Abstract
The ability to selectively modify proteins at two or more defined locations opens new avenues for manipulating, engineering, and studying living systems. As a chemical biology tool for the site-specific encoding of noncanonical amino acids into proteins in vivo, genetic code expansion (GCE) represents a powerful tool to achieve such modifications with minimal disruption to structure and function through a two-step “Dual Encoding And Labeling” (DEAL) process. In this review, we summarize the state of the field of DEAL using genetic code expansion. In doing so, we describe the basic principles of GCE-based DEAL, catalogue compatible encoding systems and reactions, explore demonstrated and potential applications, highlight emerging paradigms in DEAL methodologies, and propose novel solutions to current limitations.
Keywords: Genetic Code Expansion, Bioorthogonal Chemistry, Dual Encoding And Labeling, DEAL, Protein Bioconjugation, Protein Engineering
Introduction and Scope
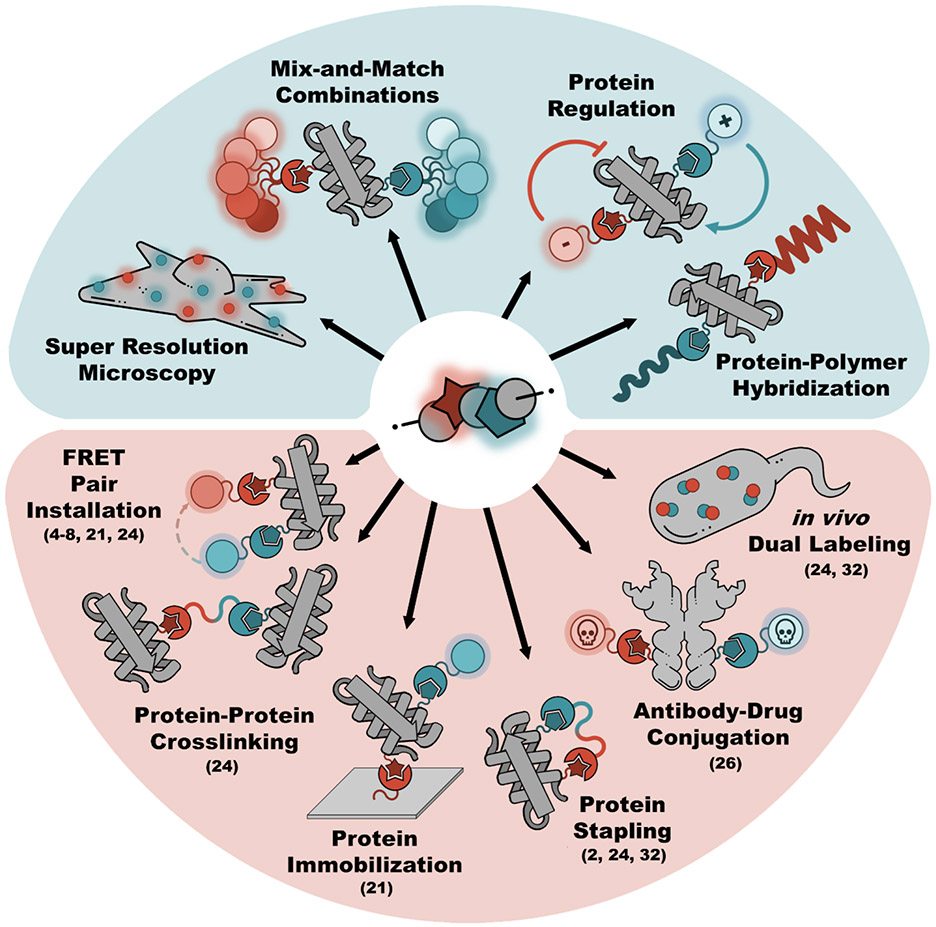
The ability to selectively attach two different labels onto biomolecules enables powerful approaches to detect, construct, characterize and study biomolecules and/or their interacting partners with great precision, even in their native contexts. In general, achieving this requires a two-step process composing of 1) biochemical encoding of two distinct bioorthogonal and mutually orthogonal reactive groups or “handles” into the targeted macromolecule(s), and 2) the subsequent selective labeling of these handles through reaction with the probes of interest (Figure 1). Here, we call this general two-step process “Dual Encoding and Labeling” (DEAL)1. DEAL has been achieved through several approaches including: metabolic labeling (i.e. metabolic oligosaccharide engineering, etc.)2,3, affinity tags (i.e. FlAsH- and ReAsH-tags, etc.)4-7, chemoenzymatic labeling (i.e. Biotin ligase, Sortase, Lipoic acid ligase, etc.)8-13, self-labeling enzymatic domains (i.e. SNAP-, CLIP-, and Halo-tags, etc.)14-16, and genetic code expansion1,17,18. Many impactful applications of DEAL include (Figure 1): simultaneous tracking and imaging of two distinct species of biomolecules9,19-22, temporal labeling of different biomolecular populations21,23,24, studying protein-protein interactions25, monitoring of protein dynamics using spectroscopic probes26-29, engineering and modifying proteins through site-specific conjugation10,30,31, protein-protein crosslinking1,11,32-34, constructing supramolecular protein complexes8,10,25,35,36, and designing novel biosensor platforms37-40.
Figure 1.
Some demonstrated (red background) and potential (blue background) applications of DEAL. Dual encoded groups are indicated by the red star and blue pentagon and the ligated compounds are indicated by the complementary shapes of matching color. For demonstrated applications, the numbers in parentheses correspond to the publications listed in Table 1.
An “ideal” approach to DEAL would be: 1) site-specific and compatible with virtually any location on a protein’s surface, permitting attachments to be non-disruptive to the protein’s structure and function; 2) modular, so it can be easily retooled for new applications, and 3) based on labeling chemistry that is bioorthogonal, biocompatible (i.e. non-toxic), biostable, and efficient in vivo. Unfortunately, most approaches listed above fall short of these criteria, as they either lack the required levels of site-specificity41-45, are non-covalent4,5,5-7,46-48, or require large and disruptive encoding approaches9-16,25,49. Recently, genetic code expansion (GCE)50-52 has emerged as a technique that meets all of the criteria for an “ideal” DEAL approach. As a chemical biology tool that allows the site-specific encoding of unobstructive noncanonical amino acids (ncAAs) into proteins, it is unparalleled in its exquisite site-specificity and its minimal invasiveness (for recent reviews see51-54). Advances in GCE have enabled the simultaneous encoding of multiple ncAAs, including those with bioorthogonal reactivity, thereby transforming the protein into a “blank slate” chassis for functionalization with virtually any imaginable modification at virtually any site, even in vivo.
Through summarizing and synthesizing the information in the 32 papers we have found that represent the GCE-based DEAL field thus far (Table 1), the purpose of this review is to highlight how GCE opens up new possibilities for powerful DEAL experiments, including those in vivo, and to provide information that lowers the barriers to its use and further development. We first outline the basic principles underlying the use of GCE for efficient DEAL. Then, based on published GCE-base DEAL experiments (Table 1), we summarize the specific compatible pairings of GCE components that are useful for DEAL. Finally, we identify aspects of GCE-based DEAL (referred to henceforth simply as “DEAL”) that need improvement and also showcasing recent advances addressing these shortcomings. As our scope is limited to in vivo GCE applications, we will not cover residue-specific GCE55-57, and cell-free GCE58-62, which have been reviewed elsewhere.
Table 1.
Publications reporting GCE-based dual-ncAA encoding that are identified in this review. Entries indicated by an “*” perform both dual encoding and dual labeling, “†” indicates that meta-azidophenylalanine (mAzF), a meta-substituted isomer of pAzF (C) was used.
| Subsystem A | Subsystem B | Subsystem C | System D | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Entry | Protein(s) | ncAA(s) | Codon | aaRS/tRNA | ncAA(s) | Codon | aaRS/tRNA | ncAA | Codon | aaRS/tRNA | ncAA | Codon | aaRS/tRNA | Reference |
| Prokaryotic Systems | ||||||||||||||
| 1 | Myoglobin | OMeY | UAG | MjTyr | hGln | AGGA | PhLys | Anderson et al., 2004182 | ||||||
| 2* | Calmodulin | pAzF (C) | AGGA | MjTyr | PrK (A) BocK |
UAG | MbPyl | Neumann et al., 201034 | ||||||
| 3 | GFPuv | pAzF (C) | UAG | MjTyr | BocK PrK (A) CycpK |
UAA | MbPyl | Wan et al., 201090 | ||||||
| 4* | QBP | pAzF | UAG | MjTyr | KetoK (F) | UAA | MmPyl | Wu et al., 201228 | ||||||
| 5* | Calmodulin | pAcF (G) | UAG | MjTyr | PrK (A) | UGA | MmPyl | Kim et al., 201326 | ||||||
| 6* | KSI GFP | pAzF (C) pAcF (G) |
UAG | MjTyr | AzK (D) BocK |
UAA | MbPyl | Chatterjee et al., 201318 | ||||||
| 7* | GST-CaM | pAzF (C) Tet1.0 (H) PrY (B) Bpa |
UAG AGGA |
MjTyr | BocK PrK (A) AlkK NbK (L) |
UAG AGUA UAGA |
MbPyl | Wang et al., 201417 | ||||||
| 8* | Calmodulin | PrY (B) | UAG | MjTyr | CpK (M) | AGUA | MbPyl | Sachdeva et al., 201491 | ||||||
| 9 | GFP GST-MBP | pAzF (C) | AGGA | MjTyr | PrK (A) BocK |
UAG | MbPyl | Lammers et al., 201427 | ||||||
| 10 | sfGFP MDH | Sep | UAG | MmSep | AcK (D) | UAA | MbPyl | Venkat et al., 2018125 | ||||||
| 11 | QBP | pAzF (C) | UAG | MjTyr | PrK (A) | UAA | MmPyl | Tharp and Liu, 201896 | ||||||
| 12 | sfGFP Histone H3 | Bpa | UGA | MjTyr | AcK | UAG | MbPyl | Zheng et al., 201892 | ||||||
| 13 | GST-Calmodulin | CbzK | UAG | MaPyl | CpK (M) | AGGA | MmPyl | Willis and Chin, 2018103 | ||||||
| 14 | TrxR1 | AcK | UAG | MbPyl | Sec | UGA | EcSec | Wright et al., 2018183 | ||||||
| 15* | sfGFP Herceptin-Fab | pAzF (C) OMeY |
UAG | MjTyr | CpK (M) BocK |
UAA | MbPyl | 5HTP (N) | UGA | EcTrp | Italia et al., 201972 | |||
| 16 | GFPuv | pAcF (G) | UAGA | MjTyr | BocK PrK (A) |
AGGA | MbPyl | Hankore et al., 201993 | ||||||
| 17 | ScFv-P3 | PrY (B) | UAG | MjTyr | CpK (M) | AGUA | MbPyl | Oller-Salvia et al., 201994 | ||||||
| 18 | sfGFP | pAzF (C) | AXC | MjTyr | AzK (D) | AGX | MmPyl | N/A | GXT | N/A | Fischer et al., 202097 | |||
| 19 | GFP | BocK | UAG | MmPyl | NmH | AGGA | Lum1Pyl | CbzK | AGUA | 1R26Pyl | Dunkelmann et al., 2020105 | |||
| 20* | sfGFP | MeF | UAG | MjTyr | BocK | UAA | MaPyl | Tharp et al., 2020130 | ||||||
| 21* | tsCA | pAzF (C) | UAG | MjTyr | Tet3.0 (I) | UAA | MbPyl | Sosa et al., 202195 | ||||||
| 22 | Ubiquitin Noncanonical polymer | pIF | UAG | AfTyr | CbzK | UCG | 1R26Pyl | BocK AllocK CpK (M) PrK (A) |
UCA | MmPyl | Robertson et al., 2021136 | |||
| AllocK PrK (A) | UCG | MmPyl | CbzK | UCA | 1R26Pyl | |||||||||
| 23 | Strep-GFP | NmH | AGGA | RumEnPyl | CbzK | AGUA | G1Pyl | AllocK | UAGA | MmPyl | pIF | CUAG | AfTyr | Dunkelmann et al., 2021131 |
| 24* | sfGFP SUMO-sfGFP sfGFP-mTagBFP2 scRPA | pAzF (C) | UAG | MjTyr | Tet3.0 (I) | UAA | MbPyl | Bednar et al., 20211 | ||||||
| 25* | sfGFP | mAzF (C†) | UAG | MaPyl | PrK (A) | UAA | MmPyl | pAcF | UAU | MjTyr | Tharp et al., 202198 | |||
| Eukaryotic Systems | ||||||||||||||
| 26* | EGFP Anti-HER2 IgG | pAcF (G) OMeY |
UAG | EcTyr | AzK (D) BocK |
UAA | MbPyl | Xiao et al., 201330 | ||||||
| 27 | CRF1R | pAzF (C) | UAG | EcTyr | CbzK | UGA | MbPyl | Serfling et al., 2017184 | ||||||
| 28 | EGFP | OMeY pAzF (C) PrY (B) pAcF (G) |
UGA UAG |
EcTyr | BocK AzK (D) PrK (A) CpK (M) |
UGA UAG |
MbPyl | Zheng et al., 201799 | ||||||
| Cap | UAG | EcLeu | ||||||||||||
| 29 | sfGFP | TCOK (J) | UAG | MaPyl | AcK | UAA | MmPyl | Meineke et al., 201880 | ||||||
| 30 | EGFP | C5Az (E) | UAG | EcLeu | CpK (M) NbK (L) |
UGA | MbPyl | Zheng et al., 201879 | ||||||
| 31 | GFP | NmH | UGA | MaPyl | BocK | UAG | MmPyl | Beranek et al., 2019104 | ||||||
| 32* | CRF1R SynNotch sfGFP | PrK (A) | UAG | G1Pyl | TCOK (J) | UAA | MmPyl | Meineke et al., 202081 | ||||||
| 33 | GFP | Anap | UAG | EcLeu | pAcF | UAA | EcTyr | AzK (D) | UGA | MmPyl | Shi et al., 2022100 | |||
Basic Principle 1: Dual encoding requires ncAA-encoding subsystems with sufficient mutual orthogonality at three key steps
GCE uses an orthogonal encoding subsystem for each ncAA to be incorporated
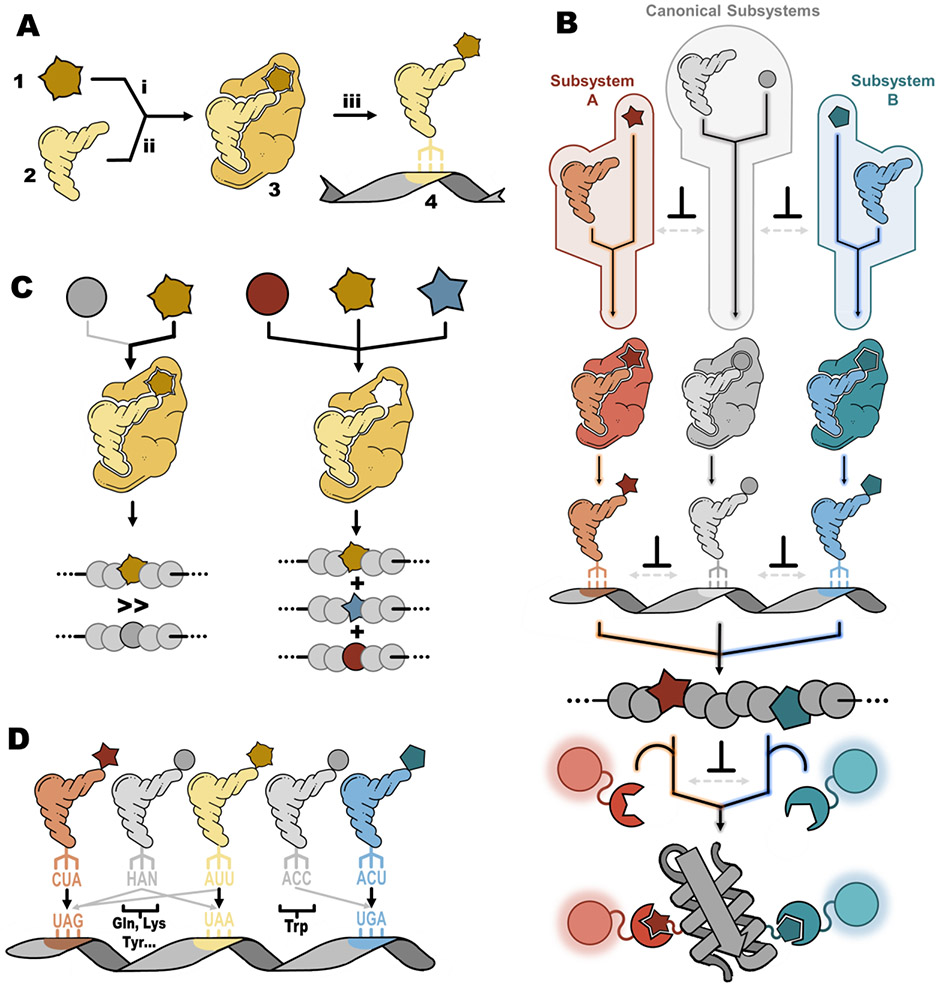
Genetic code expansion relies on the action of an exogenous aminoacyl-tRNA synthetases and their cognate tRNAs (aaRS/tRNA pairs) to outcompete endogenous decoding systems and/or release factors to encode an ncAA. This process has historically been called suppression since such a process worked to suppress the action of release factors, but we will simply refer to it as encoding. An ncAA-encoding “subsystem” is comprised of four components (Figure 2A): 1) the ncAA to be encoded, 2) the tRNA that the ncAA will be charged to, 3) the cognate aaRS that charges the ncAA to the tRNA, and 4) the blank codon that the tRNA decodes. A key requirement for faithful encoding of the targeted ncAA is that there be sufficient orthogonality63 throughout the encoding process between the exogenous ncAA-encoding subsystem and all other host organism’s subsystems. During encoding, the need for orthogonality arises at three distinct molecular recognition steps (Figure 2A). These are the (i) ncAA-aaRS, (ii) aaRS-tRNA, and (iii) tRNA anticodon-codon recognition steps, with orthogonality at each step being governed by both the concentration and nature of molecular interactions between each of the components of an exogenous subsystem and their host counterparts (Figure 2B). Failure to maintain sufficient orthogonality at any of these three steps leads to either contamination of canonical amino acids at the desired ncAA encoding site64,65, or unwanted encoding of the ncAA sporadically throughout the proteome66. Notably, maintaining orthogonality becomes more challenging when performing DEAL, because two separate and mutually orthogonal decoding subsystems are needed to encode two distinct ncAAs. In the next three subsections we discuss factors related to orthogonality at each of the three steps and highlight important considerations for combining subsystems to enable DEAL.
Figure 2. Orthogonality considerations in dual encoding and labeling.
A Schematic of the four key components of an encoding subsystem: 1) ncAA , 2) orthogonal tRNA, 3), orthogonal aaRS, and 4) codon as they participate in the three orthogonal encoding steps: i) ncAA recognition by the aaRS, ii) tRNA recognition by the aaRS, and iii) codon decoding by the charged tRNA. B Overview of DEAL indicating desired interactions (black arrows with color-coded outlines for each encoding subsystem), and undesirable interactions that violate orthogonality (hashed gray arrows). The ⊥ symbol indicates a point of essential orthogonality (i.e. where no crosstalk is required) between subsystems. C Relative fidelity (left image) is when an aaRS has sufficient preference for its cognate ncAA (black arrow) that in its presence no canonical amino acids (thin gray arrow) are incorporated. Permissivity (right image) is when an aaRS is able to efficiently incorporate multiple ncAA, depending on which ones are included in the medium. D Documented examples of near-cognate suppression due to orthogonal tRNAs (colored) and endogenous tRNAs (gray with canonical amino acid residue types involved given in bold) interacting not just with their cognate codons (black arrows) but with off-target noncognate codons that violate orthogonality (gray arrows) via near cognate suppression. This can include a UAA suppressor tRNA (yellow) decoding the UAG codon (red).
Two key considerations related to ncAA-aaRS orthogonality
The substrate specificity of aaRSs is mediated by key active-site residues that make contact with the ncAA substrate67. This specificity is often acquired through directed evolution to enrich for aaRSs that recognize the new ncAA substrate but not canonical amino acid substrates68,69. Depending on the stringency of the selection parameters, the resulting aaRSs will exhibit varying degrees of fidelity and permissivity, two key factors to consider in designing DEAL experiments. As illustrated in Figure 2C, fidelity is the ability of the aaRS to discriminate against all canonical amino acid types and permissivity is the ability of the aaRS to recognize additional ncAAs beyond the one it was selected to recognize70.
For DEAL, relative fidelity is more crucial than absolute fidelity.
Low fidelity can occur when selected aaRSs retain some level of activity towards their ancestral (or similar) canonical amino acids, as has been observed repeatedly with the commonly used tyrosyl aaRS originating from Methanocaldococcus jannashii (MjTyrRS), among others65,71,72. For example, Odoi and colleagues observed that in the absence of the preferred ncAA substrate, MjTyrRS led to incorporation of phenylalanine, which closely resembles the ancestral tyrosine substrate73. While this may seem problematic, we and others have noted that in the presence of the ncAA, homogenous ncAA incorporation can still be observed, indicating a sufficient relative preference for the ncAA18,70-73. This leads to the importance of distinguishing what we call absolute fidelity and relative fidelity: the former is the ability to avoid incorporating a canonical amino acid in the absence of ncAA, and the second is the ability to avoid incorporating a canonical amino acid in the presence of ncAA. While it may seem that lower absolute fidelity is always detrimental, a lower absolute fidelity can be advantageous when it is accompanied by sufficient relative fidelity. Indeed, as shown by Cooley and colleagues, lowering the selective stringency for absolute fidelity resulted in an aaRS with sufficient relative fidelity but with much higher overall encoding efficiency, showing that lower absolute fidelity can be associated with higher encoding efficiency and minimal fidelity penalties under conditions where ncAA is provided70. Interestingly, the commonly used pyrrolysyl aaRSs (PylRS) for DEAL originating from various archaeal and bacterial species have been shown to exhibit markedly lower relative fidelity18,71,73, possibly as a result of the unique structure of the ancestral substrate which is absent in most organisms74. Thus, in dual encoding applications, high background encoding in the absence of ncAA (i.e. low absolute fidelity) does not represent a limitation to mutual orthogonality, provided ncAA will be present during the experiments.
Permissivity.
Permissivity or “polyspecificity” (Figure 2C) refers to an aaRS exhibiting specificity for more than one ncAA75. While this property is generally useful for increasing the number of ncAAs that can be encoded by a single aaRS75-77, in the case of DEAL it can lead to a breakdown in the mutual orthogonality of the two ncAA-encoding subsystems. This may be particularly problematic as, from our experience, many aaRSs have at least some permissivity towards structurally similar ncAAs75-78. In one example, Zheng and coworkers observed that the leucyl aaRS from E. coli (EcLeuRS) and PylRS from Methanosarcina barkeri (MbPylRS) they had generated shared specificity for several ncAAs, preventing their concomitant use, a problem that was bypassed by simply using ncAAs with less structural similarity79. In general, permissivity-based orthogonality breaches appear to be more common when two closely related aaRS are used, such as two PylRSs. While developing DEAL systems, Meineke and colleagues observed that such aaRSs often have overlapping permissivity profiles, a condition that could be partially offset through active-site mutations, but one that nevertheless limited the suite of available ncAA combinations and subsequent applications80,81. It remains to be seen whether, under dual encoding contexts, such systems could exhibit sufficient relative fidelity between the ncAAs to avoid ncAA intermixing. Regardless, these examples highlight the importance of assessing aaRS-ncAA compatibility, particularly between similar ncAAs and aaRSs. As such, we recommend empirically validating the relative fidelity of any new ncAA-encoding subsystem combinations at the ncAA-aaRS step to ensure that both systems are sufficiently orthogonal to each other as well as to the host machinery.
aaRS-tRNA orthogonality and mutually-orthogonal aaRS-tRNA pairings
The recognition of tRNAs by cognate aaRSs is mediated by identity factors present on the tRNA82–features that are typically located in distal portions of the tRNA such as the acceptor-stem and anticodon83,84. Such interactions within a single organism have been fine-tuned over evolutionary timespans to provide mutual orthogonality between native encoding subsystems. However, if there are overlapping identity factors, ncAA-encoding subsystems might also charge an endogenous tRNA, leading to a breakdown in orthogonality. This phenomenon renders the commonly used MjTyrRS/tRNATyr pair orthogonal in E. coli, but not in eukaryotes because the presence of a shared C1:G72 identity factor (among other sequence similarities85) between archaeal and eukaryotic tRNATyr (86) results in mischarging of ncAA and Tyr between exogenous and endogenous tRNAs.
Common, natural, mutually-orthogonal aaRS-tRNA pairings.
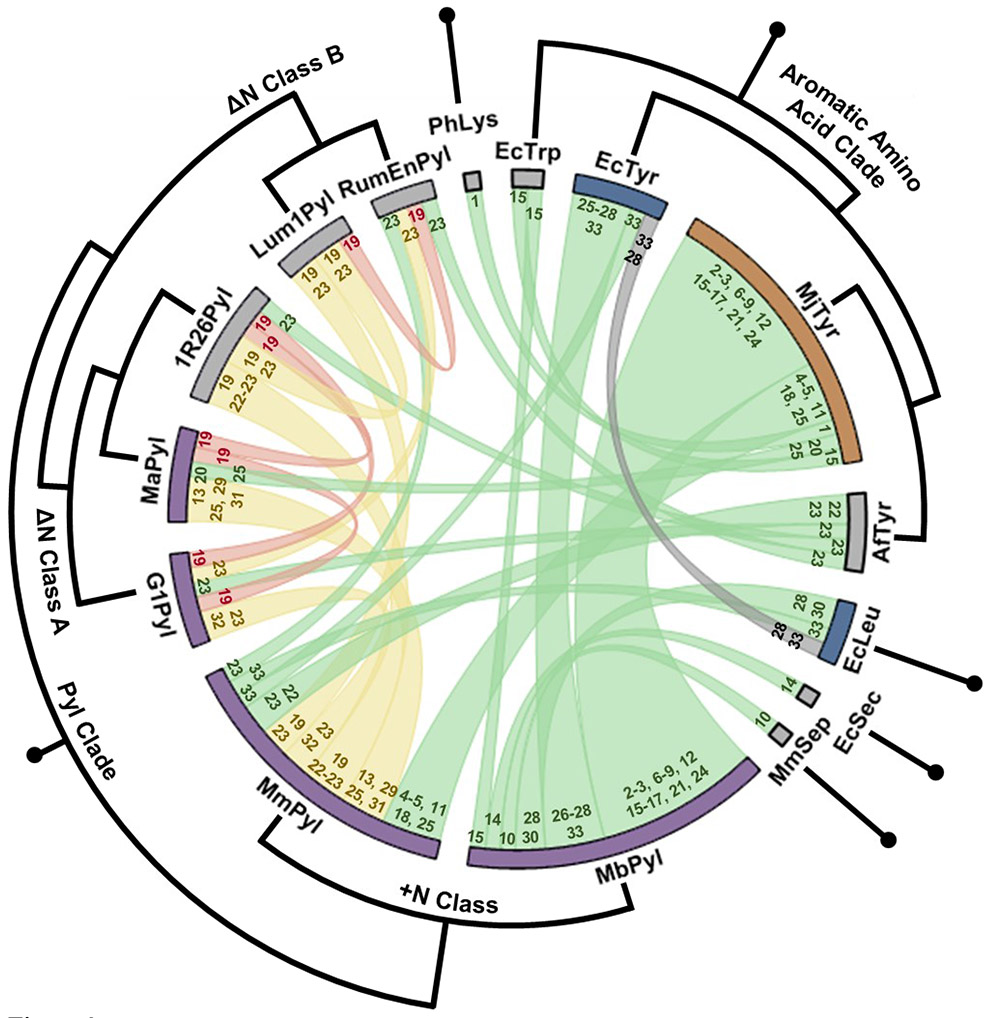
In DEAL experiments, this breakdown in orthogonality is of particular concern because it can also occur between two otherwise orthogonal ncAA-encoding subsystems, thereby violating mutual orthogonality (Figure 2D). Fortunately, in published studies (Table 1) researchers have identified numerous compatible subsystems, which we summarize in Figure 3. The majority of compatible combinations involve various PylRS/tRNAPyl encoding subsystems87, due their unique constellation of identity factors compared with most other classes of aaRS/tRNA pairs87-89. In bacteria, the compatibility of such PylRS/tRNAPyl systems with the MjTyrRS/tRNATyr pair has been extensively documented18,26-28,34,90-92,72,93-95,1,96-98. In eukaryotic systems, where the MjTyrRS/tRNATyr pair is not orthogonal, the PylRS/tRNAPyl pair has often been combined with aaRS/tRNA pairs from bacteria which, given their ancient evolutionary divergence, often possess sufficiently distinct identity factors89. Two bacterial aaRS/tRNA pairs in common use are the TyrRS/tRNATyr and LeuRS/tRNALeu pairs from E. coli79,99. Intriguingly, even though the E. coli Tyr and Leu-encoding subsystems are orthogonal in their native host, in mammalian cells the EcTyrRS can charge the EctRNALeu (99). Despite also observing this phenomenon, Shi and colleagues recently found that the EcTyr and EcLeu subsystems (along with the MmPyl subsystem) could still be paired in mammalian cells, indicating that these two systems may exhibit sufficient relative fidelity to be of functional use100. This highlights the need to be cautious in checking for orthogonality of ncAA-encoding subsystems during DEAL and to empirically check for relative fidelity of tRNA/RS needed for specific applications.
Figure 3.
Orthogonality relationships between all aaRS/tRNA subsystem pairings from the publications in Table 1. The thickness of the chord between a pair of aaRS nodes reflects the frequency that that combination has been used. The colors indicate pairs that are naturally orthogonal (green), are not orthogonal (red), or that have been engineered to be orthogonal (yellow). The gray chord indicates a discrepancy between observations in Table 1 entries 27 and 32. The numbers at the ends of the chords indicates the relevant entry numbers in Table 1. The color of the node (the bar at the edge of the circle) indicates orthogonality of that subsystem with only prokaryotic (orange), with only eukaryotic (blue) or with both (purple) host decoding systems, while gray indicates that orthogonality has only been assessed in one host. Surrounding the chord plot is a circular cladogram showing the level of similarity of aaRSs within various clades (alignments performed using Clustal Omega185).
Engineered mutually-orthogonal aaRS-tRNA pairings.
Given the limited number of available, mutually orthogonal ncAA-encoding subsystems, researchers have sought to engineer and discover new subsystems with mutual orthogonality. An innovative approach pioneered by the Chatterjee group was to replace the endogenous EcTrpRS/tRNATrp system within E. coli with a functional yeast analog71. This functional replacement for the encoding of Trp allowed the authors to engineer the EcTrpRS/tRNATrp to encode ncAAs directly in E. coli, since in this context it no longer retained the obligate role of encoding Trp. This new system, along with the MjTyrRS/tRNATyr and MbPylRS/tRNAPyl encoding subsystems, allowed successful triple encoding and labeling in E. coli72. A similar approach has since been applied to the EcTyrRS/tRNATyr system101. Others have taken a more direct approach to engineering mutually orthogonal ncAA-encoding subsystems. For example, given that the N-terminal domain of PylRS plays a major role in molecular discrimination of tRNAs, the Chin group utilized several archaeal PylRSs – including the PylRS of Methanomethylophilus alvus – that lack this N-terminal domain (so-called ΔN PylRSs, originally discovered by the Brugère group102) and so might serve as the basis of an orthogonalization approach103. By selecting tRNAPyl with an expanded and mutagenized variable loop, they found several tRNAs with specificity towards “ΔN” over “+N” PylRS, effectively orthogonalizing the two ncAA-encoding subsystems104. Taking this approach further, the “ΔN” class of PylRS could be divided into two distinct subgroups that were partially orthogonal to one another. Several further rounds of engineering generated triply mutually orthogonal PylRS/tRNAPyl pairs consisting of a “+N”, a “Class A ΔN,” and a “Class B ΔN” PylRSs, which were then used to encode three distinct ncAAs105. Together, these studies reveal that mutagenizing both the acceptor stem and variable loop of differing tRNAs is an effective means of engineering orthogonality, even between closely related aaRSs80,103,105. Another innovative approach developed by Reinkemeier and Lemke was to engineer a system in which aaRSs were physically separated in cellular space, and thus became orthogonal by proximity, despite being orthogonal only at the ncAA-aaRS step106. To do so, the authors generated two discrete, film-like, membraneless organelles comprised of localized, phase-separated proteins in different regions of a mammalian cell. By targeting two distinct PylRS and two distinct mRNA to these different organelles, Reinkemeier and Lemke were able to effectively co-opt the TAG codon to encode two distinct protein populations with different ncAAs in two different loci of the same cell. As designed, the system cannot encode two distinct ncAAs into the same protein chain. Despite this, these findings indicate that “spatial orthogonality”107 (using partitioning to bypass the requirement for molecular discrimination) is a legitimate avenue for engineering orthogonality for certain applications106.
Discovering new natural, mutually-orthogonal aaRS-tRNA pairings.
Discovering new natural mutually orthogonal aaRS/tRNA pairs from nature has also been a fruitful avenue of identifying mutually orthogonal aaRS/tRNA pairs. As exemplified, Cervettini and colleagues developed a computational pipeline to score ~3 million metagenomic tRNA sequences on the basis of known E. coli identity factors108. From these, the authors identified five new functional and orthogonal aaRS/tRNA pairs which also shared mutual orthogonality with three previously know aaRS/tRNA pairs. All the above studies demonstrate that the identity factor paradigm for explaining aaRS-tRNA molecular orthogonality is powerfully predictive and can be leveraged for the design and scalable discovery of novel, mutually orthogonal aaRS/tRNA pairs.
tRNA-Codon orthogonality and the problem of near cognate suppression
mRNA decoding occurs at the ribosome and is initiated by triplet base-pair interactions at the A-site during protein translation. Whereas there are 61 sense codons (and 3 stop codons) in the genetic code, there are only 47 unique isoacceptor tRNAs spread across 86 genes that are responsible for encoding all 21 amino acids (including selenocysteine) within the E. coli proteome109. This discrepancy is in part resolved by the occurrence of wobble base pair interactions, as predicted by Crick in 1966110. Since then, it has become apparent that a single tRNA is capable of recognizing and decoding more than one codon through a variety of non-Watson-Crick interactions, and that these can be influenced by post-transcriptional modifications111. Although such cross-talk is an essential part of proper genetic encoding, it is also the source of misincorporation through what has been called “near-cognate suppression”112. GCE relies on the use of unassigned “blank” codons–ones ideally not used for another purpose–as a component of the ncAA-encoding subsystem. The near-cognate suppression reading of these “blank” codons by near-cognate tRNAs disrupts orthogonality and is of increased concern for dual encoding, since there are two different codons that can be misread, either by endogenous or exogenous decoding systems (Figure 2D).
Near cognate suppression by canonical tRNAs.
All codons (including nonsense codons) are susceptible to near-cognate suppression from natural decoding systems (Figure 2D). In fact, the error rate of translation is estimated to be between 10−3 – 10−4, indicating that such errors are common, well-tolerated, and may even provide cellular fitness benefits under certain circumstances113. In the context of dual encoding, where protein stalling is hypothesized to be exacerbated, endogenous near-cognate suppression can become more pronounced, often in a codon-dependent manner64,114. For example, up to 10 distinct amino acids have been reported to contaminate UAG (amber) suppression sites during GCE, with the most frequently reported being Gln, Tyr, and Lys, all of which involve only a single mismatch64,73,114-119 UGA (opal) codons reportedly have the highest levels of near-cognate suppression at around 3% in E. coli, due chiefly to wobble interactions with tRNATrp (73,112,116,118) (Figure 2D) that appear to be highly dependent on codon context in the 3’ direction120,121. In contrast, UAA (ochre) codons have less endogenous near-cognate suppression relative to the commonly utilized UAG (amber) codon, presumably due to an overlap of recognition from both release factors116,119. Conversely, it should be noted that the UAG codon still enjoys higher suppression efficiency in eukaryotes, despite the presence of only a single release factor79. This observation underscores the notion that relatively little is understood about the nature of tRNA anticodon-codon interactions in eukaryotic systems, wherein additional factors such as differing post-transcriptional tRNA modifications116, and nonsense-mediated decay122 can come into play. While Indeed, the removal of release factors, as in the E. coli C321.ΔA.exp and B95 strains123,124, seems to exacerbate endogenous near-cognate suppression64,115,117,119. Furthermore, the observation that such near-cognate suppression can be overcome by improving the efficiency of an ncAARS64, further highlights that it is a relative rather than an absolute fidelity feature of ncAA-encoding subsystem that impacts its effectiveness73,115,119.
Near cognate suppression of UAG codons by UAA suppressor tRNAs.
In dual encoding contexts it has been observed that UAA-decoding tRNAs can, to a small degree, decode UAG codons18,73,80,99,125,126, presumably due to the strong U:G base pairing typical of wobble interactions127. This cross-recognition may appear to be a “DEAL breaker” for configurations using the UAG+UAA codon combination, as the UAA suppressor tRNA may insert the wrong ncAA at UAG sites. However, this is another case in which relative fidelity outweighs absolute fidelity, as we and others have observed that in the presence of a functional ncAA-charged UAG suppressor, relative fidelity is still achieved1,18,118. We suspect that the UAG suppressor tRNA simply outcompetes near-cognate suppression, but this has not been shown. Interestingly, the UAG suppressor tRNA has not been observed to decode UAA codons; while it may seem strange, this asymmetry is easily rationalized in that a C:A mispairing at the third position is known to not form a strong wobble interaction18,73,99.
Choosing “blank” codon pairs for DEAL.
For the purpose of DEAL, a wide variety of blank codons have been utilized, ranging from the standard triplet nonsense codons72, to quadruplet (also referred to as frameshift) codons17, to sense codons58, and even unnatural codons97 (Table 1). Owing to its higher suppression efficiencies18,34,79, the UAG codon remains the most widely used blank codon for dual encoding (Table 1). While this may often be the case, the Venkat and colleagues observed that within E. coli, the PylRS/tRNA pair performed roughly as well at all three stop codons125, whereas Zheng et al., noted that in a mammalian context, this same pair exhibited significantly higher encoding efficiency while decoding UAG stop codons79. These results highlight the fact that encoding efficiency depends on a myriad of factors and can be highly contextual. Nevertheless, the UAG codon frequently finds use with the UAA codon, and to a lesser extent the UGA codons during dual-ncAA encoding (Table 1). An exception to this is in mammalian settings, where the UGA codon enjoys a similar usage to the UAA codon79,99,104. Despite the allure of the vastly expanded codon space afforded by quadruplet codons (256 codons), their usage has been plagued by low suppression efficiencies125 and notable near-cognate suppression118. As reported by the Söll group, the dominant amino acid encoded at AGGA codons is actually contaminant Arg (possibly through near-cognate suppression by tRNAArg in combination with a +1 frameshift to retain the reading frame)118. To overcome this limitation, the Chin group developed a series of orthogonal ribosomes based on a unique Shine-Dalgarno sequence that substantially reduces background encoding and improves quadruplet decoding34. These systems have been used for dual encoding17,34,91,94,104,105; however, they tend to suffer from lower suppression efficiencies (yields ~1% of the wild-type protein level)27 when compared to dual triplet stop codons (yields ~5-20% of wild-type)18,99.
While the innate properties of each codon are often considered when selecting which codon to use, the choice is often limited by the aaRS, which may recognize the anticodon as a key identity factor. For example, the MjTyrRS had to be extensively engineered to recognize its UAG-decoding suppressor tRNA after codon reassignment, which has essentially shoehorned it for usage with this codon128,129. Moreover, codon reassignment of a suppressor tRNA may alter the orthogonality of the tRNA, as Italia and colleagues noted that reassigning the E. coli tRNATrp to a UAG suppressor rendered it a substrate for the native EcGlnRS71. Alternatively, as mentioned earlier PylRSs are not known to interact with the anticodon loop of their cognate tRNA, granting them immense flexibility to their codon compatibility for DEAL purposes. For a visual representation of the various combinations and frequencies of codons used for dual encoding in prokaryotes and eukaryotes, see Figure S1.
Seeking scalable solutions for multi-ncAA encoding.
Despite their popularity, the use of triplet nonsense codons is not a scalable solution for multi-codon suppression, as was illustrated by Italia et al, and Shi et al, who used all three stop codons to simultaneously encode three ncAAs in E. coli12 and mammalian cells100 respectively, leaving no codons to encode termination. In the former case, the authors needed to use a protease to produce the correct final product72. While serving to demonstrate that it is possible to encode three ncAAs, these examples represent the upper limit of triplet nonsense codons for multi-ncAA encoding. A novel approach by Tharp and colleagues was to utilize an ncAA-encoding initiator tRNA in addition to stop codons98. By reassigning a previously-generated UAG-decoding MjTyr tRNA that shares identity factors with the native E. coli fMet initiator tRNA130 to decode UAU codons, the authors were able to encode a number of ncAAs at the first position within the protein of interest, thereby generating a dual-use sense codon that encodes a ncAA at the initiation position, while retaining its Tyr-encoding assignment elsewhere in the protein98. By combining this with two other encoding subsystems, Tharp and coworkers were able to achieve triple-ncAA encoding and labeling in E. coli while still reserving a UGA stop codon to terminate translation. While limited to the N-terminus, such an approach enables effective triple encoding potential without the need for additional processing steps, or the quadruple encoding of ncAAs with such processing. Despite its drawbacks, the use of a quadruplet genetic code, on the other hand, does represent a scalable approach, as Dunkelmann et al. have demonstrated through a combination of UAG, AGGA, and AGUA codons105. The authors have continued to scale this approach to the encoding of up to four distinct ncAAs in a single protein using four quadruplet codons, exemplifying that quadruplet codons indeed represent a legitimate scalable approach to multi-ncAA encoding131. Further support for this notion was found by DeBenedictis and colleagues, who identified that all 20 E. coli isoacceptor tRNA classes can be converted to quadruplet codon suppressors, nine of which can be used to faithfully encode their respective amino acid as a mutually orthogonal set, indicating that the E. coli genetic code is tolerant towards expansion by quadruplet codon usage132. Another approach, pioneered by the Romesberg group, has been to simply expand the genetic alphabet to create novel codons based on noncanonical base-pairs133. Through this method, an E. coli genome possessing 67 codons was used to encode two different ncAAs and a serine through three new codons97. While the high levels of background render sense codons impractical for GCE58,134, the Chin group has demonstrated that these codons can be used for multi-ncAA encoding in refactored genomes that lack some redundant sense codons. In a tour de force effort, the E. coli genome was engineered so as to remove all 18,214 instances of TAG, TCG, and TCA codons, as well as their respective decoding machinery, effectively creating a 61 codon “compressed” genome with three new blank codons135. This E. coli strain (dubbed Syn61) has since been used to encode up to three ncAAs using the three liberated blank codons136. It will be interesting to see which approach will present itself as the scalable method-of-choice for multi-ncAA encoding. Regardless, we are excited at the plurality of avenues to access new vistas for ncAA encoding in the currently-limited codon landscape, and look forward to greater opportunities for developing improved multi-encoding approaches.
Basic Principle 2: Dual Labeling requires sufficiently mutually orthogonal reactive groups
The need for mutually orthogonal bioorthogonal chemistry:
Much like dual encoding, the success of a dual labeling approach depends on orthogonality. When two encoded bioorthogonal handles react with two exogenously added labels, there are six possible reactions: two desirable on-target reactions, and four undesirable off-target reactions (Figure 4A). In this case, the two labeling reactions, in addition to being bioorthogonal, need to be mutually orthogonal – having sufficiently low off-target reactions between noncognate reactants. This property is frequently referred to as chemical orthogonality or mutually orthogonal bioorthogonality. Depending on the experimental configuration, exogenous labels can sometimes be added sequentially in a step-wise manner, potentially with a washout phase, thereby reducing the number of off-target reactions that must be considered. Nevertheless, a loss in chemical orthogonality will result in scrambled labeling and thus a deterioration of site-specificity; therefore, balancing the reactivity of the participating encoded handles is key to successful labeling.
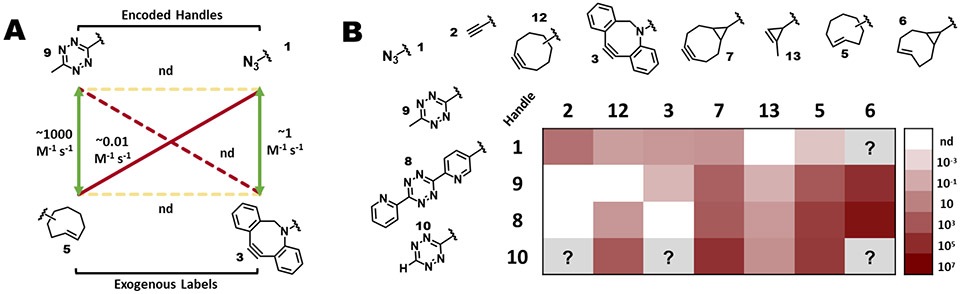
Figure 4.
Identifying chemical orthogonality for reactive groups. A Schematic overview of all possible reactions that can occur during an example dual labeling scenario involving encoded tetrazine 9 and azide 1, and exogenously added TCO 5 and DBCO 3. Depicted are the targeted cognate reactions (green arrows), and the possible off-target reactions between the encoded handles or exogenously added labels (yellow dashed arrows), and between either encoded handle and the non-cognate added label (red arrows; solid arrows indicate observed reactivity and dashed arrows indicate reaction is not detected). For this case, reaction rates (from Karver et al., 2012146) are as indicated for each reaction, with “nd” meaning not detected. As encoded handles that do not react with each other are always selected, and the exogenous labels may be added one after the other, the reactions indicated by the two yellow arrows and one of the red arrows may not be of concern. B Reaction rates of common cycloaddition bioorthogonal handles. Compounds are identified by numbers as indicated. Each panel is color coded (as indicated in the bar to the right) to convey the second order rate constant in M−1 s−1 for the reaction of that pair of compounds. White boxes indicate reaction was not detected (“nd” on the scale bar), and gray boxes “?” indicates that the reaction rate is not published.
Achieving sufficient chemical orthogonality through relative reaction rates and/or sequentiality.
The relative rate of possible reactions is what dictates whether the chemical orthogonality of the components of a system is sufficient to yield acceptably homogeneous labeled products. As Hu and colleagues note, a general recommendation is simply that the on-target reaction reactions be at least around two orders of magnitude faster than the competing off-target reactions137. (The relative rates of some commonly used bioorthogonal reactants are summarized in Figure 4B). This approach has been exploited extensively by Lemke and colleagues who leveraged the tunability of inverse electron demand Diels-Alder (IEDDA) reactions138 to enable DEAL. In this example, the researchers encoded two distinct lysine-derivatized ncAAs bearing either a cyclooctene (TCO; J, in Figure 5) or a cyclooctyne (OCT; similar to 13, in Figure 4) group into two temporally-separated populations of proteins using a single polyspecific PylRS/tRNAPyl system. The resulting proteins could be sequentially labeled using two carefully selected tetrazine labels bearing either methyl (9) or pyridyl groups (8)23. The added ring-strain on the TCO group allowed reaction with the less-reactive methyl-tetrazine group, which showed minimal reactivity towards the OCT group. Subsequently, the OCT group could be labeled through reaction with the much more reactive pyridyl-tetrazine-functionalized label, thereby permitting dual labeling in live mammalian cells. This example highlights how carefully selected reactants with differential reactivity can be leveraged, along with their order-of-addition to enable DEAL in a living system.
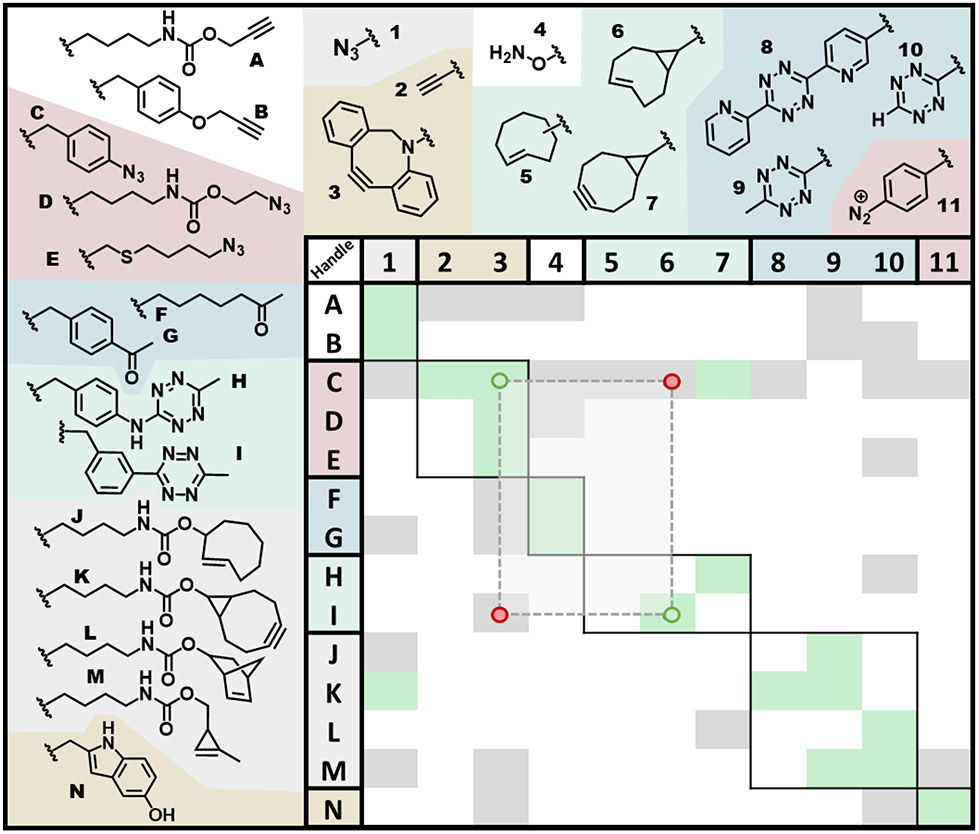
Figure 5.
Orthogonality matrix between encoded ncAAs and their exogenous cognate reaction partners used in the Table 1 DEAL papers. The matrix coloring indicates observed reactivity (green), non-reactivity (dark gray) or lack of information (white) for an encoded ncAA handle (identified by a letter) paired with a possible exogenous labeling groups (identified by a number). The background colors on which compounds are shown group them into subsets with similar reaction chemistry (e.g. azide-alkyne, and IEDDA cycloadditions, oxime ligation, and CRACR). The dashed planchette illustrates a useful way of using this matrix to select chemically orthogonal groups. A suitable chemically orthogonal pair of reactions will have two opposite corners of the rectangle in green boxes – indicating cognate handle-label pairs that react well (green dots here and green arrows in Figure 4A) – and will have the two opposite corners of the rectangle in gray boxes – indicating that the “noncognate off-target” reactions (red dots here and red arrows in Figure 4A) do not occur. The suitable pair illustrated with the rectangle shown was used by Bednar et al., 20211.
Paired reactions that have been successfully used for DEAL.
Following these basic principles, researchers have employed several mutually orthogonal bioorthogonal reactions for DEAL (Figure 5). Early demonstrations, like those by Kim and colleagues139, relied on relatively simple reactions, such as the oxime140 and copper-catalyzed azide-alkyne coupling (CuAAC)141 reactions between encoded carbonyl- (F-G)/azide (C-E) handles and their cognate alkoxylamine (4)/alkyne labels (2), respectively. Such reactions are either slow in the case of oxime labeling142, or can generate damaging reactive oxygen species in the case of the CuAAC reaction143. In pursuit of more biocompatible options, many researchers have turned to the strain-promoted azide-alkyne coupling (SPAAC) reaction18,28,30,97, which while slower than the CuAAC reaction, relies on the intrinsic ring strain of reactants such as dibenzoannulated cyclooctynes (DBCO; 3), among other reagents, to avoid the use of a metal catalyst while still retaining chemical orthogonality towards many other reactions144. Dispensing with both sluggish reaction rates and toxic catalysts, the IEDDA reaction has become, by and large, the most popular reaction for DEAL1,17,79,81,91,94,95,99, not least for its exceptional speed and tunability, excellent biocompatibility138,145, and orthogonality towards other popular cycloaddition reactions146,147. For example, we previously demonstrated that these properties of IEDDA (between I and 6) allowed us to perform DEAL, in combination with the SPAAC reaction (between C and 3), even within a living system, illustrating the ability of this approach to study proteins in their native context1. Moreover, Italia and colleagues recently showed that the SPAAC and IEDDA reactions are also triply orthogonal to a novel reaction between 5-hydroxytryptophan (N) and diazonium compounds (11) referred to as the Chemoselective and Rapid Azo-Coupling Reaction (CRACR)148, which they used to site-specifically label a protein at three distinct locations72
Paired reactions that appear to be of possible use for DEAL.
As can be implied from above, a relatively small subset of bioorthogonal reactions have been adopted for DEAL, despite there being a plethora of potential reactions that could easily be adapted for this purpose, which have been summarized in Figure 6. For example, Bruins and colleagues recently demonstrated that the recently described Strain-Promoted Cyclooctyne-Quinone reaction (between 5 and 17)149, with an exceptionally fast reaction rate, was orthogonal to the SPAAC reaction (between 1 and 7) and could be used in combination with it (although, this reaction is not orthogonal to the IEDDA reaction)13. Similarly, Cheng et al. demonstrated that a reimagined variant of the classical Staudinger ligation consisting of tetrafluoro-para-azidophenylalanine handle (15) could react rapidly enough (k2 of ~73 M−1 s−1) with triphenylphosphine (16) in an orthogonal manner to the SPAAC reaction, when order of additions were controlled inside of live HEK cells150. Orthogonality to SPAAC in live mammalian cells has also been demonstrated for the reaction between in-situ generated nitrile oxides (from chlorooximes; 20) and isonitriles (21), provided that this reaction proceeds SPAAC labeling151,152. Isonitriles have also found to be reactive towards 1,2,4,5-tetrazines. Interestingly, as the Houk and Francini groups have shown, by adding bulky substituents onto the 3,6-positions of tetrazines (e.g. 18) this handle can be tuned to adopt preferential and rapid reactivity towards tertiary isonitriles (19; k2 of ~60 M−1 s−1) over the more commonly-used strained alkenes such as TCO (5; k2 of ~0.2 M−1 s−1), providing the basis for mutually orthogonal reactivity between bulky tetrazines and isonitriles in combination with the more traditional IEDDA, and the SPAAC reactions153. Likewise, Wu and Boger recently demonstrated that unlike their 1,2,4,5- relatives, 1,2,3,5-tetrazines (22) are capable of undergoing a distinct reaction mechanism with amidines (23), which has been shown to be orthogonal to the classic IEDDA reaction involving 1,2,4,5-tetrazines (between 12 and 7)154. In pursuit of triply-orthogonal reactions, Schart and colleagues demonstrated that nitrile imines (generated in situ through photolysis from 25) are capable of chemoselectively reacting with terminal alkenes (24), even in the presence of methylcyclopropene (14) and azide handles (1) which participate in IEDDA and SPAAC reactions, respectively, thereby enabling orthogonal triple labeling in HEK cells155. Adopting a different approach, the Raines and Schomaker groups used a computational approach to rationally design a series of Sulfur-, Nitrogen-, and Oxygen-containing Cyclooctynes (SNO-OCTs; 26 and 28) that enabled differential reactivity towards either tetrazines (10) or diazoacetamides (27). Surprisingly, these handles were shown to participate in reactions that were not only orthogonal to one another, but also towards the bioorthogonal reaction between hydrazines (29) and boronic acids (30)12, creating a palette of three mutually orthogonal reactions for chemospecific triple labeling of proteins137. The orthogonality of these reactions have been summarized in supplemental Figures S3-S10. For more information on how to utilize Figure 6 for the discovery of new reaction pairings, see supplemental Figures S2 and S11.
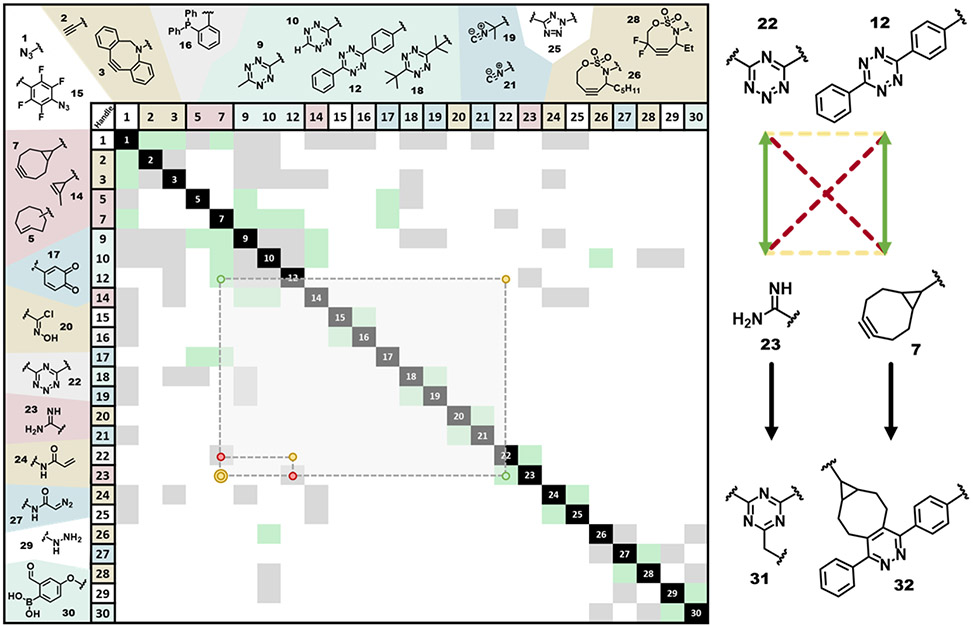
Figure 6.
Orthogonality matrix for a larger group of reactions of potential use in DEAL, including many that have not yet been used in DEAL. Coloring is as in Figure 5 (green for reactions that occur, gray for reactions that don’t or only minimally occur, and white for uncharacterized reactions). A set of two overlapping planchettes (as in Figure 5) represent information about the 6 unique reaction combinations possible for the handles and labeling agents used by Wu and Boger 2019154 as an example. These compounds and reaction combinations are shown to the right of the matrix using a scheme like Figure 4. For a more complete explanation and examples of how to use this matrix for the discovery of new chemically orthogonal reactions, see Figures S2 and S11. For a concise visual representation of the reactions that comprise this table, see Figures S3-S10.
Computational and empirical efforts to find additional compatible pairs.
As mentioned above, the use of computational screening approaches is having an increasing impact on the design of chemically orthogonal reactions. Liu et al. recently developed a distortion/interaction model to predict the impact of sterics/electronics on reaction kinetics. Using this approach, they estimated the reaction rate for 121 cycloaddition reactions, allowing the identification of putatively mutually orthogonal reactions156, demonstrating that there is a wide breadth and depth of chemical space that has yet to be explored, and that computational approaches may be powerful tools for navigating this space. Both computational and empirical approaches are continuing to uncover novel reaction modalities that can serve as a feedstock for DEAL purposes. While few of these new reactions have been adapted for GCE, we are confident that the field will continue to develop new ncAAs and their accompanying encoding subsystems. As the number of blank codons increases and opportunities for site-specific multi-ncAA encoding and labeling grow to complement them, we are eager to see which reactions will be embraced to meet these applications.
Applications of GCE-based Dual Encoding and Labeling
Genetic encoding of bioorthogonally reactive handles has enabled a number of exciting applications, ranging from protein immobilization157 to in vivo labeling158. Since bioorthogonally reactive groups essentially serve as a modular hub for the attachment of virtually any subsequent functionality, applications are far reaching. The ability to site-specifically install and subsequently label a protein at two or more locations extends the power and versatility of protein labeling by enabling the multiplexing of such abilities. Simultaneous installation of both probes and auxiliary functional moieties provides opportunities to study and manipulate proteins in an increasingly precise and sophisticated manner (Figure 1). In the below sections, we highlight published DEAL applications, as well as identify potential areas where DEAL could enhance existing approaches, and/or provide novel applications.
Förster Resonance Energy Transfer
Forster resonance energy transfer (FRET) is the nonradiative transfer of energy between an excited donor and acceptor fluorophore whose excitation spectrally overlaps with the donor’s emission, leading to a fluorescent emission from the acceptor upon excitation of the donor159. Owing to its exceptional distance-dependence (r6), FRET has been adopted as a biophysical tool to monitor intra- and inter-molecular distances and study the dynamic changes of biomolecules160. However, for the tool to be utilized to its full potential, minimally-invasive and site-specific labeling is required161, a condition that, unlike competing approaches162,163, can be uniquely satisfied by DEAL. The earliest example of dual encoding for FRET was demonstrated by the Hohsaka group, who in 2006 used chemically-acylated tRNAs bearing fluorescent ncAAs to encode a FRET pair into calmodulin164. As this example highlights, the encoding of FRET pairs using GCE has historically been limited by the relatively small size of fluorescent ncAAs that can be encoded, crowding the available spectral landscape with dim, blue-shifted dyes165; however, subsequent labeling using bioorthogonally-reactive ncAAs bypasses this limitation. In 2012, Wu and colleagues modernized the approach by encoding two bioorthogonally-reactive ncAAs, paving the way for a generalized DEAL approach that enables installation of any FRET pairs at virtually any location within a protein with minimal invasiveness28. Since then, DEAL has been used to install FRET pairs into numerous proteins such as glutamine binding protein28, ketosteroid isomerase18, and calmodulin17,139. Nevertheless, DEAL for FRET remains underutilized, despite its benefits and limited drawbacks making it the ideal tool for such applications. With the introduction of new and efficient mutually orthogonal bioorthogonal reactions, we expect to see greater adoption of DEAL for FRET applications, particularly in in vivo contexts.
In vivo Applications
Since DEAL within living cells uses mutually orthogonal bioorthogonal reactions, the approach is highly suitable for in vivo applications. Indeed, DEAL offers the most precise approach to modifying proteins in their most native environment. Despite virtually unfettered freedom to install a wide variety of modifications at practically any location, only recently have advances in bioorthogonal chemistry and improvements in encoding efficiency made the use of in vivo DEAL feasible. The first report of in vivo DEAL by Meineke and colleagues improves on their previous work80 by rationally orthogonalizing two PylRS/tRNAPyl ncAA-encoding subsystems for usage on live mammalian cells81. Polyspecificity at the ncAA-aaRS level limited the compatible subset of ncAAs leading the researchers to use CuAAC, and unfortunately, the toxicity and limited permeability of the copper catalyst limited labeling to the extracellular regions of SynNotch and the CRFR1 receptor; nevertheless, this work demonstrated the feasibility of DEAL for two-color labeling on the exterior of live cells. While the Meineke system could not be used for cytoplasmic labeling, our group has shown that through a careful configuration that maximizes both encoding and labeling efficiency, labeling inside live cells can be achieved. Our system relied on highly active ncAA-encoding subsystems that encoded two ncAAs that optimized SPAAC and IEDDA for robust in vivo labeling The abilities we demonstrated inside live E. coli cells included: two-color labeling (for the dual labeling of a single protein for i.e. FRET, or for the dual labeling of distinct populations of proteins for i.e. localization/interaction studies), protein-protein crosslinking, and protein stapling1. Despite this progress, there is much room for improvements in in vivo DEAL, and given the freedom and precision offered, we expect a rapid expansion in in vivo applications.
Additional Applications
Aside from FRET and various in vivo demonstrations, DEAL has been used in applications ranging from material sciences to biomedicine. One early application was for the construction of antibody-based theranostic agents. The Schultz group used DEAL to site-specifically install both an imaging agent (Alexa488) and an auristatin warhead onto a single antibody, the resulting dual-functionalized theranostic serving both for efficient drug-delivery and imaging30. The precision of DEAL lends itself to easier characterization compared to existing stochastic labeling approaches166,167 and greatly extends the functional space available for modification. Moreover, we can see this approach synergizing with emerging modalities, such as bispecific antibodies to provide a biologic capable of both bivalent binding, while also hosting two distinct cargos, such as drugs, probes, or conjugated proteins, greatly expanding the functionality of existing treatments.
Another potential DEAL application is protein stapling. With the potential to stabilize/control function168, protein stapling has traditionally been limited to the use of engineered disulfide bonds which are not universally employable and are susceptible to reduction169. As demonstrated by Chin and others, DEAL can provide a generalizable stapling approach that does not have those limitations1,34,81.
DEAL has also been applied to answer key questions in material sciences. The development of protein-based biomaterials such as biosensors, proteomic microarrays, and immobilized biocatalysts rely on proteins that are attached to a surface170; however, relatively little is known about how proteins interact with surfaces171. In a collaboration with the Kaar group, we used DEAL in combination with traditional cysteine labeling to generate triply-reactive proteins. This combination enabled us to use these three different handles to both monitor protein conformation by FRET while also achieving covalent immobilization at varying rates, depending on which handle was used for the immobilization reaction. This allowed us to observe that the rate of protein immobilization strongly affected the folded state – and thereby the activity – of the immobilized protein population95. These studies illustrate that dual encoding and labeling has the potential to solve meaningful problems associated with site-specific manipulation of proteins for a wide variety of applications in various fields of study.
Future Possibilities
While many uses of DEAL have already been exemplified, we have only seen the “tip of the iceberg” in terms of the potential for novel DEAL applications. For example, while the Lemke group has demonstrated that mutually orthogonal bioorthogonal reactions can be used to track two protein populations by super-resolution microscopy, their use of a single permissive suppression system and pulse-chase design limited the trackable species to two time-resolved populations of the same protein23. A more flexible DEAL system, alternatively, would enable seamless tracking of two (or more) distinct proteins, the use of multiple probe modalities, the use of FRET-based tracking, and even the tracking of multiple temporally-resolved protein subpopulations via pulse-chase approaches. Also, as we showed through our simple demonstration in E. coli1, DEAL can also be used for the construction of topologically-defined protein-protein complexes. Such an approach could be used for controlled protein dimerization, the artificial assembly of nanomaterials172, the construction of metabolon-like protein superstructures173-175, and/or the targeting of proteins for degradation through controlled, covalent “molecular glue-like” labeling176, all in vivo. Since the two encoded handles behave like a “blank-slate,” one can envision dual labeling systems using virtually any functionality. Included among them is the attachment of two distinct prosthetic groups, such as stimuli-responsive polymers177,178, tethered small-molecule regulators39,179, and/or multi-modal probes to generate multi-functional protein-based biosensors with functionalities that cannot be accessed naturally, or with current technologies. Using this DEAL approach, we envision that virtually any protein can become a chassis for extended biochemical functionality for both in vivo and in vitro applications.
Conclusions and Outlook
As the examples highlighted in this review make clear, DEAL offers unparalleled freedom and precision for the dual modification of proteins even in their most native environment. Despite these advantages, DEAL remains underutilized. We foresee a future of expanding DEAL capabilities through the development of novel, high-yielding, rapid, and mutually orthogonal bioorthogonal reactions as well as the encoding systems to support them. A greater understanding of interplay between the components involved in dual encoding will improve the development of DEAL systems and illuminate the path from “trial-and-error” approaches to more straightforward, principled approaches. In this regard, we have emphasized a “relative fidelity paradigm,” in which it is not absolute qualities, but the competition between various components that dictates the success of a DEAL experiment; and as such, we expect a systems biology approach to provide useful insights into the fine-tuning of such systems.
Even more importantly, we believe the key to greater adoption of DEAL is to substantially improve the efficiencies of ncAA-encoding subsystems – which are often ~3 orders of magnitude lower than their natural counterparts78,180,181. Although low yields can be workable for single ncAA-encoding experiments, the challenges they cause are exacerbated under DEAL conditions and severely limit the potential of the tool. We are excited to see that scalable solutions exist for many other limitations of DEAL, including approaches to designing and discovering new orthogonal aaRS/tRNA pairs, and approaches to expand the available codon space. Concordant with such advances an increasing availability of necessary ncAAs and labeling reagents, we expect to see increased interest in dual- and multi-ncAA encoding and labeling, and with this, new and innovative applications that capitalize on them. The development of such applications will contribute to a greater understanding of the roles of proteins in their biological settings and will be an important tool in the next generation of protein-based biotechnologies.
Supplementary Material
Significance.
As the chemical biology community continues to push the boundaries of protein research, there is an increasing need for tools that enable the site-selective modification of proteins at multiple defined locations. Among the top technologies for meeting this demand, genetic code expansion is well suited for such a task due to its ability to direct the site-specific encoding virtually any reactive handle at any location in any protein with minimal invasiveness. Despite this potential, the use of genetic code expansion for multiple site-specific protein labeling remains underutilized. In this review, we catalogue the available encoding systems and reactions and aim to provide a conceptual toolkit that will guide users in the basic principles and practices of utilizing genetic code expansion for Dual Encoding And Labeling (DEAL).
Acknowledgements
The authors would like to thank Dr. Richard Cooley and Alex J. Eddins for thoughtful conversations and input during the writing of this review. This review was funded partly by the GCE4All Biomedical Technology Development and Dissemination Center supported by the National Institute of General Medical Science, grant RM1-GM144227 as well as National Institutes of Health grant 1R01GM131168-01 and National Science Foundation, NSF-2054824 awarded to R.A.M.
Glossary
- Encoding
The full set of processes required for the site-specific incorporation of an amino acid into a growing peptide chain during translation of mRNA codons to proteins.
- Labeling
The conjugation of a functional group present in a biomolecule with an exogenous chemical moiety.
- Blank codon
Any codon which does not does not specify a canonical amino acid. May refer to stop codons, quadruplet codons, unnatural codons, or sense codons which lack a cognate tRNA capable of decoding the codon in question.
- Suppression
The process of outcompeting a release factor to encode an amino acid in place of translational termination.
- Mutual orthogonality
A property of two or more encoding subsystems wherein each subsystem does not interact with (i.e. is orthogonal to) all other subsystems.
- Fidelity
The faithful encoding of an amino acid in response to its cognate codon.
- Absolute fidelity
A form of fidelity wherein an encoding subsystem exhibits fidelity (i.e. does not incorporate a different amino acid) even in the absence of its cognate amino acid.
- Relative fidelity
A form of fidelity wherein an encoding subsystem exhibits fidelity when its cognate amino acid is present, but may incorporate other amino acids in the absence of its cognate amino acid.
- Permissivity
The ability of an aaRS to recognize multiple ncAAs but not canonical amino acids.
- Identity factor
A structural element or locus of a tRNA molecule that confers specific recognition by their cognate aaRSs.
- Near-cognate suppression
An error in translation wherein the wrong amino acid is encoded due to a mismatch between the mRNA codon and a similar (but non-cognate) tRNA anticodon during ribosomal decoding.
- Frameshift codon
A four-base codon; also referred to as a quadruplet codon.
- Chemical orthogonality
A property of two or more chemical reactions wherein reactions only occur between the intended cognate groups, and not with any of the other chemical groups present. Also referred to as mutually orthogonal bioorthogonality.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- (1).Bednar RM; Jana S; Kuppa S; Franklin R; Beckman J; Antony E; Cooley RB; Mehl RA Genetic Incorporation of Two Mutually Orthogonal Bioorthogonal Amino Acids That Enable Efficient Protein Dual-Labeling in Cells. ACS Chem. Biol 2021, 16 (11), 2612–2622. 10.1021/acschembio.1c00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Yang X; Qian K Protein O-GlcNAcylation: Emerging Mechanisms and Functions. Nat Rev Mol Cell Biol 2017, 18 (7), 452–465. 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gilormini P-A; Batt AR; Pratt MR; Biot C Asking More from Metabolic Oligosaccharide Engineering. Chem Sci 2018, 9 (39), 7585–7595. 10.1039/c8sc02241k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Giepmans BNG; Adams SR; Ellisman MH; Tsien RY The Fluorescent Toolbox for Assessing Protein Location and Function. Science 2006, 312 (5771), 217–224. 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- (5).Griffin BA; Adams SR; Tsien RY Specific Covalent Labeling of Recombinant Protein Molecules inside Live Cells. Science 1998, 281 (5374), 269–272. 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- (6).Lotze J; Reinhardt U; Seitz O; Beck-Sickinger AG, Peptide-Tags for Site-Specific Protein Labelling in Vitro and in Vivo. Molecular BioSystems 2016, 12 (6), 1731–1745. 10.1039/C6MB00023A. [DOI] [PubMed] [Google Scholar]
- (7).Aime S; Botta M; Fasano M; Terreno E Lanthanide(III) Chelates for NMR Biomedical Applications. Chem. Soc. Rev 1998, 27 (1), 19–29. 10.1039/A827019Z. [DOI] [Google Scholar]
- (8).Rashidian M; Kumarapperuma SC; Gabrielse K; Fegan A; Wagner CR; Distefano MD Simultaneous Dual Protein Labeling Using a Triorthogonal Reagent. J. Am. Chem. Soc 2013, 135 (44), 16388–16396. 10.1021/ja403813b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lopez Aguilar A; Meng L; Hou X; Li W; Moremen KW; Wu P Sialyltransferase-Based Chemoenzymatic Histology for the Detection of N- and O-Glycans. Bioconjugate Chem. 2018, 29 (4), 1231–1239. 10.1021/acs.bioconjchem.8b00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Harmand TJ; Bousbaine D; Chan A; Zhang X; Liu DR; Tam JP; Ploegh HL One-Pot Dual Labeling of IgG 1 and Preparation of C-to-C Fusion Proteins Through a Combination of Sortase A and Butelase 1. Bioconjug Chem 2018, 29 (10), 3245–3249. 10.1021/acs.bioconjchem.8b00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhang Y; Blanden MJ; Sudheer C; Gangopadhyay SA; Rashidian M; Hougland JL; Distefano MD Simultaneous Site-Specific Dual Protein Labeling Using Protein Prenyltransferases. Bioconjug Chem 2015, 26 (12), 2542–2553. 10.1021/acs.bioconjchem.5b00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chio TI; Gu H; Mukherjee K; Tumey LN; Bane SL Site-Specific Bioconjugation and Multi-Bioorthogonal Labeling via Rapid Formation of a Boron–Nitrogen Heterocycle. Bioconjugate Chem. 2019, 30 (5), 1554–1564. 10.1021/acs.bioconjchem.9b00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Bruins JJ; Blanco-Ania D; van der Doef V; van Delft FL; Albada B Orthogonal, Dual Protein Labelling by Tandem Cycloaddition of Strained Alkenes and Alkynes to Ortho -Quinones and Azides. Chem. Commun 2018, 54 (53), 7338–7341. 10.1039/C8CC02638F. [DOI] [PubMed] [Google Scholar]
- (14).Keppler A; Gendreizig S; Gronemeyer T; Pick H; Vogel H; Johnsson K A General Method for the Covalent Labeling of Fusion Proteins with Small Molecules in Vivo. Nature Biotechnology 2003, 21 (1), 86–89. 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- (15).Gautier A; Juillerat A; Heinis C; Corrêa IR; Kindermann M; Beaufils F; Johnsson K An Engineered Protein Tag for Multiprotein Labeling in Living Cells. Chemistry & Biology 2008, 15 (2), 128–136. 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- (16).Los GV; Encell LP; McDougall MG; Hartzell DD; Karassina N; Zimprich C; Wood MG; Learish R; Ohana RF; Urh M; Simpson D; Mendez J; Zimmerman K; Otto P; Vidugiris G; Zhu J; Darzins A; Klaubert DH; Bulleit RF; Wood KV HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem Biol 2008, 3 (6), 373–382. 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- (17).Wang K; Sachdeva A; Cox DJ; Wilf NM; Lang K; Wallace S; Mehl RA; Chin JW Optimized Orthogonal Translation of Unnatural Amino Acids Enables Spontaneous Protein Double-Labelling and FRET. Nature Chemistry 2014, 6 (5), 393–403. 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chatterjee A; Sun SB; Furman JL; Xiao H; Schultz PG A Versatile Platform for Single- and Multiple-Unnatural Amino Acid Mutagenesis in Escherichia Coli. Biochemistry 2013, 52 (10), 1828–1837. 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cole CM; Yang J; Šečkutė J; Devaraj NK Fluorescent Live-Cell Imaging of Metabolically Incorporated Unnatural Cyclopropene-Mannosamine Derivatives. Chembiochem 2013, 14 (2), 205–208. 10.1002/cbic.201200719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Niederwieser A; Späte A-K; Nguyen LD; Jüngst C; Reutter W; Wittmann V Two-Color Glycan Labeling of Live Cells by a Combination of Diels–Alder and Click Chemistry. Angewandte Chemie International Edition 2013, 52 (15), 4265–4268. 10.1002/anie.201208991. [DOI] [PubMed] [Google Scholar]
- (21).Ge J; Zhang C-W; Ng XW; Peng B; Pan S; Du S; Wang D; Li L; Lim K-L; Wohland T; Yao SQ Puromycin Analogues Capable of Multiplexed Imaging and Profiling of Protein Synthesis and Dynamics in Live Cells and Neurons. Angewandte Chemie International Edition 2016, 55 (16), 4933–4937. 10.1002/anie.201511030. [DOI] [PubMed] [Google Scholar]
- (22).Zürn A; Klenk C; Zabel U; Reiner S; Lohse MJ; Hoffmann C Site-Specific, Orthogonal Labeling of Proteins in Intact Cells with Two Small Biarsenical Fluorophores. Bioconjugate Chem. 2010, 21 (5), 853–859. 10.1021/bc900394j. [DOI] [PubMed] [Google Scholar]
- (23).Nikić I; Plass T; Schraidt O; Szymański J; Briggs JAG; Schultz C; Lemke EA Minimal Tags for Rapid Dual-Color Live-Cell Labeling and Super-Resolution Microscopy. Angew Chem Int Ed Engl 2014, 53 (8), 2245–2249. 10.1002/anie.201309847. [DOI] [PubMed] [Google Scholar]
- (24).Zhang MM; Tsou LK; Charron G; Raghavan AS; Hang HC Tandem Fluorescence Imaging of Dynamic S-Acylation and Protein Turnover. PNAS 2010, 107 (19), 8627–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mie M; Naoki T; Kobatake E Development of a Split SNAP-CLIP Double Labeling System for Tracking Proteins Following Dissociation from Protein-Protein Complexes in Living Cells. Anal Chem 2016, 88 (16), 8166–8171. 10.1021/acs.analchem.6b01906. [DOI] [PubMed] [Google Scholar]
- (26).Kim J; Seo M-H; Lee S; Cho K; Yang A; Woo K; Kim H-S; Park H-S Simple and Efficient Strategy for Site-Specific Dual Labeling of Proteins for Single-Molecule Fluorescence Resonance Energy Transfer Analysis. Anal. Chem 2013, 85 (3), 1468–1474. 10.1021/ac303089v. [DOI] [PubMed] [Google Scholar]
- (27).Lammers C; Hahn LE; Neumann H Optimized Plasmid Systems for the Incorporation of Multiple Different Unnatural Amino Acids by Evolved Orthogonal Ribosomes. ChemBloChem 2014, 15 (12), 1800–1804. 10.1002/cbic.201402033. [DOI] [PubMed] [Google Scholar]
- (28).Wu B; Wang Z; Huang Y; Liu WR Catalyst-Free and Site-Specific One-Pot Dual Labeling of a Protein Directed by Two Genetically Incorporated Noncanonical Amino Acids. Chemblochem 2012, 13 (10), 1405–1408. 10.1002/cbic.201200281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Widder P; Berner F; Summerer D; Drescher M Double Nitroxide Labeling by Copper-Catalyzed Azide–Alkyne Cycloadditions with Noncanonical Amino Acids for Electron Paramagnetic Resonance Spectroscopy. ACS Chem. Biol 2019, 14 (5), 839–844. 10.1021/acschembio.8b01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Xiao H; Chatterjee A; Choi S; Bajjuri KM; Sinha SC; Schultz PG Genetic Incorporation of Multiple Unnatural Amino Acids into Proteins in Mammalian Cells. Angewandte Chemle International Edition 2013, 52 (52), 14080–14083. 10.1002/anie.201308137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Dickgiesser S; Rasche N; Nasu D; Middel S; Hörner S; Avrutina O; Diederichsen U; Kolmar H Self-Assembled Hybrid Aptamer-Fc Conjugates for Targeted Delivery: A Modular Chemoenzymatic Approach. ACS Chem. Biol 2015, 10 (9), 2158–2165. 10.1021/acschembio.5b00315. [DOI] [PubMed] [Google Scholar]
- (32).Rutkowska A; Plass T; Hoffmann J-E; Yushchenko DA; Feng S; Schultz C T-CrAsH: A Heterologous Chemical Crosslinker. ChemBioChem 2014, 15 (12), 1765–1768. 10.1002/cbic.201402189. [DOI] [PubMed] [Google Scholar]
- (33).Worthy HL; Auhim HS; Jamieson WD; Pope JR; Wall A; Batchelor R; Johnson RL; Watkins DW; Rizkallah P; Castell OK; Jones DD Positive Functional Synergy of Structurally Integrated Artificial Protein Dimers Assembled by Click Chemistry. Commun Chem 2019, 2 (1), 1–12. 10.1038/s42004-019-0185-5. [DOI] [Google Scholar]
- (34).Neumann H; Wang K; Davis L; Garcia-Alai M; Chin JW Encoding Multiple Unnatural Amino Acids via Evolution of a Quadruplet-Decoding Ribosome. Nature 2010, 464 (7287), 441–444. 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- (35).de Montes EG; Jiménez-Moreno E; Oliveira BL; Navo CD; Cal PMSD; Jiménez-Osés G; Robina I; Moreno-Vargas AJ; Bernardes GJL Azabicyclic Vinyl Sulfones for Residue-Specific Dual Protein Labelling. Chem. Sci 2019, 10 (16), 4515–4522. 10.1039/C9SC00125E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Geissinger SE; Schreiber A; Huber MC; Stühn LG; Schiller SM Adjustable Bioorthogonal Conjugation Platform for Protein Studies in Live Cells Based on Artificial Compartments. ACS Synth. Biol 2020, 9 (4), 827–842. 10.1021/acssynbio.9b00494. [DOI] [PubMed] [Google Scholar]
- (37).Li L; Zhang S-Y; Li Y-M; Chen Y-X Dual-Labeling of Ubiquitin Proteins by Chemoselective Reactions for Sensing UCH-L3. Mol Biosyst 2016, 12 (6), 1764–1767. 10.1039/c6mb00165c. [DOI] [PubMed] [Google Scholar]
- (38).Wu Q; Zhang KY; Dai P; Zhu H; Wang Y; Song L; Wang L; Liu S; Zhao Q; Huang W Bioorthogonal “Labeling after Recognition” Affording an FRET-Based Luminescent Probe for Detecting and Imaging Caspase-3 via Photoluminescence Lifetime Imaging. J. Am. Chem. Soc 2020, 142 (2), 1057–1064. 10.1021/jacs.9b12191. [DOI] [PubMed] [Google Scholar]
- (39).Tsai Y-H; Essig S; James JR; Lang K; Chin JW Selective, Rapid and Optically Switchable Regulation of Protein Function in Live Mammalian Cells. Nat Chem 2015, 7 (7), 554–561. 10.1038/nchem.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Xue L; Prifti E; Johnsson K A General Strategy for the Semisynthesis of Ratiometric Fluorescent Sensor Proteins with Increased Dynamic Range. J. Am. Chem. Soc 2016, 138 (16), 5258–5261. 10.1021/jacs.6b03034. [DOI] [PubMed] [Google Scholar]
- (41).Ochtrop P; Hackenberger CPR Recent Advances of Thiol-Selective Bioconjugation Reactions. Current Opinion in Chemical Biology 2020, 58, 28–36. 10.1016/j.cbpa.2020.04.017. [DOI] [PubMed] [Google Scholar]
- (42).Lazar AC; Wang L; Blättler WA; Amphlett G; Lambert JM; Zhang W Analysis of the Composition of Immunoconjugates Using Size-Exclusion Chromatography Coupled to Mass Spectrometry. Rapid Commun Mass Spectrom 2005, 19 (13), 1806–1814. 10.1002/rcm.1987. [DOI] [PubMed] [Google Scholar]
- (43).Tsuchikama K; An Z Antibody-Drug Conjugates: Recent Advances in Conjugation and Linker Chemistries. Protein Cell 2018, 9 (1), 33–46. 10.1007/s13238-016-0323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ratner V; Kahana E; Eichler M; Haas E A General Strategy for Site-Specific Double Labeling of Globular Proteins for Kinetic FRET Studies. Bioconjug Chem 2002, 13 (5), 1163–1170. 10.1021/bc025537b. [DOI] [PubMed] [Google Scholar]
- (45).Kao MW-P; Yang L-L; Lin JC-K; Lim T-S; Fann W; Chen RP-Y Strategy for Efficient Site-Specific FRET-Dye Labeling of Ubiquitin. Bioconjug Chem 2008, 19 (6), 1124–1126. 10.1021/bc700480j. [DOI] [PubMed] [Google Scholar]
- (46).Yano Y; Matsuzaki K Live-Cell Imaging of Membrane Proteins by a Coiled-Coil Labeling Method—Principles and Applications. Biochimica et Biophysica Acta (BBA) - Biomembranes 2019, 1861 (5), 1011–1017. 10.1016/j.bbamem.2019.02.009. [DOI] [PubMed] [Google Scholar]
- (47).Ojida A; Honda K; Shinmi D; Kiyonaka S; Mori Y; Hamachi I Oligo-Asp Tag/Zn(II) Complex Probe as a New Pair for Labeling and Fluorescence Imaging of Proteins. J. Am. Chem. Soc 2006, 128 (32), 10452–10459. 10.1021/ja0618604. [DOI] [PubMed] [Google Scholar]
- (48).Chen B; Cao H; Yan P; Mayer MU; Squier TC Identification of an Orthogonal Peptide Binding Motif for Biarsenical Multiuse Affinity Probes. Bioconjug Chem 2007, 18 (4), 1259–1265. 10.1021/bc0603900. [DOI] [PubMed] [Google Scholar]
- (49).Rashidian M; Dozier JK; Distefano MD Chemoenzymatic Labeling of Proteins: Techniques and Approaches. Bioconjug Chem 2013, 24 (8), 1277–1294. 10.1021/bc400102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Chin JW Expanding and Reprogramming the Genetic Code. Nature 2017, 550 (7674), 53–60. 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- (51).Young DD; Schultz PG Playing with the Molecules of Life. ACS Chem Biol 2018, 13 (4), 854–870. 10.1021/acschembio.7b00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Shandell MA; Tan Z; Cornish VW Genetic Code Expansion: A Brief History and Perspective. Biochemistry 2021, 60 (46), 3455–3469. 10.1021/acs.biochem.1c00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).de la Torre D; Chin JW Reprogramming the Genetic Code. Nature Reviews Genetics 2021, 22 (3), 169–184. 10.1038/s41576-020-00307-7. [DOI] [PubMed] [Google Scholar]
- (54).Chung CZ; Amikura K; Söll D Using Genetic Code Expansion for Protein Biochemical Studies. Front. Bioeng. Biotechnol 2020, 8. 10.3389/fbioe.2020.598577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Johnson JA; Lu YY; Van Deventer JA; Tirrell DA Residue-Specific Incorporation of Non-Canonical Amino Acids into Proteins: Recent Developments and Applications. Curr Opin Chem Biol 2010, 14 (6), 774–780. 10.1016/j.cbpa.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Acevedo-Rocha CG; Budisa N Xenomicrobiology: A Roadmap for Genetic Code Engineering. Microb Biotechnol 2016, 9 (5), 666–676. 10.1111/1751-7915.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Hoesl MG; Budisa N In Vivo Incorporation of Multiple Noncanonical Amino Acids into Proteins. Angewandte Chemie International Edition 2011, 50 (13), 2896–2902. 10.1002/anie.201005680. [DOI] [PubMed] [Google Scholar]
- (58).Cui Z; Johnston WA; Alexandrov K Cell-Free Approach for Non-Canonical Amino Acids Incorporation Into Polypeptides. Front. Bioeng. Biotechnol 2020, 8. 10.3389/fbioe.2020.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Khambhati K; Bhattacharjee G; Gohil N; Braddick D; Kulkarni V; Singh V Exploring the Potential of Cell-Free Protein Synthesis for Extending the Abilities of Biological Systems. Front. Bioeng. Biotechnol 2019, 7. 10.3389/fbioe.2019.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Hirose H; Tsiamantas C; Katoh T; Suga H In Vitro Expression of Genetically Encoded Non-Standard Peptides Consisting of Exotic Amino Acid Building Blocks. Current Opinion in Biotechnology 2019, 58, 28–36. 10.1016/j.copbio.2018.10.012. [DOI] [PubMed] [Google Scholar]
- (61).Rogers JM; Suga H Discovering Functional, Non-Proteinogenic Amino Acid Containing, Peptides Using Genetic Code Reprogramming. Org. Biomol. Chem 2015, 13 (36), 9353–9363. 10.1039/C5OB01336D. [DOI] [PubMed] [Google Scholar]
- (62).Vinogradov AA; Yin Y; Suga H Macrocyclic Peptides as Drug Candidates: Recent Progress and Remaining Challenges. J. Am. Chem. Soc 2019, 141 (10), 4167–4181. 10.1021/jacs.8b13178. [DOI] [PubMed] [Google Scholar]
- (63).Arranz-Gibert P; Patel JR; Isaacs FJ The Role of Orthogonality in Genetic Code Expansion. Life (Basel) 2019, 9 (3). 10.3390/life9030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Beyer JN; Hosseinzadeh P; Gottfried-Lee I; Van Fossen EM; Zhu P; Bednar RM; Karplus PA; Mehl RA; Cooley RB Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine TRNA Synthetase. J Mol Biol 2020, 432 (16), 4690–4704. 10.1016/j.jmb.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Antonczak AK; Simova Z; Yonemoto IT; Bochtler M; Piasecka A; Czapińska H; Brancale A; Tippmann EM Importance of Single Molecular Determinants in the Fidelity of Expanded Genetic Codes. PNAS 2011, 108 (4), 1320–1325. 10.1073/pnas.1012276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Dieterich DC; Link AJ; Graumann J; Tirrell DA; Schuman EM Selective Identification of Newly Synthesized Proteins in Mammalian Cells Using Bioorthogonal Noncanonical Amino Acid Tagging (BONCAT). Proc Natl Acad Scl U S A 2006, 103 (25), 9482–9487. 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Kaiser F; Krautwurst S; Salentin S; Haupt VJ; Leberecht C; Bittrich S; Labudde D; Schroeder M The Structural Basis of the Genetic Code: Amino Acid Recognition by Aminoacyl-TRNA Synthetases. Scientific Reports 2020, 10 (1), 12647. 10.1038/s41598-020-69100-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Wang L; Brock A; Herberich B; Schultz PG Expanding the Genetic Code of Escherichia Coli. Science 2001, 292 (5516), 498–500. 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- (69).Smolskaya S; Andreev YA Site-Specific Incorporation of Unnatural Amino Acids into Escherichia Coli Recombinant Protein: Methodology Development and Recent Achievement. Biomolecules 2019, 9 (7). 10.3390/biom9070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Cooley RB; Feldman JL; Driggers CM; Bundy TA; Stokes AL; Karplus PA; Mehl RA Structural Basis of Improved Second-Generation 3-Nitro-Tyrosine TRNA Synthetases. Biochemistry 2014, 53 (12), 1916–1924. 10.1021/bi5001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Italia JS; Addy PS; Wrobel CJJ; Crawford LA; Lajoie MJ; Zheng Y; Chatterjee A An Orthogonalized Platform for Genetic Code Expansion in Both Bacteria and Eukaryotes. Nature Chemical Biology 2017, 13 (4), 446–450. 10.1038/nchembio.2312. [DOI] [PubMed] [Google Scholar]
- (72).Italia JS; Addy PS; Erickson SB; Peeler JC; Weerapana E; Chatterjee A Mutually Orthogonal Nonsense-Suppression Systems and Conjugation Chemistries for Precise Protein Labeling at up to Three Distinct Sites. Journal of the American Chemical Society 2019. 10.1021/jacs.8b12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Odoi KA; Huang Y; Rezenom YH; Liu WR Nonsense and Sense Suppression Abilities of Original and Derivative Methanosarcina Mazei Pyrrolysyl-TRNA Synthetase-TRNAPyl Pairs in the Escherichia Coli BL21(DE3) Cell Strain. PLoS One 2013, 8 (3). 10.1371/journal.pone.0057035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Gaston MA; Jiang R; Krzycki JA Functional Context, Biosynthesis, and Genetic Encoding of Pyrrolysine. Curr Opin Microbiol 2011, 14 (3), 342–349. 10.1016/j.mib.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Miyake-Stoner SJ; Refakis CA; Hammill JT; Lusic H; Hazen JL; Deiters A; Mehl RA Generating Permissive Site-Specific Unnatural Aminoacyl-TRNA Synthetases. Biochemistry 2010, 49 (8), 1667–1677. 10.1021/bi901947r. [DOI] [PubMed] [Google Scholar]
- (76).Cooley RB; Karplus PA; Mehl RA Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-Canonical Amino Acid-TRNA Synthetases. Chembiochem 2014, 15 (12), 1810–1819. 10.1002/cbic.201402180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Stokes AL; Miyake-Stoner SJ; Peeler JC; Nguyen DP; Hammer RP; Mehl RA Enhancing the Utility of Unnatural Amino Acid Synthetases by Manipulating Broad Substrate Specificity. Mol Biosyst 2009, 5 (9), 1032–1038. 10.1039/b904032c. [DOI] [PubMed] [Google Scholar]
- (78).Bryson DI; Fan C; Guo L-T; Miller C; Söll D; Liu DR Continuous Directed Evolution of Aminoacyl-TRNA Synthetases. Nat Chem Biol 2017, 13 (12), 1253–1260. 10.1038/nchembio.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Zheng Y; Mukherjee R; Chin MA; Igo P; Gilgenast MJ; Chatterjee A Expanding the Scope of Single and Dual Noncanonical Amino Acid Mutagenesis in Mammalian Cells Using Orthogonal Polyspecific Leucyl-TRNA Synthetases. Biochemistry 2018, 57 (4), 441–445. 10.1021/acs.biochem.7b00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Meineke B; Heimgärtner J; Lafranchi L; Elsässer SJ Methanomethylophilus Alvus Mx1201 Provides Basis for Mutual Orthogonal Pyrrolysyl TRNA/Aminoacyl-TRNA Synthetase Pairs in Mammalian Cells. ACS Chem. Biol 2018, 13 (11), 3087–3096. 10.1021/acschembio.8b00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Meineke B; Heimgärtner J; Eirich J; Landreh M; Elsässer SJ Site-Specific Incorporation of Two NcAAs for Two-Color Bioorthogonal Labeling and Crosslinking of Proteins on Live Mammalian Cells. Cell Reports 2020, 31 (12), 107811. 10.1016/j.celrep.2020.107811. [DOI] [PubMed] [Google Scholar]
- (82).Giegé R; Sissler M; Florentz C Universal Rules and Idiosyncratic Features in TRNA Identity. Nucleic Acids Research 1998, 26 (22), 5017–5035. 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Giegé R; Frugier M Transfer RNA Structure and Identity; Landes Bioscience, 2013. [Google Scholar]
- (84).Melnikov SV; Söll D Aminoacyl-TRNA Synthetases and TRNAs for an Expanded Genetic Code: What Makes Them Orthogonal? Int J Mol Sci 2019, 20 (8). 10.3390/ijms20081929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Krahn N; Tharp JM; Crnković A; Söll D Engineering Aminoacyl-TRNA Synthetases for Use in Synthetic Biology. Enzymes 2020, 48, 351–395. 10.1016/bs.enz.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Tsunoda M; Kusakabe Y; Tanaka N; Ohno S; Nakamura M; Senda T; Moriguchi T; Asai N; Sekine M; Yokogawa T; Nishikawa K; Nakamura KT Structural Basis for Recognition of Cognate TRNA by Tyrosyl-TRNA Synthetase from Three Kingdoms. Nucleic Acids Res 2007, 35 (13), 4289–4300. 10.1093/nar/gkm417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Tharp JM; Ehnbom A; Liu WR TRNAPyl: Structure, Function, and Applications. RNA Biol 2017, 15 (4–5), 441–452. 10.1080/15476286.2017.1356561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Ambrogelly A; Gundllapalli S; Herring S; Polycarpo C; Frauer C; Söll D Pyrrolysine Is Not Hardwired for Cotranslational Insertion at UAG Codons. Proc Natl Acad Sci U S A 2007, 104 (9), 3141–3146. 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Nozawa K; O’Donoghue P; Gundllapalli S; Araiso Y; Ishitani R; Umehara T; Söll D; Nureki O Pyrrolysyl-TRNA Synthetase–TRNA Pyl Structure Reveals the Molecular Basis of Orthogonality. Nature 2009, 457 (7233), 1163–1167. 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Wan W; Huang Y; Wang Z; Russell WK; Pai P-J; Russell DH; Liu WR A Facile System for Genetic Incorporation of Two Different Noncanonical Amino Acids into One Protein in Escherichia Coli. Angew Chem Int Ed Engl 2010, 49 (18), 3211–3214. 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- (91).Sachdeva A; Wang K; Elliott T; Chin JW Concerted, Rapid, Quantitative, and Site-Specific Dual Labeling of Proteins. J. Am. Chem. Soc 2014, 136 (22), 7785–7788. 10.1021/ja4129789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Zheng Y; Gilgenast MJ; Hauc S; Chatterjee A Capturing Post-Translational Modification-Triggered Protein–Protein Interactions Using Dual Noncanonical Amino Acid Mutagenesis. ACS Chem. Biol 2018, 13 (5), 1137–1141. 10.1021/acschembio.8b00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Hankore ED; Zhang L; Chen Y; Liu K; Niu W; Guo J Genetic Incorporation of Noncanonical Amino Acids Using Two Mutually Orthogonal Quadruplet Codons. ACS Synth Biol 2019, 8 (5), 1168–1174. 10.1021/acssynbio.9b00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Oller-Salvia B; Chin JW Efficient Phage Display with Multiple Distinct Non-Canonical Amino Acids Using Orthogonal Ribosome-Mediated Genetic Code Expansion. Angewandte Chemle International Edition 2019, 58 (32), 10844–10848. 10.1002/anie.201902658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Chaparro Sosa AF; Bednar RM; Mehl RA; Schwartz DK; Kaar JL Faster Surface Ligation Reactions Improve Immobilized Enzyme Structure and Activity. J. Am. Chem. Soc 2021, 143 (18), 7154–7163. 10.1021/jacs.1c02375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Tharp JM; Liu WR Using Amber and Ochre Nonsense Codons to Code Two Different Noncanonical Amino Acids in One Protein Gene. Methods Mol Biol 2018, 1728, 147–154. 10.1007/978-1-4939-7574-7_9. [DOI] [PubMed] [Google Scholar]
- (97).Fischer EC; Hashimoto K; Zhang Y; Feldman AW; Dien VT; Karadeema RJ; Adhikary R; Ledbetter MP; Krishnamurthy R; Romesberg FE New Codons for Efficient Production of Unnatural Proteins in a Semisynthetic Organism. Nature Chemical Biology 2020, 16 (5), 570–576. 10.1038/s41589-020-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Tharp JM; Vargas-Rodriguez O; Schepartz A; Söll D Genetic Encoding of Three Distinct Noncanonical Amino Acids Using Reprogrammed Initiator and Nonsense Codons. ACS Chem. Biol 2021, 16 (4), 766–774. 10.1021/acschembio.1c00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Zheng Y; Addy PS; Mukherjee R; Chatterjee A Defining the Current Scope and Limitations of Dual Noncanonical Amino Acid Mutagenesis in Mammalian Cells. Chem. Sci 2017, 8 (10), 7211–7217. 10.1039/C7SC02560B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Shi N; Tong L; Lin H; Zheng Z; Zhang H; Dong L; Yang Y; Shen Y; Xia Q Optimizing ERF1 to Enable the Genetic Encoding of Three Distinct Noncanonical Amino Acids in Mammalian Cells. Adv Biol (Weinh) 2022, e2200092. 10.1002/adbi.202200092. [DOI] [PubMed] [Google Scholar]
- (101).Italia JS; Latour C; Wrobel CJJ; Chatterjee A Resurrecting the Bacterial Tyrosyl-TRNA Synthetase/TRNA Pair for Expanding the Genetic Code of Both E. Coli and Eukaryotes. Cell Chem Biol 2018, 25 (10), 1304–1312.e5. 10.1016/j.chembiol.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Borrel G; Gaci N; Peyret P; O’Toole PW; Gribaldo S; Brugère J-F Unique Characteristics of the Pyrrolysine System in the 7th Order of Methanogens: Implications for the Evolution of a Genetic Code Expansion Cassette. Archaea 2014, 2014, e374146. 10.1155/2014/374146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Willis JCW; Chin JW Mutually Orthogonal Pyrrolysyl-TRNA Synthetase/TRNA Pairs. Nature Chemistry 2018, 10 (8), 831–837. 10.1038/s41557-018-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Beránek V; Willis JCW; Chin JW An Evolved Methanomethylophilus Alvus Pyrrolysyl-TRNA Synthetase/TRNA Pair Is Highly Active and Orthogonal in Mammalian Cells. Biochemistry 2019, 58 (5), 387–390. 10.1021/acs.biochem.8b00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Dunkelmann DL; Willis JCW; Beattie AT; Chin JW Engineered Triply Orthogonal Pyrrolysyl–TRNA Synthetase/TRNA Pairs Enable the Genetic Encoding of Three Distinct Non-Canonical Amino Acids. Nature Chemistry 2020, 12 (6), 535–544. 10.1038/s41557-020-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Reinkemeier CD; Lemke EA Dual Film-like Organelles Enable Spatial Separation of Orthogonal Eukaryotic Translation. Cell 2021, 184 (19), 4886–4903.e21. 10.1016/j.cell.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Van Fossen EM; Bednar RM; Mehl RA Engineering Spatial Orthogonality into Protein Translation. Biochemistry 2019, 58 (31), 3325–3327. 10.1021/acs.biochem.9b00569. [DOI] [PubMed] [Google Scholar]
- (108).Cervettini D; Tang S; Fried SD; Willis JCW; Funke LFH; Colwell LJ; Chin JW Rapid Discovery and Evolution of Orthogonal Aminoacyl-TRNA Synthetase–TRNA Pairs. Nature Biotechnology 2020, 38 (8), 989–999. 10.1038/s41587-020-0479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Blattner FR; Plunkett G; Bloch CA; Perna NT; Burland V; Riley M; Collado-Vides J; Glasner JD; Rode CK; Mayhew GF; Gregor J; Davis NW; Kirkpatrick HA; Goeden MA; Rose DJ; Mau B; Shao Y The Complete Genome Sequence of Escherichia Coli K-12. Science 1997, 277 (5331), 1453–1462. 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- (110).Crick FHC Codon—Anticodon Pairing: The Wobble Hypothesis. Journal of Molecular Biology 1966, 19 (2), 548–555. 10.1016/S0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- (111).Yokoyama S; Watanabe T; Murao K; Ishikura H; Yamaizumi Z; Nishimura S; Miyazawa T Molecular Mechanism of Codon Recognition by TRNA Species with Modified Uridine in the First Position of the Anticodon. PNAS 1985, 82 (15), 4905–4909. 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Eggertsson’ G; Soll D Transfer Ribonucleic Acid-Mediated Suppression of Termination Codons in Escherichia Coli. MICROBIOL. REV 1988, 52, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Allan Drummond D; Wilke CO The Evolutionary Consequences of Erroneous Protein Synthesis. Nature Reviews Genetics 2009, 10 (10), 715–724. 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Ma NJ; Hemez CF; Barber KW; Rinehart J; Isaacs FJ Organisms with Alternative Genetic Codes Resolve Unassigned Codons via Mistranslation and Ribosomal Rescue. eLife 2018, 7, e34878. 10.7554/eLife.34878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Aerni HR; Shifman MA; Rogulina S; O’Donoghue P; Rinehart J Revealing the Amino Acid Composition of Proteins within an Expanded Genetic Code. Nucleic Acids Res 2015, 43 (2), e8. 10.1093/nar/gku1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Blanchet S; Cornu D; Hatin I; Grosjean H; Bertin P; Namy O Deciphering the Reading of the Genetic Code by Near-Cognate TRNA. Proc Natl Acad Sci USA 2018, 115 (12), 3018–3023. 10.1073/pnas.1715578115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Lajoie MJ; Rovner AJ; Goodman DB; Aerni H-R; Haimovich AD; Kuznetsov G; Mercer JA; Wang HH; Carr PA; Mosberg JA; Rohland N; Schultz PG; Jacobson JM; Rinehart J; Church GM; Isaacs FJ Genomically Recoded Organisms Expand Biological Functions. Science 2013, 342 (6156), 357–360. 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).O’Donoghue P; Prat L; Heinemann IU; Ling J; Odoi K; Liu WR; Söll D Near-Cognate Suppression of Amber, Opal and Quadruplet Codons Competes with Aminoacyl-TRNAPyl for Genetic Code Expansion. FEBS Lett 2012, 586 (21), 3931–3937. 10.1016/j.febslet.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Johnson DBF; Wang C; Xu J; Schultz MD; Schmitz RJ; Ecker JR; Wang L Release Factor One Is Nonessential in Escherichia Coli. ACS Chem. Biol 2012, 7 (8), 1337–1344. 10.1021/cb300229q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Engelberg-Kulka H UGA Suppression by Normal TRNA Trp in Escherichia Coli: Codon Context Effects. Nucleic Acids Res 1981, 9 (4), 983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Kopelowitz J; Hampe C; Goldman R; Reches M; Engelberg-Kulka H Influence of Codon Context on UGA Suppression and Readthrough. Journal of Molecular Biology 1992, 225 (2), 261–269. 10.1016/0022-2836(92)90920-F. [DOI] [PubMed] [Google Scholar]
- (122).Wang L Engineering the Genetic Code in Cells and Animals: Biological Considerations and Impacts. Acc. Chem. Res 2017, 50 (11), 2767–2775. 10.1021/acs.accounts.7b00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Zhu P; Gafken PR; Mehl RA; Cooley RB A Highly Versatile Expression System for the Production of Multiply Phosphorylated Proteins. ACS Chem. Biol 2019, 14 (7), 1564–1572. 10.1021/acschembio.9b00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Schwark DG; Schmitt MA; Fisk JD Dissecting the Contribution of Release Factor Interactions to Amber Stop Codon Reassignment Efficiencies of the Methanocaldococcus Jannaschii Orthogonal Pair. Genes (Basel) 2018, 9 (11). 10.3390/genes9110546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Venkat S; Sturges J; Stahman A; Gregory C; Gan Q; Fan C Genetically Incorporating Two Distinct Post-Translational Modifications into One Protein Simultaneously. ACS Synth. Biol 2018, 7 (2), 689–695. 10.1021/acssynbio.7b00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Ozer E; Chemla Y; Schlesinger O; Aviram HY; Riven I; Haran G; Alfonta L In Vitro Suppression of Two Different Stop Codons. Biotechnology and Bioengineering 2017, 114 (5), 1065–1073. 10.1002/bit.26226. [DOI] [PubMed] [Google Scholar]
- (127).Varani G; McClain WH The G·U Wobble Base Pair. EMBO Rep 2000, 1 (1), 18–23. 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Kobayashi T; Nureki O; Ishitani R; Yaremchuk A; Tukalo M; Cusack S; Sakamoto K; Yokoyama S Structural Basis for Orthogonal TRNA Specificities of Tyrosyl-TRNA Synthetases for Genetic Code Expansion. Nat Struct Biol 2003, 10 (6), 425–432. 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- (129).Gan R; Perez JG; Carlson ED; Ntai I; Isaacs FJ; Kelleher NL; Jewett MC Translation System Engineering in Escherichia Coli Enhances Non-Canonical Amino Acid Incorporation into Proteins. Biotechnol Bioeng 2017, 114 (5), 1074–1086. 10.1002/bit.26239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Tharp JM; Ad O; Amikura K; Ward FR; Garcia EM; Cate JHD; Schepartz A; Söll D Initiation of Protein Synthesis with Non-Canonical Amino Acids In Vivo. Angewandte Chemie International Edition 2020, 59 (8), 3122–3126. 10.1002/anie.201914671. [DOI] [PubMed] [Google Scholar]
- (131).Dunkelmann DL; Oehm SB; Beattie AT; Chin JW A 68-Codon Genetic Code to Incorporate Four Distinct Non-Canonical Amino Acids Enabled by Automated Orthogonal MRNA Design. Nat. Chem 2021, 13 (11), 1110–1117. 10.1038/s41557-021-00764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).DeBenedictis EA; Söll D; Esvelt KM Measuring the Tolerance of the Genetic Code to Altered Codon Size. eLife 2022, 11, e76941. 10.7554/eLife.76941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Morris SE; Feldman AW; Romesberg FE Synthetic Biology Parts for the Storage of Increased Genetic Information in Cells. ACS Synth Biol 2017, 6 (10), 1834–1840. 10.1021/acssynbio.7b00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Biddle W; Schmitt MA; Fisk JD Evaluating Sense Codon Reassignment with a Simple Fluorescence Screen. Biochemistry 2015, 54 (50), 7355–7364. 10.1021/acs.biochem.5b00870. [DOI] [PubMed] [Google Scholar]
- (135).Fredens J; Wang K; de la Torre D; Funke LFH; Robertson WE; Christova Y; Chia T; Schmied WH; Dunkelmann DL; Beránek V; Uttamapinant C; Llamazares AG; Elliott TS; Chin JW Total Synthesis of Escherichia Coli with a Recoded Genome. Nature 2019, 569 (7757), 514–518. 10.1038/s41586-019-1192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Robertson WE; Funke LFH; de la Torre D; Fredens J; Elliott TS; Spinck M; Christova Y; Cervettini D; Böge FL; Liu KC; Buse S; Maslen S; Salmond GPC; Chin JW Sense Codon Reassignment Enables Viral Resistance and Encoded Polymer Synthesis. Science 2021, 372 (6546), 1057–1062. 10.1126/science.abg3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Hu Y; Roberts JM; Kilgore HR; Mat Lani AS; Raines RT; Schomaker JM Triple, Mutually Orthogonal Bioorthogonal Pairs through the Design of Electronically Activated Sulfamate-Containing Cycloalkynes. J. Am. Chem. Soc 2020, 142 (44), 18826–18835. 10.1021/jacs.0c06725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Oliveira BL; Guo Z; Bernardes GJL Inverse Electron Demand Diels–Alder Reactions in Chemical Biology. Chem. Soc. Rev 2017, 46 (16), 4895–4950. 10.1039/C7CS00184C. [DOI] [PubMed] [Google Scholar]
- (139).Kim J; Seo M-H; Lee S; Cho K; Yang A; Woo K; Kim H-S; Park H-S Simple and Efficient Strategy for Site-Specific Dual Labeling of Proteins for Single-Molecule Fluorescence Resonance Energy Transfer Analysis. Anal. Chem 2013, 85 (3), 1468–1474. 10.1021/ac303089v. [DOI] [PubMed] [Google Scholar]
- (140).Kölmel DK; Kool ET Oximes and Hydrazones in Bioconjugation: Mechanism and Catalysis. Chem Rev 2017, 117 (15), 10358–10376. 10.1021/acs.chemrev.7b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Haldón E; Nicasio MC; Pérez PJ Copper-Catalysed Azide–Alkyne Cycloadditions (CuAAC): An Update. Org. Biomol. Chem 2015, 13 (37), 9528–9550. 10.1039/C5OB01457C. [DOI] [PubMed] [Google Scholar]
- (142).Patterson DM; Nazarova LA; Prescher JA Finding the Right (Bioorthogonal) Chemistry. ACS Chem. Biol 2014, 9 (3), 592–605. 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- (143).Li S; Cai H; He J; Chen H; Lam S; Cai T; Zhu Z; Bark SJ; Cai C Extent of the Oxidative Side Reactions to Peptides and Proteins During the CuAAC Reaction. Bioconjug Chem 2016, 27 (10), 2315–2322. 10.1021/acs.bioconjchem.6b00267. [DOI] [PubMed] [Google Scholar]
- (144).Dommerholt J; Rutjes FPJT; van Delft FL Strain-Promoted 1,3-Dipolar Cycloaddition of Cycloalkynes and Organic Azides. Top Curr Chem (Cham) 2016, 374 (2), 16. 10.1007/s41061-016-0016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Blizzard RJ; Backus DR; Brown W; Bazewicz CG; Li Y; Mehl RA Ideal Bioorthogonal Reactions Using A Site-Specifically Encoded Tetrazine Amino Acid. J Am Chem Soc 2015, 137 (32), 10044–10047. 10.1021/jacs.5b03275. [DOI] [PubMed] [Google Scholar]
- (146).Karver MR; Weissleder R; Hilderbrand SA Bioorthogonal Reaction Pairs Enable Simultaneous, Selective, Multi-Target Imaging. Angew Chem Int Ed Engl 2012, 51 (4), 920–922. 10.1002/anie.201104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Liang Y; Mackey JL; Lopez SA; Liu F; Houk KN Control and Design of Mutual Orthogonality in Bioorthogonal Cycloadditions. J. Am. Chem. Soc 2012, 134 (43), 17904–17907. 10.1021/ja309241e. [DOI] [PubMed] [Google Scholar]
- (148).Addy PS; Erickson SB; Italia JS; Chatterjee A A Chemoselective Rapid Azo-Coupling Reaction (CRACR) for Unclickable Bioconjugation. J. Am. Chem. Soc 2017, 139 (34), 11670–11673. 10.1021/jacs.7b05125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Borrmann A; Fatunsin O; Dommerholt J; Jonker AM; Löwik DWPM; van Hest JCM; van Delft FL Strain-Promoted Oxidation-Controlled Cyclooctyne–1,2-Quinone Cycloaddition (SPOCQ) for Fast and Activatable Protein Conjugation. Bioconjugate Chem. 2015, 26 (2), 257–261. 10.1021/bc500534d. [DOI] [PubMed] [Google Scholar]
- (150).Cheng L; Kang X; Wang D; Gao Y; Yi L; Xi Z The One-Pot Nonhydrolysis Staudinger Reaction and Staudinger or SPAAC Ligation. Org. Biomol. Chem 2019, 17 (23), 5675–5679. 10.1039/C9OB00528E. [DOI] [PubMed] [Google Scholar]
- (151).Schäfer RJB; Monaco MR; Li M; Tirla A; Rivera-Fuentes P; Wennemers H The Bioorthogonal Isonitrile–Chlorooxime Ligation. J. Am. Chem. Soc 2019, 141 (47), 18644–18648. 10.1021/jacs.9b07632. [DOI] [PubMed] [Google Scholar]
- (152).Debets MF; van Hest JCM; Rutjes FPJT Bioorthogonal Labelling of Biomolecules: New Functional Handles and Ligation Methods. Org. Biomol. Chem 2013, 11 (38), 6439–6455. 10.1039/C3OB41329B. [DOI] [PubMed] [Google Scholar]
- (153).Tu J; Svatunek D; Parvez S; Liu ACG; Levandowski BJ; Eckvahl HJ; Peterson RT; Houk KN; Franzini RM Stable, Reactive and Orthogonal Tetrazines: Dispersion Forces Promote the Cycloaddition with Isonitriles. Angew Chem Int Ed Engl 2019, 58 (27), 9043–9048. 10.1002/anie.201903877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Wu Z-C; Boger DL Synthesis, Characterization, and Cycloaddition Reactivity of a Monocyclic Aromatic 1,2,3,5-Tetrazine. J. Am. Chem. Soc 2019, 141 (41), 16388–16397. 10.1021/jacs.9b07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Schart VF; Hassenrück J; Späte A-K; Dold JEGA; Fahrner R; Wittmann V Triple Orthogonal Labeling of Glycans by Applying Photoclick Chemistry. ChemBioChem 2019, 20 (2), 166–171. 10.1002/cbic.201800740. [DOI] [PubMed] [Google Scholar]
- (156).Liu F; Liang Y; Houk KN Bioorthogonal Cycloadditions: Computational Analysis with the Distortion/Interaction Model and Predictions of Reactivities. Acc. Chem. Res 2017, 50 (9), 2297–2308. 10.1021/acs.accounts.7b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).Bednar RM; Golbek TW; Kean KM; Brown WJ; Jana S; Baio JE; Karplus PA; Mehl RA Immobilization of Proteins with Controlled Load and Orientation. ACS Appl. Mater. Interfaces 2019, 11 (40), 36391–36398. 10.1021/acsami.9b12746. [DOI] [PubMed] [Google Scholar]
- (158).Jang HS; Jana S; Blizzard RJ; Meeuwsen JC; Mehl RA Access to Faster Eukaryotic Cell Labeling with Encoded Tetrazine Amino Acids. Journal of the American Chemical Society 2020, 7245–7249. 10.1021/jacs.9b11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (159).Lerner E; Cordes T; Ingargiola A; Alhadid Y; Chung S; Michalet X; Weiss S Toward Dynamic Structural Biology: Two Decades of Single-Molecule Förster Resonance Energy Transfer. Science 2018, 359 (6373). 10.1126/science.aan1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (160).Sahoo H Förster Resonance Energy Transfer – A Spectroscopic Nanoruler: Principle and Applications. Journal of Photochemistry and Photobiology C: Photochemistry Reviews 2011, 12 (1), 20–30. 10.1016/j.jphotochemrev.2011.05.001. [DOI] [Google Scholar]
- (161).Haney CM; Wissner RF; Petersson EJ Multiply Labeling Proteins for Studies of Folding and Stability. Curr Opin Chem Biol 2015, 28, 123–130. 10.1016/j.cbpa.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (162).Sadoine M; Cerminara M; Kempf N; Gerrits M; Fitter J; Katranidis A Selective Double-Labeling of Cell-Free Synthesized Proteins for More Accurate SmFRET Studies. Anal Chem 2017, 89 (21), 11278–11285. 10.1021/acs.analchem.7b01639. [DOI] [PubMed] [Google Scholar]
- (163).Miyawaki A; Llopis J; Heim R; McCaffery JM; Adams JA; Ikura M; Tsien RY Fluorescent Indicators for Ca2+ Based on Green Fluorescent Proteins and Calmodulin. Nature 1997, 388 (6645), 882–887. 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- (164).Kajihara D; Abe R; Iijima I; Komiyama C; Sisido M; Hohsaka T FRET Analysis of Protein Conformational Change through Position-Specific Incorporation of Fluorescent Amino Acids. Nature Methods 2006, 3 (11), 923–929. 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- (165).Harkiss AH; Sutherland A Recent Advances in the Synthesis and Application of Fluorescent α-Amino Acids. Organic & Biomolecular Chemistry 2016, 14 (38), 8911–8921. 10.1039/C6OB01715K. [DOI] [PubMed] [Google Scholar]
- (166).Akkapeddi P; Azizi S-A; Freedy AM; Cal PMSD; Gois PMP; Bernardes GJL Construction of Homogeneous Antibody–Drug Conjugates Using Site-Selective Protein Chemistry. Chem. Sci 2016, 7 (5), 2954–2963. 10.1039/C6SC00170J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Chudasama V; Maruani A; Caddick S Recent Advances in the Construction of Antibody-Drug Conjugates. Nat Chem 2016, 8 (2), 114–119. 10.1038/nchem.2415. [DOI] [PubMed] [Google Scholar]
- (168).Moore EJ; Zorine D; Hansen WA; Khare SD; Fasan R Enzyme Stabilization via Computationally Guided Protein Stapling. PNAS 2017, 114 (47), 12472–12477. 10.1073/pnas.1708907114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Gori A; Gagni P; Rinaldi S Disulfide Bond Mimetics: Strategies and Challenges. Chemistry – A European Journal 2017, 23 (60), 14987–14995. 10.1002/chem.201703199. [DOI] [PubMed] [Google Scholar]
- (170).Wong LS; Khan F; Micklefield J Selective Covalent Protein Immobilization: Strategies and Applications. Chem. Rev 2009, 109 (9), 4025–4053. 10.1021/cr8004668. [DOI] [PubMed] [Google Scholar]
- (171).Rabe M; Verdes D; Seeger S Understanding Protein Adsorption Phenomena at Solid Surfaces. Adv Colloid Interface Sci 2011, 162 (1–2), 87–106. 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- (172).Stephanopoulos N Hybrid Nanostructures from the Self-Assembly of Proteins and DNA. Chem 2020, 6 (2), 364–405. 10.1016/j.chempr.2020.01.012. [DOI] [Google Scholar]
- (173).Jørgensen K; Rasmussen AV; Morant M; Nielsen AH; Bjarnholt N; Zagrobelny M; Bak S; Møller BL Metabolon Formation and Metabolic Channeling in the Biosynthesis of Plant Natural Products. Current Opinion in Plant Biology 2005, 8 (3), 280–291. 10.1016/j.pbi.2005.03.014. [DOI] [PubMed] [Google Scholar]
- (174).Oreb M Construction of Artificial Membrane Transport Metabolons – an Emerging Strategy in Metabolic Engineering. FEMS Microbiology Letters 2020, 367 (fnaa027). 10.1093/femsle/fnaa027. [DOI] [PubMed] [Google Scholar]
- (175).Müller J; Verrijzer P Biochemical Mechanisms of Gene Regulation by Polycomb Group Protein Complexes. Current Opinion in Genetics & Development 2009, 19 (2), 150–158. 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- (176).Dong G; Ding Y; He S; Sheng C Molecular Glues for Targeted Protein Degradation: From Serendipity to Rational Discovery. J. Med. Chem 2021, 64 (15), 10606–10620. 10.1021/acs.jmedchem.1c00895. [DOI] [PubMed] [Google Scholar]
- (177).Chen C; Ng DYW; Weil T Polymer Bioconjugates: Modern Design Concepts toward Precision Hybrid Materials. Progress in Polymer Science 2020, 105, 101241. 10.1016/j.progpolymsci.2020.101241. [DOI] [Google Scholar]
- (178).Qiu Y; Askounis E; Guan F; Peng Z; Xiao W; Pei Q Dual-Stimuli-Responsive Polymer Composite with Ultrawide Tunable Stiffness Range Triggered by Water and Temperature. ACS Appl. Polym. Mater 2020, 2 (5), 2008–2015. 10.1021/acsapm.0c00181. [DOI] [Google Scholar]
- (179).Schena A; Griss R; Johnsson K Modulating Protein Activity Using Tethered Ligands with Mutually Exclusive Binding Sites. Nature Communications 2015, 6 (1), 7830. 10.1038/ncomms8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (180).Umehara T; Kim J; Lee S; Guo L-T; Söll D; Park H-S N-Acetyl Lysyl-TRNA Synthetases Evolved by a CcdB-Based Selection Possess N-Acetyl Lysine Specificity in Vitro and in Vivo. FEBS Lett 2012, 586 (6), 729–733. 10.1016/j.febslet.2012.01.029. [DOI] [PubMed] [Google Scholar]
- (181).Tanrikulu IC; Schmitt E; Mechulam Y; Goddard WA; Tirrell DA Discovery of Escherichia Coli Methionyl-TRNA Synthetase Mutants for Efficient Labeling of Proteins with Azidonorleucine in Vivo. PNAS 2009, 106 (36), 15285–15290. 10.1073/pnas.0905735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Anderson JC; Wu N; Santoro SW; Lakshman V; King DS; Schultz PG An Expanded Genetic Code with a Functional Quadruplet Codon. PNAS 2004, 101 (20), 7566–7571. 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Wright DE; Altaany Z; Bi Y; Alperstein Z; O’Donoghue P Acetylation Regulates Thioredoxin Reductase Oligomerization and Activity. Antioxid Redox Signal 2018, 29 (4), 377–388. 10.1089/ars.2017.7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Serfling R; Lorenz C; Etzel M; Schicht G; Böttke T; Möri M; Coin I Designer TRNAs for Efficient Incorporation of Non-Canonical Amino Acids by the Pyrrolysine System in Mammalian Cells. Nucleic Acids Res 2018, 46 (1), 1–10. 10.1093/nar/gkx1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Sievers F; Wilm A; Dineen D; Gibson TJ; Karplus K; Li W; Lopez R; McWilliam H; Remmert M; Söding J; Thompson JD; Higgins DG Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Molecular Systems Biology 2011, 7 (1), 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.