Abstract
Nucleotide excision repair, a general repair mechanism for removing DNA damage, is initiated by dual incisions bracketing the lesion. In procaryotes, the dual incisions result in excision of the damage in 12- to 13-nucleotide-long oligomers, and in eucaryotes they result in excision of the damage in the form of 24- to 32-nucleotide-long oligomers. We wished to find out if Archaea perform excision repair. Using cell extracts from Methanobacterium thermoautotrophicum, we found that this organism removes UV-induced (6-4) photoproducts in the form of 10- to 11-mers by incising the sixth to seventh phosphodiester bond 5′ to the damage and the fourth phosphodiester bond 3′ to the damage.
DNA repair plays an important role in the survival of organisms and the maintenance of species (5). Several DNA repair pathways work in a complementary manner to eliminate lesions from DNA. Of these repair systems, nucleotide excision repair (referred to herein as excision repair) has a unique place in cellular defense because it has a wide substrate range and the capability of removing virtually all base lesions from the genome (14–16, 22). Indeed, the presence of this rather complex enzymatic system in Mycoplasma genitalium, which is considered the minimal life form (4), has been taken as evidence of the indispensability of excision repair in the maintenance of species (14).In recent years, excision repair has been characterized in considerable detail for both procaryotes and eucaryotes. In both Bacteria and Eucarya, damage is excised by the joint actions of several proteins in an ATP-dependent reaction. The multisubunit system which makes the dual incisions is referred to as excision nuclease (excinuclease). The Escherichia coli excinuclease, comprising the UvrA, UvrB, and UvrC proteins, hydrolyzes the eighth phosphodiester bond 5′ to the damaged base(s) and the fourth to fifth phosphodiester bond 3′ to the damaged base(s) and excises the damage in the form of a 12- to 13-mer (17). Partial or complete genome sequences of about a dozen bacteria have revealed that all bacterial species analyzed so far have excinucleases similar in function to the E. coli enzyme (3). For some organisms, such as Salmonella typhimurium and Bacillus subtilis, functional assays support genomic data showing that both gram-negative and gram-positive procaryotes possess excinucleases that are highly similar in structure and function (16).
Eucaryotic excinucleases in mammalian cells and in Saccharomyces cerevisiae have been studied extensively. Those studies revealed a high degree of structural and functional similarities between the excision repair system of these two eucaryotes (5, 13, 16, 22). Furthermore, the limited data for other eucaryotes suggest a universal excision repair system in eucaryotes (15, 20). The basic mechanism of eucaryotic excinuclease, based on results for humans and yeast, is quite similar to the procaryotic prototype: a multisubunit enzyme system removes damage from DNA in an ATP-dependent reaction and by dual incisions bracketing the lesion (9, 20). However, the eucaryotic excinuclease differs from its procaryotic counterpart in two important aspects. First, the eucaryotic excinuclease removes the damage in 24- to 32-mers by hydrolyzing the 20th ± 5th phosphodiester bond 5′ to the lesion and the 6th ± 3rd phosphodiester bond 3′ to the lesion (8, 20). More significantly, none of the subunits of the eucaryotic excinuclease exhibits any significant homology to the procaryotic enzyme (5, 13), indicating that the appearance of the dual incision mechanisms in these two kingdoms can be described by the convergent-evolution model. This contrasts with all other repair systems, in which there are considerable homologies between the procaryotic and eucaryotic enzymes (5, 17).
Archaea, sometimes referred to as the third biological kingdom (21), have recently attracted considerable attention both from an evolutionary perspective and because they have novel biochemical pathways with no counterparts in the other kingdoms (12). For these reasons, we wished to investigate the mechanism of excision repair in Archaea. We chose to conduct our studies with Methanobacterium thermoautotrophicum because of the availability of cultures of this organism in the quantities required for biochemical studies.
M. thermoautotrophicum Marburg was obtained from the Oregon Collection of Methanogens (catalogue no. OCM82) and cultured on H2-CO2-H2S (80%/20%/0.1%) at 65°C in a 14-liter New Brunswick fermentor (7, 18). Media were prepared as previously described (18). M. thermoautotrophicum Marburg was harvested anaerobically during log phase (optical density of ∼3.0 at 578 nm). A cell extract was prepared in an N2-H2 (95%/5%) atmosphere in an anaerobic chamber (Coy Instruments) according to a previously described procedure (7). After removal of Ti(III) citrate and methyl viologen from the cell extract by gel filtration (Bio-Gel P-6), the cell extract was stored at −80°C until use. The substrate was a 136-bp duplex with a T-T (6-4) photoproduct (T[6-4]T) in the center of one strand and a 32P label at the fifth phosphodiester bond 5′ to the photoproduct (11). The excision assay measures the release of a radiolabeled oligomer containing the lesion from this duplex (9).
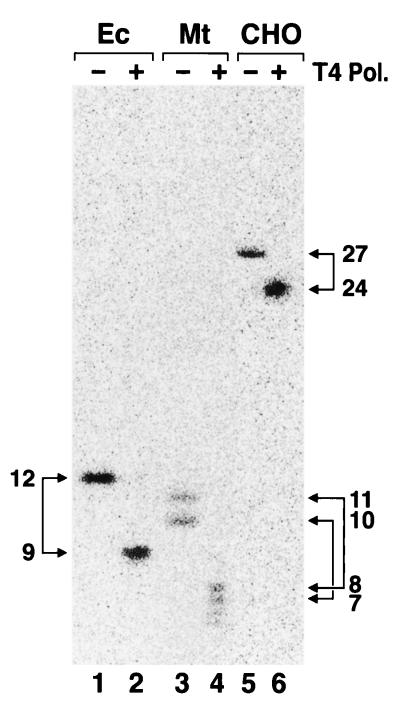
Figure 1 shows the results of excision assays conducted with cell extracts of members of the three kingdoms. E. coli excinuclease reconstituted from purified subunits (lane 1) and mammalian excinuclease in cell extracts of Chinese hamster ovary (CHO) AA8 cells (lane 3) excised the (6-4) photoproduct primarily in the form of 12- and 27-mers, respectively, in agreement with earlier results (11, 16). The cell extract of the methanogen released two oligomers 10 to 11 nucleotides (nt) in length (lane 2). Even though the efficiency of excision by the methanogen extract was rather low compared to those of reconstituted E. coli excinuclease and the CHO cell extracts, the 10- to 11-nt-long oligomer was consistently observed in reactions with the methanogen extract and hence was considered to be a bona fide repair reaction product. The low efficiency of excision is most likely due to suboptimal reaction conditions, since no systematic search to maximize excision by the methanogen extract was made. In addition to the 10- to 11-mers considered to be primary excision products, fragments of other sizes generated by nonspecific degradation of the substrate by the cell extract are seen at the background level in lane 2. In this experiment, a 17-nt oligomer was also observed in substrate treated with the methanogen extract. However, because this oligomer was not observed consistently and it contained no damage (data not shown; see below), we consider it a product of nonspecific degradation of the substrate. Thus, we conclude that the methanogen excises DNA damage by a mechanism similar to that involving the procaryotic excinuclease. Both the procaryotic and eucaryotic excinuclease systems have absolute dependence on ATP. Hence, to confirm that the 10- to 11-mers produced by the methanogen cell extract are excinuclease products, we performed the excision reaction with and without ATP. Figure 2 shows that ATP is required for excision, lending further support to the conclusion that the 10- to 11-mers are excinuclease products.
FIG. 1.
Excision assay with members of the three kingdoms. The reaction mixture (25 μl) contained 25 mM HEPES-KOH (pH 7.9), 45 mM KCl, 4.4 mM MgCl2, 2 mM ATP, a 20 μM concentration of each deoxynucleoside triphosphate, 0.5 mM dithiothreitol, 0.16 mM EDTA, and 200 μg of bovine serum albumin per ml, plus 10 nM substrate and repair protein as follows: E. coli (Ec), 180 nM (each) UvrA, UvrB, and UvrC; M. thermoautotrophicum (Mt) cell extract, 63 μg; and CHO AA8 cell extract, 60 μg. The reaction mixtures were incubated for 1 h at 37°C with the E. coli excinuclease and the methanogen extract and at 30°C with the CHO cell extracts. The products were separated on an 8% denaturing polyacrylamide gel and visualized by phosphorimaging. Oligomers in the 3- to 20-nt range with a photoproduct migrate one space slower than corresponding oligomers free of photoproduct (2). Lane 4 contains molecular size markers (M); sizes are indicated on the right, in nucleotides. The arrows on the left indicate the positions of the major excision products of the three species. The proportions of substrate excised were 12, 0.15, and 1.6% for E. coli, M. thermoautotrophicum, and CHO cells, respectively.
FIG. 2.
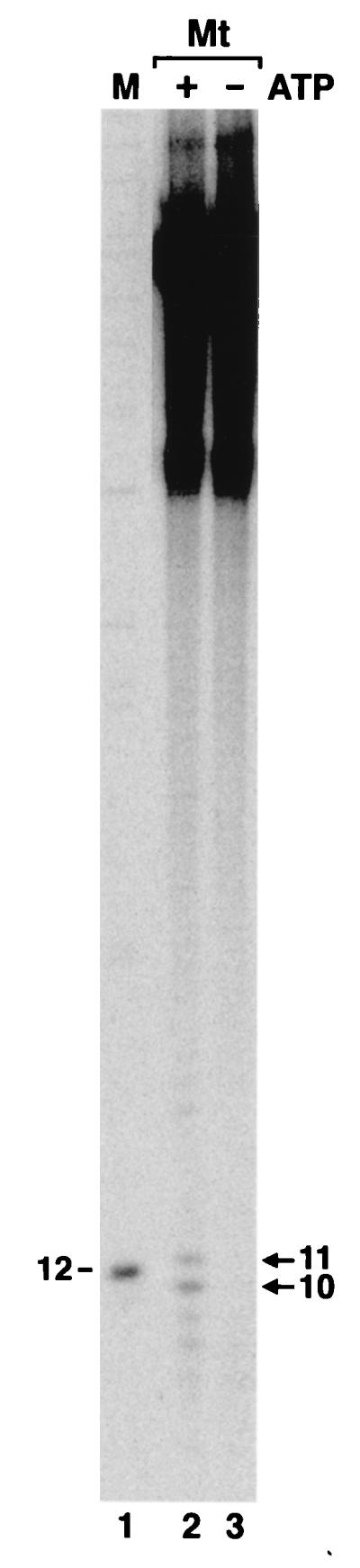
M. thermoautotrophicum (Mt) excinuclease is ATP dependent. Reactions were carried out with cell extract as for Fig. 1 in the presence and absence of ATP as indicated. The arrows indicate the two major excision products. Lane 1 contains a molecular size marker (M) in nucleotides.
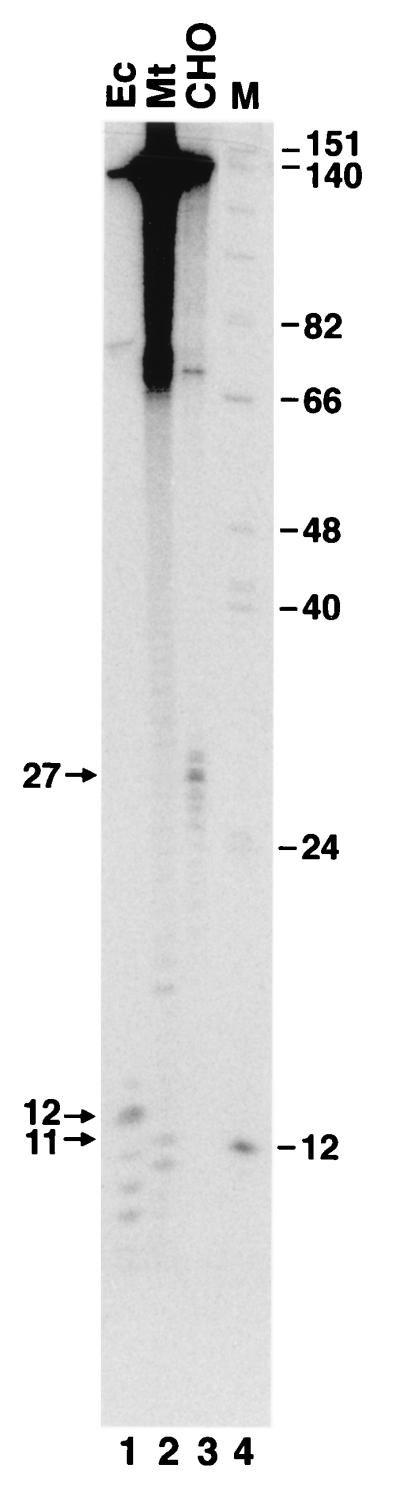
The dual incisions made by procaryotic and eucaryotic excinucleases occur at relatively precise distances from the lesion (16). To determine the incision sites of the methanogen excinuclease, the excision products generated by the cell extract were digested with T4 DNA polymerase, which, in the absence of deoxynucleoside triphosphates, acts as a potent 3′-to-5′ exonuclease, and its progress is blocked by DNA lesions (2, 6). Thus, it is possible to determine whether a DNA fragment contains a DNA lesion and the position of this lesion by digestion with T4 DNA polymerase (9, 20). Figure 3 shows the analyses of the excision fragments produced by the excinuclease systems of the three kingdoms. This figure illustrates that the 10- to 11-mers produced by methanogen extract are the products of specific incisions which, by definition, are generated by an excinuclease.
FIG. 3.
Location of the (6-4) photoproduct in the excision products of the three species. The major excision products seen in Fig. 1 were eluted from preparative-scale gels and digested with T4 DNA polymerase (T4 Pol.) 3′-to-5′ exonuclease as described by Svoboda et al. (20). The products were analyzed on an 8% denaturing polyacrylamide gel. The sizes, in nucleotides, of the fragments before and after exonuclease digestion are indicated. Ec, E. coli; Mt, M. thermoautotrophicum.
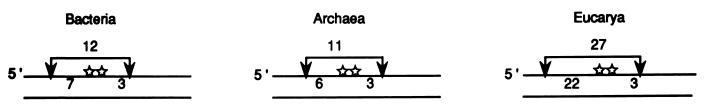
To summarize, the incision patterns on a (6-4) photoproduct substrate for the three species used in this study are as follows: for E. coli, the 8th phosphodiester bond 5′ to the lesion and the 4th phosphodiester bond 3′ to the lesion; for M. thermoautotrophicum, the 6th to 7th phosphodiester bond 5′ to the photoproduct and the 4th phosphodiester bond 3′ to the photoproduct; and for CHO cells, the 23rd phosphodiester bond 5′ to the diadduct and the 4th phosphodiester bond 3′ to the diadduct. These incision patterns are schematically shown in Fig. 4. It must be noted, however, that Fig. 3 and 4 represent the major species and that in all three systems other dual incision products are observed at lower frequencies (Fig. 1 and 2).
FIG. 4.
Incision patterns for members of the three kingdoms. The arrows indicate the incision sites, the two stars mark the position of the (6-4) photoproduct, and the numbers below the brackets represent the numbers of nucleotides from the site of incision to the first nucleotide of the diadduct. Only the major incision sites are shown for simplification.
In summary, our results show that at least one member of the third kingdom has a procaryotic-type excision pattern. This is not surprising, since the sequence of the M. thermoautotrophicum genome shows that this organism has uvrA, uvrB, and uvrC homologs (19). Considering the high degree of sequence identity of the methanogen genes to those of E. coli, it is possible that the primary excision products of the methanogen are identical to those of E. coli and that the smaller species are generated by postexcision degradation by nonspecific nucleases (20). More importantly, in contrast to members of the other two kingdoms, members of Archaea show greater variability in their gene and enzyme systems, and it is quite likely that the dual incision pattern we found in M. thermoautotrophicum is not universal to all members of Archaea. Indeed, the genome sequences of Methanococcus jannaschii (1) and Archaeoglobus fulgidus (10) do not reveal a complete set of excinuclease genes homologous to those of either Bacteria or Eucarya. Hence, whether these species possess an excinuclease in the conventional sense remains to be determined biochemically. When sufficient quantities of cultures of these organisms become available, it is possible that Archaea may yet be revealed to have a third type of excinuclease.
Acknowledgments
This work was supported by NIH grant GM32833.
We thank Joyce T. Reardon for her guidance and for comments on the manuscript, Tadayoshi Bessho for advice, and Mitsuo Wakasugi for providing some of the substrate used in this study.
REFERENCES
- 1.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 2.Doetsch P W, Chan G L, Haseltine W A. T4 DNA polymerase (3′-5′) exonuclease, an enzyme for the detection and quantitation of stable DNA lesions: the ultraviolet light example. Nucleic Acids Res. 1985;13:3285–3304. doi: 10.1093/nar/13.9.3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doolittle R F. Microbial genomes opened up. Nature. 1998;392:339–342. doi: 10.1038/32789. [DOI] [PubMed] [Google Scholar]
- 4.Fraser C M, Gocayne J D, White D, Adams M D, Clayton R A, Fleishmann R D, Bult C J, Kerlavage A R, Sutton G, Kelly J M, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: ASM Press; 1995. pp. 191–365. [Google Scholar]
- 6.Fuchs R P P. DNA binding spectrum of the carcinogen N-acetoxy-N-2-acetylaminofluorene significantly differs from the mutation spectrum. J Mol Biol. 1984;177:173–180. doi: 10.1016/0022-2836(84)90063-9. [DOI] [PubMed] [Google Scholar]
- 7.Goubeaud M, Schreiner G, Thauer R K. Purified methyl-coenzyme-M reductase is activated when the enzyme-bound coenzyme F4320 is reduced to the nickel (I) oxidation state by titanium (III) citrate. Eur J Biochem. 1997;243:110–114. doi: 10.1111/j.1432-1033.1997.00110.x. [DOI] [PubMed] [Google Scholar]
- 8.Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A and transcription factor TFIIH. J Biol Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 9.Huang J C, Svoboda D L, Reardon J T, Sancar A. Human nucleotide excision nuclease removes thymine dimers by hydrolyzing the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. The complete genome sequence of the hyperthermophilic, sulfate reducing archaeon Archaeoglobus fulgidus. Nature. 1997;310:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 11.Mu D, Tursun M, Duckett D R, Drummond J T, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol Cell Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen G J, Woese C R. Archaeal genomics: an overview. Cell. 1997;89:991–994. doi: 10.1016/s0092-8674(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 13.Prakash S, Sung P, Prakash L. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu Rev Genet. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- 14.Sancar A. Mechanisms of excision repair. Science. 1994;266:1954–1956. doi: 10.1126/science.7801120. [DOI] [PubMed] [Google Scholar]
- 15.Sancar A. Excision repair in mammalian cells. J Biol Chem. 1995;270:22008–22016. doi: 10.1074/jbc.270.37.22008. [DOI] [PubMed] [Google Scholar]
- 16.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 17.Sancar A, Rupp W D. A novel repair enzyme: UvrABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 18.Schonheit P, Moll J, Thauer R K. Growth parameters (D, μmax, Ys) of Methanobacterium thermoautotrophicum. Arch Microbiol. 1980;127:59–65. doi: 10.1007/BF00403508. [DOI] [PubMed] [Google Scholar]
- 19.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svoboda D L, Taylor J S, Hearst J E, Sancar A. DNA repair by eukaryotic nucleotide excision nuclease: removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by Xenopus laevis oocytes. J Biol Chem. 1993;268:1931–1936. [PubMed] [Google Scholar]
- 21.Woese C R, Fox G E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood R D. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]