Abstract
We identified in Streptococcus mutans six new genes (rgpA through rgpF), whose disruption results in a loss of serotype-specific antigenicity, specified by the glucose side chains of rhamnose-glucose polysaccharide from the cell wall. Rhamnose and glucose content of the cell wall decreased drastically in all these disruption mutants, except that in the rgpE mutant only the glucose content decreased. RgpC and RgpD are homologous to ATP-binding cassette transporter components and may be involved in polysaccharide export, whereas RgpE may be a transferase of side chain glucose.
The rhamnose-glucose polysaccharides (RGPs) of Streptococcus mutans (serotypes c, e, and f) have a backbone structure of α1,2- and α1,3-linked rhamnosyl polymers with glucose side chains (11, 20). These polysaccharide antigens have received much attention because in vitro stimulation of human monocytes with the serotype f-specific polysaccharide was reported to induce the release of inflammatory cytokines, such as tumor necrosis factor alpha and interleukin-1β (24), and provoke nitric oxide production in the rat aorta (13).
Little is known about the biosynthesis of these polysaccharide antigens. We recently demonstrated (26, 27) that in S. mutans serotype c four genes (rmlA, rmlB, rmlC, and rmlD) are involved in the synthesis of dTDP-l-rhamnose, which serves as an immediate precursor of the backbone of the RGP (27). In addition to cloning the rml genes, we cloned a gene (gluA) encoding glucose-1-phosphate uridylyltransferase involved in side chain formation of RGP (31).
Cloning of additional genes for RGP synthesis.
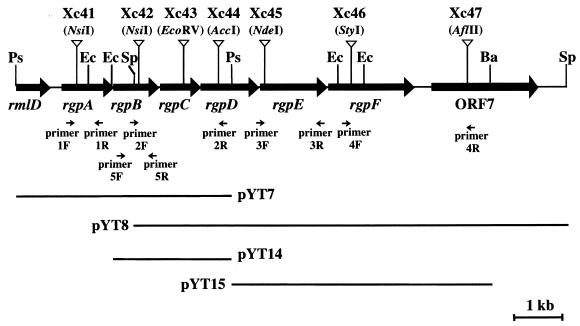
We identified additional genes involved in RGP synthesis in the region downstream from the rmlD gene (27). An approximately 10-kb fragment was cloned from the region downstream from the rmlD gene of S. mutans Xc by a marker rescue method (16). Standard DNA recombinant procedures were carried out as described previously (30). pResYT10, which is composed of P15A replicon (23) and the erythromycin resistance gene (23) with the unique PvuII site immediately downstream of this gene, was linearized with PvuII and inserted into the unique NsiI site downstream from rmlD on the insert fragment of pYT5, which was previously constructed (27). The resultant plasmid, designated pYT41, in which both the erythromycin resistance gene and the rmlD gene were oriented in the same direction (in order to avoid the polarity effects [see below]), was selected, digested with BssHII, and introduced into the chromosome of S. mutans Xc by a double crossover recombination. The correct insertion of pResYT10 into the target site on the chromosome was confirmed by PCR amplification with primers 1F and 1R (Fig. 1). One transformant was designated strain Xc41, and its chromosome was digested with PstI, self-ligated, and used to transform Escherichia coli DH5 to erythromycin resistance. The plasmid carrying the 3.1-kb downstream region of rmlD was isolated from one transformant and designated pYT7 (Fig. 1). The 2.3-kb EcoRI-PstI fragment from pYT7 was subcloned into pBluescriptII KS+, and the resultant plasmid was designated pYT14 (Fig. 1).
FIG. 1.
Restriction map of the region downstream from the rmlD gene of S. mutans Xc. The arrows indicate the locations of the eight ORFs including the carboxy-terminal half of rmlD. The pResYT10 integration sites for the insertional inactivation of these ORFs are indicated by the inverted open triangles. The locations of the insertional fragments are indicated in the lower portion of the diagram. Restriction enzyme abbreviations and primer sequences are as follows: Ba, BamHI; Ec, EcoRI; Ps, PstI; Sp, SpeI; 1F, 5′-ATGGTCACGTCCAGTGCA-3′; 1R, 5′-GTCCAATACCGTGCAGCA-3′; 2F, 5′-AGCTAAGCAGTGGAAGCAG-3′; 2R, 5′-CTGCATCCCACTAGAATAG-3′; 3F, 5′-GAAGCTGGATTGGTGAAGC-3′; 3R, 5′-TGTAGGAATGGTCCAACG-3′; 4F, 5′-AGGTGATTGACCAGTATG-3′; 4R, 5′-ATCTGTCACTAGCAGAGG-3′; 5F, 5′-TGTCCACCTACAATGGTC-3′; and 5R, 5′-GAAAGGCACGATTCTTAG-3′.
Essentially the same procedure was repeated to clone DNA further downstream, by inserting pResYT10 into pYT14 and by introducing the fragment into the chromosome, finally generating strain Xc42. The chromosome of strain Xc42 was digested with SpeI, self-ligated, and used to transform E. coli DH5. A plasmid carrying an additional 6.8-kb downstream region, which was isolated from one transformant, was designated pYT8 (Fig. 1).
Sequence analysis.
The cloned fragments were subcloned into pBluescriptII KS+ or SK+, and the nucleotide sequence was determined with a 373 STRETCH automated sequencer (Applied Biosystems, Inc., Foster City, Calif.) (30). Sequence analysis revealed the presence of seven open reading frames (ORFs) in the 10-kb region, as shown in Fig. 1. These ORFs were designated rgpA, rgpB, rgpC, rgpD, rgpE, rgpF, and ORF7. Possible Shine-Dalgarno sequences were identified just upstream of the potential initiation codons of all seven ORFs. rgpA and ORF7 were located 173 and 182 bp downstream from the immediately adjacent upstream gene, respectively. The region between rmlD and rgpA contained a stem-loop structure (positions 680 to 695) with a free energy of −15.6 kcal/mol followed by a poly(T) sequence, which may act as a transcriptional terminator for rmlD, and a consensus −10 and −35 E. coli promoter-like sequence (TCGCAAN17TATAAT; positions 705 to 733) for rgpA. Another hairpin-loop structure (positions 8110 to 8122) without the features of a typical transcription terminator and with a free energy of −14.4 kcal/mol was located between rgpF and ORF7. The putative initiation codons for rgpB, rgpC, rgpD, and rgpF overlapped the stop codons of the preceding genes, and rgpE was located 22 bp downstream from rgpD.
The nucleotide database was searched for homologous genes by using the program FASTA (12) on the DDBJ e-mail server at the National Institute of Genetics, Mishima, Japan. The amino acid sequence deduced from rgpE showed 14% identity with the amino acid sequence of the glycosyltransferase of Neisseria gonorrhoeae (5). Hydrophobic cluster analysis (HCA) with the program HCA-Plot (10) at the Web site of Systèmes Moléculaires & Biologie Structurale (http://www.lmcp.jussieu.fr/∼mornon/) revealed a conserved structural feature, domain A, in the N-terminal portion of the amino acid sequence of RgpE as well as in those of other bacterial glycosyltransferases. Domain A consists of four β-sheets and three α-helices, and there are at least one Asp residue in the loop at the C-terminal end of the β2-region and DXDD at the C-terminal end of the β4-region (7, 22).
The amino acid sequences deduced from rgpC and rgpD showed significant homology with those of components of the ATP-binding cassette (ABC) transport system of several bacterial polysaccharides (3, 9, 18). rgpC encoded a polypeptide with a predicted size of 30.7 kDa, and its hydrophobic profile is similar to the profiles of the KpsM (18), BexB (9), and CtrC (3) proteins, which are integral membrane components (2). Each member of this family of proteins has a molecular mass of ∼30 kDa, similar to that of the RgpC protein. The RgpD protein was relatively hydrophilic compared to the RgpC protein and showed 27.8 to 29.5% identity with proteins such as BexA, KpsT, and CtrD, which are peripheral inner membrane proteins that contain the ATP-binding domain (3, 9, 18, 19). Although the molecular mass of the RgpD protein was much higher than that of the BexA, KpsT, and CtrD proteins (45.5 kDa versus 24.5 to 24.9 kDa), the structure of the central part of the protein was very similar to the complete structures of the BexA, KpsT, and CtrD proteins, and the ATP-binding motifs consisting of the A and B sites were conserved (Fig. 2).
FIG. 2.
Multiple sequence alignment of RgpC with KpsT (18), BexA (9), and CtrD (3). Multiple alignments of the amino acid sequences were generated with the program CLUSTAL V (6). Identical amino acids in all four sequences are indicated by asterisks. Dashes indicate gaps.
Creation of nonpolar insertions in rgp genes.
To study the functions of the rgp genes in S. mutans, the rgp genes were insertionally inactivated by homologous recombination. Procedures for the insertional inactivation of the rgpA gene in strain Xc41 and the rgpB gene in strain Xc42 have been described above. rgpC, rgpD, rgpE, rgpF, and ORF7 were insertionally inactivated by homologous recombination using the respective gene fragments from pYT8, which were interrupted by linearized pResYT10 at the restriction sites indicated in Fig. 1. The fragments on which both the erythromycin resistance gene and the inactivated gene were oriented in the same direction were used for the insertional inactivation. Their insertional inactivation was confirmed by detecting an amplified fragment 1.6 kb larger than the corresponding fragment detected by PCR with the appropriate pair of primers: primers 2F and 2R, primers 3F and 3R, and primers 4F and 4R (Fig. 1). The resultant mutant strains were designated Xc43, Xc44, Xc45, Xc46, and Xc47, respectively, as shown in Fig. 1.
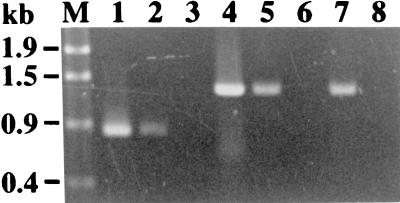
The rgp genes are located close to each other, suggesting the polycistronic transcription of these genes. Therefore, we used pResYT10 to insertionally inactivate each gene in order to reduce the likelihood of a polar effect on the transcription of the downstream genes. To confirm the transcription of rgpB in strain Xc41 and rgpE in strains Xc43 and Xc44, reverse transcriptase-mediated PCR (RT-PCR) was utilized. Total RNA was prepared from strains Xc41, Xc43, and Xc44 by using the FastPrep Device (Bio 101, Vista, Calif.) in combination with a FastPrep BLUE tube (Bio 101) in accordance with the protocol of the supplier. An RT-PCR kit, the SuperScript One-Step RT-PCR system (Gibco/BRL Life Technologies, Cleveland, Ohio), was used to amplify cDNA synthesized from rgpB- or rgpE-specific mRNA. A set of primers was added to the RT-PCR mixture in advance. Primers 5F and 5R as well as primers 3F and 3R were used to detect rgpB and rgpE transcription, respectively. cDNA was transcribed from 100 ng of total RNA at 50°C for 30 min and heated at 94°C for 2 min. Successive PCR was performed under the following condition: 25 cycles at 94°C for 15 s, 55°C for 30 s, and 72°C for 1 min. In the control reaction, the final heating at 94°C after the RT reaction was omitted. A 0.8-kb RT-PCR fragment was observed in the total RNA preparation from strain Xc41, while 1.3-kb RT-PCR fragments were observed in the total RNA preparations from strains Xc43 and Xc44 (Fig. 3). No RT-PCR product was observed in the control reaction mixture that was heated at 94°C for 2 min prior to the addition of the primers, suggesting that the RT-PCR products were derived from mRNA but not from contaminating chromosomal DNA. pResYT10 integrated into the chromosome thus did not interrupt transcription of the downstream genes.
FIG. 3.
Detection of rgpB transcription in strain Xc41 and rgpE transcription in strains Xc43 and Xc44 by RT-PCR. Lanes 1 and 4, PCR using the chromosomal DNA of Xc as a template; lanes 2 and 3, RT-PCR using total RNA from strain Xc41 as a template; lanes 5 and 6, RT-PCR using total RNA from strain Xc43 as a template; and lanes 7 and 8, RT-PCR using total RNA from strain Xc44 as a template. Lanes 1 to 3, amplification with primers 5F and 5R; lanes 4 to 8, amplification with primers 4F and 4R; lanes 3, 6, and 8, negative control reaction mixtures that were initially heated at 94°C for 2 min.
Chemical analysis of cell wall polysaccharide.
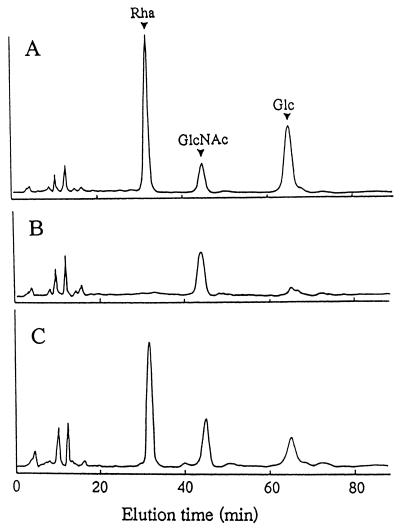
The sugar composition of the cell wall preparations from the mutant strains was analyzed by high-pressure liquid chromatography (HPLC) (Fig. 4) (26). The ratio of rhamnose to glucose in the cell wall preparation from strains Xc (data not shown) and Xc47 (Fig. 4A) was nearly 2. On the other hand, both the rhamnose and glucose contents were drastically reduced in the cell wall preparations from strains Xc41 (Fig. 4B) and Xc42, Xc43, Xc44, and Xc46 (data not shown). Only the glucose content was reduced in the cell wall preparation from strain Xc45 (Fig. 4C). There was no significant difference in the hexosamine content among these cell wall preparations (data not shown) as determined by the method of Strominger et al. (25). The intracellular UDP-d-glucose concentration in strain Xc45 was not different from that in Xc, when determined as previously described (31).
FIG. 4.
HPLC patterns of monosaccharides obtained by acid hydrolysis of cell wall preparations of strains Xc47 (A), Xc41 (B), and Xc45 (C). The pyridylamino sugars were analyzed by HPLC using a PALPAK type A column. Arrowheads labeled Rha, GlcNAc, and Glc indicate the elution times of pyridylaminated l-rhamnose, pyridylaminated N-acetylglucosamine, and pyridylaminated d-glucose, respectively.
Immunological analysis of polysaccharide antigens.
The polysaccharide antigen was extracted from whole cells of S. mutans strains and Streptococcus pyogenes A486var (kindly provided by V. A. Fischetti, Laboratory of Bacterial Pathogenesis, Rockefeller University, New York, N.Y.) by using the formamide extraction procedure of Fuller (4), which is an effective method for extracting RGP (28). Immunodiffusion analysis was carried out with serotype c-specific antiserum (26) as well as rhamnan-specific rabbit antiserum raised against whole cells of S. mutans Xc31 (31), which is a mutant that is deficient in glucose-1-phosphate uridylyltransferase and produces a poly-l-rhamnose backbone without a glucose side chain. The latter antiserum was raised by three subcutaneous injections of cell suspension at 2-week intervals in incomplete Freund’s adjuvant and then absorbed with whole cells of S. mutans MT703 (serotype e). The serotype c-specific antiserum did not react with the extracts from any of the mutant strains other than strain Xc47 (data not shown). The presumed rhamnan-specific antiserum formed a single precipitin line between the extracts from strains Xc43, Xc44, and Xc45 (Fig. 5). However, the amount of poly-l-rhamnose produced in Xc43 and Xc44 was probably small because we needed extracts from Xc43 and Xc44 much more concentrated than those from Xc31 and Xc45 in order to produce precipitin lines of the same intensity. This precipitin line fused with the line produced by the extract of S. pyogenes A486var, a variant of group A producing solely a poly-l-rhamnose that is identical to the backbone structure of RGP (1). The antiserum did not react with the extract from strains Xc (Fig. 5).
FIG. 5.

Immunodiffusion analysis of polysaccharide extracts with antiserum against a poly-l-rhamnose polymer of RGP. Immunodiffusion was performed in 1% (wt/vol) Noble agar in saline (17). The center well contains rabbit antiserum against whole cells of strain Xc31. The outer wells contain the formamide polysaccharide extracts from S. mutans Xc (well 1), S. mutans Xc31 (well 2), S. pyogenes A486var (well 3), S. mutans Xc45 (well 4), S. mutans Xc44 (well 5), and S. mutans Xc43 (well 6).
Probable function of RgpE.
The cell wall polysaccharide extracts from strains Xc45 and Xc31 shared the same antigenicity with the extract from S. pyogenes A486var (Fig. 5). Thus, the cell wall polysaccharide of the rgpE mutant (Xc45) consists of an alternating α1,2- and α1,3-linked l-rhamnosyl polymer without glucose side chains, and RgpE is involved in glucose side chain formation. Indeed, the RgpE sequence has features typical of glycosyltransferases.
Probable function of RgpC and RgpD.
The RgpC and RgpD proteins, apparent ABC transporter components, are likely to transport polysaccharide across the cell membrane. Both the rhamnose and glucose contents were drastically reduced in the cell wall fractions of rgpC and rgpD mutants (Fig. 4B), even though a poly-l-rhamnose backbone structure appeared to have been synthesized at very low levels (Fig. 5). There are two pathways for assembling the O antigen of lipopolysaccharide in gram-negative bacteria (29). These pathways differ in the process used to export polysaccharide across the cytoplasmic membrane. In the Rfc-dependent pathway, each O unit is transported across the cytoplasmic membrane by a flippase encoded by rfbX. O-antigen heteropolymers are then synthesized by block polymerization, which is believed to faithfully reproduce the complex repeating unit structure of the heteropolysaccharides. In the other pathway, Rfc-independent polymerization, the polymerized product is then exported across the cytoplasmic membrane by an ABC transporter. Although the latter pathway is limited to homopolymeric O antigen, it is used for the export of heteropolymeric capsular polysaccharides in gram-negative bacteria (21). Recently a third pathway for O polysaccharide synthesis, involving a protein with dual transferase-transport function, was proposed in the biosynthesis of the O:54 antigen of Salmonella enterica LPS (7). It seems likely that RGP, a heteropolysaccharide, is exported by RgpCD transporter, somewhat similarly to the capsular heteropolysaccharide. In contrast, type 14, 19B, and 19F pneumococcal capsular polysaccharides, which have rather complicated repeat units, appear to be synthesized by an Rfc-dependent mechanism (8, 14, 15), and we are not aware of any other streptococcal cell wall polysaccharides exported by ABC transporters.
The rml gene mutants that are completely unable to synthesize RGP (26, 27) showed a characteristic colony morphology (32), circular and convex with a dull surface but not rough even in the presence of sucrose and smaller than that of the parental strain. This morphology is clearly distinct also from that of strain Xc31 in which glucose side chains in the cell wall polysaccharide are defective (Fig. 5). The colony morphology of strains Xc43 and Xc44 was identical to that of rml gene mutants (data not shown).
Possible functions of other Rgp proteins.
At present, it is difficult to speculate on the functions of RgpA, RgpB, and RgpF. However, the biochemical and immunological properties of strains Xc41, Xc42, and Xc46 strongly support the hypothesis that all of rgpA, rgpB, and rgpF are required for RGP synthesis in S. mutans in addition to rgpC, rgpD, and rgpE.
Nucleotide sequence accession number.
The 11,085-bp nucleotide sequence described in this work has been submitted to the EMBL/GenBank/DDBJ data bank under accession number AB010970.
Acknowledgments
We thank J.-P. Mornon for his helpful suggestion on HCA and T. Shiroza for his useful advice on the construction of pResYT10.
This work was supported in part by Grant-in-Aid for Developmental Scientific Research (B)09470474 (to Y.Y.) from the Ministry of Education, Science, Sports and Culture of Japan and grants from the Takeda Science Foundation (to Y.Y.) and the Uehara Memorial Foundation (to T.K.).
REFERENCES
- 1.Coligan J E, Kindt T J, Krause R M. Structure of the streptococcal groups A, A-variant and C carbohydrates. Immunochemistry. 1978;15:755–760. doi: 10.1016/0161-5890(78)90105-0. [DOI] [PubMed] [Google Scholar]
- 2.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frosch M, Edwards U, Bousset K, Krauße B, Weisgerber C. Evidence for a common molecular origin of the capsule gene loci in Gram-negative bacteria expressing group II capsular polysaccharides. Mol Microbiol. 1991;5:1251–1263. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 4.Fuller A T. Formamide method for the extraction of polysaccharides from hemolytic streptococci. Br J Exp Pathol. 1938;19:130. [Google Scholar]
- 5.Gotschlich E C. Genetic locus for the biosynthesis of the variable portion of Neisseria gonorrhoeae lipooligosaccharide. J Exp Med. 1994;180:2181–2190. doi: 10.1084/jem.180.6.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 7.Keenleyside W J, Whitfield C. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J Biol Chem. 1996;271:28581–28592. doi: 10.1074/jbc.271.45.28581. [DOI] [PubMed] [Google Scholar]
- 8.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 9.Kroll J S, Loynds B, Brophy L N, Moxon E R. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol Microbiol. 1990;4:1853–1862. doi: 10.1111/j.1365-2958.1990.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 10.Lemesle-Varlot L, Henrissat B, Gaboriaud C, Bissery V, Morgat A, Mornon J-P. Hydrophobic cluster analysis: procedures to derive structural and functional information from 2-D representation of protein sequences. Biochimie. 1990;72:555–574. doi: 10.1016/0300-9084(90)90120-6. [DOI] [PubMed] [Google Scholar]
- 11.Linzer R, Reddy M S, Levine M J. Immunochemical aspects of serotype carbohydrate antigens of Streptococcus mutans. In: Hamada S, Michalek S M, Kiyono H, Menaker L, McGhee J R, editors. Molecular microbiology and immunobiology of Streptococcus mutans. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. pp. 29–38. [Google Scholar]
- 12.Lipman D J, Pearson W R. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 13.Martin V, Kleschyov A L, Klein J-P, Beretz A. Induction of nitric oxide production by polyosides from the cell walls of Streptococcus mutans OMZ 175, a gram-positive bacterium, in the rat aorta. Infect Immun. 1997;65:2074–2079. doi: 10.1128/iai.65.6.2074-2079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 15.Morona J K, Morona R, Paton J C. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 19B. J Bacteriol. 1997;179:4953–4958. doi: 10.1128/jb.179.15.4953-4958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niaudet B, Goze A, Ehrlich S D. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene. 1982;19:277–284. doi: 10.1016/0378-1119(82)90017-8. [DOI] [PubMed] [Google Scholar]
- 17.Ouchterlony Ö. Diffusion-in-gel methods for immunological analysis. Prog Allergy. 1958;5:1–78. [PubMed] [Google Scholar]
- 18.Pavelka M S, Jr, Wright L F, Silver R P. Identification of two genes, kpsM and kpsT, in region 3 of the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1991;173:4603–4610. doi: 10.1128/jb.173.15.4603-4610.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavelka M S, Jr, Hayes S F, Silver R P. Characterization of KpsT, the ATP-binding component of the ABC-transporter involved with the export of capsular polysialic acid in Escherichia coli K1. J Biol Chem. 1994;269:20149–20158. [PubMed] [Google Scholar]
- 20.Pritchard D G, Gregory R L, Michalek S M, McGhee J R. Biochemical aspects of serotype carbohydrate antigens of Streptococcus mutans. In: Hamada S, Michalek S M, Kiyono H, Menaker L, McGhee J R, editors. Molecular microbiology and immunobiology of Streptococcus mutans. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. pp. 39–49. [Google Scholar]
- 21.Roberts I S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 22.Saxena I M, Brown R M, Jr, Fevre M, Geremia R A, Henrissat B. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiroza T, Kuramitsu H K. Construction of a model secretion system for oral streptococci. Infect Immun. 1993;61:3745–3755. doi: 10.1128/iai.61.9.3745-3755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soell M, Lett E, Holveck F, Schöller M, Wachsmann D, Klein J-P. Activation of human monocytes by streptococcal rhamnose glucose polymers is mediated by CD14 antigen, and mannan binding protein inhibits TNF-α release. J Immunol. 1995;154:851–860. [PubMed] [Google Scholar]
- 25.Strominger J L, Park J T, Thompson R E. Composition of the cell wall of Staphylococcus aureus: its relation to the mechanism of action of penicillin. J Biol Chem. 1959;234:3263–3268. [PubMed] [Google Scholar]
- 26.Tsukioka Y, Yamashita Y, Oho T, Nakano Y, Koga T. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J Bacteriol. 1997;179:1126–1134. doi: 10.1128/jb.179.4.1126-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsukioka Y, Yamashita Y, Oho T, Nakano Y, Koga T. Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J Bacteriol. 1997;179:4411–4414. doi: 10.1128/jb.179.13.4411-4414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wetherell J R, Jr, Bleiweis A S. Antigen of Streptococcus mutans: characterization of a polysaccharide antigen from walls of strain GS-5. Infect Immun. 1975;12:1341–1348. doi: 10.1128/iai.12.6.1341-1348.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;178:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita Y, Takehara T, Kuramitsu H K. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J Bacteriol. 1993;175:6220–6228. doi: 10.1128/jb.175.19.6220-6228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamashita Y, Tsukioka Y, Nakano Y, Tomihisa K, Oho T, Koga T. Biological functions of UDP-glucose synthesis in Streptococcus mutans. Microbiology. 1998;144:1235–1245. doi: 10.1099/00221287-144-5-1235. [DOI] [PubMed] [Google Scholar]
- 32.Yamashita, Y., K. Tomihisa, Y. Tsukioka, and T. Koga. Unpublished data.