Abstract
Zinc is an essential nutrient for all cells, but remarkably little is known regarding bacterial zinc transport and its regulation. We have identified three of the key components acting to maintain zinc homeostasis in Bacillus subtilis. Zur is a metalloregulatory protein related to the ferric uptake repressor (Fur) family of regulators and is required for the zinc-specific repression of two operons implicated in zinc uptake, yciC and ycdHIyceA. A zur mutant overexpresses the 45-kDa YciC membrane protein, and purified Zur binds specifically, and in a zinc-responsive manner, to an operator site overlapping the yciC control region. A similar operator precedes the ycdH-containing operon, which encodes an ABC transporter. Two lines of evidence suggest that the ycdH operon encodes a high-affinity zinc transporter whereas YciC may function as part of a lower-affinity pathway. First, a ycdH mutant is impaired in growth in low-zinc medium, and this growth defect is exacerbated by the additional presence of a yciC mutation. Second, mutation of ycdH, but not yciC, alters the regulation of both the yciC and ycdH operons such that much higher levels of exogenous zinc are required for repression. We conclude that Zur is a Fur-like repressor that controls the expression of two zinc homeostasis operons in response to zinc. Thus, Fur-like regulators control zinc homeostasis in addition to their previously characterized roles in regulating iron homeostasis, acid tolerance responses, and oxidative stress functions.
Despite the essential role of zinc as a structural and catalytic cofactor in numerous metalloproteins, mechanisms of zinc homeostasis in bacteria are poorly understood (33, 34). As for other essential metal ions, it is likely that both the uptake and efflux of zinc are tightly regulated in response to availability. Zinc uptake and efflux proteins have recently been identified in several bacteria, including Streptococcus pneumoniae (11), Haemophilus influenzae (21), Staphylococcus aureus (40), Synechocystis strain PCC 6803 (38), and Escherichia coli (3, 28, 30). Expression of these transporters, where known, seems to be regulated by zinc-sensing metalloregulatory proteins (28, 38).
Metalloregulatory proteins sense the intracellular levels of specific metal ions and mediate a transcriptional or translational response. For example, the E. coli ferric uptake repressor (Fur) protein regulates the iron-dependent repression of iron uptake pathways (2) and is the prototype for a large family of regulators (17). Remarkably, Bacillus subtilis contains three distinct Fur homologs (5). Although many Fur homologs regulate iron uptake, Fur-like regulators control other functions as well. A B. subtilis Fur homolog designated PerR regulates the peroxide stress response (5), while E. coli Zur controls the transcription of a zinc uptake operon (28).
Zinc-specific metalloregulation has been documented for yeast, mammals, and bacteria. Much of this work has centered on the induction of metallothionein by zinc. For example, a zinc-sensitive inhibitor protein prevents the interaction of MTF-1 with metal response elements preceding mammalian metallothionein genes (27). In Synechococcus, metallothionein expression is induced when the SmtB repressor dissociates from its operator in response to zinc (8, 14, 39). A similar repressor, ZiaA, regulates the zinc-inducible expression of a zinc efflux pump in Synechococcus strain PCC 6803 (38). The regulation of zinc transport in Saccharomyces cerevisiae has also been well characterized. In that organism, Zap1p activates the transcription of both low- and high-affinity zinc transporters under conditions of zinc limitation (13).
Here we describe a B. subtilis Fur homolog, Zur, that regulates two operons implicated in zinc transport. Zur mediates the zinc-dependent repression of the ycdH-containing operon, encoding a putative high-affinity zinc transport system, and yciC, encoding an integral membrane protein of unknown function.
MATERIALS AND METHODS
Bacterial strains.
E. coli DH5α was used for routine DNA cloning (31). B. subtilis strains are all derivatives of ZB307A (W168 SPβc2Δ2::Tn917-lacZ::pSK10Δ6 MLSr) (44) or HB1000 (ZB307A attSPβ). The zur (yqfV) mutant strain, HB6542, contains a zur::spc gene disruption and has been described elsewhere (5).
Growth conditions.
B. subtilis was grown on LB plates or a defined morpholinepropanesulfonic acid-buffered minimal medium (MM) (6) prepared by using Milli-Q-treated water and supplemented with trace metals including 30 nM Co(II), 10 nM Cu(II), 10 nM Zn(II), 80 nM Mn(II), and 5 μM Fe(III) except as noted. Zinc-limited MM (LZMM) was prepared by omission of zinc from MM. Erythromycin (1 μg/ml) and lincomycin (25 μg/ml) (for testing macrolide-lincosamide-streptogramin B resistance), spectinomycin (100 μg/ml), kanamycin (10 μg/ml), neomycin (10 μg/ml), and chloramphenicol (5 μg/ml) were used for the selection of various B. subtilis strains.
Membrane protein preparation and analysis.
Cell wall- and membrane-associated proteins were isolated from the zur mutant and the wild type as described previously (15). Briefly, strains were grown in MM, and the cells were collected by centrifugation, resuspended in 50 mM Tris-HCl (pH 7.5)–10 mM MgCl2–0.5 mM phenylmethylsulfonyl fluoride, and disrupted in a French press. The walls were recovered by centrifugation at 20,000 × g for 5 min. The supernatant was centrifuged at 100,000 × g for 1 h to collect the cell membranes. Cell wall and membrane preparations were resuspended in the same buffer, mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and boiled for 10 min prior to analysis by SDS-PAGE on a 12% gel. For amino-terminal sequencing of the 45-kDa protein, samples were electroblotted to polyvinylidene difluoride membranes prior to gas-phase microsequencing by automated Edman degradation at the Cornell Biotechnology facility. The resulting amino-terminal sequence, MKKIPVTVLSGYLGA, is identical to the predicted amino-terminal sequence of YciC.
Construction of yciC and ycdH transcriptional fusions.
Promoter regions were amplified from the B. subtilis genome by PCR using primers 5′-CTGAAGCTTCCAGATGCGAAATGGGTATA-3′ and 5′-CGGGATCCAATGCTGTTCAGCAATGTTGTTT-3′ for yciC and primers 5′-CCGAAGCTTCCGGACGATCCGGACT-3′ and 5′-GCGGATCCTTTTGTTGATGAGTTGCC-3′ for ycdH. The resulting products were cloned as HindIII-to-BamHI fragments (sites underlined) into pJPM122 (35) to generate yciC′-cat-lacZ and ycdH′-cat-lacZ operon fusions, respectively. The resulting plasmids were linearized with ScaI and transformed into ZB307A with selection for neomycin resistance to generate strains HB8008 and HB8009, respectively. SPβ transducing lysates (SPβ8008 and SPβ8009) were prepared by heat induction and transduced to HB1000 and HB6542 to generate reporter strains designated HB8010 (HB1000 SPβ8008), HB8011 (HB6542 SPβ8008), HB8012 (HB1000 SPβ8009), and HB8003 (HB6542 SPβ8009).
Construction of yciC and ycdH mutant strains.
Chromosomal DNA from the yciC region was amplified by using PCR primers 5′-CTGGGTACCCCAGATGCGAAATGGGTATAT-3′ and 5′-CTGGATCCGATGATTACAGTGCCGGAATT-3′ and then digested with BamHI and KpnI (sites underlined); the resulting fragment was cloned in pBKS+ (Stratagene) to generate pAG34. A gene cassette coding for kanamycin resistance (Kmr) was isolated from pJM114 (29) as a BamHI-to-HindIII fragment and treated with the Klenow fragment of DNA polymerase I to generate blunt ends. This fragment was then ligated into the unique PstI site within yciC, after treatment with mung bean nuclease to generate blunt ends, to generate pAG34Km. PCR amplification of the disrupted gene was done with the same primers, and the resulting fragment was transformed to B. subtilis HB1000 with selection for Kmr. To construct a ycdH mutant, the ycdH-ycdI region was amplified with primers 5′-GCGAAGCTTTGTTTGTGATTGGAGCGTG-3′ and 5′-CGTCTAGATGGCAATGGACACTTCTG-3′, digested with HindIII and XbaI (sites underlined), and cloned in pBKS+ (Stratagene) to generate pAG2023. An internal PstI-to-EcoRV fragment, spanning the last 130 codons of ycdH and the first 7 codons of ycdI, was replaced with a chloramphenicol resistance (Cmr) gene cassette, isolated as a PstI-to-HincII fragment from pJM105A (29), to generate pAG2023Cm. The disrupted genes were amplified with the same primers, and the DNA fragment was transformed to HB1000, selecting for Cmr. Genomic DNA was prepared from the yciC and ycdH mutants, and the gene disruption was confirmed by PCR. A yciC ycdH double mutant was constructed by transforming DNA containing the ycdH mutation to the yciC mutant and selecting for Kmr and Cmr.
β-Galactosidase assays.
To induce zinc deficiency, overnight cultures were diluted 1:100 in LZMM and grown to mid-log phase. Cells were collected by centrifugation, washed once with 10 mM EDTA, diluted 1:10 in LZMM containing 1 mM Mn2+ and Zn2+ as indicated, and grown overnight at 37°C with vigorous shaking. Samples of 1 ml were harvested and assayed for β-galactosidase as described elsewhere (6, 22). If the EDTA wash step was omitted, as little as 10 nM Zn was sufficient to repress transcription of yciC, consistent with the absence of YciC protein from membrane fractions prepared from wild-type cells grown in MM.
For examining the zinc ion concentration dependence of gene expression, the data were fit, using the DeltaGraph Professional reiterative curve-fitting algorithm, to an equation of the form y = max −{(max − min)(1/1 + (K2/[Zn]2))}, where y is the measured β-galactosidase activity (Miller units), max is the maximal (fully induced) level of expression, min is the fully repressed level of expression, K is the apparent binding constant for the interaction of zinc with Zur, and [Zn] is the concentration of added zinc. Note that by this equation, repression varies as the square of the added zinc concentration, since this was found to give a better fit for every data set. Correlation coefficients are >0.99 for the wild-type strains and >0.97 in all other cases. Essentially identical equilibrium dissociation constants are also obtained by using a linear dependence on zinc.
Overproduction and purification of Zur.
The coding region of zur was amplified from a previously described PCR product (5, 23), characterized as part of the genome sequencing project, using the primers 5′-CTGGCTAGGTACCGTTCAATG-3′ and 5′-GGGACCCCATATGAACGTCC-3′. The resulting fragment was digested with KpnI and NdeI and cloned into pET17b (Novagen) to generate pHB6506 (4). pHB6506 was transformed into E. coli BL21(DE3)(pLysS) (37). For purification of Zur, a single colony was grown overnight in 2 ml of LB containing ampicillin (200 μg/ml), chloramphenicol (34 μg/ml), and 0.4% (wt/vol) glucose. The overnight culture was used to inoculate a 30-ml culture, which was then used to inoculate 2 liters of 2× LB, and the cells were incubated at 37°C with vigorous shaking to an optical density at 600 nm of 0.8. Isopropyl-β-d-thiogalactopyranoside was added to 1 mM (final concentration), and the cells were harvested after further incubation for 2.5 h.
For purification of Zur apoprotein, the cell pellet was resuspended in buffer A (50 mM Tris-HCl [pH 8], 50 mM NaCl, 0.1 mM EDTA, 2 mM dithiothreitol [DTT], 5% glycerol) containing 2% sodium deoxycholate and lysed by sonication. Inclusion bodies were recovered by centrifugation and washed twice with buffer A containing 2% sodium deoxycholate. The inclusion bodies were dissolved in buffer A containing 0.4% Sarkosyl and 100 mM EDTA and incubated at 20°C for 30 min. Zur was diluted 10-fold slowly, by 2-fold dilutions with buffer A at 4°C, and dialyzed overnight against buffer A at 4°C. The solution was applied to a 10-ml Sepharose Q column, washed with buffer A, washed with 2 volumes of buffer A containing 0.1 M NaCl, and then washed with 2 volumes of buffer A containing 0.3 M NaCl. Zur was eluted with 0.6 M NaCl. Peak fractions were pooled and dialyzed against buffer A containing 50% glycerol and stored at −20°C. For some experiments, Zur was further purified by chromatography on a 25-ml FPLC Superdex-75 column, using buffer A modified to contain 150 mM NaCl. Zur elutes after about 11.5 ml, which by comparison with protein standards suggests that Zur is a dimer in solution. All glassware and the columns were washed with 10 mM EDTA and Milli-Q-treated water prior to use. Initial preparations, leading to Zur that was active even in the absence of added metal ions, were prepared as described above except that buffer A contained 0.1 mM DTT and the inclusion bodies were denatured, and solubilized protein was renatured, in the absence of EDTA.
Electrophoretic mobility shift and restriction enzyme protection assays.
PCR fragments containing the promoter regions of yciC or a control fragment (e.g., the yknW promoter region) were purified and labeled with [γ-32P]ATP. Then 300 ng of Zur protein was incubated at room temperature for 20 min in 20 μl of binding buffer (20 mM Tris-HCl [pH 8], 50 mM KCl, 1 mM DTT, 5% glycerol, 0.1 mg of bovine serum albumin per ml, 5 μg of sheared salmon sperm DNA per ml) containing metal ions or EDTA as indicated; 1 fmol of labeled DNA was added to each tube, and incubation continued for an additional 20 min. Samples were loaded on a 4% polyacrylamide gel prepared and run in 40 mM Tris-acetate buffer (with no added EDTA), pH 8.0. The gel was dried and exposed to a phosphorimager screen.
For restriction enzyme protection assays, 1 pmol of DNA (330 ng) was mixed with 800 ng of protein in RE buffer (10 mM Tris-HCl [pH 7], 50 mM KCl, 5% glycerol, 50 μg of bovine serum albumin per ml, 1 mM β-mercaptoethanol, 0.05% Nonidet P-40), and samples were incubated at room temperature for 1 h in the presence or absence of 10 U of DraI (or control endonuclease). Control reaction mixtures contained no Zur or Zur and 25 mM EDTA. The preparation of Zur used for these experiments was active for DNA binding without added zinc. The resulting DNA fragments were separated by PAGE on a 6% gel, stained with ethidium bromide, and visualized under UV light.
Computer analysis.
All predicted protein sequences were compared against the nonredundant protein databases by using BLAST 2.0 (1). Protein searches against the B. subtilis genome were executed with either BLAST or FASTA as available on the SubtiList web site (24) at http://www.pasteur.fr/Bio/SubtiList.html. Protein localization was predicted with both PSORT (25) and TMPred (18) programs, available at http://psort.nibb.ac.jp:8800/form.html and http://www.isrec.isb-sib.ch/software/TMPRED_form.html, respectively.
RESULTS
The sequence of the B. subtilis genome reveals three genes encoding Fur homologs (Fig. 1): fur (yqkL), perR (ygaG), and zur (yqfV) (5, 20). We have previously demonstrated that Fur controls iron transport functions whereas PerR regulates a peroxide stress regulon (5). Since we herein demonstrate that yqfV encodes a Zn(II)-uptake regulator, we will refer to this gene as zur. Zur is quite dissimilar relative to Fur proteins from either B. subtilis (23% identity) or Escherichia coli (26% identity [32]) but shows nearly 50% identity with a Fur-like regulator of unknown function from Staphylococcus epidermidis (16). This similarity is even higher in regions presumed important for either DNA- or metal-binding selectivity (Fig. 1). Interestingly, of the three Fur homologs in B. subtilis, Zur is the one least similar to the recently described E. coli Zur protein (28). However, since these proteins are both Fur homologs, and they appear to control similar functions, we propose to use the same designation, Zur.
FIG. 1.
Multiple sequence alignment of Fur-like regulatory proteins. B. subtilis encodes three Fur-like regulatory proteins: Fur (BsuFur), PerR (BsuPerR), and Zur (BsuZur) (5, 23). These proteins are aligned against the E. coli Fur (EcoFur) (32) and Zur (EcoZur) (33) proteins and a Fur-like regulatory protein from S. epidermidis (SepFur) (16) that is closely related to B. subtilis Zur. The amino-terminal domain contains a proposed helix-turn-helix motif with a conserved recognition helix. The carboxyl-terminal metal-binding domain contains a cluster of conserved His and Cys residues.
Zur controls the expression of an integral membrane protein, YciC.
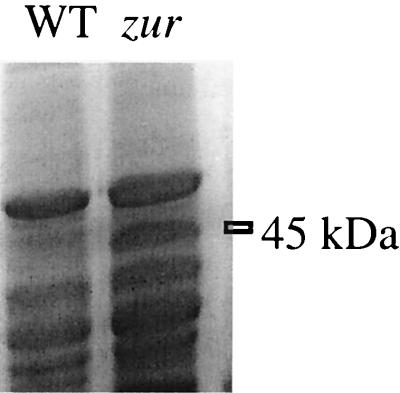
We hypothesized that zur might affect a metal homeostasis system involving one or more integral membrane proteins. When we compared the membrane protein profiles from wild-type and zur mutant cells grown in MM, we noted an abundant ∼45-kDa protein in the zur mutant that was absent from the wild type (Fig. 2) and the fur and perR mutants (data not shown). Expression of this protein is therefore controlled, either directly or indirectly, by the zur-encoded metalloregulatory protein.
FIG. 2.
Regulation of YciC by Zur. Membrane protein fractions from wild-type (WT) and zur mutant strains grown in minimal medium were fractionated by SDS-PAGE (12% gel) and stained with Coomassie blue. The abundant 45-kDa protein present in the zur mutant strain is indicated. Apart from YciC, no other changes in protein expression were visible on this gel.
Amino-terminal sequencing identified the 45-kDa protein as the product of the yciC gene. YciC is a 45.145-kDa protein with 43% identity to the P47K protein of Pseudomonas chlororaphis (26) and with similarity to CobW of Pseudomonas denitrificans (10). The roles of these proteins are not known. YciC is predicted to be an ATP-binding, integral membrane protein with at least one membrane-spanning region.
Transcription of yciC is repressed by zinc in the wild type but not in the zur mutant.
The promoter region of yciC was used to construct a lacZ transcriptional fusion on an SPβ prophage that was then transduced into isogenic wild-type and zur mutant backgrounds. On rich (LB) medium containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), the wild-type strain is white while the zur mutant is dark blue. We postulated that the Zur-dependent repression on LB required a metal ion. To identify the relevant regulatory ion(s), we first investigated yciC-lacZ expression on defined MM–X-Gal plates overlaid with an EDTA-soaked filter. In the wild-type strain, yciC-lacZ expression is strongly induced in the region flanking the zone of growth inhibition due to EDTA. This induction was blocked by the juxtaposition of filters containing Zn ion but not other metal ions. Interestingly, on LB plates yciC-lacZ is not induced around the zone of inhibition due to EDTA, indicating that chelation of a metal ion besides zinc limits growth on LB medium.
To quantify these effects of metal ion addition on yciC-lacZ regulation, we resuspended EDTA-washed, mid-exponential-phase cells in LZMM supplemented with 5 μM iron, manganese, cobalt, nickel, cadmium, copper, or zinc. Zinc was the most efficient at eliciting repression of the yciC-lacZ fusion (Fig. 3), although cadmium and nickel were also partially effective. In the zur mutant, yciC transcription was constitutive (Fig. 3).
FIG. 3.
Regulation of yciC as determined by using a yciC-cat-lacZ transcriptional fusion. Cells were grown overnight in MM containing either no additional metal ions or 5 μM indicated divalent cation, and β-galactosidase (Beta-gal) activity was determined. Expression of the yciC-cat-lacZ fusion is constitutive in the zur mutant strain HB8011 (right).
Zur binds specifically to the yciC promoter region.
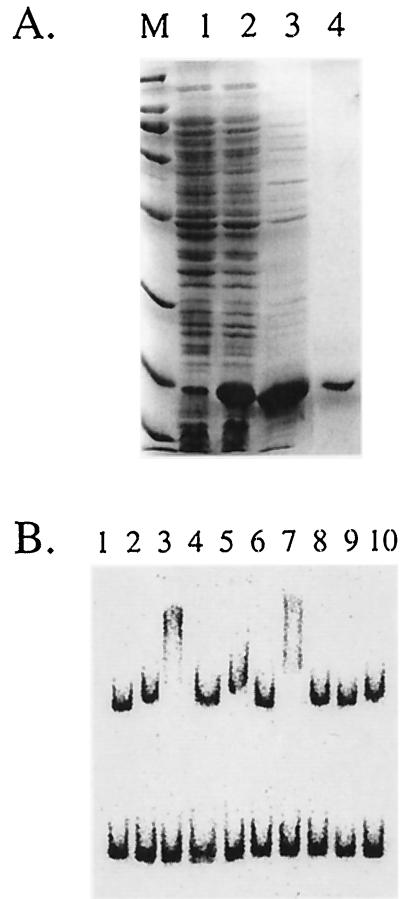
Zur was overproduced in E. coli under the control of T7 RNA polymerase and purified to homogeneity (Fig. 4A). Zur, as initially purified, bound to the yciC promoter region either with or without the addition of zinc but did not bind to control DNA fragments. Addition of 25 mM EDTA abolished binding, suggesting that a metal ion is required for binding. To further investigate this metal ion requirement, we prepared apoprotein by renaturation of protein in the presence of high concentrations of EDTA. The resulting protein binds to the yciC promoter region in the presence of either Zn2+ or Mn2+, as evidenced by the much slower migration of the yciC-containing DNA fragment (Fig. 4B). In contrast, addition of several other metal ions did not lead to the appearance of the slower-migrating complex. Zur, in either the presence or absence of metal ions, did not shift any of three other promoter fragments (Fig. 4B and data not shown). These data suggest that the interaction of Zur with the yciC promoter region is sequence specific.
FIG. 4.
Purification and DNA-binding selectivity of Zur. (A) SDS-PAGE analysis of Zur apoprotein purified from E. coli. Lanes: M, molecular weight markers (the last five bands are 66, 45, 31, 21, and 14 kDa; Zur runs just below the 21-kDa marker); 1, uninduced cell extract; 2, induced cell extract; 3, pooled fractions from QAE-Sepharose; 4, fraction from Superdex-75. (B) Electrophoretic mobility shift analysis of Zur apoprotein in the presence of yciC promoter fragments (top band) and an unrelated DNA fragment (from the B. subtilis yknW gene). Lane 1, DNA alone. Lanes 2 to 10 contain DNA plus 200 ng of Zur with 100 μM metal ions as follows: lane 2, none; lane 3, Zn2+; lane 4, Zn2+ plus 25 mM EDTA; lane 5, Cd2+; lane 6, Cu2+; lane 7, Mn2+; lane 8, Ni2+; lane 9, Co2+; lane 10, Fe3+. The formation of a protein-DNA complex is evident from slower migration of the yciC promoter fragment in lanes amended with Zn or Mn.
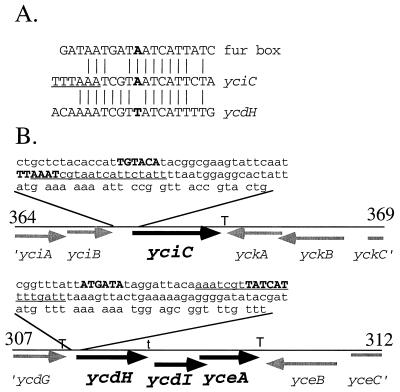
We used a restriction enzyme protection assay to localize the Zur-binding site within the yciC control region. Bound Zur specifically impedes cleavage at a unique DraI site located 36 bp upstream of the yciC start codon but does not affect cleavage at several other restriction sites within this fragment (data not shown). Upon inspection, we noticed a 12-of-19 match to the Fur box consensus sequence that overlaps this DraI site and is therefore a candidate for the Zur operator (Fig. 5A). Though low, this level of similarity is expected since Zur coexists in B. subtilis with Fur, which recognizes operators that closely match the Fur consensus and thereby regulates iron homeostasis (5). This putative operator site overlaps a candidate ςA-type promoter sequence (Fig. 5B).
FIG. 5.
(A) The yciC promoter region contains a Fur box-like sequence overlapping a DraI site (TTTAAA; site underlined) that is protected against digestion in presence of Zur. A similar operator-like sequence is found in the promoter region of the ycdH operon. (B) The genomic context of the yciC and ycdH ycdI yceA transcription units is illustrated. Each line represents a 5-kbp segment of the B. subtilis genome with boundaries indicated in kilobase pairs, based on the complete genome sequence (20). The DNA sequences preceding the yciC and ycdH genes are shown to illustrate putative ςA-like promoter elements (in bold) and the relative positions of the operator-like sequences (underlined). Proposed transcription terminator sites (T) and a possible termination site within the ycdH-containing operon (t) are indicated.
The ycdH-containing operon encodes an ABC transporter regulated by Zur.
When the sequence of the Zur operator region was used to search the B. subtilis genome (20, 24), a closely related site was identified immediately preceding the ycdH gene (Fig. 5A). The ycdH-containing operon (Fig. 5B) contains three genes (ycdH, ycdI, and yceA) that encode an ABC transporter most closely related to the Streptococcus pneumoniae AdcABC system involved in high-affinity zinc transport (11, 12). Specifically, YcdH is 42% identical to AdcA, a Zn(II)-binding lipoprotein, while YcdI is 50% identical to AdcC, the ATP-binding protein. YceA is 36% identical to the hydrophobic membrane protein AdcB. This suggests that the ycdH operon also encodes a Zn(II)-translocating ATPase.
Regulation of the ycdH operon was measured by using a ycdH-lacZ transcriptional fusion in wild-type and zur mutant strains grown in zinc-limited and zinc-replete MM. As shown for yciC, ycdH is repressed by zinc (see below) and this repression requires Zur (data not shown). The regulation of ycdH expression was almost identical to that of yciC expression: repression was achieved with 5 μM zinc, and partial repression was observed with 5 μM cadmium. In addition, Zur, in the presence of zinc, binds to the promoter region of the ycdH operon in an electrophoretic mobility shift assay, as noted for yciC (data not shown).
ycdH and yciC mutations affect zinc-limited growth.
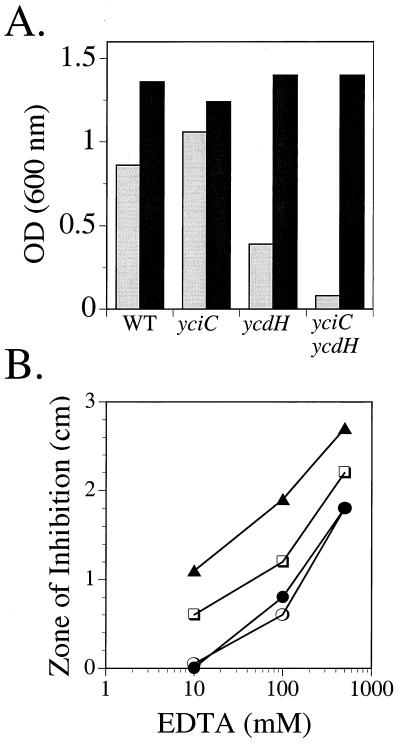
Growth studies suggest that the ycdH-encoded ABC transporter is important for zinc transport. In a zinc-deficient medium (LZMM), the ycdH mutant grew more slowly and with lower cell yield than the isogenic wild-type strain (Fig. 6A). This defect could be completely suppressed by addition of 10 μM Zn(II) but not by the addition of other metal ions, including Mn(II), Co(II), and Fe(III). In contrast, the yciC mutant grew as well as the wild type under these conditions.
FIG. 6.
Effects of Zn and EDTA on growth of wild-type (WT) and yciC, ycdH, and yciC ycdH mutant strains. (A) Cell optical density (OD) after overnight growth in LZMM (light bars) or LZMM supplemented with 10 μM Zn2+ (dark bars) is indicated for each strain. The results are representative of experiments performed three or more times. (B) Growth inhibition by EDTA. For each strain, the diameter of the zone of growth inhibition was determined on MM plates surrounding a 0.6-cm-diameter filter paper disk containing 10 μl of EDTA at either 10, 100, or 500 mM. Strains are wild type (○), yciC (•), ycdH (□), and yciC ycdH (▴). Values represent the overall diameter of inhibition minus the diameter of the filter and are reproducible to within 0.2 cm.
To determine if YciC plays a role in a lower-affinity zinc transport pathway, we compared the growth of the ycdH mutant with that of a ycdH yciC double mutant (Fig. 6). The yciC mutation clearly exacerbates the growth defect of the ycdH single mutant in LZMM, and this defect can be suppressed by added zinc (Fig. 6A). When zinc limitation was imposed by using EDTA, we again observed an increased sensitivity in the presence of both the ycdH and yciC mutations (Fig. 6B). Therefore, we suggest that YciC is a component of a low-affinity pathway whose role can be revealed in the absence of the high-affinity ABC transporter.
A ycdH mutation affects Zur-mediated repression.
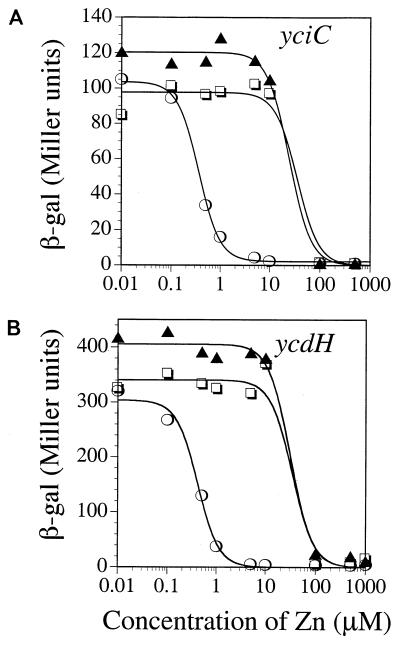
Zur is postulated to sense intracellular zinc to mediate the transcriptional repression of both the yciC and ycdH operons. This provides an indirect but powerful method of monitoring the intracellular zinc levels. Indeed, we find that mutants deficient in zinc transport are altered in Zur-mediated transcriptional control, as seen previously in analogous studies of high- and low-affinity zinc transporters in S. cerevisiae (13, 41, 42). In the case of yciC-lacZ (Fig. 7A), the concentration of Zn(II) needed for half-maximal repression is approximately 0.36 μM in the wild-type strain (determined by curve fitting; see Materials and Methods) but increases to 34 μM in the absence of the putative high-affinity system encoded by the ycdH operon (ycdH or ycdH yciC double mutant). Very similar results are seen with the ycdH-lacZ reporter fusion (Fig. 7B). This suggests that when zinc is internalized via the low-affinity pathway (in a ycdH mutant), 100-fold-higher levels of added zinc are needed to reach intracellular levels of zinc sufficient to activate Zur for DNA binding.
FIG. 7.
Effects of yciC and ycdH mutations on regulation. β-Galactosidase (β-gal) assays were performed for strains containing the yciC-cat-lacZ (A) or ycdH-cat-lacZ (B) transcriptional fusion after overnight growth in LZMM supplemented with Zn at the indicated concentration. The fusions were assayed in the wild-type (○) and the ycdH (□) and yciC ycdH (▴) mutant backgrounds. In the yciC mutant background, the results were indistinguishable from those for the wild type (not shown). The curves shown are best fits to the experimental data as described in Materials and Methods.
DISCUSSION
Although the essential role of zinc in cell growth is unquestioned, the regulation and mechanism of bacterial zinc transport have received little attention. We have now identified three of the key components mediating zinc homeostasis in B. subtilis. Zur is a Fur homolog that is necessary for the zinc-dependent repression of yciC and the ycdH-containing operon. The ycdH operon encodes an ABC transporter necessary for growth under conditions of zinc limitation. YciC appears to be important for growth only under the conditions of severe zinc limitation achieved by growth of a ycdH mutant on low-zinc medium. Since YciC is an integral membrane protein, we suggest that it may be part of an as yet uncharacterized zinc transport pathway.
Zur, one of three distinct Fur homologs in B. subtilis, is 35% identical to PerR and 24% identical to the ferric uptake repressor Fur (5). All three of these metalloregulators are also related to various Fur proteins from both gram-negative and gram-positive bacteria (Fig. 1). Zur is most similar (50% identity) to a Fur-like repressor of unknown function from S. epidermidis (16). Although Zur is functionally analogous to the recently described E. coli Zur protein (28), it is more closely related to E. coli Fur (29% identity) than to Zur (<25% identity).
It is likely that all members of the Fur family have a conserved structure with an amino-terminal DNA-binding domain and a carboxyl-terminal metal-binding domain (9, 36). Recent results indicate the presence of two metal-binding sites per monomer: a tetrahedral zinc site involving two of the conserved cysteine residues and at least one histidine, and a second regulatory site acting to sense divalent cation levels (19). The metal ion selectivity of Fur-like repressors varies and is presumably determined by the precise spatial arrangement of potential metal ligands around the regulatory site. In B. subtilis, the Fur-dependent repression of iron uptake genes is elicited only by iron, whereas the PerR-mediated repression of peroxide stress genes is elicited by either iron or manganese and weakly by other metal ions (5–7).
Zur has evolved the ability to sense zinc in vivo (Fig. 3), and both zinc and manganese can activate DNA binding in vitro (Fig. 4B). Presumably, intracellular concentrations of manganese are regulated such that manganese is not an effective corepressor in the cell. Zur may have evolved the ability to sense zinc by modification of the iron-sensing site characteristic of Fur repressor proteins. Alternatively, Zur may sense zinc by monitoring occupancy of the recently described zinc site that apparently plays a structural, rather than a regulatory, role in E. coli Fur (19).
Our analysis of Zur-mediated repression has identified the ycdH-containing operon as encoding a zinc-repressible ABC transporter important for growth in low-zinc medium. Both the regulation of the ycdH operon by zinc and the growth defect of the mutant suggest that this operon encodes a Zn(II)-translocating ATPase. The products of the ycdH operon are most similar to the AdcABC transporter of Streptococcus pneumoniae, which is also implicated in Zn(II) transport (11).
Zur also mediates the zinc-dependent repression of YciC, an abundant membrane protein. Regulation of yciC is at the transcriptional level and Zur binds specifically to the yciC regulatory region. YciC is closely related to a protein from P. chlororaphis (26) that is essential for nitrile hydratase production. In the absence of this protein, nitrile hydratase accumulates in inclusion bodies. This suggests a defect in protein folding, possibly due to the lack of a required metal cofactor. The amino-terminal region of YciC also displays significant similarity to P. denitrificans CobW, a protein involved in cobalamine biosynthesis (10). In each case, these proteins possess an ATP-binding motif and one predicted membrane-spanning region. A yciC mutation clearly impairs growth in cells lacking the high-affinity zinc transport system, and this growth defect is reversed by adding zinc. This finding indicates that YciC may be part of a low-affinity Zn(II) transport system, although direct measurements of zinc transport in wild-type and mutant strains will be required to test this hypothesis.
Zinc homeostasis in B. subtilis bears a number of striking similarities with that in S. cerevisiae. The yeast zinc-sensing metalloregulator, Zap1p, mediates the transcriptional activation (under low-zinc conditions) of both high-affinity (ZRT1) and low-affinity (ZRT2) zinc transporters (43). A ZRT2 mutation does not by itself affect zinc-limited growth, but it exacerbates the growth defect of a ZRT1 mutant (42). Finally, a ZRT1 mutation leads to a 75-fold increase in the level of external zinc necessary to mediate transcriptional control (41). In this case, it was demonstrated that the level of cell-associated (presumably internal) zinc necessary to inactivate Zap1p was unchanged. This is similar to the effect of the ycdH mutation on transcriptional repression by Zur (Fig. 7).
Mechanisms contributing to zinc homeostasis in bacteria are receiving increasing attention. While bacterial zinc uptake has long been known to be an energy-dependent process, the corresponding transport machinery has only recently been identified. In several bacteria, zinc uptake appears to be mediated by an ABC-type transporter. Examples include the AdcABC transporter in S. pneumoniae (11) and the recently described E. coli ZnuACB transporter (28). It seems likely that the periplasmic zinc-binding protein found in H. influenzae (21) is part of a similar system. Excess zinc also leads to the induction of homeostasis mechanisms. Well-characterized examples include zinc-inducible efflux systems, such as the E. coli ZntA (3, 30) and the Synechocystis strain PCC 6803 ZiaA P-type ATPases or, in the case of cyanobacteria, metallothionein (14, 39). It is interesting that zinc uptake seems to be regulated by Fur-like repressor proteins (reference 28 and this study), while zinc efflux or sequestration is controlled by proteins of the SmtB (ArsR) family (8, 38, 39). Further characterization of Zur, and its interactions with both operator DNA and metal ions, will help clarify the molecular basis of zinc homeostasis in B. subtilis.
ACKNOWLEDGMENTS
We thank N. Bsat for construction of the zur mutant and overproducing plasmid, T. Santangelo for assistance with Zur purification, and A. Herbig, N. Bsat, and Q. Que for helpful suggestions and comments on the manuscript.
This work was supported by grant MCB9630411 from the National Science Foundation.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagg N, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard S J, Hashim R, Membrillo-Hernandez J, Hughes M N, Poole R K. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol Microbiol. 1997;25:883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x. [DOI] [PubMed] [Google Scholar]
- 4.Bsat N. Regulation of iron uptake systems in Bacillus subtilis by an iron-specific Fur protein. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1998. [Google Scholar]
- 5.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtilis contains multiple Fur homologs: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, James L P, Helmann J D. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially regulated by metal ions. J Bacteriol. 1993;175:5428–5437. doi: 10.1128/jb.175.17.5428-5437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Keramati L, Helmann J D. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci USA. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook W J, Kar S R, Taylor K B, Hall L M. Crystal structure of the cyanobacterial metallothionein repressor SmtB: a model for metalloregulatory proteins. J Mol Biol. 1998;275:337–346. doi: 10.1006/jmbi.1997.1443. [DOI] [PubMed] [Google Scholar]
- 9.Coy M, Neilands J B. Structural dynamics and functional domains of the Fur protein. Biochemistry. 1991;30:8201–8210. doi: 10.1021/bi00247a016. [DOI] [PubMed] [Google Scholar]
- 10.Crouzet J, Levy-Schil S, Cameron B, Cauchois L, Rigault S, Rouvez M C, Blanche F, Debussche L, Thibaut D. Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanyltransferase. J Bacteriol. 1991;173:6074–6087. doi: 10.1128/jb.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dintilhac A, Alloing G, Granadel C, Claverys J P. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement of Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 12.Dintilhac A, Claverys J P. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res Microbiol. 1997;148:119–131. doi: 10.1016/S0923-2508(97)87643-7. [DOI] [PubMed] [Google Scholar]
- 13.Eide D. Molecular biology of iron and zinc uptake in eukaryotes. Curr Opin Cell Biol. 1997;9:573–577. doi: 10.1016/s0955-0674(97)80036-1. [DOI] [PubMed] [Google Scholar]
- 14.Erbe J L, Taylor K B, Hall L M. Metalloregulation of the cyanobacterial smt locus: identification of SmtB binding sites and direct interaction with metals. Nucleic Acids Res. 1995;23:2472–2478. doi: 10.1093/nar/23.13.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster S J. Analysis of autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1992;174:464–470. doi: 10.1128/jb.174.2.464-470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidrich C, Hantke K, Bierbaum G, Sahl H-G. Identification and analysis of a gene encoding a Fur-like protein of Staphylococcus epidermidis. FEMS Microbiol Lett. 1996;140:253–259. doi: 10.1111/j.1574-6968.1996.tb08345.x. [DOI] [PubMed] [Google Scholar]
- 17.Helmann J D. Metal cation regulation in gram positive bacteria. In: Walden W E, Silver S, editors. Metal ions in gene regulation. New York, N.Y: Chapman & Hall; 1997. pp. 45–76. [Google Scholar]
- 18.Hofmann K, Stoffel W. TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 19.Jacquamet L, Aberdam D, Adrait A, Hazemann J-L, Latour J-M, Michaud-Soret I. X-ray absorption spectroscopy of a new zinc site in the Fur protein from Escherichia coli. Biochemistry. 1998;37:2564–2571. doi: 10.1021/bi9721344. [DOI] [PubMed] [Google Scholar]
- 20.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borris R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Duesterhoeft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo M F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee S M, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Mellado R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S H, Parro V, Pohl T M, Portetelle D, Porwollik S, Prescott A M, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror S J, Serror P, Shin B S, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 21.Lu D, Boyd B, Lingwood C A. Identification of the key protein for zinc uptake in Haemophilus influenzae. J Biol Chem. 1997;272:29033–29038. doi: 10.1074/jbc.272.46.29033. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. Experiments in molecular genetics, p. 352–355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Mizuno M, Masuda S, Takemaru K-I, Hosono S, Sato T, Takeuchi M, Kobayashi Y. Systematic sequencing of the 283 kb 210°–232° region of the Bacillus subtilis genome containing the skin element and many sporulation genes. Microbiology. 1996;142:3103–3111. doi: 10.1099/13500872-142-11-3103. [DOI] [PubMed] [Google Scholar]
- 24.Moszer I, Glaser P, Danchin A. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology. 1995;141:261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 25.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins Struct Funct Genet. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 26.Nishiyama M, Horinouchi S, Kobayahi M, Nagasawa T, Yamaka H, Beppu T. Cloning and characterization of genes responsible for metabolism of nitrile compounds from Pseudomonas chlororaphis B23. J Bacteriol. 1991;173:2465–2472. doi: 10.1128/jb.173.8.2465-2472.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmiter R D. Regulation of metallothionein genes by heavy metals appears to be mediated by a zinc-sensitive inhibitor that interacts with a constitutively active transcription factor, MTF-1. Proc Natl Acad Sci USA. 1994;91:1219–1223. doi: 10.1073/pnas.91.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patzer S I, Hantke K. The ZnuABC high affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 29.Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- 30.Rensing C, Mitra B, Rosen B P. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Schäffer S, Hantke K, Braun V. Nucleotide sequence of the iron regulatory gene fur. Mol Gen Genet. 1985;200:110–113. doi: 10.1007/BF00383321. [DOI] [PubMed] [Google Scholar]
- 33.Silver S. Transport of inorganic cations. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1091–1102. [Google Scholar]
- 34.Silver S, Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992;56:195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slack F J, Mueller J P, Sonenshein A L. Mutations that relieve nutritional repression of the Bacillus subtilis dipeptide permease operon. J Bacteriol. 1993;175:4605–4614. doi: 10.1128/jb.175.15.4605-4614.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stojilkovic I, Hantke K. Functional domains of the Escherichia coli ferric uptake regulator protein (Fur) Mol Gen Genet. 1995;247:199–205. doi: 10.1007/BF00705650. [DOI] [PubMed] [Google Scholar]
- 37.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 38.Thewell C, Robinson N J, Turner-Cavet J S. An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc Natl Acad Sci USA. 1998;95:10728–10733. doi: 10.1073/pnas.95.18.10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner J S, Glands P D, Samson A C R, Robinson N J. Zn2+-sensing by the cyanobacterial metallothionein repressor SmtB: different motifs mediate metal-induced protein-DNA dissociation. Nucleic Acids Res. 1996;24:3714–3721. doi: 10.1093/nar/24.19.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong A, Jayaswal R K. Molecular characterization of a chromosomal determinant conferring resistance to zinc and cobalt ions in Staphylococcus aureus. J Bacteriol. 1998;180:4024–4029. doi: 10.1128/jb.180.16.4024-4029.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci USA. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao H, Eide D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem. 1996;271:23203–23210. doi: 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- 43.Zhao H, Eide D J. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:5044–5052. doi: 10.1128/mcb.17.9.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuber P, Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987;169:2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]