Abstract
The comparison of Receiver Operating Characteristic (ROC) curves is frequently used in the literature to compare the discriminatory capability of different classification procedures based on diagnostic variables. The performance of these variables can be sometimes influenced by the presence of other covariates, and thus they should be taken into account when making the comparison. A new non-parametric test is proposed here for testing the equality of two or more dependent ROC curves conditioned to the value of a multidimensional covariate. Projections are used for transforming the problem into a one-dimensional approach easier to handle. Simulations are carried out to study the practical performance of the new methodology. The procedure is then used to analyse a real data set of patients with Pleural Effusion to compare the diagnostic capability of different markers.
KEYWORDS: Bootstrap, covariates, hypothesis testing, projections, ROC curves
1. Introduction
In any classification problem such as a diagnostic method – in which the aim is to discriminate between two populations, usually identified as the healthy population and the diseased population – the main concern is to minimize the number of subjects that are misclassified. Receiver Operating Characteristic (ROC) curves are commonly used in this context for studying the behaviour of the classification variables [see, for example, the monograph of 19, as an introduction to the topic]. They combine the notions of sensitivity (the ability of classifying a diseased patient as diseased) and specificity (the ability of classifying a healthy individual as healthy), two measurements that can be expressed in terms of the cumulative distribution functions of the diagnostic variables of the diseased and the healthy populations.
When there is more than one variable for diagnosing a certain disease one can compare their respective ROC curves in order to decide whether their discriminatory capability is different or not. This is what happens in the medical example that we will be analysing in this paper, a real data set containing the information of patients with pleural effusion. In this data set there are two variables (the carbohydrate antigen 152 and the cytokeratin fragment 21-1) that can be used for deciding whether that pleural effusion is due to the presence of a malignant tumour or not. The objective of the analysis will be to compare the diagnostic capability of those markers.
There are several methodologies discussed in the literature for making that sort of comparisons [for a review of such methodologies, see 6], although most of them do not consider the possible effect that the presence of covariates can have in the performance of the test. In the example provided, apart from the diagnostic variables there are other covariates such as the age or the neuron-specific enolase of the patients. It is important to take this information into account, because the diagnostic capability of a marker may change with the value of a covariate [17].
In this paper the aim is to propose a test to compare ROC curves that includes the presence of a multidimensional covariate in the analysis. With this methodology, given a new patient waiting for a diagnosis on his or her pleural effusion, we could compare the different diagnosis mechanisms taking into account the covariate values of this particular individual and see which one would be the most appropriate.
One way of introducing the effect of the covariates into the study is by using the conditional ROC curve. If we consider and as the continuous diagnostic markers in the diseased and healthy populations, respectively, as the continuous dimensional covariate of the diseased population and as the continuous dimensional covariate of the healthy population, then, given a fixed value (where is the intersection of and , the supports of and , and is assumed to be non-empty), the conditional ROC curve is defined as
| (1) |
where , and .
By comparing these conditional ROC curves instead of the standard ROC curves it is possible to incorporate the potential effect of the covariates in the analysis of the equivalence of two or more methods of diagnosis. A test for performing this comparison is proposed in [7] for the case of a continuous one-dimensional covariate. The objective here is to extend that methodology to the case in which we have a multidimensional covariate. Thus, the aim is to test, given a certain ,
| (2) |
where K is the number of diagnostic markers (and thus, ROC curves) that are being compared. In this context we would have K diagnostic variables and one dimensional covariate in the healthy population, , and similar variables in the diseased population, . In practice this kind of test could help to design a more personalized diagnostic method based on the covariate values of each patient. With this methodology, in the medical example at hand we could determine whether the carbohydrate antigen 152 and the cytokeratin fragment 21-1 are equally suitable for the diagnosis of a patient with a certain age and a certain enolase value.
In order to be able to make this comparison, we are going to rely on the estimation of the corresponding conditional ROC curves. There is a wide range of estimation methods in the literature: some of them estimate the conditional distribution functions involved in the definition of the conditional ROC curve, others use regression functions to include the effect of the covariates (following direct or indirect approaches). See [17] for a further review of this topic.
In [7] the estimation of the conditional ROC curve that is used is based on the indirect (or induced) regression methodology, which incorporates the covariate information through regression models by considering the effect of those covariates in the diagnostic marker in each population of healthy or diseased separately. However, this method was originally designed for one single covariate. One could think of extending that methodology by changing the estimator of the conditional ROC curve for another capable of handling multidimensional covariates. Nevertheless, there are not many methods in the literature capable of considering more than one covariate when estimating the conditional ROC curve, and most of them have some parametric assumptions that we would like to avoid making. See [11] as an example of a non-parametric Bayesian model to estimate the conditional distribution functions involved in the ROC curves, [13], [21] or [22] as examples of a direct ROC regression model (where generalized additive models are used to directly regress the ROC curve) or [20] as an example of induced methodology (framed in a Bayesian setting). In our case we will be following a frequentist approach.
The tests related to multidimensional data tend to become less powerful when the dimension of the problem increases. This is why, in this paper, the problem of comparing conditional ROC curves is first transformed using projections in such a way that the multidimensional problem becomes a unidimensional problem easier to handle. This idea has been applied several times in the literature for reducing the dimension in goodness-of-fit problems [see, for example, 5,8,18], but, to the best of our knowledge, it is the first time that it is applied on an ROC curve setting. In the last few years random projections are increasingly being used as a way to overcome the curse of dimensionality. The characterization of the multidimensional distribution of the original data by the distribution of the randomly projected unidimensional data is what allows for the reduction of the dimension.
To that end, in Section 2, which is dedicated to the exposition of the methodology introduced in this paper, we begin by showing how (2) can be transformed in a test with one-dimensional covariates using projections. In Section 2.2 we show how to compare ROC curves in that simplified setting, with a unidimensional covariate and in Section 2.3, which contains the major contribution of this paper, we propose a methodology for testing that equivalent hypothesis with a multi-dimensional covariate. This includes the proposal of a bootstrap algorithm to approximate the distribution of the statistic under the null hypothesis. In Section 3 the results from a simulation study show the practical performance of the test in terms of level approximation and power. The procedure is illustrated in Section 4 by analysing the real data set containing information of patients with pleural effusion.
2. Methodology
This section is divided in three subsections. In the first one, Section 2.1, we present a result that allows us to transform the problem discussed in (2) into an equivalent one, easier to handle, by using projections to reduce the multidimensional role of the covariate to a unidimensional one.
In Section 2.2 we show a methodology to test the equality of conditional ROC curves on a unidimensional problem [based on the one proposed in 7]. Finally, in Section 2.3, we combine that methodology with the result obtained in Section 2.1 to solve our original problem with multidimensional covariates. Both Sections 2.2 and 2.3 include the statistic proposed to perform the test and a bootstrap algorithm to approximate its distribution.
2.1. An equivalent problem
In order to present the transformation of the problem, first we need to introduce the definition of the ROC curve conditioned to a pair :
| (3) |
This concept is very similar to the conditional ROC curve (1): the only difference is that this new definition allows us to condition on different values for the diseased and healthy populations. In this case and are unidimensional, but the definition could be applied on a multidimensional case. Even if the interpretability of this new ROC curve is not very clear in practice, theoretically it does not present any problems (as it will not do its estimation), as the population of healthy and diseased are always considered to be independent.
The following result is the base for developing the test for comparing ROC curves with multidimensional covariates. It borrows the ideas in [5] of using projections for reducing the dimension of the covariate in a regression context. Since here we are dealing with ROC curves, the dimension reduction is less straightforward and some adjustments are required, as each ROC depends on two cumulative distribution functions. To the best of our knowledge, the idea of using projections has not been considered in the context of ROC curves.
Given , denotes the scalar product of the vectors and . For now on, all the vectors representing the projections will be considered to be contained in the d-dimensional unit sphere . This way we ensure that all possible directions are equally important.
Lemma 2.1
Assume and for every . Then, given a certain , and assuming dependence among the ROC curves (meaning the covariate is common for all the K curves considered), then
if and only if
where and are dimensional coordinates in that represent the directions of the projections.
The proof of this Lemma can be found in the Appendix. Note that and are one-dimensional values. By using these ROC curves conditioned to a pair of projected covariates (as defined in 3), the problem is reduced to a one-dimensional covariate conditional ROC curve comparison test for each possible direction and .
Thus, taking advantage of the result in Lemma 2.1, instead of testing for the null hypothesis (2), we may use this equivalent formulation to develop a methodology that, given a certain , tests
| (4) |
against the general alternative is not true. The notation ∀ will be used instead of ‘for any’ to shorten the expression (this applies mainly in the proofs found in the Appendix).
Note that values of the projections do not have a meaning on their own: there is not optimal direction of the coefficients to be found, they are all equally important. This methodology should not be confused with the search for the best linear combination of markers for developing new diagnostic tests, like the ones proposed in [23] or in [12]. Here we are combining the components of a multidimensional covariate to perform a test about the performance of two or more markers.
In a first step, a statistic for testing the equivalence of these ROC curves is presented for a certain pair of fixed projections, and then that statistic is adapted to include all possible directions.
2.2. Test for a one-dimensional covariate
The objective in this section is to develop a test for the equivalent problem presented in Lemma 2.1 for a fixed pair of projections and . Here a test is presented for comparing two or more dependent ROC curves conditioned to two one-dimensional values. Given the pair , the aim is then to test
| (5) |
against the general alternative is not true.
The samples available in this context are:
an i.i.d. sample from the distribution of ,
an i.i.d. sample from the distribution of ,
with and the sample sizes of the diseased and healthy populations, respectively. Define as the total sample size used for the estimation of each conditional ROC curve (that will be the same for all ). Note that both and are here one-dimensional covariates.
The method used for the estimation of the conditional ROC curves is based on the one proposed in [9], which relies on non-parametric location-scale regression models. To be more precise, for each , assume that
| (6) |
| (7) |
where, for , and are the conditional mean and the conditional variance functions (both of them unknown smooth functions), and the error is independent of . The dependence structure between the K diagnostic variables is modelled by introducing a dependence structure between the errors: will follow a multivariate distribution function with zero mean and a covariance matrix with ones in the diagonal.
Given this location-scale regression model structure for the diagnostic variables, the k-th ROC curve conditioned to a pair of values can be expressed in terms of the marginal cumulative distribution functions of the errors, and :
| (8) |
where
Thus, this k-th conditional ROC curve can be estimated by
| (9) |
where, for ,
,
, with ,
is a non-parametric estimator of based on local weights depending on a bandwidth parameter ,
is a non-parametric estimator of . For simplicity we take the same bandwidth parameter that is used for the estimation of the regression function ,
are Nadaraya–Watson-type weights, where and κ is a probability density function symmetric around zero,
and ,
is a bandwidth parameter responsible for the smoothness of the estimator. Its value does not seem to have a significant effect on the conditional ROC curve estimation.
This way of estimating the conditional ROC curve is similar to the one proposed in [9], with the difference that they condition the ROC curve on a single value x and here we have a pair of values and , each one of them related to the diseased and the healthy population, respectively. As both populations are independent, the adaptation of the methodology of [9] to this case is straightforward.
Once we know how to estimate this doubly conditional ROC curve we can propose a test statistic for the test (5):
| (10) |
where:
for , , where and are bandwidth parameters involved in the estimation of the kth conditional ROC curve.
for , is the estimated conditional ROC curve given , as seen in (9),
is a sort of weighted average of the K conditional ROC curves.
- ψ is a real-valued function that measures the difference between each estimated conditional ROC curve and the weighted average of all of them. This function may be similar to the ones used for the comparison of cumulative distribution functions (after all, a ROC curve can be viewed as a cumulative distribution function). For example, if one considers the -measure, then the resulting test statistic is
On the other hand, when using the Kolmogorov–Smirnov criteria the resulting test statistic is
The null hypothesis will be rejected for large values of . In order to obtain the distribution of this statistic, a bootstrap algorithm is proposed. This bootstrap algorithm is adapted from the procedure proposed in [14] and has been already used by Martínez-Camblor et al. [15] and by Fanjul-Hevia et al. [7] in the context of ROC curves. The key of this algorithm is that
coincides with the statistic as long as the null hypothesis holds, where
The quantity can be rewritten as
| (11) |
where . Note that, in general, cannot be computed from the data, as it depends on the unknown theoretical conditional ROC curves, but it is useful when applying the bootstrap algorithm.
The bootstrap algorithm suggested to approximate a p-value for this test is the following:
-
A.1
From the original samples, and , compute the test statistic value (10), that we will denote by .
-
A.2For , generate the bootstrap samples and as follows:
- For each , let be an i.i.d. sample from the empirical cumulative multivariate distribution function of the original residuals.
- Reconstruct the bootstrap samples for each , where .
-
A.3Compute the test statistic based on the bootstrap samples, for using (11) as
where is the estimated j-th conditional ROC curve of the b-th bootstrap sample. -
A.4The distribution of under the null hypothesis (and thus, the distribution of ) is approximated by the empirical distribution of the values and the p-value is approximated by
In contrast with the usual bootstrap algorithms in testing setups, in this case the null hypothesis is not employed when generating of the bootstrap samples (Step A.2), because replicating the null hypothesis of equal ROC curves is not a straightforward problem. Instead, it is used in the computation of the bootstrap statistic (Step A.3) by using instead of , that are equal under the null hypothesis. This particularity also appears in the bootstrap algorithm of the next section.
There are two kind of bandwidth parameters that appear in the estimation of the th conditional ROC curve (9), with . The first one, , is taken as , and the second ones, and , are selected by least-squares cross-validation. Note that, for each bootstrap iteration, the bandwidth parameters could change, as their selection depends on the sample. However, remains constant, as we are choosing it in terms of the sample size, and that is the same for each bootstrap iteration. As for and , for computational issues we have decided to compute them on step A.1 using the original sample, and then apply the same bandwidths for all the bootstrap estimations. The cross-validation method can be very time-consuming, and this simplification prevents the simulations to become infeasible.
2.3. Test for a multidimensional covariate
In the previous subsections we have shown how to transform our original multidimensional problem into a one-dimensional one by using projections and how to test the equality of ROC curves conditioned to a given pair (and more particularly, to a fixed pair of directions). It is time now to resume our main objective, which was to compare ROC curves conditioned to a multidimensional covariate.
Once having seen a strategy for testing (4) for only one pair of fixed directions, the idea now is to modify the previous procedure so the new statistic takes into account all the possible directions that and can take. For that purpose, consider the test statistic
| (12) |
where and represent the uniform density on the sphere of dimension d, . This ensures that all directions are equally important.
The expression is equal to the statistic used in (10) for testing the equality of K ROC curves when conditioned to the value of the pair , that is,
Note that, in this context with dimensional covariates, the samples are and , with and .
In practice, as it is done in [1], to compute the test statistic random directions and are drawn uniformly from , where is the number of random directions considered (the same number of directions is taken for and for ). With them, the approximated statistic is
| (13) |
In order to obtain the distribution of the statistic, a bootstrap algorithm (similar to the one described in the previous section) is proposed. To do so, the following expression is introduced:
| (14) |
where is the same as in (11), but for the conditioning values of :
As it happened in (11), cannot be computed without knowing the true distribution of the diagnostic markers. However, it can be computed in the bootstrap algorithm below, and there is approximated by
| (15) |
As happened before, for two given projections and , and coincide as long as the null hypothesis holds, and thus the same happens with and .
Taking into account these approximations, the resulting bootstrap algorithm goes as follows:
-
B.1Draw random directions and uniformly from . This can be done by using the method proposed by Muller [16] to generate points in a multidimensional sphere. For each random direction , with and :
- Generate d values independently from a normal standard distribution: .
- Consider the projection obtained by normalizing the vector :
-
B.2
For each random directions and (with ), consider the sample and and the conditioning values . With them, following steps A.1–A.3 of the bootstrap algorithm of the previous subsection, compute the value of and the B corresponding .
- B.3
-
B.4Approximate the p-value of the test by:
Remark 2.1
Note that represents the number of random directions drawn from considered for the approximation of (13) and (15), but that, in fact, we are using different combination of pairs to make that approximation. This could become a problem from the computational point of view, as the complexity of the problem increases very fast when increasing the value of .
As an alternative, we could consider using
instead of statistic (12), where represents the uniform density on the torus of dimension d, . This ensures, as before, that all pairs of directions are equally important. Thus, in practice, instead of using the approximation (13) we could consider
where are pairs of random directions drawn uniformly from , and where would represent here the same as before, with the advantage that it allows for more flexibility because it can assume non-squared values. A similar adaptation could be applied for the approximation of in (14).
Remark 2.2
In the literature we can find papers, like for example [2] or [3], that use only one random projection. The main idea is to perform the test at hand for a randomly selected projection instead of for all possible projections. The use of projections results in a dimension reduction (as desired), and, despite being a procedure that may produce less powerful tests, the use of one single projection results in a reduction of the computational cost.
Following that idea, instead of testing the equality of covariate-projected ROC curves for all possible projections, we could test the equality of covariate-projected ROC curves for some random pair of projections given a certain , meaning:
(16) The equivalence between this hypothesis and the one of interest in this paper given in (2) still needs theoretical justification. However, it is a possibility worth studying, if only for computational reasons. A way of perform this approach could be to consider the proposed methodology for .
Additionally, in a practical situation we should take into account that the different magnitudes of the covariates that we are projecting may obscure the effect of some of them. In order to prevent this from happening we suggest standardizing the multidimensional covariate before we start the analysis. Note that an ROC curve conditioned to a certain value is the same as the ROC curve in which the covariate is modified by a one-to-one transformation and the ROC curve is conditioned to the corresponding transformed value.
Given a non-degenerate multidimensional covariate the standardization proposed here is to consider the multidimensional covariate , with a diagonal matrix with in the diagonal and . Then, for a given variable Y, a given and a certain value of the covariate ,
with and, thus,
The standardization that takes place here does not care for the covariance between the covariates that conform , as we are only interested on obtaining covariates with similar magnitudes. Also, in practice the standardization is made considering the sample mean and the sample standard deviation of the covariates at hand.
3. Simulations
In order to analyse the performance of the proposed methodology, simulations were run for the comparison of several dependent conditional ROC curves. On a first stage, these simulations were focused on analysing the behaviour of the unidimensional test described in Section 2.2, but we do not display them here, as they are very similar to the ones that can be found in [7]. Instead, we show the results for several scenarios (first under the null hypothesis and then under the alternative) in which we compare K ROC curves (with ) conditioned to a dimensional covariate (with ).
All the curves used in the simulation study were drawn from location-scale regression models similar to the ones presented in (6) and (7), only that, in this case, the regression and the conditional standard deviation functions are for d-dimensional covariates. The construction of those curves is summarized in Table 1, were all the different conditional mean and conditional standard deviation functions are displayed.
Table 1.
Conditional mean and conditional standard deviation functions of the conditional ROC curves considered in the simulation study.
| Covariate | ROC curves | Regression functions | Conditional standard deviation functions |
|---|---|---|---|
The regression errors were considered to have multivariate normal distribution with zero mean, variance one and correlation ρ for all the models.
In all scenarios the covariates , , , , and are uniformly distributed in the unit interval. Thus, the value of the multidimensional covariate at which the conditional ROC curves should be compared is contained in . Particularly, the comparisons are made for and for , for d = 2 and d = 3, respectively.
The study contains simulations for different sample sizes , , and different values of ρ that represent different possible degrees of correlation between the diagnostic variables under comparison ( ).
Moreover, two different functions ψ were considered for the construction of : one based on the measure and the other one based on the Kolmogorov-Smirnov criterion (from now on denoted by and KS, respectively). The number of iterations used in the bootstrap algorithm was 200, and 500 data sets were simulated to compute the proportion of rejection in each scenario.
Furthermore, the number of directions that was used for approximating the test statistic was taken as (as mentioned in Remark 2.1, notice that this means that different pairs of directions were considered).
3.1. Level of the test
The scenarios that were considered for calibrating the level of the test (by comparing the same conditional ROC curves) are represented in Table 2. The results of the simulations obtained for are summarized in Figures 1 (for d = 2) and 2 (for ). Each subfigure represents the test of one scenario for a particular sample size. The nominal level considered is 0.05. The estimated proportion of rejections over 500 replications of the data sets is represented along with the rejection region of such nominal level. For the test to be well calibrated the estimated proportions should fall between the gray lines.
Table 2.
Scenarios under the null hypothesis considered for calibrating the level of the test.
 |
Figure 1.
Estimated proportion of rejection under the null hypothesis and the corresponding limits of the critical region (in gray) for the level 0.05 (dotted black line) with d = 2 and for different sample sizes and different ρ.
Figure 2.
Estimated proportion of rejection under the null hypothesis and the corresponding limits of the critical region (in gray) for the level 0.05 (dotted black line) with d = 3 and for different sample sizes and different ρ.
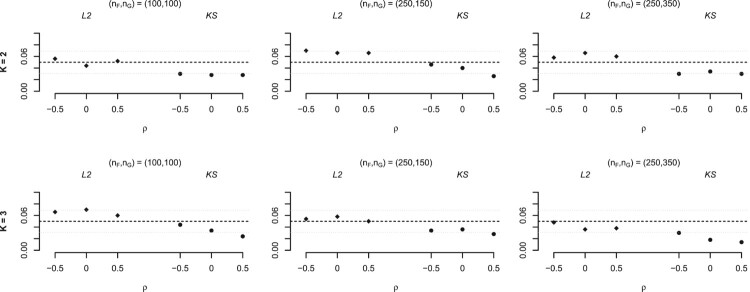
In general it can be said that the expected nominal level is reached, as most of the estimated proportions are close to the corresponding nominal level. The statistic seems to overestimate the level in a few scenarios, but its behaviour improves when increasing the sample size. The KS statistic is a little more conservative.
3.2. Power of the test
On the other hand, the scenarios that were considered for studying the power of the test (by comparing different conditional ROC curves) are represented in Table 3. The results of the simulations are summarized in Figure 3 (for ). In those figures the first and second row represent the simulation results for the scenarios with K = 2 and K = 3, respectively, and the first and the second column represent the simulation results for d = 2 and for d = 3, respectively. In this case, only was considered.
Table 3.
Scenarios under the alternative hypothesis considered for calibrating the power of the test.
 |
Note: and are represented in purple, and in green, and and in yellow.
Figure 3.
Estimated proportion of rejection under the alternative hypothesis for different sample sizes and different ρ, for ( ).
It can be seen that the power of the test grows with the considered sample sizes. The statistic yields higher power than the KS statistic, which is consistent with KS being more conservative. Moreover, the difference between the conditional ROC curves considered for the case of d = 2 is bigger than the difference between the ROC curves in the scenarios with d = 3, which translates in higher power for the cases in which d = 2.
We can also observe that for each scenario, the highest power is always obtained for the cases in which the correlation of the diagnostic variables is , and the lowest for .
Note that for the scenario with d = 3 and the power of the test does not increase significantly from the first sample size to the second (in fact, for K = 3 it even decreases a little), but this can be due to the fact that the lower sample size has balanced data, ( , ) being (100,100). whereas for the second sample size considered ( , ) take the value (250,150). The highest sample size is also unbalanced, but not so much.
Remark 3.1
In order to evaluate the modification of the method proposed in Remarks 2.1 and 2.2 we have run simulations for the same scenarios previously described. We show here the results for the scenarios with K = 2 and d = 2 under the null and the alternative hypotheses for assessing the level and the power of the test, respectively. Similar conclusions were obtained with the rest of the scenarios. The parameters that are used here are the same as before, with the exception that now 1000 data-sets were simulated instead of 500.
Figure 4 shows the results of the simulations when considering the modification of Remark 2.1 for (first row) and (second row), and the results for considering only one random projection (Remark 2.2), i.e. (third row). Note that taking is comparable with used in the previous simulations (see first row of Figure 1), and that the results are very similar: the estimated proportion or rejections is a little overestimated for the statistic for the smaller sample size and otherwise close to the nominal level, and the KS statistic is always more conservative. Increasing from 25 to 50 does not seem to affect the results significantly, and neither does reducing it to a single random projection ( ).
In Figure 5 we can observe the results for the simulations under the alternative hypothesis, once again for , and . The firs two graphics are very similar to the one obtained for (see the first graphic of Figure 3), but from the last graphic it is obvious that by using only one random projection the power of the test decreases considerably (as it was expected).
In the light of these results it seems that the alternative methodology proposed in Remark 2.1 yields similar conclusions than the first proposal, with no noticeable gain when increasing the number used to approximate the value of the statistic from 25 to 50. It remains an open problem to determine an optimal value for that parameter.
As for the idea mentioned in Remark 2.2, using only one random projection seems to produce a well calibrated test, despite having considerably lower power.
Figure 4.
Estimated proportion of rejection under the null hypothesis and the corresponding limits of the critical region (in gray) for the level 0.05 (dotted black line) with K = 2, d = 2 and for different sample sizes and different ρ.
Figure 5.
Estimated proportion of rejection under the alternative hypothesis for different sample sizes and different ρ, for and for the scenarios with K = 2 and d = 2.
3.3. Some extra simulations: changing the distribution of the covariates
The previous simulations were all obtained for scenarios in which the covariates are uniformly distributed in the unit interval. The point at which the comparisons where made was selected ensuring that it has enough data around. However, in practice the covariates may not follow a uniform distribution and the may even behave differently in the diseased and the helathy populations. This may result in scenarios where the point is closer to the boundaries of the supports of the covariates or in scenarios where it would be advisable to standardize the covariates with different magnitudes.
In this section we repeat the simulations for two of the scenarios considered previously, changing just the distribution of the covariates. The first one is the scenario under the null hypothesis where we compare two ROC curves equal to (defined in Table 1) for d = 2, with and , at . The second one is the scenario under the alternative hypothesis where we compare two curves, and , also for d = 2, with and and at . Both scenarios are represented in Tables 2 and 3. Note that it does not matter if we change the distribution of the covariates: the conditional ROC curve remains the same. The simulations were run in similar conditions as before, with , 200 bootstrap iterations and 500 replications of the data sets.
We considered a total of 16 new models for the covariates, with different combinations of the distributions of , , and . Models A.1–A.4 follow uniform distributions; the difference between them is that the point is almost at the centre of the support of the first model A.1, but as we move from A.1 to A.4 it gets closer and closer to the boundary of the support. The same applies for models B.1–B.4, but now with normal distributions. In models C.1–C.4 we add the feature that the the four variables have different distributions, and that the healthy and the diseased populations grow apart for each model. The distributions of the covariates in each case are described in Table 4. We also consider the models D.1–D.4, that used the same distributions than in C.1–C.4 but for which we applied the standardization recommended at the end of Section 2.
Table 4.
Distributions of the covariates considered for the simulations.
| A.1 | ||||
| A.2 | ||||
| A.3 | ||||
| A.4 | ||||
| B.1 | ||||
| B.2 | ||||
| B.3 | ||||
| B.4 | ||||
| C.1 | ||||
| C.2 | ||||
| C.3 | ||||
| C.4 |
Note: Models D.1–D.4 are not included as they are a standardization of models C.1–C.4.
The results for the scenario under the null hypothesis are summarized in Figure 6, considering the 16 different combinations of covariate distribution for both the and the KS type of statistic. The estimated proportion of rejections and its rejection region is presented as before for nominal level 0.05.
Figure 6.
Estimated proportion of rejection under the null hypothesis and the corresponding limits of the critical region (in gray) for the level 0.05 (dotted black line) for different combinations of distribution for the bidimensional covariates (models A.1–A.4–D.1–D4) for both the and the KS statistics.
The simulations show that the test is well calibrated regardless of the distribution of the covariates. It shows some problems when we take the point too close to the boundaries or in a place where there is limited data (models .4), but it is also expected considering that the sample size that we are using is not too large, . Models .1 and .2 are comparable with the ones studied in previous sections, and only A.3 of the .3 models overestimates the level of the test for the statistic. Also, the good behaviour of the test for models D.1–D.4 shows that the standardization proposed in this methodology does not affect the result of the test.
The results of the simulations for the scenario under the alternative hypothesis are collected in Table 5. Leaving aside the differences between the results for the and the KS statistic, that were observed also in the previous sections, the power of the test does not change significantly when the covariate distribution changes, and it is comparable to the power obtained when we compared the same conditional ROC curves in the prevoius section. As it could be expected, it decreases when we move from the models with more data (.1) to the models with less data (.4) around the point . Moreover, the power experiments some increasing from models in C to models in D for the statistic, which means that the standardization does not worsen the behaviour of the test. Note that in this case the models in C do not need the standardization for the test to work, as the difference of magnitude of the covariates involved is minimal.
Table 5.
Estimated proportion of rejection under the alternative hypothesis for different combinations of distribution for the bidimensional covariates (models A.1–A.4–D.1–D4) for both the and the KS statistics.
| KS | ||||||||
|---|---|---|---|---|---|---|---|---|
| Models | ||||||||
| A | 0.670 | 0.636 | 0.590 | 0.530 | 0.520 | 0.454 | 0.426 | 0.386 |
| B | 0.650 | 0.614 | 0.580 | 0.538 | 0.470 | 0.472 | 0.420 | 0.386 |
| C | 0.642 | 0.626 | 0.602 | 0.588 | 0.464 | 0.458 | 0.440 | 0.424 |
| D | 0.652 | 0.638 | 0.614 | 0.600 | 0.452 | 0.458 | 0.438 | 0.424 |
4. Application
An illustration of the proposed test is displayed in this section through the analysis of the previously mentioned data set concerning 463 patients with pleural effusion. This data set has been provided by Dr. F. Gude, from the Unidade de Epidemioloxía Clínica of the Hospital Clínico Universitario de Santiago (CHUS), and it has been used for a previous study in [24].
From a medical perspective, the goal is to find a way to discriminate the patients in which the pleural effusion (PE) has a malignant origin (MPE) from those in which the PE is due to other non-cancer-related causes. 200 individuals form the sample had MPE (the diseased population in this context), against 263 who did not (healthy population). For that matter, two diagnostic markers were considered, the carbohydrate antigen 152 (ca125) and the cytokeratin fragment 21-1 (cyfra). Moreover, the information of two different covariates is also available: the age and the neuron-specific enolase (nse). Due to the characteristics of the data (positive values, most of them close to zero, with some extreme high values), logarithms of those variables – excluding the variable age – were considered for the study. Being the logarithm a monotone transformation, its use does not have an effect on the estimation of the common ROC curve. However, it does affect the estimation of the conditional ROC curves, as it reduces the effect of the more extreme values of the variables. A representation of the relationship of each one of those biomarkers with the two covariates is depicted in Figure 7, for both MPE (green) and the non-MPE (blue) patients.
Figure 7.
Scatterplots of the two different diagnostic biomarkers in function of the two covariates considered: age and . Contour plots were added in the bidimensional marginal scatterplots. The healthy subjects are represented in blue and the diseased ones in green (in the printed version, the healthy subjects appear in a darker colour than the diseased subjects).
It can be observed that the shape of the point clouds of the two populations changes with the values of the covariates, specially in the case of the diseased population.
In order to evaluate whether the discriminatory capability of those markers ( and as the variables containing the information of , and and as the variables containing the information of ) is the same when the covariates age and are taken into account, the methodology explained in previous sections is applied, comparing their respective ROC curves conditioned to different values of the bidimensional covariate with age and . In order to explore the advantages of using this method over the ones that do not consider multidimensional covariates, we also test the equivalence of the ROC curves of those diagnostic markers for the case in which no covariates are taken into account and for the case in which only one of the covariates is included in the analysis. Figure 8 shows how those two covariates are distributed in the diseased and healthy populations. The scatterplot included there highlights also the pair of values of the bidimensional covariate at which we are going compare the ROC curves in this analysis. Note that the covariates have different magnitudes: the values that the variable age takes are always going to be bigger than the values of . Thus, if we were to use the procedure directly over these variables, when projecting the multidimensional covariate on any direction, the effect of the second component will be overshadowed by the first component's. To prevent this from happening we decided to use the standardized variables of and instead of the originals. This also affects the value at which the conditional ROC curves are being compared. So in this case, instead of the multidimensional covariate and the conditional value , we consider the standardized version and , with and the sample standard deviation of the marginal covariates.
Figure 8.
Histograms, boxplots and scatterplots (with the corresponding contour plots) of the two covariates considered (age and ). The healthy subjects are represented in blue and the diseased ones in green (in the printed version, the healthy subjects appear in a darker colour than the diseased subjects). The black histogram lines and the white boxplot correspond to the two populations of the healthy and the diseased patients combined. The red points in the scatterplot correspond to the values of at which the ROC curves are compared.
We start the analysis of the performance of the two diagnostic markers by comparing their respective ROC curves without taking into account any covariate information. For that matter we use the method proposed by DeLong et al. [4]. The estimated ROC curves for both markers are depicted in Figure 9. The p-value obtain for that comparison was 0.138. Similar results were obtained when using other ways of comparing ROC without covariates (like [15] or [25]). Thus, we do not find significant differences between the two diagnostic variables in terms of diagnostic accuracy.
Figure 9.
ROC curve estimation for both diagnostic variables (log(ca125) and log(cyfra), represented by the solid and the dashed line, respectively) without covariates and conditioned to different values of the covariates age and .
Next, we compare the two diagnostic markers taking into account a unidimensional covariate using the test proposed in [7] for dependent diagnostic markers. We consider the covariates age and , each one at a time. We test the equality of the ROC curves conditioned to the values of in the case of age and the values of in the case of . The corresponding ROC curve for every case is estimated in Figure 9. For each considered covariate and each value of the covariate we obtain a p-value of the test, summarized in Table 6. The test is made considering two types of statistics, one based on the -measure and the other in the Kolmogorov-Smirnov criteria, although both of them yield similar results. When comparing the ROC curves conditioned on different values of the age, the results are in line with the obtained for the previous case, in which no covariates where taken into account: the equality of the two curves is not rejected. However, when considering the covariate , we see that for a certain value (1.14) the null hypothesis is rejected (for a significance level of 5 ). This matches the representation of the conditional ROC curves depicted in Figure 9.
Table 6.
Results for the comparison of the ROC curves of the diagnostic markers and when considering a unidimensional covariate, that covariate being the age or the .
| age | 51 | 67 | 83 |
| p-values ( ) | 0.454 | 0.218 | 0.936 |
| age | 51 | 67 | 83 |
| p-values (KS) | 0.512 | 0.202 | 0.762 |
| -0.92 | 1.14 | 3.20 | |
| p-values ( ) | 0.844 | 0.012 | 0.470 |
| -0.92 | 1.14 | 3.20 | |
| p-values (KS) | 0.900 | 0.008 | 0.412 |
Finally, we compare the performance of the two diagnostic variables considering the effect of both the age and the at the same time. This is where we use the methodology proposed in this paper. We test the equality of their respective ROC curves conditioned to nine pairs of values of the two covariates: the ones obtained by making all the possible combinations of and . As before, two different type of statistics were considered: and KS (and once again, the results are similar in both cases). The results obtained are summarized in Table 7.
Table 7.
Results for the comparison of the ROC curves of the diagnostic markers log(ca125) and log(cyfra) when considering the multidimensional covariate (age, ) for the and the KS statistics (to the left and to the right, respectively).
| age | 51 | 67 | 83 |
|---|---|---|---|
| 3.20 | 0.026 | 0.056 | 0.010 |
| 1.14 | 0.152 | 0.070 | 0.004 |
| -0.92 | 0.000 | 0.030 | 0.258 |
| age | 51 | 67 | 83 |
|---|---|---|---|
| 3.20 | 0.066 | 0.196 | 0.032 |
| 1.14 | 0.212 | 0.050 | 0.016 |
| -0.92 | 0.004 | 0.048 | 0.424 |
Note that in this case we did not represent the estimated ROC curves conditioned to the bidimensional covariate . This is to stress the fact that, with this methodology, (with bidimensional) does not need to be computed at all.
The obtained p-values show that, depending on the pair of values of the covariate considered, we can find significative differences between the ROC curves of the and the markers, including pairs of values that when considered separately in the previous test did not rejected the null hypothesis. Likewise, finding differences between the ROC curves conditioned to marginal covariates at certain values does not mean that those differences will be significant when considering the multidimensional covariates (for example, when we conditioned the ROC curves marginally to the value of 1.14 we find differences, but when considering both covariates this difference between the ROC curves only remains significant for the age of 83).
This means that if two patients with pleural effusion entered the doctors office, patient A with 1.14 level of and 67 years old, and patient B also with 1.14 level of but 83 years old, we could apply this methodology and take into account those covariate values to personalize their diagnostic. Without including the covariates in the analysis both diagnostic methods (based on the and the ) would seem equally effective to detect MPE. However, when we take into account that multidimensional covariate information, we would see that for patient B (and not for patient A) there are significant differences between the two diagnostic markers.
5. Discussion
In this work a new non-parametric methodology has been presented for comparing two or more dependent ROC curves conditioned to the value of a continuous multidimensional covariate. This method combines existing techniques for reducing the dimension in goodness-of-fit tests and for estimating and comparing ROC curves conditioned to a one-dimensional covariate. Although in this paper we have used a induced regression models to include the covariate effect in the ROC curves, we believe that this test could be adapted and extended to other estimation techniques. This opens the door to future research that could include longitudinal or functional covariates, using, for example, the approach presented in [10] for extending the induced ROC methodology to functional covariates.
A simulation study was carried out in order to analyse the practical performance of the test. Two different functions were proposed for the construction of the statistic, the and the KS, the second one being a little more conservative. Different correlations between the diagnostic variables and different sample sizes have been considered, including uneven ones without any appreciable effect on the test performance.
The behaviour of the test was also studied for different distributions of the covariates. It showed that, even if those distribution do not seem to affect the test, the lack of data around the point at which we perform the test does reduce its power, so the points taken too close to the boundary of the sample range should be analysed with precaution.
Finally, the methodology was illustrated by means of an application to a data set: with this new test it was possible to detect differences on the discriminatory ability of two diagnostic variables conditioned to two different covariates without the need of an estimator of an ROC curve conditioned to a multidimensional covariate. With this application it becomes clear the importance of being able to include the effect of multidimensional covariates to the ROC curves analysis, as different conclusions could be drawn of the comparison of those curves when considering a multidimensional covariate, when considering unidimensional covariates or when excluding the covariates from the study. It can also be an important tool for the personalized medicine, as it allows us to compare different diagnostic methodologies using the personal information of each patient.
Acknowledgements
The authors would like to thank the Associate Editor and the anonymous reviewers for their constructive comments and suggestions on an earlier version of this manuscript. The Supercom puting Center of Galicia (CESGA) is acknowledged for providing the computational resources that allowed to run most of the simulations. Dr. F. Gude (Unidade de Epidemioloxía Clínica, Hospital Clínico Universitario de Santiago) is thanked for providing the data set analysed in this article.
Appendix A. Proofs
The proofs needed for Lemma 2.1 are presented below.
Lemma A.1
[5] or [3]: Given a random variable Y such that ,
From now on it will be assumed that all directions considered satisfy .
Lemma A.2
Let be K dependent random variables with cumulative distribution functions , respectively, such that for every . Let be a multidimensional covariate. Then, given ,
(A1) with and where for .
Proof.
It is proven for K = 2.
Seeing that is the same as proving that , which, given that the random variables are dependent in the sense that they are conditioned to the same covariate , is equivalent to Now, because of Lemma A.1, this is the same as saying that
Using again the dependence between the random variables, that is equivalent to , which in turn is equivalent to .
Definition A.3
The inverted conditional ROC curve IROC is defined as:
Related to the previous definition, the inverted conditional ROC curve ( ), given the pair , is defined as:
Lemma A.4
The equality of ROC curves is equivalent to the equality of the inverted ROC curves, i.e.
Moreover, the same property holds when talking about conditional ROC curves. Given the pair , holds if and only if .
Proof.
It is proven for the unconditional case, and for K = 2. The conditional case is similar.
Take (and hence, ). q will take all the values in , and thus, . Then,
Proof Proof of Lemma 2.1 —
It is proven for K = 2. For ,
Using Lemma A.2, that is equivalent to
which in turn is the same as saying that Now, using Lemma A.4 we know that this is equivalent to
By the definition of the inverted conditional ROC curve, that is the same as saying that
Using again the result of Lemma A.2, the previous statement is equivalent to
By definition, that is the same as saying that and, using again Lemma A.4, that is equivalent to
where and for i = 1, 2.
Funding Statement
The research of A. Fanjul-Hevia is supported by the Ministerio de Educación, Cultura y Deporte (fellowship FPU14/05316), as well as by the Spanish Ministerio de Educación y Formación Profesional (Mobility Grant EST18/00673). A. Fanjul-Hevia, W. González-Manteiga and I. Van Keilegom acknowledge the support by the Grant PID2020-116587GB-I00 from Spanish Ministerio de Ciencia e Innovación (MCIN/AEI/ 10.13039/501100011033). J.C. Pardo-Fernández acknowledges financial support by the Grant PID2020-118101GB-I00 from Spanish Ministerio de Ciencia e Innovación (MCIN/AEI/10.13039/501100011033). I. Van Keilegom is financially supported by the European Research Council (2016-2021, Horizon 2020 / ERC grant agreement No. 694409).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Colling B. and Van Keilegom I., Goodness-of-fit tests in semiparametric transformation models using the integrated regression function, J. Multivar. Anal. 160 (2017), pp. 10–30. [Google Scholar]
- 2.Cuesta-Albertos J.A., Fraiman R., and Matrán C., The random projection method in goodness of fit for functional data, Comput. Stat. Data. Anal. 51 (2007), pp. 4814–4831. [Google Scholar]
- 3.Cuesta-Albertos J.A., García-Portugués E., Febrero-Bande M., and González-Manteiga W., Goodness-of-fit tests for the functional linear model based on randomly projected empirical processes, Ann. Stat. 47 (2019), pp. 439–467. [Google Scholar]
- 4.DeLong E.R., DeLong D.M., and Clarke-Pearson D.L., Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach, Biometrics 44 (1988), pp. 837–845. [PubMed] [Google Scholar]
- 5.Escanciano J.C., A consistent diagnostic test for regression models using projections, Econ. Theory. 22 (2006), pp. 1030–1051. [Google Scholar]
- 6.Fanjul-Hevia A. and González-Manteiga W., A comparative study of methods for testing the equality of two or more ROC curves, Comput. Stat. 33 (2018), pp. 357–377. [Google Scholar]
- 7.Fanjul-Hevia A., González-Manteiga W., and Pardo-Fernández J.C., A non-parametric test for comparing conditional ROC curves, Comput. Stat. Data. Anal. 157 (2021), pp. 107146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García-Portugués E., González-Manteiga W., and Febrero-Bande M., A goodness-of-fit test for the functional linear model with scalar response, J. Comput. Graph. Stat. 23 (2014), pp. 761–778. [Google Scholar]
- 9.González-Manteiga W., Pardo-Fernández J.C., and Van Keilegom I., ROC curves in non-parametric location-scale regression models, Scand. J. Stat. 38 (2011), pp. 169–184. [Google Scholar]
- 10.Inácio V., González-Manteiga W., Febrero-Bande M., Gude F., Alonzo T.A., and Cadarso-Suárez C., Extending induced ROC methodology to the functional context, Biostatistics 13 (2012), pp. 594–608. [DOI] [PubMed] [Google Scholar]
- 11.Inácio de Carvalho V., Jara A., Hanson T.E., and de Carvalho M., Bayesian nonparametric ROC regression modeling, Bayesian Anal. 8 (2013), pp. 623–646. [Google Scholar]
- 12.Kim E., Zeng D., and Zhou X.-H., Semiparametric transformation models for multiple continuous biomarkers in ROC analysis, Biom. J. 57 (2015), pp. 808–833. [DOI] [PubMed] [Google Scholar]
- 13.Li J., Applications of the bootstrap in roc analysis, Commun. Stat. Simul. Comput. 41 (2012), pp. 865–877. [Google Scholar]
- 14.Martínez-Camblor P. and Corral N., A general bootstrap algorithm for hypothesis testing, J. Stat. Plan. Inference. 142 (2012), pp. 589–600. [Google Scholar]
- 15.Martínez-Camblor P., Carleos C., and Corral N., General nonparametric ROC curve comparison, J. Korean. Stat. Soc. 42 (2013), pp. 71–81. [Google Scholar]
- 16.Muller M.E., A note on a method for generating points uniformly on n-dimensional spheres, Commun. ACM. 2 (1959), pp. 19–20. [Google Scholar]
- 17.Pardo-Fernández J.C., Rodríguez-Álvarez M.X., and Van Keilegom I., A review on ROC curves in the presence of covariates, Revstat Stat. J. 12 (2014), pp. 21–41. [Google Scholar]
- 18.Patilea V., Sánchez-Sellero C., and Saumard M., Testing the predictor effect on a functional response, J. Amer. Statist. Assoc. 111 (2016), pp. 1684–1695. [Google Scholar]
- 19.Pepe M.S., The Statistical Evaluation of Medical Tests for Classification and Prediction, Oxford University Press, Oxford, 2003. [Google Scholar]
- 20.Rodríguez A. and Martínez J.C., Bayesian semiparametric estimation of covariate-dependent ROC curves, Bioestatistics 15 (2014), pp. 353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez-Álvarez M.X., Roca-Pardiñas J., and Cadarso-Suárez C., A new flexible direct ROC regression model: Application to the detection of cardiovascular risk factors by anthropometric measures, Comput. Stat. Data. Anal. 55 (2011), pp. 3257–3270. [Google Scholar]
- 22.Rodríguez-Álvarez M.X., Roca-Pardiñas J., Cadarso-Suárez C., and Tahoces P.G., Bootstrap-based procedures for inference in nonparametric receiver-operating characteristic curve regression analysis, Stat. Methods. Med. Res. 27 (2018), pp. 740–764. [DOI] [PubMed] [Google Scholar]
- 23.Su J.Q. and Liu J.S., Linear combinations of multiple diagnostic markers, J. Amer. Statist. Assoc. 88 (1993), pp. 1350–1355. [Google Scholar]
- 24.Valdés L., San-José E., Ferreiro L., González-Barcala F.-J., Golpe A., Álvarez-Dobaño J.M., Toubes M.E., Rodríguez-Núñez N., Rábade C., Lama A., and Gude F., Combining clinical and analytical parameters improves prediction of malignant pleural effusion, Lung 191 (2013), pp. 633–643. [DOI] [PubMed] [Google Scholar]
- 25.Venkatraman E.S. and Begg C.B., A distribution-free procedure for comparing receiver operating characteristic curves from a paired experiment, Biometrika 83 (1996), pp. 835–848. [Google Scholar]