Abstract
A recombinant plasmid isolated from a Mycobacterium fortuitum genomic library by selection for gentamicin and 2-N′-ethylnetilmicin resistance conferred low-level aminoglycoside and tetracycline resistance when introduced into M. smegmatis. Further characterization of this plasmid allowed the identification of the M. fortuitum tap gene. A homologous gene in the M. tuberculosis H37Rv genome has been identified. The M. tuberculosis tap gene (Rv1258 in the annotated sequence of the M. tuberculosis genome) was cloned and conferred low-level resistance to tetracycline when introduced into M. smegmatis. The sequences of the putative Tap proteins showed 20 to 30% amino acid identity to membrane efflux pumps of the major facilitator superfamily (MFS), mainly tetracycline and macrolide efflux pumps, and to other proteins of unknown function but with similar antibiotic resistance patterns. Approximately 12 transmembrane regions and different sequence motifs characteristic of the MFS proteins also were detected. In the presence of the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP), the levels of resistance to antibiotics conferred by plasmids containing the tap genes were decreased. When tetracycline accumulation experiments were carried out with the M. fortuitum tap gene, the level of tetracycline accumulation was lower than that in control cells but was independent of the presence of CCCP. We conclude that the Tap proteins of the opportunistic organism M. fortuitum and the important pathogen M. tuberculosis are probably proton-dependent efflux pumps, although we cannot exclude the possibility that they act as regulatory proteins.
Mycobacteria cause several diseases, including tuberculosis (Mycobacterium tuberculosis), leprosy (M. leprae), systemic infections in AIDS patients (M. avium), and nosocomial and opportunistic infections (M. fortuitum). The treatment of mycobacterial infections is often difficult because these bacteria are intrinsically resistant to most common antibiotics and chemotherapeutic agents (21). Furthermore, the emergence of multidrug-resistant strains has become an additional handicap in the control of tuberculosis.
In recent years, considerable work has been done on the characterization of genes involved in acquired drug resistance in mycobacteria, specifically following the emergence of clinical isolates of multidrug-resistant M. tuberculosis (26). This work has led to the identification of the molecular basis of resistance: structural or metabolic genes determine a high level of resistance to a single drug when altered. In most cases, multidrug-resistant isolates have accumulated independent mutations in several of these genes (14).
There are several examples in other bacterial species of a multidrug resistance phenotype determined by a single gene product (29, 35). Such products are typically membrane transport proteins which can remove toxic compounds by active transport after they have entered the cytoplasm by diffusion. These bacterial membrane efflux pumps form a large and heterogeneous family of energy-dependent membrane proteins capable of extruding either a single antibiotic, such as tetracycline, or a wide variety of chemically and structurally unrelated substances. In these pumps, mutations can increase the level of resistance (18) or broaden the range of substances transported. On the other hand, an inhibitor of such pumps can make the organism more susceptible to antimicrobial agents (27). The characterization of efflux pumps may thus allow the design of new therapeutic strategies.
Recently, the efflux pump LfrA, conferring low-level resistance to fluoroquinolones and other compounds (22, 42), Tet(V), conferring resistance to tetracycline (9), and Emb, conferring resistance to ethambutol (43), were found in nonpathogenic M. smegmatis. However, in a characterization of the efpA gene, encoding a putative efflux protein from M. tuberculosis (11), the associated resistance phenotype could not be detected.
In this work, we describe the characterization of a gene encoding a putative multidrug efflux pump which confers low-level resistance to aminoglycosides and tetracycline in the opportunistic pathogen M. fortuitum (the terms tapfor and Tapfor are used here to indicate the gene and the protein, respectively, isolated from M. fortuitum). Its homologue in the important pathogen M. tuberculosis confers resistance to tetracycline (the corresponding gene and protein are designated taptub and Taptub, respectively). These proteins show a significant homology to some proton drug antiporters, especially tetracycline and macrolide transporters, suggesting they are proton-motive-force-dependent efflux pumps capable of exporting tetracycline and some aminoglycosides.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
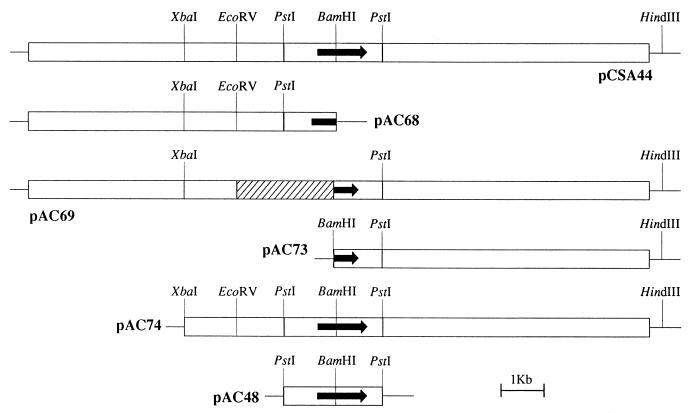
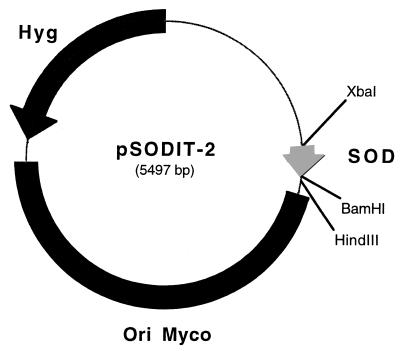
The reference strains and plasmids are listed in Table 1. The subclones constructed from plasmid pCSA44 are shown in Fig. 1. The expression vector pSODIT2 was constructed as follows. The M. tuberculosis superoxide dismutase promoter (46) was amplified with the oligonucleotides 5′-GCTCTAGACAGCCTGGGGGCGTCCTG-3′ and 5′-CGGGATCCCACGGCATTCCTTCCTTCGAT-3′ as primers, M. tuberculosis H37Rv DNA as a template, and Deep Vent polymerase (New England Biolabs). The primers provide artificial XbaI and BamHI sites at the ends of the superoxide dismutase promoter. The PCR product was digested with XbaI and BamHI and ligated in the shuttle vector pOLYG (32) cut with the same enzymes to produce pSODIT-2 (Fig. 2).
TABLE 1.
Relevant characteristics of the bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| M. smegmatis mc2155 | Efficient plasmid transformation mutant | 41 |
| E. coli XL1-Blue | Host strain for plasmid propagation | 39 |
| E. coli DH5α | Host strain for plasmid propagation | 39 |
| Plasmids | ||
| pSUM36 and pSUM38 | KmrMycobacterium and E. coli shuttle vectors | 2 |
| pCSA44 | pSUM36 with a 15-kb fragment from M. fortuitum containing the tapfor gene | This work |
| pCSA44* | pCSA44 BamHI digested, blunt ended, and religated | This work |
| pAC68 | pCSA44 with BamHI-HindIII deleted | This work |
| pAC69 | pCSA44 with BamHI-EcoRV deleted | This work |
| pAC73 | pSUM36 with BamHI-HindIII fragment from pCSA44 | This work |
| pAC74 | pSUM38 with XbaI-HindIII fragment from pCSA44 | This work |
| pAC48 | pSUM36 with a 2.3-kb PstI fragment from pCSA44 | This work |
| pOLYG | Hygr mycobacterial expression vector | 32 |
| pSODIT-2 | Hygr pOLYG with the superoxide dismutase promoter | This work |
| pefpD | pSODIT-2 with a 1.2-kb BamHI-HindIII fragment containing the taptub gene | This work |
FIG. 1.
Physical map of pCSA44 and its derivatives. pCSA44 contains the 15-kb DNA fragment with the tapfor gene (indicated by arrow) from M. fortuitum. Only the PstI sites delimiting the 2.3-kb fragment containing the tapfor gene are shown. The hatched area in plasmid pAC69 represents the deleted fragment.
FIG. 2.
Map of the expression vector pSODIT-2. The thick black line denotes the region containing the mycobacterial origin of replication; the thin line denotes the E. coli origin of replication and ampicillin selection marker. Hyg, hygromycin selection marker; SOD, superoxide dismutase promoter.
All media were obtained from Difco Laboratories, Detroit, Mich. Middlebrook 7H9 broth with 0.05% Tween 80, Middlebrook 7H10 agar, or Luria broth (LB) with 0.5% Tyloxapol (Sigma Chemical Co., St. Louis, Mo.) was used to culture the mycobacterial strains. Escherichia coli XL1-Blue or E. coli DH5α was cultured in LB or brain heart infusion. All the cultures were incubated at 37°C. Kanamycin A (Sigma) at 20 μg/ml or hygromycin (Boehringer GmbH, Mannheim, Germany) at 50 μg/ml for mycobacteria or 250 μg/ml for E. coli was added when necessary to maintain the plasmids.
Antibiotic susceptibility testing.
The MICs of the antibiotics were determined by serial dilution of the antibiotics either by plate dilution or by microdilution in liquid medium. Mueller-Hinton agar (Difco) plates containing antibiotic were inoculated with cultures diluted to 105 CFU/ml and incubated at 37°C for up to 5 days. For liquid cultures, 100 μl of medium with antibiotic was serially diluted in 100-μl aliquots of medium without antibiotic in microtiter plate wells. Subsequently, the microtiter plate wells were inoculated with 10 μl of cultures diluted to 106 CFU/ml and incubated at 37°C for up to 4 days. The Alamar Blue assay (13) was used in order to precisely determine the lowest concentration that inhibited growth. All MIC determinations were repeated at least three times.
The MICs of the antibiotics were determined with and without the membrane deenergizer carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma) at a concentration of 0.8 μg/ml in order to evaluate the effects of alterations in the proton gradient on resistance levels (20).
Assay of tetracycline accumulation.
The accumulation of tetracycline was monitored as described previously (22, 24). Briefly, M. smegmatis mc2155 was cultured in Middlebrook 7H9 broth until the exponential phase was reached. Cells were then pelleted, washed with 50 mM KPO4–10 mM MgSO4 (pH 7.0), and resuspended to 5 mg/ml in the same buffer. For each experiment, 5 mg of cells was incubated at 37°C for 10 min, and 20 μl of tritiated tetracycline (American Radiolabeled Chemicals Inc., St. Louis, Mo.) at 125 μCi/ml (1 Ci/mmol) was added. Samples of 50 μl were removed every 4 min, diluted into 1 ml of 100 mM KPO4–100 mM LiCl (pH 7.0), filtered through 0.45-μm-pore-size filters (Millipore), and quickly washed with 8 ml of the same buffer. Finally, filters were dried and counts were determined with a liquid scintillation counter (Beckman LS-6000 IC). When the effect of protonophores was studied, CCCP was added to a final concentration of 20 μg/ml 10 min after the addition of tetracycline. In a different experiment, cells were preincubated with CCCP 60 min before the addition of tetracycline. In both cases, the cells were treated as described above.
Electroporation of M. smegmatis.
M. smegmatis mc2155 was transformed by electroporation. Briefly, competent cells were prepared by culturing strains to an optical density (OD) of 0.75 and were washed three times with ice-cold 10% glycerol. Aliquots were rapidly frozen in a dry ice-ethanol bath and stored at −80°C. Electroporation was performed with a Gene Pulser (Bio-Rad Laboratories Inc., Richmond, Calif.) at 2.5 kV, 25 μF, and 1,000 Ω. Plates containing 20 μg of kanamycin per ml or 50 μg of hygromycin per ml were used to select the transformants.
Molecular biology procedures.
Electrophoresis, digestion, ligation, dephosphorylation, plasmid extraction, transformation, and electroporation were performed as described elsewhere (39) or according to supplier recommendations (Boehringer). Double-stranded DNA sequencing was performed by the dideoxynucleotide chain termination method with M13 universal primers, a Cy5 Autocycle sequencing kit (Pharmacia), and an ALFexpress DNA analysis system (Pharmacia) in accordance with manufacturer instructions.
Computer analysis.
Nucleotide and amino acid sequences were analyzed and compared with Genetics Computer Group software (10) either at the Centro Nacional de Biotecnología, Madrid, Spain, or at the Human Genome Mapping Project, Cambridge, United Kingdom. Databases were searched with the programs BLAST and PSI-BLAST (3a) at the National Center for Biological Information. The TMpred program (16) was used to estimate the number of transmembrane segments (TMS). Clustal W (44) was used to generate the phylogenetic tree, and TreeView (33) was used to display it.
Nucleotide sequence accession number.
The nucleotide sequence of the M. fortuitum tapfor gene has been deposited in GenBank under accession no. AJ000283.
RESULTS
Isolation of an M. fortuitum multidrug resistance gene, tapfor.
We previously described the construction of an M. fortuitum genomic library in M. smegmatis mc2155 and the selection of 80 clones that conferred resistance to 4 μg of gentamicin per ml (2). An analysis of some of these clones allowed the characterization of the aac(2′)-Ib gene (3), which encodes the enzyme aminoglycoside 2′-N-acetyltransferase enzyme [AAC(2′)]. The 80 gentamicin-resistant clones were then screened for resistance to other aminoglycosides, and 7 clones were found to be resistant to 2-N′-ethylnetilmicin, an aminoglycoside that lacks the 2′ amino group and therefore is not a substrate for the AAC(2′) enzyme. The plasmids contained in the seven clones were transferred to E. coli XL1-Blue and analyzed by endonuclease restriction. Six of the seven plasmids were found to contain the aac(2′)-Ib gene; plasmid pCSA44 did not contain the aac(2′)-Ib gene. When transformed into M. smegmatis mc2155, plasmid pCSA44 conferred gentamicin and 2′-N-ethylnetilmicin resistance. This plasmid was further studied.
The insert of pCSA44 is 15 kb long. Three unique sites (XbaI, EcoRV, and BamHI) are located within the insert. Initially, these sites and the HindIII site of the polylinker of pSUM36 were used to construct four subclones: pAC68, pAC69, pAC73, and pAC74 (see Fig. 1 and Table 1 for further details on the constructs). The three constructs in which the BamHI site was affected (pAC68, pAC69, and pAC73) abolished the phenotype of 2′-N-ethylnetilmicin resistance. Only plasmid pAC74, which contained the BamHI site unaffected, produced the same phenotype as parental plasmid pCSA44.
In order to confirm that the region around the BamHI site was responsible for the resistance phenotype, two further plasmids were constructed. First, pCSA44 was digested with BamHI, filled in, and religated, producing plasmid pCSA44*; the latter plasmid was unable to confer the resistance phenotype to M. smegmatis mc2155 (Table 2). Second, the 2.3-kb PstI fragment containing the BamHI site was cloned in vector pSUM36, resulting in pAC48; the latter plasmid conferred the same phenotype to M. smegmatis mc2155 as pCSA44 and pAC74.
TABLE 2.
Relative MICs for M. smegmatis mc2155 carrying diverse constructs containing the M. fortuitum tapfor gene and the M. tuberculosis taptub genea
| Compound(s) | Relative MICs (actual MICs), in μg/ml, for M. smegmatis mc2155 harboring the following plasmid:
|
||||
|---|---|---|---|---|---|
| None or pSUM36 | pCSA44 or pAC48 (tapfor) | pCSA44*(tapfor disrupted) | pSODIT-2 | pefpD (taptub) | |
| 2′-N-Ethylnetilmicin | 1 (8) | 16 | 1 | 2 | 2 |
| 2′-N-Ethylnetilmicin + CCCP | 1 | 4 | 1 | 1 | 1 |
| 6′-N-Ethylnetilmicin | 1 (16) | 8 | 0.5 | 1 | 1 |
| 6′-N-Ethylnetilmicin + CCCP | 1 | 1 | 1 | 1 | 1 |
| Tetracycline | 1 (0.5) | 8–16 | 1 | 1 | 4 |
| Tetracycline + CCCP | 1 | 4 | 1 | 1 | 2 |
| Gentamicin | 0.5–1 (2) | 2 | 0.5 | 1 | 1 |
| Gentamicin + CCCP | 1 | 0.5–1 | 0.5 | 1 | 0.5 |
| Streptomycin | 0.5–1 (2) | 2 | 0.5 | 0.5 | 0.5 |
| Streptomycin + CCCP | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
See Table 1 and Fig. 1 for details on the constructs. The MICs were determined by plate dilution and microdilution and are expressed as relative resistances, i.e., the MIC for each strain divided by the MIC for wild-type M. smegmatis mc2155. The actual MICs shown were those for wild-type M. smegmatis mc2155.
The sequence of both strands of the 2.3-kb PstI DNA fragment in pAC48 was determined. A search for coding regions revealed the presence of an open reading frame (ORF) with a G+C content of 68.1%, in agreement with values previously described for mycobacterial genes and genomes (7), as well as the G+C content of the third base of the codons (88.5%). This ORF spans from nucleotides 620 to 1849 and contains the BamHI site mentioned above at position 736. Preceding the ATG start codon, two putative ribosome binding site sequences, GATA and AGA, were found. No putative transcription signals were detected by inspection of the sequence, but since pAC48 and pAC49 contain the same insert in opposite orientations and confer the same level of resistance (data not shown), the upstream region probably contains the necessary transcription signals. This ORF has been designated tapfor (tetracycline-aminoglycoside resistance) and encodes a protein that confers low-level resistance to aminoglycosides and tetracycline (see below).
Cloning of the M. tuberculosis taptub gene.
Homology searches of databases with the M. fortuitum tapfor nucleotide sequence found the ORF Rv1258 in the M. tuberculosis genome (8), which showed 71% nucleotide identity. This ORF was amplified by PCR from M. tuberculosis H37Rv DNA with the primers 5′-AAGGATCCATGAGAAACAGCAACCGC-3′ and 5′-AGCTAAGCTTCAGGCCAGCCAGCAC-3′, which contain, respectively, the start and stop codons proposed for the ORF (indicated in bold). The 1.26-kb product was digested with BamHI and HindIII and ligated to pSODIT-2. The resulting plasmid was designated pefpD, following the nomenclature used to designate efflux pumps in M. tuberculosis (11). This plasmid was electroporated into M. smegmatis mc2155, and changes in susceptibilities to a wide range of compounds were determined. The homologous gene found in M. tuberculosis has been designated taptub.
Further sequence analysis of the putative Tap proteins.
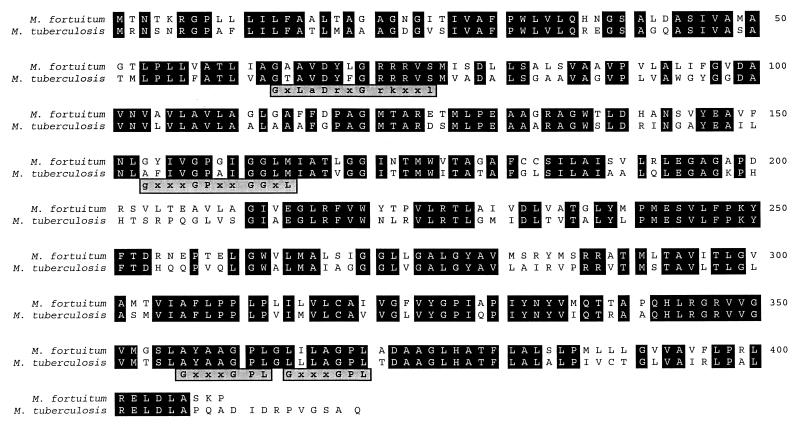
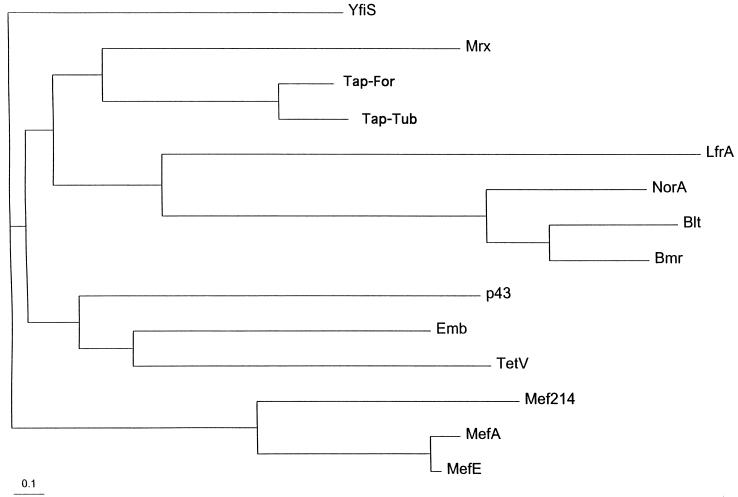
The deduced protein sequences of the Tapfor and Taptub proteins showed 68% amino acid identity and 83% amino acid similarity. The alignment of both proteins is shown in Fig. 3. Protein databases were searched with the deduced Tapfor sequence in order to find other, similar proteins. The homologous proteins identified with the program BLAST are summarized in Table 3. Proteins showing the highest scores for amino acid identity to Tapfor over the whole protein sequence are Mrx from E. coli, a protein with unknown function but involved in macrolide resistance (31); another E. coli protein closely related to Mrx and encoded by an ORF located close to the macrolide phosphotransferase K gene (17); and a putative multidrug transporter (YfiS) identified in the sequencing of the Bacillus subtilis genome (19). In addition, three M. smegmatis proteins were found to be homologous to Tapfor: a putative transport protein encoded by a gene adjacent to (but probably not involved in) the ethambutol resistance operon (43), a tetracycline efflux pump (9), and the quinolone transporter LfrA (42). Lower, but significant, homology was found with the p43 protein of E. coli, whose gene is adjacent to but independent from the ferric enterobactin transport operon (5, 40). Approximately 20% amino acid identity was found with other well-known multidrug resistance proteins, such as the macrolide efflux pumps (Mef) from Lactobacillus lactis, Streptococcus pyogenes (6), and Streptococcus pneumoniae; the B. subtilis transporters Bmr (27) and Blt (1); and the NorA protein from Staphylococcus aureus (45). The latter proteins belong to the major facilitator superfamily (MFS) of proton antiporter proteins and are multidrug efflux pumps (35). In addition, some homology was found with diverse metabolite transporters (data not shown). A phylogenetic tree showing the relationships among the proteins mentioned is shown in Fig. 4, in which it can be appreciated that the E. coli Mrx protein is the closest homologue of the Tap proteins.
FIG. 3.
Alignment of the deduced M. fortuitum and M. tuberculosis Tap protein sequences. Identical amino acids are shown in black boxes. Various sequence motifs are shown in grey boxes under the alignment: motif GxLaDrxGrkxxl is characteristic of the MFS and is also described in the Prosite database as a sugar transport protein signature (accession no. PS00216); motif gxxxGPxxGGxl has been described as characteristic of drug export proteins; and motif GxxxGPL is characteristic of the 12-TMS family.
TABLE 3.
Amino acid sequence identity between the deduced amino acid sequence of the M. fortuitum Tapfor protein and the sequences of other, homologous proteins of different origins, as identified with the program BLAST
| Protein | Organism | % Identity to Tapfora | Reference or source |
|---|---|---|---|
| Mrx | Escherichia coli | 29.7 | 31 |
| Product of gene adjacent to mphK gene | Escherichia coli | 28.9 | 17 |
| YfiS | Bacillus subtilis | 27.2 | 19 |
| Emb | Mycobacterium smegmatis | 26.6 | 43 |
| TetV | Mycobacterium smegmatis | 25.7 | 9 |
| LfrA | Mycobacterium smegmatis | 22.6 | 42 |
| p43 | Escherichia coli | 22.4 | 5, 40 |
| Mef214 | Lactobacillus lactis | 22.3 | V. Perreten et al.; GenBank accession no. X92946 |
| MefA | Streptococcus pyogenes | 21.6 | 6 |
| Bmr | Bacillus subtilis | 21.5 | 27 |
| MefE | Streptococcus pneumoniae | 20.2 | A. Tait-Kamradt et al.; GenBank accession no. U83667 |
| Blt | Bacillus subtilis | 19.7 | 1 |
| NorA | Staphylococcus aureus | 19.5 | 45 |
The degree of identity was determined with the program Gap from the Genetics Computer Group software package.
FIG. 4.
Phylogenetic tree showing the relationships among different proteins homologous to the mycobacterial Tap proteins. A list of proteins analyzed is given in Table 3.
The program PSI-BLAST was used for a further search of the databases and identified many tetracycline efflux pumps, macrolide efflux pumps, and other multidrug resistance proteins as homologs (data not shown).
Several sequence motifs have been described as important for the structure or function of the MFS proteins (34, 35), although it is difficult to specify roles for specific regions. Pao et al. (34) described a sequence motif characteristic of most protein families in the MFS, and the same motif is described in the Prosite database as a sugar transport protein signature (accession no. PS00216). This motif is present in both Tap proteins, as are other motifs characteristics of drug export proteins and the 12-TMS family (Fig. 3). Other motifs such as the ones described by Pao et al. (34) for drug efflux systems possessing 12 or 14 TMS could not be clearly identified.
Members of the MFS contain either 12 or 14 TMS (35). Transmembrane regions of both Tap proteins were determined with the TMpred algorithm (16), which suggested the presence of at least 10 TMS (data not shown), although it was not possible to unambiguously determine the number of TMS from the primary sequence (37). Most likely, these proteins have 12 TMS because of their homology to other proteins of the MFS with 12 TMS and the presence of the sequence motif characteristic of the 12-TMS family.
Substrate profile, resistance levels, and proton motive force dependence of the Tap proteins.
Sensitivity tests to determine the substrate profile, resistance levels, and proton dependence of the Tap proteins were carried out by plate dilution and by microdilution. Both methods gave the same results.
The clone containing the tapfor gene was initially identified by its ability to confer an increase in the levels of resistance of M. smegmatis mc2155 to the aminoglycosides gentamicin and 2-N′-ethylnetilmicin. However, the screening of other compounds showed that Tapfor also confers an increase in the levels of resistance to 6-N′-ethylnetilmicin, streptomycin, and tetracycline (Table 2) but not netilmicin (data not shown). The MICs for the aminoglycosides neomycin, amikacin, and kanamycin could not be tested with the existing plasmid constructions because of the specificity of the resistance marker kan of the pSUM36 vector used. There was no increase in the levels of resistance to other compounds, such as chloramphenicol, ciprofloxacin, and ethidium bromide (data not shown).
The Taptub protein has a more restricted specificity, conferring resistance to tetracycline (Table 2) but not streptomycin, gentamicin, netilmicin, 2-N′-ethylnetilmicin, 6-N′-ethylnetilmicin, or any other aminoglycoside tested (including neomycin, kanamycin, amikacin, dibekacin, sisomycin, puromycin, and tobramycin). It also has no effect on resistance to fluoroquinolones (ciprofloxacin and ofloxacin); antituberculosis drugs (isoniazid, rifampin, ethionamide, and pyrazinamide); or other compounds, such as chloramphenicol, ethidium bromide, acridine orange, capreomycin, chlortetracycline, crystal violet, daunomycin, doxycycline, doxorubicin, erythromycin, ethambutol, fusidic acid, lincomycin, minocycline, p-aminosalicylic acid, phosphomycin, thiacetazone, vancomycin, clarithromycin, cycloserine, or rhodamine.
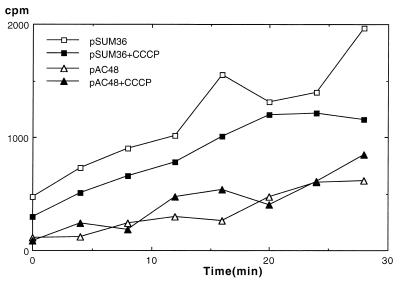
The resistance levels conferred by either of the Tap proteins to M. smegmatis are low. In general, membrane efflux pumps also confer low levels of resistance, which contrast with the high levels that enzymatic inactivation mechanisms can confer. In addition membrane efflux pumps can confer resistance to unrelated antibiotics. These findings, together with the sequence homology to some multidrug efflux pumps, led us to consider the hypothesis that the Tap proteins act as proton-motive-force-dependent efflux pumps. In order to test this hypothesis, two related experiments were performed. First, the levels of resistance conferred by the Tap proteins were compared in the presence and absence of the membrane energy uncoupler CCCP, which is known to disperse the proton gradient across membranes. The MICs (expressed as relative resistance) for the different constructs studied are shown in Table 2. The MICs for wild-type M. smegmatis mc2155 carrying no plasmid were not altered by the presence of CCCP; however, the MICs for cells carrying either the tapfor gene or the tapfub gene were notably reduced by the presence of CCCP. Second, the time course of the accumulation of tritiated tetracycline was determined for M. smegmatis mc2155 carrying either the vector pSUM36 or the plasmid pAC48, which contains the tapfor gene. The results of these experiments (Fig. 5) showed that M. smegmatis mc2155 carrying the vector pSUM36 accumulated more tetracycline than M. smegmatis mc2155 carrying the plasmid pAC48. However, the accumulation levels were not changed by preincubation with CCCP (Fig. 5) or by the addition of CCCP during the time course (data not shown), showing that the mechanism of resistance in cells expressing the tapfor gene apparently was not affected by the proton motive force.
FIG. 5.
Time course of tetracycline accumulation (ordinate) for M. smegmatis cells (open symbols) carrying the plasmid pSUM36 or pAC48 (tapfor gene). The closed symbols represent the same time course when the cells were preincubated for 60 min in the presence of CCCP.
DISCUSSION
Mycobacteria are intrinsically resistant to a wide range of antibiotics. The thick, waxy mycobacterial cell wall often has been implicated as one of the reasons for this resistance (21, 30). Although the cell wall considerably slows down the diffusion of antibiotics into the bacterium, it cannot completely prevent it. Other active mechanisms of resistance therefore must be present. In the last few years, considerable work has been done on the characterization of such resistance mechanisms in mycobacteria (26), especially as the emergence of multidrug-resistant strains has become an important problem. In other bacteria, genes conferring resistance to a range of different and unrelated antibiotics usually encode membrane efflux pumps (35). The recent publication of the complete M. tuberculosis genome (8) has revealed the presence of genes for 20 putative multidrug transporters, as in other genomes of the same size (38). One of these genes, efpA, has been characterized, but no resistance phenotype could be detected (11).
The sequences of the putative membrane efflux pumps Tapfor from M. fortuitum and Taptub from M. tuberculosis, described in the present work, showed homology to those of a wide range of membrane proteins (Table 3). Some of these proteins are efflux proteins that have been shown to recognize antibiotics as substrates; these proteins include the products of the lfrA, tet(V), emb, norA, bmr, and blt genes. When the program PSI-BLAST was used, proton-dependent tetracycline and macrolide efflux pumps were identified as homologous to the Tap proteins. The presence of TMS and the high percentage of hydrophobic amino acids strongly suggest a membrane location of the Tap proteins. In addition, sequence motifs characterizing the cluster of 12 TMS in members of the MFS family (35) were found (Fig. 3).
Tap proteins conferred low-level resistance to tetracycline and aminoglycosides to M. smegmatis. When studying the tapfor gene, we detected slight differences in the resistance levels conferred by plasmids pCSA44 and pAC48. This finding could reflect either a difference in the stabilities of the plasmids due to insert size or, more likely, the presence of a gene homologous to the M. tuberculosis putative regulatory ORF Rv1255, which would be present in pCSA44 but not in pAC48.
The specificities of the two Tap proteins are different: Taptub confers resistance only to tetracycline, but Tapfor confers resistance to tetracycline and the structurally unrelated aminoglycosides streptomycin, gentamicin, 2′-N-ethylnetilmicin, and 6′-N-ethylnetilmicin. The genes encoding the S. aureus QacA and QacB efflux proteins differ by only 7 nucleotides, but this difference is enough to confer different specificities (36). Therefore, the difference in the nucleotide sequences of the tap genes (29%) would explain the difference in the specificities of the Tap proteins.
We determined the effect of the protonophore CCCP on the resistance levels conferred by the Tap proteins. First, as CCCP is toxic for bacteria, we determined the MICs of CCCP in LB (2 μg/ml) and Middlebrook 7H9 broth (4 μg/ml). Our results contrast with the data found by other authors, who used 15 μg/ml to study the LfrA transporter (42) in the same strain (M. smegmatis mc2155).
The resistance levels in the presence of the protonophore CCCP are shown in Table 2. CCCP reduced the MICs of all five antibiotics used (2′-N-ethylnetilmicin, 6′-N-ethylnetilmicin, tetracycline, gentamicin, and streptomycin) in cells containing the plasmid-carried tapfor gene. For plasmid pefpD (which carries the taptub gene), CCCP reduced the MIC of tetracycline. Aminoglycosides are known to enter cells by an energy-dependent mechanism (4, 25); as a result, CCCP can affect the levels of resistance to aminoglycosides by decreasing both the uptake of aminoglycosides and extrusion through Tap proteins. Our results showed that the MICs of aminoglycosides were reduced in the presence of CCCP, suggesting that a decrease in the extrusion of aminoglycosides was the predominant effect.
The accumulation experiments with radiolabelled tetracycline showed that the cells containing the tapfor gene accumulated only one-half the tetracycline accumulated by the same strain harboring only the vector pSUM36 (Fig. 5). However, these levels were not substantially changed by preincubation of the cells with CCCP (Fig. 5) or by the addition of CCCP during the time course of tetracycline accumulation (data not shown). With proton-dependent membrane efflux pumps, the addition of CCCP should increase the accumulation of antibiotic in cells overexpressing the pumps. We do not have a plausible explanation for our results.
In summary, the majority of the results in our work support the idea that the Tap proteins are membrane efflux pumps. First, both Tap proteins have sequence similarities to other proteins associated with multidrug-resistance phenotypes, especially tetracycline and macrolide proton-dependent efflux pumps, and a sequence motif characteristic of drug export proteins. Second, the presence of a number of TMS suggests a membrane location. Third, the Tap proteins confer low-level resistance and have a broad substrate specificity. Fourth, the resistance levels of tap-expressing cells are decreased in the presence of CCCP. Fifth, cells expressing Tap proteins accumulate less tetracycline than control cells. The fact that the Tap proteins could export aminoglycosides is an interesting topic for future work, since only the E. coli efflux pump MdfA has been described as a pump for aminoglycosides (12).
Regulatory functions have been described as well for the ABC family of transporters (15). The idea of the Tap proteins regulating other specific resistance mechanisms is an interesting hypothesis, especially because a recent publication has indicated that the expression of the E. coli multiple-antibiotic-resistance marA gene in M. smegmatis mc2155 produces an increase in the levels of resistance to multiple drugs (23). We cannot completely rule out the possibility that the Tap proteins, either directly or indirectly, act as regulators. The lack of effect of CCCP in the tetracycline accumulation experiments is consistent with such a hypothesis. The production of the Tap proteins may alter membrane structure or permeability, resulting in different levels of antibiotic resistance.
We consider as well the possibility that the Tap proteins transport other substrates, as it has been postulated that the ability to remove antibiotics may not be the primary physiological role for the multidrug efflux pumps (28) and the transporter Blt is also involved in the transport of the polyamine spermidine (44a). In this connection, the presence of a sugar transport signature and the low but significant homology to some sugar transporters (data not shown) may provide some clues for the primary function of the Tap proteins.
ACKNOWLEDGMENTS
We thank Carmen Lafoz and Santiago Uranga for technical assistance and Mervyn J. Bibb for enthusiastic help in the elaboration of the phylogenetic tree. Three unknown reviewers are acknowledged for useful and interesting comments and suggestions. 2′-N-Ethylnetilmicin and 6′-N-ethylnetilmicin were kindly provided by Schering Plough Research Institute.
This work was supported by Ministry of Health, Spain (Fondo Investigaciones Sanitarias de la Seguridad Social), grant FIS97-0042 and by European Commission BIOMED grant CT96-1241. K.A.L.D. was supported by the Glaxo Wellcome Action TB Program.
REFERENCES
- 1.Ahmed M, Lyass L, Markham P N, Taylor S S, Vázquez-Laslop N, Neyfakh A A. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J Bacteriol. 1994;177:3904–3910. doi: 10.1128/jb.177.14.3904-3910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aínsa J A, Martín C, Cabeza M, De la Cruz F, Mendiola M V. Construction of a family of Mycobacterium/Escherichia coli shuttle vectors derived from pAL5000 and pACYC184: their use for cloning an antibiotic resistance gene from Mycobacterium fortuitum. Gene. 1996;176:23–26. doi: 10.1016/0378-1119(96)00202-8. [DOI] [PubMed] [Google Scholar]
- 3.Aínsa J A, Martín C, Gicquel B, Gómez-Lus R. Characterization of the chromosomal aminoglycoside 2′-N-acetyltransferase gene from Mycobacterium fortuitum. Antimicrob Agents Chemother. 1996;40:2350–2355. doi: 10.1128/aac.40.10.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryan L E, van den Elzen H M. Streptomycin accumulation in susceptible and resistant strains of Escherichia coli and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1976;9:928–938. doi: 10.1128/aac.9.6.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chenault S S, Earhart C F. Organization of genes encoding membrane proteins of the Escherichia coli ferrienterobactin permease. Mol Microbiol. 1991;5:1405–1413. doi: 10.1111/j.1365-2958.1991.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 6.Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 7.Clark-Curtiss J E. Genome structure of mycobacteria. In: McFadden J, editor. Molecular biology of the mycobacteria. London, England: Surrey University Press; 1990. pp. 77–96. [Google Scholar]
- 8.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 9.De Rossi E, Blokpoel M C J, Cantoni R, Branzoni M, Riccardi G, Young D B, De Smet K A L, Ciferri O. Molecular cloning and functional analysis of a novel tetracycline resistance determinant, tet(V), from Mycobacterium smegmatis. Antimicrob Agents Chemother. 1998;42:1931–1937. doi: 10.1128/aac.42.8.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereux J P, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doran J L, Pang Y, Mdluli K E, Moran A J, Victor T C, Stokes R W, Mahenthiralingam E, Kreiswirth B N, Butt J L, Baron G S, Treit J D, Kerr V J, van Helden P D, Roberts M C, Nano F E. Mycobacterium tuberculosis efpA encodes an efflux protein of the QacA transporter family. Clin Diagn Lab Immunol. 1997;4:23–32. doi: 10.1128/cdli.4.1.23-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzblau S G, Witzig R S, McLaughlin J C, Torres P, Madico G, Hernandez A, Degnan M T, Cook M B, Quenzer V K, Ferguson R M, Gilman R H. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol. 1998;36:362–366. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heym B, Honoré N, Truffot-Pernot C, Banerjee A, Schurra C, Jacobs W R, van Embden J D A, Grosset J H, Cole S T. Implications of multidrug resistance for the future of short course chemotherapy of tuberculosis: a molecular study. Lancet. 1994;344:293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 15.Higgins C F. The ABC of channel regulation. Cell. 1995;82:693–696. doi: 10.1016/0092-8674(95)90465-4. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann K, Stoffel W. TMbase, a database of membrane spanning protein segments. Biol Chem Hoppe Seyler. 1993;347:166. [Google Scholar]
- 17.Kim S K, Baek M C, Choi S S, Kim B K, Choi E C. Nucleotide sequence, expression and transcriptional analysis of the Escherichia coli mphK gene encoding macrolide-phosphotransferase K. Mol Cells. 1996;6:153–160. [Google Scholar]
- 18.Klyachko K A, Neyfakh A A. Paradoxical enhancement of the activity of a bacterial multidrug transporter caused by substitutions of a conserved residue. J Bacteriol. 1998;180:2817–2821. doi: 10.1128/jb.180.11.2817-2821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 20.Levy S B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992;36:695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Rosemberg E Y, Nikaido H. Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc Natl Acad Sci USA. 1995;92:11254–11258. doi: 10.1073/pnas.92.24.11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Takiff H E, Nikaido H. Active efflux of fluoroquinolones in Mycobacterium smegmatis mediated by LfrA, a multidrug efflux pump. J Bacteriol. 1996;178:3791–3795. doi: 10.1128/jb.178.13.3791-3795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDermott P F, White D G, Podglajen I, Alekshun M N, Levy S B. Multidrug resistance following expression of the Escherichia coli marA gene in Mycobacterium smegmatis. J Bacteriol. 1998;180:2995–2998. doi: 10.1128/jb.180.11.2995-2998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMurray L, Petrucci R E, Jr, Levy S B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller M H, Edberg S C, Mandel L J, Behar C F, Steigbigel N H. Gentamicin uptake in wild-type and aminoglycoside-resistant small-colony mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1980;18:722–729. doi: 10.1128/aac.18.5.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neyfakh A A, Bidnenko V E, Chen L B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci USA. 1991;88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neyfakh A A. Natural functions of bacterial multidrug transporters. Trends Microbiol. 1997;5:309–313. doi: 10.1016/S0966-842X(97)01064-0. [DOI] [PubMed] [Google Scholar]
- 29.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 31.Noguchi N, Emura A, Matsuyama H, O’Hara K, Sasatsy M, Kono M. Nucleotide sequence and characterization of erythromycin resistance determinant that encodes macrolide 2′-phosphotransferase I in Escherichia coli. Antimicrob Agents Chemother. 1995;39:2359–2363. doi: 10.1128/aac.39.10.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Gaora P, Barnini S, Hayward C, Filley E, Rook G, Young D, Thole J. Mycobacteria as immunogens: development of expression vectors for use in multiple mycobacterial species. Med Principles Practice. 1997;6:91–96. [Google Scholar]
- 33.Page R D M. TreeView: an application to display phylogenetic trees on personal computers. CABIOS. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 34.Pao S S, Paulsen I T, Saier M H., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulsen I T, Brown M H, Littlejohn T G, Mitchell B A, Skurray R A. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci USA. 1996;93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulsen I T, Skurray R A. Topology, structure and evolution of two families of proteins involved in antibiotic and antiseptic resistance in eukaryotes and prokaryotes—an analysis. Gene. 1993;124:1–11. doi: 10.1016/0378-1119(93)90755-r. [DOI] [PubMed] [Google Scholar]
- 38.Saier M H, Jr, Paulsen I T, Sliwinski M K, Pao S S, Skurray R A, Nikaido H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–274. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Shea C M, Mcintosh M A. Nucleotide sequence and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein-dependent systems in Escherichia coli. Mol Microbiol. 1991;5:1415–1428. doi: 10.1111/j.1365-2958.1991.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 41.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 42.Takiff H E, Cimino M, Musso M C, Weisbrod T, Martínez R, Delgado M B, Salazar L, Bloom B R, Jacobs W R., Jr Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc Natl Acad Sci USA. 1996;93:362–366. doi: 10.1073/pnas.93.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telenti A, Philipp W, Sreevatsan S, Bernasconi C, Stockbauer K E, Wieles B, Musser J M, Jacobs W R. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med. 1997;3:567–570. doi: 10.1038/nm0597-567. [DOI] [PubMed] [Google Scholar]
- 44.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix code. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Woolridge D P, Vazquez-Laslop N, Markman P N, Chevalier M S, Gerner E W, Neyfakh A A. Efflux of the natural polyamine spermidine facilitated by the Bacillus subtilis multidrug transporter Blt. J Biol Chem. 1997;272:8864–8866. doi: 10.1074/jbc.272.14.8864. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida H, Bogaki M, Nakamura S, Ubutaka K, Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990;173:6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, García M J, Lathigra R, Allen B, Moreno C, van Embden J D A, Young D. Alterations in the superoxide dismutase gene of an isoniazid resistance strain of Mycobacterium tuberculosis. Infect Immun. 1992;60:2160–2165. doi: 10.1128/iai.60.6.2160-2165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]