||||
| LFR1 | |||||||||
| 0.505 | 0.506 | 0.509 | 0.513 | 0.513 | 0.512 | 0.519 | 0.518 | 0.516 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| LFR2 | |||||||||
| 0.501 | 0.493 | 0.500 | 0.500 | 0.501 | 0.500 | 0.503 | 0.507 | 0.501 | |
| 1.033 | 1.000 | 0.996 | 0.984 | 1.009 | 0.987 | 1.005 | 0.988 | 0.994 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.984 | 0.994 | 0.994 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.526 | 0.728 | 0.824 | 0.952 | 0.984 | 0.996 | 1.000 | 1.000 | 1.000 | |
Figure 9.
The average AIC scores (left panel) and average 10-fold cross-validation RMSE (right panel) over 500 datasets simulated from the LFR2 method, with the second stage model being normal linear regression.
The results demonstrate the profound implications involved with these analytic choices. As shown in the results based on the naive model, by including all the highly correlated explanatory variables in the model, the existence of multicollinearity can seriously bias the estimation of the covariate effect. Under the PCR approach, the regression coefficients for the PCs as the explanatory variables in the multiple linear regression model were estimated to be negative on average. The LFR2 method offers a solution to disentangle unique from shared contributions of the exposure effects on the outcome variable. The advantage of LFR2 in comparison to LFR1 is more pronounced when the VIF is not extremely high.
5.3.4. Additional simulation study results
In the simulation study, the exposure variables are all positively correlated; however, in some cases, the explanatory variable might be highly negatively correlated. To investigate the robustness of the performance of the proposed method, an additional set of the simulated study was carried out, where the explanatory variables are negatively correlated. The setting for this additional simulation study is very similar to the previous setting for positively correlated exposures. The results gave consistent conclusions about the ones when the explanatory variables were positively correlated. Moreover, in reality, the explanatory variables may have a negative effect on the outcome, so we conducted an additional simulation study by setting the regression coefficient, i.e. , as a negative value. Consistent conclusions were obtained. We also increased the sample size from n = 300 to n = 1000 in the simulation studies. The results and conclusions for comparing the methods remained similar.
6. Discussion
Using simulated datasets and real-life data, we showed that although PCR method can help reduce the dimensionality of the data into lower space, the interpretation of the effect of principal component scores on the outcome variable can be unrealistic and untenable. The other key finding of our study was to reveal the limitation of the conventional LFR method, i.e. LFR1 method, which regresses the outcome variable only on the common latent factor(s) and omits the unique effect of the exposure variables. In the circumstances when the exposure variables have an additional effect on the outcome variable in addition to the common effect driven by the latent factor(s), our study showed that ignoring the exposure-specific residual components resulted in an inferior model fit.
In contrast to the PCR and LFR1 methods of handling the multicollinearity problem of explanatory variables, we demonstrated the advantage of using LFR2 method to uncover the common and exposure-specific residual components of multiple correlated exposure variables, allowing the multiple highly correlated explanatory variables to be investigated simultaneously in the same model to predict the outcome. The common factor helps us understand how the outcome is simultaneously affected by the multiple covariates. The exposure-specific residual components capture any additional effect of the exposure variables on the outcome variable after accounting for the common effect of the exposure variables. For example, the real data analysis showed that the three pollutants PM2.5, PM10, and NO2 jointly increased the respiratory disease risk, and NO2 exhibited an additional effect on increasing the respiratory disease risk.
Several future directions of this study could be pursued. First, as environmental health data are often collected over geographical regions, spatial autocorrelation may exist underlying these data. However, when modeling the effects of exposures on the disease outcome, the highly correlated exposures and residual spatial autocorrelation could be confounded [29]. In this case, assessing the impact of multicollinearity among the explanatory variables on the disease risk can be challenging in the presence of residual spatial autocorrelation. Therefore, an improved method to appropriately separate the spatial effect from the effects of highly correlated exposure variables warrants future investigation.
Further, the latent common factor scores in the LFR method are assumed to be spatially independent. However, the common factor scores may be spatially correlated, with nearby locations having similar neighborhood environments. Incorporating spatial correlation in the latent factors could increase precision for parameter estimation gained by ‘borrowing of information’ from neighboring regions, especially when the data are sparse. In literature, common spatial factor models have been developed in the literature for studying the spatial variations of multivariate disease outcomes [6,18,40]. However, few studies have applied the spatial factor model to address the multicollinearity issue. Updating the LFR method presented in this article to properly account for this spatial correlation warrants future research.
In addition, in the proposed two-stage LFR method, the latent common factor and residuals are assumed to have a linear relationship with all the exposure variables, which are valid assumptions in our two case studies; however, the linearity assumption might not be true in other applications. The conventional factor analysis model has been extended to address non-linear structure underlying multivariate data [3], which could be used to tackle multicollinearity problems. More flexible non-parametric spline functions [41] could also be considered to model the non-linear effect between the exposure and the common factor. Similarly, in the second stage of the two-stage LFR method, the latent factor and exposure-specific residuals are assumed to be linearly related to the outcome variable. Such linearity assumption should also be carefully examined.
In our investigation, the LFR is a two-stage approach. In the first stage, the common and residual components are extracted based on factor analysis method. The estimated mean structures of the common and residual components are then used as the input variables in modeling the response variable in the second stage. Ideally, the uncertainty of the estimated common and residual components should also be accounted for. A single-stage model could be developed to achieve this goal by simultaneously modeling the exposure and response variables in one model with multiple equations. This type of model can be regarded as an extension of the structural equation model (SEM) [5], a multivariate statistical analysis technique used to analyze structural relationships. SEM is the combination of factor analysis and multiple regression analysis, which allows the simultaneous analysis of a set of regression equations to determine the relationships between latent and measured variables. However, in SEM, only the common factor(s) of the exposure variables are linked to the response variable(s) [30]. Our goal is not only to study the effects of the common factor(s) but also the residual components of the exposure variables on the response variable. Therefore, we are interested in extending the SEM to link the common factor(s) and the residual components of the exposure variables to the response variable. Further, when the data are measured repeatedly over time in the longitudinal setting, dynamic factor models [8,33,34] could be used to extract the underlying, latent, unobserved process. The latent structures from repeated measurements could then be linked to the response variable.
Acknowledgements
The authors would like to thank the Editor, Associate Editor, and two anonymous reviewers for their very insightful and valuable comments, which greatly helped improve the quality of the manuscript. The authors would also like to thank Dr. George Kephart, Professor at Dalhousie University for his very insightful comments and suggestions. The authors would like to thank the financial support from the Natural Sciences and Engineering Research Council of Canada for this research. The authors are also very grateful to Dr. Duncan Lee for providing the air pollution dataset in the real data analysis.
Funding Statement
This research was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant [RGPIN/70212 -2019].
Disclosure statement
No conflict of interests.
References
- 1.Agresti A., Foundations of Linear and Generalized Linear Models, Wiley, New Jersey, 2015. [Google Scholar]
- 2.Ambler G. and Benner A., mfp: Multivariable Fractional Polynomials (2022). Available at https://CRAN.R-project.org/package=mfp, R package version 1.5.2.2
- 3.Amemiya Y. and Yalcin I., Nonlinear factor analysis as a statistical method, Stat. Sci. 16 (2001), pp. 275–294. [Google Scholar]
- 4.Artigue H. and Smith G., The principal problem with principal components regression, Cogent Math. Stat. 6 (2019), p. 1622190. [Google Scholar]
- 5.Bollen K., Structural Equations with Latent Variables, John Wiley & Sons, New York, 1989. [Google Scholar]
- 6.Christensen W.F. and Amemiya Y., Latent variable analysis of multivariate spatial data, J. Am. Stat. Assoc. 97 (2002), pp. 302–317. [Google Scholar]
- 7.De Marco J.P. and Nóbrega C.C., Evaluating collinearity effects on species distribution models: an approach based on virtual species simulation, PLoS ONE 13 (2018), pp. 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engle R. and Watson M., A one-factor multivariate time series model of metropolitan wage rates, J. Am. Stat. Assoc. 76 (1981), pp. 774–781. [Google Scholar]
- 9.Feng C., Li J., Sun W., Zhang Y., and Wang Q., Impact of ambient fine particulate matter (PM2.5) exposure on the risk of influenza-like-illness: a time-series analysis in Beijing, China, Environ. Health Perspect. 15 (2016), pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox J., Applied Regression Analysis and Generalized Linear Models, 2nd ed., Sage, Los Angeles, CA, 2008. [Google Scholar]
- 11.Graham M.H., Factors determining the upper limit of giant kelp, Macrocystis pyrifera Agardh, along the Monterey Peninsula, central California, USA, J. Exp. Mar. Biol. Ecol. 218 (1997), pp. 127–149. [Google Scholar]
- 12.Graham M.H., Confronting multicollinearity in ecological multiple regression, Ecology 84 (2003), pp. 2809–2815. [Google Scholar]
- 13.Gunst R.F., Regresion analysis with multicollinear predictor variables: definition, derection, and effects, Commun. Stat. Theory Methods 12 (1983), pp. 2217–2260. [Google Scholar]
- 14.Gunst R.F., Comment: toward a balanced assessment of collinearity diagnostics, Am. Stat. 38 (1984), pp. 79–82. [Google Scholar]
- 15.Hadi A.S. and Ling R.F., Some cautionary notes on the use of principal components regression, Am. Stat. 52 (1998), pp. 15–19. [Google Scholar]
- 16.Hanspach J., Kühn I., Schweiger O., Pompe S., and Klotz S., Geographical patterns in prediction errors of species distribution models, Glob. Ecol. Biogeogr. 20 (2011), pp. 779–788. [Google Scholar]
- 17.Hoffmann J. and Shafer K., Linear Regression Analysis: Applications and Assumptions, NASW Press, North Carolina, 2015. [Google Scholar]
- 18.Hogan J.W. and Tchernis R., Bayesian factor analysis for spatially correlated data, with application to summarizing area-level material deprivation from census data, J. Am. Stat. Assoc. 99 (2004), pp. 314–324. [Google Scholar]
- 19.Hotelling H., Analysis of a complex of statistical variables into principal components, J. Educ. Psychol. 24 (1933), pp. 417–441. [Google Scholar]
- 20.Huang G., Lee D., and Scott E.M., Multivariate space-time modelling of multiple air pollutants and their health effects accounting for exposure uncertainty, Stat. Med. 37 (2018), pp. 1134–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson R.A. and Wichern D.W., Applied Multivariate Statistical Analysis, Pearson Prentice Hall, New Jersey, 2007. [Google Scholar]
- 22.Johnson R.W., Fitting percentage of body fat to simple body measurements, J. Stat. Educ. 4 (1996), p. null. [Google Scholar]
- 23.Jolliffe I.T., Principal Component Analysis, 2nd ed., Springer Series in Statistics, Springer, New York, 2002. [Google Scholar]
- 24.Kalnins A., Multicollinearity: how common factors cause type 1 errors in multivariate regression, Strat. Manag. J. 39 (2018), pp. 2362–2385. [Google Scholar]
- 25.Kutner M., Nachtsheim C., and Neter J., Applied Linear Statistical Models, McGraw-Hill/Irwin, New York, 2004. [Google Scholar]
- 26.Laden F., Neas L.M., Dockery D.W., and Schwartz J., Association of fine particulate matter from different sources with daily mortality in six U.S. cities, Environ. Health Perspect. 108 (2000), pp. 941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafi S. and Kaneene J., An explanation of the use of principal-components analysis to detect and correct for multicollinearity, Prev. Vet. Med. 13 (1992), pp. 261–275. [Google Scholar]
- 28.Lavery M.R., Acharya P., Sivo S.A., and Xu L., Number of predictors and multicollinearity: what are their effects on error and bias in regression?, Commun. Stat. Simul. Comput. 48 (2019), pp. 27–38. [Google Scholar]
- 29.Lee D., Rushworth A., and Sahu S.K., A Bayesian localized conditional autoregressive model for estimating the health effects of air pollution, Biometrics 70 (2014), pp. 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Wall M.M., and Hodges J.S., Generalized spatial structural equation models, Biostatistics 6 (2005), pp. 539–557. [DOI] [PubMed] [Google Scholar]
- 31.Mansfield E.R. and Helms B.P., Detecting multicollinearity, Am. Stat. 36 (1982), pp. 158–160. [Google Scholar]
- 32.Massy W.F., Principal components regression in exploratory statistical research, J. Am. Stat. Assoc. 60 (1965), pp. 234–256. [Google Scholar]
- 33.Molenaar P., A dynamic factor model for the analysis of multivariate time series, Psychometrika 50 (1985), pp. 181–202. [Google Scholar]
- 34.Molenaar P., De Gooijer J.G., and Schmitz B., Dynamic factor analysis of nonstationary multivariate time series, Psychometrika 57 (1982), pp. 333–349. [Google Scholar]
- 35.Myers R.H., Classical and Modern Regression with Applications, 2nd ed., Duxbury Advanced Series in Statistics and Decision Sciences, PWS-KENT PubCo, Boston, 1990. [Google Scholar]
- 36.Pituch K. and Stevens J., Applied Multivariate Statistics for the Social Sciences Analyses with SAS and IBM's SPSS, Wiley, New Jersey, 2015. [Google Scholar]
- 37.Powell H. and Lee D., Modelling spatial variability in concentrations of single pollutants and composite air quality indicators in health effects studies, J. R. Stat. Soc. Ser. A (Stat. Soc.) 177 (2014), pp. 607–623. [Google Scholar]
- 38.Song Z., Chomicki K.M., Drouillard K., and Weidman R.P., A statistical framework for testing impacts of multiple drivers of surface water quality in nearshore regions of large lakes, Sci. Total Environ. 811 (2022), p. 152362. [DOI] [PubMed] [Google Scholar]
- 39.Vatcheva K.P., Lee M., McCormick J.B., and Rahbar M.H., Multicollinearity in regression analyses conducted in epidemiologic studies, Epidemiology 6 (2016), pp. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F. and Wall M.M., Generalized common spatial factor model, Biostatistics 4 (2003), pp. 569–582. [DOI] [PubMed] [Google Scholar]
- 41.Wood S.N., Inference and computation with generalized additive models and their extensions, Test 29 (2020), pp. 307–339. [Google Scholar]