Abstract
Kidney function‐adjusted drug dosing is currently based solely on the estimated glomerular filtration rate (GFR), however, kidney drug handling is accomplished by a combination of filtration, tubular secretion, and re‐absorption. Mechanistic physiologically‐based pharmacokinetic (PBPK) models recapitulate anatomic compartments to predict elimination from estimated perfusion, filtration, secretion, and re‐absorption, but clinical applications are limited by a lack of empiric individual‐level measurements of these functions. We adapted and validated a PBPK model to predict drug clearance from individual biomarker‐based estimates of kidney perfusion and secretory clearance. We estimated organic anion transporter‐mediated secretion via kynurenic acid clearance and kidney blood flow (KBF) via isovalerylglycine clearance in human participants, incorporating these measurements with GFR into the model to predict kidney drug clearance. We compared measured and model‐predicted clearances of administered tenofovir and oseltamivir, which are cleared by both filtration and secretion. There were 27 outpatients (age 55 ± 15 years, mean iohexol‐GFR [iGFR] 76 ± 31 mL/min/1.73 m2) in this drug clearance study. The mean observed and mechanistic model‐predicted tenofovir clearances were 169 ± 102 mL/min and 163 ± 80 mL/min, respectively; estimated mean error of the mechanistic model was 37.1 mL/min (95% confidence interval [CI]: 24–52.9), compared to a mean error of 41.8 mL/min (95% CI: 25–61.6) from regression model. The mean observed and model‐predicted oseltamivir carboxylate clearances were 183 ± 104 mL/min and 179 ± 89 mL/min, respectively; estimated mean error of the mechanistic model was 42.9 mL/min (95% CI: 29.7–56.4), versus error of 48.1 mL/min (95% CI: 31.2–67.3) from the regression model. Individualized estimates of tubular secretion and KBF improved the accuracy of PBPK model‐predicted tenofovir and oseltamivir kidney clearances, suggesting the potential for biomarker‐informed measures of kidney function to refine personalized drug dosing.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Kidney dosing of drugs clinically is based on estimated glomerular filtration rate (GFR) alone, despite the importance of proximal tubular secretion and reabsorption in determining drug clearance. Although typically linked, these functions may not decline proportionately to GFR. Physiologic‐based pharmacokinetic (PBPK) models can incorporate multiple elimination mechanisms in a population but are not responsive to individual variation in filtration versus secretion for clinical drug dosing.
WHAT QUESTION DID THIS STUDY ADDRESS?
Can clearance predictions be improved for drugs eliminated by both secretion and filtration using a previously validated PBPK model informed by individualized estimates of secretory clearance and kidney blood flow (KBF), in addition to GFR, compared to more standard linear models using GFR alone?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
We show that drug clearance estimates of tenofovir and oseltamivir carboxylate are improved when utilizing the PBPK model trained on KBF and secretory clearance, in addition to measured GFR. We estimated KBF by measuring isovalerylglycine clearance, an endogenous solute with rapid elimination by filtration and secretion, and OAT‐mediated secretory clearance by measuring the clearance of kynurenic acid, an endogenous solute that is highly protein bound and eliminated by OAT1/3. This methodology was applied to 27 participants across a wide range of kidney function in a dedicated pharmacokinetic study.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
These methods highlight the potential to better personalize drug dosing using individual biomarker‐based functional estimates of secretory clearance and KBF for drugs that have complex kidney handling.
INTRODUCTION
The kidneys eliminate hundreds of prescribed drugs and their metabolites from the circulation. Dose adjustment for kidney function represents standard clinical practice for achieving optimal drug exposure. Yet, prevailing methods for kidney dose‐adjustment do not account for the complex mechanisms of drug clearance. Specifically, kidney drug handling is determined by blood flow, plasma protein binding, glomerular filtration, tubular re‐absorption, and tubular secretion. In many cases, tubular secretory clearance represents the primary mechanism for eliminating protein bound drugs that are inefficiently filtered due to size/charge selectivity of the glomerular basement membrane. Tubular secretion is theoretically linked with glomerular filtration but this assumption is untested in real‐world settings and fails to account for individual‐level variation in renin‐angiotensin‐aldosterone activation, tubular transporter expression/function, and the underlying etiology of kidney disease. 1 , 2 , 3 , 4 Despite these considerations, clinical dose adjustments for decreased kidney function is currently based on only estimates of the glomerular filtration rate (eGFR), which are calculated from serum concentrations of creatinine and/or cystatin C. 5 , 6
Many drugs have mixed routes of elimination including both filtration and secretion, and individualized dosing tailored to the specific drug and patient is important in many clinical situations. 7 , 8 For instance, critically ill patients typically experience a complex milieu of malperfusion and subsequent multi‐organ failure, compounded by polypharmacy and a high likelihood of interactions as well as impaired metabolism and elimination; often, acute kidney injury can further catalyze nephrotoxicity from unadjusted dosing/overdosing. 9 In addition, oncologic medications have narrow therapeutic indices with decades of established clinical practices seeking optimal adjusted dosing to avoid inadvertent toxicity. In both scenarios, there is also a morbid risk from lost efficacy in underdosing further highlighting a need for responsive and personalized adjustments.
Mechanistic physiologically‐based pharmacokinetic (PBPK) models capture multiple aspects of kidney drug clearance by dividing the kidney into separate compartments and modeling passage across vascular, cellular, and tubular spaces using physiological parameters and drug‐specific in vitro data. 10 , 11 Mixed routes of drug elimination can be modeled using different functional parameters to permit more accurate dosing and facilitate the best clinical practice by predicting drug disposition, even in untested situations where formal pharmacokinetic data are not available. 10 , 11 PBPK models incorporate testable anatomic and biochemical determinants in its design therefore striving to align with the underlying physiologic process; however, remaining accurate for the single patient requires individual‐level functional and physiological information to separately capture filtration, re‐absorption, and tubular secretion to generate patient‐specific predictions in various clinical circumstances. Herein, we tested the feasibility of integrating individual, biomarker‐based estimates of kidney blood flow (KBF) and organic anion transporter (OAT)‐mediated tubular secretion for individualized predictions of kidney drug clearance. We estimated KBF and OAT‐mediated secretion in 27 participants from a kidney pharmacokinetic study via measured kidney clearance of isovalerylglycine and kynurenic acid (KA), respectively, and incorporated these with iohexol‐measured GFR into a mechanistic kidney PBPK model. We then compared measured and model‐predicted kidney clearances of administered tenofovir and oseltamivir carboxylate (the active metabolite of oseltamivir), which are cleared by both glomerular filtration and tubular secretion. 12 , 13 , 14 , 15
METHODS
Study design
This study was ancillary to the Proximal Tubular Clearance of Renal Medications (PROCLAIM) study, a pharmacokinetic study of kidney drug metabolism and excretion. 16 Participants were recruited from 2017 to 2019 by medical record screening at the University of Washington (UW) hospital clinics. Recruitment was stratified by categories of normal and impaired kidney function based on eGFR. Patients were ineligible if possessing a history of nephrotic syndrome, cirrhosis, requiring renal replacement therapy, or actively using any of the study medications. A total of 54 participants completed the primary PROCLAIM study visit assessing furosemide and famciclovir clearance. All were invited to a secondary visit to investigate the pharmacokinetics of tenofovir and oseltamivir carboxylate; half completed the second visit, which is the subject of this investigation (Table 1). All participants provided voluntary informed consent and all study procedures were overseen by the UW Institutional Review Board.
TABLE 1.
Patient characteristics.
| Characteristic | N = 27 a |
|---|---|
| Age, years | 54 (15) |
| Male | 17 (63%) |
| Race/ethnicity | |
| Black | 11 (41%) |
| White | 15 (56%) |
| Other/prefer not to answer | 1 (3.7%) |
| Body mass index, kg/m2 | 29.0 (5.7) |
| History of diabetes | 3 (11%) |
| History of hypertension | 4 (15%) |
| Relevant laboratory measurements | |
| iGFR, mL/min/1.73 m2 | 76 (30) |
| Kynurenic acid clearance, mL/min b | 226 (138) |
| Isovalerylglycine clearance, mL/min b | 575 (387) |
| Creatinine clearance, mL/min b | 105 (50) |
| Urine albumin, mg/24 h c | 7.9 (3.5–26) |
| Serum albumin, g/dL | 4.03 (0.24) |
| Medications | |
| RAAS inhibitor | 7 (26%) |
| Thiazide diuretic | 1 (3.7%) |
| K sparing diuretic | 2 (7.4%) |
| Non‐steroid anti‐inflammatory | 1 (3.7%) |
| Proton pump inhibitor | 3 (11%) |
Abbreviations: iGFR, measured GFR based on renal clearance of iohexol; RAAS, renin‐angiotensin‐aldosterone system.
Mean (SD); n (%).
Derived from 10‐h urine collection and averaged plasma levels collected at 0, 60, 300, and 480 min.
Median (interquartile range); standardized to 24 h from supervised daytime urine collection.
Drug clearance measurements
Study personnel administered oral tenofovir alafenamide 50 mg and oral oseltamivir 30 mg, then collected 13 plasma samples (15, 30, 45, 60, 90, 120, 150, 180, 210, 240, 300, 480, and 600 min) and a concurrent 10‐h timed urine collection after drug administration. Study drug concentrations of tenofovir and oseltamivir carboxylate, the active metabolite of oseltamivir, were quantified by liquid chromatography/tandem mass spectrometry analysis using validated assays. 16 A standard noncompartmental analysis was conducted to determine pharmacokinetic parameters of each study drug. The area under the plasma concentration versus time curve (AUC) was calculated using the trapezoidal method. We calculated kidney clearance of each drug as:
where Ae0 → t represents the drug quantity recovered in the 10‐h urine sample and AUCplasma0 → t represents the 10‐h plasma AUC.
Secretory solute measurements
We estimated effective KBF by the kidney clearance of isovalerylglycine, an endogenously produced solute, due to its rapid clearance (>4‐fold higher than GFR), low protein binding, and high extraction ratio. We estimated OAT‐mediated tubular secretory clearance by the kidney clearance of KA, an endogenously produced avid substrate of OAT1/3 that is highly (>90%) protein bound, limiting filtration. 17 , 18 , 19 To increase the precision we measured plasma concentrations of isovalerylglycine and KA at 0, 60, 300, 480 min, and in the supervised 10‐h urine collection, using our validated mass‐spectroscopic assay. 16 Briefly, solvent‐precipitated plasma underwent solid‐phase extraction (Phree phospholipid removal plate; Phenomenex). Urine samples underwent two solid‐phase extractions (HLB μElution plates; Waters). Dried extracts were reconstituted in acetonitrile/formic acid and passed through a large‐pore filter plate (MSBVN1210; Millipore). The resulting samples underwent liquid chromatography/tandem mass spectrometry (Shimadzu/Sciex 6500 q‐Trap) and quantification by internal/external standard curves.
The kidney clearance (CLR) of solutes were calculated as:
where U x represents the urinary solute concentration, V represents urine flow (mL/min), n is the number of plasma measurements, and P x the time‐weighted plasma concentration. We measured GFR by plasma iohexol disappearance with concentrations measured by the University of Minnesota Research and Diagnostic Laboratory. 20
Mechanistic kidney model
In this study, we leveraged our previously published physiologically‐based mechanistic kidney model to first estimate intrinsic secretory clearance via KA clearance, then to predict the kidney clearance of tenofovir and oseltamivir carboxylate for individual patients. Model equations and codes were previously provided. 10 , 11 This model consists of 35 compartments that resemble the physiology and anatomy of human kidneys, including glomerulus, peritubular blood vessels, renal tubular epithelial cells, tubules, and bladder. The vasculature, cells, and tubules are longitudinally divided into Bowman's capsule, proximal tubule, loop of Henle, distal tubule, and collecting duct. This allows the model to simulate drug concentration in different sections of the kidney at any timepoint following drug administration. To mechanistically describe kidney handling, the model includes unbound filtration, transporter‐mediated active secretion, and pH‐dependent passive re‐absorption. For patients with varying levels of kidney function (i.e., GFR), adaptive/compensatory physiological changes of tubular flow were incorporated, whereas other parameters, such as peritubular blood flow and tubular volume, all decreased proportionally with GFR. 10 , 11 , 21 This mechanistic kidney model has been verified with clearance data of 46 drugs in healthy subjects and 20 compounds in subjects across a range of normal and abnormal GFRs. 10 , 11 In the present study, we incorporated individual‐level markers of KBF and proximal tubular secretory clearance into the model for each participant that was subsequently used for kidney drug clearance prediction as described below. This approach accounts for the intrinsic interindividual variability in kidney physiology and drug handling parameters and separates secretory and filtration mechanisms where they may not decline proportionally with GFR. The mechanistic kidney model structure and the modeling and simulation workflow is shown in Figure 1. All parameter values for the biomarker and drugs used in the physiologically‐based mechanistic kidney model are summarized in Table 2.
FIGURE 1.
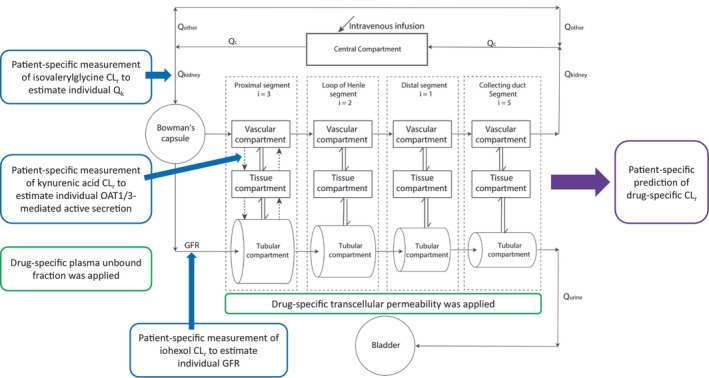
Diagram of the mechanistic kidney model structure and the modeling and simulation workflow of leveraging individual measurement of CLr of isovalerylglycine, iohexol, and kynurenic acid to estimate intrinsic active secretion individually and to predict renal drug clearance for each subject. CLr, renal clearance; GFR, glomerular filtration rate; i, the number of subsegment each segment is divided into; OAT1/3, organic anion transporter 1/3; Qc, blood flow in the central compartment; Qkidney/Qk, blood flow to the kidney; Qurine, urine formation flow; single solid arrow, the fluid flow; single dash arrow, active secretion or active reabsorption; double arrow, bidirectional passive diffusion.
TABLE 2.
Parameter values for the biomarker and drugs in the physiologically‐based mechanistic kidney model.
Estimation of individual intrinsic proximal tubular secretory clearance using kynurenic acid
To estimate the OAT1/3‐mediated intrinsic secretion capacity, we used the iohexol‐GFR (iGFR) and kidney clearance of isovalerylglycine from each individual as model input values for GFR and KBF, respectively. We set the plasma unbound fraction of KA at 7% for all subjects, derived as the average from previous measurements in the literature of protein binding using different methodologies and also based on our finding that KA binding does not vary among patients with normal and impaired kidney function. 17 , 22 , 23 , 24
After inputting GFR and KBF to the model, we simulated KA clearance using the mechanistic model allowing for a range (10–1000 L/h) of secretory clearance values applied to both basolateral uptake and apical efflux clearance. We selected the model parameters from which the simulated intrinsic secretory clearance was numerically closest to the measured KA kidney clearance. We then quantified the degree of optimality (prediction/observation ratio) in which a ratio of 1 indicates an optimal prediction. The optimized unbound intrinsic secretion clearance of KA for each subject is summarized in Table 3.
TABLE 3.
Measured CLr of iohexol, isovalerylglycine, and KA as the model inputs, and the model output of the optimized unbound intrinsic secretion clearance of KA for each subject.
| ID | Iohexol CLr (mL/min) | Isovalerylglycine CLr (mL/min) | KA CLr (mL/min) | Optimized unbound intrinsic secretion clearance (L/h) |
|---|---|---|---|---|
| 1 | 128 | 1145 | 313 | 320 |
| 2 | 143 | 735 | 288 | 340 |
| 3 | 146 | 1212 | 409 | 460 |
| 5 | 116 | 468 | 156 | 160 |
| 6 | 40 | 134 | 38 | 40 |
| 13 | 48 | 292 | 81 | 85 |
| 14 | 55 | 237 | 159 | 280 |
| 15 | 63 | 413 | 150 | 160 |
| 16 | 66 | 551 | 157 | 160 |
| 18 | 38 | 369 | 99 | 100 |
| 22 | 139 | 315 | 143 | 180 |
| 23 | 110 | 416 | 195 | 240 |
| 24 | 134 | 1692 | 386 | 380 |
| 25 | 95 | 746 | 211 | 220 |
| 30 | 84 | 567 | 199 | 220 |
| 31 | 110 | 359 | 174 | 220 |
| 33 | 123 | 667 | 203 | 220 |
| 34 | 96 | 639 | 249 | 280 |
| 35 | 98 | 912 | 235 | 240 |
| 36 | 91 | 496 | 164 | 180 |
| 38 | 74 | 192 | 202 | 1000 |
| 41 | 111 | 493 | 501 | 1000 |
| 42 | 138 | 1327 | 565 | 700 |
| 45 | 89 | 416 | 398 | 1000 |
| 46 | 54 | 533 | 296 | 420 |
| 49 | 20 | 76 | 53 | 95 |
| 51 | 33 | 161 | 50 | 55 |
Abbreviations: CLr, renal clearance; KA, kynurenic acid.
Mechanistic prediction of kidney clearance of tenofovir and oseltamivir
We used iGFR and KBF from each individual as above as model input values to predict kidney clearance of tenofovir and oseltamivir carboxylate for each participant. The plasma unbound fractions of tenofovir and oseltamivir carboxylate were set as 99.3% 25 and 97% 26 for all subjects based on literature, and the estimated clearance of tenofovir and oseltamivir carboxylate in each participant was scaled based on the estimated intrinsic secretory clearance of KA as described above. Scalars were necessary to translate biomarker‐derived secretory clearance to a specific drug, which accounts for differences in substrate‐specific transporter maximum velocity and affinity (i.e., K d or K m) between KA and study drugs. We calculated optimized scalars of 0.033 for tenofovir and 0.038 for oseltamivir carboxylate when comparing to observed clearance.
Assessing prediction accuracy
We assessed the mechanistic model by generating goodness‐of‐fit plots comparing the individual prediction with individual observation alongside adjusted R‐square and Pearson and Spearman r. We generated a linear regression empirical model, fitting iGFR to observed drug clearance, as a prevailing method for determining function‐adjusted kidney drug clearance without use of a mechanistic model. We calculated absolute prediction error for each individual by subtracting the observed kidney clearance from the predicted kidney clearance, as well as mean and average‐fold error. Given small sample sizes, which make point estimates of error unreliable, we used bootstrapping to estimate the 95% confidence interval of prediction errors.
RESULTS
Patient characteristics
The mean age of the 27 study participants was 54 ± 15 years, 63% were men, and 56% identified as White. The mean iGFR was 76 ± 31 mL/min/1.73 m2 with the following distribution: six patients (22%) had iGFR less than 45, four (15%) with iGFR 45–60, nine (33%) with iGFR 60–90, and eight (30%) with iGFR greater than 90. Median urinary albumin was 7.9 mg/24 h (interquartile range: 3.5–26; Table 1).
Fitting intrinsic secretory clearance with kynurenic acid clearance
The intrinsic secretory clearance of KA was determined by fitting the mechanistic model using observed KA kidney clearance (Figure 2, left panel). Predicted KA kidney clearance using the optimized intrinsic secretory clearance differed from the observed KA kidney clearance by less than 5% for all but three patients (Table 2, Figure 3). In those participants (38, 41, and 45), the measured kidney clearance of KA was greater than KBF estimated from isovalerylglycine clearance. The model follows the physiologic principle that secretory clearance cannot exceed KBF, leading to spurious predictions when violated by these imperfect functional estimates. For instance, measured isovalerylglycine kidney clearance in participant 38 was 192 mL/min, lower than the observed KA kidney clearance of 202 mL/min. Hence, the maximum simulated kidney clearance of KA would asymptotically approach the measured kidney clearance of isovalerylglycine of 192 mL/min as it represents the KBF but could not reach the observed kidney clearance of KA of 202 mL/min as secretory clearance could not exceed KBF (Figure S1B). Therefore, the estimated unbound intrinsic secretory clearance of KA (Table 3) for the affected participants reached the predefined maximum of 1000 L/h. The mechanistic reason for this phenomenon is unclear and warrants further investigation, one hypothesis is that, in the case of these three patients, KA is more efficiently extracted by the kidneys than isovalerylglycine which serves as the biomarker for renal blood flow. This may be due to differences in OAT1/OAT3 expression level of specific variants that transport KA and isovalerylglycine with different capacity and efficiency.
FIGURE 2.
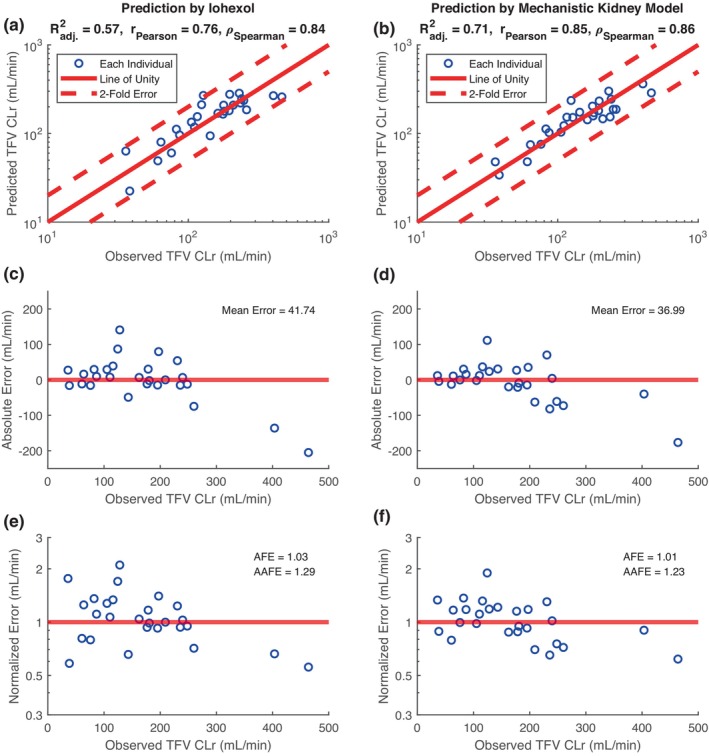
Assessment of mechanistic kidney model in predicting tenofovir (TFV) renal clearance (CLr). Linear regression model (a, c, e) using CLr of iohexol (iGFR) alone is compared to the mechanistic kidney model (b, d, f) which incorporates iGFR, kidney blood flow (via isovalerylglycine clearance), and OAT‐mediated secretory clearance (via kynurenic acid clearance). Goodness‐of‐fit plots show the comparison between individual prediction of TFV CLr and individual observation of TFV CLr, with adjusted R‐square, Pearson r, and Spearman ρ displayed (a, b). Error plots show the individual absolute prediction error (c, d) and individual normalized prediction error (e, f) with mean error, average fold error, and absolute average fold error displayed. Blue circles represent individual data, red dashed lines represent twofold error, and red solid lines represent error‐free.
FIGURE 3.
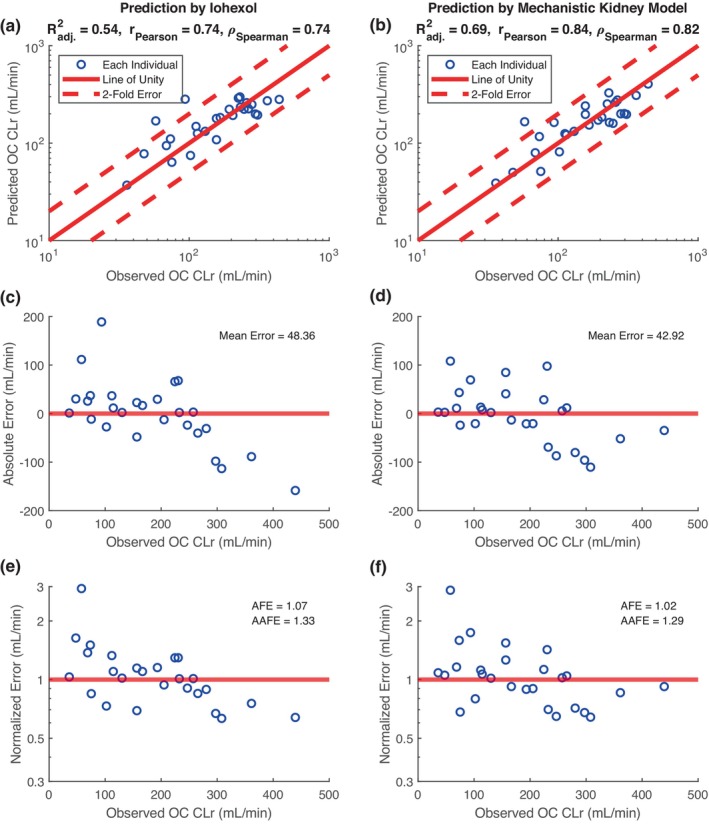
Assessment of mechanistic kidney model in predicting oseltamivir carboxylate (OC) renal clearance (CLr). Linear regression model (a, c, e) using CLr of iohexol (iGFR) alone is compared to the mechanistic kidney model (b, d, f) which incorporates iGFR, kidney blood flow (via isovalerylglycine clearance), and OAT‐mediated secretory clearance (via kynurenic acid clearance). Goodness‐of‐fit plots show the comparison between individual prediction of TFV CLr and individual observation of TFV CLr, with adjusted R‐square, Pearson r, and Spearman ρ displayed (a, b). Error plots show the individual absolute prediction error (c, d) and individual normalized prediction error (e, f) with mean error, average fold error, and absolute average fold error displayed. Blue circles represent individual data, red dashed lines represent twofold error, and red solid lines represent error‐free.
Drug clearance prediction using mechanistic kidney model
Tenofovir
The mean observed tenofovir clearance in the study participants was 168.7 ± 101.7 mL/min, approximately two‐times higher than iGFR. The mean predicted tenofovir clearance using the mechanistic PBPK model was 162.8 ± 79.8 mL/min. Using linear regression and iGFR as the independent variable, the predicted tenofovir clearance exhibited a mean absolute error of 41.7 mL/min (R 2 = 0.57; Figure 2a,c,e). Using the mechanistic kidney model trained with individual iGFR, isovalerylglycine derived blood flow, and KA clearance, tenofovir clearance prediction exhibited the lowest mean error point estimate of 37 mL/min (R 2 = 0.71; Figure 2b,d,f). Using bootstrapping, the mechanistic PBPK model prediction had a mean error of 37.1 mL/min (95% CI: 24.0–52.9) compared to the iGFR linear regression model which had a mean error of 41.8 mL/min (95% CI: 25.0–61.6). All individual predictions and observations of tenofovir are summarized in Table 4.
TABLE 4.
Observed versus predicted kidney clearance for TFV and OS for each subject.
| ID | Observed TFV CLr | Predicted TFV CLr | P/O ratio of TFV CLr | Observed OC CLr | Predicted OC CLr | P/O ratio of OC CLr |
|---|---|---|---|---|---|---|
| 1 | 240 | 244 | 1.02 | 257 | 263 | 1.02 |
| 2 | 197 | 233 | 1.18 | 225 | 253 | 1.12 |
| 3 | 231 | 301 | 1.30 | 230 | 328 | 1.43 |
| 5 | 236 | 154 | 0.65 | 232 | 163 | 0.70 |
| 6 | 36 | 48 | 1.33 | 48 | 50 | 1.04 |
| 13 | 64 | 75 | 1.17 | 69 | 80 | 1.16 |
| 14 | 87 | 102 | 1.17 | 74 | 117 | 1.58 |
| 15 | 82 | 113 | 1.38 | 115 | 122 | 1.06 |
| 16 | 111 | 123 | 1.11 | 130 | 132 | 1.02 |
| 18 | 76 | 76 | 1.00 | 102 | 81 | 0.79 |
| 22 | 128 | 152 | 1.19 | 94 | 163 | 1.73 |
| 23 | 179 | 158 | 0.88 | 193 | 172 | 0.89 |
| 24 | 464 | 287 | 0.62 | 361 | 309 | 0.86 |
| 25 | 181 | 171 | 0.94 | 205 | 184 | 0.90 |
| 30 | 116 | 153 | 1.32 | 58 | 166 | 2.86 |
| 31 | 209 | 146 | 0.70 | 247 | 160 | 0.65 |
| 33 | 248 | 187 | 0.75 | 281 | 200 | 0.71 |
| 34 | 195 | 181 | 0.93 | 308 | 198 | 0.64 |
| 35 | 260 | 187 | 0.72 | 297 | 201 | 0.68 |
| 36 | 163 | 143 | 0.88 | 167 | 153 | 0.92 |
| 38 | 105 | 103 | 0.98 | 112 | 125 | 1.12 |
| 41 | 124 | 236 | 1.90 | 265 | 277 | 1.05 |
| 42 | 404 | 363 | 0.90 | 440 | 405 | 0.92 |
| 45 | 177 | 204 | 1.15 | 157 | 241 | 1.54 |
| 46 | 143 | 174 | 1.22 | 157 | 197 | 1.25 |
| 49 | 38 | 34 | 0.89 | 36 | 39 | 1.08 |
| 51 | 61 | 48 | 0.79 | 75 | 51 | 0.68 |
Abbreviations: CLr, renal clearance; OC, oseltamivir; P/O ratio, prediction‐over‐observation ratio of renal drug clearance; TFV, tenofovir.
Oseltamivir carboxylate
The mean observed oseltamivir clearance carboxylate was 182.8 ± 103.7 mL/min, compared to predicted clearance using the mechanistic PBPK model of 178.9 ± 88.5 mL/min. Using linear regression and iGFR, the mean absolute error in predicting oseltamivir clearance carboxylate was 48.4 mL/min (R 2 = 0.54; Figure 3a,c,e). The mechanistic model trained with iGFR, isovalerylglycine derived blood flow, and KA clearance, resulted in the lowest mean absolute error of 42.9 mL/min (R 2 = 0.69; Figure 3b,d,f). Using bootstrapping, the mean error of the mechanistic PBPK model was 42.9 mL/min (95% CI: 29.7–56.4), compared to the mean error of 48.1 mL/min (95% CI: 31.2–67.3) from linear regression. All individual predictions and observations of oseltamivir carboxylate are summarized in Table 4.
DISCUSSION
In this study, we demonstrate the ability to predict the kidney clearance of tenofovir and oseltamivir carboxylate, two drugs with complex kidney elimination pathways, using a physiologically‐based mechanistic kidney model informed by biomarker‐based representations of KBF and OAT‐mediated tubular secretory clearance, as well as GFR, with improved prediction accuracy over regression by GFR. The mechanistic model allows simultaneous consideration of glomerular filtration, passive re‐absorption, and active secretion with various levels of kidney function; in prior usage the relative changes in glomerular versus tubular hemodynamics were generalized to all patients, and to our knowledge this is the first use of a PBPK to incorporate individualized markers of such characteristics to enhance the prediction of different drugs' kidney clearances. Considering that tenofovir and oseltamivir carboxylate are cleared by both glomerular filtration and tubular secretion, this study highlights the ample potential for PBPK models to refine individualized drug clearance prediction and dosing recommendations by incorporating markers of both functions simultaneously. 12 , 14 This may have implications for dosing of drugs with complex kidney disposition as well as in clinical scenarios expected to result in disparate alterations between glomerular and tubular dynamics.
It is important to note the potential benefits and drawbacks of modeling kidney function using biomarkers of limited scope. We expect that elimination of freely filtered drugs would best align with GFR whether by endogenous or exogenous markers, however, GFR may not accurately predict the clearance of a predominantly secreted drug. 27 Our previously developed assay of multiple endogenously produced metabolites was intended to summarily reflect the action of the proximal tubules, however, each metabolite varies significantly in clearance, affinity for transporters, and protein binding. 16 Given these inherent differences, each solute likely reflects different aspects of clearance and may therefore best predict clearance of biochemically similar compounds. Specificity to secretory clearance differs between each solute, and we continue to search for more optimal and generalizable markers of secretion. 28 This study estimated secretory clearance by fitting the model to patient‐specific KA measurements, a metabolite cleared by OAT1/3; whereas tenofovir and oseltamivir carboxylate are also largely cleared by OAT1/3, more data are needed to understand how it would apply to other drugs. Pharmacogenomic studies reveal that variation at individual transporters can significantly modify drug clearance even at the same gross level of organ function, indicating the need for more specific markers. 29 , 30 Apart from predicting clearance, some metabolites may approximate the risk for drug–drug interactions which could aid medication choice and dosing. 31 Ideally, incorporating multiple solute measurements of different mechanistic specificity could provide the most universal predictive capacity limited only by laboratory practicality. We therefore expect that combining more inclusive information covering different kidney clearance pathways will perform better in predicting kidney pharmacokinetics for the individual, particularly for medications with complex kidney handling.
There are several strengths of this study. The PBPK model has been previously validated in a large dataset combining kidney perfusion, glomerular filtration, secretion, and passive re‐absorption. 10 , 11 Here, we compared predicted and observed drug clearance in a diverse cohort of 27 patients with different stages of kidney impairment, highlighting model versatility. The measurements of isovalerylglycine and KA have been specifically linked to dysfunction of the proximal tubules in other studies, most recently showing tubulointerstitial fibrosis in proportion to clearance reduction. 32 , 33 Several limitations should be noted. Some participants were on stable, pharmacologic doses of routine medications that may impact clearance of study drugs or metabolites. KA serves as a reasonable secretory clearance marker for tenofovir and oseltamivir carboxylate owing to commonalities in OAT1/3 clearance, however, as described above, this many not necessarily apply to drugs not cleared by OAT1/3. It is unknown whether different transporters' activity declines differentially or in parallel in chronic kidney disease, and so may limit the generalizability of KA clearance to all routes of tubular secretory clearance; this could be ameliorated by incorporating other secreted endogenous solutes of different transporter specificity. Measurement of KA and isovalerylglycine clearance requires paired plasma and timed urine sampling which can be cumbersome, subject to imprecision, and may complicate clinical adaptation. 34 Variation in KA and isovalerylglycine clearance could occur due to metabolite or drug interaction, or changes in extra‐renal clearance, that would hinder model accuracy. Furthermore, functional estimates that violate the physiological principles underpinning the mechanistic model (such as KA clearance exceeding isovalerylglycine clearance) may be challenging to reconcile.
In summary, this study highlights the potential for improved accuracy and individual‐level prediction of drug clearance using a PBPK model informed by markers of KBF, glomerular filtration, and tubular secretion. Glomerular filtration alone may fail to capture the function of the tubules; by co‐measuring KA and isovalerylglycine clearance, markers of tubular secretory clearance, and KBF, respectively, we have shown the ability to predict the kidney clearance of tenofovir and oseltamivir carboxylate across a cohort of patients with varying degrees of kidney impairment. Co‐measuring GFR with these secretory markers may be a reasonable strategy for future PBPK models to capture added physiologic information about kidney function and, therefore, with further work, allow for improved and more personalized drug dosing for patient use.
AUTHOR CONTRIBUTIONS
M.L.G. and W.H. wrote the manuscript. All authors designed the research. M.L.G. and W.H. performed the research. All authors analyzed the data. C.K.Y., N.I., and B.K. contributed new analytical tools.
FUNDING INFORMATION
No funding was received for this work.
CONFLICT OF INTEREST STATEMENT
The authors declared no competing interests for this work.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
M.L.G. was supported by grants from the American Society of Nephrology as a Ben J. Lipps Award recipient, as well as a Pilot and Feasibility Award granted by the University of Michigan O'Brien Kidney Translational Core Center (NIDDK P30DK081943). N.I. was supported in part by National Institutes of Health grant P01 DA032507 and UW School of Pharmacy's Milo Gibaldi Endowed Chair. C.K.Y. was supported by National Institutes of Health grant R01 GM121354. W.H. is currently a salaried employee and stockholder of Genentech, Inc./Roche. Genentech, Inc./Roche was not involved in the study design and publication. The authors would like to acknowledge and thank all study participants.
Granda ML, Huang W, Yeung CK, Isoherranen N, Kestenbaum B. Predicting complex kidney drug handling using a physiologically‐based pharmacokinetic model informed by biomarker‐estimated secretory clearance and blood flow. Clin Transl Sci. 2024;17:e13678. doi: 10.1111/cts.13678
Michael L. Granda and Weize Huang contributed equally to the manuscript as co‐first authors.
REFERENCES
- 1. Thomson SC, Blantz RC. Glomerulotubular balance, tubuloglomerular feedback, and salt homeostasis. J Am Soc Nephrol. 2008;19:2272‐2275. [DOI] [PubMed] [Google Scholar]
- 2. Kurokawa K. Tubuloglomerular feedback: its physiological and pathophysiological significance. Kidney Int Suppl. 1998;67:S71‐S74. [DOI] [PubMed] [Google Scholar]
- 3. Vallon V, Thomson SC. The tubular hypothesis of nephron filtration and diabetic kidney disease. Nat Rev Nephrol. 2020;16:317‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han YF, Fan XH, Wang XJ, et al. Association of intergenic polymorphism of organic anion transporter 1 and 3 genes with hypertension and blood pressure response to hydrochlorothiazide. Am J Hypertens. 2011;24:340‐346. [DOI] [PubMed] [Google Scholar]
- 5. Matzke GR, Aronoff GR, Atkinson AJ, et al. Drug dosing consideration in patients with acute and chronic kidney disease—a clinical update from Kidney Disease: Improving Global Outcomes (KDIGO). Kidney Int. 2011;80:1122‐1137. [DOI] [PubMed] [Google Scholar]
- 6. Lowenstein J, Grantham JJ. The rebirth of interest in renal tubular function. Am J Physiol Renal Physiol. 2016;310:F1351‐F1355. [DOI] [PubMed] [Google Scholar]
- 7. Ruiz S, Minville V, Asehnoune K, et al. Screening of patients with augmented renal clearance in ICU: taking into account the CKD‐EPI equation, the age, and the cause of admission. Ann Intensive Care. 2015;5:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Udy AA, Jarrett P, Stuart J, et al. Determining the mechanisms underlying augmented renal drug clearance in the critically ill: use of exogenous marker compounds. Crit Care. 2014;18:657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perazella MA. Drug use and nephrotoxicity in the intensive care unit. Kidney Int. 2012;81:1172‐1178. [DOI] [PubMed] [Google Scholar]
- 10. Huang W, Isoherranen N. Novel mechanistic PBPK model to predict renal clearance in varying stages of CKD by incorporating tubular adaptation and dynamic passive reabsorption. CPT Pharmacometrics Syst Pharmacol. 2020;9:571‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang W, Isoherranen N. Development of a dynamic physiologically based mechanistic kidney model to predict renal clearance. CPT Pharmacometrics Syst Pharmacol. 2018;7:593‐602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calcagno A, Cusato J, Marinaro L, et al. Clinical pharmacology of tenofovir clearance: a pharmacokinetic/pharmacogenetic study on plasma and urines. Pharmacogenomics J. 2016;16:514‐518. [DOI] [PubMed] [Google Scholar]
- 13. Kohler JJ, Hosseini SH, Green E, et al. Tenofovir renal proximal tubular toxicity is regulated by OAT1 and MRP4 transporters. Lab Investig J Tech Methods Pathol. 2011;91:852‐858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He G, Massarella J, Ward P. Clinical pharmacokinetics of the prodrug oseltamivir and its active metabolite Ro 64‐0802. Clin Pharmacokinet. 1999;37:471‐484. [DOI] [PubMed] [Google Scholar]
- 15. Hill G, Cihlar T, Oo C, et al. The anti‐influenza drug oseltamivir exhibits low potential to induce pharmacokinetic drug interactions via renal secretion—correlation of in vivo and in vitro studies. Drug Metab Dispos. 2002;30:13‐19. [DOI] [PubMed] [Google Scholar]
- 16. Chen Y, Zelnick LR, Hoofnagle AN, et al. Prediction of kidney drug clearance: a comparison of tubular secretory clearance and glomerular filtration rate. J Am Soc Nephrol. 2021;32:459‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Y, Zelnick LR, Wang K, et al. CRIC Study Investigators: association of tubular solute clearances with the glomerular filtration rate and complications of chronic kidney disease: the chronic renal insufficiency cohort study. Nephrol Dial Transplant. 2020;36:1271‐1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mor A, Kalaska B, Pawlak D. Kynurenine pathway in chronic kidney disease: what's old, what's new, and what's next? Int J Tryptophan Res. 2020;13:1178646920954882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uwai Y, Honjo H, Iwamoto K. Interaction and transport of kynurenic acid via human organic anion transporters hOAT1 and hOAT3. Pharmacol Res. 2012;65:254‐260. [DOI] [PubMed] [Google Scholar]
- 20. Delanaye P, Ebert N, Melsom T, et al. Iohexol plasma clearance for measuring glomerular filtration rate in clinical practice and research: a review. Part 1: how to measure glomerular filtration rate with iohexol? Clin Kidney J. 2016;9:682‐699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang W, Czuba LC, Isoherranen N. Mechanistic PBPK modeling of urine pH effect on renal and systemic disposition of methamphetamine and amphetamine. J Pharmacol Exp Ther. 2020;373:488‐501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jansen J, Jankowski J, Gajjala PR, Wetzels JFM, Masereeuw R. Disposition and clinical implications of protein‐bound uremic toxins. Clin Sci. 2017;131:1631‐1647. [DOI] [PubMed] [Google Scholar]
- 23. Ma Y, Xin M, Li K, et al. An LC‐MS/MS analytical method for the determination of uremic toxins in patients with end‐stage renal disease. J Pharm Biomed Anal. 2020;191:113551. [DOI] [PubMed] [Google Scholar]
- 24. Kumar R, Adiga A, Novack J, et al. The renal transport of hippurate and protein‐bound solutes. Physiol Rep. 2020;8:e14349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Drugs@FDA: FDA‐Approved Drugs. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&ApplNo=021752. Accessed April 19, 2022.
- 26. Drugs@FDA: FDA‐Approved Drugs. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&applno=021087. Accessed April 19, 2022.
- 27. Hall AM, Trepiccione F, Unwin RJ. Drug toxicity in the proximal tubule: new models, methods and mechanisms. Pediatr Nephrol. 2022;37:973‐982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Granda ML, Prince DK, Fiehn O, et al. Metabolomic profiling identifies new endogenous markers of tubular secretory clearance. Kidney360. 2022;4(1):23‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Evans WE, McLeod HL. Pharmacogenomics — drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538‐549. [DOI] [PubMed] [Google Scholar]
- 30. Taube SE, Clark GM, Dancey JE, McShane LM, Sigman CC, Gutman SI. A perspective on challenges and issues in biomarker development and drug and biomarker codevelopment. J Natl Cancer Inst. 2009;101:1453‐1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang J, Shen H, Zhao X, et al. Endogenous plasma kynurenic acid in human: a newly discovered biomarker for drug‐drug interactions involving organic anion transporter 1 and 3 inhibition. Drug Metab Dispos Biol Fate Chem. 2021;49:1063‐1069. [DOI] [PubMed] [Google Scholar]
- 32. Granda ML, Zelnick LR, Prince DK, Hoofnagle A, Young BA, Kestenbaum B. Tubular secretion and eGFR decline in the Jackson Heart Study. Kidney Int Rep. 2022;7(12):2668‐2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garimella PS, Katz R, Waikar SS, et al. Kidney tubulointerstitial fibrosis and tubular secretion. Am J Kidney Dis. 2022;79:709‐716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kestenbaum B, Ix JH, Gansevoort R, et al. Population‐based limits of urine creatinine excretion. Kidney Int Rep. 2022;7(11):2474‐2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang S‐Y, Huang W, Chapron A, et al. Incorporating uremic solute‐mediated inhibition of OAT1/3 improves PBPK prediction of tenofovir renal and systemic disposition in patients with severe kidney disease. Pharm Res. 2023. 10.1007/s11095-023-03594-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Instiaty I, Lindegardh N, Jittmala P, et al. Comparison of oseltamivir and oseltamivir carboxylate concentrations in venous plasma, venous blood, and capillary blood in healthy volunteers. Antimicrob Agents Chemother. 2013;57:2858‐2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morimoto K, Nakakariya M, Shirasaka Y, et al. Oseltamivir (Tamiflu) efflux transport at the blood‐brain barrier via P‐glycoprotein. Drug Metab Dispos Biol Fate Chem. 2008;36:6‐9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


