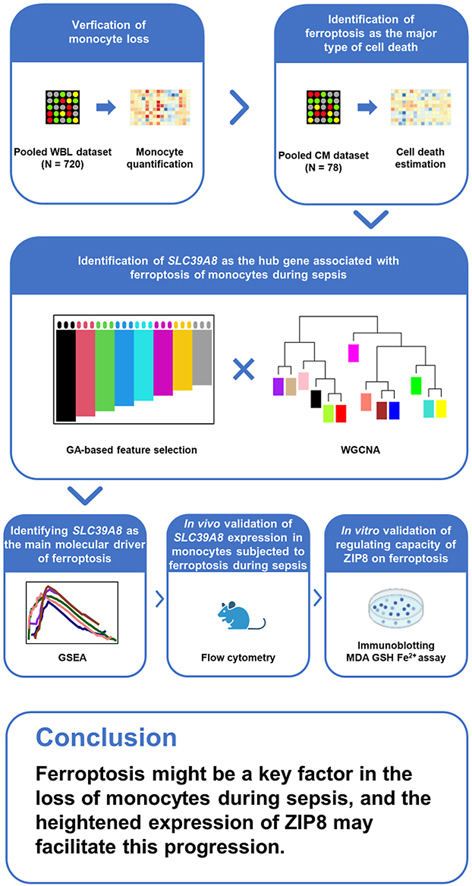
Abstract
Sepsis is a heterogenous syndrome with concurrent hyperinflammation and immune suppression. A prominent feature of immunosuppression during sepsis is the dysfunction and loss of monocytes; however, the major type of cell death contributing to this depletion, as well as its underlying molecular mechanisms, are yet to be identified. In this study, we confirmed the monocyte loss in septic patients based on a pooled gene expression data of periphery leukocytes. Using the collected reference gene sets from databases and published studies, we identified ferroptosis with a greater capacity to distinguish between sepsis and control samples than other cell death types. Further investigation on the molecular drivers, by a genetic algorithm-based feature selection and a weighted gene co-expression network analysis, revealed that zrt-/irt-like protein 8 (ZIP8), encoded by SLC39A8, was closely associated with ferroptosis of monocytes during sepsis. We validated the increase of ZIP8 of monocytes with in vivo and in vitro experiments. The in vitro studies also showed that downregulation of ZIP8 alleviated the lipopolysaccharide-induced lipid peroxidation, as well as restoring the reduction of GPX4, FTH1 and xCT. These findings suggest that ferroptosis might be a key factor in the loss of monocytes during sepsis, and that the heightened expression of ZIP8 may facilitate this progression.
Keywords: Sepsis, Monocyte, Cell death, Ferroptosis, ZIP8
Graphical abstract
Highlights
-
•
Peripheral monocytes encounter cell death and loss during sepsis.
-
•
Ferroptosis, rather than other types of cell death, may act as the leading cause to the loss of monocytes.
-
•
The increased ZIP8 may facilitate the progression of ferroptosis of monocytes.
1. Introduction
Sepsis is now acknowledged as one of the leading causes of mortality and critical illness of hospitalized patients worldwide [1]. A recent study from the Global Burden of Disease Study revealed that a total of 48.9 million cases of sepsis had been reported across the globe in 2017, yielding an alarming mortality rate of 22.5 %, accounting for nearly 20 % of all global deaths [2]. Besides, sepsis survivors are accompanied by a long-term morbidity, which entails a 15 % mortality rate in the first-year post-discharge and 6–8% in the following five years [3,4]. As the prevalence of sepsis continues to rise, it is likely to have substantial impacts on public health and cost.
Sepsis is defined as an organ dysfunction syndrome caused by an aberrant host response to an infection [5]. However, this definition fails to portray the complexity of the immune state present during sepsis, which is triggered by persistent unremovable pathogens and damage-associated molecular patterns and may comprise concurrent hyperinflammation and immune suppression resulting in a final immune collapse and mortality [6]. Excessive release of pro-inflammatory cytokines and activation of complement and clotting systems could contribute to tissue damage and organ dysfunction in septic patients; however, anti-inflammation therapies delivered in clinical trials were met with limited success [[7], [8], [9], [10]]. Therefore, the role of sustained immunosuppression in the progression of sepsis is gaining more interest. Although yet to be fully elucidated, it is postulated that the mechanisms of immunosuppression involve different cell types and pathophysiological processes.
More recently, the indicator value of monocyte distribution width in the detection of sepsis has been demonstrated, thus highlighting the early involvement of monocytes in sepsis progression [11]. The dysfunction of monocytes is a prominent feature of immunosuppression, which is characterized by the abnormal expression of major histocompatibility complex, class II, DR (HLA-DR) and programmed cell death 1 ligand 1 (PDL1) and diminished capacity to release pro-inflammatory cytokines. Compared to functional abnormalities, loss of monocytes may have a more drastic impact on the development of sepsis. Studies focusing on circulating immunocytes have found a strong inverse correlation between absolute monocyte counts and the degree of severity and mortality of septic patients [12,13]. Furthermore, apoptosis, necrosis, as well as pyroptosis of monocytes have all been found to be related to development and severity of sepsis [[14], [15], [16]]. A recent study has revealed apoptosis, autophagy and ferroptosis are main types of cell death in monocytes [17]; however, the limitations, such as sequencing-based identification of monocytes from peripheral blood mononuclear cell (PBMC), single source-derived cell death gene sets, and small sample size, demand necessity of further exploration on the predominant type of cell death signaling in monocytes during acute sepsis.
In this study, we leveraged population-level transcriptomic data in humans with sepsis to determine the principal types of cell death pathways expressed in monocytes during sepsis and found that monocytes from patients with sepsis displayed significant upregulation of ferroptosis-related pathways. To further explore specific underlying mechanisms, we utilized a feature selection method based on genetic algorithm and performed a weighted gene co-expression network analysis (WGCNA) to filter out significant genes. Consequently, zrt-/irt-like protein 8 (ZIP8), encoded by SLC39A8, was identified and validated for its critical role in monocyte ferroptosis during sepsis. Our findings could potentially shed light on the molecular pathogenesis of sepsis and subsequently assist in optimizing therapies for septic patients.
2. Material and methods
2.1. Dataset inclusion criteria
Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) public gene expression repositories were systematically searched for any relevant gene expression microarray or next-generation sequencing (NGS/RNAseq) datasets of sepsis with the following terms: sepsis, septic, SIRS, acute infection, trauma, shock, surgery, infection, pneumonia, critical, ICU, inflammatory, nosocomial. Upon further scrutiny, datasets containing transcriptomic profiling of leukocytes and/or monocytes from whole blood sample of Homo sapiens, regardless of sex, were considered potentially eligible and retained. Datasets simply pertaining to SIRS, acute infection or pneumonia, which did not meet sepsis diagnostic criteria, were excluded. In addition, datasets were also excluded if they solely comprised pediatric or neonatal patients, utilized endotoxin as a model for sepsis, were in vitro experiments, focused on diseases with special pathogen (HIV, HBV, etc.), were derived from sorted cells (without monocytes), or merely contained non-coding RNA profiling.
2.2. Ethics statement
Gene expression data used in this research were obtained from publicly available sources with preexisting ethics approval from original studies. Informed consent was obtained for each participant. The study was conducted in accordance to the Declaration of Helsinki.
All animal experiments were carried out in accordance with the Guides for the Care and Use of Laboratory Animals (National Academy of Sciences, China) and with approval from the Ethics Committee of Shanghai Pulmonary Hospital (ethical approval number: K21-181Z), adhered to the recommendations in the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health.
2.3. Dataset normalization and pooling
All Affymetrix datasets were procured and re-normalized according to the original studies. The remaining array datasets were normalized using R package ‘limma’ [18]. During processing, probes with the maximum mean expression intensity across all samples were retained when multiple probes were mapped to the same symbol, whilst probes mapped to multiple symbols or having no mapping were removed. Expression from NGS datasets were processed as counts per million total reads and were normalized using the ‘voom’ method in R package ‘limma’, for compatibility with microarray studies. Dataset pooling was performed via a batch effect correction by an empirical Bayes framework [19], with a principal component analysis for quality inspection.
2.4. Monocyte quantification
Monocyte quantification was implemented using gene expression data of leukocytes through computational approaches based on sets of monocyte-specific marker genes or overall expression signatures. We calculated the enrichment score (ES) of monocyte in each leukocyte sample leveraging on the single sample gene set enrichment analysis (ssGSEA) method (R package ‘gsva’). The aforementioned monocyte-specific marker genes were extracted from CellMarker and PanglaoDB databases [20,21]. Additionally, the relative fraction of monocyte was also quantitatively estimated using the CIBERSORT and quanTIseq methods (R package ‘immunedeconv’) [22,23]. The enrichment scores and relative fractions obtained from different methods were normalized by deviation standardization for presentation.
2.5. Cell death estimation
Extents of various cell death types were estimated using ssGSEA method (R package ‘gsva’). The renamed reference gene sets of multiple cell death forms, including apoptosis, necrosis, necroptosis, pyroptosis, ferroptosis and cuproptosis, were curated from published studies or public databases and listed in Table S1. In brief, defined cell death associated gene sets were retrieved from MSigDB HALLMARK and GOBP collections [24], REACTOME, WIKI, and KEGG databases [[25], [26], [27]]. Each cell death term was also searched in GeneCards database [28], and genes with relevant score greater than 95 % quantile were retained as members of corresponding gene sets. Subsequently, the enrichment scores of each cell death pathway were compared between septic patients and healthy control subjects, and potential significant cell death types were further evaluated via receiver operating characteristic (ROC) curve analysis (R package ‘pROC’).
2.6. K-means clustering analysis
The scaled enrichment scores (z-scores) generated from selected cell death pathways were employed for performing k-means clustering analysis. The optimal number of clusters was determined by applying elbow method and average silhouette method [29].
2.7. Feature selection through multivariate method
Feature selection was performed to screen genes representative of characteristics of sepsis samples. The procedure was implemented through a multivariate method, which was based on a 5-gene neural network (NN) model (R package ‘nnet’) for classification and a genetic algorithm (GA) approach (R package ‘GALGO’) coupled with ROC analysis (R package ‘pROC’) for variable search and evaluation [30]. GA methods, a computing technique commonly applied to address optimization issues through simulating natural selection, operate by iterating sets of random variables (‘chromosomes’) to fit designated criteria from an initialized random population. In this study, a number of random 5-gene combinations were established as the initial chromosomes. The chromosomes were then adopted for the construction of a class prediction model using a neural network. The NN model was trained and tested with 4-fold cross validation (automatically detected splits), and the corresponding testing area under operating characteristic curve (AUC) values delineated the ability of the chromosomes to discern the group affiliation of each sample. The chromosomes with a higher fitness score, a parameter proportional to the AUC value, were underwent replication, cross-over, and at times, mutation, for generating more offspring until one of them achieved the predetermined ‘goalFitness’ or until 200 generations had occurred. The chromosomes with a fitness score greater than 0.99 were stored and the attendances of the corresponding genes were recorded for frequency ranking. The convergence of the GA was detected by monitoring the stability of the gene rankings. The aforementioned process was defined as one evolution cycle, which was performed 2000 times to guarantee the steadiness of genes with the most attendance frequency in this study.
2.8. Gene co-expression network construction
Prior to network construction, sample quality control was conducted to eliminate outlier samples or samples with missing values through ‘hclust’ and ‘goodSamplesGenes’ functions in R package ‘WGCNA’ [31]. Legitimate samples were then employed for co-expression network construction using the R package ‘WGCNA’. The soft-threshold power β was detected by ‘pickSoftThreshold’ function to construct a standard scale-free network. The constructed adjacency matrix was then transformed into a topological overlap matrix with the β value set as 9 (scale-free R2 = 0.85). Finally, the identification of co-expression gene modules was implemented through dynamic tree cut method with the cut height of 0.25 and minimal module size set as 50.
2.9. Identification of the hub gene
In order to identify the hub gene, we employed a two-step approach combining feature selection and WGCNA analysis. We first selected genes with a frequency greater than 15 in the feature selection procedure to obtain a list of hub gene candidates. The fold-change values of the candidates were calculated using R package ‘limma’ and visualized in a volcano plot. In WGCNA procedure, after identifying gene co-expression modules, we quantified the correlations, which were defined as ‘geneTraitSignificance’ (GS) values, between the module eigengene value of each module and z-scores of significant cell death pathways with high AUC values. A p value < 0.05 was considered as statistically significant. The mean GS values of each legitimate module were then arranged to screen out the hub gene module. In the hub gene module, genes with respective module membership (MM) value and GS value greater than 0.8 and 0.3 were selected as candidates of the hub gene. The intersected candidates from feature selection procedure and WGCNA process were then re-arranged according to their GS values, with the one having the highest GS value being considered as the hub gene.
2.10. Functional enrichment analysis
Pathway enrichment analyses were performed through DAVID tools (https://david.ncifcrf.gov/) based on KEGG, REACTOME and WIKI databases [32]. Gene set enrichment analysis (GSEA) was conducted using GSEA software (version 4.2.1) [33]. Based on the median expression of the hub gene, samples were divided into high-expression and low-expression groups. The reference gene set, ‘c2.cp.v2023.1.Hs.symbols.gmt’, was acquired from the Molecular Signature Database. The NORM-p value < 0.05 was considered as statistically significant.
2.11. Animals
Male adult C57BL/6 wild-type mice, 8-10-week-old, obtained from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China), were used in this study. All animals were housed under specific pathogen-free conditions and had access to water and food pellets ad libitum.
2.12. Cecal ligation and puncture model
Cecal ligation and puncture (CLP) was performed to induce sepsis as described previously [34]. Briefly, animals were anesthetized with ketamine (100 mg/kg i. p.) and xylazine (5 mg/kg i. p.) prior to disinfection of abdominal skin. A 1.5-cm midline abdominal incision was performed to allow for complete exposure of the cecum. The cecum was then ligated at half the distance between the distal pole and the base of the cecum and punctured with a 22-gauge needle. Thereafter, the cecum was slightly compressed to extrude a small amount of feces. Once this procedure was completed, the cecum was relocated, and the incision was then sutured. In sham controls, the cecum was exposed but not ligated or punctured. Following surgical procedures, all mice were given subcutaneous injections of prewarmed normal saline (50 μl/g) for fluid resuscitation and buprenorphine (0.05 mg/kg) for analgesia.
2.13. Flow cytometry
Twenty-four or 48 h post-CLP, mice were anesthetized and whole blood was collected via cardiac puncture for flow cytometry analysis. After lysis of erythrocytes (eBioscience Cat# 00–4300) and Fc receptor blocking (BioLegend Cat# 156604, RRID:AB_2783138), blood samples were stained with anti-CD45 (BioLegend Cat# 103114, RRID:AB_312979), anti-CD11b (BioLegend Cat# 101212, RRID:AB_312795) and anti-Ly6G (BioLegend Cat# 127608, RRID:AB_1186099). 7-AAD (BioLegend Cat# 420404) was simultaneously used to differentiate between live and dead cells. For counterstaining of ZIP8, GPX4, FTH1, and xCT, leukocytes were fixed (eBioscience Cat# 00–8222) and permeabilized (eBioscience Cat# 00–8333) before antibody incubation. Alexa Fluor 488 labeled anti-ZIP8, anti-GPX4, and anti-xCT were respectively produced by modifying anti-ZIP8 (Proteintech Cat# 20459-1-AP, RRID:AB_10697830), anti-GPX4 (Proteintech Cat# 30388-1-AP), and anti-xCT (Invitrogen Cat# PA1-16893, RRID:AB_2286208) using a conjugation kit (Abcam Cat# ab236553). FTH1 was detected using primary antibody (Abcam Cat# ab75973, RRID:AB_1310222), followed by staining with Alexa Fluor 488 conjugated anti-Rabbit IgG (Abcam Cat# ab150077, RRID:AB_2630356). All samples were then tested in a FACSCanto II flow cytometer (BD Biosciences) and analyzed using FlowJo software.
2.14. Cell culture, treatment and transfection
The mouse monocyte/macrophage cell line RAW264.7 was obtained from American type culture collection, and was cultured in Dulbecco's modified Eagle's medium (Gibco Cat# C11995500BT) with 10 % fetal bovine serum (Gibco Cat# 10091–148) and 1 % penicillin-streptomycin (Gibco Cat# 15070–063) in a 37 °C incubator with 5 % CO2. When necessary, cells were treated with lipopolysaccharide (LPS, Sigma Cat# L2880) after 48 h of transfection. Cells were cultured in each well of a 6-well plate, adjusted to a density of approximately 6 × 105 cells per well before being transfected with small interfering RNA (target sequences CGACACTGTCAGCGTTGTA, 5′-CGACACUGUCAGCGUUGUA-3′ and 5′-UACAACGCUGACAGUGUCG-3′) for the purpose of Slc39a8 expression knockdown using RNAiMAX reagent (Invitrogen Cat# 13778).
2.15. Western blotting assay
Cells were washed with phosphate buffer saline (PBS) and ultrasonically disrupted before lysed in high strength radioimmunoprecipitation assay lysis reagent (Epizyme Cat# PC101) containing protease inhibitors (NCM Cat# P002). The protein concentration in the cell supernatants was determined using the BCA protein assay kit (Thermo Cat# 23225). Subsequently, 15 μg protein was loaded and separated by 12.5 % sodium dodecyl sulphonate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane. Following blocking, the membrane was incubated with primary antibody, including anti-ZIP8 (Proteintech Cat# 20459-1-AP, RRID:AB_10697830), anti-GPX4 (ABclonal Cat# A1933, RRID:AB_2763960), anti-FTH1 (Affinity Biosciences Cat# DF6278, RRID:AB_2838244; ABclonal Cat# A19544, RRID:AB_2862659), anti-xCT (ABclonal Cat# A2413, RRID:AB_2863004) and anti-GAPDH (Abcam Cat# ab9485, RRID:AB_307275). The membranes were then incubated with horseradish peroxidase-conjugated IgG (Jackson ImmunoResearch Labs Cat# 111-035-003, RRID:AB_2313567) and signals were detected using an enhanced electrochemiluminescence kit (Epizyme Cat# SQ201).
2.16. Intracellular iron measurement
Intracellular ferrous ions were measured using a commercial kit (Elabscience Cat# E-BC-K881-M). In brief, cells were washed with PBS prior to collection and separated for cell counting and next analysis. Then, 200 μl of buffer solution was added per 1 × 106 cells for lysis. The cells were lysed on ice for a total of 30 min and vortexed every 10 min. After centrifugation at 15000 g and 4 °C for 10 min, the supernatant was collected and mixed with 80 μl of chromogenic solution and incubated for 90 min at 37 °C in the dark. All mixtures were then measured for absorbance at OD593 nm. The total intracellular ferrous ion concentration was calculated according to the standard curve and normalized by cell counts.
2.17. Lipid peroxidation assay
Total reduced glutathione (GSH) and malondialdehyde (MDA) were measured respectively using a GSH assay kit (Elabscience Cat# E-BC-K030-M) and an MDA assay kit (Elabscience Cat# E-BC-K028-M). The experimental procedures were conducted according to the manufacturer's instructions.
2.18. Statistical analysis
The normalized enrichment scores and relative fractions of monocytes, as well as normalized enrichment scores of cell death, were analyzed with a two-tailed, unpaired t-test method between two groups. The relationship between the generated class by k-means clustering and disease identity was determined by conducting a chi-square test. The correlations between normalized enrichment scores of ferroptosis and gene modules were assessed via ‘corPvalueStudent’ function in R package ‘WGCNA’. The correlations between normalized enrichment scores of ferroptosis and expression of genes in the blue module were analyzed using Pearson correlation analysis. The correlations between normalized enrichment scores of ferroptosis and expression of SLC39A8 were analyzed through Pearson correlation analysis. The percentages of ZIP8, GPX4, FTH1, and xCT positive monocytes and the mean fluorescence intensities (MFIs) of ZIP8, GPX4, FTH1, and xCT in monocytes were analyzed using a two-tailed, unpaired t-test method between two groups. Expression level analyses between seven groups were performed using one-way analysis of variance (ANOVA) followed by a Dunnett's multiple comparisons test. Analyses of MDA, GSH, ferrous ions, and protein levels between four groups were analyzed using a two-way ANOVA followed by a Holm-Sidak multiple comparisons test. All statistics were completed using R 4.1.1 and GraphPad Prism 9.5.0 software. All data were presented as the mean ± standard error. Statistical significance was considered to occur at p < 0.05/adjusted p < 0.05.
3. Results
3.1. Included datasets
After initial search, 64 datasets from GEO and 12 datasets from ArrayExpress were screened for further analysis. Datasets derived in patients with sepsis (rather than endotoxemia, acute infection, SIRS, etc.) from whom whole blood specimens were collected and mRNA profiling performed (microarray or bulk RNAseq) were included for analysis. In total, 56 datasets from GEO and ten datasets from ArrayExpress were excluded due to exclusion criteria, including using whole blood sample without leukocyte isolation or employing sorted immunocytes (without monocytes), providing placebo treatment to septic patients, failing to enroll healthy controls, or recruiting pediatric or neonatal patients. The remaining of ten datasets were finally included. Among them, six datasets utilizing whole blood leukocyte samples were combined for monocyte quantification, while the other four datasets analyzing mRNA profiles of circulating monocytes were pooled for further analysis. General features of included datasets are provided in Table S2. The further analyses and whole study design are presented in Fig. S1.
3.2. Quantification of circulating monocytes in sepsis
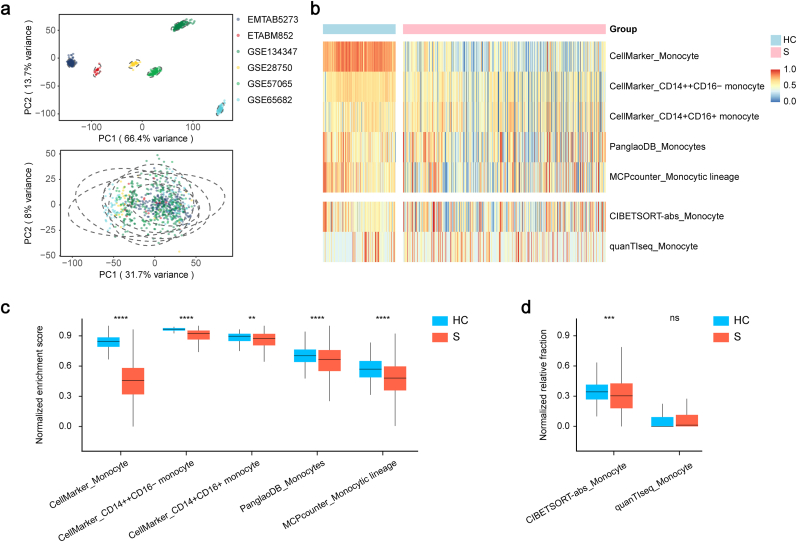
To investigate the variance of circulating monocyte counts between septic patients and healthy volunteers, we conducted a quantitative analysis based on a combined leukocyte dataset. As shown in Fig. 1a, batch effect from non-biological technical biases was reduced after correction between the six leukocyte datasets. Quantification of circulating monocytes was implemented using both marker-based methods, such as MCPcounter and ssGSEA, and deconvolution methods, including CIBERSORT and quanTIseq (Fig. 1b). A total of three and one reference gene sets were curated from CellMarker and PanglaoDB databases, respectively. Compared to healthy controls, the normalized enrichment scores of septic patients were all downregulated (Fig. 1c). Furthermore, the CIBERSORT analysis revealed that the normalized relative fraction of peripheral monocytes in the sepsis group was lower than that in the healthy control group (Fig. 1d). However, quanTIseq analysis demonstrated no statistical difference in the fraction between two groups (Fig. 1d). On the whole, the above findings suggest that circulating monocytes are significantly reduced in septic patients compared to healthy controls.
Fig. 1.
Quantification of circulating monocytes in sepsis.
a. The first two principal components of the clustering results using all 8267 genes present in the pooling whole blood leukocyte profiling before (up) and after (down) batch effect correction. b. Heatmap of the landscape of both NESs calculated based on distinct monocyte reference gene sets and NRFs generated by deconvolutional methods of monocytes. c, d. Distribution of the NES (c) and NRF (d) of monocytes exhibited a decline in the septic patients compared with healthy controls. Two-tailed Student's t-test was performed in c and d. nsp > 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Abbreviations: PC: principal component, HC: healthy control, S: septic patient, NES: normalized enrichment score, NRF: normalized relative fraction.
3.3. Identification of primary cell death mode of monocytes
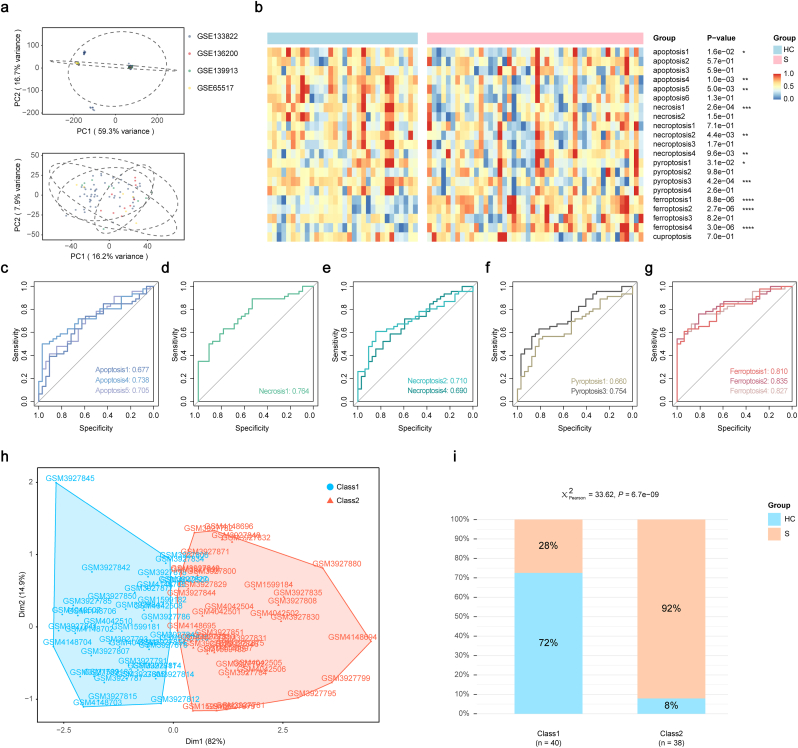
To probe the predominant type of cell death forms associated with the decreased circulating monocyte counts, we implemented a ssGSEA analysis based on the pooled monocyte dataset. Batch effects were corrected with an empirical Bayes framework in dataset integration (Fig. 2a). The enrichment analysis showed that all cell death pathways, except cuproptosis, had distinct regulation between the two groups, of which three pathways of apoptosis, one of necrosis, two of necroptosis, two of pyroptosis and three of ferroptosis were significantly regulated (Fig. 2b).
Fig. 2.
Identification of primary cell death mode of circulating monocytes in sepsis.
a. The first two principal components of the clustering results using all 9279 genes present in the pooling circulating monocyte profiling before (up) and after (down) batch effect correction. b. Heatmap of the landscape of NESs calculated based on various reference gene sets of different cell death types. Apoptosis 1/4/5, necrosis 1, necroptosis 2/4, pyroptosis 1/3 and ferroptosis1/2/4 were abnormally regulated in septic patients compared with healthy controls. c-g. The ROC curve analyses of NESs of apoptosis (c), necrosis (d), necroptosis (e), pyroptosis (f), and ferroptosis (g) exhibited the NESs of ferroptosis possessed the greatest area under operating characteristic curve values. h. Distribution of subjects in the two most important dimensions in k-means clustering analysis based on NESs of ferroptosis1, 2, and 4. i. Proportion of disease identity in the two classes generated from the k-means clustering analysis. Two-tailed Student's t-test was performed in b. Chi-square test was performed in i. Abbreviations: PC: principal component, HC: healthy control, S: septic patient, Dim: dimension, ROC: receiver operating characteristic, NES: normalized enrichment score.
We next operated ROC analyses to further investigate the contribution order of these cell death types. The AUCs of three significant ferroptosis pathways all exceeded 0.8, which were greater than those for other cell death types (Fig. 2c–g). To verify the role of ferroptosis of monocytes in the development of sepsis, we conducted a k-means clustering analysis using the normalized enrichment scores of the significantly regulated ferroptosis pathways. Elbow and average silhouette methods were employed to determine the optimal number of clusters (Fig. S2), following which monocytes from septic patients and healthy volunteers were categorized into two classes (Fig. 2h). Notably, this ferroptosis-based classification could exactly distinguish the identity of sepsis (Fig. 2i). There were 40 subjects in class 1 (51.3 %) and 38 subjects in class 2 (48.7 %). Subjects in class 2 showed significantly higher sepsis sample rate as compared to class 1. Additionally, compared to ferroptosis, the reclassification based on the normalized enrichment scores of other significantly regulated cell death types showed insufficient correlations with sepsis identity (Fig. 2i, Fig. S3). Taken together, the present results demonstrate that ferroptosis could be a major contributor in the development of sepsis.
3.4. Identification of SLC39A8 as the hub gene related to monocyte ferroptosis in sepsis
We took a two-step approach to identify candidate genes related to both sepsis and ferroptosis. We first investigated specifically regulated genes in monocytes during sepsis by a multivariate method. In the 2000 cycles of variable search procedure, the attendance frequencies of selected genes in the 5-gene classification NN model attaining the goal fitness were all tallied and used for the association assessment of sepsis (Fig. S4a). The genes with higher attendance frequency were colored and labeled (Fig. S4b and c). The GA was converged before reaching 2000 cycles, reflected by the stability of gene ranks, which indicated no new solutions were found (Fig. S4c). Genes with an attendance frequency greater than 15 were thought to be closely linked with sepsis development and were selected for further investigation (Table S3, Fig. S4d and e).
We then identified ferroptosis-related candidates by implementing a WGCNA analysis, which partitions genes into distinct modules based on their expression patterns, and can be used for functional analysis. Before gene network construction, we performed quality control of the samples by sequentially eliminating samples with missing values and outlier samples, resulting in 76 qualified samples (Fig. S5a). Then a scale-free network was created based on the legitimate samples using a soft thresholding power set as 9, which was detected according to a network topology analysis (Fig. S5b and c). Thirteen gene modules were identified by average linkage clustering method, with non-clustering genes grouped in the gray module (Fig. S5d). To identify the ferroptosis-related gene module, we calculated the mean value of correlations between the z-scaled ferroptosis enrichment scores and each module's module eigengene value. Genes in the blue module were found to be most statistically related to ferroptosis scores (Fig. S5e-i). To validate the function of genes in the blue module, we performed functional enrichment analyses based on reference gene sets from three public databases. The results revealed that genes from the blue module were enriched in the ‘Ferroptosis’ pathway and lipid metabolism related pathways, which suggested their potential roles in ferroptosis induction (Fig. S5j-l). Ninety-one candidate ferroptosis-related genes were then screened for further analysis by setting a threshold of MM values greater than 0.8 and GS values to z-scaled ferroptosis enrichment scores greater than 0.3 (Table S4).
To pinpoint the hub gene related to both sepsis and ferroptosis, we intersected the candidates from GA-based feature selection analysis and WGCNA analysis. Twenty-six overlapped genes were identified and then ranked according to their mean GS values to investigate their contribution ordering to ferroptosis (Fig. S6a). SLC39A8, which encodes ZIP8, emerged as the candidate gene hub given its possession of the highest mean GS value and a high level of expression in sepsis (Table S4, Fig. S6).
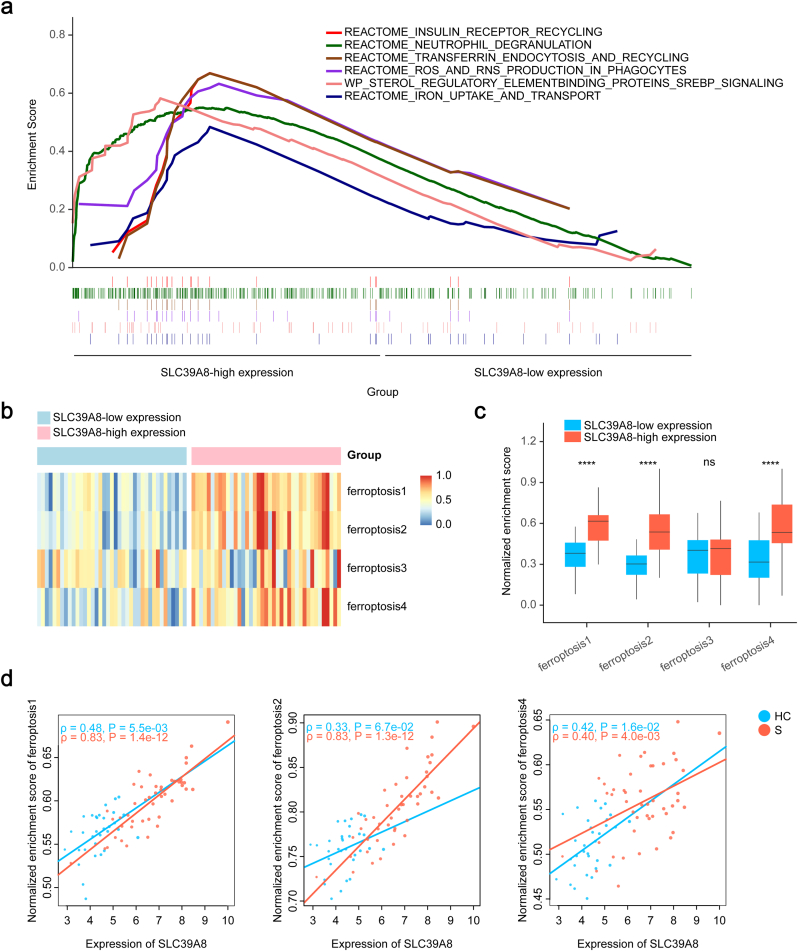
To further investigate the role of ZIP8 in ferroptosis and sepsis, we divided the samples from the pooled monocyte dataset into two groups according to the median expression of SLC39A8 and conducted a GSEA analysis (Table S5). The top six enriched pathways are presented in Fig. 3a, where ferroptosis-related ‘IRON_UPTAKE_AND_TRANSPORT’, ‘TRANSFERRIN_ENDOCYTOSIS_AND_RECYCLING’ and ‘ROS_AND_RNS_PRODUCTION_IN_PHAGOCYTES’ pathways were all upregulated in the SLC39A8-high expressed group. Furthermore, z-scaled enrichment scores of ferroptosis 1/2/4 pathways were all markedly higher in SLC39A8-high expressed group (Fig. 3b and c). Additionally, correlation analyses demonstrated that the normalized ferroptosis scores all had strong associations with the expression of SLC39A8 in septic patients (Fig. 3d). Taken together, these data suggest ZIP8 is closely associated with monocyte ferroptosis during sepsis, and may play a potential role in ferroptosis induction.
Fig. 3.
Association between SLC39A8 expression and ferroptosis NESs in circulating monocytes in sepsis.
a. The top six pathways with higher NESs in monocytes of the SLC39A8 high-expression group. b. Heatmap of the landscape of ferroptosis NESs of the SLC39A8 high-expression and low-expression groups. c. Distribution of the NESs of ferroptosis exhibited an increase of ferroptosis1/2/4 NESs in the SLC39A8 high-expression group compared to the SLC39A8 low-expression group. d. Correlation analysis revealed a strong relationship between the expression of SLC39A8 and NESs of ferroptosis1/2/4. Two-tailed Student's t-test was performed in c. Pearson correlation analysis was performed in d. nsp > 0.05, ****p < 0.0001. Abbreviations: HC: healthy control, S: septic patient, NES: normalized enrichment score.
3.5. ZIP8 is upregulated in monocytes subjected to ferroptosis under sepsis conditions in vivo
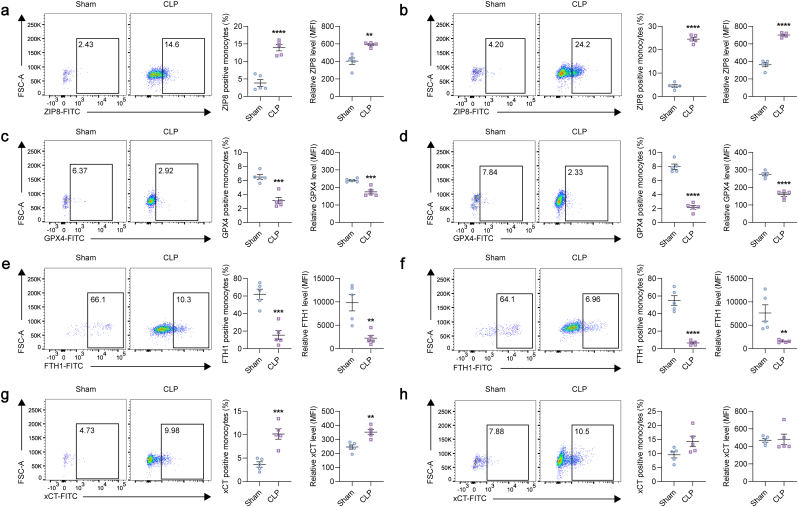
To validate the increased expression of ZIP8 in monocytes subjected to ferroptosis under sepsis conditions, we performed flow cytometry analyses, using whole blood from mice subjected to sepsis induced by CLP model. The CD45high/CD11bhigh/Ly6Gnega-int population was characterized as circulating monocytes (Fig. S7). The two main components of the key antioxidant GSH system, GPX4 and xCT, and the constituent of the major iron storage protein ferritin, FTH1, were used as ferroptosis indicators. It is revealed that, at 24 h and 48 h post-CLP, ZIP8 was significantly upregulated (Fig. 4a and b), reflected by both the increased positive cell fraction and MFI. As for ferroptosis indicators, GPX4 and FTH1 were decreased both at 24 h and 48 h post-CLP (Fig. 4c–f). However, compared to sham controls, xCT was upregulated at 24 h post-CLP (Fig. 4g), and had no differential expression between two groups at 48 h post-CLP (Fig. 4h).
Fig. 4.
ZIP8 is upregulated in monocytes subjected to ferroptosis from mice treated with CLP.
a-h. Quantification of ZIP8 (a, b), GPX4 (c, d), FTH1 (e, f), and xCT (g, h) in circulating CD45high/CD11bhigh/Ly6Gnega-int monocytes isolated from 8 to 10 weeks old male C57BL/6 mice at 24 h (a, c, e, g) and 48 h (b, d, f, h) post-CLP treatment (n = 5/group). Two-tailed Student's t-test was performed in a-h. **p < 0.01, ***p < 0.001, ****p < 0.0001. Abbreviations: CLP: cecal ligation and puncture, MFI: mean fluorescence intensity.
3.6. ZIP8 regulates ferroptosis of monocytes under sepsis conditions in vitro
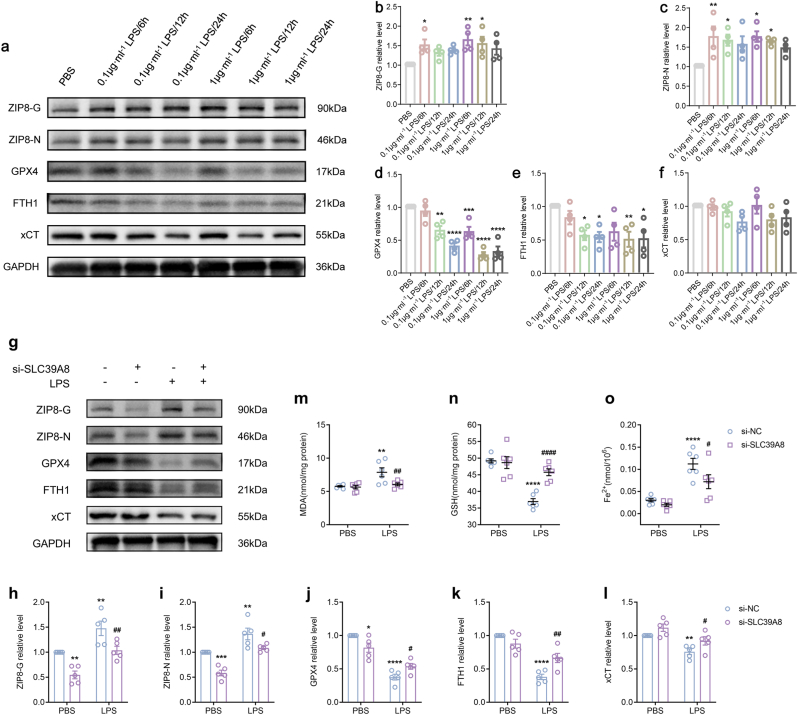
We also verified our in silico analyses with in vitro experiments by evaluating the effect of LPS on the expression of ferroptosis markers in RAW264.7 monocyte/macrophage cell lines. We examined both native and glycosylated ZIP8 as glycosylation is essential for ZIP8 trafficking to the plasma membrane. The results showed a peak in partially glycosylated ZIP8 (ZIP8-G) expression at 6 h for both concentrations of LPS, followed by a time-dependent decrease (Fig. 5a and b). The regulation of native ZIP8 (ZIP8-N) showed a similar pattern (Fig. 5a, c). Additionally, the ferroptosis indicators, GPX4 and FTH1, notably decreased in response to 1 μg/ml LPS at 12 h (Fig. 5a, d, e). However, the expression of xCT was not statistically regulated (Fig. 5a, f). These results suggest that ZIP8 may be associated with ferroptosis development of circulating monocytes during sepsis.
Fig. 5.
ZIP8 regulates ferroptosis of monocytes under sepsis conditions in vitro.
a. Representative immunoblots of ZIP8-G, ZIP8-N, GPX4, FTH1 and xCT in RAW264.7 cells treated with different concentrations of LPS and durations. b-f. Immunoblot analyses of ZIP8-G (b, n = 4/group), ZIP8-N (c, n = 4/group), GPX4 (d, n = 4/group), FTH1 (e, n = 4/group), and xCT (f, n = 4/group) exhibited a sequential pattern of expression changes between ZIP8 (peaked at 6 h post-treatment) and ferroptosis related GPX4 and FTH1 (hit the lowest at 12 h post-treatment). g. Representative immunoblots of ZIP8-G, ZIP8-N, GPX4, FTH1 and xCT in RAW264.7 cells with/without decreased Slc39a8 expression and/or LPS treatment. h-l. Immunoblot analyses of ZIP8-G (h, n = 5/group), ZIP8-N (i, n = 5/group), GPX4 (j, n = 5/group), FTH1 (k, n = 5/group), and xCT (l, n = 5/group) exhibited that suppressing ZIP8 attenuated the decrease of GPX4, FTH1 and xCT in response to LPS. m-o. Quantification analysis showed that decreasing ZIP8 alleviated the LPS-induced upregulations of MDA (m, n = 6/group) and ferrous ions (o, n = 6/group) and reversed the decreased GSH level (n, n = 6/group). One-way ANOVA was performed in b-f. Two-way ANOVA was performed in h-o. Compared to PBS treated si-NC group: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; Compared to LPS treated si-NC group: #p < 0.05, ##p < 0.01, ####p < 0.0001.
Abbreviations: NC: normal control.
To verify its involvement in the ferroptosis of monocytes during sepsis, we knocked down the expression of ZIP8 by transfecting si-Slc39a8 sequences to RAW264.7 cells. As shown in Fig. 5g–i, ZIP8-G and ZIP8-N were effectively repressed in the absence of LPS challenge, and compared to si-NC group, the expressions of ZIP8-G and ZIP8-N were also decreased under LPS treatment in the si- Slc39a8 group. As anticipated, suppressing ZIP8 attenuated the decreases of GPX4, FTH1 and xCT in response to LPS (Fig. 5g, j, k, l). Interestingly, we noticed that, even without LPS stimulation, the downregulation of ZIP8 could directly cause a slight reduction in GPX4 (Fig. 5g, j). Additionally, knocking down ZIP8 remarkably alleviated the LPS-induced upregulations of concentrations of MDA and ferrous ions and reversed the decreased GSH level (Fig. 5m-o). These results suggest that suppressing ZIP8 could protect RAW264.7 cells from LPS-induced ferroptosis.
4. Discussion
In this study, we employed both six and four independent mRNA profiles of whole blood leukocytes and monocytes from two separate databases, respectively [[35], [36], [37], [38], [39], [40], [41], [42], [43], [44]], to explore the predominant cell death type of circulating monocytes in sepsis and to further identify the most prominent gene with significant regulating capacity in ferroptosis of monocytes. The results indicated that ferroptosis may serve as a leading factor in the decreased counts of circulating monocytes in sepsis. Additionally, we have revealed that ZIP8 could modulate the ferroptosis of monocytes in vitro, which might underline the ferroptosis of circulating monocytes in septic patients.
Sepsis is characterized by an imbalance of immune response to infection, with an exaggerated immune activation and immune paralysis or suppression, which could occur either sequentially or synchronously [45]. To date, early antimicrobial administration and support of organ function remain the mainstay therapeutic strategies against this enigmatic disease [46,47]. Early diagnosis and proper therapies to rectify the dysregulated immune response may prove instrumental in improving global morbidity and mortality from sepsis. Evidence reveals that the dominating driver of sepsis is the dysregulated host response to infection, rather than the invading organisms themselves [48]. Specifically, monocytes, which serve as pathogen detectors and pro-inflammatory drivers, play a critical role in the defense against both foreign pathogens and nosocomial opportunistic pathogens associated with secondary infections. Dysfunction or depletion of these cells leads to prolonged immune dysfunctions and consequent organ failure and mortality. Our in silico analyses showed septic patients admitted to ICU had a drastic depletion of monocytes when compared to healthy controls. Although our study was unable to investigate the association between the decrease of monocyte counts and mortality due to the limited counts of accessible samples in the public database, numerous studies have reported the specific prognostic impact of monocyte absolute numbers on the overall survival of septic patients [[12], [13], [14]]. In the in silico analyses, we employed both GSEA-based algorithms and complete deconvolution methods to investigate the changing counts of monocytes. The GSEA-based methods enabled the computation of the ES of monocytes, which is a semi-quantitative score describing the enrichment of monocytes in certain samples [49]. We used reference gene sets of monocytes collected from diverse databases to address potential technical biases. Further, the complete deconvolution methods, including quanTIseq and the absolute mode of CIBERSORT, enabled us to calculate the relative cell fraction of circulating monocytes in whole blood leukocytes [50]. The joint application of these two distinct types of methods could provide a more comprehensive view of the specific changing trends of the monocyte transcriptome in the sepsis condition.
After quantifying the number of monocytes, we examined the levels of different modes of cell death using ssGSEA method based on reference gene sets curated from diverse databases and published studies. Ferroptosis scores had a much greater discriminating ability between septic patients and healthy volunteers rather than the other cell death modes, indicating that ferroptosis may play a more prominent role in the decrease of circulating monocytes. On the other hand, although the monocyte was decreased, the apoptosis related pathways presented a counterintuitive downregulation, which might be explained by the sampling timing. It has been reported by a previous study investigating the role of apoptosis and necrosis in monocyte exhaustion that the sampling timing could directly influence the results. The dead monocytes had a significant increase only within the first 72 h of the onset of sepsis, but not subsequently between day 7 and day 10, nor at discharge [14]. The sampling timings of monocyte mRNA profiles included in this study were as followed: GSE133822-day 2 to day 7 after ICU admission; GSE136200-within 24 h after ICU admission; GSE139913-day 1, day 3, and day 7 after ICU admission; GSE65517-upon diagnosis [[41], [42], [43], [44]]. Considering the timings of admission and diagnosis were later than sepsis onset, the overall sampling time points of datasets used in this study were similar to the reported sampling timing, at which the apoptosis activity remained unchanged or even decreased.
In the procedure of hub gene identification, we opted to utilize a multivariate approach rather than relying on univariate methods for searching sepsis-related genes. Currently used univariate statistical approaches all aim to identify differentially expressed genes with ability to discriminate between two or more groups of samples. However, the inherent conceptual restriction of univariate methods, which assumes genes work independently and neglects the connectivity between pathways, can lead to incorrect evaluations regarding gene predictivity. Multivariate approaches, which contemplate the interactions between genes as they are tested in combinations, have the potential to alleviate this limitation to a certain degree. However, the extremely large number of combinations cannot be exhaustively evaluated using currently available computational resources. To address this issue, we adopted a GA method as the variable searching strategy, which could expeditiously explore the ‘combination library’ for the optimal set of genes.
ZIP8, a member of fourteen zinc import proteins, was predicted to have seven transmembrane domains (TMDs), in which the EExxH motif located between TMD IV and V, and the histidine-rich cluster (Hx)n located between TMD III and IV, have been suggested to confer an ability to bind/transport metal ions [51,52]. ZIP8 was found to be able to transport several divalent metals, including Zn2+, Mn2+, as well as Fe2+, Co2+, and Cd2+ [51,52]. Uptake kinetic studies found Mn2+ and Zn2+ to be the best physiological substrate for ZIP8 in comparison with other divalent cations [53,54]. In our study, ZIP8 was detected with two molecular sizes in western blotting experiments, around 100 kDa and 50 kDa, corresponding to the partially glycosylated and native forms, respectively [52]. ZIP8 was substantially increased in both in vivo and in vitro experiments conducted under sepsis conditions, in accordance with the results of our in silico analyses. Due to the essential regulation function of manganese and zinc in cell responses to infection, the role of ZIP8 in the host defense already had been studied in numbers of researches. In fact, the discovery of ZIP8 was precisely due to its evident induction in monocytes in response to Mycobacteria infection [55]. Moreover, a previous clinical study found that the expression of SLC39A8 in circulating monocytes was closely associated with the severity of sepsis where SLC39A8 expression was highest in patients with the lowest plasma zinc concentrations and the highest severity of illness [56]. Consistent with that, in our study, ZIP8 of monocytes dramatically increased at 24 h and 48 h post-CLP, due to a polymicrobial infection. Previous in vitro studies found ZIP8 can be induced by LPS in bone marrow derived macrophages in a time-dependent manner, where it peaked at 4 h post-treatment then decreased at 24 h. Similar ZIP8 induction trend was observed in our in vitro experiments, which were performed using RAW264.7 monocyte/macrophage cell line, where the expression of ZIP8 culminated at 6 h after LPS administration in both concentrations, however, instead of decreasing, ZIP8 still underwent a slight increase in comparison with the PBS control group at 24 h after LPS treatment.
To detect the ferroptosis induction, we probed the expression of ferroptosis indicators, GPX4, FTH1, and xCT with in vivo and in vitro experiments. GPX4 and xCT are components of the xc−-GSH-GPX4 system, which provides main antioxidant defense against ferroptosis [57]. GPX4 is a recognized ferroptosis gatekeeper and plays a central role in limiting lipid peroxidation by directly reducing lipid hydroperoxide to nontoxic lipid alcohol in the membrane, and its proper function depends on the availability of cellular GSH [57]. xCT is one of the two subunits of antiporter xc−, which is a cystine/glutamate antiporter and contributes to the synthesis of GSH through importing extracellular cystine [58]. The decrease of GPX4 and xCT indicates an impairment of antioxidant capacity, which is a marker event in ferroptosis [57]. On the other hand, FTH1 is a constituent of ferritin, which is the main iron storage protein. The abundance of FTH1 is closely associated with the concentration of intracellular irons. The decrease of FTH1 is normally a consequence of autophagy (defined as ferritinophagy), which indicates an iron release leading to a sharp increase of intracellular iron concentration [57]. In our in vivo study, GPX4 and FTH1 presented a significant decreasing pattern in circulating monocytes both at 24 h and 48 h post-CLP, reflected by the decrease of both positive cell percentage and MFI. Besides, our in vitro results showed that GPX4 and FTH1 were decreased under LPS treatment regardless of concentrations and stimulation durations, which was consistent with published studies [[59], [60], [61]], indicating an induction of ferroptosis in RAW264.7 cells. On the other hand, the expression of xCT under sepsis conditions was not decreased from beginning. Our in vivo results showed that, at 24 h post-CLP, xCT in circulating monocytes was significantly induced, while at 48 h post-CLP, this induction was weakened and there was no difference of xCT expression between two groups. In our in vitro study, under conditions of 0.1 μg⋅ml−1/6 h, 0.1 μg⋅ml−1/12 h, 0.1 μg⋅ml−1/24 h, 1 μg⋅ml−1/6 h and 1 μg⋅ml−1/24 h, xCT abundance had no difference between groups; however, under the condition of 1 μg⋅ml−1/12 h, xCT was decreased. Previous published studies showed both up and down regulation patterns of xCT in monocytes under LPS treatment [[62], [63], [64]]. These seemingly contradictory results might be explained by the complicated regulation mechanisms involving transcription and degradation activities and variance of experimental conditions. Firstly, under sepsis conditions, or in a broader sense, stress conditions, the positive and negative transcription factors (TFs) of xCT could be both induced, mainly including activating transcription factor 4 (ATF4) and NFE2 like bZIP transcription factor 2 (NFE2L2) (positive), tumor protein p53 (TP53) and activating transcription factor 3 (ATF3) (negative) [65]. The proteasomal degradation may also take part in the regulation, reflected by the contrary regulations of xCT mRNA and protein under the stimulation of 1 μg⋅ml−1 LPS/24 h, in which mRNA of xCT was upregulated while protein was decreased [63,66]. On the other hand, the treatment conditions may be another factor. In previous studies, the upregulation of xCT was induced under a low concentration of LPS (50 ng⋅ml−1), while at concentrations of 1 μg⋅ml−1 and 10 μg⋅ml−1, expression of xCT was significantly inhibited [[62], [63], [64]]. Considering the reported inconsistency in mRNA and protein levels of xCT, we speculated the specific mechanism of xCT expression may be associated with the balance between transcription and degradation regulations. And the expression patterns of xCT may reflect the changing states of activation to collapse of the antioxidant system reacted to a stress stimulation.
In our in vitro experiments, we also found that the GPX4 and FTH1 levels had a significant decrease at 12 h after 1 μg/ml LPS administration, which was later than the timing of obvious increase of ZIP8. Combined with our GSEA results, the sequential presentation of ZIP8 upregulation and ferroptosis induction wondered us whether there existed a causal relationship between them. Actually, the critical roles of ZIP8 in modulating cellular processes in immunocytes have been widely studied. Loss of ZIP8 could lead to impaired phagocytosis of macrophages in bacterial pneumonia [67]. Additionally, under inflammatory conditions, ZIP8 could dampen LPS-induced pro-inflammatory cytokine production and block IL-10 release in macrophages by a zinc-dependent manner [68,69]. In adaptive immunity, the zinc-dependent T cell activation is also regulated by ZIP8 [70]. In terms of controlling cell death, it was revealed that ZIP8-mediated Zn2+ transport into primary human lung epithelia could protect against apoptosis [71]. Interestingly, contrary to impairing cellular viability by promoting apoptotic cell death, knocking down ZIP8 was identified as inhibiting ferroptosis in our in vitro study, indicated by the reversals of increased MDA and intracellular ferrous ions. Considering the critical functions of ZIP8 in the uptake of several metal ions, its regulation capacity of ferroptosis might be associated with the intracellular ion dyshomeostasis. Although zinc at physiological levels can work as an antioxidant [72], an excessive amount of zinc or ‘iron-free’ zinc oxide nanoparticles could boost generation of mitochondrial reactive oxygen species and induce lipid peroxidation [[73], [74], [75]]. Moreover, a recent study illustrated that, in the presence of iron chelators, zinc could still trigger ferroptosis [76]. Likewise, excessive manganese could induce ferroptosis in a type-I IFN-dependent manner [77]. We observed that reducing ZIP8 could reverse the decreases of xCT, GSH, GPX4, and FTH1, and decrease the intracellular iron, which suggested that ferroptosis induced by ZIP8 may be associated with both the impairment of antioxidant GSH system and the boost of iron overload. Considering the critical roles of ZIP8 in ion transport, these dysregulations may be induced by the intracellular ion disturbance. The abundance of GPX4, xCT and FTH1 embodies a competition of their transcription and degradation, which could be both regulated by ions to a great extent. The expression of the main TFs of GPX4, ATF4 and NFE2L2, both could be increased by the metal ions transported by ZIP8, including Zn2+, Mn2+, Fe2+, Cd2+, and Co2+ [[78], [79], [80], [81], [82], [83], [84], [85], [86], [87]]. However, several inducers of the degradation pathways of GPX4 could also be increased. Specifically, Mn2+ could enhance the expression of NEDD4 like E3 ubiquitin protein ligase (NEDD4L) [88], an E3 ligase, which was recently found to mediate GPX4 degradation through ubiquitin proteasome system (UPS) [89]. Heat shock protein family A (Hsp70) member 8 (HSPA8), lysosomal associated membrane protein 2 (LAMP2A), and heat shock protein 90 (HSP90), promoting chaperone-mediated autophagy (CMA)-mediated GPX4 degradation, all could be increased by ZIP8 transported ions [[90], [91], [92], [93], [94], [95], [96], [97]]. The increases of ATF4 and NFE2L2 could also promote the expression of xCT; however, through ATF3 and TP53, ZIP8 transported ions can induce the transcription inhibition of xCT as well [[98], [99], [100], [101], [102], [103], [104]]. Besides, the decrease of xCT may be caused by the UPS-mediated degradation. Fascin actin-bundling protein 1 (FSCN1) was recently found to promote xCT degradation in a UPS-dependent way [105], and OTU deubiquitinase, ubiquitin aldehyde binding 1 (OTUB1) could deubiquitinate and stabilize xCT [106]. The above two UPS activator and inhibitor could be increased and decreased by Co2+ and Zn2+, respectively [107,108]. The xCT disturbance could inhibit the synthesis of GSH, which could further lead to the dysfunction of GPX4. Finally, the decrease of FTH1 may be a result of enhanced ferritinophagy activity, which could be induced by excess intracellular Zn2+ and Cd2+ [83,109]. And the released iron by ferritinophagy, together with the iron directly transported by ZIP8 could boost the concentration of intracellular irons, excessively promoting ferroptosis. The future work aiming to figure out the occurrence and importance orders of the mechanisms concerning ZIP8 induced xCT/GSH/GPX4 dysregulation and iron overload could be better for learning the roles of ZIP8 in ferroptosis. Besides, we observed that decreased ZIP8 expression in PBS-treated RAW264.7 cells could directly induce a down-regulation of GPX4, which may be explained by the inhibited expression of ATF4 and NFE2L2 caused by zinc deficiency.
Our manuscript has limitations that need to be mentioned. Due to the limited accessible resources and our inclusion criteria, only 78 monocyte samples were included, which was not quite wealthier compared to the sample size of whole blood leukocyte samples. However, genes capable to distinguish between sepsis and control samples and gene modules associated with ferroptosis were exactly identified from respective feature selection and WGCNA analysis, and responding results had robust statistical significance. This might be interpreted by the small sample threshold allowed in both analyses [30,31]. Actually, the GA-based feature selection was designed to dispose the ‘big data’ problems [30], where the number of variables commonly far exceeds that of the observations. Besides, although we used reference gene sets of cell death curated from distinct sources to minimize the technical bias, the estimation of cell death implemented by the computational methods requires further experimental validation. We also acknowledge that the in vitro validation of the regulation ability of ZIP8 on ferroptosis, conducted using a murine monocyte/macrophage cell line, may not fully recapitulate human primary monocytes, and more work with primary monocytes should be implemented in future study.
In conclusion, through computational approaches, we identified ferroptosis as the major contributor to the decrease of circulating monocytes in sepsis. The raised ZIP8 played an essential role in the ferroptosis induction of monocytes in vitro. However, experimental validations are needed to verify the role of ferroptosis in monocyte depletion, and the mechanisms underlining ZIP8 functions require further investigations to be explored. Overall, we provided supplementary information that ZIP8 induced ferroptosis could potentially serve as a therapeutic target for the monocyte exhaustion in sepsis.
Funding
This work was supported by the Program of Shanghai Academic Research Leader [grant number 21XD1402800]; the Natural Science Foundation of Shanghai [grant number 22ZR1452200]; and the National Natural Science Foundation of China [grant number 82100094].
Role of the funding source
The funders had no role in the study design, data collection, analyses, or interpretation of data, and were not involved in the writing or submission process.
Contributions
Conceptualization: Xin Lv, Tong Zhang, and Shuqing Jin. Methodology: Tong Zhang, Sheng Wang, and Dongsheng Hua. Investigation: Tong Zhang, Sheng Wang, and Dongsheng Hua. Data curation and software: Tong Zhang, Sheng Wang, Xuan Shi, and Huimin Deng. Visualization: Tong Zhang, and Sheng Wang. Formal analysis: Tong Zhang, Sheng Wang, and Dongsheng Hua. Drafting, review and/or revision of the manuscript: Tong Zhang, Sheng Wang, Dongsheng Hua, Xuan Shi, Huimin Deng, Shuqing Jin, and Xin Lv. Material support: Shuqing Jin, Xuan Shi, and Huimin Deng. Project administration: Xin Lv, Shuqing Jin, and Tong Zhang. Supervision: Xin Lv, and Shuqing Jin. All authors have read and approved the final version of the manuscript. Tong Zhang, Sheng Wang, and Dongsheng Hua have accessed and verified the data. All authors have full access to all the data and accept responsibility to submit for publication. Xin Lv was responsible for the decision to submit the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Program of Shanghai Academic Research Leader (21XD1402800), Shanghai Natural Science Foundation (22ZR1452200), and National Natural Science Foundation of China (82100094).
Abbreviations
- 7-AAD
7-aminoactinomycin D
- ANOVA
analysis of variance
- ATF3
activating transcription factor 3
- ATF4
activating transcription factor 4
- AUC
area under operating characteristic curve
- CD11b
integrin alpha M
- CD45
protein tyrosine phosphatase receptor type C
- CLP
cecal ligation and puncture
- CMA
chaperone-mediated autophagy
- ES
enrichment score
- FTH1
ferritin heavy chain 1
- FSCN1
fascin actin-bundling protein 1
- GA
genetic algorithm
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GEO
gene expression omnibus
- GPX4
glutathione peroxidase 4
- GS
gene trait significance
- GSEA
gene set enrichment analysis
- GSH
glutathione
- HBV
hepatitis B virus
- HIV
human immunodeficiency virus
- HLA-DR
major histocompatibility complex, class II, DR
- HSPA8
heat shock protein family A (Hsp70) member 8
- HSP90
heat shock protein 90
- ICU
intensive care unit
- IFN
interferon
- IgG
immunoglobulin G
- IL-10
interleukin 10
- kDA
kilodalton
- LAMP2A
lysosomal associated membrane protein 2
- LPS
lipopolysaccharide
- Ly6G
lymphocyte antigen 6 family member G
- MDA
malondialdehyde
- MFI
mean fluorescence intensity
- MM
module membership
- mRNA
messenger RNA
- NC
normal control
- NEDD4L
NEDD4 like E3 ubiquitin protein ligase
- NES
normalized enrichment score
- NFE2L2
NFE2 like bZIP transcription factor 2
- NGS/RNA-seq
next-generation sequencing
- NN
neural network
- NRF
normalized relative fraction
- OTUB1
OTU deubiquitinase, ubiquitin aldehyde binding 1
- PBMC
peripheral blood mononuclear cell
- PBS
phosphate buffer saline
- PDL1
programmed cell death 1 ligand 1
- RNA
ribonucleic acid
- ROC
receiver operating characteristic
- SIRS
system inflammatory reaction syndrome
- SLC39A8
solute carrier family 39 member 8
- ssGSEA
single sample gene set enrichment analysis
- TF
transcription factor
- TMD
transmembrane domain
- TP53
tumor protein p53
- UPS
ubiquitin proteasome system
- WGCNA
weighted gene co-expression network analysis
- xCT
light-chain subunit of the cystine/glutamate antiporter
- z-score
standard score
- ZIP8
zrt-/irt-like protein 8
- ZIP8-G
glycosylated ZIP8
- ZIP8-N
native ZIP8
- ZIP1–14
zrt-/irt-like proteins 1-14
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102985.
Contributor Information
Shuqing Jin, Email: jinshuqing1@163.com.
Xin Lv, Email: xinlvg@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Fleischmann C., Scherag A., Adhikari N.K., Hartog C.S., Tsaganos T., Schlattmann P., Angus D.C., Reinhart K. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am. J. Respir. Crit. Care Med. 2016;193(3):259–272. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 2.Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R., Colombara D.V., Ikuta K.S., Kissoon N., Finfer S., Fleischmann-Struzek C., Machado F.R., Reinhart K.K., Rowan K., Seymour C.W., Watson R.S., West T.E., Marinho F., Hay S.I., Lozano R., Lopez A.D., Angus D.C., Murray C.J.L., Naghavi M. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet (London, England) 2020;395(10219):200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angus D.C., Opal S. Immunosuppression and secondary infection in sepsis: Part, not all, of the story. JAMA. 2016;315(14):1457–1459. doi: 10.1001/jama.2016.2762. [DOI] [PubMed] [Google Scholar]
- 4.Prescott H.C., Osterholzer J.J., Langa K.M., Angus D.C., Iwashyna T.J. Late mortality after sepsis: propensity matched cohort study. Br. Med. J. 2016;353:i2375. doi: 10.1136/bmj.i2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., Hotchkiss R.S., Levy M.M., Marshall J.C., Martin G.S., Opal S.M., Rubenfeld G.D., van der Poll T., Vincent J.L., Angus D.C. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Poll T., Shankar-Hari M., Wiersinga W.J. The immunology of sepsis. Immunity. 2021;54(11):2450–2464. doi: 10.1016/j.immuni.2021.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Torres L.K., Pickkers P., van der Poll T. Sepsis-induced immunosuppression. Annu. Rev. Physiol. 2022;84:157–181. doi: 10.1146/annurev-physiol-061121-040214. [DOI] [PubMed] [Google Scholar]
- 8.van der Poll T., van de Veerdonk F.L., Scicluna B.P., Netea M.G. The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol. 2017;17(7):407–420. doi: 10.1038/nri.2017.36. [DOI] [PubMed] [Google Scholar]
- 9.Venet F., Monneret G. Advances in the understanding and treatment of sepsis-induced immunosuppression. Nat. Rev. Nephrol. 2018;14(2):121–137. doi: 10.1038/nrneph.2017.165. [DOI] [PubMed] [Google Scholar]
- 10.Kerris E.W.J., Hoptay C., Calderon T., Freishtat R.J. Platelets and platelet extracellular vesicles in hemostasis and sepsis. J. Invest. Med. : the official publication of the American Federation for Clinical Research. 2020;68(4):813–820. doi: 10.1136/jim-2019-001195. [DOI] [PubMed] [Google Scholar]
- 11.Hausfater P., Robert Boter N., Morales Indiano C., Cancella de Abreu M., Marin A.M., Pernet J., Quesada D., Castro I., Careaga D., Arock M., Tejidor L., Velly L. Monocyte distribution width (MDW) performance as an early sepsis indicator in the emergency department: comparison with CRP and procalcitonin in a multicenter international European prospective study. Crit. Care. 2021;25(1):227. doi: 10.1186/s13054-021-03622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S.H., Park B.G., Park C.J., Kim S., Kim D.H., Jang S., Hong S.K., Chi H.S. An extended leukocyte differential count (16 types of circulating leukocytes) using the CytoDiff flow cytometric system can provide information for the discrimination of sepsis severity and prediction of outcome in sepsis patients. Cytometry B Clin. Cytometry. 2014;86(4):244–256. doi: 10.1002/cyto.b.21123. [DOI] [PubMed] [Google Scholar]
- 13.Liu S., Li Y., She F., Zhao X., Yao Y. Predictive value of immune cell counts and neutrophil-to-lymphocyte ratio for 28-day mortality in patients with sepsis caused by intra-abdominal infection. Burns & trauma. 2021;9 doi: 10.1093/burnst/tkaa040. tkaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adrie C., Bachelet M., Vayssier-Taussat M., Russo-Marie F., Bouchaert I., Adib-Conquy M., Cavaillon J.M., Pinsky M.R., Dhainaut J.F., Polla B.S. Mitochondrial membrane potential and apoptosis peripheral blood monocytes in severe human sepsis. Am. J. Respir. Crit. Care Med. 2001;164(3):389–395. doi: 10.1164/ajrccm.164.3.2009088. [DOI] [PubMed] [Google Scholar]
- 15.Venet F., Pachot A., Debard A.L., Bohe J., Bienvenu J., Lepape A., Powell W.S., Monneret G. Human CD4+CD25+ regulatory T lymphocytes inhibit lipopolysaccharide-induced monocyte survival through a Fas/Fas ligand-dependent mechanism. J. Immunol. 2006;177(9):6540–6547. doi: 10.4049/jimmunol.177.9.6540. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., Liu Y., Liu Q., Zheng Q., Dong X., Liu X., Gao W., Bai X., Li Z. Caspase-1-Dependent pyroptosis of peripheral blood mononuclear cells is associated with the severity and mortality of septic patients. BioMed Res. Int. 2020;2020 doi: 10.1155/2020/9152140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S., Duan C., Xie J., Zhang J., Luo X., Wang Q., Liang X., Zhao X., Zhuang R., Zhao W., Yin W. Peripheral immune cell death in sepsis based on bulk RNA and single-cell RNA sequencing. Heliyon. 2023;9(7) doi: 10.1016/j.heliyon.2023.e17764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C., Luo T., Xu L., Liao G., Yan M., Ping Y., Li F., Shi A., Bai J., Zhao T., Li X., Xiao Y. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–d728. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franzén O., Gan L.M., Björkegren J.L.M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database : the journal of biological databases and curation. 2019:2019. doi: 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finotello F., Mayer C., Plattner C., Laschober G., Rieder D., Hackl H., Krogsdam A., Loncova Z., Posch W., Wilflingseder D., Sopper S., Ijsselsteijn M., Brouwer T.P., Johnson D., Xu Y., Wang Y., Sanders M.E., Estrada M.V., Ericsson-Gonzalez P., Charoentong P., Balko J., de Miranda N., Trajanoski Z. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11(1):34. doi: 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., Griss J., Sevilla C., Matthews L., Gong C., Deng C., Varusai T., Ragueneau E., Haider Y., May B., Shamovsky V., Weiser J., Brunson T., Sanati N., Beckman L., Shao X., Fabregat A., Sidiropoulos K., Murillo J., Viteri G., Cook J., Shorser S., Bader G., Demir E., Sander C., Haw R., Wu G., Stein L., Hermjakob H., D'Eustachio P. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50(D1):D687–d692. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pico A.R., Kelder T., van Iersel M.P., Hanspers K., Conklin B.R., Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6(7):e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., Kaplan S., Dahary D., Warshawsky D., Guan-Golan Y., Kohn A., Rappaport N., Safran M., Lancet D. The GeneCards suite: from gene data mining to disease genome sequence analyses. Current protocols in bioinformatics. 2016;54:1. doi: 10.1002/cpbi.5. 30.1-1.30.33. [DOI] [PubMed] [Google Scholar]
- 29.Charrad M., Ghazzali N., Boiteau V., Niknafs A. NbClust: an R package for determining the relevant number of clusters in a data set. J. Stat. Software. 2014;61(6):1–36. [Google Scholar]
- 30.Trevino V., Falciani F. GALGO: an R package for multivariate variable selection using genetic algorithms. Bioinformatics. 2006;22(9):1154–1156. doi: 10.1093/bioinformatics/btl074. [DOI] [PubMed] [Google Scholar]
- 31.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rittirsch D., Huber-Lang M.S., Flierl M.A., Ward P.A. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 2009;4(1):31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burnham K.L., Davenport E.E., Radhakrishnan J., Humburg P., Gordon A.C., Hutton P., Svoren-Jabalera E., Garrard C., Hill A.V.S., Hinds C.J., Knight J.C. Shared and distinct aspects of the sepsis transcriptomic response to fecal peritonitis and pneumonia. Am. J. Respir. Crit. Care Med. 2017;196(3):328–339. doi: 10.1164/rccm.201608-1685OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koh G.C., Maude R.R., Schreiber M.F., Limmathurotsakul D., Wiersinga W.J., Wuthiekanun V., Lee S.J., Mahavanakul W., Chaowagul W., Chierakul W., White N.J., van der Poll T., Day N.P., Dougan G., Peacock S.J. Glyburide is anti-inflammatory and associated with reduced mortality in melioidosis. Clin. Infect. Dis. : an official publication of the Infectious Diseases Society of America. 2011;52(6):717–725. doi: 10.1093/cid/ciq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scicluna B.P., Uhel F., van Vught L.A., Wiewel M.A., Hoogendijk A.J., Baessman I., Franitza M., Nürnberg P., Horn J., Cremer O.L., Bonten M.J., Schultz M.J., van der Poll T. The leukocyte non-coding RNA landscape in critically ill patients with sepsis. Elife. 2020;9 doi: 10.7554/eLife.58597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutherland A., Thomas M., Brandon R.A., Brandon R.B., Lipman J., Tang B., McLean A., Pascoe R., Price G., Nguyen T., Stone G., Venter D. Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Crit. Care. 2011;15(3):R149. doi: 10.1186/cc10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cazalis M.A., Lepape A., Venet F., Frager F., Mougin B., Vallin H., Paye M., Pachot A., Monneret G. Early and dynamic changes in gene expression in septic shock patients: a genome-wide approach. Intensive care medicine experimental. 2014;2(1):20. doi: 10.1186/s40635-014-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scicluna B.P., Klein Klouwenberg P.M., van Vught L.A., Wiewel M.A., Ong D.S., Zwinderman A.H., Franitza M., Toliat M.R., Nürnberg P., Hoogendijk A.J., Horn J., Cremer O.L., Schultz M.J., Bonten M.J., van der Poll T. A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am. J. Respir. Crit. Care Med. 2015;192(7):826–835. doi: 10.1164/rccm.201502-0355OC. [DOI] [PubMed] [Google Scholar]
- 41.Washburn M.L., Wang Z., Walton A.H., Goedegebuure S.P., Figueroa D.J., Van Horn S., Grossman J., Remlinger K., Madsen H., Brown J., Srinivasan R., Wolf A.I., Berger S.B., Yi V.N., Hawkins W.G., Fields R.C., Hotchkiss R.S. T cell- and monocyte-specific RNA-sequencing analysis in septic and nonseptic critically ill patients and in patients with cancer. J. Immunol. 2019;203(7):1897–1908. doi: 10.4049/jimmunol.1900560. Baltimore, Md. : 1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan H.N., Brands X., Aufiero S., Hoogendijk A.J., Klarenbeek A.M., van Engelen T.S.R., Haak B.W., van Vught L.A., Horn J., Schultz M.J., Zwinderman A.H., van der Poll T., Scicluna B.P. The circular RNA landscape in specific peripheral blood mononuclear cells of critically ill patients with sepsis. Crit. Care. 2020;24(1):423. doi: 10.1186/s13054-020-03146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liepelt A., Hohlstein P., Gussen H., Xue J., Aschenbrenner A.C., Ulas T., Buendgens L., Warzecha K.T., Bartneck M., Luedde T., Trautwein C., Schultze J.L., Koch A., Tacke F. Differential gene expression in circulating CD14(+) monocytes indicates the prognosis of critically ill patients with sepsis. J. Clin. Med. 2020;9(1) doi: 10.3390/jcm9010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergenfelz C., Larsson A.M., von Stedingk K., Gruvberger-Saal S., Aaltonen K., Jansson S., Jernström H., Janols H., Wullt M., Bredberg A., Rydén L., Leandersson K. Systemic monocytic-MDSCs are generated from monocytes and correlate with disease progression in breast cancer patients. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0127028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cecconi M., Evans L., Levy M., Rhodes A. Sepsis and septic shock. Lancet (London, England) 2018;392(10141):75–87. doi: 10.1016/S0140-6736(18)30696-2. [DOI] [PubMed] [Google Scholar]
- 46.Marik P.E., Linde-Zwirble W.T., Bittner E.A., Sahatjian J., Hansell D. Fluid administration in severe sepsis and septic shock, patterns and outcomes: an analysis of a large national database. Intensive Care Med. 2017;43(5):625–632. doi: 10.1007/s00134-016-4675-y. [DOI] [PubMed] [Google Scholar]
- 47.Evans L., Rhodes A., Alhazzani W., Antonelli M., Coopersmith C.M., French C., Machado F.R., McIntyre L., Ostermann M., Prescott H.C., Schorr C., Simpson S., Wiersinga W.J., Alshamsi F., Angus D.C., Arabi Y., Azevedo L., Beale R., Beilman G., Belley-Cote E., Burry L., Cecconi M., Centofanti J., Coz Yataco A., De Waele J., Dellinger R.P., Doi K., Du B., Estenssoro E., Ferrer R., Gomersall C., Hodgson C., Møller M.H., Iwashyna T., Jacob S., Kleinpell R., Klompas M., Koh Y., Kumar A., Kwizera A., Lobo S., Masur H., McGloughlin S., Mehta S., Mehta Y., Mer M., Nunnally M., Oczkowski S., Osborn T., Papathanassoglou E., Perner A., Puskarich M., Roberts J., Schweickert W., Seckel M., Sevransky J., Sprung C.L., Welte T., Zimmerman J., Levy M. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Intensive Care Med. 2021;47(11):1181–1247. doi: 10.1007/s00134-021-06506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas L. Germs, The New England journal of medicine. 1972;287(11):553–555. doi: 10.1056/NEJM197209142871109. [DOI] [PubMed] [Google Scholar]
- 49.Hackl H., Charoentong P., Finotello F., Trajanoski Z. Computational genomics tools for dissecting tumour-immune cell interactions. Nat. Rev. Genet. 2016;17(8):441–458. doi: 10.1038/nrg.2016.67. [DOI] [PubMed] [Google Scholar]
- 50.Sturm G., Finotello F., Petitprez F., Zhang J.D., Baumbach J., Fridman W.H., List M., Aneichyk T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35(14):i436–i445. doi: 10.1093/bioinformatics/btz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkitkasemwong S., Wang C.Y., Mackenzie B., Knutson M.D. Physiologic implications of metal-ion transport by ZIP14 and ZIP8, Biometals : an international journal on the role of metal ions in biology. biochemistry, and medicine. 2012;25(4):643–655. doi: 10.1007/s10534-012-9526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zang Z.S., Xu Y.M., Lau A.T.Y. Molecular and pathophysiological aspects of metal ion uptake by the zinc transporter ZIP8 (SLC39A8) Toxicology research. 2016;5(4):987–1002. doi: 10.1039/c5tx00424a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He L., Girijashanker K., Dalton T.P., Reed J., Li H., Soleimani M., Nebert D.W. ZIP8, member of the solute-carrier-39 (SLC39) metal-transporter family: characterization of transporter properties. Mol. Pharmacol. 2006;70(1):171–180. doi: 10.1124/mol.106.024521. [DOI] [PubMed] [Google Scholar]
- 54.Wang C.Y., Jenkitkasemwong S., Duarte S., Sparkman B.K., Shawki A., Mackenzie B., Knutson M.D. ZIP8 is an iron and zinc transporter whose cell-surface expression is up-regulated by cellular iron loading. J. Biol. Chem. 2012;287(41):34032–34043. doi: 10.1074/jbc.M112.367284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Begum N.A., Kobayashi M., Moriwaki Y., Matsumoto M., Toyoshima K., Seya T. Mycobacterium bovis BCG cell wall and lipopolysaccharide induce a novel gene, BIGM103, encoding a 7-TM protein: identification of a new protein family having Zn-transporter and Zn-metalloprotease signatures. Genomics. 2002;80(6):630–645. doi: 10.1006/geno.2002.7000. [DOI] [PubMed] [Google Scholar]
- 56.Besecker B.Y., Exline M.C., Hollyfield J., Phillips G., Disilvestro R.A., Wewers M.D., Knoell D.L. A comparison of zinc metabolism, inflammation, and disease severity in critically ill infected and noninfected adults early after intensive care unit admission. Am. J. Clin. Nutr. 2011;93(6):1356–1364. doi: 10.3945/ajcn.110.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X., Li J., Kang R., Klionsky D.J., Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021;17(9):2054–2081. doi: 10.1080/15548627.2020.1810918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J., Xia X., Huang P. xCT: a critical molecule that links cancer metabolism to redox signaling. Mol. Ther. : the journal of the American Society of Gene Therapy. 2020;28(11):2358–2366. doi: 10.1016/j.ymthe.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong C., Ni X., Wang Y., Zhang A., Zhang Y., Lin F., Li S., Lv Y., Zhu J., Yao X., Dai Q., Mo Y., Wang J. ICA69 aggravates ferroptosis causing septic cardiac dysfunction via STING trafficking. Cell death discovery. 2022;8(1):187. doi: 10.1038/s41420-022-00957-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao Y., Yuan Y., Yang Y., Liu B., Ding Z., Luo J., Chen S., Yu L. GCH1 reduces LPS-induced alveolar macrophage polarization and inflammation by inhibition of ferroptosis. Inflamm. Res. : official journal of the European Histamine Research Society ... [et al.] 2023;72(10–11):1941–1955. doi: 10.1007/s00011-023-01785-1. [DOI] [PubMed] [Google Scholar]
- 61.Luo L., Huang F., Zhong S., Ding R., Su J., Li X. Astaxanthin attenuates ferroptosis via Keap1-Nrf2/HO-1 signaling pathways in LPS-induced acute lung injury. Life Sci. 2022;311(Pt A) doi: 10.1016/j.lfs.2022.121091. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y., Wan R., Peng W., Zhao X., Bai W., Hu C. Quercetin alleviates ferroptosis accompanied by reducing M1 macrophage polarization during neutrophilic airway inflammation. Eur. J. Pharmacol. 2023;938 doi: 10.1016/j.ejphar.2022.175407. [DOI] [PubMed] [Google Scholar]
- 63.Li X., Qi H., Zhang X., Liang H., Zeng N. Jing-Fang n-butanol extract and its isolated JFNE-C inhibit ferroptosis and inflammation in LPS induced RAW264.7 macrophages via STAT3/p53/SLC7A11 signaling pathway. J. Ethnopharmacol. 2023;316 doi: 10.1016/j.jep.2023.116689. [DOI] [PubMed] [Google Scholar]
- 64.Lai K., Song C., Gao M., Deng Y., Lu Z., Li N., Geng Q. Uridine alleviates sepsis-induced acute lung injury by inhibiting ferroptosis of macrophage. Int. J. Mol. Sci. 2023;24(6) doi: 10.3390/ijms24065093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koppula P., Zhuang L., Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein & cell. 2021;12(8):599–620. doi: 10.1007/s13238-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Srisook K., Cha Y.N. Biphasic induction of heme oxygenase-1 expression in macrophages stimulated with lipopolysaccharide. Biochem. Pharmacol. 2004;68(9):1709–1720. doi: 10.1016/j.bcp.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 67.Hall S.C., Smith D.R., Dyavar S.R., Wyatt T.A., Samuelson D.R., Bailey K.L., Knoell D.L. Critical role of zinc transporter (ZIP8) in myeloid innate immune cell function and the host response against bacterial pneumonia. J. Immunol. 2021;207(5):1357–1370. doi: 10.4049/jimmunol.2001395. (Baltimore, Md. : 1950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu M.J., Bao S., Gálvez-Peralta M., Pyle C.J., Rudawsky A.C., Pavlovicz R.E., Killilea D.W., Li C., Nebert D.W., Wewers M.D., Knoell D.L. ZIP8 regulates host defense through zinc-mediated inhibition of NF-κB. Cell Rep. 2013;3(2):386–400. doi: 10.1016/j.celrep.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pyle C.J., Akhter S., Bao S., Dodd C.E., Schlesinger L.S., Knoell D.L. Zinc modulates endotoxin-induced human macrophage inflammation through ZIP8 induction and C/EBPβ inhibition. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0169531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aydemir T.B., Liuzzi J.P., McClellan S., Cousins R.J. Zinc transporter ZIP8 (SLC39A8) and zinc influence IFN-gamma expression in activated human T cells. J. Leukoc. Biol. 2009;86(2):337–348. doi: 10.1189/jlb.1208759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Besecker B., Bao S., Bohacova B., Papp A., Sadee W., Knoell D.L. The human zinc transporter SLC39A8 (Zip8) is critical in zinc-mediated cytoprotection in lung epithelia. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;294(6):L1127–L1136. doi: 10.1152/ajplung.00057.2008. [DOI] [PubMed] [Google Scholar]
- 72.Zago M.P., Oteiza P.I. The antioxidant properties of zinc: interactions with iron and antioxidants. Free Radic. Biol. Med. 2001;31(2):266–274. doi: 10.1016/s0891-5849(01)00583-4. [DOI] [PubMed] [Google Scholar]
- 73.Sensi S.L., Ton-That D., Sullivan P.G., Jonas E.A., Gee K.R., Kaczmarek L.K., Weiss J.H. Modulation of mitochondrial function by endogenous Zn2+ pools. Proc. Natl. Acad. Sci. U.S.A. 2003;100(10):6157–6162. doi: 10.1073/pnas.1031598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palmer L.D., Jordan A.T., Maloney K.N., Farrow M.A., Gutierrez D.B., Gant-Branum R., Burns W.J., Romer C.E., Tsui T., Allen J.L., Beavers W.N., Nei Y.W., Sherrod S.D., Lacy D.B., Norris J.L., McLean J.A., Caprioli R.M., Skaar E.P. Zinc intoxication induces ferroptosis in A549 human lung cells. Metallomics : integrated biometal science. 2019;11(5):982–993. doi: 10.1039/c8mt00360b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang C., Liu Z., Zhang Y., Ma L., Song E., Song Y. "Iron free" zinc oxide nanoparticles with ion-leaking properties disrupt intracellular ROS and iron homeostasis to induce ferroptosis. Cell Death Dis. 2020;11(3):183. doi: 10.1038/s41419-020-2384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen P.H., Wu J., Xu Y., Ding C.C., Mestre A.A., Lin C.C., Yang W.H., Chi J.T. Zinc transporter ZIP7 is a novel determinant of ferroptosis. Cell Death Dis. 2021;12(2):198. doi: 10.1038/s41419-021-03482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang S., Kang L., Dai X., Chen J., Chen Z., Wang M., Jiang H., Wang X., Bu S., Liu X., Zhang G., Tang H. Manganese induces tumor cell ferroptosis through type-I IFN dependent inhibition of mitochondrial dihydroorotate dehydrogenase. Free Radic. Biol. Med. 2022;193(Pt 1):202–212. doi: 10.1016/j.freeradbiomed.2022.10.004. [DOI] [PubMed] [Google Scholar]
- 78.Doi T., Hara H., Kajita M., Kamiya T., Adachi T. Zinc regulates expression of IL-23 p19 mRNA via activation of eIF2α/ATF4 axis in HAPI cells, Biometals : an international journal on the role of metal ions in biology. biochemistry, and medicine. 2015;28(5):891–902. doi: 10.1007/s10534-015-9874-4. [DOI] [PubMed] [Google Scholar]
- 79.Liu C., Yan D.Y., Wang C., Ma Z., Deng Y., Liu W., Xu B. Manganese activates autophagy to alleviate endoplasmic reticulum stress-induced apoptosis via PERK pathway. J. Cell Mol. Med. 2020;24(1):328–341. doi: 10.1111/jcmm.14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y., Wang M., Liu Y., Tao H., Banerjee S., Srinivasan S., Nemeth E., Czaja M.J., He P. Integrated regulation of stress responses, autophagy and survival by altered intracellular iron stores. Redox Biol. 2022;55 doi: 10.1016/j.redox.2022.102407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu F., Inageda K., Nishitai G., Matsuoka M. Cadmium induces the expression of Grp78, an endoplasmic reticulum molecular chaperone, in LLC-PK1 renal epithelial cells. Environ. Health Perspect. 2006;114(6):859–864. doi: 10.1289/ehp.8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao C., Yu D., He Z., Bao L., Feng L., Chen L., Liu Z., Hu X., Zhang N., Wang T., Fu Y. Endoplasmic reticulum stress-mediated autophagy activation is involved in cadmium-induced ferroptosis of renal tubular epithelial cells. Free Radic. Biol. Med. 2021;175:236–248. doi: 10.1016/j.freeradbiomed.2021.09.008. [DOI] [PubMed] [Google Scholar]
- 83.Qin X., Zhang J., Wang B., Xu G., Yang X., Zou Z., Yu C. Ferritinophagy is involved in the zinc oxide nanoparticles-induced ferroptosis of vascular endothelial cells. Autophagy. 2021;17(12):4266–4285. doi: 10.1080/15548627.2021.1911016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dodd C.A., Filipov N.M. Manganese potentiates LPS-induced heme-oxygenase 1 in microglia but not dopaminergic cells: role in controlling microglial hydrogen peroxide and inflammatory cytokine output. Neurotoxicology. 2011;32(6):683–692. doi: 10.1016/j.neuro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang J., Zhang L., Yao G., Zhao H., Wu S. 2023. NRF2 Is Essential for Iron-Overload Stimulated Osteoclast Differentiation through Regulation of Redox and Iron Homeostasis, Cell Biology and Toxicology. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y., Mandal A.K., Son Y.O., Pratheeshkumar P., Wise J.T.F., Wang L., Zhang Z., Shi X., Chen Z. Roles of ROS, Nrf2, and autophagy in cadmium-carcinogenesis and its prevention by sulforaphane. Toxicol. Appl. Pharmacol. 2018;353:23–30. doi: 10.1016/j.taap.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang B., Cheng H., Wang L., Fu J., Zhang G., Guan D., Qi R., Gao X., Zhao R. Protective roles of NRF2 signaling pathway in cobalt chloride-induced hypoxic cytotoxicity in human HaCaT keratinocytes. Toxicol. Appl. Pharmacol. 2018;355:189–197. doi: 10.1016/j.taap.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 88.Sidoryk-Wegrzynowicz M., Lee E.S., Ni M., Aschner M. Manganese-induced downregulation of astroglial glutamine transporter SNAT3 involves ubiquitin-mediated proteolytic system. Glia. 2010;58(16):1905–1912. doi: 10.1002/glia.21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang H., Jiang X., Hua Y., Li H., Zhu C., Hao X., Yi M., Li L. NEDD4L facilitates granulosa cell ferroptosis by promoting GPX4 ubiquitination and degradation. Endocrine connections. 2023;12(4) doi: 10.1530/EC-22-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zaman M.S., Barman S.K., Corley S.M., Wilkins M.R., Malladi C.S., Wu M.J. Transcriptomic insights into the zinc homeostasis of MCF-7 breast cancer cells via next-generation RNA sequencing. Metallomics : integrated biometal science. 2021;13(6) doi: 10.1093/mtomcs/mfab026. [DOI] [PubMed] [Google Scholar]
- 91.Pinsino A., Roccheri M.C., Matranga V. Manganese overload affects p38 MAPK phosphorylation and metalloproteinase activity during sea urchin embryonic development. Mar. Environ. Res. 2014;93:64–69. doi: 10.1016/j.marenvres.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 92.Timblin C.R., Janssen Y.M., Goldberg J.L., Mossman B.T. GRP78, HSP72/73, and cJun stress protein levels in lung epithelial cells exposed to asbestos, cadmium, or H2O2. Free Radic. Biol. Med. 1998;24(4):632–642. doi: 10.1016/s0891-5849(97)00325-0. [DOI] [PubMed] [Google Scholar]
- 93.Ghosh R., Gillaspie J.J., Campbell K.S., Symons J.D., Boudina S., Pattison J.S. Chaperone-mediated autophagy protects cardiomyocytes against hypoxic-cell death. Am. J. Physiol. Cell Physiol. 2022;323(5):C1555–c1575. doi: 10.1152/ajpcell.00369.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu Y., Lu X., Wu D., Cai S., Li S., Teng X. The effect of manganese-induced cytotoxicity on mRNA expressions of HSP27, HSP40, HSP60, HSP70 and HSP90 in chicken spleen lymphocytes in vitro. Biol. Trace Elem. Res. 2013;156(1–3):144–152. doi: 10.1007/s12011-013-9817-2. [DOI] [PubMed] [Google Scholar]
- 95.Sidarovich V., Adami V., Gatto P., Greco V., Tebaldi T., Tonini G.P., Quattrone A. Translational downregulation of HSP90 expression by iron chelators in neuroblastoma cells. Mol. Pharmacol. 2015;87(3):513–524. doi: 10.1124/mol.114.095729. [DOI] [PubMed] [Google Scholar]
- 96.Shinkai Y., Masuda A., Akiyama M., Xian M., Kumagai Y. Cadmium-mediated activation of the HSP90/HSF1 pathway regulated by reactive persulfides/polysulfides. Toxicol. Sci. : an official journal of the Society of Toxicology. 2017;156(2):412–421. doi: 10.1093/toxsci/kfw268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu K., Xu W., You Q., Guo R., Feng J., Zhang C., Wu W. Increased expression of heat shock protein 90 under chemical hypoxic conditions protects cardiomyocytes against injury induced by serum and glucose deprivation. Int. J. Mol. Med. 2012;30(5):1138–1144. doi: 10.3892/ijmm.2012.1099. [DOI] [PubMed] [Google Scholar]
- 98.Wei S., Li T., Xie R., Ye B., Xiang J., Liu K., Chen Z., Gao X. The role of ATF3 in ZnO nanoparticle-induced genotoxicity and cytotoxicity in bronchial epithelial cells. Int. J. Biochem. Cell Biol. 2019;113:95–102. doi: 10.1016/j.biocel.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 99.Zhang S., Chen L., Chen T., Zhang Y., Ma J., Ji H., Guo C., Xiao Z., Li J., Niu P. Transcriptome evidence reveals mitochondrial unfolded protein response participate in SH-SY5Y cells exposed to manganese. J. Integr. Neurosci. 2022;21(5):127. doi: 10.31083/j.jin2105127. [DOI] [PubMed] [Google Scholar]
- 100.He C.H., Gong P., Hu B., Stewart D., Choi M.E., Choi A.M., Alam J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J. Biol. Chem. 2001;276(24):20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 101.Fanzo J.C., Reaves S.K., Cui L., Zhu L., Wu J.Y., Wang Y.R., Lei K.Y. Zinc status affects p53, gadd 45, and c-fos expression and caspase-3 activity in human bronchial epithelial cells, American journal of physiology. Cell physiology. 2001;281(3):C751–C757. doi: 10.1152/ajpcell.2001.281.3.C751. [DOI] [PubMed] [Google Scholar]
- 102.Zhou Y., Que K.T., Zhang Z., Yi Z.J., Zhao P.X., You Y., Gong J.P., Liu Z.J. Iron overloaded polarizes macrophage to proinflammation phenotype through ROS/acetyl-p53 pathway. Cancer Med. 2018;7(8):4012–4022. doi: 10.1002/cam4.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luo H., Gu R., Ouyang H., Wang L., Shi S., Ji Y., Bao B., Liao G., Xu B. Cadmium exposure induces osteoporosis through cellular senescence, associated with activation of NF-κB pathway and mitochondrial dysfunction. Environ. Pollut. 2021;290 doi: 10.1016/j.envpol.2021.118043. (Barking, Essex : 1987) [DOI] [PubMed] [Google Scholar]
- 104.Paladini F., Cocco E., Potolicchio I., Fazekasova H., Lombardi G., Fiorillo M.T., Sorrentino R. Divergent effect of cobalt and beryllium salts on the fate of peripheral blood monocytes and T lymphocytes. Toxicol. Sci. : an official journal of the Society of Toxicology. 2011;119(2):257–269. doi: 10.1093/toxsci/kfq328. [DOI] [PubMed] [Google Scholar]
- 105.Chen C., Xie B., Li Z., Chen L., Chen Y., Zhou J., Ju S., Zhou Y., Zhang X., Zhuo W., Yang J., Mao M., Xu L., Wang L. Fascin enhances the vulnerability of breast cancer to erastin-induced ferroptosis. Cell Death Dis. 2022;13(2):150. doi: 10.1038/s41419-022-04579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu T., Jiang L., Tavana O., Gu W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 2019;79(8):1913–1924. doi: 10.1158/0008-5472.CAN-18-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bu M., Liu X., Liu X., Xu W. Upregulation of fascin-1 is involved in HIF-1α-dependent invasion and migration of hypopharyngeal squamous cell carcinoma. Int. J. Oncol. 2019;55(2):488–498. doi: 10.3892/ijo.2019.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qiao H., Mei J., Yuan K., Zhang K., Zhou F., Tang T., Zhao J. Immune-regulating strategy against rheumatoid arthritis by inducing tolerogenic dendritic cells with modified zinc peroxide nanoparticles. J. Nanobiotechnol. 2022;20(1):323. doi: 10.1186/s12951-022-01536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He Z., Shen P., Feng L., Hao H., He Y., Fan G., Liu Z., Zhu K., Wang Y., Zhang N., Hu X., Fu Y., Wu J. Cadmium induces liver dysfunction and ferroptosis through the endoplasmic stress-ferritinophagy axis. Ecotoxicol. Environ. Saf. 2022;245 doi: 10.1016/j.ecoenv.2022.114123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.