Abstract
Objective
The consequences of mutations in genes associated with monogenic forms of diabetes on human pancreas development cannot be studied in a time-resolved fashion in vivo. More specifically, if recessive mutations in the insulin gene influence human pancreatic endocrine lineage formation is still an unresolved question.
Methods
To model the extremely reduced insulin levels in patients with recessive insulin gene mutations, we generated a novel knock-in H2B-Cherry reporter human induced pluripotent stem cell (iPSC) line expressing no insulin upon differentiation to stem cell-derived (SC-) β cells in vitro. Differentiation of iPSCs into the pancreatic and endocrine lineage, combined with immunostaining, Western blotting and proteomics analysis phenotypically characterized the insulin gene deficiency in SC-islets. Furthermore, we leveraged FACS analysis and confocal microscopy to explore the impact of insulin shortage on human endocrine cell induction, composition, differentiation and proliferation.
Results
Interestingly, insulin-deficient SC-islets exhibited low insulin receptor (IR) signaling when stimulated with glucose but displayed increased IR sensitivity upon treatment with exogenous insulin. Furthermore, insulin shortage did not alter neurogenin-3 (NGN3)-mediated endocrine lineage induction. Nevertheless, lack of insulin skewed the SC-islet cell composition with an increased number in SC-β cell formation at the expense of SC-α cells. Finally, insulin deficiency reduced the rate of SC-β cell proliferation but had no impact on the expansion of SC-α cells.
Conclusions
Using iPSC disease modelling, we provide first evidence of insulin function in human pancreatic endocrine lineage formation. These findings help to better understand the phenotypic impact of recessive insulin gene mutations during pancreas development and shed light on insulin gene function beside its physiological role in blood glucose regulation.
Keywords: Insulin, Endocrinogenesis, β cell, Monogenic diabetes, iPSC differentiation, Islet composition
Highlights
-
•
Insulin-deficient SC-islets exhibit low insulin receptor signaling but display increased insulin receptor sensitivity.
-
•
Insulin shortage has no effects on human endocrine lineage induction.
-
•
Lack of insulin alters the endocrine cell composition of SC-islets.
-
•
Insulin deficiency reduces the proliferation of SC-β cells.
1. Introduction
The endocrine pancreas regulates glucose homeostasis, and its malfunction leads to diabetes mellitus. The pancreatic endocrine mini-organs, called the islet of Langerhans, comprise different hormone-producing cell types, including α cells (glucagon+), β cells (insulin+), ẟ cells (somatostatin+), PP cells (pancreatic polypeptide+) and ε cells (ghrelin+). The overall balance of the levels and function of the hormones secreted from these cells regulates blood glucose. Therefore, the composition of distinct endocrine cell types is critical for the optimal function of islets that is defined during endocrine lineage formation or endocrinogenesis [[1], [2], [3]]. During pancreas development, multipotent pancreatic progenitors (PPs) give rise to endocrine progenitors marked by transient expression of the key transcription factor (TF) neurogenin-3 (NEUROG3; NGN3) [[4], [5], [6], [7]]. A combination of signaling pathways and gene regulatory networks drive the differentiation of endocrine progenitors towards hormone-expressing endocrine cell types (Figure 1A), which form the islets of Langerhans [[8], [9], [10], [11], [12]]. Yet, how endocrine lineages are segregated from human PPs is not well understood. Several upstream signals that regulate endocrine cell induction have been reported [8,13,14], however, how islet cell composition is determined during human development is less well understood. Elucidating signals and factors that drive α and β cell fate specification will not only help to bioengineer islet clusters from pluripotent stem cells in vitro but also allows to decipher the pathomechanisms of monogenic forms of diabetes by iPSC disease modelling.
Figure 1.
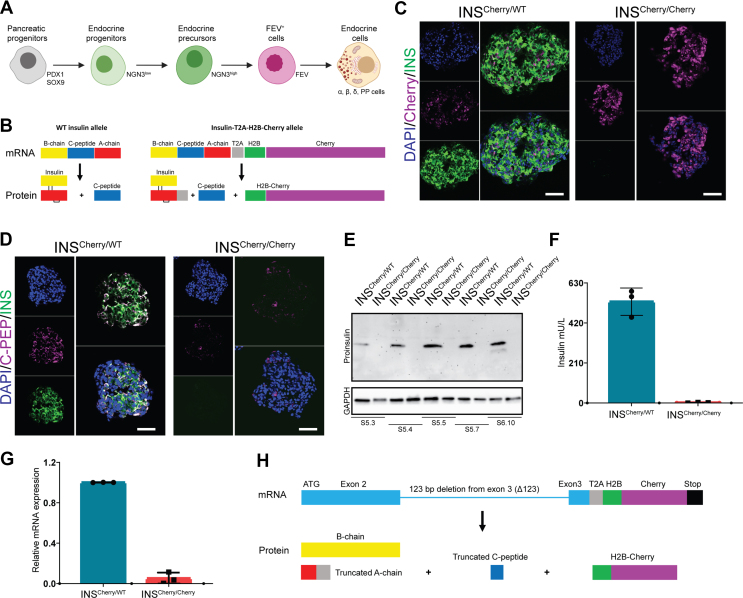
Generation and characterization of the INSCherry/Cherry(INSΔ123/Δ123) reporter iPSC line. A) Schematic picture of stepwise pancreatic endocrine lineage formation. B) Schematic representation of proinsulin mRNA and the expected translated and processed protein from WT INS and INS-T2A-H2B-Cherry alleles. C) INS and H2B-Cherry expression in INSCherry/WT and INSCherry/Cherry SC-islets. Scale bar, 50 μm. D) INS and C-peptide expression in INSCherry/WT and INSCherry/Cherry SC-islets. Scale bar, 50 μm. E) Western blot showing the expression of proinsulin in INSCherry/WT and INSCherry/Cherry SC-islets from S5.3 to S6.10. F) Total insulin from INSCherry/WT vs INSCherry/Cherry SC-islets at S6.21. G) qPCR analysis of INS mRNA expression in SC-islets at S5.7. H) Schematic picture of 123 base pair (bp) deletion in exon 3 of INS mRNA from INSCherry/Cherry cell line, and the expected translated peptides. Schemes were created with BioRender.com. Data are represented as mean ± SD.
Mutations in single genes involved in endocrine cell formation and/or function cause monogenic diabetes, which accounts for 1–5% of diabetic individuals [15]. Two major types of monogenic diabetes are neonatal diabetes mellitus (NDM) and maturity-onset diabetes of the young (MODY) [16,17]. Mutations in more than 30 genes, such as pancreas/duodenum homeobox protein 1 (PDX1), potassium channel, inwardly rectifying subfamily J member 11 (KCNJ11), hepatocyte nuclear factor 4-alpha (HNF4A) and insulin (INS) have been identified to cause monogenic diabetes [[18], [19], [20], [21]]. Among these, insulin is the main physiological regulator of blood glucose homeostasis and thus carries a central role in the pathophysiology of all types of diabetes. β cells synthesize the prohormone proinsulin in the endoplasmic reticulum (ER), which then undergoes proper folding in the ER and enzymatic processing in immature secretory granules (SGs) to produce insulin and C-peptide that are secreted from mature SGs upon glucose stimulation [22,23]. A variety of mutations in the INS locus have been identified as a major cause of monogenic diabetes [21]. Large numbers of these mutations are autosomal dominant that cause mutant INS-gene-induced diabetes of youth (MIDY). MIDY patients often carry dominant negative heterozygous missense mutations that generate misfolded proinsulin, which accumulates in β cells and trigger ER-stress and eventually lead to ER stress-mediated β cell failure or death [24,25]. Another set of INS mutations are recessive homozygous mutations, which frequently result in reduced insulin biosynthesis [26]. Because the decreased insulin synthesis initiates upon the birth of β cells during pancreas development and endocrine cell formation, the patients carrying these mutations exhibit reduced growth and birth weight and are diagnosed earlier compared to patients with MIDY [21,26]. The impact of autosomal dominant and recessive INS mutations on the prevalence of early onset of diabetes after birth has been relatively well studied. However, it is not well described how such mutations autonomously affect pancreas development, endocrine cell formation and β cell characteristics.
Here we employed a human iPSC in vitro differentiation system to explore the impact of loss of the insulin gene product on human endocrinogenesis. We generated a reporter iPSC line producing no insulin when differentiated towards stem cell-derived β cells (SC-β). This iPSC model enabled us to study the effects of insulin and insulin signaling deficiency on human endocrine lineage formation and SC-β cell characteristics in vitro. Our results uncovered that the lack of insulin blunts insulin receptor (IR) signaling in stem cell-derived islets (SC-islets). However, upon administration of exogenous insulin, insulin-deficient SC-islets showed increased IR sensitivity. Importantly, insulin shortage elevated the number of SC-β cells at the expense of SC-α cells and reduced SC-β cell proliferation. These findings not only demonstrate the effect of altered insulin levels on human endocrinogenesis but also implicate the possible existence of insulin signaling feedback loops to determine islet cell composition during human development.
2. Materials and methods
2.1. Cell sources
Episomal reprogrammed HMGUi001 [27] and the heterozygous hiPSC-INS-T2A-H2B-Cherry reporter (INSCherry/WT) [28] iPSC line were used. The homozygous hiPSC-INS-T2A-H2B-Cherry reporter iPSC line (INSCherry/Cherry) was generated with the similar strategy according to Blöchinger et al., 2020 [28]. All the iPSC lines were confirmed to be mycoplasma-free using the Lonza MycoAlert Mycoplasma Detection Kit (Lonza, catalog no. LT07-418).
2.2. Characterization of INSCherry/Cherry iPSCs
Karyotyping of the INSCherry/Cherry iPSC clones was executed during cell growth in a logarithmic phase. Cells at passage number 29 were incubated with colcemid for 2 h, and then trypsinized and treated with hypotonic solution (0.075 M KCL) for 20 min and finally were fixed with methanol/acetic acid (3:1). Metaphase chromosomes from INSCherry/Cherry cells were classified using the standard G banding technique. The final karyotype is based on the average of 85 % of around 20 methaphases. To test the multipotency of the INSCherry/Cherry iPSC clone, cells were differentiated in monolayers to endodermal, mesodermal and ectodermal cells with the StemMACS™ Trilineage Differentiation Kit (Miltenyi Biotec, Cat# 130-115-660). Immunohistochemistry was used to analyze the differentiated cells (antibody list is provided in Table S1). The obtained clones (HMGUi001-A-43) were registered at the European Human Pluripotent Stem Cell Registry (hPSCreg®): https://hpscreg.eu/cell-line/HMGUi001-A-43.
2.3. In vitro differentiation of iPSCs toward pancreatic endocrine cells
We cultured iPSCs on 1:30 diluted Geltrex (Invitrogen, catalog no. A1413302) in StemMACS iPS-Brew medium (Miltenyi Biotec, catalog no. 130-104-368). At around 70 % confluency, cultures were rinsed with PBS without Mg2+ and Ca2+ (Invitrogen, catalog no. 14190) followed by incubation with accutase (Gibco) for 3 min at 37 °C. Single cells were then rinsed with iPS-Brew and centrifuged at 1200 rpm for 3 min. The pellets were suspended in iPS-Brew medium supplemented with Y-27632 (10 μM; Sigma-Aldrich, catalog no. Y0503) and the single cells were seeded at ∼1.5–2 × 105 cells per cm2 on Geltrex-coated surfaces for maintenance. For 3D differentiation, cells were seeded at 4-5 × 106 cells per well in ultra-low attachment (ULA) plates, placed in a shaking platform at 60 rpm or 1 × 106 cells per ml in a stirring spinner flask (ABLE corporation) stirring at 60 rpm. We fed the cells every day with iPS-Brew medium. 3D differentiations in ULA plates were started 24 h following cell seeding. For spinner flask, differentiation was started as soon as ∼80 % or more aggregates achieved a size of ∼150 μM–∼200 μM that usually was obtained 72 h after seeding.
To differentiate iPSC into SC-islets, the protocol from Velazco-Cruz et al., 2019 was used [29]. Briefly, cells were subjected to differentiate to definitive endoderm (DE) using 500 ml MCDB131 medium, using a stage 1 (S1) base media for 3 days. The medium was supplemented with 2 % bovine serum albumin (BSA) (Sigma, catalog no. 10775835001), 1x glutamax (Gibco, catalog no. A12860-01), 0.25 mM ascorbic acid (Sigma, catalog no. A4544-25G), 1 % P/S, 0.22 g glucose (MilliporeSigma; G7528), 1.23 g sodium bicarbonate (MilliporeSigma; S3817) and, ITS-X (Invitrogen; 51500056). S1 media was also supplemented with 100 ng/ml Activin A (R&D Systems; 338-AC) and 3 μM Chir99021 (Stemgent; 04-0004-10) on the first day. For the next 2 days, the S1 media was supplemented with 100 ng/ml activin A. Afterwards, cells were differentiated into primitive gut tube using the stage 2 (S2) media for 3 days. This includes 500 ml of MCDB131 with 2 % BSA, 1x glutamax (Gibco, catalog no. A12860-01), 0.25 mM ascorbic acid (Sigma, catalog no. 120-14-300), 0.22 g glucose, 0.615 g sodium bicarbonate, 10 μL ITS-X, 50 ng/ml KGF (Peprotech; AF-100-19) and 1 % P/S. Next, cells were differentiated toward pancreatic endoderm using the stage 3 media (S3) for 1 day. This media includes 500 ml of MCDB131 supplemented with 1x Glutamax, 0.22 g of glucose, 0.615 g sodium bicarbonate, 2 % BSA, 2.5 mL ITS-X, 0.25 mM ascorbic acid, 1 % P/S, 50 ng/ml KGF, 200 nM LDN193189, (Reprocell; 040074), 500 nM PdBU (MilliporeSigma; 524390), 2 μM retinoic acid (MilliporeSigma; R2625), 0.25 μM Sant1 (MilliporeSigma; S4572) and 10 μM Y27632. To generate pancreatic progenitors, the stage 4 (S4) media was used for 5 days. This includes 500 ml of MCDB131 supplemented with 1x glutamax, 0.22 g of glucose, 0.615 g sodium bicarbonate, 2 % BSA, 2.5 mL ITS-X, 0.25 mM ascorbic acid, 1 % P/S, 5 ng/mL activin A, 50 ng/mL KGF, 0.1 μM retinoic acid, 0.25 μM SANT1, and 10 μM Y27632. To generate endocrine progenitors and lineages the stage 5 media was used for 7 days. This includes 500 mL MCDB131 with 0.877 g sodium bicarbonate, 2 % BSA, 1.8 g glucose, 2.5 mL ITS-X, 5 mL glutaMAX, 22 mg vitamin C, 1 % P/S, and 5 mg heparin (MilliporeSigma; A4544). This media was also supplemented with 10 μM ALK5i II (Enzo Life Sciences; ALX-270-445-M005), 1 μM XXI (MilliporeSigma; 595790), 20 ng/mL betacellulin (R&D Systems; 261-CE-050), 1 μM T3 (Biosciences; 64245), 0.1 μM retinoic acid and 0.25 μM SANT1. To further differentiate endocrine cells the stage 6 media (ESFM media) was utilized for up to 35 days. This includes 500 mL MCDB131 supplemented with 2 % BSA, 5.2 mL glutaMAX, 1 % P/S, 5 mg heparin, 0.23 g glucose, 5.2 mL MEM nonessential amino acids (Corning; 20-025-CI), 84 μg ZnSO4 (MilliporeSigma; 10883), 523 μL trace elements A (Corning; 25-021-CI), and 523 μL trace elements B (Corning; 25-022-CI).
For the insulin-depleted experiment, cells were cultured in a modified S5 media. The S5 I + T + S–E media included 500 mL MCDB131 with 0.877 g sodium bicarbonate, 2 % BSA, 1.8 g glucose, 861 nM insulin (sigma-aldrich), 30 nM transferrin human (sigma; T8158-100 MG), 19 nM sodium selenite (sigma; S5261 – 10G) and 16.4 mM ethanolamines (sigma-aldrich), 5 mL glutaMAX, 22 mg vitamin C, 1 % P/S, and 5 mg heparin (MilliporeSigma; A4544). This media was also supplemented with 10 μM ALK5i II (Enzo Life Sciences; ALX-270-445-M005), 1 μM XXI (MilliporeSigma; 595790), 20 ng/mL betacellulin (R&D Systems; 261-CE-050), 1 μM T3 (Biosciences; 64245), 0.1 μM retinoic acid and 0.25 μM SANT1.The second modified S5 media (S5 T + S + E) was prepared as the above media but without 861 nM insulin (sigma-aldrich). To reaggregate SC-islets, clusters at S5.7 were incubated with accutase (Gibco) for 5–8 min, followed by the inactivation and removal of accutase by adding ESFM media and centrifuged at 1200 rpm for 3 min. Pellets were resuspended with ESFM and passed through a 40 μm nylon cell strainer (Corning; 431752), and cultured in ESFM in 6-well plates on an orbi-shaker (Benchmark) set at 100 RPM. Some assessment assays were performed between 11 and 21 days of S6 unless otherwise stated.
2.4. Magnetic activated cell sorting (MACS) of SC-β cells
SC-islets at S6 were collected and stained for CD49a antibody. 10 μl of CD49a antibody (conjugated to PE) was added per 1 × 106 cells in a 100-μl volume of ESFM media. Samples were incubated in the dark for 30 min at RT. For magnetic labeling of the antibody, stained cells were washed 2× with PBS and then suspended in 80 μl of ESFM media with 20 μl of anti-PE microbeads per 10 × 106 cells for 15 min at 4 °C. Next, cells were washed with 5 ml PBS (2x) and resuspended at 10 × 106 cells in 500 μl of ESFM media before proceeding with magnetic sorting. For magnetic sorting of CD49a+ cells, the LS column was placed in the magnetic sorter. The column was rinsed with 3 ml PBS and 500 μl cell suspension was applied where the flow-through containing unlabeled cells was discarded after washing 3x with 3 ml PBS. The antibody-positive cells were collected by removing the column from the separator, adding the final 5 ml of S5 media and pushing the CD49a+ cells through by using the top part of the LS columns. Cells were quantified and seeded with pancreatic progenitor cells in S5 media supplemented with 10 μM Y-compound at a seeding density of 2–3 × 106 cells per well of a 6 well ultra-low plate to form aggregates.
2.5. Electron microscopy
SC-β cell-enriched aggregates were fixed in 4 % formaldehyde (freshly prepared from paraformaldehyde prills) in 100 mM phosphate buffer pH 7.4 for 2 h at RT. For embedding into epoxy resin, the samples were processed according to a modified protocol using osmium tetroxide (OsO4), thiocarbohydrazide (TCH), and again OsO4 to generate enhanced membrane contrast [30,31]. In brief, samples were postfixed overnight in modified Karnovsky fixative (2 % glutaraldehyde/2% formaldehyde in 50 mM HEPES, pH 7.4), followed by post-fixation in a 2 % aqueous OsO4 solution containing 1.5 % potassium ferrocyanide and 2 mM CaCl2 (30 min on ice), washed in water, 1 % TCH in water (20 min at RT), washed in water and a second osmium contrasting step in 2 % OsO4/water (30 min on ice). Samples were washed in water and en-bloc contrasted with 1 % uranyl acetate/water for 2 h on ice, washed again in water, and dehydrated in a graded series of ethanol/water mixtures (30 %, 50 %, 70 %, 90 %, 96 %), followed by three changes in pure ethanol on molecular sieve. Samples were infiltrated into a 1/1 mixture of the epon substitute EMBed 812 and araldite. Samples were incubated in resin/ethanol mixtures (1:3, 1:1, 3:1) for 1 h each, followed by pure resin overnight and for another 5 h, finally embedded in flat embedding molds, and cured at 65 °C overnight. Ultrathin sections (70 nm) were prepared with a Leica UC6 ultramicrotome (Leica Microsystems, Wetzlar, Germany) using a diamond knife (Diatome, Nidau, Switzerland), collected on formvar-coated slot grids, and stained with lead citrate [32] and uranyl acetate. All sections were analyzed on a JEM 1400Plus transmission electron microscope (JEOL, Freising, Germany) at 80 kV and images were taken with a Ruby digital camera (JEOL).
2.6. Proteomics analysis
2.6.1. Sample preparation
Differentiated human stem cells/islets were washed 2x in PBS and prepared for liquid chromatography-mass spectrometry (LC-MS/MS) as described previously [33]. In brief, samples were lysed in SDC lysis buffer (2 % SDC, 100 mM Tris-HCl pH 8.5) at 95 °C for 10 min at 1000 rpm, sonicated in high mode (30 s OFF, 30 s ON) for 10 cycles (Bioruptor® Plus; Diagenode). Bicinchoninic acid (BCA) assay was used to determine the protein concentration and 25 μg of protein were used for further analysis. Samples were incubated with TCEP and CAA (final concentrations of 10 mM and 40 mM, respectively) at 45 °C for 10 min with 1000 rpm shake in dark. Protein digestion was done with trypsin and LysC (1:40, protease:protein ratio) overnight at 37 °C, 1000 rpm shake. Later, peptides were acidified with isopropanol including 2 % TFA with 1:1 volume-to-volume ratio. Desalting and purification of the acidified samples were done in custom-made StageTips, which were prepared with three layers of styrene divinylbenzene reversed-phase sulfonate (SDB-RPS; 3 M Empore) membranes. Peptides were loaded on the activated (100 % ACN, 1 % TFA in 30 % Methanol, 0.2 % TFA, respectively) StageTips, run through the SDB-RPS membranes, and washed with EtOAc including 1 % TFA, isopropanol including 1 % TFA, and 0.2 % TFA, respectively. Peptides eluted from the membranes with 60 μl elution buffer (80 % ACN, 1.25 % NH4OH) were dried by vacuum centrifuge (40 min at 45 °C) and reconstituted in 6 μl of loading buffer (2 % ACN, 0.1 % TFA). Peptide concentration was estimated via optical measurement at 280 nm (Nanodrop 2000; Thermo Scientific).
2.6.2. LC-MS/MS analysis
0.5 μg of peptides were analyzed using the EASY-nLC 1200 (Thermo Fisher Scientific) connected to Orbitrap Exploris 480 Mass Spectrometer (Thermo Fisher Scientific) and a nano-electrospray ion source (Thermo Fisher Scientific). FAIMS Pro™ with Orbitrap Fusion ™ module was used with two different compensation voltages (CV) (−50V and −70V). Peptides were loaded at 60 °C onto a 50 cm, 75 μm inner diameter, in-house packed HPLC column with 1.9 μm with C18 Reprosil particles (Dr. Maisch GmbH). A binary buffer system [0.1 % formic acid (buffer A) and 80 % can, 0.1% formic acid (buffer B)] at a flow rate of 300 nl/min was used over a 60 min gradient: 5–20 % buffer B over 30 min, 20–29 % buffer B over 9 min, 29–45 % buffer B over 6 min, 45–95 % buffer B over 5 min, wash with 95 % buffer B for 5 min and 95-5% buffer B over 5 min. MS data were acquired in DIA (data-independent acquisition) mode using a 2 s cycle time scan method. Full scan MS targets were in a 300–1650 m/z scan range with 3 × 106 charges and data was acquired in 120000 at m/z 200 resolution with 45 ms maximum injection time. Precursor ions for MS/MS scans were fragmented by higher-energy C-trap dissociation (HCD) with a normalized collision energy of 30. MS/MS scan sets were 15000 at m/z 200 resolution with an ion target value of %1000 and a maximum injection time of 22 ms.
2.6.3. Proteome data processing and analysis
The raw data were searched with Spectronaut version 15.7 in directDIA mode (library-free approach) and all searches were performed against the human Uniprot FASTA database (2020). MaxLFQ settings enabled protein level label-free quantification. MaxQuant contaminant fasta file was included in the search. Default settings were kept if not stated otherwise. Perseus (version 1.6.14.0) was used for bioinformatics analysis. Data were visualized using Perseus, Adobe Illustrator or GraphPad Prism (version 9.0).
In Perseus, the raw data were filtered for “Maxquant contaminants”. Quantified proteins were filtered for at least two valid values among biological replicates in at least one condition. Missing values were imputed (Gaussian normal distribution with a width of 0.3 and a downshift of 1.8) in order to obtain PCA, hierarchical clusters, volcano plots and to apply the t-test. Two-sample test with student t-test statistics (permutation-based FDR = 0.1; s0 = 0) was performed in Perseus and significant proteins were hierarchical clustered by applying Euclidean as a distance measure for row clustering after normalization of median protein abundances of biological replicates by z-score. Fisher's Exact GOBP, GOCC, GOMF and KEGG Term enrichment of significant proteins was performed against human database (gene list with 21846 entries) in Perseus (Benjamini Hochberg FDR truncation with 0.05 threshold value).
2.7. RNA isolation, cDNA preparation and qPCR analysis
We extracted total RNA using miRNeasy mini kit (Qiagen). Extracted RNA was reverse transcribed utilizing the SuperScript Vilo cDNA and cDNA synthesis kit (Life Technologies-Thermofisher Scientific). qPCR was performed using predesigned TaqMan™ probes (Life Technologies, Table S1) and 15 ng of cDNA per reaction. Each reaction consisted of 4.5 μL cDNA in nuclease-free water, 5 μL TaqMan™ Advanced master mix (Life Technologies) and 0.5 μL TaqMan probe™ (Life Technologies). qPCR was performed using Viia7 (Thermo Fisher Scientific). Ct-values were normalized among samples, transformed to linear expression values, normalized on reference genes and on control samples. Samples were normalized to the housekeeping gene GAPDH.
2.8. RT-PCR analysis of XBP-1 mRNA splicing
RNA was isolated from whole cells and reverse transcribed to obtain total cDNA. XBP-1 primers (antisense, 5′-TCCTTCTGGGTAGACCTCTGG-3′ and sense, 5′-AAACAGAGTAGCAGCTCAGACTGC-3) were used to amplify an XBP-1 amplicon spanning the 26-nt intron in a regular 3-step PCR. The PCR products were then digested overnight using PstI-HF overnight and samples were run on agarose gel and were analyzed.
2.9. Flow cytometry
Cell aggregates were dissociated to a single cell suspension with accutase for 10–20 min at 37 °C. The accutase was inactivated and discarded by washing with iPSC Brew media and then centrifugation at 1200 rpm for 3 min. Pellets were washed with PBS (1x) and fixed with 4 % paraformaldehyde for 10 min. Samples were permeabilized with donkey blocking solution (0.1 % tween-20, 10 % FBS, 0.1 % BSA, 0.2 % Triton-X100 and 3 % donkey serum), followed by staining with primary antibodies (Table S1) diluted in the same permeabilization solution for 1 h at RT or at 4 °C overnight. When no conjugated antibody was used, the protocol continued with incubation of appropriate secondary antibodies (Table S1) for 30 min for 1 h at RT. Stained cells were then washed with PBS (3x) followed by performing flow cytometry using FACS-Aria III (BD Bioscience). FACS gating was determined using isotype, secondary only antibody and stained hiPSCs. FACS data were analyzed using FlowJo. For quantification of median fluorescence intensity (MFI) for H2B-Cherry, the H2B-Cherry-positive population was first gated and then MFI was calculated using BD FACS software.
2.10. Western blotting
For western blotting, aggregates were homogenized in RIPA buffer containing protease and phosphatase inhibitors. Cell lysates were then resolved by SDS-PAGE, transferred to PVDF membranes (BioRad), and incubated with blocking solution for 30 min at RT while rotating. Membranes were incubated with primary antibodies (Table S1) overnight at 4 °C. Next day, samples were washed 3x with PBS and they were incubated with HRP-conjugated secondary antibodies (Table S1) for 1–2 h at RT while rotating. After washing with PBS (3x), protein bands were resolved using a chemiluminescence reagent (Bio-rad, Cat #1705061) reacting with HRP conjugated antibodies. Bands were processed and quantified with ImageJ.
2.11. Immunostaining, confocal microscopy and imaging
To prepare cryosections, 20–30 SC-islets were harvested and washed with PBS followed by fixation using 4 % paraformaldehyde (PFA) for 20–30 min at RT. Samples underwent a dehydration process by sequentially incubation in 10 % and 30 % sucrose solutions for 2 h each. Next, cells were incubated in a mixed solution of 30 % sucrose and OCT (1:1) overnight at 4 °C. The following day, SC-islets were placed in a plastic mold embedded in OCT, to slow freezing on dry ice to generate appropriate size OCT blocks that were stored at 80 °C. Cryosections were generated by using a Leica cryostat. Three sections of 10 μm were placed on a glass slide (Thermo Fisher Scientific), dried at RT and stored at −20 °C. For immunostaining, cryosections were rehydrated by washing 3x with PBS. Sections were next permeabilized with 0.2 % Triton X-100 in H2O for 30 min and then blocked in blocking solution (PBS, 0.1 % Tween-20, 1 % donkey serum, 5 % FCS) for 1 h. The samples were incubated with the primary antibody (Table S1) diluted in the same blocking solution O/N at 4 °C. After 3x washing with PBS, samples were incubated with secondary antibodies (Table S1) diluted in blocking solution for 2–3 h at RT. Sections were then stained for DAPI (1:500 in PBS) for 30 min, rinsed and washed 3x with PBS and mounted.
For attached cell monolayer, dispersed cells at different stages were first prepared as single cell solutions and plated utilizing μ-Slide 8-well plates (ibidi), left O/N for their attachment to the surface and form a cell monolayer. Samples were fixed and stained as described for the cryosections. Pictures were captured using a Leica DMI 6000 microscopy using LAS AF software. Images were analyzed and quantified using LAS AF and ImageJ software programs.
2.12. Aggresome detection
To detect misfolded and aggregated proteins, we utilized the PROTEOSTAT® Aggresome Detection Reagent (Enzo life sciences). SC-islets were cultured in μ-Slide 8-well plates (ibidi) in the absence or presences of 5 μM MG132 for 6 h. After fixing the cells with 4 % PFA (10 min at RT), samples were permeabilized (0.5 % Triton X-100, 3 mM EDTA, pH 8.0) followed by incubation with the PROTEOSTAT® dye (Enzo life science) for 30 min. Samples were then washed 2x with PBS and were stained with primary and secondary antibodies and embedded as described.
2.13. Statistical analysis
Comparison of three or more datasets was performed using ordinary one-way analysis of variance (ANOVA) with multiple comparison test. For two sample comparisons two-tailed unpaired t-test (Student's t-test) assuming equal standard deviation were used. All statistics were performed using GraphPad Prism software 9.
2.14. Data availability
The mass spectrometry proteomics data produced in this study have been deposited to the ProteomeXchange Consortium via the PRIDE [34] partner repository with the dataset identifier PXD045366.
3. Result
3.1. Generation of an insulin knock-out H2B-Cherry knock-in iPSC reporter cell line
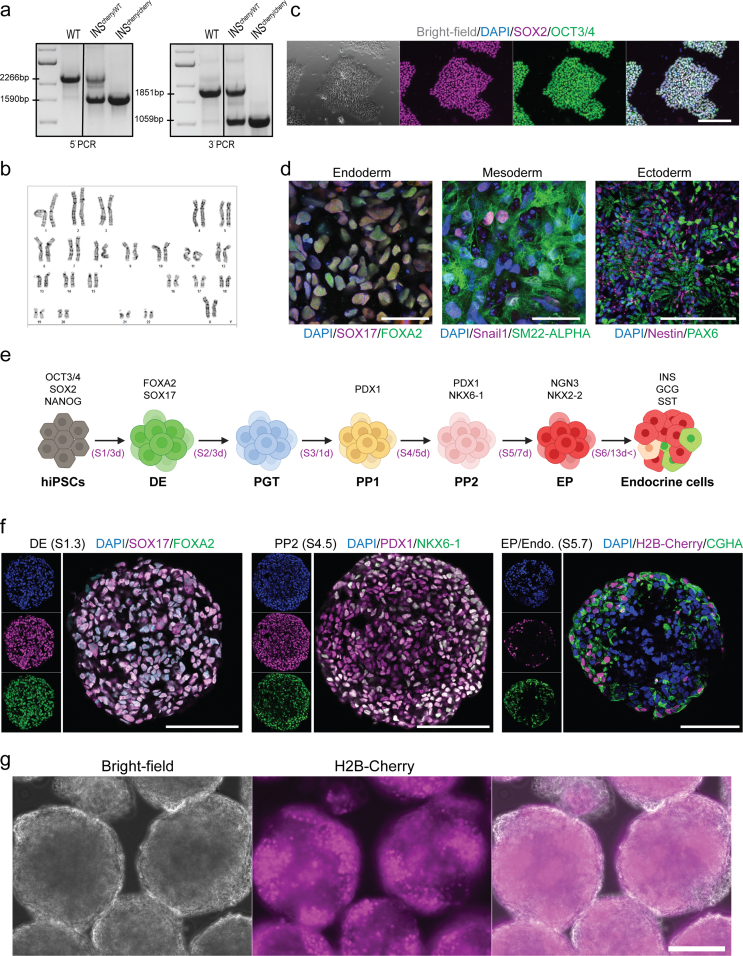
To track human β-cell formation and function we previously generated an iPSC line using an insulin T2A co-translational H2B-Cherry reporter [28]. In this iPSC line, the stop codon of terminal exon 3 of one of the endogenous insulin (INS) allele was replaced by a T2A-histone 2B(H2B)-Cherry sequence (INSCherry/WT) in order to produce equimolar of INS-T2A and H2B-Cherry transcripts and proteins (Figure 1B). Therefore, SC-β cells were fluorescently labeled due to the expression of nuclear Cherry [28]. We applied the CRISPR/Cas9 strategy and generated a line with homozygous H2B-Cherry reporter activity with a normal karyotype and the capability to differentiate into all three germ layers, i.e., endoderm, mesoderm and ectoderm (Figs. S1a–d). The homozygous clone (INSCherry/Cherry) efficiently gave rise to pancreatic endocrine lineages in a multi-step in vitro differentiation system and showed strong H2B-Cherry reporter activity (Figs. S1e–g). Surprisingly, H2B-Cherry+ cells from the INSCherry/Cherry clone revealed no detectable insulin immunoreactivity in endocrine cells (Figure 1C). However, very low levels of C-peptide (C-PEP) were detectable in a few H2B-Cherry+ cells (Figure 1D). We further confirmed the lack of insulin protein using Western blot analysis (Figure 1E). Measuring total insulin content by a very sensitive enzyme-linked immunosorbent assay (ELISA) showed no detectable signal in endocrine clusters from the INSCherry/Cherry clone (Figure 1F). Together, these data indicate that we generated a functional insulin knock-out H2B-Cherry knock-in reporter iPSC line.
To identify the possible genetic alteration underlying the lack of insulin protein production in the INSCherry/Cherry stem cell-derived islets (SC-islets), we first sequenced the genomic DNA and confirmed the correct sequence and integration of the T2A-H2B-Cherry reporter cassette. We then checked if the non-mutated INS-T2A-H2B-Cherry genomic sequence produces the intact INS mRNA containing the exon 1, 2 and 3. qPCR analysis of differentiated endocrine cells at S6 uncovered a striking reduction in INS mRNA levels in the INSCherry/Cherry clone compared to the INSCherry/WT SC-islets (Figure 1G). Next, we performed reverse transcription PCR (RT-PCR) using primer pairs against different regions of INS mRNA and applied the resulting products to Sanger sequencing. This analysis identified a deletion of 123 bp from exon 3 of the INS mRNA (while having intact genomic sequence) in the INSCherry/Cherry clone (hereafter INSΔ123/Δ123). The deleted sequence encodes for a major part of C-peptide and A-chain of proinsulin, resulting in a predicted truncated and likely non-functional unstable version of insulin that cannot be detected using insulin antibodies (Figure 1H). However, the deletion was predicted to not cause a frameshift mutation, and thus the H2B-Cherry co-translation was not affected.
3.2. Insulin-deficient SC-islets contain SC-β cells with no mature insulin secretory granules (SGs)
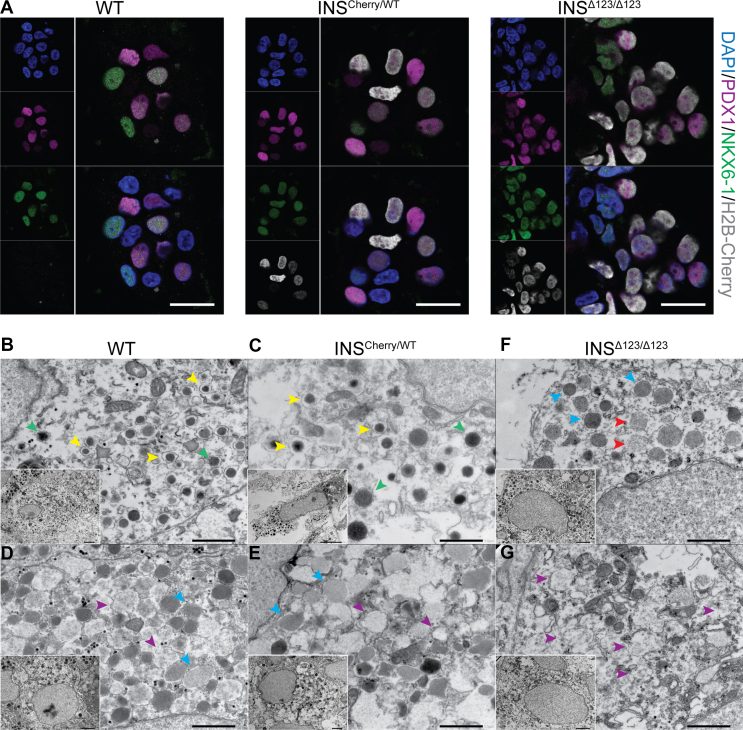
We next characterized the SC-islets derived from the INSΔ123/Δ123 cells. Immunostaining against Chromogranin A (CHGA) as the pan-endocrine marker disclosed the endocrine cell identity of H2B-Cherry+ cells in both INSCherry/WT and INSΔ123/Δ123 SC-islets at S6.14 (Fig. S2a). Furthermore, all H2B-Cherr+ cells from INSCherry/WT and INSΔ123/Δ123 SC-islets were also positive for both PDX1 and NKX6-1 TFs (labeling differentiated SC-β cells at this stage), confirming their β cell identity (Figure 2A).
Figure 2.
Characteristics of SC-β cells derived from the INSΔ123/Δ123iPSC line. A) Representative immunofluorescence pictures showing the presence of cells co-expressing PDX1, NKX6-1 and H2B-Cherry in SC-islets at S6.14. Scale bar, 20 μm. B-G) Electron microscopy imaging depicting WT and INSCherry/WT SC-β cells containing mature SGs (yellow arrowheads) and TSGs (green arrowheads) and INSΔ123/Δ123 SC-β-cells containing low-dense SGs with scattered electron density (red arrowheads). Immature SGs (blue arrowheads) and vesicles with very low/no electron density (purple arrowheads) are also shown. Scale bars: 1 μm (for inserts 2 μm).
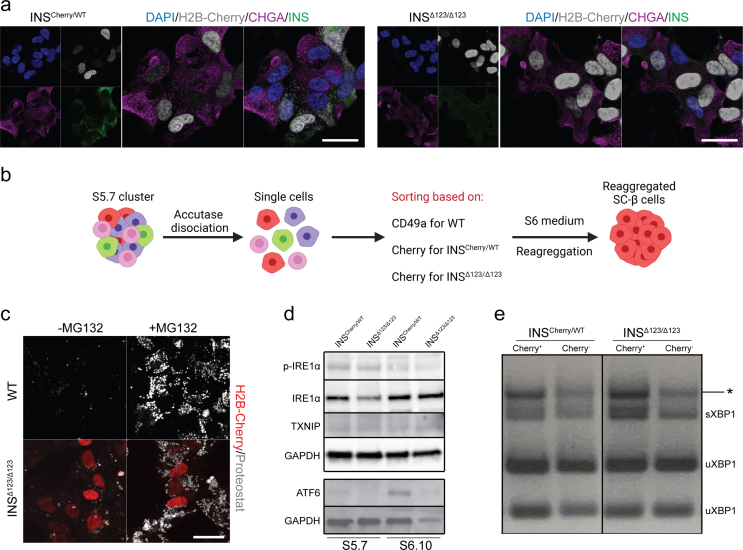
After enzymatic processing, insulin monomers bind to Zn2+ to form insulin hexamers and form mature secretory granules (SGs). Mature SGs are dense cores vesicles surrounded by wide electron-lucent halos when analyzed by transmission electron microscopy (TEM). In comparison, immature SGs containing proinsulin have no electron-lucent halos. During the processing of proinsulin to insulin by convertases, immature SGs exhibit thin electron-lucent halos and are referred to immature transforming SGs (TSGs) [35,36]. To explore the formation of SGs in the in vitro differentiated SC-β cells lacking the intact insulin protein, we performed TEM. We sorted SC-β cells at endocrine stage (S) 5.7 using Cherry fluorescent activity (for INSCherry/WT and INSΔ123/Δ123 cells) or CD49a antibody (for WT cells), which enriches for SC-β cells [37] and reaggregated the isolated cells for two weeks in S6 culture medium (Fig. S2b). A high number of WT SC-β cells enclosed mature SGs (yellow arrowheads) and TSGs (green arrowheads) (Figure 2B). Compared to the WT cells, the SC-β cells from the INSCherry/WT clone contained slightly lower number of mature SGs and TSGs (Figure 2C). Moreover, a fraction of SC-β cells from the WT and INSCherry/WT clones contained high numbers of immature SGs (blue arrowheads) and vesicles with very low electron density (purple arrowheads) (Figure 2D,E). In comparison, SC-β cells from the INSΔ123/Δ123 iPSC clone contained no mature SGs and only few cells encompassed immature SGs and vesicles with weak scattered electron density (red arrowheads) (Figure 2F). However, majority of the SC-β cells from the INSΔ123/Δ123 clone comprised no SGs but only vesicles with very weak electron density (Figure 2G). These data demonstrate that lack of insulin in SC-β cells leads to absence of mature dense core SGs.
Numerous missense mutations in the insulin gene have been identified, leading to proinsulin misfolding and aggregation that induce ER-stress in β cells [24,25]. To test whether the putative truncated insulin products in INSΔ123/Δ123 SC-islets accumulate intracellularly, we employed the thioflavin T-derivative Proteostat (intercalating with protein aggregate-associated quaternary structures) to detect aggresomes, which are inclusion bodies formed by assembly of misfolded proteins [38,39]. As a positive control, we treated SC-islets with the proteasome inhibitor MG132 to trigger protein misfolding. Laser confocal microscopy analysis of WT and INSΔ123/Δ123 SC-islets revealed comparable levels of aggresomes in both cell types, irrespective of the presence or absence of MG132 (Fig. S2c). We also assessed the expression levels of several ER stress-related proteins through Western blotting. Our results demonstrated no discernible differences between INSCherry/WT and INSΔ123/Δ123 SC-islets (Fig. S2d). Consistently, analysis of X-box binding protein 1 (XBP1) splicing revealed no alterations in the INSΔ123/Δ123 SC-islets compared to those from INSCherry/WT clone (Fig. S2e). Collectively, these analyses demonstrate that there is neither residual proinsulin aggregation nor increased ER stress in the INSΔ123/Δ123 SC-β cells.
3.3. Insulin-depleted SC-islets show reduced insulin signaling but exhibit increased insulin receptor sensitivity
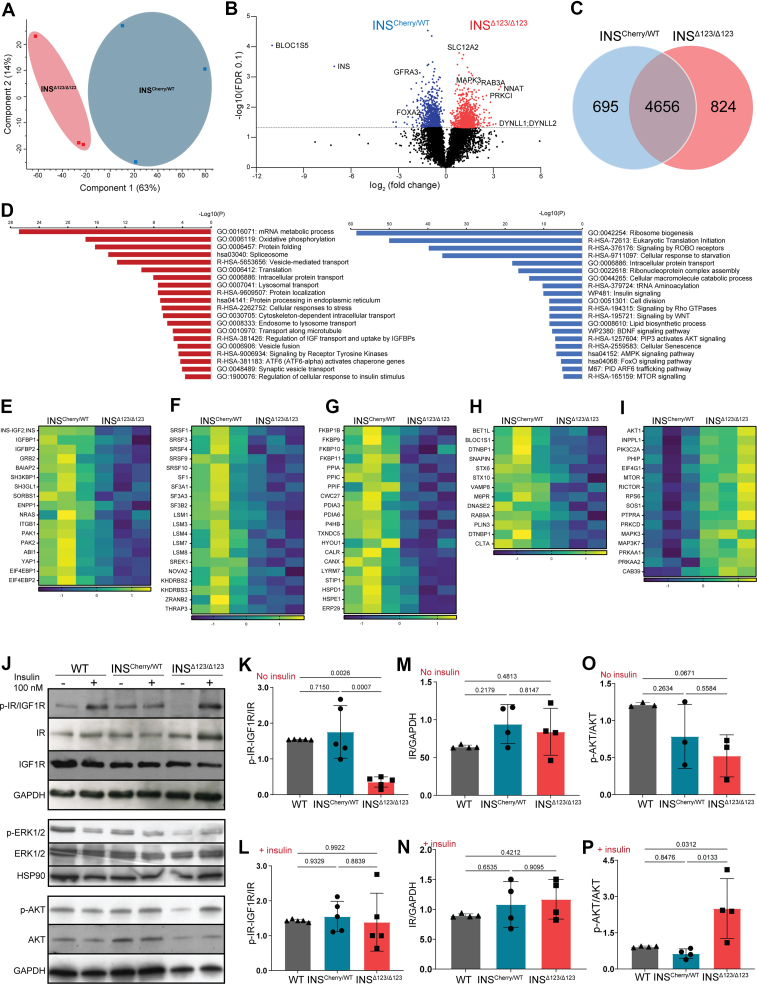
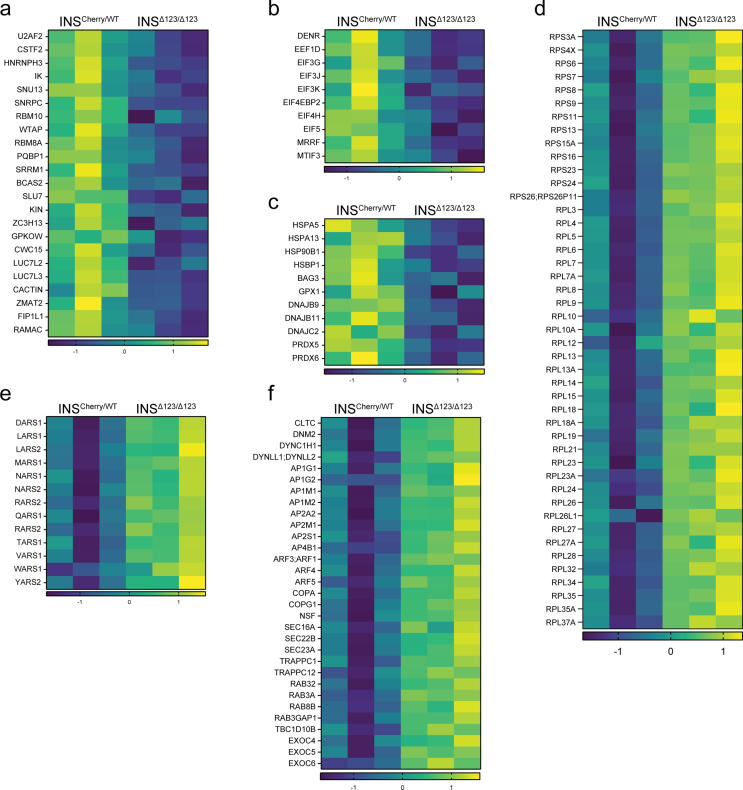
To unravel the molecular impact of insulin deficiency from early born SC-β cells on endocrine differentiation, we performed quantitative proteomics analysis of SC-islets at end of endocrine induction at S5.7. We subjected INSCherry/WT and INSΔ123/Δ123 samples from three independent differentiations to liquid chromatography-tandem mass spectrometry (LC-MS/MS) and quantified more than 5900 proteins. Principal component analysis (PCA) revealed a clear separation of the INSCherry/WT and INSΔ123/Δ123 SC-islets along the first principal component plane (accounting for 63 % of the variance), indicating a clear difference in their proteome. The second component also showed the expected sample variability in both cell lines, likely due to differences in differentiation rates of independent experiments (Figure 3A). We identified 1519 significantly different proteins with permutation-based at a false discovery rate (FDR) of 0.1 using both-sided Student's t-test, with 695 proteins downregulated and 824 proteins upregulated when comparing INS Δ123/Δ123 to INSCherry/WT SC-islets (Figure 3B,C and Table S1). As expected, one of the most downregulated proteins was INS, further confirming that the INSΔ123/Δ123 SC-islets do not produce intact insulin. We conducted pathway enrichment analysis of differentially synthesized proteins using the Metascape annotation and analysis resource [40]. Most of the downregulated proteins were involved in molecular and cellular processes associated with mRNA splicing and translation, protein processing and folding, intracellular transport, as well as insulin/receptor tyrosine kinase signaling (Figure 3D and Table S1). We observed downregulation of several proteins involved in insulin and receptor tyrosine kinase pathways, including IGFBP1/2, GRB2, SORBS1, ENPP1, PAK1/2 and YAP1 in the INSΔ123/Δ123-derived SC-islets (Figure 3E). Furthermore, several proteins involved in mRNA splicing and processing were downregulated, including the serine/arginine-rich splicing factors (SRSF1/3/4/9/10), U6 snRNA-associated Sm-like proteins (LSM3/4/7/8), and splicing factors (SF3A1/A3/B2, U2AF2) (Figure 3F and S3a). This indicates the reduced necessity for spliceosome activity likely resulting from the decreased levels of mutant INS transcripts coding for the main protein product in INSΔ123/Δ123 SC-β cells. Additionally, we found the downregulation of translation-associated proteins, such as DENR, EIF3J, EIF4H and EIF5, indicating the decline in protein translation machinery due to the lack of properly processed and spliced INS mRNA (Fig. S3b). In support of reduced protein synthesis, we also detected lower expression levels of several protein folding enzymes, including peptidyl-prolyl cis-trans isomerases (FKBP/9/10/11/1B, PPIA/C/F, CWC27) and protein disulfide-isomerases (PDIA3/6, P4HB, TXNDC5), as well as several chaperones (HYOU1, CALR, CANX, LYRM7, STIP1, HSPD1/E1, ERP29) (Figure 3G). Consistently, we identified decreased levels of proteins related to cellular response to baseline stress, including HSPA5/13, HSP90B1, BAG3, GPX1, DNAJB9/11, DNAJC2, and PRDX5/6, highlighting the reduced ER stress in INSΔ123/Δ123 SC-islets resulting from the decline in insulin synthesis (Fig. S3c), in line with what was previously reported [41]. Finally, we found downregulation of several proteins that coordinate the trafficking of insulin granules, including SNAREs (BET1l, VAMP8, STX6/10) and SNARE-associated proteins (BLOC1S1, DTNBP1, SNAPIN) (Figure 3H). Overall, these analyses demonstrate reduced mRNA processing, protein synthesis and folding, and granule trafficking in the INSΔ123/Δ123 SC-islets.
Figure 3.
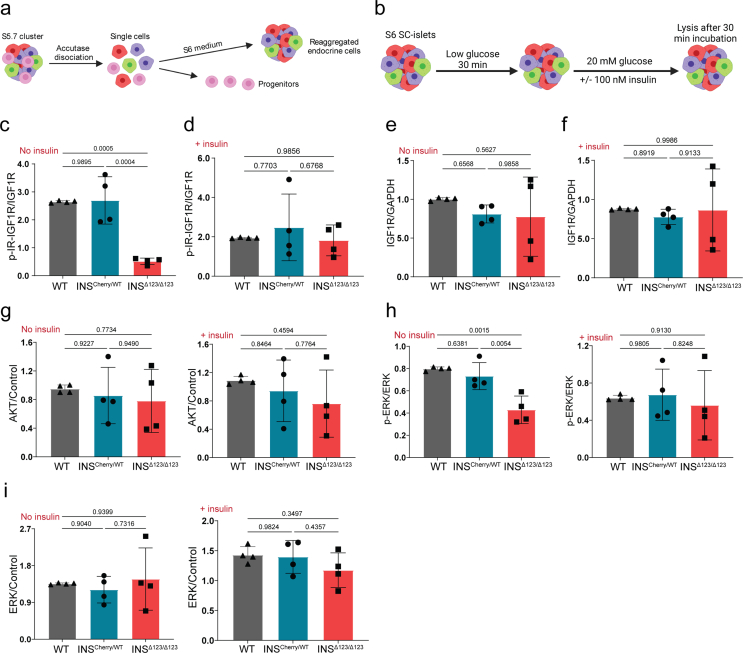
INSΔ123/Δ123SC-islets exhibit reduced IR signaling. A) Principal component analysis (PCA) of INSCherry/WT and INSΔ123/Δ123 SC-islets at S5.7. B) Volcano plot showing the log2 fold changes of proteins with respect to the log of FDR (0.1). C) Differentially upregulated and downregulated proteins in INSΔ123/Δ123 vs INSCherry/WT SC-islets. D) Selected terms from the pathway analysis of downregulated and upregulated proteins in INSΔ123/Δ123 SC-islets. E-H) Heat maps of significantly downregulated proteins associated with insulin signaling, spliceosome, protein folding and vesicle trafficking in INSΔ123/Δ123 compared INSCherry/WT SC-islets s. I) Heat map of significantly upregulated proteins associated with intracellular signaling proteins in INSΔ123/Δ123 compared INSCherry/WT SC-islets. J) Representative Western blot picture showing the phosphorylation of IR, IGF1R, and AKT from WT, INSCherry/WT and INSΔ123/Δ123 SC-islets at S6.21 treated with 20 mM glucose in the absence and presence of 100 nM exogenous insulin. K-N) Quantifications of phosphorylated IR-IGF1R and the total IR in SC-islets treated with 20 mM glucose in the absence (No insulin) and presence (+insulin) of exogenous insulin. O, P) Quantifications of phosphorylated AKT in SC-islets treated with 20 mM glucose in the absence (No insulin) and presence (+insulin) of exogenous insulin. Data are normalized to WT (K–P). All statistics have been done using one-way ANOVA multiple comparisons test. Data are represented as mean ± SD.
The lack of insulin synthesis resulted in an overexpression of proteins prominently involved in protein translation, protein transport, and several signaling pathways (Figure 3D and Table S1). We observed increased expression levels of a large number of 40S and 60S ribosomal proteins (RPSs and RPLs) (Fig. S3d). Moreover, the expression levels of several amino acid-tRNA ligases, including LARS, TARS1, VARS1 and YARS1 were increased (Fig. S3e). These enzymes ligate tRNA to leucine, threonine, valine and tyrosine, which are among the most frequently used amino acids generating proinsulin. The increase in the levels of RPSs, RPLs and amino acid-tRNA ligases likely stems from the activation of compensatory mechanisms in response to the loss of intact insulin synthesis in the INSΔ123/Δ123 SC-β cells. We also found elevated levels of intracellular trafficking proteins, including those involved in endocytosis and retrograde transport (AP2A2/M1/S1, CLTC, DNM2, DYNC1H1), trans-Golgi and endosomes sorting (AP1G1/M1/M2, AP4B1, ARF1/3/4/5), ER-Golgi transport (COPA/G1, NSF, SEC16A/22B/23A, TRAPPC1/12), and several RABs and their regulatory proteins (RAB3A/8B/32, RAB3GAP1, TBC1D10B) (Fig. S3f). Furthermore, we identified increased levels of several intracellular signaling proteins, including INPPL1, AKT1, PIK3C2A, PRKAA1, PRKAA2, MTOR, RICTOR, MAPK3, and MAP3K7 (Figure 3I). These proteins are involved with pathways associated or downstream of insulin receptor (IR) signaling, including PI3K, MTOR, MAPK, and AMPK. This proteomic analysis suggests that INSΔ123/Δ123 SC-islets respond to the loss of insulin signaling by overexpressing the downstream effector proteins and increased insulin sensitivity and protein translation, potentially trying to compensate for insulin loss.
The proteomics analysis suggests that INSΔ123/Δ123 SC-islets respond to the loss of insulin signaling by overexpressing the downstream effector proteins and increased insulin sensitivity. To test this directly, we performed Western blot analysis of the IR and IGF1R signaling and the downstream signaling effectors on SC-islets that were reaggregated at S5.7 (Fig. S4a). We starved the reaggregated cells at S6.21 in low glucose for 30 min and then incubated them with 20 mM glucose for 30 min followed by sample analysis (Fig. S4b). In WT and INSCherry/WT SC-islets, this resulted in auto- and paracrine insulin signaling assessed by IR/IGF1R phosphorylation. In comparison, we found a striking reduction of the phosphorylated IR/IGF1R in the INSΔ123/Δ123 SC-islets (Figure 3J,K and S4c). Administration of exogenous insulin led to comparable levels of phosphorylated IR/IGF1R in the INSΔ123/Δ123 SC-islets compared to the WT and INSCherry/WT cells (Figure 3J, L and S4d). Importantly, we found a tendency of increased levels of total IR but not IGF1R in the INSCherry/WT and INSΔ123/Δ123 SC-islets compared to the WT cells (Figure 3J, M, N and S4e, f). Consistently, we detected lower levels of phosphorylated AKT in the INSΔ123/Δ123 SC-islets compared to the WT cells. However, adding exogenous insulin resulted in striking increased in AKT phosphorylation in the INSΔ123/Δ123 SC-islets compared to the WT and INSCherry/WT cells (Figure 3J, O, P and S4g). Furthermore, the phosphorylation levels of the downstream effector ERK1/2 were also reduced in the INSΔ123/Δ123 SC-islets (Figure 2J and S4h, i), confirming the lack/decline of IR/IGF1R signaling in the INSΔ123/Δ123 cells. This analysis validated the proteomics data confirming that the insulin knock-out cells exhibit reduced IR signaling in response to glucose but show increased IR sensitivity when treated with exogenous insulin.
3.4. Insulin is dispensable for human endocrine lineage induction
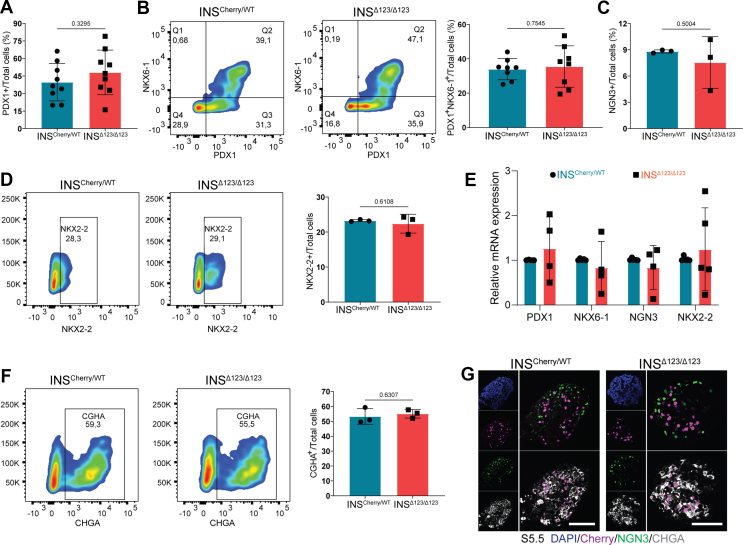
The functional role of insulin and IR signaling deficiency during human endocrinogenesis has not been studied so far. Therefore, we first investigated the impact of loss of insulin on the induction of human pancreatic and endocrine progenitor cells. Analysis of differentiated cells at S3, corresponding to pancreatic progenitor 1 (PP1) stage, revealed a comparable number of PDX1-expressing progenitors between INSCherry/WT and INSΔ123/Δ123 cells (Figure 4A). Consistently, we found a similar number of PP2 cells expressing both PDX1 and NKX6-1 in INSCherry/WT and INSΔ123/Δ123 clusters at S4.5 (Figure 4B). Next, we assessed the rate of endocrine progenitor formation. FACS analysis disclosed equal numbers of cells expressing endocrine progenitor markers, NGN3 and NKX2-2, between INSCherry/WT and INSΔ123/Δ123 clones at S5.5 (Figure 4C,D), demonstrating that insulin is dispensable for human endocrine lineage induction. This conclusion was further supported by detecting comparable levels of PDX1, NKX6-1, NGN3 and NKX2-2 mRNA in S5 clusters (Figure 4E). Additionally, we analyzed the differentiation potential of endocrine progenitors toward endocrine cells. We found a similar percentage of cells expressing pan-endocrine marker CHGA in the INSΔ123/Δ123 cells compared to the INSCherry/WT SC-islets at the end of S5 (Figure 4F,G). Collectively, these analyses demonstrate that there are no significant alterations in human pancreatic and endocrine cell induction, as well as endocrine progenitor differentiation toward endocrine cells, in the absence of insulin.
Figure 4.
Lack of insulin has no impact on human endocrine lineage induction. A) FACS quantification of total PDX1-positive cells at PP1 stage. B) Representative FACS plots and quantification of total PDX1 and NKX6-1 double positive-cells at S4.5. C) FACS quantification of total NGN3+ cells from INSΔ123/Δ123 vs INSCherry/WT SC-islets at S5.5. D) Representative FACS plots and quantification of total NKX2-2+ cells at S5.5. E) qPCR analysis of SC-islets at S5. F) Representative FACS plots of total CHGA cells at endocrine cell induction stage S5.5 from INSΔ123/Δ123 vs INSCherry/WT SC-islets. G) Immunofluorescence pictures showing NGN3 and CHGA-positive cells at S5.5. Scale bar, 80 μm. Data are normalized to INSCherry/WT (C, D). All statistics have been done using two-sided unpaired t-test. Data are represented as mean ± SD.
3.5. Lack of insulin skews human endocrine cell composition in vitro
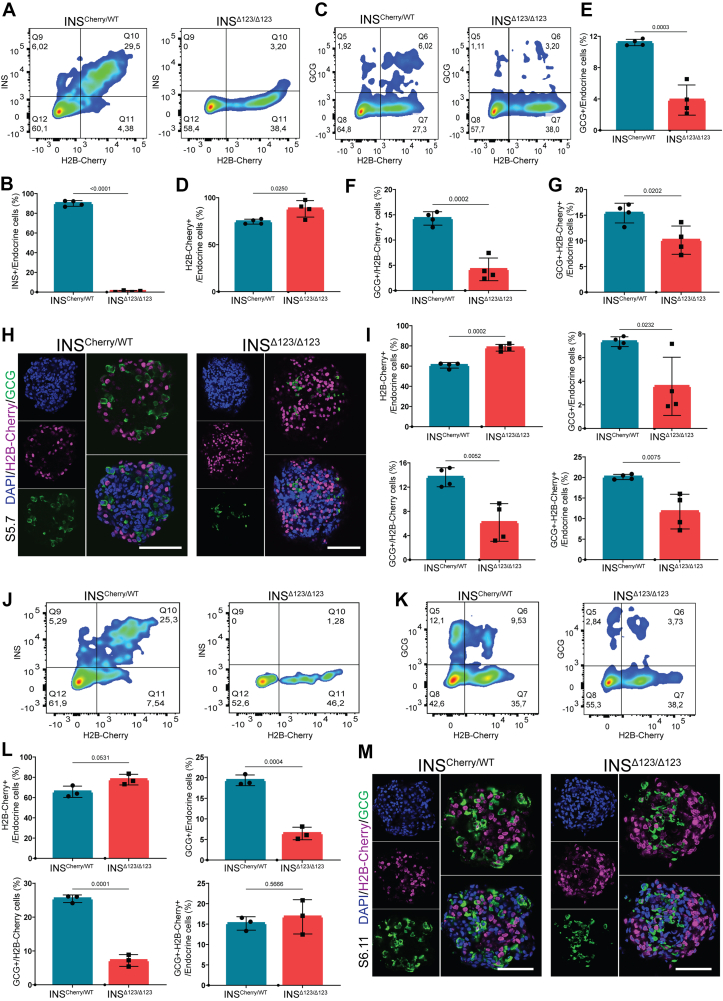
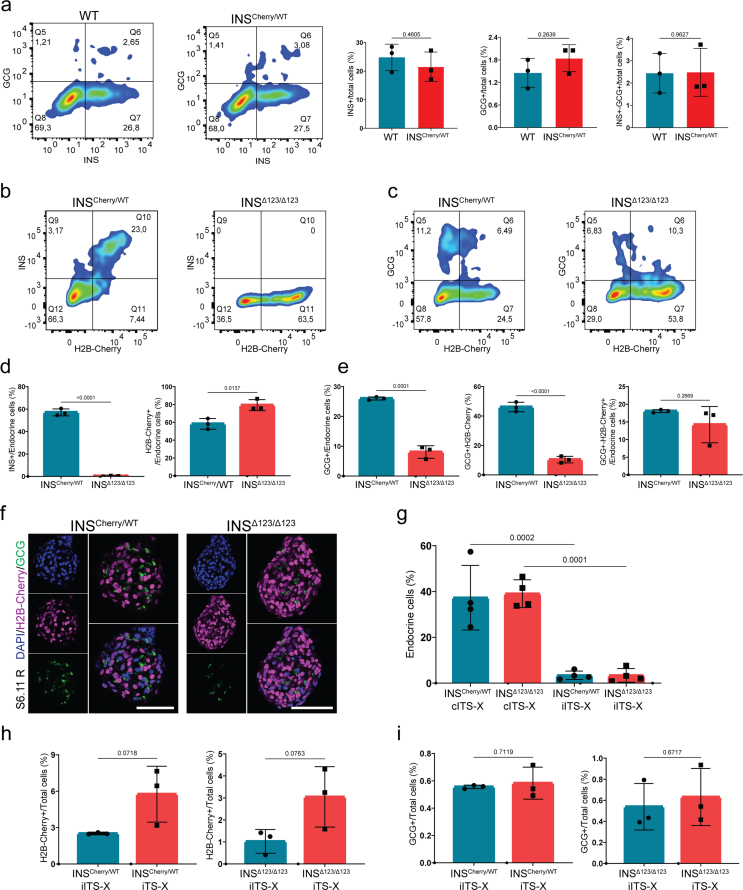
We next explored whether the loss of insulin affects the rate of differentiation of distinct hormone-producing endocrine cell types. First, we compared the endocrine cell composition between WT and INSCherry/WT SC-islets at S5.7, indicating comparable numbers of INS+, GCG+ and INS-GCG double-positive cells between the two clones (Fig. S5a). Thus, for the rest of analysis we used the INSCherry/WT clone as the control. As expected, FACS analysis showed no INS signal in the INSΔ123/Δ123 cells compared to the high number of INS+ cells in the INSCherry/WT clusters (Figure 5A,B). Conversely to this, we found a higher percentage of H2B-Cherry+ cells in the SC-islets from the INSΔ123/Δ123 cell line (Figure 5A,C, D). Importantly, we detected a lower percentage of GCG+ mono-hormonal cells in the INSΔ123/Δ123 cells compared to the INSCherry/WT SC-islets (Figure 5C,E, F). Moreover, the percentage of cells expressing both GCG and H2B-Cherry (GCG+-H2B-Cherry+) was also reduced in the INSΔ123/Δ123 SC-islets (Figure 5C,G). Similar results were obtained using immunostaining of SC-islets at S5.7 (Figure 5H–I), indicating that lack of insulin enhances the number of H2B-Cherry+ SC-β cells at the expense of formation of GCG+ SC-α cells. To search whether these changes depend on the stage of differentiation, we also analyzed the endocrine cell composition of SC-islets at S6. FACS analysis showed no insulin signals in the INSΔ123/Δ123 SC-islets at S6.11. Additionally, we observed an increased percentage of H2B-Cherry+ cells, a decreased percentage of GCG+ cells, and a comparable percentage of GCG+-H2B-Cherry+ cells in INSΔ123/Δ123 clusters compared to the INSCherry/WT SC-islets (Figure 5J-L). This analysis was also supported by IHC analysis, which showed an elevated number of H2B-Cherry+ cells along with a reduced number of GCG+ cells in the INSΔ123/Δ123 SC-islets at S6.11 (Figure 5M). Similar analysis was conducted on reaggregated SC-islets (Fig. S4a) at S6.11 that also disclosed increased H2B-Cherry+ cells accompanied by a decline in GCG+ cells in INSΔ123/Δ123 SC-islets compared to the INSCherry/WT clusters (Figs. S5b–f). Together, these analyses demonstrate that lack of insulin leads to increased number of SC-β cells and reduced differentiation of SC-α cells during human endocrinogenesis in vitro.
Figure 5.
Insulin shortage alters human endocrine cell composition. A) Representative FACS plots of INS+ and H2B-Cherry+ cells at S5.7. B) FACS quantification of INS+ cells, relative to the total endocrine cell population at S5.7. C) Representative FACS plots of GCG+ and H2B-Cherry+ cells at S5.7. D, E) FACS quantification of H2B-Cherry+ and GCG+ cells, relative to the total endocrine cell population at S5.7. F) Ratio of GCG+ to H2B-Cherry+ at S5.7. G) FACS quantification of GCG+-H2B-Cherry+ cells, relative to the total endocrine cell population at S5.7. H) Immunofluorescence pictures showing the H2B-Cherry+ and GCG+ cells at S5.7. Scale bar, 80 μm. I) Quantification of the percentage of H2B-Cherry+, GCG+ and GCG+-H2B-Cherry+ cells to the total endocrine cells, as well as the ratio of GCG+ to H2B-Cherry+ cells at S5.7. J) Representative FACS plots of INS+ and H2B-Cherry+ cells in SC-islets at S6.11. K) Representative FACS plots of GCG+ and H2B-Cherry+ cells in SC-islets at S6.11. L) Quantification of H2B-Cherry+, GCG+ and GCG+-H2B-Cherry+ cells, as well as the ratio of GCG+ to H2B-Cherry+ cells in SC-islets at S6.11. M) Immunofluorescence pictures showing the H2B-Cherry+ and GCG+ cells in the SC-islets at S6.11. Scale bar, 80 μm. Data are normalized to INSCherry/WT. All statistics have been done using two-sided unpaired t-test. Data are represented as mean ± SD.
The current differentiation protocol requires adding a commercial supplement comprising insulin-transferrin-selenium and ethanolamine (ITS-X) to the entire S5 to support cell survival at this stage [29]. Therefore, we asked whether this exogenous insulin, albeit in low concentrations, impacts the endocrine cell composition. To test this, we initially compared the differentiation rates of cells cultured in conventional media containing commercial (cITS-X) versus those cultured in media containing individual compounds of insulin, transferrin, selenium and ethanolamine (iITS-X). We observed a significant reduction in endocrine differentiation of cells cultured in iITS-X compared to cITS-X (Fig. S5g). Next, we used the media containing individual compounds in the presence (iITS-X) or absence (iTS-X) of insulin. Consistent with our earlier findings, the removal of exogenous insulin led to increased percentage of H2B-Cherry+ cells in both INSCherry/WT and INSΔ123/Δ123 SC-islets (Fig. S5h). However, we detected no differences in the rate of GCG+ cell formation in both cell lines, as the numbers of GCG+ cells were very low (Fig. S5i). This analysis provides further evidence that insulin shortage promotes SC-β cell differentiation.
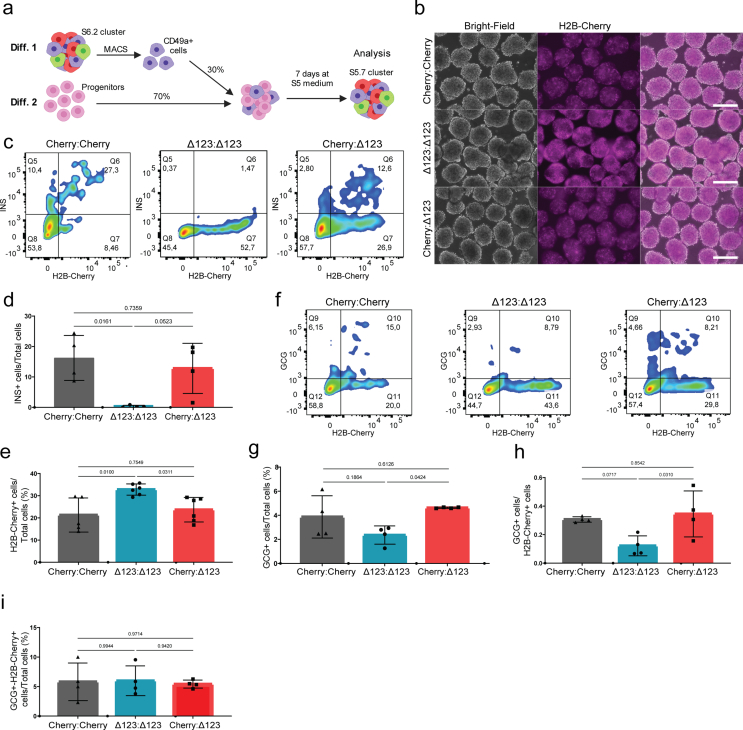
Next, we asked whether recovering insulin signaling restores the cell composition in INSΔ123/Δ123 SC-islets. To do so, we isolated and enriched SC-β cells at S6.2 using CD49a antibody and mixed them with the S5.1 progenitors and allowed the mixed culture to differentiate until S5.7 (Fig. S6a). We set up three different conditions including INSCherry/WT SC-β with INSCherry/WT progenitors (Cherry:Cherry), INSΔ123/Δ123 SC-β with INSΔ123/Δ123 progenitors (Δ123:Δ123) and INSCherry/WT SC-β with INSΔ123/Δ123 progenitors (Cherry:Δ123). Importantly, coculture of insulin-producing INSCherry/WT SC-β cells with INSΔ123/Δ123 progenitors reduced the number of H2B-Cherry+ cell formation but increased the generation of GCG+ cells compared to the insulin deficient Δ123:Δ123 mixture (Figs. S6b–i). This data shows that exogenous insulin reestablishes the proper islet cell composition in INSΔ123/Δ123 progenitors and proposes that insulin signaling rather than insulin synthesis/storage regulates human endocrine cell composition.
3.6. Insulin deficiency reduces human SC-β cell proliferation
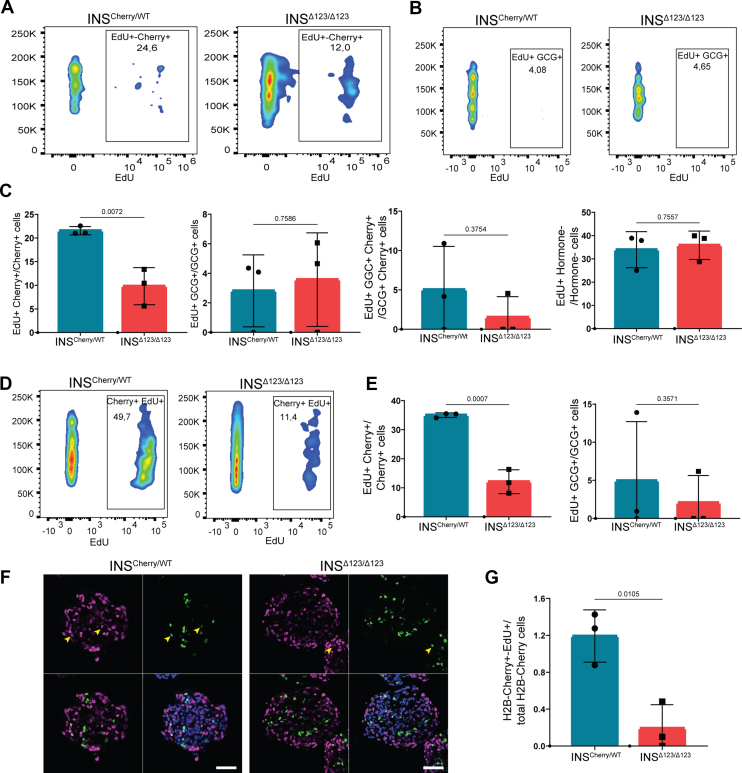
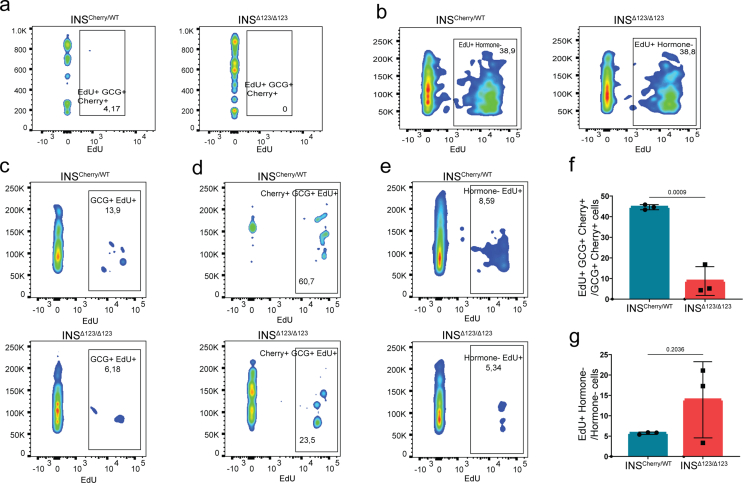
Due to the well-known function of insulin as a growth hormone, we next explored the possible contribution of cell division to the effect of insulin insufficiency on the endocrine cell composition. To this end, we applied EdU pulse-chase analysis on clusters at S5.5 for 48 h and conducted FACS analysis at S5.7. INSΔ123/Δ123-derived H2B-Cherry+ cells exhibited around 50 % reduction in proliferation compared to those originated from the INSCherry/WT line (Figure 6A,C). However, no significant differences in the proliferation rate of GCG+, GCG+-H2B-Cherry+, and non-hormone-producing cells were observed between the two clones at this stage (Figure 6B,C and S7a, b). It should be noted that the rates of proliferation of GCG+ and GCG+-H2B-Cherry+ cells were low and exhibited a high variation between independent experiments. To test if the effect of insulin shortage on SC-β proliferation depends on the stage of differentiation, we performed a similar EdU pulse-chase analysis on reaggregated SC-islets at S6.13 followed by FACS analysis at S6.15. Consistently, we found a significant reduction in the proliferation of H2B-Cherry+ cells (Figure 6D,E) and no prominent changes in the cell division rates of GCG+ and non-hormone-producing cells in the INSΔ123/Δ123 SC-islets. However, the number of proliferating GCG+-H2B-Cherry+ cells was reduced (Figure 6E and S7c–g). Moreover, immunostaining analysis of SC-islets at S5.7 confirmed the FACS data and showed a reduced proliferation of H2B-Cherry+ cells in the INSΔ123/Δ123 clusters compared to the INSCherry/WT cells (Figure 6F,G). Altogether, these analyses demonstrate the reduced SC-β cell proliferation in the absence of insulin.
Figure 6.
Insulin deficiency reduces SC-β cell proliferation. A, B) Representative FACS plots of proliferative H2B-Cherry+ and GCG+ cells at S5.7. C) FACS quantification of proliferative H2B-Cherry+, GCG+, GCG+-H2B-Cherry+ and non-hormone-expressing cells at S5.7. D) Representative FACS plots of proliferative H2B-Cherry+ cells from reaggregated SC-islets at S6.15. E) FACS quantification of proliferative H2B-Cherry+ and GCG+ cells from reaggregated SC-islets at S6.15. F, G) Representative immunofluorescence pictures and the respective quantification exhibiting the proliferating H2B-Cherry+ cells at S5.7. Scale bar, 50 μm. Data are normalized to INSCherry/WT (EdU+ Cherry+ graphs in C, E). All statistics have been done using two-sided unpaired t-test. Data are represented as mean ± SD.
4. Discussion
Here, we studied the effects of the reduced levels of insulin on human endocrine lineage segregation during development. We used an insulin insufficiency iPSC differentiation system to model INS gene mutations that have been reported in patients diagnosed with NDM [21]. Some of these mutations have been referred to “loss of INS function mutations”, meaning that the cellular processes at the levels of insulin transcription and/or translation are affected [21,26]. Due to the complete absence of intact insulin in the INSΔ123/Δ123-derived SC-β cells, this cell line resembles recessive INS mutations producing no or low levels of insulin [26]. However, unlike the so far reported INS mutations in NDM patients, INSΔ123/Δ123 iPSCs exhibited no mutations in the genomic sequence of INS gene. Additionally, the expression of H2B-Cherry that was fused to the 3′ of INS sequence indicates a functional INS promoter and therefore confirms active transcription and translation. However, we found a 123 bp deletion in the exon 3 of the INS mRNA. This indicates that fusion of H2B-Cherry sequence to INS gene impairs proper INS mRNA processing. Interestingly, the 123 bp deletion corresponds to the start sequence of exon 3, implicating the possible defect in mRNA splicing. This conclusion is supported by finding the misexpression of several proteins involved in mRNA processing and spliceosome in the proteomics analysis. The dysregulation of some of these proteins such as SR proteins and NOVA1 has been associated with β cell loss and diabetes [42,43]. These findings suggest the possible contribution of non-coding mutations of INS 3′sequence to the reduced insulin levels in NDM patients due to the impairment of proper mRNA splicing.
Our results illustrated that insulin deficiency reduces IR signaling when SC-islets were stimulated with glucose, implying impairment of autocrine and paracrine insulin signaling. However, administration of exogenous insulin caused increased IR sensitivity upon chronic insulin shortage. The degree of IR sensitization directly impacts β cell function and contributes to the development of T2D [44]. In patients with insulin resistance, elevated levels of insulin and prolonged exposure to this hormone eventually desensitizes IR and attenuates the downstream signaling affecting β cells health and function [45,46]. Therefore, IR sensitization is considered as one possible strategy for diabetes therapy [47]. In this context, the INSΔ123/Δ123-derived SC-β cells present a hypersensitized IR model due to the lack of insulin biosynthesis and signaling. Yet, the consequence of prolonged insulin hypersensitization in a such model is not so clear. Furthermore, if NDM patients with reduced insulin levels also exhibit IR hypersensitivity still needs to be explored.
We found that insulin deficiency alters SC-islet cell composition in favor of SC-β cells and at the expense of SC α-cell formation. This finding is aligned with the previous studies in which near to complete ablation of β cells in adult mouse pancreas resulted in conversion of α and δ cells into β cells [48,49]. Thus, it is likely that insulin and insulin signaling deficiency triggers a compensatory mechanism to ensure generation of enough β cells in both embryonic and adult pancreas. The elevated SC-β cell number in the INSΔ123/Δ123 SC-islets could be resulting from the enhanced specification of endocrine progenitors towards SC-β cell fate, increased reprogramming of newly differentiated SC α-cells towards SC-β cells or elevated proliferation of existing SC-β cells. The reduced numbers of poly-hormonal (GCG+-H2B-Cherry+) and cycling SC-β cells at the early stage of endocrine cell formation in INSΔ123/Δ123 cells exclude the possible contribution of SC-α to SC-β cell transdifferentiation and SC-β cell proliferation to the increased number of SC-β cells. Therefore, it is possible that lack of insulin primes endocrine progenitor towards β cell fate. In support of this, deletion of insulin or IR in zebrafish also enhanced β cell formation and reduced the differentiation of α cells. The increased β cell differentiation was attributed to the elevated expression levels of Pdx1 as one of the key β cell formations in the progenitor cells [50]. In this context, our results propose that lack of insulin determines SC-islet cell composition likely through a feedback loop. Further mechanistic analyses are needed to elucidate how insulin signaling changes the β cell program versus α cell fate in endocrine progenitors. Additionally, future studies need to explore the impact of recessive homozygous INS mutations on α cell mass and function as well as blood glucagon levels in NDM patients.
Our data revealed that the lack of insulin reduces human SC-β cell proliferation. Contrary to this, deletion of both alleles of Ins1 and Ins2 genes in mice has been shown to enhance embryonic β cell expansion. This increased β cell division was postulated to be a consequent result from increased proto-islet vasculature in the Ins1/2 double KO embryos [51]. Additionally, Szabat et al., showed that reduced levels of insulin biosynthesis impact adult mouse β cell expansion in a cell-autonomous manner independently of extracellular insulin. According to their model, reduced insulin production results in decreased ER stress that consequently activates Akt phosphorylation to trigger cell cycle [41]. Although, we cannot exclude the possible contribution of cell-autonomous effects of insulin deficiency to cell division, we found reduced phosphorylation of AKT in INSΔ123/Δ123 SC-islets, ruling out the possible involvement of the similar mechanism reported by Szabat et al. [41]. This suggests that the decreased SC-β cell proliferation in INSΔ123/Δ123 SC-islets is likely mediated by the lack of IR signaling. Indeed, administration of insulin has been shown to increase mouse β cell mass and proliferation [52,53]. However, the impact of loss of IR on β cell replication is still a matter of debate. Although some studies showed no or positive effects of loss of IR on murine β cell expansion [54,55], several other studies showed that IR deficiency reduces mouse β cell proliferation and mass [[56], [57], [58], [59]]. Additionally, deletion of insulin inhibitory receptor (inceptor), which desensitizes IR, also enhances β cell division further supporting the direct involvement of IR signaling in β cell propagation [60]. Future works should dissect the distinct modes of action of cell-autonomous effect of insulin synthesis and autocrine insulin signaling on β cell proliferation in humans. We also did not find any changes in the expansion of SC-α cells in the absence of insulin, supporting the dispensable function of insulin/insulin signaling in regulating α-cell proliferation [41]. These data also exclude the possible contribution of reduced α-cell division to the decreased α-cell number in the INSΔ123/Δ123 SC-islets, further highlighting the possible fate decision function of insulin during human endocrinogenesis.
Despite the interesting and novel findings, this study still faces several limitations. Certain analyses display high variations across independent experiments, likely attributed to the inherent nature of the differentiation protocol, resulting in fluctuations in endocrine differentiation efficiency. Furthermore, our results were obtained using a single iPSC clone, and analyzing more clones in future studies is needed to further support and strengthen these findings. In summary, we found that lack of insulin enhances sensitization of IR in SC-islets and alters their cell composition as well as reduces SC-β cell proliferation. These findings implicate the developmental impact of mutations in INS gene associated with NDM on human endocrinogenesis and β cell function. Furthermore, they highlight the possible physiological function of insulin/insulin signaling in regulating endocrine cell segregation during human development. Accordingly, insulin might exert feedback loop signaling to ensure generation of appropriate number of endocrine cells during human endocrinogenesis. These findings provide novel insights into the function of insulin during human development and further highlight the potential of insulin signaling targeting for islet cell regeneration for diabetes treatment.
CRediT authorship contribution statement
Perla Cota: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. Özüm Sehnaz Caliskan: Data curation, Formal analysis, Methodology, Software, Writing – review & editing. Aimée Bastidas-Ponce: Methodology, Writing – review & editing. Changying Jing: Methodology, Writing – review & editing. Jessica Jaki: Methodology, Writing – review & editing. Lama Saber: Methodology, Writing – review & editing. Oliver Czarnecki: Methodology, Writing – review & editing. Damla Taskin: Methodology, Writing – review & editing. Anna Karolina Blöchinger: Methodology, Writing – review & editing. Thomas Kurth: Methodology, Writing – review & editing. Michael Sterr: Methodology, Writing – review & editing. Ingo Burtscher: Methodology, Writing – review & editing. Natalie Krahmer: Resources, Supervision, Validation, Writing – review & editing. Heiko Lickert: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. Mostafa Bakhti: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Supervision, Validation, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Mara Catani, and Elke Schlüssel for the helpful comments and discussion. We thank Kerstin Diemer and Susanne Kretschmar for technical support. We thank Donna Marie Thomson for administrative support. We thank G. Lederer, G. Eckstein and T. Meitinger for the karyotyping. The work was supported by the Helmholtz Society, Helmholtz Portfolio Theme’Metabolic Dysfunction and Common Disease, and German Center for Diabetes Research (DZD e.V.). P.C. and L.S. were supported by the Helmholtz Research School for Diabetes (HRD), which is funded by the Helmholtz Association - Initiative and Networking Fund (IVF). C.J. was supported by a scholarship under the State Scholarship Fund by the China Scholarship Council (File No. 202208080154). This project was supported by DZD NEXT Young Talent Program (M.B.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101853.
Contributor Information
Heiko Lickert, Email: heiko.lickert@helmholtz-munich.de.
Mostafa Bakhti, Email: mostafa.bakhti@helmholtz-munich.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Supplementary figure 1. Characterization of INSCherry/Cherry (INSΔ123/Δ123) reporter iPSC line. a) PCR analyses for genotyping WT, INScherry/wt and INScherry/cherry clones. b) INScherry/cherry is characterized by a normal female karyotype (46, XX). c) SOX2 and OCT3/4 expression in INScherry/cherry clone. Scale bar, 80 μm. d) IHC of directed differentiation of INScherry/cherry in-vitro towards the three germ layers, endoderm, mesoderm and ectoderm. Scale bar 50 μm for endoderm and mesoderm and 100 μm for ectoderm. e) Schematic representation of human in-vitro endocrine differentiation stages. f) IHC showing definitive endoderm (DE), pancreatic progenitor (PP2) and endocrine cells, confirms in-vitro differentiation capacity of INScherry/cherry clone. Scale bar, 80 μm. g) Live imaging detecting H2B-Cherry signal in INSCherry/cherry cells at S6 confirming insulin promoter activity. Schemes were created with BioRender.com.
Supplementary figure 2. Analysis of ER stress levels in INSΔ123/Δ123 SC-β cells. a) Representative confocal pictures showing the presence of cells co-expressing CHGA and H2B-Cherry in SC-islets at S6.14. Scale bar, 20 μm. b) Schematic picture showing the sorting and reaggregation procedure of SC-β cells at S5.7. c) Representative confocal pictures indicating the levels of aggresome (Proteostat staining) in the absence and presence of MG132 in WT and INSΔ123/Δ123 SC-islets at S6.10. Scale bar, 20 μm. d) Western blot analysis of ER stress-related markers in SC-islets. e) RT-PCR analysis depicting XBP1 splicing in sorted H2B-Cherry-positive (Cherry+) and -negative (Cherry−) SC-islets at S6.15. Schemes were created with BioRender.com.
Supplementary figure 3. Proteomics analysis of INSΔ123/Δ123 SC-islets at S5.7. a-c) Heat maps of significantly downregulated proteins involved in spliceosome and mRNA homeostasis, translation, and ER-stress in INSΔ123/Δ123 vs INSCherry/WT SC-islets at S5.7. d-f) Heat maps of significantly upregulated ribosomal, intracellular transport and amino acid-tRNA ligase proteins in INSΔ123/Δ123 vs INSCherry/WT SC-islets at S5.7.
Supplementary figure 4. Quantification of downstream IR signaling in SC-islets. a) Schematic picture showing the reaggregation procedure to eliminate progenitor cells and enrich endocrine cells at S5.7. b) Schematic of the SC-islet treatment with glucose and insulin to prepare samples for Western blotting. c-f) WB quantifications of phosphorylated IR-IGF1R and total IGF1R treated with 20 mM glucose in the absence and presence of exogenous insulin. g) WB quantifications of total AKT in SC-islets treated with 20 mM glucose in the absence and presence of exogenous insulin. h, i) WB quantifications of phosphorylated ERK1/2 and total ERK1/2 in SC-islets treated with 20 mM glucose in the absence and presence of exogenous insulin. Schemes were created with BioRender.com. Data are normalized to WT (c-i). All statistics have been done using one-way ANOVA multiple comparisons test. Data are represented as mean ± SD.
Supplementary figure 5. Impact of insulin deficiency on SC-islet cell composition. a) Representative FACS plots and quantifications of endocrine cell types between WT and INScherry/WT SC-islets at S5.7. b) Representative FACS plots of total INS+ and H2B-Cherry+ cells in reaggregated SC-islets at S6.11. c) Representative FACS plots of total GCG+ and H2B-Cherry+ cells in reaggregated SC-islets at S6.11. d) FACS quantification of INS+ cells and H2B-Cherry+ cells, relative to total endocrine cell population in reaggregated SC-islets at S6.11. e) FACS quantification of GCG+ cells and GCG+-H2B-Cherry+ cells relative to total endocrine cell population, as well as the ratio of GCG+ to H2B-Cherry+ cells in reaggregated SC-islets at S6.11. Data are normalized to INSCherry/WT (d, e). f) Representative immunofluorescence pictures showing H2B-Cherry+ and GCG+ cells at S6.11 from reaggregated SC-islets at S6.11. Scale bar, 80 μm. g) FACS quantification of the percentage of endocrine cell differentiation in INScherry/WT and INSΔ123/Δ123 SC-islets cultured in media supplemented with cITS-X and iITS-X. h) FACS quantification of H2B-Cherry+ cells relative to total cells in INScherry/WT and INSΔ123/Δ123 SC-islets differentiated in media containing iITS-X or iTS-X. i) FACS quantification of GCG+ cells relative to total cells in INScherry/WT and INSΔ123/Δ123 SC-islets differentiated in media containing iITS-X or iTS-X. Data are normalized to iTS-X in INSCherry/WT graphs (h, i). All statistics have been done using two-sided unpaired t-test. Data are represented as mean ± SD.
Supplementary figure 6. Reinstalling insulin signaling restores endocrine cell composition in INSΔ123/Δ123 SC-islets. a) Schematic picture depicting that the sorted and enriched SC-β cells from differentiation (diff.) 1 were mixed with progenitors from diff. 2 at ratio of 30:70 % and the differentiation proceeds for 7 days. b) Live microscopy imaging of H2B-Cherry+ cells in INSCherry/WT SC-β cells reaggregated with INSCherry/WT progenitors (Cherry:Cherry), INSΔ123/Δ123 SC-β with INSΔ123/Δ123 progenitors (Δ123:Δ123) and INSCherry/WT SC-β with INSΔ123/Δ123 progenitors (Cherry:Δ123). An increased Cherry signals in the Δ123:Δ123 mixture is observed compared to the Cherry:Cherry condition. Notably, the Cherry:Δ123 mixture exhibits reduced Cherry signals at the levels of the Cherry:Cherry culture. Scale bar, 300 μm. c) Representative FACS plots of INS+ and H2B-Cherry+ cells in Cherry:Cherry, Δ123:Δ123 and Cherry:Δ123 SC-islets. d) FACS quantification of total INS+ cells. Data are normalized to Δ123:Δ123. e) FACS quantification of H2B-Cherry+ cells to total cells. Data are normalized to Δ123:Δ123. f) Representative FACS plots of GCG+ and H2B-Cherry+ cells. g) FACS quantification of GCG+ cells to total cells. Data are normalized to Cherry:Δ123. h) FACS quantification of the ratio of GCG+ cells to H2B-Cherry+ cells. Data are normalized to Cherry:Cherry. i) FACS quantification of GCG+-H2B-Cherry+ cells to total cells. Data are normalized to Cherry:Δ123. Schemes were created with BioRender.com. All statistics have been done using one-way ANOVA and Tukey's multiple comparisons test. Data are represented as mean ± SD.
Supplementary figure 7. The impact of insulin deficiency on pancreatic and endocrine cell proliferation. a) Representative FACS plots of proliferative GCG+ cells at S5.7. b) Representative FACS plots of proliferative hormone− cells at S5.7. c) Representative FACS plots of proliferative GCG+ cells in reaggregated SC-islets at S6.15. d) Representative FACS plots of proliferative GCG+-H2B-Cherry+ cells in reaggregated SC-islets at S6.15. e) Representative FACS plots of proliferative hormone− cells in reaggregated SC-islets at S6.15. f) FACS quantification of GCG+-H2B-Cherry+-EdU+ cells relative to total GCG+-H2B-Cherry+ cells in reaggregated SC-islets at S6.15. g) Quantification of hormone−-EdU+ cells relative to total hormone− cells in reaggregated SC-islets All statistics have been done using two-sided unpaired t-test. Data are normalized to INSCherry/WT (f, g). All statistics have been done using two-sided unpaired t-test. Data are represented as mean ± SD.
Data availability
Data will be made available on request.
References
- 1.Brissova M., Fowler M.J., Nicholson W.E., Chu A., Hirshberg B., Harlan D.M., et al. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J Histochem Cytochem. 2005;53(9):1087–1097. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- 2.Cabrera O., Berman D.M., Kenyon N.S., Ricordi C., Berggren P.-O., Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc Natl Acad Sci USA. 2006 doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim A., Miller K., Jo J., Kilimnik G., Wojcik P., Hara M. Islet architecture: a comparative study. Islets. 2009;1(2):129–136. doi: 10.4161/isl.1.2.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gradwohl G., Dierich a., LeMeur M., Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA. 2000;97(4):1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu G., Dubauskaite J., Melton D.a. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–2457. doi: 10.1146/annurev.cellbio.20.012103.094648. [DOI] [PubMed] [Google Scholar]
- 6.Jennings R.E., Berry A.A., Strutt J.P., Gerrard D.T., Hanley N.A. Human pancreas development. Development. 2015;142(18):3126–3137. doi: 10.1242/dev.120063. [DOI] [PubMed] [Google Scholar]
- 7.Jennings R.E., Berry A.a., Kirkwood-Wilson R., Roberts N.a., Hearn T., Salisbury R.J., et al. Development of the human pancreas from foregut to endocrine commitment. Diabetes. 2013;62(10):3514–3522. doi: 10.2337/db12-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Löf-Öhlin Z.M., Nyeng P., Bechard M.E., Hess K., Bankaitis E., Greiner T.U., et al. EGFR signalling controls cellular fate and pancreatic organogenesis by regulating apicobasal polarity. Nat Cell Biol. 2017;19(11):1313–1325. doi: 10.1038/ncb3628. [DOI] [PubMed] [Google Scholar]
- 9.Bakhti M., Bastidas-ponce A., Tritschler S., Tarquis-medina M., Nedvedova E., Scheibner K., et al. Synaptotagmin 13 orchestrates pancreatic endocrine cell egression and islet morphogenesis. Nat Commun. 2022;13:4540. doi: 10.1038/s41467-022-31862-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bankaitis E.D., Bechard M.E., Wright C.V.E. Feedback control of growth, differentiation, and morphogenesis of pancreatic endocrine progenitors in an epithelial plexus niche. Gene Dev. 2015;29(20):2203–2216. doi: 10.1101/gad.267914.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arda H.E., Benitez C.M., Kim S.K. Gene regulatory networks governing pancreas development. Dev Cell. 2013;25(1):5–13. doi: 10.1016/j.devcel.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouzi M., Kim Y.H., Katsumoto K., Johansson K., Grapin-Botton A. Neurogenin3 initiates stepwise delamination of differentiating endocrine cells during pancreas development. Dev Dynam. 2011;240(3):589–604. doi: 10.1002/dvdy.22544. [DOI] [PubMed] [Google Scholar]
- 13.Shih H.P., Kopp J.L., Sandhu M., Dubois C.L., Seymour P.a., Grapin-Botton A., et al. A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development. 2012;139(14):2488–2499. doi: 10.1242/dev.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mamidi A., Prawiro C., Seymour P.A., Lichtenberg K.H. De., Jackson A., Serup P., et al. Mechanosignalling via integrins directs fate decisions of pancreatic progenitors. Nature. 2018;564:114–118. doi: 10.1038/s41586-018-0762-2. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y., Chan L. Monogenic diabetes: what it teaches us on the common forms of type 1 and type 2 diabetes. Endocr Rev. 2016;37(3):190–222. doi: 10.1210/er.2015-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greeley S.A.W., Naylor R.N., Philipson L.H., Bell G.I. Neonatal diabetes: an expanding list of genes allows for improved diagnosis and treatment. Curr Diabetes Rep. 2011;11(6):519–532. doi: 10.1007/s11892-011-0234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fajans S.S., Bell G.I. MODY: history, genetics, pathophysiology, and clinical decision making. Diabetes Care. 2011:1878. doi: 10.2337/dc11-0035. 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hani E.H. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J Clin Invest. 1999;104(9):R41–R48 104. doi: 10.1172/JCI7469. R41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gloyn A.L., Pearson E.R., Antcliff J.F., Proks P., Bruining G.J., Slingerland A.S., et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 20.Yamagata K., Furuta H., Oda N., Kaisaki P.J., Menzel S., Cox N.J., et al. Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384(6608):458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 21.Støy J., De Franco E., Ye H., Park S.Y., Bell G.I., Hattersley A.T. In celebration of a century with insulin – update of insulin gene mutations in diabetes. Mol Metabol. 2021;52(June):101280. doi: 10.1016/j.molmet.2021.101280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokarz V.L., MacDonald P.E., Klip A. The cell biology of systemic insulin function. JCB (J Cell Biol) 2018;217(7):1–17. doi: 10.1083/jcb.201802095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu M., Weiss M.A., Arunagiri A., Yong J., Rege N., Sun J., et al. Biosynthesis, structure, and folding of the insulin precursor protein. Diabetes Obes Metabol. 2018:28–50. doi: 10.1111/dom.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M., Hodish I., Haataja L., Lara-Lemus R., Rajpal G., Wright J., et al. Proinsulin misfolding and diabetes: mutant INS gene-induced diabetes of youth. Trends Endocrinol Metabol. 2010;21(11):652–659. doi: 10.1016/j.tem.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahin G.S., Lee H., Engin F. An accomplice more than a mere victim: the impact of β-cell ER stress on type 1 diabetes pathogenesis. Mol Metabol. 2021:101365. doi: 10.1016/j.molmet.2021.101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garin I., Edghill E.L., Akerman I., Rubio-Cabezas O., Rica I., Locke J.M., et al. Recessive mutations in the INS gene result in neonatal diabetes through reduced insulin biosynthesis. Proc Natl Acad Sci USA. 2010;107(7):3105–3110. doi: 10.1073/pnas.0910533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Sterr M., Burtscher I., Chen S., Hieronimus A., Machicao F., et al. Genome-wide analysis of PDX1 target genes in human pancreatic progenitors. Mol Metabol. 2018;9:57–68. doi: 10.1016/j.molmet.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blöchinger A.K., Siehler J., Wißmiller K., Shahryari A., Burtscher I., Lickert H. Generation of an INSULIN-H2B-Cherry reporter human iPSC line. Stem Cell Res. 2020;45(April):101797. doi: 10.1016/j.scr.2020.101797. [DOI] [PubMed] [Google Scholar]
- 29.Velazco-Cruz L., Song J., Maxwell K.G., Goedegebuure M.M., Augsornworawat P., Hogrebe N.J., et al. Acquisition of dynamic function in human stem cell-derived β cells. Stem Cell Rep. 2019;(12):351–365. doi: 10.1016/j.stemcr.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanker J.S., Deb C., Wasserkrug H.L., Seligman A.M. Staining tissue for light and electron microscopy by bridging metals with multidentate ligands. Science. 1966;152(3729):1631–1634. doi: 10.1126/science.152.3729.1631. [DOI] [PubMed] [Google Scholar]
- 31.Völkner M., Wagner F., Steinheuer L.M., Carido M., Kurth T., Yazbeck A., et al. HBEGF-TNF induce a complex outer retinal pathology with photoreceptor cell extrusion in human organoids. Nat Commun. 2022;13(1):6183. doi: 10.1038/s41467-022-33848-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venable J.H., Coggeshall R. A simplified lead citrate stain for use in electron microscopy. J Cell Biol. 1965;25:407–408. doi: 10.1083/jcb.25.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11(3):319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill C.P., Dodson E.J., Dodson G.G., Dauter Z., Dauter Z., Dunn M.F. X-Ray structure of an unusual Ca2+ site and the roles of Zn2+ and Ca2+ in the assembly, stability, and storage of the insulin hexamer. Biochemistry. 1991;30(4):917–924. doi: 10.1021/bi00218a006. [DOI] [PubMed] [Google Scholar]
- 36.Rao A., McBride E.L., Zhang G., Xu H., Cai T., Notkins A.L., et al. Determination of secretory granule maturation times in pancreatic islet β-cells by serial block-face electron microscopy. J Struct Biol. 2020;212(1):107584. doi: 10.1016/j.jsb.2020.107584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veres A., Faust A.L., Bushnell H.L., Engquist E.N., Kenty J.H.-R., Harb G., et al. Charting cellular identity during human in vitro β-cell differentiation. Nature. 2019;569(7756):368–373. doi: 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen D., Coleman J., Chan E., Nicholson T.P., Dai L., Sheppard P.W., et al. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem Biophys. 2011;60(3):173–185. doi: 10.1007/s12013-010-9138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kahraman S., Dirice E., Basile G., Diegisser D., Alam J., Johansson B.B., et al. Abnormal exocrine–endocrine cell cross-talk promotes β-cell dysfunction and loss in MODY8. Nat Metab. 2022;4(1):76–89. doi: 10.1038/s42255-021-00516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szabat M., Page M.M., Panzhinskiy E., Skovsø S., Mojibian M., Fernandez-Tajes J., et al. Reduced insulin production relieves endoplasmic reticulum stress and induces β cell proliferation. Cell Metabol. 2016;23:179–193. doi: 10.1016/j.cmet.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 42.Jeffery N., Richardson S., Chambers D., Morgan N.G., Harries L.W. Cellular stressors may alter islet hormone cell proportions by moderation of alternative splicing patterns. Hum Mol Genet. 2019;28(16):2763–2774. doi: 10.1093/hmg/ddz094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juan-Mateu J., Rech T.H., Villate O., Lizarraga-Mollinedo E., Wendt A., Turatsinze J.V., et al. Neuron-enriched RNA-binding proteins regulate pancreatic beta cell function and survival. J Biol Chem. 2017;292(8):3466–3480. doi: 10.1074/jbc.M116.748335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldfine A.B., Kulkarni R.N. Modulation of β-cell function: a translational journey from the bench to the bedside. Diabetes Obes Metabol. 2012;14(SUPPL.3):152–160. doi: 10.1111/j.1463-1326.2012.01647.x. [DOI] [PubMed] [Google Scholar]
- 45.Boucher J., Kleinridders A., Ronald Kahn C. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harbor Perspect Biol. 2014;6(1):a009191. doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zick Y. Science’s STKE : Signal Transduction Knowledge Environment; 2005. Ser/Thr phosphorylation of IRS proteins: a molecular basis for insulin resistance. pe4. [DOI] [PubMed] [Google Scholar]
- 47.Jain C., Ansarullah, Bilekova S., Lickert H. Targeting pancreatic β cells for diabetes treatment. Nat Metab. 2022;4(9):1097–1108. doi: 10.1038/s42255-022-00618-5. [DOI] [PubMed] [Google Scholar]
- 48.Chera S., Baronnier D., Ghila L., Cigliola V., Jensen J.N., Gu G., et al. Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature. 2014;514:503–507. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thorel F., Népote V., Avril I., Kohno K., Desgraz R., Chera S., et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye L., Robertson M.A., Mastracci T.L., Anderson R.M. An insulin signaling feedback loop regulates pancreas progenitor cell differentiation during islet development and regeneration. Dev Biol. 2016;409(2):354–369. doi: 10.1016/j.ydbio.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duvillié B., Currie C., Chrones T., Bucchini D., Jami J., Joshi R.L., et al. Increased islet cell proliferation, decreased apoptosis, and greater vascularization leading to β-cell hyperplasia in mutant mice lacking insulin. Endocrinology. 2002;143(4):1530–1537. doi: 10.1210/endo.143.4.8753. [DOI] [PubMed] [Google Scholar]
- 52.Beith J.L., Alejandro E.U., Johnson J.D. Insulin stimulates primary β-cell proliferation via Raf-1 kinase. Endocrinology. 2008;149(5):2251–2260. doi: 10.1210/en.2007-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Movassat J., Saulnier C., Portha B. Insulin administration enhances growth of the β-cell mass in streptozotocin-treated newborn rats. Diabetes. 1997;46(9):1445–1452. doi: 10.2337/diab.46.9.1445. [DOI] [PubMed] [Google Scholar]
- 54.Skovsø S., Panzhinskiy E., Kolic J., Cen H.H., Dionne D.A., Dai X.Q., et al. Beta-cell specific Insr deletion promotes insulin hypersecretion and improves glucose tolerance prior to global insulin resistance. Nat Commun. 2022;13(1):1–22. doi: 10.1038/s41467-022-28039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trinder M., Zhou L., Oakie A., Riopel M., Wang R. β-cell insulin receptor deficiency during in utero development induces an islet compensatory overgrowth response. Oncotarget. 2016;7(29):44927–44940. doi: 10.18632/oncotarget.10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kulkarni R.N., Brüning J.C., Winnay J.N., Postic C., Magnuson M.A., Ronald Kahn C. Tissue-specific knockout of the insulin receptor in pancreatic β cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96(3):329–339. doi: 10.1016/S0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 57.Ueki K., Okada T., Hu J., Chong W.L., Assmann A., Dahlgren G.M., et al. Total insulin and IGF-I resistance in pancreatic β cells causes overt diabetes. Nat Genet. 2006;38(5):583–588. doi: 10.1038/ng1787. [DOI] [PubMed] [Google Scholar]
- 58.Okada T., Chong W.L., Hu J., Hinault C., Michael M.D., Krützfeldt J., et al. Insulin receptors in β-cells are critical for islet compensatory growth response to insulin resistance. Proc Natl Acad Sci USA. 2007;104(21):8977–8982. doi: 10.1073/pnas.0608703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otani K., Kulkarni R.N., Baldwin A.C., Krutzfeldt J., Ueki K., Stoffel M., et al. Reduced β-cell mass and altered glucose sensing impair insulin-secretory function in βIRKO mice. Am J Physiol Endocrinol Metabol. 2004;286(1 49–1):E41–E49. doi: 10.1152/ajpendo.00533.2001. [DOI] [PubMed] [Google Scholar]
- 60.Ansarullah, Jain C., Far F.F., Homberg S., Wißmiller K., von Hahn F.G., et al. Inceptor counteracts insulin signalling in β-cells to control glycaemia. Nature. 2021;590(7845):326–331. doi: 10.1038/s41586-021-03225-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data produced in this study have been deposited to the ProteomeXchange Consortium via the PRIDE [34] partner repository with the dataset identifier PXD045366.
Data will be made available on request.