Abstract
In Acinetobacter sp. strain ADP1, alkane degradation depends on at least five essential genes. rubAB and xcpR are constitutively transcribed. Here we describe inducible transcription of alkM, which strictly depends on the presence of the transcriptional activator AlkR. alkR itself is expressed at a low level, while a chromosomally located alkM::lacZ fusion is inducible by middle-chain-length alkanes from heptane to undecane, which do not support growth of ADP1, and by long-chain-length alkanes from dodecane to octadecane, which are used as sources of carbon and energy. The putative AlkM substrate 1-dodecene is also an effective inducer. Products of alkane hydroxylase activity like 1-dodecanol prevent induction of alkM expression. alkM is expressed only in stationary phase, suggesting its dependence on at least one other regulatory mechanism.
Degradation of n-alkanes is a widespread trait among bacteria, but little is known about the regulation of genes encoding alkane utilization. Acinetobacter sp. strain ADP1 is able to use long-chain-length alkanes with at least 12 carbon atoms as the sole source of carbon and energy. This requires at least five essential genes: rubAB, encoding rubredoxin and rubredoxin reductase (6); alkM, encoding the alkane hydroxylase; alkR, encoding a protein with similarity to AraC-XylS-like transcriptional regulators (19); and xcpR, which is part of the general secretory pathway (18).
The crucial step is the initial oxidation of the inert alkane to the respective primary alcohol, which is achieved by a three-component alkane monooxygenase complex composed of AlkM, RubA, and RubB (6, 19), as primarily characterized for Pseudomonas oleovorans (24).
The biochemistry and genetics of this ω-hydroxylation pathway have been studied extensively for the P. oleovorans-borne alk system (24). It confers growth on medium-chain-length alkanes from hexane to dodecane. The alk genes identified so far are located in two different regions of the OCT plasmid. The alkBFGHJKL operon encodes the alkane hydroxylase, two rubredoxins, an aldehyde dehydrogenase, an alcohol dehydrogenase, an acyl coenzyme A synthetase, and an outer membrane protein with unknown function (24). The second locus contains alkS and alkT, which encode a LuxR-UhpA-like regulator and rubredoxin reductase (24). AlkS is necessary for activation of expression of the alkBFGHJKL operon (3). The organization of alk genes in Acinetobacter sp. strain ADP1 is completely different. They are neither contained in large operons nor clustered or localized on a plasmid but occur in apparent disorder on the Acinetobacter chromosome (9). Expression of rubAB and xcpR is constitutive (6, 18).
We describe here the regulation and expression of alkR and alkM. Transcription of alkM depends strictly on AlkR and is inducible by hydrocarbons of various chain lengths. It seems to be repressed by oxidized alkane derivatives, while alkR is transcribed at a low level.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Acinetobacter sp. strain ADP1 is synonymously called Acinetobacter sp. strain BD413 and was formerly classified as Acinetobacter calcoaceticus ADP1 (13, 23).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| Acinetobacter sp. strain ADP1 | Wild type | 13, 23 |
| Acinetobacter sp. strain WH404 | alkR::lacZ Kmr | This study |
| Acinetobacter sp. strain WH405 | alkM::lacZ Kmr | 19 |
| Acinetobacter sp. strain WH407 | alkR(ΔMluI-StyI) CmralkM::lacZ Kmr | This study |
| E. coli DH5α | recA1 endA1 supE44 gyrA96 thi hsdR17(rK− mK+) relA1 φ80ΔlacZΔM15 Δ(lacZYA-argF)U169 | 20 |
| Plasmids | ||
| pKOK6.1 | Apr Kmr; promoterless lacZ | 14 |
| pWH767 | Apr; shuttle vector, contains original gene library insert, including alkR and alkM | 19 |
| pWH775 | ApralkR::lacZ Kmr; part of alkM (up to HindIII site in Fig. 1 bottom) | This study |
| pWH777 | ApralkR(ΔMluI-StyI) Cmr; part of alkM (up to HindIII site in Fig. 1 bottom) | 19 |
| pWH785 | ApralkR part of alkM (up to HindIII site in Fig. 1 bottom) | 19 |
| pWH1274 | Apr, Tcr; shuttle vector | 12 |
General methods.
Escherichia coli and Acinetobacter sp. strain ADP1 were transformed as described previously (17, 20). Electroporation of Acinetobacter was performed with a Gene Pulser (Bio-Rad Laboratories, Munich, Germany). Preparation of total DNA and small-scale preparations of plasmids were done as described before (6); large-scale preparations of plasmids were done with the Nucleobond kit (Macherey-Nagel, Düren, Germany). Southern hybridizations were done as described before (7). All other general methods and DNA manipulations were performed as described previously (20).
Media, growth conditions, and β-galactosidase assays.
Acinetobacter sp. strains were grown at 28°C on Luria broth (LB) (20) plates. Selectivity was achieved by adding ampicillin (300 mg/liter), kanamycin (10 mg/liter), or chloramphenicol (5 mg/liter). Selection for growth on specific carbon sources was performed on minimal medium supplemented with metal solution 44 and solidified with 1.5% agar (Noble agar; Difco, Detroit, Mich.) as described before (18). The carbon sources were supplied through the gas phase by spotting 200 μl (>99% pure) onto the center of a sterile filter paper disk placed in the lid of an inverted petri dish. E. coli was grown at 37°C in LB and under selective conditions with ampicillin (100 mg/liter), kanamycin (30 mg/liter), or chloramphenicol (20 mg/liter). Cultures for preparation of total or plasmid DNA were grown in LB supplemented with an antibiotic if appropriate. Overnight cultures for β-galactosidase assays were grown in LB for at least 15 h to stationary phase in the presence of an appropriate antibiotic. When growth-dependent β-galactosidase activity was determined, 100 ml of LB medium was inoculated to an optical density at 600 nm (OD600) of 0.01 or 0.02 and the OD600 was monitored for at least 27 h. Samples for β-galactosidase assays were taken at times indicated in the respective results and treated as described before (7). Alternatively, β-galactosidase activity in cultures grown in 4 ml of LB was assayed by using sealed tubes to prevent evaporation of volatile compounds. The medium was inoculated to an OD600 of 0.02, and the cultures were grown as specified in the respective results. The OD600 was determined, and 50 μl was used directly as described by Miller (16). The data (OD600, Miller units, and percent expression) given in the results were determined from three independent cultures.
Primer extension.
Total RNA was prepared with the RNeasy Mini Kit from Qiagen (Hilden, Germany). Primer extension reactions were performed as described before (22). Twelve micrograms of total RNA was used; the sequence of the 5′-labelled, alkM-specific primer (PSK1-3) is 5′-GTACAGGTGCATTCATAGTG-3′. The corresponding sequence ladder was obtained by sequencing pWH785 with the same primer by the dideoxy chain termination method (21) using Sequenase (U.S. Biochemicals, Cleveland, Ohio) and [α-32P]dATP.
RESULTS
Transcription initiation site of alkM and sequence analysis of the intergenic region.
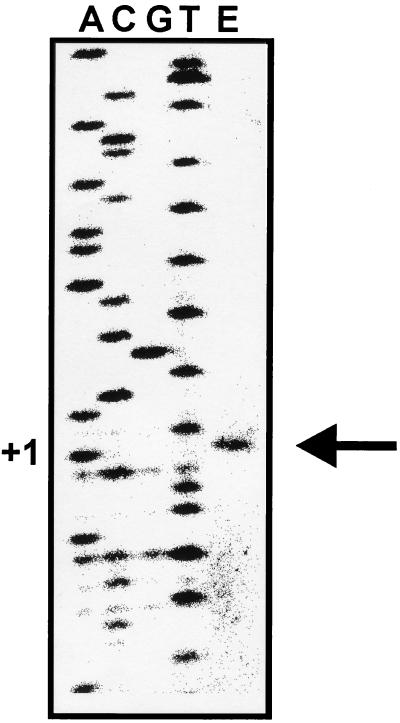
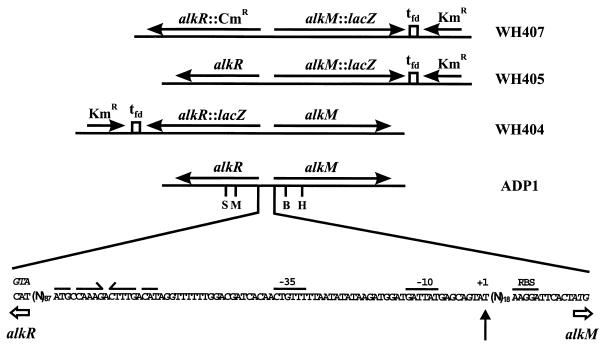
Primer extension analysis with RNA from hexadecane-grown Acinetobacter sp. strain ADP1 was performed with an alkM-specific primer. The primer binding site overlaps the putative start codon of alkM. Figure 1 top shows the result revealing the start site of transcription located 30 nucleotides upstream of the ATG. Analysis of the preceding sequence (Fig. 1 bottom) shows only weakly conserved −10 and −35 promoter consensus sequences. A weakly conserved −12, −24 consensus element for a ς54 promoter is located 5 bp upstream from the transcription start. The rpoN-negative strain WH396 (gift of B. Argauer) was able to grow with hexadecane and dodecane as sole carbon and energy sources, revealing that degradation of these substrates is not ς54 dependent. No direct repeat, which would represent the typical binding sequence of AraC or XylS (5), is found within the intergenic region between alkR and alkM. Instead, a nearly perfect inverted repeat is located 31 bp upstream from the putative −35 element (Fig. 1 bottom). Inverted repeats have been described as target sites for RhaS and MelR, which are also members of the AraC-XylS family (5).
FIG. 1.
(Top) Primer extension of the alkM mRNA from Acinetobacter sp. strain ADP1. The experiment was done with total RNA and primer PSK1-3. Lanes A, C, G, and T show the respective sequencing products. Lane E represents the signal obtained from the experiment using 12 μg of total RNA from cells grown with hexadecane as the sole carbon source. The position of the signal is indicated by an arrow; the corresponding sequencing product is designated with +1. (Bottom) Genetic organization of the alkR-alkM region in ADP1 and reporter strains WH404, WH405, and WH407. The arrangements of genes and markers and their transcriptional directions are shown. tfd, transcriptional terminator sequence of phage fd. Restriction sites within alkR and alkM used for constructions were StyI (S), MluI (M), BclI (B), and HindIII (H). The intergenic region between alkR and alkM is shown at the bottom of this panel. The putative start codons of alkM and alkR are given in italics, and the putative ribosome binding site (RBS) of alkM is indicated above the sequence. The nucleotide corresponding to the primer extension signal is highlighted by a vertical arrow. Putative −10 and −35 promoter elements are indicated. An inverted repeat with one mismatch is marked by arrows above the sequence.
Transcription of alkM is growth phase dependent and inducible.
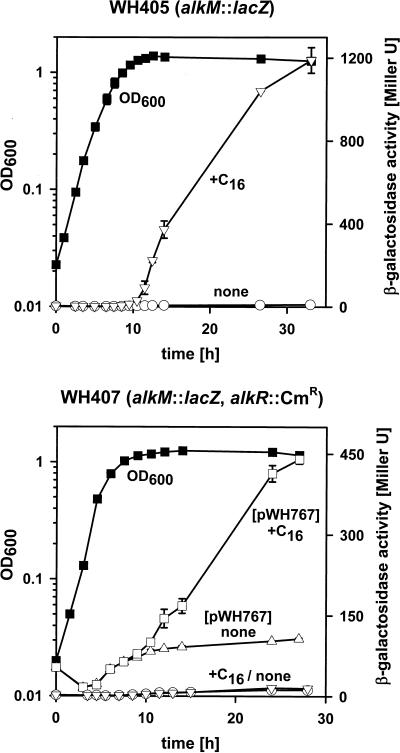
Expression analysis of alkM was performed by measuring the β-galactosidase activity expressed from a chromosomally located alkM::lacZ transcriptional fusion in mutant strain WH405 (19) (see Fig. 1 bottom). Acinetobacter sp. strain ADP1 has no endogenous β-galactosidase activity (data not shown). WH405 was grown in LB with and without hexadecane and showed only basal β-galactosidase expression at all growth phases in the absence of hexadecane. Induction of alkM occurred during the transition from exponential growth to stationary phase in the presence of hexadecane (Fig. 2 top). The same result was obtained when the precultures were grown in LB with or without hexadecane or when additional carbon sources like glucose, ethanol, or glutamate were added to the broth. These results demonstrate growth phase- and hexadecane-dependent alkM expression.
FIG. 2.
Growth phase- and inducer-dependent expression of the alkM::lacZ transcriptional fusion. Growth patterns in LB were identical with (10 mM) and without hexadecane (filled squares). β-Galactosidase activities were determined at the indicated times. Values represent the averages of three independent determinations with the indicated error ranges. (Top) alkM expression of WH405 with (open inverted triangles) and without (open circles) hexadecane; (bottom) expression of alkM in the alkR mutant strain WH407 with (open inverted triangles) and without (open circles) hexadecane and expression pattern of alkM in WH407(pWH767) with (open squares) and without (open triangles) hexadecane. C16, hexadecane.
alkR is transcribed at a low level.
A suitable single-copy reporter strain for analysis of alkR expression was constructed by insertion of the lacZ-Kmr cassette of pKOK6.1 into alkR on the chromosome. The promoterless lacZ gene is preceded by stop codons in all reading frames (15). pWH785 was cleaved with MluI and StyI (see Fig. 1 bottom), and the protruding ends were filled in. The lacZ-Kmr cassette, excised with BamHI from pKOK6.1, was blunt ended and inserted to yield pWH775. pWH775 was digested with SacII and ApaI, and the linear fragments were transformed into Acinetobacter sp. strain ADP1. Selection for kanamycin resistance resulted in strain WH404 (Fig. 1 bottom). The chromosomal integration was confirmed by Southern blotting (data not shown). The mutant strain is alkane negative but grows on 1-dodecanol. The expression pattern of alkR was analyzed by determining the β-galactosidase expression of strain WH404 grown in LB with or without hexadecane. The β-galactosidase activity increased about fivefold during exponential growth but remained at a low level (17 ± 1 Miller U [mean ± standard error] after 27 h). The presence of hexadecane led to a twofold increase of β-galactosidase activity in the late stationary phase (30 ± 1 Miller U after 27 h). To test autoregulation of alkR, we determined expression of alkR::lacZ in strain WH404 with pWH767 providing AlkR in trans. The expression patterns were nearly the same with and without pWH767; only the β-galactosidase activity was slightly increased during the entire experiment (31 ± 1 Miller U after 27 h). No increase of β-galactosidase activity was detectable in the strain containing pWH767 in the presence of hexadecane (32 ± 2 Miller U after 27 h). Control experiments with WH404 after electroporation of the shuttle vector pWH1274 gave the same results as those shown for WH404 without plasmid. Since we frequently observed recombination after electroporation of pWH767 into WH404, the presence of intact pWH767 was confirmed in the cells at the end of the expression experiments by restriction analysis (data not shown). We conclude from these results that alkR is transcribed at a low level.
alkR encodes an activator for alkM transcription.
To clarify the role of alkR in alkM regulation, we inactivated alkR in WH405. For that purpose, pWH777, containing alkR disrupted by a Cmr cassette (19), was cleaved with SacII and ApaI and the linear fragments were transformed into WH405. Screening for Cmr and Kmr yielded strain WH407. Figure 1 bottom shows the chromosomal organization of alkR and alkM in the double mutant as confirmed by Southern hybridization (data not shown). β-Galactosidase assays of strain WH407 showed no alkM transcription at any growth phase (Fig. 2 bottom). Electroporation of the alkR-containing plasmid pWH767 into WH407 restored regulation of alkM expression (Fig. 2 bottom), even though the expression level in the induced state is lower, and the noninduced expression level is higher, than that found in the single-copy situation of alkR in Fig. 2 top. Electroporation of the shuttle vector pWH1274 into WH407 yielded the same β-galactosidase expression pattern as that shown for WH407 without plasmid (Fig. 2 bottom). The presence of intact pWH767 in the cultures was confirmed at the end of the expression experiments by restriction analysis (data not shown). These results demonstrate that alkR encodes an inducer-dependent activator of alkM transcription.
Inducer specificity of alkM expression.
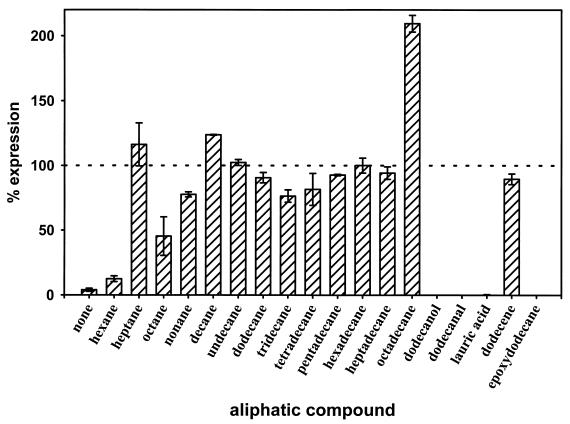
We analyzed the inducibility of alkM expression in strain WH405 with n-alkanes of various chain lengths from dodecane to octadecane, which all serve as growth substrates for Acinetobacter sp. strain ADP1, and with medium-chain-length n-alkanes from hexane to undecane, which do not support growth of ADP1. To avoid different growth rates, which might result from the proposed toxicity of medium-chain-length alkanes (1), the cultures were grown to stationary phase without alkanes, which were then added to a final concentration of 10 mM. β-Galactosidase activities were determined after continued incubation for 9 h (Fig. 3). Alkanes of chain length C7 to C17 induce transcription of alkM to about the same level. Hexane does not induce, and octadecane induces to about twofold, the hexadecane level. The inducing capacity of alkanes, which do not support growth of ADP1, is surprising and different from alk regulation in P. oleovorans, where the substrate specificity of AlkS seems to limit utilization of various alkanes (24). In contrast, alkane utilization in Acinetobacter sp. strain ADP1 seems to be limited by the specificity of AlkM.
FIG. 3.
alkM expression in response to alkanes with different chain lengths and putative intermediates from the alkane oxidation pathway. Expression induced with hexadecane in strain WH405 was defined as 100% and corresponds to a β-galactosidase activity of 736 Miller U. All chemicals were added to a final concentration of 10 mM. Values represent the averages of three independent determinations with the indicated error ranges.
We also determined the inducibility of alkM transcription in WH405 by putative substrates and intermediates from the alkane oxidation pathway. Dodecane, 1-dodecanol, dodecanal, lauric acid, 1-dodecene, and 1,2-epoxydodecane were added to the cultures to a final concentration of 10 mM in stationary phase. After continued incubation for 9 h, β-galactosidase activities were determined and revealed a clear discrimination between inducers and noninducers (Fig. 3). None of the oxidized compounds containing a hydrophilic modification induced alkM, whereas the hydrophobic compound 1-dodecene induced alkM as efficiently as dodecane did. We then wanted to determine whether Acinetobacter sp. strain ADP1 and the mutant strains WH405 and WH404 are able to use these compounds as sole sources of carbon and energy. In contrast to the wild type, neither mutant grows on dodecane or 1-dodecene, while 1-dodecanol and 1,2-epoxydodecane, the products of alkane hydroxylase activity (25), are utilized by all three strains.
A potential inhibitory effect of an oxidized alkane derivative on alkM expression was elucidated by adding 1-dodecanol and dodecane to a final concentration of 10 mM each to WH405 in stationary phase. β-Galactosidase activities determined after continued incubation for 9 h demonstrate that 1-dodecanol strictly prevents induction by dodecane. The mixture of dodecane and 1-dodecanol gave the same response as 1-dodecanol alone (Fig. 3).
To address the sensitivity of induction, the minimal hexadecane concentration necessary to induce alkM transcription in strain WH405 was determined. Hexadecane was added to various concentrations in stationary phase, and β-galactosidase activities were determined after continued incubation for 7 h. While 10 mM (100.0% ± 5.6% β-galactosidase expression, corresponding to 231 Miller U) and 1 mM (106.1% ± 2.7% β-galactosidase expression) hexadecane gave maximal expression, 100 μM (8.1% ± 0.8% β-galactosidase expression) hexadecane yielded only partial induction. Concentrations of 10 μM (3.9% ± 0.1% β-galactosidase expression) and lower (2.8% ± 0.0% β-galactosidase expression) of the alkane did not induce expression.
DISCUSSION
alkM is the only one of five presently characterized genes necessary for alkane degradation, which is regulated by alkanes (6, 18). The expression of a respective alkM::lacZ transcriptional fusion is induced about 100-fold by hexadecane in stationary phase, whereas only a basal expression level is detected in the absence of an appropriate inducer (Fig. 2 top). AlkM, RubA, and RubB supposedly form a three-component alkane monooxygenase complex (6, 19), as previously described for P. oleovorans (24). Rubredoxins are also involved in various other electron transfer reactions (4, 8, 11) and should, therefore, not depend on alkanes for expression.
The identification of AlkR as the transcriptional activator of alkM (Fig. 2) explains that both genes are needed for growth of ADP1 on alkanes (19). AlkR belongs to the family of AraC-XylS-like transcriptional regulators (19), which are characterized by a conserved C-terminal domain mediating DNA recognition, while effector molecules bind to their nonhomologous N-terminal and central regions (5). The homology of AlkR to AraC-XylS-like proteins implies a different regulatory mechanism than that proposed for AlkS, which activates alkBFGHJKL operon expression in P. oleovorans (3). AlkS is a LuxR-UhpA-like transcriptional regulator and contains an ATP or GTP binding motif, suggesting that ATP binding might be necessary for induction of the alkBFGHJKL operon (24). There is no evidence for ATP binding to AlkR by virtue of its primary structure.
AlkR requires the presence of an appropriate effector molecule to activate alkM expression (Fig. 3). The great variety of medium- and long-chain-length alkanes and the clear cutoff between the inducer heptane and the noninducer hexane suggest a broad specificity of AlkR for inducer binding with a sharp limitation in size. Since there is also a discrimination between hydrophobic inducers like dodecane and 1-dodecene and their noninducing oxidized derivatives 1-dodecanol and 1,2-epoxydodecane (Fig. 3), two basic requirements must be fulfilled: the inducer must exceed a minimum chain length and it must be hydrophobic. AlkR and AlkS recognize different inducers, because AlkS leads to induction of the alkBFGHJKL operon in response to alkanes, alkenes, and the respective primary alcohols (10). This correlates with the fact that alkBFGHJKL encodes the alcohol dehydrogenase in addition to the alkane hydroxylase (24).
Full expression of the alkM::lacZ fusion was observed when WH405 cells were exposed to 1 mM hexadecane, and partial induction was observed in the presence of 100 μM hexadecane. By virtue of its physical properties, we cannot assume an equal distribution of hexadecane between the bulk liquid and the bacterial cytoplasm. Therefore, the concentration directly available to AlkR might be much lower. With respect to the broth, the sensitivity to hexadecane is nearly 1,000-fold lower than that for p-hydroxybenzoate-induced pobA expression in ADP1 (2). Alkanes may not be a preferred substrate for this organism. Limitation of inducibility to stationary phase (Fig. 2 top) indicates an influence of general starvation or some form of catabolite repression. However, various carbon sources, as different as glucose, ethanol, and glutamate, prolong the exponential growth phase of WH405 in LB to a higher OD600 and delay the inducibility by hexadecane accordingly. This stationary-phase effect is most probably not due to a lack of AlkR in exponential phase, because transcription of alkR seems to be approximately constitutive. Expression of alkM is regulated by the presence of alkanes through AlkR and by at least one other unidentified mechanism.
ACKNOWLEDGMENT
This work was supported by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Asperger O, Kleber H-P. Metabolism of alkanes by Acinetobacter. In: Towner K J, Bergogne-Berezin E, Fewson C A, editors. The biology of Acinetobacter. New York, N.Y: Plenum Press; 1991. pp. 323–351. [Google Scholar]
- 2.DiMarco A A, Averhoff B, Ornston L N. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J Bacteriol. 1993;175:4499–4506. doi: 10.1128/jb.175.14.4499-4506.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eggink G, Engel H, Meijer W G, Otten J, Kingma J, Witholt B. Alkane utilization in Pseudomonas oleovorans. Structure and function of the regulatory locus alkR. J Biol Chem. 1988;263:13400–13405. [PubMed] [Google Scholar]
- 4.Fu C, Maier R J. Organization of the hydrogenase gene cluster from Bradyrhizobium japonicum: sequences and analysis of five more hydrogenase-related genes. Gene. 1994;145:91–96. doi: 10.1016/0378-1119(94)90328-x. [DOI] [PubMed] [Google Scholar]
- 5.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geissdörfer W, Frosch S C, Haspel G, Ehrt S, Hillen W. Two genes encoding proteins with similarities to rubredoxin and rubredoxin reductase are required for conversion of dodecane to lauric acid in Acinetobacter calcoaceticus ADP1. Microbiology. 1995;141:1425–1432. doi: 10.1099/13500872-141-6-1425. [DOI] [PubMed] [Google Scholar]
- 7.Geissdörfer W, Ratajczak A, Hillen W. Transcription of ppK from Acinetobacter sp. strain ADP1 encoding a putative polyphosphate kinase is induced by phosphate starvation. Appl Environ Microbiol. 1998;64:896–901. doi: 10.1128/aem.64.3.896-901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes C M, Silva G, Oliveira S, LeGall J, Liu M Y, Xavier A V, Rodrigues-Pousada C, Teixeira M. Studies on the redox centers of the terminal oxidase from Desulfovibrio gigas and evidence for its interaction with rubredoxin. J Biol Chem. 1997;272:22502–22508. doi: 10.1074/jbc.272.36.22502. [DOI] [PubMed] [Google Scholar]
- 9.Gralton E M, Campbell A L, Neidle E L. Directed introduction of DNA cleavage sites to produce a high-resolution genetic and physical map of the Acinetobacter sp. strain ADP1 (BD413UE) chromosome. Microbiology. 1997;143:1345–1357. doi: 10.1099/00221287-143-4-1345. [DOI] [PubMed] [Google Scholar]
- 10.Grund A, Shapiro J, Fennewald M, Bacha P, Leahy J, Markbreiter K, Nieder M, Toepfer M. Regulation of alkane oxidation in Pseudomonas putida. J Bacteriol. 1975;123:546–556. doi: 10.1128/jb.123.2.546-556.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugenholtz J, Ljungdahl L G. Metabolism and energy generation in homoacetogenic clostridia. FEMS Microbiol Rev. 1990;7:383–389. doi: 10.1111/j.1574-6968.1990.tb04941.x. [DOI] [PubMed] [Google Scholar]
- 12.Hunger M, Schmucker R, Veerebrahma K, Hillen W. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene. 1990;87:45–51. doi: 10.1016/0378-1119(90)90494-c. [DOI] [PubMed] [Google Scholar]
- 13.Juni E, Janik A. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 15.Lotz, W. 1997. Personal communication.
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 17.Palmen R, Vosman B, Buijsman P, Breek C K, Hellingwerf K J. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J Gen Microbiol. 1993;139:295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 18.Parche S, Geissdörfer W, Hillen W. Identification and characterization of xcpR encoding a subunit of the general secretory pathway necessary for dodecane degradation in Acinetobacter calcoaceticus ADP1. J Bacteriol. 1997;179:4631–4634. doi: 10.1128/jb.179.14.4631-4634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratajczak A, Geissdörfer W, Hillen W. Alkane hydroxylase from Acinetobacter sp. strain ADP1 is encoded by alkM and belongs to a new family of bacterial integral-membrane hydrocarbon hydroxylases. Appl Environ Microbiol. 1998;64:1175–1179. doi: 10.1128/aem.64.4.1175-1179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schirmer F, Ehrt S, Hillen W. Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J Bacteriol. 1997;179:1329–1336. doi: 10.1128/jb.179.4.1329-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strätz M, Mau M, Timmis K N. System to study horizontal gene exchange among microorganisms without cultivation of recipients. Mol Microbiol. 1996;22:207–215. doi: 10.1046/j.1365-2958.1996.00099.x. [DOI] [PubMed] [Google Scholar]
- 24.van Beilen J B, Wubbolts M G, Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994;5:161–174. doi: 10.1007/BF00696457. [DOI] [PubMed] [Google Scholar]
- 25.Witholt B, de Smet M J, Kingma J, van Beilen J B, Kok M, Lageveen R G, Eggink G. Bioconversions of aliphatic compounds by Pseudomonas oleovorans in multiphase bioreactors: background and economic potential. Trends Biotechnol. 1990;8:46–52. doi: 10.1016/0167-7799(90)90133-i. [DOI] [PubMed] [Google Scholar]