Abstract
We describe a small family of proteins, CHR, which contains members that function in chromate and/or sulfate transport. CHR proteins occur in bacteria and archaea. They consist of about 400 amino acyl residues, appear to have 10 transmembrane α-helical segments in an unusual 4+6 arrangement, and arose by an intragenic duplication event.
Several families of bacterial transport proteins are involved in solute translocation as secondary carriers (e.g., transporters of the major facilitator superfamily [MFS] [6, 10, 19], the cation diffusion facilitator [CDF] family [15, 21], and the resistance-Nodulation-cell division [RND] family [13, 25, 27]). Such active transporters are generally believed to form a transmembrane pathway that allows the substrate to cross the cytoplasmic membrane (18). This channel is usually composed of amphipathic α-helices (18). Carrier and channel forms of some permeases seem to be interconvertible (3, 4, 28, 29), at least in some cases. The number of transmembrane α-helical segments (TMSs) may vary from 2 to 14. Twelve or 14 TMSs are observed exclusively for secondary carriers of the MFS (20), and many transport proteins seem to consist of two units composed of five to seven transmembrane α-helices. Transporter families with six transmembrane spanners have also been described (e.g., the cation diffusion facilitators [CDFs] [15, 20]).
The MFS carriers (26) and RND family permeases (27), each with 12 to 14 TMSs, appear to have originated by gene duplication events, and the term “6+6 spanner transporter” has been proposed for these proteins (18). In this communication, we describe a small family of transport proteins, most members of which apparently consist of two structurally similar repeat units. The N-terminal repeat unit appears to possess four TMSs; the C-terminal repeat unit, however, appears to possess six TMSs. We show that these two topologically dissimilar repeat units probably arose from a single primordial polypeptide unit by an internal gene duplication event.
The CHR protein family.
Table 1 presents the fully sequenced proteins of the CHR family currently available in the databases. While all of these sequences are clearly homologous, with comparison scores well in excess of 10 standard deviations (RDF2 program with 500 random shuffles when the entireties of the protein sequences were compared [22]), they show no significant similarity to other proteins in the databases. The two functionally characterized proteins are the plasmid-encoded ChrA proteins from Alcaligenes eutrophus [ChrA(Aeu)] (12) and Pseudomonas aeruginosa [ChrA(Pae)] (2). Both proteins seem to catalyze the energy-dependent extrusion of chromate. One other protein, SrpC, from the cyanobacterium Synechococcus, is sulfur regulated and may function in sulfate uptake (11). The other CHR family homologs have not been functionally characterized. Since one of these homologs is from the archaeon Methanococcus jannaschii (1), the family is not restricted to the Bacteria domain. Both this protein and the one found in Synechocystis are chromosomally encoded (7, 11).
TABLE 1.
The chromate resistance (CHR) protein family
| Abbreviation | Description | Source | Length (amino acids) | Accession no. |
|---|---|---|---|---|
| ChrA(Pae) | Chromate resistance protein A | Pseudomonas aeruginosa | 416 | spP14285 |
| Orf1(Ssp) | ChrA homolog | Synechocystis sp. | 399 | gbD90916 |
| SrpC(Ssp) | ChrA homolog, sulfur regulated | Synechococcus sp. | 393 | pirC56274 |
| ChrA(Aeu) | Chromate resistance protein A | Alcaligenes eutrophus | 401 | gbP17551 |
| Orf1(Mja) | ChrA homolog | Methanococcus jannaschii | 402 | gbU67518 |
| Orf1(Bsu) | ChrA homolog (YwrA) | Bacillus subtilis | 197 | gbZ93767 |
| Orf2(Bsu) | ChrA homolog (YwrB) | Bacillus subtilis | 178 | gbZ93767 |
All of these proteins are about the same size (393 to 416 amino acyl residues). In contrast, two homologs of Bacillus subtilis, Orf1 (YwrA) and Orf2 (YwrB), are about half this size (197 and 178 amino acyl residues, respectively) (24). These two putative Bacillus proteins are encoded by two adjacent overlapping genes that may be translationally coupled. Evidence suggesting that a sequencing error is responsible for an apparent splice site within a single gene is lacking. The genomes of Mycoplasma genitalium, Mycoplasma pneumoniae, Helicobacter pylori, Escherichia coli, Haemophilus influenzae, Methanobacterium thermautotrophicum, Archaeoglobus fulgidus, and Saccharomyces cerevisiae do not encode ChrA homologs.
Operons encoding CHR family proteins and their possible physiological function.
Among genes surrounding those encoding CHR family proteins, there appear to be some that are involved in resistance to oxidative stress, such as chrC in A. eutrophus, a chrC-homologous open reading frame (ORF) following chrA in P. aeruginosa, and the srpA putative periplasmic catalase from Synechococcus sp. (2, 11, 23). When chromate is reduced, toxic oxygen species are produced. The presence of the periplasmic SrpA catalase suggests that chromate reduction may occur in the periplasm, most likely catalyzed by a membrane-bound redox enzyme system.
Insertions into the ChrA-like srpC gene of Synechococcus lead to an increase, rather than a decrease, in chromate resistance (11). In P. aeruginosa and A. eutrophus, chromate is accumulated by sulfate uptake systems, and expression of ChrA leads to reduced accumulation of chromium (2, 12, 16, 17). In Alcaligenes, the chrA gene is preceded by chrB. Expression of Alcaligenes chrA without chrB leads to hyperaccumulation of chromium and an increase in sensitivity (12), as in Synechococcus. Thus, the ChrA protein of A. eutrophus may be a chromate uptake system when expressed alone.
These contradictory observations can be explained if CHR family proteins catalyze chromate/sulfate antiport, thereby correcting the deficiency of the nonspecific sulfate uptake system by exporting the erroneously accumulated chromate, which would be exchanged for sulfate. Since the sulfate concentration is usually high compared to the chromate concentration, the former might be sufficient to drive chromate/sulfate antiport. In Synechococcus, the effect of the srpC mutation was tested at low sulfate concentrations (11), when the antiporter should work as a chromate uptake system.
Multiple alignment of the CHR family proteins.
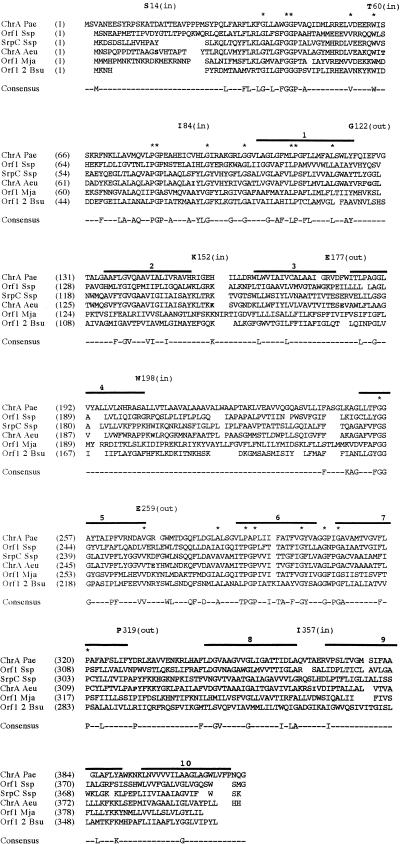
Figure 1 presents a multiple alignment of the CHR family proteins. In the top line, two adjacent G’s are fully conserved, and these are followed by several additional fully conserved residues, producing the following motif: GGX12VX4WX16PGPX10GX7G (X = any residue). Starting at the end of line 4, another conserved motif was found: GGX12VX4WX16PGPX8GX7G. A comparison of the two halves of the CHR family proteins by using the RDF2 program (22) revealed that the two halves are homologous, suggesting that they (as well as the two ORFs from B. subtilis) may have been derived from a common ancestral sequence. Based on the multiple alignment shown in Fig. 1, average similarity and hydropathy plots were derived (data not shown). Following a short, poorly conserved region containing the first 30 positions, the next 120 residues of both halves are better conserved. The hydropathy plot of all proteins (shown for ChrA(Aeu) in Fig. 2A [all other proteins not shown]) revealed two N-terminal peaks of moderate hydrophobicity, followed by 10 peaks of a significantly hydrophobic character.
FIG. 1.
Multiple alignment of six full-length members of the CHR family. The two Bacillus ORFs (Orf1 and Orf2) were artificially fused to generate a full-length protein. The multiple alignment was generated with the TREE program (5). Protein abbreviations are as indicated in Table 1. Asterisks above the alignment indicate fully conserved residues. A residue appears in the consensus sequence if it occurs in a majority (four of six) of the aligned sequences at any one position. Solid bars above the alignment indicate the putative TMSs. The numbers above these lines indicate the number of the TMS in the proposed topological model (Fig. 2). The residue number of the first residue in each line is presented in parentheses following the abbreviation of that protein. Residues to which LacZ and PhoA fusion domains were attached in the topological analyses reported in this paper are underlined and indicated above the alignment with the demonstrated orientation (in or out) indicated in parentheses.
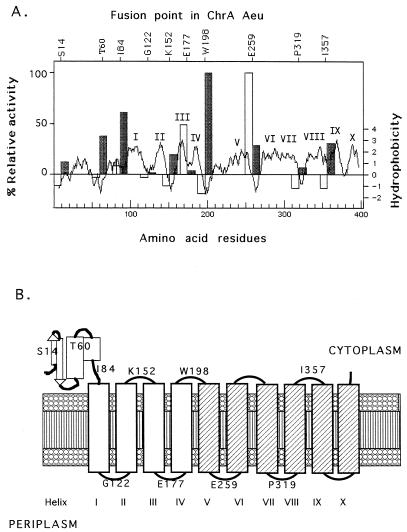
FIG. 2.
Localization score, hydrophobicity, and model of ChrA(Aeu). (A) Parts of the chrA gene of A. eutrophus were fused with the phoA (light grey bars) or lacZ (dark grey bars) topological reporter gene. The chrA parts were from the 5′ end of the gene up to the point indicated by the amino acid residue written above the x axis. After expression in E. coli CC118 (9), the specific activities were determined in triplicate, and mean values were calculated. The results for the respective negative controls (vector plasmids with no insert) were subtracted from the specific activities, and the resulting corrected values from both assays were divided by the highest specific activity obtained, which was 0.55 U/mg of protein (ChrA-E259) for phoA fusions or 10.7 U/mg of protein (ChrA-W198), respectively. The light grey PhoA bars and the dark grey LacZ bars were drawn in the hydrophobicity plot of ChrA(Aeu) with the roman numerals indicating the predicted transmembrane spans. (B) Proposed topological model for the ChrA(Aeu). The four assumed membrane-spanning α-helices in the first half of the protein are shown as open boxes; the six membrane-spanning helices in the second half are presented as shaded boxes. S14, T60, I84, G122, K152, E177, W198, E259, P319, and I357 are the amino acid residues that were used as the sites of construction of the LacZ- and PhoA-reporter fusions. The model is based on the assumption that ChrA functions alone and as a monomer.
Phylogenetic tree for the CHR family proteins and their halves.
ChrA of A. eutrophus and SrpC of Synechococcus are closely related, but all other homologs are about equally distant from each other and from these two proteins (data not shown). This observation is surprising, because Methanococcus, being an archaeon, is not closely related to the other organisms. These facts suggest either the appearance of paralogous isoforms of the CHR proteins early during evolution or horizontal transmission of genetic material.
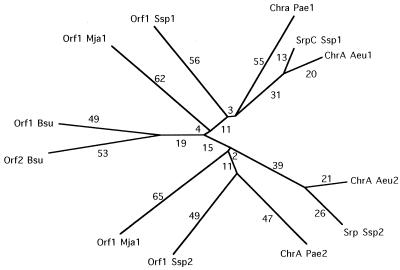
In the multiple alignment of the 12 homologous 200-residue protein halves of the CHR family members, 6 residues are fully conserved: 3 G’s, 2 P’s, and a V (data not shown). These and other residues that appear in the consensus sequence are likely to be of structural significance. The phylogenetic tree for all of the halves of the CHR family proteins is shown in Fig. 3. Branch lengths connecting halves 1 are approximately the same as those for halves 2, suggesting that the two halves of these proteins have undergone sequence divergence at about the same rate. Most surprising is the fact that the two Bacillus ORFs are closer to each other than they are to any other segment analyzed. This fact strongly suggests that they arose by an extragenic duplication event that occurred later than the intragenic duplication event that gave rise to the larger members of the CHR family. These Bacillus ORFs represent the structural equivalent of the primordial protein that presumably gave rise to the members of the CHR family. Although we have analyzed nearly 150 families of transport proteins (8), several of which arose by intragenic duplication events, this is the first instance in which both extant primordial polypeptide unit-equivalent and full-length duplicated proteins have been identified.
FIG. 3.
Phylogenetic tree of the halves of the CHR family proteins, based on the TREE program (5) and the multiple alignment of the CHR protein halves. Protein abbreviations are as indicated in Table 1. Branch lengths are assumed to be approximately proportional to phylogenetic distance. Numbers 1 and 2 at the ends of abbreviations represent first and second halves, respectively.
Topological analyses of the CHR family proteins.
The structure of the ChrA protein from A. eutrophus was investigated by using translational lacZ and phoA fusions. Various fragments of the chrA gene were cloned into the fusion vectors pECD499 and pECD500 (25). The activities of alkaline phosphatase (9) and β-galactosidase (14) were determined with E. coli CC118 (9) as published previously (25). In fusions at positions S14, T60, and I84 (Fig. 1 and 2A), the PhoA activities were negligible, but LacZ activities were significant (Fig. 2A). Thus, the amino-terminal 84 residues of A. eutrophus ChrA are probably located in the cytoplasm.
Following the first 90 residues are four peaks of hydrophobicity, and four fusions were constructed in this region at residues G122, K152, E177, and W198 (Fig. 1). The high PhoA and low LacZ activities of the fusions at E177 indicated a periplasmic location (Fig. 2A). Two other fusions, in contrast, indicated a cytoplasmic location of residue W198 and residue K152. Based on the very low LacZ activity, residue G122, between peaks I and II, is probably localized to the periplasmic side of the membrane. In agreement with these data, there is a cluster of positively charged amino acids following peak II in ChrA(Aeu), as well as in all of the ChrA homologs (Fig. 1).
As noted above, the first halves of the ChrA homologs are homologous to the second halves. However, the ChrA(Aeu)-PhoA fusion at residue E259 displayed an extraordinarily high PhoA activity (Fig. 2A), and E259 must be located on the periplasmic side. Thus, between W198 (on the cytoplasmic side) and E259, an odd number of transmembrane spans must exist. We suggest that a single transmembrane spanner is present.
A third hydrophobic region is localized to the carboxy terminus of the large CHR family proteins, starting at alignment position 290 (Fig. 2). There are four or five hydrophobic peaks C terminal to alignment position 290. Only two fusion points could be obtained in this region, at residues P319 and I357 (Fig. 1). The high LacZ activity of fusion I357 (Fig. 2A) indicates a cytoplasmic location, similar to the fusion K152 in the first half of this protein. The result for P319 is not so clear, but the low LacZ activity and the similarity to the fusion point G122 should mean a periplasmic location for P319.
Summing up the predicted structure of A. entrophus ChrA(Aeu) and the average hydrophobicity of all CHR family proteins, there appear to be four transmembrane helices in the first halves of these proteins, with the amino termini up to alignment position 90 localized to the cytoplasmic side of the membrane. The C-terminal halves of the proteins start with two spans which are at the same position as the nonspanning, relatively hydrophilic regions of the amino-terminal halves. Finally, the proteins exhibit four C-terminal transmembrane helices which are homologous to the four spans in the hydrophobic region of the N-terminal halves of these proteins. The total number of transmembrane spans in CHR proteins is therefore probably 10 (Fig. 2B).
Since all of the large CHR family proteins may be composed of two homologous halves, the CHR family ancestor would probably have been a six-span transporter, probably involved in oxyanion transport. The primordial fusion protein may well have had 12 spans. Presumably, the first two evolved an increased degree of hydrophilicity and became localized to the cytoplasmic side of the membrane.
Acknowledgments
We are grateful to Milda Simonaitis for her assistance with the preparation of the manuscript and to an unknown reviewer for boiling down the manuscript into a note.
Work in the laboratory of M.H.S. was supported by USPHS grants 5RO1 AI21702 from the National Institutes of Allergy and Infectious Diseases and 9RO1 GM55434 from the National Institute of General Medical Sciences, as well as by the M. H. Saier, Sr., Memorial Research Fund. Work in the laboratory of D.H.N. was supported by Forschungsmittel des Landes Sachsen-Anhalt and by Fonds der Chemischen Industrie.
REFERENCES
- 1.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 2.Cervantes C, Ohtake H, Chu L, Misra T K, Silver S. Cloning, nucleotide sequence, and expression of the chromate resistance determinant of Pseudomonas aeruginosa plasmid pUM505. J Bacteriol. 1990;172:287–291. doi: 10.1128/jb.172.1.287-291.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dierks T, Salentin A, Krämer R. Pore-like and carrier-like properties of the mitochondrial aspartate/glutamate carrier after modification by SH-reagents: evidence for a preformed channel as a structural requirement of carrier-mediated transport. Biochim Biophys Acta. 1990;1028:281–288. doi: 10.1016/0005-2736(90)90177-p. [DOI] [PubMed] [Google Scholar]
- 4.Dierks T, Saletin A, Heberger C, Krämer R. The mitochondrial aspartate/glutamate and ADP/ATP carrier switch from obligate counterexchange to unidirectional transport after modification by SH-reagents. Biochim Biophys Acta. 1990;1028:268–280. doi: 10.1016/0005-2736(90)90176-o. [DOI] [PubMed] [Google Scholar]
- 5.Feng D-F, Doolittle R F. Progressive alignment and phylogenetic tree construction of protein sequences. Methods Enzymol. 1990;183:375–387. doi: 10.1016/0076-6879(90)83025-5. [DOI] [PubMed] [Google Scholar]
- 6.Griffith J K, Baker M E, Rouch D A, Page M G P, Skurray G A, Paulsen I T, Chater K F, Baldwin S A, Henderson P J F. Membrane transport proteins: implications of sequence comparisons. Curr Opin Cell Biol. 1992;4:684–695. doi: 10.1016/0955-0674(92)90090-y. [DOI] [PubMed] [Google Scholar]
- 7.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 8.Le, T., T.-T. Tseng, and M. H. Saier, Jr. Flexible programs for the estimation of average amphipathicity of multiply aligned homologous proteins: application to integral membrane transport proteins. Submitted for publication. [DOI] [PubMed]
- 9.Manoil C. Analysis of protein localization by use of gene fusions with complementary properties. J Bacteriol. 1990;172:1035–1042. doi: 10.1128/jb.172.2.1035-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marger M D, Saier M H., Jr A major superfamily of transmembrane facilitators catalyzing uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 11.Nicholson M L, Laudenbach D E. Genes encoded on a cyanobacterial plasmid are transcriptionally regulated by sulfur availability and CysR. J Bacteriol. 1995;177:2143–2150. doi: 10.1128/jb.177.8.2143-2150.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nies A, Nies D H, Silver S. Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus. J Biol Chem. 1990;265:5648–5653. [PubMed] [Google Scholar]
- 13.Nies D H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J Bacteriol. 1995;177:2707–2712. doi: 10.1128/jb.177.10.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nies D H. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J Bacteriol. 1992;174:8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nies D H, Silver S. Ion efflux systems involved in bacterial metal resistances. J Ind Microbiol. 1995;14:186–199. doi: 10.1007/BF01569902. [DOI] [PubMed] [Google Scholar]
- 16.Nies D H, Silver S. Metal ion uptake by a plasmid-free metal-sensitive Alcaligenes eutrophus strain. J Bacteriol. 1989;171:4073–4075. doi: 10.1128/jb.171.7.4073-4075.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nies D H, Silver S. Plasmid-determined inducible efflux is responsible for resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989;171:896–900. doi: 10.1128/jb.171.2.896-900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikaido H, Saier M H., Jr Transport proteins in bacteria: common themes in their design. Science. 1992;258:936–942. doi: 10.1126/science.1279804. [DOI] [PubMed] [Google Scholar]
- 19.Pao S S, Paulsen I T, Saier M H., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulsen I T, Saier M H., Jr A novel family of ubiquitous heavy metal ion transport proteins. J Membr Biol. 1997;156:99–103. doi: 10.1007/s002329900192. [DOI] [PubMed] [Google Scholar]
- 22.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peitzsch, N., S. Juhnke, and D. H. Nies. Unpublished data.
- 24.Presecan E, Moszer I, Boursier L, Cruz Ramos H, de la Fuente V, Hullo M-F, Lelong C, Schleich S, Sekowska A, Song B H, Villani G, Kunst F, Danchin A, Glaser P. The Bacillus subtilis genome from gerBC (311°) to licR (334°) Microbiology. 1997;143:3313–3328. doi: 10.1099/00221287-143-10-3313. [DOI] [PubMed] [Google Scholar]
- 25.Rensing C, Pribyl T, Nies D H. New functions for the three subunits of the CzcCBA cation-proton-antiporter. J Bacteriol. 1997;22:6871–6879. doi: 10.1128/jb.179.22.6871-6879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saier M H., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saier M H, Jr, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11:841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 28.Schroers A, Krämer R, Wohlrab H. The reversible antiport-uniport conversion of the phosphate carrier from yeast mitochondria depends on the presence of a single cysteine. J Biol Chem. 1997;272:10558–10564. doi: 10.1074/jbc.272.16.10558. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz M, Gross A, Steinkamp T, Flügge U-I, Wagner R. Ion channel properties of the reconstituted chloroplast triose phosphate/phosphate translocator. J Biol Chem. 1994;269:29481–29489. [PubMed] [Google Scholar]