Abstract
The pyrazinamidase from Mycobacterium smegmatis was purified to homogeneity to yield a product of approximately 50 kDa. The deduced amino-terminal amino acid sequence of this polypeptide was used to design an oligonucleotide probe for screening a DNA library of M. smegmatis. An open reading frame, designated pzaA, which encodes a polypeptide of 49.3 kDa containing motifs conserved in several amidases was identified. Targeted knockout of the pzaA gene by homologous recombination yielded a mutant, pzaA::aph, with a more-than-threefold-reduced level of pyrazinamidase activity, suggesting that this gene encodes the major pyrazinamidase of M. smegmatis. Recombinant forms of the M. smegmatis PzaA and the Mycobacterium tuberculosis pyrazinamidase/nicotinamidase (PncA) were produced in Escherichia coli and were partially purified and compared in terms of their kinetics of nicotinamidase and pyrazinamidase activity. The comparable Km values obtained from this study suggested that the unique specificity of pyrazinamide (PZA) for M. tuberculosis was not based on an unusually high PZA-specific activity of the PncA protein. Overexpression of pzaA conferred PZA susceptibility on M. smegmatis by reducing the MIC of this drug to 150 μg/ml.
Pyrazinamide (PZA) is one of the most important drugs in tuberculosis chemotherapy, but in contrast to other first-line drugs, relatively little is known about its mechanism of action (12, 27). PZA is converted in the mycobacterium to pyrazinoic acid (POA) by the action of the enzyme pyrazinamidase. POA is thought to function as the toxic metabolite by virtue of its activity as a weak acid and/or by specifically inhibiting a metabolic process (12). In a recent seminal study, point mutations in the M. tuberculosis pncA gene that abrogated the pyrazinamidase/nicotinamidase activity of this organism were shown to confer resistance to PZA and the natural resistance of M. bovis to this drug was shown to be attributable to an inactivating point mutation in pncA (27). Subsequent studies have shown that 72 to 95% of PZA-resistant clinical isolates of M. tuberculosis are associated with pncA mutations (26, 29), suggesting that pyrazinamidase-catalyzed hydrolysis of PZA is essential for the activity of this drug in M. tuberculosis. This conclusion is consistent with the fact that POA esters show good antimycobacterial activity against PZA-susceptible and -resistant isolates of M. tuberculosis (4, 35). Paradoxically, other mycobacterial species such as M. smegmatis (5, 33) and M. avium (31) are known to possess pyrazinamidase activity yet are resistant to PZA. We therefore decided to investigate the PZA-hydrolyzing activity of M. smegmatis. We report the purification to homogeneity of the major pyrazinamidase from this organism and the subsequent cloning and targeted knockout of its encoding gene. We also show that overexpression of this enzyme conferred PZA susceptibility on M. smegmatis.
(Preliminary reports on the purification of this activity have been published elsewhere [2, 18]).
MATERIALS AND METHODS
Materials, bacterial strains, media, and growth conditions.
Enzymes were from Boehringer Mannheim or New England Biolabs; radiochemicals were from Amersham; the Sequenase 2.0 sequencing kit was from United States Biochemicals; and PZA, POA, nicotinamide, and other fine chemicals were from Sigma. The bacterial strains and plasmids used in this study are listed in Table 1, and the oligonucleotides are given in Table 2. Escherichia coli strains were grown in Luria-Bertani broth (LB) or agar (LA) for all plasmid isolations or in 2YT broth for expression of recombinant proteins (24). E. coli DH5α was used for plasmid manipulations, JM101 was used for M13 cloning, and BL21(DE3)pLysS was used for expression. Ampicillin, chloramphenicol, and kanamycin were used at 100, 34, and at 50 μg/ml, respectively, for E. coli. M. smegmatis was maintained in MADC-Tw (Middlebrook 7H9 broth [Difco] supplemented with 0.085% NaCl, 0.2% glucose, and 0.05% Tween 80) and on Middlebrook 7H10 supplemented with 0.085% NaCl and 0.2% glucose as the solid medium.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| M. smegmatis strains | ||
| mc2155 | High-frequency transformation mutant of M. smegmatis ATCC 607 | 28 |
| pzaA::aph | pzaA knockout mutant of M. smegmatis mc2155; Kmr | This work |
| [pzaA::aph](pOam) | pzaA::aph carrying pzaA on pOam; Kmr, Hygr | This work |
| mc2155(pOam) | mc2155 carrying pzaA on pOam; Hygr | This work |
| Plasmids | ||
| pGEM3Z(+)f | E. coli cloning vector; Apr | Promega |
| pET15b | E. coli expression vector; Apr | Novagen |
| p45E5 | pBluescript SK+ construct containing complete M. smegmatis pzaA with ca. 200 bp of upstream sequence | M. Everett |
| p38O20 | pBluescript SK+ construct containing partial M. smegmatis pzaA with 380 bp of upstream sequence | M. Everett |
| pTam | pET15b derivative carrying pzaA | This work |
| pTncA | pET15b derivative carrying pncA | This work |
| pGam | pGEM3Zf(+) derivative carrying M. smegmatis pzaA with 382 bp of upstream sequence cloned as a HindIII-XbaI fragment | This work |
| pMam | M13mp19 construct containing the 1,824-bp HindIII-XbaI fragment from pGam | This work |
| pMamB | pMam derivative in which the internal BamHI site in pzaA was mutated to BglII | This work |
| pGamB | pGEM3Zf(+) construct carrying the EcoRV-XbaI fragment from pMamB | This work |
| pGamk | pGamB derivative carrying the aph gene in the BglII site engineered in pzaA | This work |
| pOLYG | Multicopy E. coli-Mycobacterium shuttle vector; Hygr | P. O’Gaora |
| pOam | pOLYG derivative carrying the HindIII-XbaI fragment from pGam | This work |
| pHINT | E. coli-Mycobacterium integrating shuttle vector; Hygr | P. O’Gaora |
| pHam | pHINT derivative carrying the HindIII-XbaI fragment from pGam | This work |
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′-3′) | Strand | Position and description |
|---|---|---|---|
| M. smegmatis pzaA primersa | |||
| PZA-P1 | ATCGAGGTCGCGGAGAAGATCCGCACGAAGGAGGTCAGCCCGGTCGAGGT | + | Guessmer used to clone the pzaA gene. |
| AMD-F | GGGGCCATGGAACTCTACGAA | + | Positions 382–396. G/C clamp and NcoI site are italic; start codon is underlined. |
| AMD-R | GGGGCATATGTCAGCCCACCTT | − | Positions 1777–1791. G/C clamp and NdeI site are italic; stop codon is underlined. |
| muAMD | GGATGCGGATAGATCTGCCCGTATCGCTGC | − | Positions 881–910. Used to mutate internal BamHI to BglII (underlined). |
| M. tuberculosis pncA primersb | |||
| PncA-F | GGGGCCATGGGACGGGCGTTGATCATC | + | Positions 1–18. G/C clamp and NcoI site are italic; start codon is underlined; extra Gly2 codon is in bold. |
| PncA-R | GGGGGGATCCTCAGGAGCTGCAAAC | − | Positions 548–561. G/C clamp and BamHI site are in italic; stop codon is underlined. |
Purification of the M. smegmatis pyrazinamidase.
M. smegmatis ATCC 607 was grown and harvested as described by Scherman et al. (25). Cell pellets (35 g) were resuspended in 70 ml of buffer A (20 mM sodium phosphate [pH 6.3], 1 mM EDTA, 1 mM dithiothreitol [DTT]) containing 1 mM phenylmethylsulfonyl fluoride, 1 μM leupeptin, and 0.2 μM aprotinin and sonicated at 4°C for 15 cycles of a 15-s pulse followed by a 90-s cooling. The extract was clarified by centrifugation at 25,400 × g for 20 min at 4°C followed by 148,000 × g for 60 min at 4°C. Extracts (50 ml) were loaded on a 2.5- by 10-cm Q Sepharose Fast Flow column (Pharmacia LKB) equilibrated in buffer A and eluted with a linear gradient of 0 to 0.5 M NaCl in buffer A (1,500 ml). Pyrazinamidase-containing fractions were pooled, concentrated to 8 ml by ultrafiltration through a 30-kDa exclusion limit membrane (Amicon), and fractionated on a HiLoad 26/60 Superdex (Pharmacia LKB) column in buffer A (0.8 ml/min). Peak fractions were loaded on a Mimetic Blue 1 A6XL column (0.5 by 3 cm; Affinity Chromatography Ltd.) equilibrated in buffer A, and the enzyme was eluted in 50 mM KCl in buffer A (15 ml). A second anion-exchange separation was performed on a MonoQ HR5/5 column (Pharmacia LKB) eluting with a 0 to 0.5 M NaCl linear gradient in buffer A (30 ml). Ammonium sulfate (1 M) was added to peak fractions, and the protein was fractionated on a phenyl-Superose HR5/5 column (Pharmacia LKB), eluting with a linear gradient of 1 to 0 M (NH4)2SO4 in buffer A (30 ml). Protein concentrations were determined by a Bradford assay (Bio-Rad kit II), and proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Protein sequencing.
Cysteine residues were derivatized with 4-vinylpyridine, and the proteins were subsequently separated by electrophoresis on a 10% Tricine gel. After being blotted to a polyvinylidene difluoride membrane, the proteins were subjected to Edman degradation and analysis on an Applied Biosystems ABI476A protein sequencer.
Cloning and sequencing of the pzaA gene.
The probe PZA-P1 (Table 2), designed from the amino-terminal sequence of the purified protein (MELYELPLIEVAEKIRTKEVSPVEVTES), was used to screen a gridded plasmid library of M. smegmatis provided by M. Everett (Glaxo Wellcome, Stevenage, United Kingdom). Two overlapping clones (p45E5 and p38O20) thus identified were subcloned in M13mp18/19 for DNA sequencing. The nucleotide sequence was analyzed with the Lasergene suite of programs (DNASTAR, Madison, Wis.).
Expression of M. smegmatis PzaA and M. tuberculosis PncA in E. coli.
The M. smegmatis pzaA gene was amplified by PCR with Vent polymerase (Promega) and the AMD-F and AMD-R primer pair. The product was cloned in pET15b to create pTam, which directed the overexpression of a recombinant protein with the authentic PzaA sequence. Overexpression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to cultures at an optical density at 600 nm of 0.6. After 2 h at 37°C, the cells were harvested, washed, and resuspended at 0.2 g/ml in buffer B (20 mM sodium phosphate [pH 5.9], 1 mM EDTA, 1 mM DTT, 0.04% NaN3) containing protease inhibitors (Complete; Boehringer Mannheim), freeze-thawed once, and lysed by sonication. Clarified extracts were obtained by centrifugation at 20,000 × g for 60 min. Most of the recombinant PzaA was precipitated in inclusion bodies, but sufficient activity remained in the supernatant for further analysis. The protein was partially purified by loading a 50-ml extract on a 2.5- by 12-cm Q Sepharose Fast Flow column equilibrated in buffer B and eluting with a linear gradient of 0 to 0.5 M NaCl in buffer B (600 ml).
The M. tuberculosis pncA gene was amplified by PCR with the PncA-F and PncA-R primer pair (Table 2). The product was cloned in pET15b to form pTncA. Since PncA-F introduced a Gly codon between the first and second codons of pncA, the recombinant form of PncA was designated PncA-G2+. Overexpression and lysis were carried out as described above, and the protein was partially purified by fractionating a 50-ml extract on a 2.5- by 12-cm Q Sepharose Fast Flow column equilibrated in buffer B and eluting with a linear gradient of 0 to 0.5 M NaCl in buffer B (600 ml). Peak fractions were treated with (NH4)2SO4 (1.6 M) and loaded on a Toyopearl phenyl 650M 2- by 16-cm column [equilibrated in 1.6 M (NH4)2SO4–buffer C] and eluted with a 300-ml linear gradient of 1.6 to 0 M (NH4)2SO4 in buffer C (10 mM Tris · HCl [pH 7.5], 1 mM DTT, 0.04% NaN3). Peak fractions were loaded on a 2- by 16-cm hydroxyapatite column (Fast Flow; Calbiochem) equilibrated in buffer D (1 mM potassium phosphate, 10 mM Tris · HCl [pH 8], 0.2 mM CaCl2, 0.04% NaN3), and the protein was eluted with a 300-ml linear gradient of 0 to 100% buffer E (0.3 M potassium phosphate [pH 7.4], 0.01 mM CaCl2, 0.04% NaN3).
Enzyme assays.
The pyrazinamidase assay was a modification of the Wayne test (34). The activity was assayed by adding a 50-μl aliquot of test sample to 50 μl of a solution containing 40 mM PZA, 100 mM sodium phosphate (pH 6.5), and 2 mM DTT. After incubation at 37°C for 5 to 10 min, 10 μl of 20% FeNH4(SO4)2 and 890 μl of 0.1 M Gly · HCl (pH 3.4) were added, precipitates were removed by centrifugation (13,000 × g for 10 min), and the OD480 of the supernatant was determined. The POA concentration was determined from a standard curve. Since this modified Wayne test was limited in its sensitivity, a coupled amidase enzyme assay based on ammonia release was developed to determine the Km values for nicotinamide and PZA. The 750-μl assay mixture consisted of 30 mM Tris · HCl (pH 7.5), 15 U of l-glutamate dehydrogenase per ml, 800 μM α-ketoglutarate, 160 μM NADPH, and PZA (0; 70 μM to 5 mM) or nicotinamide (0; 10 μM to 5 mM) and the appropriate units of amidase, with the substrate being added last. The reaction mixtures were incubated at room temperature (21 to 25°C), and the optical density at 340 nm was monitored with a Shimadzu UV1601 spectrophotometer during the course of the reaction.
Construction of a pzaA knockout mutant of M. smegmatis.
Plasmid pGam was constructed by cloning the 282-bp HindIII-StuI upstream fragment from p38O20 and the 1,542-bp StuI-XbaI fragment from p45E5 in pGEM-3Zf(+). The 1,824-bp HindIII-XbaI fragment from pGam was cloned in M13mp19 to produce pMam, which was used as the template for site-directed mutagenesis with a Muta-Gene kit (Bio-Rad) and the oligonucleotide muAMD (Table 2) to yield pMamB. The EcoRV-XbaI fragment from pMamB was cloned in pGEM3Zf(+) to yield pGamB. The Tn903 aph (kanamycin resistance) gene, carried on a 1.3-kb BamHI fragment (10), was cloned in the BglII site of pGamB to form pGamk. M. smegmatis was electroporated with 1 μg of alkali-denatured pGamk (19) as described by Jacobs et al. (13), except that the cells were washed and prepared in 1% glycerol. Cells were plated on LA containing 10 μg of kanamycin per ml, and colonies were grown in LB containing 10 μg of kanamycin per ml. Genomic DNA was isolated as previously described (10), and Southern blots were probed with the pzaA PCR product described above. Cell extracts were prepared by resuspension of cell pellets from 10-ml cultures in 0.7 ml of 20 mM sodium phosphate (pH 6), disruption in a Bio 101/Savant FastPrep cell disruptor as previously described (7), and removal of cells and debris by centrifugation at 13,000 × g for 30 min.
Construction of PzaA expression vectors.
The M. smegmatis pzaA gene was excised as a HindIII-XbaI fragment from pGam and cloned in the hygromycin resistance vectors pOLYG and pHINT (9) to form pOam and pHam, respectively (Table 1). The constructs were electroporated into M. smegmatis, DNA was extracted from the recombinants, and Southern blotting was performed as described above. The presence of pOLYG-based vectors was also confirmed by electroduction into E. coli (10). The pyrazinamidase activity of recombinant clones was determined as previously described (10).
Determination of PZA susceptibility.
Dilutions of cultures were prepared in sterile water and plated on Middlebrook 7H10 adjusted to pH 5.2, supplemented with NaCl and glucose as described above, and containing a range of PZA concentrations (0, 50, 100, 150, 200, 300, 500, and 2,200 μg/ml). Colonies were counted after the appropriate growth period, and the MIC was defined as the lowest PZA concentration that caused a 90% reduction in the colony count. Nicotinamide susceptibility studies were carried out in a similar manner.
Nucleotide sequence accession number.
The pzaA gene sequence was deposited in GenBank under accession no. AF058285.
RESULTS AND DISCUSSION
Purification of the M. smegmatis pyrazinamidase and cloning of its encoding gene.
The pyrazinamidase activity from M. smegmatis was purified 3,270-fold by a combination of anion-exchange, gel filtration, dye affinity, and hydrophobic interaction chromatography (Table 3). SDS-PAGE of the purified protein revealed a major species at ca. 50 kDa (Fig. 1). The amino-terminal sequence was used to design a 50-mer oligonucleotide probe (PZA-P1 [Table 2]) for screening a gridded plasmid library of M. smegmatis DNA. An open reading frame (ORF) encoding 468 amino acids (49.3 kDa) was identified in positive clones and designated pzaA. Mycobacterial promoter elements (1) could not be identified in the region 382 nucleotides upstream of the putative start codon, although a ribosome binding site (GAAAAGGAA) was found 7 nucleotides upstream of this site. The ORF had 52 and 64% amino acid identity and similarity, respectively, to an enantiomer-selective amidase from Rhodococcus (17) but showed little homology to the M. tuberculosis (27), M. avium (31), and M. kansasii PncA proteins. This observation is consistent with the fact that M. avium and M. tuberculosis pncA probes failed to hybridize to M. smegmatis genomic DNA (27, 31). PzaA belongs to a large family of enzymes which bear a conserved amidase signature (16, 17) and which act on a wide range of substrates. Although this family includes a putative amidase from M. tuberculosis, it is unlikely to hydrolyze nicotinamide or PZA, since loss of the pncA-encoded amidase function is sufficient to abrogate the pyrazinamidase/nicotinamidase activity of this organism (26, 27).
TABLE 3.
Summary of the purification of the M. smegmatis pyrazinamidase activity
| Step | Vol (ml) | Concn of protein (mg/ml) | Total amt of protein (mg) | Sp act (U/mg)a | Total activity (U) | Yield (%) | Fold purifi-cation |
|---|---|---|---|---|---|---|---|
| Crude cell extract | 70 | 12 | 840 | 0.45 | 378 | 100 | 1 |
| Q Sepharose | 190 | 0.39 | 75 | 3.53 | 265 | 71 | 8 |
| Superdex 200 | 20 | 0.089 | 1.8 | 35.3 | 64 | 17 | 80 |
| Blue 1 A6XL | 3 | 0.083 | 0.25 | 201 | 50 | 13 | 448 |
| Mono Q HR5/5 | 2.5 | 0.029 | 0.072 | 607 | 44 | 11 | 1,350 |
| Phenyl-Superose | 2 | 0.0085 | 0.017 | 1,471 | 25 | 6 | 3,270 |
A unit (1 U) is defined as the number of micromoles of product formed per minute at 37°C.
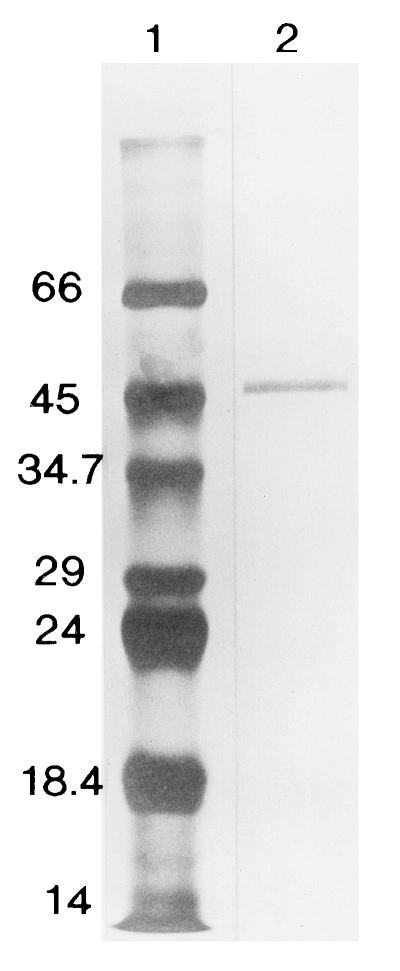
FIG. 1.
Purification of the pyrazinamidase from M. smegmatis. Lanes: 1, molecular weight markers (sizes shown in thousands); 2, phenyl-Superose fraction that was subjected to N-terminal amino acid sequencing. Samples were fractionated by SDS-PAGE in a 10% gel, and bands were visualized by silver staining.
Biochemical characterization of M. smegmatis PzaA and comparison with M. tuberculosis PncA.
The pyrazinamidase specific activity of a cell extract of M. tuberculosis H37Rv (0.02 U/mg) was found to be considerably lower than that of M. smegmatis (0.45 U/mg). To determine whether the difference in susceptibilities of M. tuberculosis and M. smegmatis to PZA could be ascribed to different specificities of the amidases for their substrates, PncA and PzaA were expressed as nonfusion recombinant proteins in E. coli and were partially purified for use in steady-state kinetic studies (Fig. 2). The Km values for nicotinamide and PZA were not significantly different for the two enzymes (33 and 300 μM for PncA-G2+ and 25 and 400 μM for PzaA). The Km values for nicotinamide were within the ranges reported for other bacterial and yeast nicotinamidases (3, 20, 32, 36), suggesting that these mycobacterial amidases are theoretically competent for the incorporation of nicotinamide as a precursor of NAD synthesis via the Preiss-Handler pathway (22). The high Km values for PZA suggested that PzaA and PncA have an equally low specificity for this drug. The biochemical data thus ruled out the possibility that the 20-fold higher apparent pyrazinamidase activity observed in crude extracts of M. smegmatis was an artifact of the assay conditions and confirmed that M. smegmatis is at least as proficient as M. tuberculosis in producing POA. On the basis of these observations, we therefore concluded that the relative resistance of M. smegmatis to PZA must be due to other factors such as the nature of the downstream target(s) of POA.
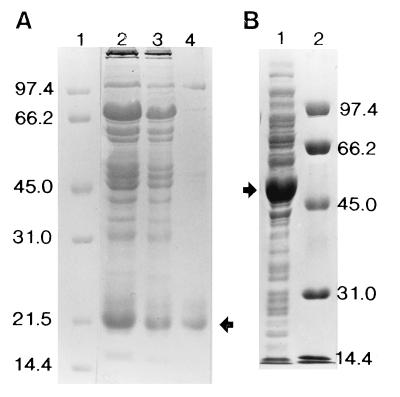
FIG. 2.
Partial purification of recombinant forms of M. smegmatis PzaA and M. tuberculosis PncA (PncA-G2+) expressed in E. coli. (A) PncA-G2+. Lanes: 1, molecular weight markers; 2, Q Sepharose fraction; 3, Toyopearl phenyl fraction; 3, hydroxyapatite fraction. (B) PzaA. Lanes: 1, Q Sepharose fraction; 2, molecular weight markers. Marker sizes are as indicated (in thousands), and the positions of the recombinant amidases are shown by arrows. Samples were fractionated by SDS-PAGE in a 10% gel which was stained with Coomassie brilliant blue.
Construction and phenotypic characterization of a pzaA knockout mutant of M. smegmatis.
Allelic exchange mutagenesis was used to confirm that pzaA encodes the major pyrazinamidase of M. smegmatis. The gene was disrupted by insertion of a drug resistance marker at a locus corresponding to the proposed active site of the encoded amidase (Fig. 3). Allelic exchange was detected in 4 of the 40 recombinants, with the remainder corresponding to products of site-specific single crossover (Fig. 3). The M. smegmatis pzaA::aph mutant displayed a 3-fold reduction in pyrazinamidase specific activity (Table 4). The residual pyrazinamidase activity of the mutant could be inactivated by heat (data not shown), suggesting that in addition to PzaA, M. smegmatis contains other minor, PZA-hydrolyzing enzymes. Comparison of the growth rate of cultures of the pzaA::aph mutant grown in MADC-Tw to that of wild-type controls suggested that loss of the amidase activity conveyed no obvious growth disadvantage (data not shown). Since disruption of a gene required in the recycling pathway of an essential cofactor might cause a decreased survival rate under conditions of nutrient limitation, the viability of stationary-phase cultures of the pzaA::aph mutant was also compared to the wild-type control, but no differences were detected (data not shown). This observation argues against the salvage of nicotinamide in the NAD biosynthetic pool by the sequential action of nicotinamidase and nicotinic acid phosphoribosyltransferase (PncB [8]) during stationary phase in M. smegmatis and is consistent with the inability to demonstrate the presence of PncB in M. tuberculosis by using nicotinic acid as a precursor for the synthesis of intermediates of the pyridine nucleotide cycle (6, 14). Therefore, continued de novo synthesis and/or bypass of this arm of the recycling pathway by the action of nicotinamide mononucleotide deamidase would be the preferred routes for the formation of NAD during the stationary phase of M. smegmatis.
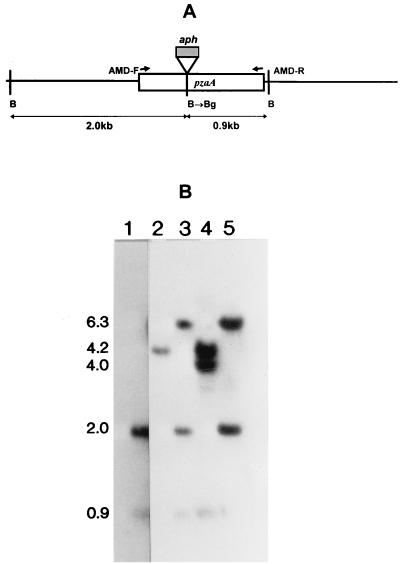
FIG. 3.
Targeted knockout of the M. smegmatis pzaA gene. (A) Restriction map of the pzaA locus showing the site of insertion of the aph marker (hatched box) in the pzaA gene (open box). The positions of the primers used to amplify the PzaA ORF, and the BamHI (B) and BglII (Bg) sites are indicated. (B) Southern blot of recombination products. Lanes: 1, wild-type M. smegmatis mc2155; 2, representative double-crossover product, pzaA::aph; 3 and 5, single crossover (downstream); 4, single crossover (upstream). The gel was probed with the pzaA PCR product, and the sizes of the hybridizing bands are as indicated (in kilobases).
TABLE 4.
Pyrazinamidase specific activities and MICs for M. smegmatis strainsa
| Parental strain of M. smegmatis | Sp act (U/mg)b | MIC of PZA (μg/ml) | MIC of Nam (μg/ml) |
|---|---|---|---|
| Wild type | |||
| mc2155 | 0.45 ± 0.04 | ≥2,200 | >2,200 |
| mc2155(pOam) | 10 ± 1 | 150 | 150 |
| mc2155::pHam | 1.12 ± 0.05 | ≥2,000 | 500 |
| Mutant | |||
| pzaA::aph | 0.15 ± 0.03 | ≥2,000 | >2,200 |
| [pzaA::aph](pOam) | 9 ± 1 | 150 | 200 |
| [pzaA::aph]::pHam | 1.04 ± 0.03 | NDc | ND |
MIC determinations were performed at pH 5.2.
Mean and standard deviation from three independent experiments.
ND, not determined.
PZA susceptibility testing in M. smegmatis and in recombinants with altered pyrazinamidase activity.
The fact that the pyrazinamidase activity of the pzaA::aph knockout mutant was restored by complementation with a single copy of the pzaA gene carried on pHam confirmed that pzaA encodes the major pyrazinamidase of M. smegmatis (Table 4). However, the level of pyrazinamidase activity was unexpectedly high (threefold higher than in the wild type). Overexpression of PzaA in M. smegmatis from the multicopy plasmid, pOam, resulted in a 25-fold increase in the specific activity of the pyrazinamidase enzyme in cell lysates, which was also considerably higher than expected from the estimated copy number of pAL5000-based plasmids (30). Therefore, the pzaA expression cassette contained in pHam and pOam appeared to direct a higher level of expression of pzaA than the corresponding, chromosomally encoded gene. One possible explanation is that the chromosomal copy of pzaA is negatively regulated by flanking sequences which were not present in the pzaA cassette in pOam and pHam (382 nucleotides of upstream sequence). Consistent with this notion is the fact that regulation of amidase gene expression is a commonly observed phenomenon (11, 15, 17, 21). However, on the basis of the available data, we cannot exclude the possibility that the high-level expression directed by pOam and pHam was instead the result of a cloning artifact.
Overexpression of pzaA resulted in a 10-fold increase in the susceptibility of M. smegmatis to PZA (Table 4), suggesting that when produced at sufficiently high levels, POA can overwhelm relatively insensitive downstream target(s) and render the organism susceptible to this drug. The fact that nicotinamide and PZA inhibited the PzaA overexpressor with similar MICs suggests that nicotinic acid and POA have similar inhibitory effects, although it is unclear whether the toxicity is attributable to general, intracytoplasmic acidification or to inhibition of a specific cellular target(s). We note that the site of production of POA might also play an important role in determining the susceptibility of a mycobacterium to PZA. Indeed, Raynaud et al. (23) showed that 27% of the nicotinamidase activity of M. tuberculosis is secreted into the extracellular medium and that the cell-associated activity could be detected in its outermost capsule whereas M. smegmatis nicotinamidase was not detected in the extracellular medium but was buried much deeper within the outer capsule.
PZA-resistant revertants of mc2155(pOam) that were able to grow at 2 mg of PZA per ml were isolated at a frequency of 3 × 10−3 to 4 × 10−3. Of the 16 clones initially isolated, 4 were subsequently shown to be genuinely resistant to this concentration of PZA. In all four of the revertants, the pyrazinamidase activity was reduced to wild-type levels as a result of gross rearrangements within the pzaA region of pOam, as detected by restriction analysis of the recovered plasmid (data not shown). We note that such a mechanism for PZA resistance caused by abrogation of pyrazinamidase overproduction in M. smegmatis precisely mirrors the dominant role of PncA inactivation in PZA resistance in M. tuberculosis (27, 29).
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support of the Glaxo Wellcome Action TB Initiative, the South African Medical Research Council, and the South African Institute for Medical Research Foundation.
We are deeply indebted to Neil Freeman, Glaxo Wellcome, for carrying out the protein sequencing, to Steven Martin and Martin Everett for providing the gridded library and for recovering clones, and to Ken Duncan for advice, encouragement, and constructive criticisms. We thank F. Kiepela and W. Sturm for advice during the early part of this project, P. O’Gaora for providing pOLYG and pHINT, and Bill Jacobs, Oren Zimhony, and Jeffery Cox for providing mc2155 and for critically reviewing the manuscript.
REFERENCES
- 1.Bashyam M D, Kaushal D, Dasgupta S K, Tyagi A K. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J Bacteriol. 1996;178:4847–4853. doi: 10.1128/jb.178.16.4847-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boshoff H I, Mizrahi V. Abstracts of the ASM Conference on Tuberculosis: Past, Present and Future 1997. Washington, D.C: American Society for Microbiology; 1997. Characterization of the pyrazinamidase activity of Mycobacterium smegmatis, abstr. B-72; p. 42. [Google Scholar]
- 3.Calbreath D F, Joshi J G. Inhibition of nicotinamidase by nicotinamide adenine dinucleotide. J Biol Chem. 1971;246:4334–4339. [PubMed] [Google Scholar]
- 4.Cynamon M H, Klemens S P, Chou T S, Gimi R H, Welch J T. Antimycobacterial activity of a series of pyrazinoic acid esters. J Med Chem. 1992;35:1212–1215. doi: 10.1021/jm00085a007. [DOI] [PubMed] [Google Scholar]
- 5.Damato J J, Collins M T, McClatchy J K. Niacin, nitrate and pyrazinamide studies using Middlebrook 7H10 broth. Eur J Clin Microbiol. 1984;3:546–549. doi: 10.1007/BF02013616. [DOI] [PubMed] [Google Scholar]
- 6.Dudley M A, Willett H P. Nicotinamide adenine dinucleotide synthesis by Mycobacterium tuberculosis var. bovis. Proc Soc Exp Biol Med. 1965;119:807–812. doi: 10.3181/00379727-119-30307. [DOI] [PubMed] [Google Scholar]
- 7.Durbach S I, Andersen S J, Mizrahi V. SOS induction in mycobacteria. Analysis of the DNA binding activity of a LexA-like repressor and its role in the DNA damage induction of the recA gene from Mycobacterium smegmatis. Mol Microbiol. 1997;26:643–653. doi: 10.1046/j.1365-2958.1997.5731934.x. [DOI] [PubMed] [Google Scholar]
- 8.Foster J W, Moat A G. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev. 1980;44:83–105. doi: 10.1128/mr.44.1.83-105.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garbe T R, Barathi J, Barnini S, Zhang Y, Abou-Zeid C, Tang D, Mukherjee R, Young D B. Transformation of mycobacterial species using hygromycin resistance as a selectable marker. Microbiology. 1994;140:133–138. doi: 10.1099/13500872-140-1-133. [DOI] [PubMed] [Google Scholar]
- 10.Gordhan B G, Andersen S J, De Meyer A R, Mizrahi V. Construction by homologous recombination and phenotypic characterization of a polA mutant of Mycobacterium smegmatis. Gene. 1996;178:125–130. doi: 10.1016/0378-1119(96)00350-2. [DOI] [PubMed] [Google Scholar]
- 11.Hashimoto Y, Nishiyama M, Honirouchi S, Beppu T. Nitrile hydratase gene from Rhodococcus sp. N-774 requirement for its downstream region for efficient expression. Biosci Biotechnol Biochem. 1994;58:1859–1865. doi: 10.1271/bbb.58.1859. [DOI] [PubMed] [Google Scholar]
- 12.Heifets L B, Flory M A, Lindholm-Levy P J. Does pyrazinoic acid as an active moiety of pyrazinamide have specific activity against Mycobacterium tuberculosis? Antimicrob Agents Chemother. 1989;33:1252–1254. doi: 10.1128/aac.33.8.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs W R, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Genetic systems for mycobacteria. Methods Enzymol. 1991;381:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 14.Kasãrov L B, Moat A G. Metabolism of nicotinamide adenine dinucleotide in human and bovine strains of Mycobacterium tuberculosis. J Bacteriol. 1972;110:600–603. doi: 10.1128/jb.110.2.600-603.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komeda H, Kobayashi M, Shimizu S. A novel gene cluster including the Rhodococcus rhodochrous J1 nhlBA genes encoding low molecular mass nitrile hydratase (L-Nhase) induced by its reaction product. J Biol Chem. 1996;271:15796–15802. doi: 10.1074/jbc.271.26.15796. [DOI] [PubMed] [Google Scholar]
- 16.Mayaux J F, Cerebelaud E, Soubrier F, Faucher D, Petre D. Purification, cloning and primary structure of an enantiomer-selective amidase from Brevibacterium sp. strain R312: structural evidence for genetic mapping with nitrile reductase. J Bacteriol. 1990;172:6764–6773. doi: 10.1128/jb.172.12.6764-6773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayaux J F, Cerbelaud E, Soubrier F, Yeh P, Blanche F, Petre D. Purification, cloning, and primary structure of a new enantiomer-selective amidase from a Rhodococcus strain: structural evidence for a conserved genetic coupling with nitrile reductase. J Bacteriol. 1991;173:6694–6704. doi: 10.1128/jb.173.21.6694-6704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller L P, Marcinkeviciene J A, Blanchard J S, Jacobs W R., Jr . Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Purification and characterization of the M. smegmatis pyrazinamidase, abstr. U-79; p. 115. [Google Scholar]
- 19.Oh S-H, Chater K. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardee A B, Benz E J, Jr, St. Peter D A, Krieger J N, Meuth M, Trieshmann H W., Jr Hyperproduction and purification of nicotinamide deamidase, a microconstitutive enzyme of Escherichia coli. J Biol Chem. 1971;246:6792–6796. [PubMed] [Google Scholar]
- 21.Parish T, Mahenthiralingam E, Draper P, Dabis E, Colston M J. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology. 1997;143:2267–2276. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- 22.Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotides. I. Identification of the intermediates. J Biol Chem. 1958;233:483–492. [PubMed] [Google Scholar]
- 23.Raynaud C, Etienne G, Peyron P, Laneelle M A, Daffe M. Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology. 1998;144:577–587. doi: 10.1099/00221287-144-2-577. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Scherman M, Weston A, Duncan K, Whittington A, Upton R, Deng L, Comber R, Friedrich J D, McNeil M. Biosynthetic origin of mycobacterial cell wall arabinosyl residues. J Bacteriol. 1995;177:7125–7130. doi: 10.1128/jb.177.24.7125-7130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:540–543. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 28.Snapper S B, Melton R E, Kieser T, Mustafa S, Jacobs W R., Jr Isolation and characterization of high efficiency plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 29.Sreevatsan S, Pan X, Zhang Y, Kreiswirth B N, Musser J M. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41:636–640. doi: 10.1128/aac.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stolt P, Stoker N G. Functional definition of regions necessary for replication and incompatibility in the Mycobacterium fortuitum plasmid pAL5000. Microbiology. 1996;142:2795–2802. doi: 10.1099/13500872-142-10-2795. [DOI] [PubMed] [Google Scholar]
- 31.Sun Z, Scorpio A, Zhang Y. The pncA gene from naturally pyrazinamide-resistant Mycobacterium avium encodes pyrazinamidase and confers pyrazinamide susceptibility to resistant M. tuberculosis complex organisms. Microbiology. 1997;143:3367–3373. doi: 10.1099/00221287-143-10-3367. [DOI] [PubMed] [Google Scholar]
- 32.Tanigawa Y, Shimoyama M, Dohi K, Ueda I. Purification and properties of nicotinamide deamidase from Flavobacterium peregrinum. J Biol Chem. 1972;247:8036–8042. [PubMed] [Google Scholar]
- 33.Tarnok I, Rohrscheidt E. Biochemical background of some enzymatic tests used for the differentiation of mycobacteria. Tubercle. 1976;57:145–150. doi: 10.1016/0041-3879(76)90052-0. [DOI] [PubMed] [Google Scholar]
- 34.Wayne L G. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am Rev Respir Dis. 1974;109:147–151. doi: 10.1164/arrd.1974.109.1.147. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto S, Toida I, Watanabe N, Ura T. In vitro antimycobacterial activities of pyrazinamide analogs. Antimicrob Agents Chemother. 1995;39:2088–2091. doi: 10.1128/aac.39.9.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan C, Sloan D L. Purification and characterization of nicotinamide deamidase from yeast. J Biol Chem. 1987;262:9082–9087. [PubMed] [Google Scholar]