Summary
Phosphorylation of α-synuclein at the Serine-129 site (α-syn Ser129P) is an established pathologic hallmark of synucleinopathies and a therapeutic target. In physiologic states, only a fraction of α-syn is phosphorylated at this site, and most studies have focused on the pathologic roles of this post-translational modification. We found that unlike wild-type (WT) α-syn which is widely expressed throughout the brain, the overall pattern of α-syn Ser129P is restricted, suggesting intrinsic regulation. Surprisingly, preventing Ser129P blocked activity-dependent synaptic attenuation by α-syn – thought to reflect its normal function. Exploring mechanisms, we found that neuronal activity augments Ser129P, which is a trigger for protein-protein interactions that are necessary for mediating α-syn function at the synapse. AlphaFold2-driven modeling and membrane-binding simulations suggest a scenario where Ser129P induces conformational changes that facilitate interactions with binding partners. Our experiments offer a new conceptual platform for investigating the role of Ser129 in synucleinopathies, with implications for drug-development.
eTOC
Phosphorylation of α-synuclein at the Serine-129 site is an established pathologic hallmark of Parkinson’s disease and related ‘synucleinopathies’, but new findings from Parra-Rivas et al. suggest a physiologic role for this post-translational modification in triggering α-synuclein function by facilitating protein-protein interactions at synapses.
Introduction
Aggregation of α-syn into Lewy bodies and Lewy neurites is a hallmark of Parkinson’s disease (PD) and related disorders, collectively called ‘synucleinopathies’. Almost all pathologically aggregated α-syn is phosphorylated at the Ser129-site1–3, and antibodies to Ser129 α-syn are the most sensitive marker for neuropathologic diagnosis of all synucleinopathies4. Given the correlation between aggregation, phosphorylation, and human pathology, studies in animal models and cell-based systems routinely use this modification as a surrogate marker for pathology. Assuming a pathologic role for Ser129P, inhibitors against Polo-like Kinase 2 (PLK2) – the enzyme phosphorylating α-syn5 – have also been explored as a drug target in PD6. However, contrary to expectations, PLK2 over-expression suppresses dopaminergic neurodegeneration and functional deficits in vivo7, and experiments seeking to establish a pathologic role for α-syn Ser129P have repeatedly failed to assign a clear link between phosphorylation, aggregation, and toxicity8–11.
Though Ser129P is produced during normal metabolism, biochemical estimates show that under physiologic conditions, only a small fraction (~ 4%) of α-syn is phosphorylated at this site1, which has further reinforced the idea that Ser129P has an exclusive role in pathology. However, low-frequency events such as serine phosphorylation are known to regulate numerous biological properties12. It also seems reasonable to speculate that pathologic modifications that are also seen in physiologic states – regardless of their absolute levels – would have initially evolved to perform some normal function. Indeed, the constitutive nature of Ser129P in brains suggests a physiologic role involving repeated phosphorylation and dephosphorylation; nevertheless, almost all studies to date have examined α-syn Ser129 in the context of pathology. A very recent study showed that increasing neuronal activity augmented α-syn phosphorylation at the Ser129-site13, suggesting physiologic roles, but mechanistic events following phosphorylation, functional consequences of this phosphorylation at a single-synapse level, and even the overall localization of α-syn Ser129P in the brain is unclear.
Here, we systematically examined α-syn Ser129P using various cell/molecular assays, including optical probes to visualize synaptic vesicle recycling, in vivo studies, cell-free assays with purified proteins, mass-spectrometry, electron microscopy, and dynamic simulations with Alphafold2-driven structures. We propose a model where Ser129P induces conformational changes at the α-syn C-terminus, facilitating its association with functional interacting partners and eliciting α-syn function. Our findings may have broad implications for understanding the pathophysiologic transition of α-syn in PD and other synucleinopathies.
Results
Restricted expression of Ser129P α-syn in brains
In mammals, α-syn robustly localizes to presynapses throughout the brain, but studies have also reported α-syn in the nucleus14,15. While previous work examined α-syn Ser129P levels in the brain by biochemistry, the spatial distribution of post-translationally modified α-syn in brains is unclear. Given the overall low abundance of this post-translational modification in the brain, we had assumed that all α-syn-positive synapses would have small amounts of the phosphorylated protein. However, we found that while WT-α-syn was broadly distributed throughout synapses in the brain as expected, Ser129P α-syn was restricted to a subset of brain regions, with prominent staining in the superficial cortex, hippocampus, olfactory bulb, parts of the basal ganglia, and cerebellum (Fig. 1A–B, Supp. Fig. 1A–B; also see optimization of immunohistochemistry in methods). A similar staining pattern was reported in a recent study using a signal-amplification technique to detect Ser129P α-syn16. In addition to dense synaptic staining of Ser129P, we also saw nuclear staining in several brain regions, including granule cells of the olfactory bulb and stratum pyramidale of the hippocampus (arrowheads in Fig. 1B’ and Supp. Fig. 1A/a’). Nuclear localization of α-syn Ser129P in the brain was also reported by a recent study using phospho-specific antibodies17. However, nuclear α-syn has not been consistently observed, and variabilities in tissue fixation and preservation of the phosphorylation state might play a role in detecting the nuclear α-syn fraction18. Regardless, the discrete pattern of brain α-syn Ser129P expression suggests a tight post-translational regulation and putative physiologic roles.
Figure 1: Localization of α-syn Ser129P and effects of phosphorylation on synaptic targeting.
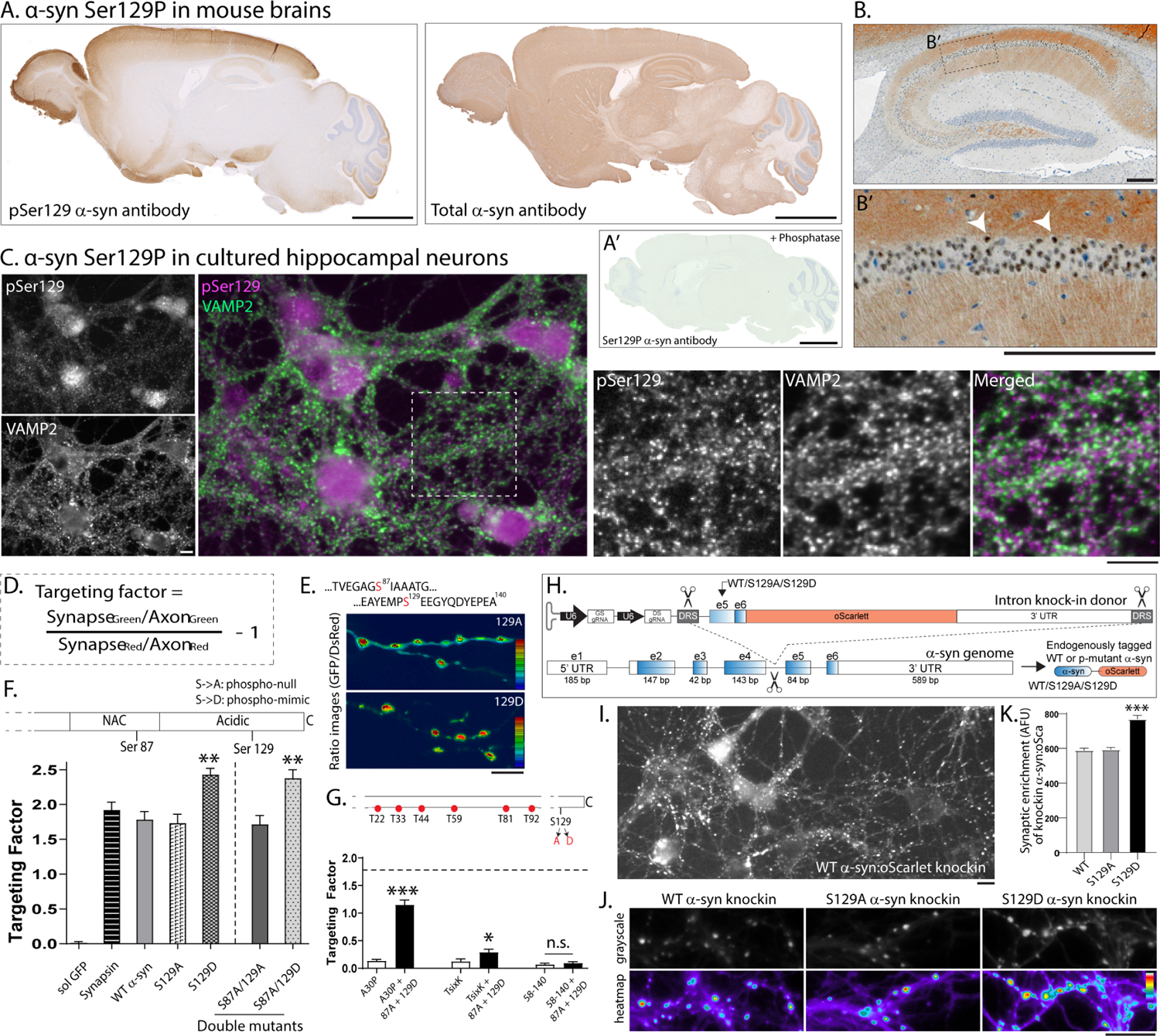
(A) Staining for α-syn Ser29P (left) or total α-syn (right) in mouse brains (representative). Unlike broad expression of total α-syn, Ser129P α-syn staining is restricted to subsets of brain regions. Section in A’ was pre-treated with phosphatases, which eliminated the Ser129P staining. See Supp. Fig. 1A–B for further characterization. Scale bar = 2 mm.
(B) Hippocampus section: Note α-syn Ser129P staining in both synapses and nuclei – white arrowheads mark some pSer129+ nuclei (B’ zoomed image from B). Scale bar = 200 μm.
(C) Cultured hippocampal neurons stained with antibodies to α-syn Ser129P and VAMP2 (to mark synapses). Note α-syn Ser129P is localized to nuclei and synapses (also see Supp. Fig. 1C). Scale bar = 10 μm. Quantitative analyses (see methods) showed that 50.95 +/− 3.32% (mean +/− SD) VAMP2-positive puncta were colocalized with α-syn Ser129P. Several Ser129P α-syn puncta were immediately adjacent to synapses or in processes resembling axons.
(D) Formula for evaluating presynaptic targeting of transfected h-α-syn in α-syn −/− neurons.
(E) Top: Serine phosphorylation sites in α-syn. Bottom: Representative images from synaptic targeting assays showing increased targeting of phospho-mimic (129D) α-syn. Scale bar = 10 μm.
(F) Quantification of synaptic targeting for phospho-incompetent (129A) and phospho-mimic (129D) mutants (also see Supp. Fig. 2). Note targeting was substantially increased by mimicking phosphorylation at the 129-site, and this was independent of 87A phosphorylation status (~ 500–1000 boutons from 6–15 coverslips were analyzed for each condition; **p<0.001, one-way ANOVA followed by Kruskal-Wallis test).
(G) Synaptic targeting of h-α-syn phospho-mutants relative to WT h-α-syn (horizontal dashed line marks targeting of WT h-α-syn). Note decreased presynaptic targeting of the familial PD mutant A30P and TsixK (mutant that disrupts membrane-binding) was augmented by the 129D (but not 129A) mutant, and as expected, the N-terminus was required for targeting (~ 600–900 boutons from 8–20 coverslips were analyzed for each condition; **p<0.001, Mann-Whitney nonparametric test).
(H) Knock-in strategy to replace the last two exons (exon 5 and exon 6) and insert a fluorescent tag (oScarlet) at the C-terminus of endogenous mouse α-syn. Note that the A/D phospho-mutants can be inserted into exon 5 using this strategy (also see Supp. Fig. 3A–D and methods).
(I) Representative image of cultured neurons expressing the knockin construct (Ser at 129-residue, more examples in Supp. Fig. 3E).
(J) Exemplary images from cultured neurons expressing knock-in WT and S129A/D tagged to mScarlet, synaptic enrichment quantified in (K). Note augmented targeting of S129D α-syn (mean ± SEM ***p<0.0001 Mann-Whitney nonparametric test).
Ser129P augments the presynaptic targeting of α-syn
Ser129P α-syn in cultured mouse hippocampal neurons was also localized to presynapses and nuclei, though punctate staining in neurites (presumably axons) was also seen (Fig. 1C and Supp. Fig. 1C–E). Since many studies have shown that protein levels of α-syn at synapses is linked to its function19–22, we first asked if Ser129P alters the presynaptic abundance of α-syn. Towards this, we transiently transfected GFP-tagged h-α-syn in cultured hippocampal neurons from α-syn null mice – to eliminate potential confounding effects of the endogenous mouse α-syn – and used a ratio-metric imaging paradigm that accounts for variabilities in transgene expression and robustly reports presynaptic accumulation23 (Fig. 1D; also see methods and Supp. Fig. 2). Serine phosphorylation of α-syn occurs at two sites – Ser87 and Ser129 (Fig. 1E – top), although only the 129-site is predominantly phosphorylated in synucleinopathies. To test the effects of phosphorylation on presynaptic targeting, we transfected neurons with GFP-tagged human-α-syn (h-α-syn) phospho-mutants where Ser129 was either mutated to Alanine (S129A), which cannot be phosphorylated, or Aspartate (S129D), a residue that can mimic phosphorylation and has been used in previous studies to simulate α-syn Ser129P8–10,24–27. Since the Ser129P-site is located very close to the C-terminus of full-length α-syn, which has 140 amino-acids, we placed the GFP-tag at the N-terminus of h-α-syn. Note that both N- and C-terminus tagged h-α-syn were targeted equivalently to synapses, and their abundance was quantitatively comparable to another cytosolic presynaptic protein synapsin (Supp. Fig. 2C).
While presynaptic targeting of phospho-incompetent S129A resembled WT h-α-syn, synaptic targeting of the phospho-mimic (S129D) h-α-syn was substantially increased (Fig. 1E–F). This augmented enrichment was specifically due to phosphorylation at the Ser129-site and not the Ser87-site because mutating the latter to alanine (S87A) did not lower the augmented targeting seen by mimicking Ser129 (see “double-mutants” in Fig. 1F). Thus our data suggest that phosphorylation at the 129-site may act as a switch that promotes presynaptic α-syn abundance. In further support of this, mimicking Ser129P even augmented the synaptic targeting of α-syn A30P and TsixK mutants that normally show diminished synaptic targeting28,29 (Fig. 1G). Expectedly, Ser129P did not augment targeting of the deletion-mutant lacking the N-terminus (residues 58–140, Fig. 1G), since membrane binding via the N-terminus is essential for presynaptic targeting30,31. To evaluate synaptic targeting in a more physiologic setting, we developed a strategy to knock-in the Ser129WT or Ser129A/D phospho-mutants – and a fluorescent reporter optimized to decrease aggregation in neurons (oScarlet, see32) – at the C-terminus of endogenous mouse α-syn (Fig. 1H, also see Supp. Fig. 3A–D and methods). Using this knock-in strategy in cultured neurons, we saw a punctate synaptic staining Resembling α-syn (Fig. 1I and Supp. Fig. 3E). Quantification of synaptic fluorescence in this knock-in setting also showed that mimicking Ser129P augmented the synaptic targeting of α-syn (Fig. 1J–K). Sequence alignment indicates that while Ser87 is variably conserved across species, Ser129 is highly conserved in mammals (Supp. Fig. 3F), consistent with an important role for this post-translational modification. Collectively, the data suggest that dynamic Ser129 phosphorylation and dephosphorylation alters the targeting of α-syn to synapses – with augmented phosphorylation leading to enhanced targeting – though Ser129P is not required for presynaptic targeting of α-syn (since phospho-incompetent mutants target normally).
Blocking Ser129P prevents physiologic effects of α-syn on synapses
Next, we used optical pHluorin assays to visualize the effects of Ser129P on SV recycling33,34. In our assay, the pHluorin – a pH-sensitive GFP that acts as a sensor for pH changes – is tagged to the transmembrane presynaptic protein vGlut1 and localized inside SVs35. While vGlut1-pHluorin is quenched in resting SVs (pH ~ 5.5); upon stimulation, SVs fuse with the presynaptic plasma membrane, resulting in an increase in intra-luminal pH and rise in fluorescence, which is subsequently quenched as SVs are endocytosed and reacidified (Fig. 2A–B, also see Supp. Movie 1). The rise and fall of fluorescence in this assay is largely a measure of the SV exo/endocytic cycle, respectively, and at the end of the experiment, all vesicles are alkalinized by adding NH4Cl to the bath to normalize datasets. Previous studies using pHluorins have shown that modest over-expression of h-α-syn in cultured hippocampal neurons suppresses activity-dependent SV-recycling21,31,36,37, leading to the concept that α-syn is a physiologic attenuator of activity-dependent neurotransmitter release. We developed a lentiviral-based system for expressing vGLUT1:pHluorin and h-α-syn:mScarlet in cultured hippocampal neurons – where essentially all synapses on a coverslip expressed both transgenes – and tested the effects of modest (~ 1.5-fold) synaptic over-expression of WT and phospho-mutant h-α-syn on SV-recycling (Supp. Fig. 4A–B and methods). Since h-α-syn suppresses SV-recycling in pHluorin-assays, and phospho-mimic (S129D) h-α-syn increases synaptic α-syn levels, we expected that over-expressing α-syn S129D would further attenuate SV-recycling, and this was indeed the case (Fig. 2C). Surprisingly however, even more than two-fold synaptic over-expression of phospho-incompetent Ser129A h-α-syn did not have any effect on activity-induced SV-recycling (Fig. 2D – all pHluorin data quantified in Fig. 2E). Experiments with transiently transfected vGlut:pHl and untagged α-syn constructs – introduced into neurons by nucleofection (see methods and21,31,36) – gave similar results (Supp. Fig. 4C–E). Taken together, these experiments imply an unexpected physiologic role for this post-translational modification.
Figure 2: Ser129P is required for α-syn-induced synaptic attenuation in pHluorin assays.
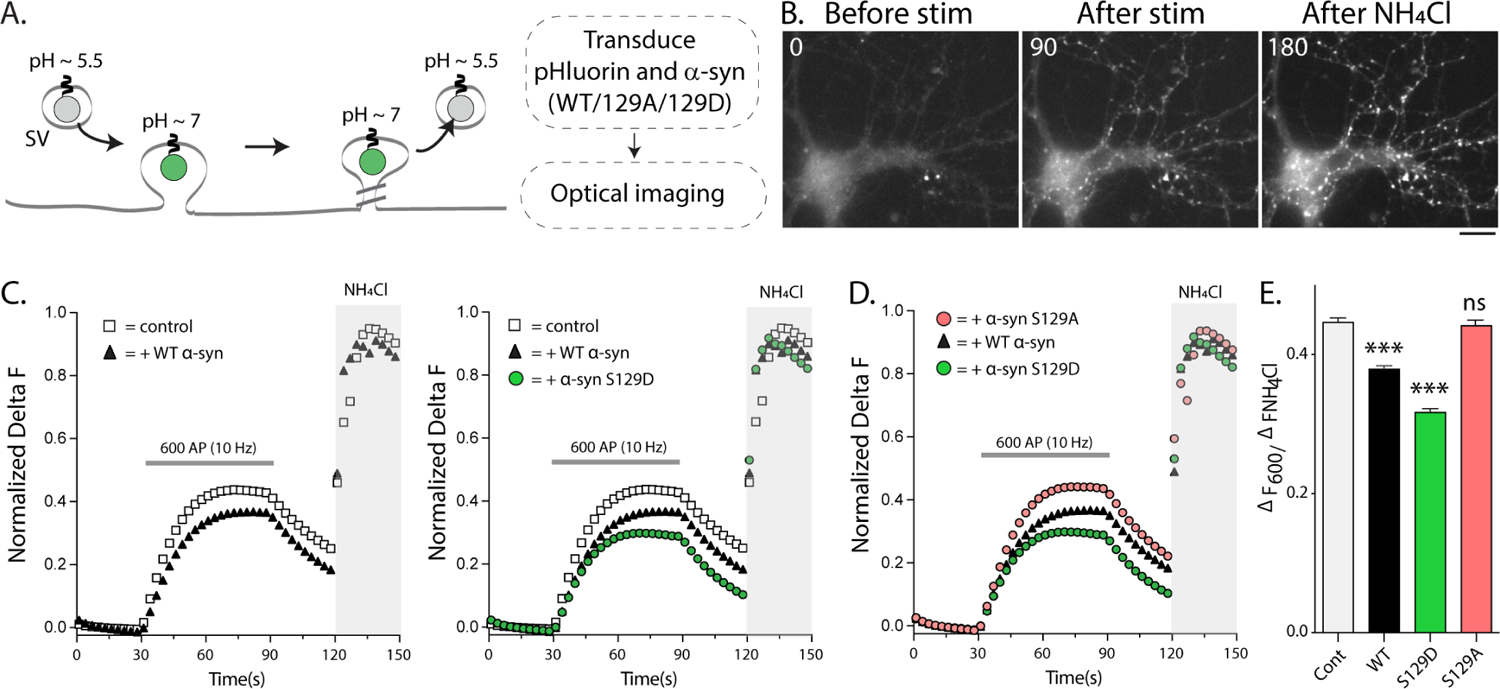
(A) Schematic of the pHluorin assay with experimental design on right.
(B) Representative pre/post-stimulation images from visualizing vGLUT:pHl fluorescence in cultured mouse hippocampal neurons. Elapsed time in seconds is shown on the upper left (also see Supp. Movie 1). Scale bar = 5 μm.
(C) Fluorescence fluctuations of vGLUT:pHl in presence of WT and phospho-mutant α-syn. WT-α-syn attenuates SV-recycling in pHluorin assays as previously reported, and Ser129 phospho-mimic mutant (S129D) further augments this suppression.
(D) Preventing α-syn Ser129P (S129A) abolished the α-syn-induced synaptic attenuation.
(E) Quantification of the recycling pool size in all pHluorin experiments (data from 12–15 coverslips, 30–50 boutons per coverslips and 4 independent cultures for each condition; ***p<0.0001 one-way ANOVA followed by a nonparametric Kruskal-Wallis; ns = not significant).
Neuronal activity augments α-syn Ser129P
The α-syn-induced synaptic attenuation in pHluorin assays is activity-dependent, and neuronal activity and calcium influx are known to induce phosphorylation of a number of synaptic proteins38,39. Very recent studies have also shown that neuronal activity triggers α-syn Ser129P in cultured neurons13, and we first asked if neuronal activity also augmented α-syn Ser129P in cells and in-vivo. First, cultured hippocampal neurons were activated by chemical [4-aminopyridine or 4-AP, a voltage-gated K+ channel blocker40], or electrical stimulation (Fig. 3A and Supp. Fig. 4F–K), and α-syn Ser129P levels were determined biochemically. As shown in Fig. 3B, 4-AP treatment led to a gradual increase in Ser129P α-syn levels without a change in total α-syn. Similarly, electrical activity (600AP at 10Hz – resembling the pHluorin experiments) also led to a rapid and selective increase in Ser129P levels, which was blocked by the sodium channel blocker tetrodotoxin (TTX, Fig. 3C). Previous studies have implicated PLK2 in Ser129P of α-syn5,24,41, and the PLK2 inhibitor BI2536 also prevented the α-syn phosphorylation induced by electrical activity (Fig. 3C). To test the activity-dependent rise of α-syn Ser129P in-vivo, we injected mice intraperitoneally with 4-aminopyridine (4-AP) – widely used to induce neuronal activity in rodents42 – and examined brains for changes in phosphorylation levels (Fig. 3D). Indeed, 4-AP treatment led to an increase in Ser129P levels without altering total brain α-syn levels (Fig. 3E). Immunohistochemical examination of brains from control and 4-AP injected mice also showed a marked increase in Ser129P levels (Fig. 3F).
Figure 3: Neuronal activity augments Ser129 phosphorylation.
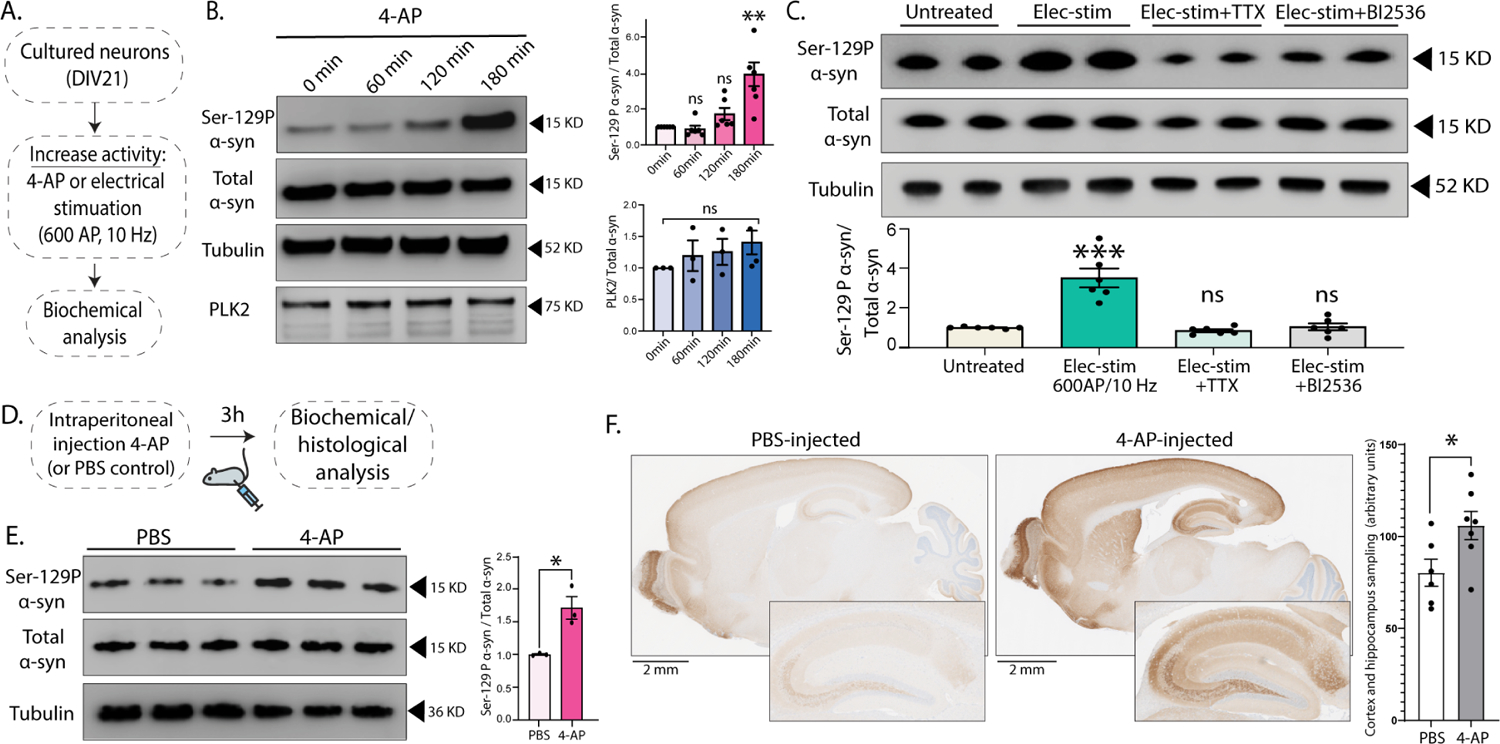
(A) Experimental design to induce activity in cultured neurons by chemical (4-AP) or electrical (600AP) stimulation.
(B) Western blots showing time-dependent increase in α-syn Ser129P after 4-AP treatment; quantified o right (n=6 for Ser129-P α-syn/total α-syn and n=3 for PLK2/total α-syn **p<0.001 unpaired Student’s t-test).
(C) Top: Western blots showing increased Ser129P after 600AP/10Hz stimulation (resembling vGLUT:pHl assays, see Methods). Note that pre-incubated with 1 μM TTX or 50 nM PLK2 inhibitor BI2536 (putative kinase for Ser129P) for 15min blocked Ser129P (two biological replicates shown). Bottom: Quantification of the western blots.
(D) Schematic for inducing neuronal activity in vivo by intraperitoneal 4-AP injections.
(E) Western blots from 4-AP-injected mouse brains show increase in Ser29P α-syn without a change in total α-syn levels (three biological replicates shown); quantified on the right (n=3 mice, all data are means ± SEM - *p < 0.01, unpaired Student’s t-test).
(F) Spatial distribution of increased Ser129P in 4-AP injected mouse brains, shown in representative sections stained with the Ser129P α-syn antibody (zoomed insets show hippocampal staining). Bar graph on the right shows quantification of intensities sampled from the cortex and hippocampus (*p<0.05, unpaired Student’s t-test, see methods for details).
Ser129P is required for binding of α-syn to functional interacting partners VAMP2 and synapsin
How does Ser129P trigger α-syn-mediated functionality? In previous studies, we found that the binding of α-syn to two presynaptic proteins – VAMP2 (synaptobrevin-2) and synapsin – was essential for the α-syn mediated synaptic attenuation seen in pHluorin assays36,37. Specifically, mutating the C-terminus VAMP2-binding site in α-syn36 or transgenic elimination of all synapsins37 abolished the α-syn-induced dampening of SV-recycling seen in pHluorin experiments. Interestingly, there are other examples at synapses where activity-induced phosphorylation or dephosphorylation regulates protein-protein interactions and functions. One of the best-studied examples is dynamin, a protein with critical roles in endocytic membrane fission. Neuronal activity induces dephosphorylation at Ser-774/778 of dynamin, which augments its association with syndapin and promotes SV endocytosis at times of elevated neuronal activity [reviewed in43]. In other examples, serine phosphorylation of the SNARE SNAP25 accelerates SV recruitment and SNARE-complex formation44, and phosphorylation of syntaxin at Ser14/188 regulates its interaction with Munc18–145,46. Thus, we wondered if Ser129P of α-syn also regulated the interaction of α-syn with its functional binding partners.
To test this idea, we first used standard co-immunoprecipitation assays to determine the binding of WT or phospho-mutant α-syn to VAMP2 or synapsin-1a (henceforth called synapsin). Neuro2a cells were transfected with myc-tagged WT/129A/129D h-α-syn and Green-Lantern (GL)-tagged VAMP2 or synapsin; VAMP2/synapsin was immunoprecipitated using an antibody to GL; and levels of h-α-syn was evaluated in the precipitates (Fig. 4A). While WT h-α-syn bound to VAMP2 and synapsin as expected36,37,47, strikingly, these interactions were essentially eliminated when the Ser129-site was rendered phospho-incompetent (S129A), and more binding was seen with phospho-mimic (S129D) α-syn (Fig. 4B–D; also see Supp. Fig. 5A–B). To visualize these associations at synapses, we designed a proximal ligation assay (PLA) to determine the interaction of α-syn and VAMP2 in cultured neurons. Based on a previous study48, the principle and overall experimental design is shown in Figure 4E. Neurons were transduced with h-α-syn (WT or S129A/D) tagged to mScarlet; widespread transduction was confirmed by red fluorescence; and proximity of h-α-syn (WT or S129A/D) to endogenous mouse VAMP2 was determined by visualizing fluorescent PLA-signals. As expected, WT h-α-syn:mScarlet expression led to a punctate PLA-signal that was restricted to synapses (Fig. 4F – left panels). However, the PLA-signal was essentially absent in neurons expressing phospho-null (S129A) h-α-syn:mScarlet (Fig. 4F – middle panels), while an augmented signal was seen with phospho-mimic (S129D) h-α-syn (Fig. 4F – right panels). In control experiments, the PLA-signal was absent when only untagged mScarlet was expressed, or when either of the two antibodies was missing (all data quantified in Fig. 4G).
Figure 4: Ser129P regulates association of α-syn with functional binding partners VAMP2 and synapsin.
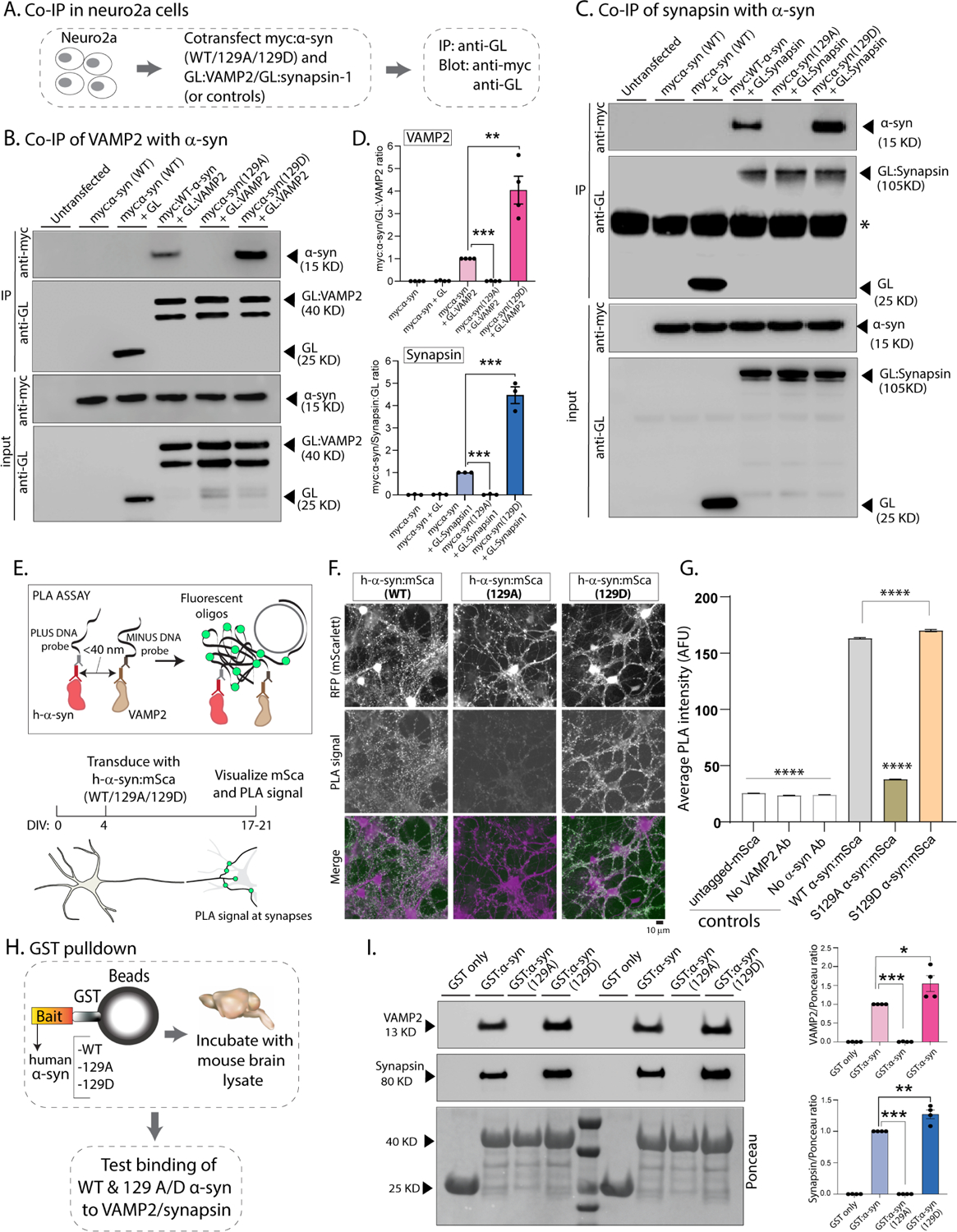
(A) Workflow for co-immunoprecipitation experiments in neuro2a cells.
(B, C) Western blots from co-immunoprecipitation experiments show that both VAMP2 (B) and synapsin (C) co-immunoprecipitated with WT-h-α-syn but not phospho-incompetent (129A) h-α-syn. Mimicking Ser129P (S129D) augmented this interaction – quantified on the right (n=4 for myc α-syn/GL-VAMP2 and n=3 for myc α-syn/GL-Synapsin co-IP). All western-blot data quantified in (D, mean ± SEM ***p<0.0001, unpaired Student’s t-test).
(E) Principle of our PLA assay and experimental design (neuron cartoon courtesy of Christophe Leterrier, Marseille). Note that a fluorescent signal is expected if transduced h-α-syn:mScarlet (WT or 129A/D) and endogenous mouse VAMP2 are <40 nm apart.
(F) Representative images of fluorescent mScarlet and PLA signals. Note that neurons transduced with WT h-α-syn show punctate PLA signal at synapses (left column), while essentially no signal is seen with phospho-incompetent (S129A) h-α-syn (middle column). Increased PLA-signal is seen with S129D h-α-syn (right column); all data quantified in (G, mean ± SEM ****p<0.0001 one-way ANOVA followed by a nonparametric Kruskal-Wallis test).
(H) Workflow for pulldown of GST-tagged WT/129A/129D h-α-syn after incubation with mouse brain lysates. Equivalent amounts of immobilized GST h-α-syn (or its phospho variants) were used.
(I) Samples from GST-pulldown were analyzed by NuPAGE and immunoblotted with antibodies against VAMP2 (top panel) and synapsin (middle panel); two biological replicates are shown. Ponceau staining (bottom panel) shows equivalent loading of fusion proteins. Note that preventing Ser129P (S129A) blocked interaction with VAMP2 and synapsin; blots quantified below (n=4, mean ± SEM *p<0.01,**p<0.001,***p<0.0001, unpaired Student’s t-test). Also note that the GST-h-α-syn in our experiments appears to be innately phosphorylated at Ser129 (see results and further characterization in Supp. Fig. 5).
To evaluate the interaction of phospho-incompetent and phospho-mimic h-α-syn in the context of the brain, we incubated GST-tagged recombinant WT/S129A/S129D h-α-syn with mouse brain lysates, and analyzed proteins bound to GST:h-α-syn by western blotting (see strategy in Fig. 4H). The overall idea here is that if the binding of α-syn to VAMP2 and synapsin is dependent on Ser129P, altering the phosphorylation status of the GST-bound h-α-syn would also alter its interaction with VAMP2/synapsin in the brain lysates. While performing these experiments, we noticed that under our experimental conditions, recombinant GST-tagged WT-α-syn expressed in bacteria appeared to be innately phosphorylated at the 129D-site (see characterization of purified proteins in Supp. Fig. 5C–F and Supp. Table 1). Consistent with the immunoprecipitation and PLA experiments, brain VAMP2 and synapsin also failed to bind phospho-incompetent (S129A) h-α-syn. In contrast, robust binding was seen with both WT and S129D h-α-syn, with relatively more phospho-mimetic α-syn binding to both proteins (Fig. 4I).
If phosphorylation and dephosphorylation of α-syn at the Ser129-site regulates the binding and unbinding of VAMP2 and synapsin – as predicted by our experiments – acutely altering the phosphorylation status of GST-tagged h-α-syn in the GST-pulldown assay would also change its binding to VAMP2 and synapsin. To test this, we performed on-bead dephosphorylation or phosphorylation of GST-tagged human α-syn – using alkaline phosphatase or recombinant PLK2, the kinase involved in Ser129P – and asked if these manipulations also altered the interaction of GST:h-α-syn with brain VAMP2/synapsin (see experimental plan in Fig. 5A). Indeed, on-bead dephosphorylation greatly attenuated interaction of h-α-syn with VAMP2 and synapsin (Fig. 5B), whereas on-bead phosphorylation augmented these interactions (Fig. 5C – all on-bead data quantified in Fig. 5D); further supporting the notion that this post-translational event is critical for protein-protein interactions. Since neuronal activity augments α-syn Ser129P, which facilitates α-syn binding to both VAMP2 and synapsin, a prediction is that under conditions of enhanced neuronal activity, a higher fraction of Ser129P α-syn would associate with VAMP2 and synapsin. To test this prediction in an in-vivo setting, we injected mice with 4-AP to induce neuronal activity (or PBS injections as controls); immunoprecipitated endogenous VAMP2 or synapsin from the mouse brains; and determined levels of Ser129P associating with the two synaptic proteins (see workflow in Fig. 5E). Indeed, a relatively higher amount of Ser129P α-syn was associated with VAMP2 and synapsin in 4-AP injected mouse brains compared to controls (Fig. 5F–G), suggesting that the phosphorylation-triggered association of α-syn with VAMP2 and synapsin is also seen in-vivo (note that a phospho-insensitive antibody was used to detect synapsin in these experiments, see methods). Taken together, data from a variety of techniques – co-immunoprecipitation in neuro2a cells, PLA experiments, GST-bead pulldown with brain lysates (including on-bead α-syn dephosphorylation and phosphorylation in this setting), and in vivo pulldowns after 4-AP injections – support a model where activity-dependent phosphorylation at the 129-site of α-syn triggers its association with VAMP2 and synapsin, which is required for mediating effects of α-syn on SV-recycling.
Figure 5: Activity-dependent Ser129P triggers α-syn association with VAMP2 and synapsin in vivo.
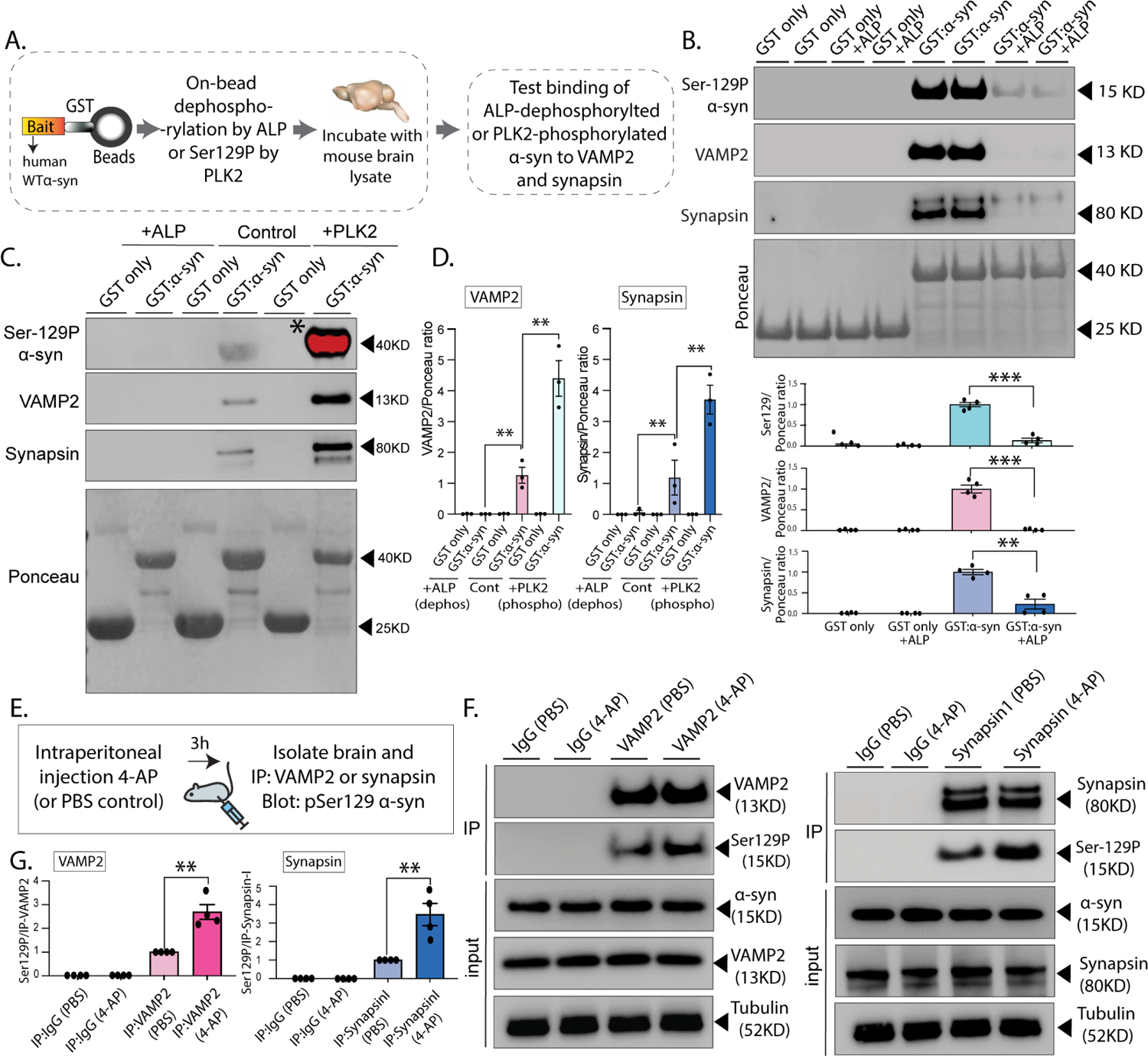
(A) Workflow for on-bead dephosphorylation and phosphorylation. GST-tagged WT-h-α-syn (or S129A/D) were pretreated with alkaline phosphatase for 3h before pulldown, or with recombinant PLK2 overnight at 30 °C (see methods for details).
(B, C) Samples from GST-pulldowns were analyzed by NuPAGE and immunoblotted with antibodies against α-syn Ser129P, VAMP2, and synapsin. Ponceau staining (bottom panels) show equivalent loading of fusion proteins. Note that α-syn dephosphorylation attenuates VAMP2 and synapsin interaction with α-syn, while Ser129P by PLK2 augments these associations (note that PLK2-induced phosphorylation leads to a substantial increase in phosphorylation that saturated the band marked with an asterisk). Quantification (n=3) of all pulldown experiments in bottom (B – mean ± SEM **p<0.001,***p<0.0001, unpaired Student’s t-test) and (D – mean ± SEM **p<0.001, unpaired Student’s t-test).
(E) Workflow for chemically inducing activity (4-AP injections), followed by co-immunoprecipitation..
(F) Note that after 4-AP injections, a relatively higher fraction of Ser129P α-syn co-immunoprecipitated with VAMP2 (left) and synapsin (right), indicating that neuronal activity augmented these associations in vivo.
(G) Quantification of western blots (n=4 and n=5 for Ser129P/VAMP2 and Ser129P/synapsin experiments, respectively). All data presented as mean ± SEM (*p < 0.01, **p < 0.001, ***p < 0.0001, t-test).
A network of synaptic proteins preferentially associating with Ser129P α-syn
Previous studies using mass-spectrometry have identified several putative binding partners of α-syn49–52. Interestingly, using a combination of peptide pulldown and mass-spectrometry with solubilized synaptosomal preparations from mouse brains, one such study (McFarland et al., ref #52) reported differential sets of proteins binding to Ser129-phosphorylated/dephosphorylated h-α-syn, suggesting that Ser129P may regulate a protein-protein interaction network. However, this study has always been viewed in the context of pathology; and moreover, the α-syn bait used in ref #52 (residues 101–140) did not contain the entire VAMP2/synapsin binding region that starts at residue 96 (Supp. Fig. 6A). In the case of VAMP2, our previous alanine-scanning experiments have specifically identified residues ~ 96–104 in α-syn as the critical site for VAMP2 binding36. Nevertheless, in hindsight, the McFarland et al. paper suggested to us that Ser129P α-syn might be physiologically binding to a broader repertoire of synaptic proteins besides VAMP2 and synapsin. To explore this possibility, we combined GST-pulldowns (using the protocol outlined in Fig. 4H) and mass-spectrometry to identify proteins from mouse brain lysates that were preferentially associated with full-length S129D α-syn, compared to S129A. Indeed, in addition to VAMP2 and synapsin, many other synaptic proteins also showed preferential affinity for WT/phospho-mimic α-syn, compared to phospho-incompetent α-syn. This interactome included several key regulators of synaptic function, including proteins involved in SNARE-assembly and exo/endocytosis (Figs. 6A–C and Supp. Tables 2–3), and we validated some candidates by western blots from mouse brain fractions (Fig. 6D). Note that both VAMP2 and synapsins (synapsin-1 and synapsin-2) preferentially associated with WT/S129D as expected, and several proteins in this group were also common with ref #52 (Supp. Table 4–5). Other proteins preferentially associating with S129D include β-spectrin, which was recently shown to bind to α-syn and play a role in pathology53, and proteins related to calcium-regulation/sensing and mitochondria, which have been long implicated in α-syn pathophysiology52,54. Collectively, the data suggest that α-syn Ser129P triggers the association of α-syn with a host of synaptic proteins involved in exo/endocytosis.
Figure 6: Ser129P Is a trigger for protein-protein interactions.
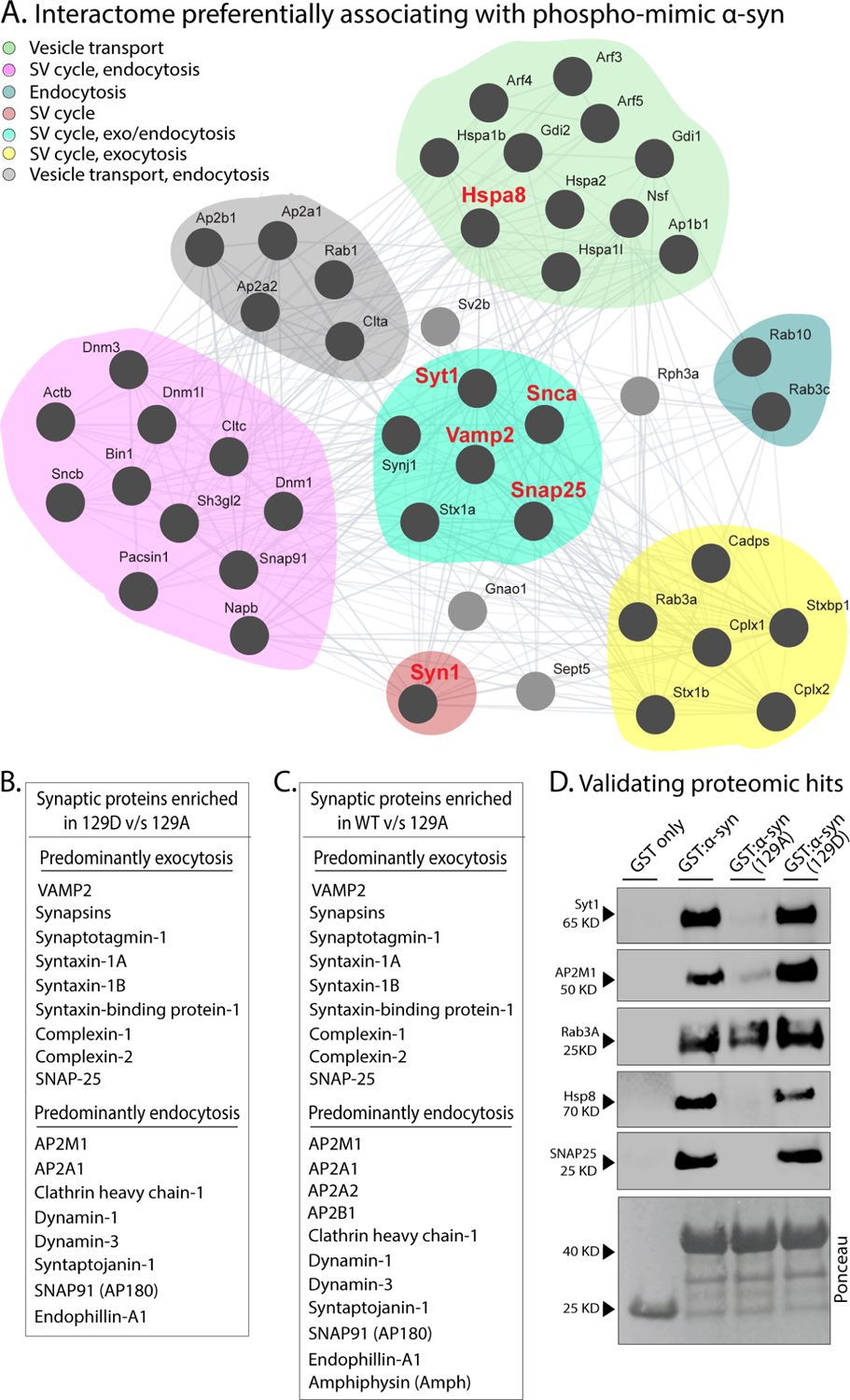
(A) Interactome of synapse-related proteins in mouse brains preferentially associating with WT or phospho-mimic (129D) h-α-syn, compared to phospho-incompetent (129A) h-α-syn (see results for experimental details). GST-tagged h-α-syn (WT/129A/12D) was incubated with mouse brain lysates, and bound proteins were detected by mass-spectrometry. Note that a host of synaptic proteins preferentially associated with S129D α-syn.
(B, C) List of some synaptic proteins preferentially associating with S129 and WT h-α-syn, compared to S129A.
(D) Western-blot validation of selected candidates from the mass-spectrometry experiments above. Ponceau staining (bottom panel) shows equivalent loading of fusion proteins.
Ser129P triggers α-syn binding to a subset of synaptic vesicles and facilitates vesicle clustering
The α-syn C-terminus (residues 96–140) has emerged as an important site for protein-protein interactions30,36,47,52, and both VAMP2 and synapsin bind to this region (ref #36, and Supp. Fig. 6A). Previous in-vitro experiments with purified proteins found that α-syn facilitates the association and clustering of small synaptic-like vesicles, and that VAMP2-binding at the C-terminus of α-syn was required for this clustering36,55. Direct tethering of adjacent vesicles by α-syn has also been proposed54,56. In previous studies, we found that h-α-syn can cluster SVs in cultured neurons31, and in-vivo ultrastructural studies have also shown that a loss of α-syn decreases SV clustering, while α-syn over-expression augments clustering57. Thus, α-syn-dependent binding and mobilization of specific SVs may be important clues to its function. To test a putative role of Ser129P in SV organization, we turned to our GST-beads/brain-lysate assay, but used intact SV preparations instead of brain lysates; incubating GST-tagged h-α-syn C-terminus-96–140 (WT/129A/129D) with SVs isolated from mouse brains (Fig. 7A–B, see fractionation protocol in Supp. Fig. 6B). GST:h-α-syn pulldown samples were blotted for VAMP2 and synapsin, as well as SV2, a ubiquitous membrane glycoprotein found on all SVs, with minimal variability amongst vesicles58. Note that synucleins are absent from these SV-fractions as described previously59 – presumably due to its relatively weaker association with SVs – eliminating potential confounding effects of mouse α-syn in these experiments (Fig. 7C). Interestingly, levels of all three SV proteins – including ubiquitous SV2 – were greatly attenuated in the GST-h-α-syn pulldown samples (Fig. 7D), suggesting that SVs enriched in VAMP2 and synapsin were unable to bind to phospho-incompetent α-syn (quantified in Fig. 7E).
Figure 7: Ser129P α-syn binds to a subset of synaptic vesicles and facilitates vesicle clustering.

(A) Workflow for pulldown of C-terminus of h-α-syn (residues 96–140) tagged to GST(WT/129A/129D) after incubation with intact SV-fractions (LP2) from mouse brain lysates.
(B) Schematic for isolating SVs; see fractionation protocol in Supp. Fig. 5B and methods.
(C) Western blots from the pellet (LP2, 15 μg) and supernatant (S4) were analyzed by western blots. Note that mouse α-syn is not present in these enriched fractions, eliminating potential confounding factors.
(D) Western blots from GST h-α-syn C-terminus pulldown experiments. Samples were analyzed by NuPAGE and immunoblotted with antibodies against VAMP2 (top), synapsin (middle), and SV2 (bottom). Note that SVs robustly associate with WT (innately phosphorylated) or S129D α-syn 96–140. However, the binding of SVs enriched in VAMP2, synapsin, and the ubiquitous glycoprotein SV2 to the α-syn C-terminus was attenuated when Ser129 was rendered phospho-incompetent (S129A). These findings suggest that Ser129P triggers association of α-syn with a subset of SVs enriched in VAMP2 and synapsin (also see results and discussion).
(E) Quantification of blots: VAMP2 (n=4), synapsin (n=4), and SV2 (n=4). All data presented as mean ± SEM (*p < 0.01,**p < 0.001, unpaired Student’s t test).
(F) Top: Schematic of experimental design to evaluate WT or S129A/D h-α-syn effects on SV clustering. Bottom: Representative electron microscopy images of synapses. Note that while WT h-α-syn over-expression increased SV clustering, phospho-incompetent (S129A) h-α-syn decreased clustering, while phospho-mimic S129D h-α-syn further augmented SV clustering. Inter-vesicular distances are quantified in (G – mean ± SEM ***p<0.001 one-way ANOVA followed by a nonparametric Kruskal-Wallis test).
In the experiments above, we asked if α-syn phosphorylation at the 129-residue affected the binding of intact SVs (specifically, integral membrane proteins on SVs) to the α-syn C-terminus. Since the α-syn N-terminus can also bind to SVs, we used a C-terminal fragment in these experiments (residues 96–140, that cannot bind to vesicles30,31) to exclude potentially confounding effects of N-terminus binding. Indeed, when we used full-length GST:h-α-syn as bait, there were equivalent levels of the ubiquitous SV2 glycoprotein in the pulldown samples (Supp. Fig. 6C – top panel), indicating that a similar number of SVs were binding to GST:h-α-syn in this setting, regardless of phosphorylation-status (presumably many of these vesicles are binding to the N-terminus). Nevertheless, more VAMP2/synapsin enriched SVs were associated with S129D h-α-syn, compared to S129A (Supp. Fig. 6C – middle and bottom panels; quantified in Supp. Fig. 6D), suggesting that despite N-terminus associations, phosphorylation status of the C-terminus was still influencing the binding of VAMP2/synapsin-enriched SVs in these experiments. Notably, there was less binding of synapsin-enriched SVs to WT h-α-syn in this setting, which may relate to the probabilistic nature of these experiments, where vesicles can associate with either N- and/or C-terminus of α-syn. While precise characterization of the composition of SVs is beyond the scope of this study, our data suggest that α-syn may preferentially associate with a subset of SVs that are enriched in VAMP2 – which is one of the most abundant proteins on SVs60,61 – and that this binding is required for activity-dependent neurotransmitter release62. In this context, while bulk biochemical approaches have estimated average numbers of proteins on SVs60 and all synaptic vesicles look similar at the ultrastructural level, numerous lines of evidence indicate that SVs within a single synapse are not homogenous, and the molecular underpinnings of SV-pool heterogeneity is an active area of investigation [reviewed in63,64].
Given the emerging evidence linking α-syn Ser129P to VAMP2-binding and vesicle clustering, we next asked if the phosphorylation status of α-syn influenced vesicle clustering at bona-fide synapses. For these experiments, we transduced cultured neurons with WT or S129A/D h-α-syn tagged to mScarlet, confirmed broad transduction by red fluorescence, and performed electron microscopy to visualize and quantify SV clusters within single synapses. While SVs were clustered upon over-expression of WT h-α-syn as expected31, interestingly, quantification of inter-vesicular distance (see methods) indicated that there was a reciprocal effect of Ser129P on vesicle clustering. While inter-vesicular distances were increased in S129A-expressing synapses – with more spacing between individual SVs within a synapse – augmented SV-clustering was seen in S129D-expressing neurons (Fig. 7F–G). Taken together, these findings suggest a scenario where phosphorylation of α-syn at the 129-site triggers protein-protein interactions at synapses that regulate SV clustering and mobilization, eventually influencing SV recycling and neurotransmission.
Ser129-P mediated electrostatic interactions may stabilize the α-syn C-terminus and facilitate protein-protein interactions
How does a single phosphorylation event modulate α-syn interactions? Since the Ser129P-dependent differential binding of α-syn with its interacting partners is solely mediated by residues 96–140 (Fig. 7D), one possibility is that α-syn Ser129P induces conformational changes within the C-terminus that stabilize this region, facilitating its binding to interacting partners. Indeed, there are many examples in biology where phosphorylation alters protein conformation and protein-protein binding65. To explore the effects of Ser129P on α-syn structure, we used AlphaFold2-driven modeling and dynamic membrane-binding atomic scale simulations. AlphaFold2 uses machine learning algorithms that incorporate both bioinformatics and physical knowledge of protein structures – leveraging multi-sequence alignments – to accurately predict three-dimensional structures at atomic resolution; and has emerged as a valuable tool in predicting protein structures66. To model putative changes in α-syn structure upon Ser129P, we used ColabFold, a free and accessible platform based on AlphaFold2 and RoseTTAFold, which reproduces and implements many of the ideas incorporated by AlphaFold267. Since regulation of α-syn Ser129P is dynamic24,68, we considered two states in our simulations – one where the Ser129 h-α-syn remains unphosphorylated, and another where h-α-syn is constitutively phosphorylated (phospho-mimic S129D); employing models that had the highest confidence values in ColabFold (Fig. 8A and Supp. Fig. 7A). Finally, we set up dynamic simulations where resultant ColabFold structures were allowed to associate with artificial bilayers resembling SV membranes under physiologic conditions, and penetration of h-α-syn into the bilayers (upper and lower membrane leaflets) was computationally determined (Fig. 8B; see methods for details). For each scenario, the near-final stabilized, membrane-embedded conformation from the simulations was considered (last 20 ns, ~10% of the simulation, averaging 107 time-points at 2 femto-second resolution).
Figure 8: Ser129-P mediated interactions may stabilize α-syn C-terminus and facilitate protein-protein interactions.
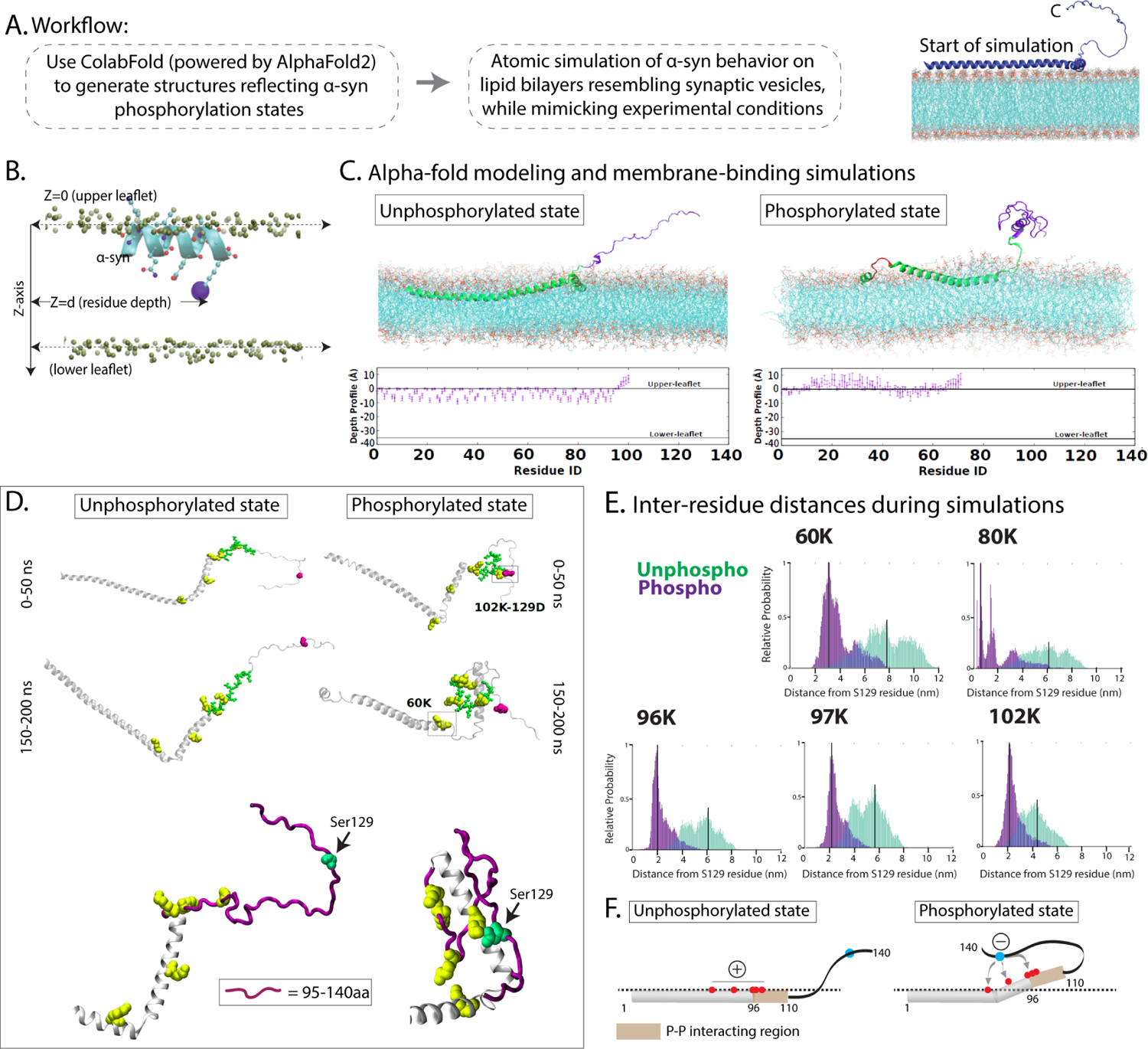
(A) Workflow for generating α-syn structures and molecular simulations. Illustration on the right shows overall principle of the membrane-binding simulations (see results and methods for more details).
(B) Schematic showing boundary-definitions of membrane leaflets and protein residue depths used to calculate membrane penetrance of α-syn; viewed from the side. Note that the depths to which the alpha-helices can embed the membrane can be precisely calculated.
(C) Results from membrane-binding simulations. Top: Near-final snapshots from simulations showing depth profiles of membrane-embedded unphosphorylated (left) phosphorylated (right) states of α-syn. Bottom: Quantitative residue-wise depth-profile (mean +/− SEM of each residue) shows membrane-embedding in the two states. Note that the C-terminus (magenta) appears folded in the phosphorylated state due to intramolecular electrostatic interactions.
(D) Representative snapshots from early (0–50 ns) and late (150–200 ns) stages of the simulation show that in the phosphorylated state, five positively charged lysine residues within the 95–140 residues (60K, 80K, 96K, 97K, and 102K – marked in yellow) crowd around the Ser129-site (marked by an arrow in the zoomed images below). The distances between these five lysine residues and the Ser129-site across the simulations is quantified in (E). Note the relatively greater proximity of the five lysines to Ser129 in the phosphorylated state.
(F) Schematic depicts a possible scenario where electrostatic interactions between the negatively charged S129-site and positive lysines along the C-terminus expose the α-syn protein-protein interaction site (residues 96–140), allowing functional associations with other synaptic proteins. Dashed line represents upper membrane leaflet.
In the unphosphorylated state, our simulations showed that the N-terminus of α-syn was embedded into the membrane bilayer, as reported by previous studies69–71. Although mimicking phosphorylation induced some changes in membrane-binding, the N-terminus was largely embedded into the membrane and there was little change in the overall conformation of the vesicle-binding surface (Fig. 8C and Supp. Fig. 7B). Since membrane-binding of α-syn is thought to mainly rely on the first ~ 20 residues72, these changes are not expected to have a significant effect on SV-binding. However, there were substantial changes in the folding of the C-terminus when we mimicked the phosphorylated state, triggered by electrostatic associations between the negatively charged Ser129P with positively charged lysine residues within the VAMP2/synapsin-binding region (60K, 80K, 96K, 97K, and 102K – see snapshots from simulation in Fig. 8D, and folding-kinetics in Supp. Movie 2). These interactions tended to crowd the five lysines around the 129-site – see a zoomed example from simulation, Fig. 8D (bottom panel) – and we reason that such associations may expose and/or stabilize the binding region of VAMP2 and synapsin (96–140 residues) and facilitate protein-protein interactions. Quantitative analyses of inter-residue distances during simulations highlight the proximity of C-terminus lysine residues to the 129-site when α-syn is in the phosphorylated state (Fig. 8E). A hypothetical scenario is shown in Figure 8F, where transient electrostatic interactions between the Ser129P residue and the positively charged lysines at the C-terminus exposes the protein-protein interaction region (residues 96–110), allowing association of α-syn with its functional interacting partners.
Discussion
Phosphorylation of α-syn at the Ser129-site is a pathologic hallmark of synucleinopathies that is often synonymously associated with disease, but our studies suggest that this post-translational modification has a physiologic role. Our main findings are: (1) Although α-syn is widely expressed in synapses throughout the brain, Ser129P is only seen in a subset of brain regions – suggesting intrinsic regulation of this post-translational modification – and this phosphorylation augments the synaptic targeting of α-syn. (2) Ser129P is required for α-syn-mediated synaptic attenuation in optical SV-recycling (pHluorin) assays, implying a functional role for this modification. (3) Neuronal activity augments Ser129P in cultured neurons and in-vivo. (4) Ser129P is required for α-syn binding to VAMP2 and synapsin – two functional interacting partners required for mediating effects of α-syn on SV-recycling – and mass-spectrometry data suggest that a wider repertoire of synaptic proteins preferentially associate with Ser129P α-syn. (5) Ser129P triggers the binding of α-syn to a subset of SVs enriched in VAMP2 and synapsin, which may dynamically facilitate the physiologic clustering of SVs. (6) Alphafold-2-driven modeling and dynamic simulations suggest that Ser129P stabilizes the C-terminus via local interactions, which may allow conformations that favor protein-protein interactions. Taken together, the data reveal a cascade of molecular events by which Ser129P of α-syn regulates synaptic function, and offers a new basis for understanding the role of this post-translational modification in pathophysiology.
What is the evidence that α-syn Ser129P triggers pathology?
If Ser129P triggered pathology, the most straightforward prediction is that increasing phosphorylation at this site would induce more neurodegeneration and cell death, but data from studies looking at this is inconsistent. AAV injections of phospho-mimic (S129D) h-α-syn into rat brains or transgenic expression of S129D h-α-syn in mouse brains lacking endogenous mouse α-syn did not show an additive pathologic effect of the phospho-mimic mutant8–10, though degeneration initially occurred at a slightly faster rate with S129D in one study27. Raising levels of PLK2/3 – kinases implicated in Ser129P – by AAV injections increased levels of Ser129P but did not augment neurodegeneration11. On the other hand, transgenic expression of S129D h-α-syn in fly models showed a faster rate of dopaminergic neuron loss with S129D26. Expression of WT h-α-syn is also toxic in yeast models73,74, and Ser129 phospho-mimic mutants did not enhance this toxicity75. Based on the extensive Ser129P seen in Lewy bodies and Lewy neurites, another prediction is that this phosphorylation would facilitate aggregation. However, the opposite has been noted, where the introduction of Ser129P suppresses α-syn aggregation7,25. Though more work is needed to draw definitive conclusions, the available data does not readily support the view that this post-translational modification makes α-syn more toxic.
A physiologic role for α-syn Ser129P in synaptic attenuation
The normal function of α-syn has been debated for many years, and the clearest insights have come from optical pHluorin assays that can quantitatively evaluate the exo/endocytic recycling of SVs in living cultured hippocampal neurons at a single-synapse level. In this system, a modest (~ 1.5-fold) over-expression of h-α-syn attenuates activity-dependent SV recycling21,31,36,37, advocating a role for α-syn in physiologically dampening neurotransmitter release. Remarkably, blocking the association of α-syn with endogenous VAMP2 or synapsin – two synaptic proteins known to bind to α-syn – abolishes the ability of α-syn to attenuate SV recycling in this assay36,37, indicating that the synaptic attenuation in these experiments occurs within the context of a functional interactome. Converse effects are seen in α-syn knockout mice, where striatal dopamine stores are reduced, consistent with increased release76. A more rapid facilitation of neurotransmitter release is also seen in these mice77,78. The α-syn gene belongs to a family of three genes α/β/γ-synucleins; and α/β-null or α/β/γ-null synuclein mice show substantial increase in striatal dopamine release in-vivo79,80 as well as enhanced neurotransmission in hippocampal slices, at least in young mice81.
While exploring the role of Ser129P, we were surprised to find that preventing phosphorylation at this site also abolished the α-syn-mediated synaptic attenuation seen in pHluorin assays (Fig. 2), which led us to investigate underlying mechanisms. This synaptic attenuation is activity-dependent, and neuronal activity is also known to phosphorylate other synaptic proteins – which suggested links between activity and α-syn Ser129P. Our data and a recently published study13, show that neuronal activity augments α-syn Ser129P. The increase in Ser129P can be seen within minutes of activity induction, and can also be elicited in an in vivo setting (Fig. 3D–F). One caveat is that in previous studies, we found that α-syn residues 1–110 can also attenuate SV recycling36. Though this fragment is expected to bind to both VAMP2 and synapsin36, it obviously lacks the Ser129-site, so it is unclear how the synaptic attenuation is achieved in this scenario. Perhaps C-terminus truncation changes the conformation of the protein-protein binding region (residues 96–110) and allows persistent interactions. The restricted pattern of normal Ser129P in the brain (Fig. 1A–B) suggests a scenario where Ser129P-dependent α-syn functionality may be constitutively required in specific brain circuits. It is also possible that dephosphorylated and phosphorylated α-syn have different functions, and more work is needed to clarify these issues. Regarding histology, brain Ser129P staining was seen in both synapses and nuclei, with clear nuclear staining in several groups of neurons, including hippocampal, cortical, and olfactory neurons (Fig. 1A and Supp. Fig. 1A–B). Similar staining was also seen in cultured hippocampal neurons (Fig. 1C and Supp. Fig. 1C). The nuclear localization of Ser129P α-syn in selected neurons within the brain might be an important clue to its function.
A model linking α-syn Ser129P, protein-protein interaction, and α-syn function
What are the molecular events connecting α-syn Ser129P to α-syn-induced synaptic attenuation? Serine phosphorylation is a known trigger for protein-protein interactions in numerous cellular contexts (reviewed in82–84). Our candidate-based biochemical data showed that Ser129P was necessary for α-syn binding to both VAMP2 and synapsin, and unbiased proteomics suggests that α-syn Ser129P facilitates association of α-syn with a wider repertoire of proteins, including several that play key roles in SV recycling (Figs. 4–7). A straightforward conclusion is that the C-terminus of α-syn is a major protein-protein binding hub, and that Ser129P is required for triggering these interactions. While the binary effect of Ser129P on α-syn binding to its interacting partners is somewhat surprising, there are many examples in biology where protein phosphorylation regulates the stability, activity, or cellular localization of proteins84. Though this has primarily been studied in the context of cell-signaling, there are a few clear examples where serine phosphorylation regulates protein-protein interaction at synapses, and interestingly, some of these effects are also activity-dependent43–46.
Previous studies have established that protein phosphorylation can influence function by directly recognizing functional domains or interfering with protein-protein binding (orthosteric). Alternatively, phosphorylation can induce conformational changes that can alter protein-protein interactions (allosteric). A common mode of allosteric interaction is when phosphorylation leads to the formation of hydrogen bonds and salt bridges between the phosphate oxygens and arginine or lysine side chains that change the conformation of the protein84. Our modeling data and membrane-binding simulations suggest a scenario where Ser129P triggered substantial changes in the conformation of the unstructured C-terminus of α-syn due to the formation of salt bridges between the negatively charged 129-site and positively charged lysines within this region (Fig. 8D–F and Supp. Movie 2). Since both VAMP2 and synapsin bind within amino-acids 96–140 of α-syn (Supp. Fig. 6A), we propose that the electrostatic interactions triggered by Ser129P stabilize this region and facilitate its association with interacting partners. Additionally, the negatively charged Ser129P-site may also form long-range electrostatic interactions with the N-terminus of α-syn on an adjacent vesicle. In this scenario, α-syn may facilitate vesicle clustering – as seen in minimally reconstituted in vitro systems36,55, and also in our experiments in cultured neurons (Fig. 7F – also see31,85). The broader repertoire of protein-protein interaction upon S129P may also be the basis for augmented synaptic targeting seen with S129D α-syn (Fig. 1D–G).
Taken together, our experiments support a physiologic role for α-syn Ser129P at synapses, advocating a model where activity-induced Ser129P triggers the interaction of α-syn with a network of synaptic proteins that eventually leads to physiologic attenuation of neurotransmitter release. Precise mechanisms by which α-syn and its functional partners dampen neurotransmission are still unclear, though we posit that Ser129P-induced physiologic changes in SV mobilization and membrane-binding are important. The invariably high levels of Ser129P α-syn in synucleinopathies imply an important role of this post-translational modification in disease. However, most disease models have viewed pathologic events as a direct consequence of Ser129P, and a possible physiologic role of this modification has not been considered. In this context, our data from mouse brains suggest that there is an age-related increase in Ser129P (Supp. Fig. 1B), which may have disease relevance. Our collective data – and the proposed new model for the role of Ser129P in α-syn function – offer a fresh conceptual platform that can serve as the foundation for a deeper understanding of the pathophysiologic transition of α-syn in synucleinopathies.
STAR METHODS TEXT
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Subhojit Roy (sroy@ucsd.edu).
Material availability
Backbone plasmids generated in this study have been deposited to Addgene. These vectors include pCCL-mScarlet (Addgene #209889), pCCL-Green Lantern (Addgene #209890), and pGEX-KG myc (Addgene#209891).
Data and code availability
All data to reproduce figure panels has been deposited and archived by Zenodo, DOI is listed in the key resource table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Ser129P α-syn | Abcam | Cat#ab51253,RRID:AB_869973 |
| NeuN | Abcam | Cat#ab236870,RRID:AB_2927651 |
| VAMP2 | Synaptic Systems | Cat#104211BT,RRID:AB_2619758 |
| α-syn | BD Biosciences | Cat# 610787,RRID:AB_398108 |
| β-syn | Abcam | Cat#ab76111,RRID:AB_1309981 |
| Tubulin | Abcam | Cat#ab7291,RRID:AB_2241126 |
| PLK2 | Thermo Fisher Scientific | Cat# PA5–14094,RRID:AB_2167742 |
| Synaptophysin-1 | Synaptic Systems | Cat#101011C2,RRID:AB_10890165 |
| SV2A | Abcam | Cat#ab32942,RRID:AB_778192 |
| Synapsin-1 | Abcam | Cat#ab254349,RRID:AB_2920663 |
| Synapsin-1 | Synaptic Systems | Cat#106104,RRID:AB_2721082 |
| c-myc | Sigma-Aldrich | Cat#M4439,RRID:AB_439694 |
| GFP | Abcam | Cat#ab290,RRID:AB_303395 |
| Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A32723,RRID:AB_2633275 |
| Alexa Fluor 594 | Thermo Fisher Scientific | Cat#A32740,RRID:AB_2762824 |
| Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A32733,RRID:AB_2633282 |
| α-syn(Syn 204) | Santa Cruz Biotechnology | Cat#sc-32280 |
| VAMP-2 | Abcam | Cat#ab214590 |
| DuoLink PLA technology probe | Sigma-Aldrich | Cat#DUO92002 |
| DuoLink PLA technology probe | Sigma-Aldrich | Cat#DUO92004 |
| DuoLink PLA technology probe | Sigma-Aldrich | Cat#DUO82049 |
| DuoLink PLA technology probe | Sigma-Aldrich | Cat#DUO92008 |
| DuoLink PLA technology probe | Sigma-Aldrich | Cat#DUO92014 |
| α-syn | Synaptic Systems | Cat#128211 |
| Ser129P α-syn | Cell Signaling | Cat #23706 |
| Goat anti-Rabbit HRP | Abcam | Cat#ab205718,RRID:AB_2819160 |
| Goat anti-mouse HRP | Abcam | Cat#ab205719,RRID:AB_2755049 |
| Bacterial and Virus Strains | ||
| Escherichia coli BL21(DE3) | New England Biolabs | Cat#C2530H |
| One Shot™ Stbl3™ Chemically Competent E. coli | Invitrogen | Cat#C737303 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) | TOCRIS bioscience | Cat#0190 |
| DuoLink mounting media | Sigma-Aldrich | Cat#DUO82040–5 |
| Recombinant human α-syn phospho-Ser129 protein | Proteos | Cat#RP-004 |
| Recombinant human α-syn | RnD systems | Cat#SP-485–500 |
| GST-α-syn | NovoPro | Cat#505565 |
| pGEX-KG myc | Addgene | Cat#209891 |
| Human recombinant PLK2 | Abcam | Cat#ab102108 |
| Lipofectamine 2000 | Invitrogen | Cat#11668019 |
| ProFection | Promega | Cat#E1200 |
| LentiX Concentrator | Takara | Cat#631232 |
| Isoflurane | VetOne | Cat#502017 |
| 4% Paraformaldehyde (PFA) | Thermo Scientific | Cat#J19943-K2 |
| Discovery Wash buffer | Ventana | Cat#07311079001 |
| CC1 buffer | Ventana | Cat#06414575001 |
| inhibitor CM buffer | Ventana | Cat#760–159 |
| HRP polymer | Ventana | Cat#05266548001 |
| Hematoxylin II | Ventana | Cat#05277965001 |
| Bluing reagent | Ventana | Cat#05266769001 |
| Lambda Protein Phosphatase | New England Biolab | Cat#P0753 |
| NotI restriction enzyme | New England Biolabs | Cat#R3189L |
| In-Fusion Snap Assembly Master Mix | Takara | Cat#638947 |
| DNA Ligation Kit Mighty Mix | Takara | Cat#6023 |
| PEI Max | Polysciences | Cat#24765 |
| Lucigen QuickExtract DNA extraction solution | Biosearch Technologies | Cat#QE09050 |
| Neuronal protein extraction reagent (N-PER) | Thermo Scientific | Cat#87792 |
| Protease/phosphatase inhibitors | Cell Signaling | Cat#5872 |
| Protein G-agarose beads | Thermo Scientific | Cat#20397 |
| NuPAGE LDS sample buffer | Thermo Scientific | Cat#NP007 |
| NuPAGE 4 to 12% Bis-Tris polyacrylamide gels | Thermo Scientific | Cat#NP0335BOX |
| 0.2 μM PVDF membrane | Thermo Scientific | Cat#LC2002 |
| Terrific Broth | Thermo Scientific | Cat# BP9728–2 |
| EDTA-free protease cocktail inhibitor | Roche | Cat#11836170001 |
| Glutathione-Sepharose 4B | Sigma | Cat#17-0756-01 |
| TEV protease | New England Biolabs | Cat#P8112 |
| Alkaline Phosphatase (ALP) | Thermo Scientific | Cat# EF0651 |
| Dithiothreitol (DTT) | Thermo Scientific | Cat#R0861 |
| Iodoacetamide (IAA) | Sigma | Cat#I1149 |
| Lysyl endopeptidase | Wako | Cat#125–05061 |
| Trypsin | Promega | Cat#VA9000 |
| Asp-N | Promega | Cat#VA1160 |
| HLB column | Waters | Cat#186002034 |
| Poly-D-lysine | Sigma | Cat#P6407–5MG |
| Critical Commercial Assays | ||
| Ventana Discovery ChromoMap DAB kit | Ventana | Cat#05266645001 |
| AAVpro Purification Kit (All Serotypes) | Takara | Cat#6666 |
| P3 Primary Cell 4D Nucleofector X Kit S | Lonza | Cat#V4XP-3032 |
| Monarch DNA gel extraction Kit | New England Biolabs | Cat#T1020S |
| High-select Fe-NTA Phosphopeptide enrichment kit | Thermo Scientific | Cat#A32992 |
| Deposited Data | ||
| ColabFold Protein model | This paper | Zenodo: https://zenodo.org/records/8059967 |
| Depth profile molecular simulation | This paper | Zenodo: https://zenodo.org/records/8059967 |
| Tabular data | This paper | Zenodo: https://zenodo.org/records/8059967 |
| Supplemental movies | This paper | Zenodo: https://zenodo.org/records/8059967 |
| Experimental Models: Cell Lines | ||
| HEK293T | Cellosaurus | RRID:CVCL_0063 |
| Neuro2A | Cellosaurus | Cat#TKG0509, RRID:CVCL_0470 |
| Experimental Models: Organisms/Strains | ||
| CD-1 IGS mouse (ICR) | Charles River Laboratories | Outbred Cat# 0220CD-1 |
| C57BL/6J | Jackson Labs | RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| α-syn forward primer (PCR#1): TGTGCTTTCTCTTCCCT CTCTG | IDT | N/A |
| α-syn forward primer (PCR#2): ATAACACTTCGTGCAG CACC | IDT | N/A |
| Reverse oScarlet primer (PCR#1 and PCR#2): ACAGGATGTCCCAGGA GAAG. | IDT | N/A |
| Recombinant DNA | ||
| pCCL-mScarlet | This paper | Addgene plasmid# 209889 |
| pCCL-Green Lantern | This paper | Addgene plasmid# 209890 |
| pGEX-KG myc | This paper | Addgene plasmid# 209891 |
| psPAX2 | Addgene | RRID:Addgene_12260 |
| pMD2.G | Addgene | RRID:Addgene_12259 |
| LentiCRISPR v2 | Addgene | Addgene plasmid#52961 |
| PX552 | Addgene | Addgene plasmid# 60958 |
| pAAV-SpCas9 | Addgene | Addgene plasmid# 60957 |
| pPHP.S | Addgene | Addgene plasmid #103005 |
| Software and Algorithms | ||
| GraphPad Prism Software | http://www.graphpad.com/ | RRID:SCR_002798 |
| MetaMorph Microscopy Automation and Image Analysis Software | Molecular Devices https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysissoftware/metamorph-microscopy#gref | RRID:SCR_002368 |
| MATLAB | MATLAB http://www.mathworks.com/products/matlab/ | RRID:SCR_001622 |
| CRISPick | Broad Institute https://portals.broadinstitute.org/gppx/crispick/public | N/A |
| Image Lab software version 6.1 | BioRad | RRID:SCR_014210 |
| STRING database version 11.5 | STRING | RRID:SCR_005223 |
| PANTHER database | PANTHER | RRID:SCR_004869 |
| AxIS software version 2.4 | Axion Biosystems | RRID:SCR_016308 |
| ColabFold | https://colabfold.mmseqs.com/ | N/A |
| GROMACS v.2021.3 | https://www.gromacs.org/ [doi: 10.5281/zenodo.5053201] | RRID:SCR_014565 |
| CHARMM36 force field | https://www.charmm.org/archive/charmm/resources/charmm-force-fields/ | RRID:SCR_014892 |
| LINCS algorithm | http://www.lincsproject.org/ | RRID:SCR_016486 |
| Jalview sequence fetcher | Jalview | RRID:SCR_006459 |
| Clustal Omega Webserv er | http://www.ebi.ac.uk/Tools/msa/clustalo/ | RRID:SCR_001591 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice and cell-lines
Only wild-type healthy animals were used in our studies, and all procedures were performed following University of California guidelines. Mice were housed in group cages according to standard guidelines under a 12-hour:12-hour light-dark cycle. Primary hippocampal cultures (DOI: 10.1007/978-1-0716-1990-2_8) were obtained from timed-pregnant female CD-1(RRID:MGI:5649524) mice [(CD-1 IGS mouse (ICR) Outbred 0220) https://www.criver.com/products-services/find-model/cd-1r-igs-mouse?region=3611; biochemical and in vivo experiments were performed in C57BL/6J mice [(Jackson Labs RRID:IMSR_JAX:000664). For drug-induced neuronal activity studies, 6–8 week-old C57BL/6J mice were intraperitoneally (IP) injected with 2mg/Kg 4-aminopyridine (Sigma-Aldrich Cat #A78403) and analyzed after 3hrs (DOI: dx.doi.org/10.17504/protocols.io.5qpvo38dxv4o/v1). Both females and males were used in this study in equal numbers at the age described in the text and figures. Neuro2a cells (used for biochemistry) were obtained from ATCC that has standardized and established internal authentication protocols. Cells were cultured using standard recommended protocols and tested for mycoplasma contamination.
METHODS DETAILS
Reagents and DNA constructs
The following antibodies were used for immunofluorescence experiments: Ser129P α-syn (Abcam #ab51253, RRID:AB_869973) (1:100), NeuN (Abcam # ab236870, RRID:AB_2927651)(1:1000), VAMP2 (Synaptic Systems Cat# 104 211BT, RRID:AB_2619758) (1:1000). The following antibodies were used for biochemistry experiments: Ser129P α-syn (Abcam #ab51253, RRID:AB_869973) (1:800), total α-syn (BD Biosciences Cat# 610787, RRID:AB_398108) (1:400), β-syn (Abcam Cat# ab76111, RRID:AB_1309981)(1:500), tubulin (Abcam Cat# ab7291, RRID:AB_2241126)(1:10000), PLK2 (Thermo Fisher Scientific Cat# PA5–14094, RRID:AB_2167742) (1:10000), synaptophysin-1 (Synaptic Systems Cat# 101 011C2, RRID:AB_10890165)(1:500), VAMP2 (Synaptic Systems Cat# 104 211BT, RRID:AB_2619758) (1:500), SV2A (Abcam Cat# ab32942, RRID:AB_778192) (1:500), synapsin-1(Abcam Cat# ab254349, RRID:AB_2920663)(1:500), synapsin-1 (Synaptic Systems Cat# 106 104, RRID:AB_2721082)(1:500)], c-myc (Sigma-Aldrich Cat# M4439, RRID:AB_439694) (1:500), GFP (Abcam Cat# ab290, RRID:AB_303395) (1:5000). Chemicals used: 4-Aminopyridine (4-AP, Sigma #A78403), tetrodotoxin (TTX) (Cayman#14964), 6-cyano-7-nitroquinoxaline-2,3- dione (CNQX, TOCRIS bioscience #0190), D, L-2-amino-5-phosphonovaleric acid (AP5, TOCRIS bioscience #0105), and BI2536 (Selleckchem #S1109). Antibodies used for tissue immunohistochemistry include total α-syn (Synaptic Systems, 1:15,000, Cat #128211) and Ser129P α-syn (Cell Signaling, 1:3000, Cat #23706). Commercial recombinant human α-syn phospho-Ser129 protein (Proteos #RP-004), α-syn (RnD systems #SP-485–500), and GST-α-syn (NovoPro #505565) were used in biochemical and proteomic experiments. For tagged proteins, synthetic DNA blocks (IDT) or PCR products coding for human WT α-syn, α- syn S129A, α-syn S129D, VAMP2, and synapsin1 (gift from George Augustine, Nanyang Technological University, Singapore) were subcloned into the pCCL-mScarlet (Addgene _209889), pCCL-Green Lantern (Addgene_209890), or pGEX-KG myc (Addgene_209891) vectors. Synthetic DNA blocks (IDT) coding for the truncated α-syn variants were subcloned into the pGEX-KG myc vector (Addgene_209891). Lentiviral and AAV plasmids were transformed with One Shot Stbl3 Chemically Competent E. coli (Invitrogen Cat# C737303), whereas protein expression plasmids used Escherichia coli BL21(DE3) (New England Biolabs Cat# C2530H). All cloning was performed using Gibson cloning (NEB). Constructs obtained from other laboratories are noted in the acknowledgements. All constructs were verified by sequencing.
Hippocampal Cultures, lentivirus production, vGLUT1-pHluorin imaging and analysis
Most experiments were either done in primary neuronal cultures from mice, or in vivo in mice. All animal studies were performed in accordance with University of California guidelines. Primary hippocampal cultures were obtained from postnatal (P0–P1) mouse pups using standard procedures as described previously86,87. In brief, cells from P0–P1 mice were plated (30,000–60,000 cells/cm2) and cultured to maturity – days in vitro (DIV) 17–21. For synaptic targeting experiments, neurons were transiently transfected on DIV-13 using Lipofectamine 2000 (Invitrogen Cat# 11668019) and imaged at DIV 17–21 as described previously31. HEK293T cells (RRID: CVCL_0063) were maintained in DMEM+Glutamax supplemented with 10% FBS and 1% penicillin-streptomycin for lentivirus production. Before transfection, 6×106 cells were plated in 15cm dishes (Genessee Scientific) for 14–16 hrs. HEK cells were transfected with the targeting plasmid and two helper plasmids (psPAX2 (RRID: Addgene_12260) and pMD2.G (RRID: Addgene_12259) at a 2:1.5:1 molar ratio, using ProFection (Promega Cat#E1200). The supernatant was collected and concentrated using LentiX Concentrator (Takara Cat#631232) two to three days later. The viral pellet was resuspended in 1/100th of the supernatant volume of sterile HBSS and stored at −80°C. Lentiviral packaging of vGLUT1:pHluorin (construct was gift from Shigeki Watanabe, Johns Hopkins) was carried out by GenScript (Piscataway, NJ, USA). For viral transduction, lentiviruses were added to each well of neurons at DIV 3 at MOI=5. In all cases, almost 100% transduction was achieved with the lentiviruses, as confirmed by immunostaining.
For the lentiviral vGLUT1:pHluorin experiments, cultured neurons were plated at 60,000 cells/cm2 density. DIV-3 neurons were infected with lentiviruses carrying α-syn tagged at the C-terminus to mScarlet, or mScarlet alone (multiplicity of infection or MOI=2.5). Subsequently, lentiviruses carrying vGLUT1:pHluorin (MOI=2.5) were added to DIV-5, and the transduced neurons were cultured to maturity (DIV17-DIV21) before imaging. Near 100% infection efficiency of the mScarlet-tagged constructs was confirmed before using the coverslip for pHluorin experiments. For transfected vGLUT1:pHluorin assays, P0–P1 neurons were electroporated with the respective constructs using an Amaxa 4D-Nucleofector™ System (Lonza Inc., Walkersville, MD, USA) with the P3 Primary Cell 4D-Nucleofector X Kit S (Lonza Cat# V4XP-3032) and program CL-133. The volume of the cell suspension was 20 μl per reaction, and the cell density ranged from 1×107 to 1.5×107 cells/ml. Cells were plated at a 60,000 cells/cm2 density onto poly D-lysine-coated coverslips after electroporation and cultured to maturity (DIV14-DIV17) before imaging. Neurons were imaged live using an inverted motorized epifluorescence microscope (Olympus, IX81) fitted with a Prime 95B camera. The coverslips were mounted into a Chamlide EC magnetic chamber (Live cell Instrument, ON, Canada) in Tyrode solution (pH 7.4) containing (in mM): 119 NaCl, 2.5 KCl, 2 CaCl2, 2 MgCl2, 25 HEPES, 30 glucose, 10 μM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX, TOCRIS bioscience #0190), and 50 μM D,L-2-amino-5-phosphonovaleric acid (AP5, TOCRIS bioscience #0105). NH4Cl perfusions were done with 50 mM NH4Cl in substitution of 50 mM NaCl (pH 7.4). For field stimulation, 10 V/cm pulses were applied at 10 Hz for 60 seconds using a Model 4100 Isolated High-Power Stimulator (A-M systems, Sequim, WA). Incident excitation (Lumencor LED, Spectra X) was attenuated 10-fold, and images were acquired with 500 ms exposures at three-second intervals for three minutes. For analysis, regions of interest (ROIs) were placed on each bouton, and average intensities were obtained for each frame within the time-lapse. Fmax was defined as the maximal fluorescence after NH4Cl perfusion. Baseline F0 was defined as the average fluorescence of the initial 10 frames before stimulation [(F0 = average (F1:F10)]. Fluorescence intensity of a bouton at a given time point (F) was normalized to F0 and Fmax and expressed as (F-F0) / (Fmax-F0). DOI: dx.doi.org/10.17504/protocols.io.q26g7pdkkgwz/v1.
Immunofluorescence and object-based colocalization analysis
For immunostaining, DIV17–21 neurons were fixed in 100% ice-cold methanol solution for 15 min at room temperature, followed by extraction in 0.1 % Triton X-100 for 10 min, blocked in 10% BSA for 1 h at room temperature, and incubated overnight at 4°C with primary antibodies. We found that the methanol fixation was better than paraformaldehyde fixation for pSer129 α-syn staining. After washing with 1x PBS, neurons were blocked again for 30 min at room temperature and incubated with secondary antibodies (Alexa Fluor antibodies from Invitrogen (RRID:AB_2633275, RRID:AB_2762824, RRID:AB_2633282) (1:500) for 1 h at room temperature. Images were acquired at 40X magnification. Z-stack images were obtained as previously described87, and all images were acquired and processed using the MetaMorph Microscopy Automation and Image Analysis Software (RRID:SCR_002368)(https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref). For α-syn Ser129 and VAMP2 colocalization analysis in cultured neurons, 5–6 regions of interest (ROIs) of 200×200 pixels were placed on each image. A total of 26 ROIs were used for this analysis. Object-based colocalization analysis was done using MATLAB (RRID:SCR_001622)(http://www.mathworks.com/products/matlab/). First, automatic puncta detection was done using local maxima. Next, detected puncta are said to have co-localized if the distance between their centers is less than the maximum radius of the two particles. DOI: dx.doi.org/10.17504/protocols.io.8epv5xdodg1b/v1.
Proximity Ligation Assay (PLA)
The PLA was used to detect α-syn interactions at synapses. PLA was performed as described previously with minor modifications48. The following antibodies were used for the PLA experiments: Syn 204 against h-αSyn (Santa Cruz Biotechnology Cat# sc-32280, RRID:AB_628319)(1:100) and EPR12790 against VAMP-2 (Abcam Cat# ab214590)(1:300). The in-situ PLA was performed on fixed primary neurons with DuoLink PLA technology probes and reagents (Sigma-Aldrich Cat# DUO92002, DUO92004, DUO82049, DUO92008, and DUO92014), following the manufacturer’s protocol. First, the neurons were permeabilized with PBS + 0.4% Triton X-100 for 10 min. After two PBS washes, the cells were incubated with a blocking solution for 2 hours at 37 °C and then incubated with the primary antibodies for 30 min at room temperature. The coverslips were washed twice for 5 min with buffer A, followed by incubation with the PLA probes (secondary antibodies against two different species bound to two oligonucleotides: anti-mouse MINUS (Sigma-Aldrich Cat# DUO92004 (also DUO92004–30RXN, DUO92004–100RXN), RRID:AB_2713942) and anti-rabbit PLUS) (Bethyl Cat# OLK-92002–0100, RRID:AB_10950581)in antibody diluent for 30 min at 37 °C. After two washes of 5 min with buffer A, the ligation step was performed with ligase diluted in ligation stock for 30 min at 37 °C. The coverslips were washed with buffer A twice for 2 min before incubation for 50 min with amplification stock solution at 37 °C. After two washes of 10 min with buffer B. Finally, the coverslips were washed with PBS and mounted with Duolink in situ mounting medium (Sigma-Aldrich Cat# DUO82040–5 ML). A negative control experiment was performed for every antibody, where only one antibody was incubated with the PLA probes. The experiments were performed 2 times. The experiments were performed 2 times. Average signal intensities were measured using the MetaMorph Microscopy Automation and Image Analysis Software (RRID:SCR_002368) (https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref) and plotted using GraphPad Prism software [(RRID:SCR_002798) http://www.graphpad.com/]. DOI: dx.doi.org/10.17504/protocols.io.j8nlko6ydv5r/v1.
Immunohistochemistry
Mice were euthanized using isoflurane (VetOne Cat#502017) inhalation followed by cervical dislocation. At autopsy, mouse brains were removed from the calvarium and rapidly placed in a 4% paraformaldehyde (PFA) solution (Thermo Scientific, Cat# J19943-K2) for drop fixation. For immunohistochemistry, 5 μm sagittal tissue sections were stained with antibodies against total α-syn (Synaptic Systems, 1:15,000, Cat #128211) and Ser129P α-syn (Cell Signaling, 1:3000, Cat #23706). Slides were stained using Ventana Discovery Ultra (Ventana Medical Systems, Tucson, AZ, USA). The sections were deparaffinized by three cycles of heating to 72°C in the presence of Discovery Wash (Ventana, Ref# 07311079001) for 4 min with a rinse between each heating step. Subsequently, antigen retrieval was performed using CC1 buffer (Tris-EDTA; pH 8.6 Ventana, Ref# 06414575001) for 40 min (5 cycles of 8 min each with fresh CC1 buffer for each incubation) at 100°C. Endogenous peroxidase was quenched by incubation with inhibitor CM (Ventana, Cat# 760–159) for 8 min. The sections were incubated with primary antibodies for 32 minutes at 37 °C, mixing the samples every 4 min to ensure an even distribution of the antibody. Primary antibodies were detected using the OmniMap system, with an anti-rabbit secondary antibody coupled to an HRP polymer (Roche Cat# 760–4311, RRID:AB_2811043) (Ventana, Ref# 05266548001). Antibody presence was visualized using the Ventana Discovery ChromoMap DAB kit (Ventana, Ref# 05266645001) as a chromagen followed by hematoxylin II (Ventana, Ref#05277965001) for 4 min followed by a 4 min treatment with bluing reagent (Ventana, Ref# 05266769001) as a counterstain. The slides were rinsed, dehydrated with alcohol and xylene, and mounted on coverslips. To confirm the specificity of the pSer129 antibody, the epitope was dephosphorylated using Lambda Protein Phosphatase (New England Biolab, Cat # P0753). Briefly, 2400 units of the Lambda Protein Phosphatase were supplemented with a 10x NEBuffer and 10x MnCl2 and added to the slides for 2h at 37°C. Antigen retrieval and peroxidase quenching were performed on the slides before incubation with the pSer129 antibody. For quantification of Ser129P staining, after background correction, the following ROIs were placed to sample the frontal lobe and hippocampus – ROI measuring ~ 2.5 mm × 1 mm flanking the frontal pole, and an irregular ROI over the CA3 region (which was strongly positive for Ser129P and easy to identify). Average intensities were considered for quantification, and data were analyzed in GraphPad Prism software [(RRID:SCR_002798) http://www.graphpad.com/]. DOI: dx.doi.org/10.17504/protocols.io.bp2l6xmzdlqe/v1.
Electron microscopy (EM) and quantification
Hippocampal neurons were plated at a density of 60,000 cells/cm2. DIV-3 neurons were infected with lentiviruses carrying α-syn tagged at the C-terminus to mScarlet, or mScarlet alone (multiplicity of infection or MOI=2.5). The transduced neurons were cultured to maturity (DIV17-DIV21) before imaging. The ~100 % infection efficiency of the mScarlet-tagged constructs was confirmed before using the coverslip for EM experiments. EM embedding protocol for monolayer cultured cells was performed on ice. The coverslips were fixed in 2 % glutaraldehyde and in 0.1 M sodium cacodylate (SC buffer) buffer for at least 60min at 4 °C, washed 3 min with 0.1 M SC buffer (5 times), and postfixed in 1% osmium tetroxide (OsO4) diluted in 0.1 M SC buffer for 30 min to 1hr. The coverslips were then washed for 3 min (5 times) with 0.1 M SC buffer, followed by a water rinse, sequential ethanol dehydration (20%, 50%, 70%, 90%, 2×100%) 1min each, and subsequent incubation with dry acetone for 1 min at room temperature (2 times). The samples were then incubated with a Durcupan:Acetone solution (50:50%) for 1hr in rotation, followed by incubation with 100% Durcupan for 1 hr (2 times). The cells were then embedded using foil plates along with wood sticks in the embedding oven (overnight incubation). 60 nm ultrathin sections were cut with a diamond knife and mounted on 300 mesh grids. Grids were post-stained with 2% of uranyl acetate for 5 min and lead for 1 min. JEOL 1400 Electron Microscopy and Gatan digital Camera were used to obtain images. For quantification of inter-vesicular (IV) distances between vesicles, we considered all the vesicles in the synapse encompassing 1μmx1μm. To mark each vesicle, a line was drawn across the center of the vesicle, recording its diameter and its position. These parameters were then fed into our algorithm on MATLAB (RRID:SCR_001622)(http://www.mathworks.com/products/matlab/), which gave the output of IV distances for a particular image. The IV distance for all images per condition was pooled together. This algorithm is available upon request. DOI: dx.doi.org/10.17504/protocols.io.yxmvm3zm6l3p/v1.
CRISPR nuclease (CRISPRn) genome editing
CRISPRn technology was used to attenuate α-syn at synapses. LentiCRISPR v2 plasmid (RRID:Addgene_52961) was used for α-syn genome editing. All sgRNAs were designed using CRISPick (https://portals.broadinstitute.org/gppx/crispick/public) and examined for genome-wide sequence specificity using the National Center for Biotechnology Information’s (NCBI) Basic Local Alignment Search Tool NCBI BLAST (RRID:SCR_004870)(http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sgRNAs (Control 5’ GACGGAGGCTAAGCGTCGCAA3’ and α-syn CRISPRn 5’ GGTTCATGAAAGGACTTTCAA3’) were cloned as described previously88. To validate the sgRNAs constructs, DIV-3 hippocampal primary neurons were infected with lentiviruses per 6hrs (MOI=5). Transduced neurons were cultured to maturity (DIV17-DIV21) and then lysate for western blotting analysis and genomic analysis. DOI: dx.doi.org/10.17504/protocols.io.dm6gp35qpvzp/v1.
CRISPR knock-in (endogenous tagging)
Plasmid construction
CRISPR knock-in technology was used to tag endogenous mouse α-syn at synapses, to evaluate targeting of α-syn in living synapses. For the C-terminus knock-in experiment, the sgRNAs and the homology-independent donor templates were generated following strategies similar to those described previously89–91. Briefly, the coding sequence of SNCA1 – the gene encoding α-syn at exon 6 was targeted by a sgRNA, and the oScarlet tag (a variant of mScarlet that is engineered to reduce aggregation; a gift from Karl Deisseroth, Addgene plasmid #137135) was knocked in using a homology-independent mechanism92. The donor sequence was designed following the SATI knock-in vector93 with slight modifications for the intron knock-in experiment. Briefly, the following sequences were directly conjugated as a donor tag; SNCA1 intron 4 (only the sequence after the gRNA targeting site), SNCA1 exon 5 (with wild type or mutation sequences; S129A or S129D), SNCA1 exon 6 (coding site until stop codon), 3X GGGGS linker, oScarlet (without a start codon and with a stop codon at the end), and SNCA1 3’ UTR. The SNCA1 intron 4 was targeted by a sgRNA (TTCTAAGTGTACCAAACCAC), and the donor tag was homology-independently knocked in. The plasmid PX552 (RRID:Addgene_60958) (a gift from Feng Zhang) was digested with a NotI restriction enzyme (New England Biolabs, Cat# R3189L) and used as a plasmid backbone. Oligonucleotides were ordered from Sigma. The genomic sequence of SNCA1 was partially synthesized by Twist Bioscience, and for the rest, it was PCR amplified from a purified genome of a C57BL/6J mouse. DNA fragments were ligated together using the In-Fusion Snap Assembly Master Mix (Takara, Cat# 638947) or DNA Ligation Kit Mighty Mix (Takara, Cat# 6023). The sgRNA sequence for knock-in is listed in the supplemental materials. The AAV-SpCas9 plasmid PX551 RRID:Addgene_60957 (a gift from Feng Zhang) was modified by removing the HA tag.
Adeno-associated virus (AAV) production, neuronal transduction, and quantification of synapses
Small-scale AAV cell lysates were produced using the AAVpro Purification Kit (All Serotypes) (Takara, Cat# 6666) with slight modifications. Briefly, HEK293T cells (RRID:CVCL_0063) were triple transfected with the AAV targeting plasmid, helper plasmid (Agilent Technologies, Cat # 240071), and serotype PHP.S plasmid pUCmini-iCAP-PHP.eB (RRID:Addgene_103005, a gift from Viviana Gradinaru) with PEI Max (Polysciences, Cat#24765). The medium was changed the next day of transfection, and cells were incubated for 3 days after transfection. HEK cells were then collected and lysed with the AAV Extraction Solution A plus. The extracted solution was centrifuged at 10,000 × g for 10 min to remove debris and mixed with Extraction Solution B. This small-scale AAV solution was stored at −80 until use. For viral transduction, hippocampal neurons were plated at 60,000 cells/cm2 density and infected 4 hours later with 10ul of the AAVs expressing SpCas9 and sgRNAs/donor. The media was replaced 2 days after infection. Before imaging or genomic analysis, the transduced neurons were cultured to maturity (DIV-17-DIV-21). For quantification of the knock-in α-syn:o-Scarlet fluorescence at synapses, images were first background-corrected, small ROIs were manually placed over ~20–30 synapses on each image, and average intensities were calculated – all using dropdown menus in MetaMorph Microscopy Automation and Image Analysis Software (RRID:SCR_002368)(https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref). The resulting datasets were statistically analyzed using GraphPad Prism [(RRID:SCR_002798) http://www.graphpad.com/]. DOI: dx.doi.org/10.17504/protocols.io.e6nvwdmqwlmk/v1.
CRISPR knock-in validation
Cultured hippocampal neurons tagged with oScarlet were prepared with Lucigen QuickExtract DNA extraction solution (Biosearch Technologies, cat# QE09050). Briefly, neurons were lysed by adding 50 ul of QuickExtract solution to each sample. The samples were mixed by pipetting and incubated at 68 °C for 15 min followed by 95 °C for 10 min in a thermocycler before being stored at −20 °C for downstream analysis. Two different PCR reactions were performed to amplify DNA products of ~500 bp corresponding to the 5’ and 3’ integration junctions. PCR#1 used the α-syn forward primer: TGTGCTTTCTCTTCCCTCTCTG and the reverse oScarlet primer CCGTCCTCGAAGTTCATCAC, whereas PCR#2 used the α-syn forward primer: ATAACACTTCGTGCAGCACC and the reverse oScarlet primer ACAGGATGTCCCAGGAGAAG. PCR products were extracted from the agarose gel using the Monarch DNA gel extraction kit (New England Biolabs, cat# T1020S), and samples were submitted for sequencing for analysis at MCLAB. DOI: dx.doi.org/10.17504/protocols.io.n2bvj3qb5lk5/v1.
Preparation and Administration of Systemic 4-AP
4-Aminopyridine (4-AP) was used to augment neuronal activity in mouse brains in vivo. 4-AP was obtained from Sigma-Aldrich (Cat #A78403), and a stock solution was prepared in saline. Mice (8 weeks old) were given intraperitoneal injections of soluble 4-AP at a low dose of 2 mg/Kg or equivalent saline. Animals were monitored every 20 min and presented no sign of convulsive seizure behavior. After 3 hrs, animals were anesthetized with isoflurane (VetOne Cat#502017), followed by cervical dislocation. Brains were rapidly removed and drop-fixed in 4% PFA solution for immunohistochemistry or placed on ice for biochemical assays. DOI: dx.doi.org/10.17504/protocols.io.5qpvo38dxv4o/v1.
Biochemical assays and evaluation
Preparation of Brain and Neuro2A Lysates
Neuro2A cells were used to evaluate protein-protein interactions in a cellular context using co-immunoprecipitation. Whole mouse brains were homogenized with a Dounce tissue grinder in neuronal protein extraction reagent (N-PER) (Thermo Scientific, Cat#87792) containing protease/phosphatase inhibitors (Cell Signaling, Cat #5872). Triton X-100 was added at a final concentration of 1%, and the samples were incubated with rotation for 1 hour at 4 °C. Samples were centrifuged at 10,000 × g for 10 min at 4 °C, and the supernatant was collected. For Neuro2A lysates, cells were washed with 1X PBS 3 times and incubated for 5 min on ice in the presence of N-PER reagent supplemented with protease inhibitors. Samples were centrifuged at 10,000 × g for 10 min at 4 °C to remove cellular debris. Total protein concentration was measured in brain and Neuro2A (Cat#TKG0509, RRID:CVCL_0470) lysates (DC™ Protein Assay Kit II, Biorad), and samples were used in subsequent experiments. DOI: dx.doi.org/10.17504/protocols.io.5jyl8pey7g2w/v1.
Preparation of an Enriched Synaptic Vesicle Fraction
Synaptic vesicle fractions were used to evaluate association of GST-tagged recombinant α-syn with synaptic vesicles. Mouse brain tissue was homogenized in a buffer containing 0.32 M sucrose in PBS, supplemented with protease/phosphatase inhibitors. The homogenate was centrifuged at 1,000 × g for 10 min at 4 °C to remove nuclei and cellular debris (P1). The supernatant (S1) was centrifuged at 15,000 × g for 15 min at 4 °C to yield a crude synaptosomal pellet (P2). This pellet was hypo-osmotically lysed in water containing protease inhibitors for 5 min at 4 °C and passed through both a 22- and 27½-gauge needle (10 times each). This suspension was centrifuged at 23,000 × g for 22 min at 4 °C, and the resulting supernatant LS1(S3) was centrifuged again at 174,900 × g for 2 hours at 4 °C in a STi32 Beckman rotor. The final pellet (LP2 or P4) containing an enriched fraction of synaptic vesicles was used for subsequent experiments. DOI: dx.doi.org/10.17504/protocols.io.3byl4qr62vo5/v1.
Immunoprecipitations and Western blots analysis
Immunoprecipitations were performed using 1–2 mg of total protein. Samples were incubated overnight with the indicated antibody at 4° C, followed by the addition of 50 μl of protein G-agarose beads (Thermo Scientific, Cat#20397). Immunoprecipitated proteins were recovered by centrifugation at 2,500× rpm for 2 min, washed three times with a buffer containing PBS and 0.15% Triton X-100. The resulting pellets were resuspended in 20 μl of 1X NuPAGE LDS sample buffer (Thermo Scientific #NP007) and incubated at 95 °C for 10 min. Samples were separated by NuPAGE 4 to 12% Bis-Tris polyacrylamide gels (Thermo Scientific #NP0335BOX), and transferred to a 0.2 μM PVDF membrane (Thermo Scientific #LC2002), using the Mini Blot Module system (Thermo Scientific). PVDF membranes were first fixed with 0.2% PFA 1x PBS per 30 min at room temperature. Then, membranes were washed three times for 10 min in PBS with 0.1% Tween 20 Detergent (TBST) and blocked for 1 h in TBST buffer containing 5% dry milk, and then incubated with the indicated primary antibody for 1 h in blocking solution, washed three times for 10 min each and incubated with HRP-conjugated secondary antibodies (RRID:AB_2819160), (RRID:AB_2755049). After antibody incubations, membranes were again washed three times with TTBS buffer, and protein bands were visualized using the ChemiDoc Imaging System (BioRad) and quantified with Image Lab software version 6.1 from BioRad (RRID:SCR_014210) http://www.bio-rad.com/en-us/sku/1709690-image-lab-software. DOI: dx.doi.org/10.17504/protocols.io.36wgq3ep5lk5/v1.
GST fusion protein production
Full-length recombinant human WT α-syn, α-syn S129A, and S129D were expressed in Escherichia coli BL21 (DE3) (New England Biolabs, Cat # C2530H) using the bacterial expression vector pGEX-KG myc. Following transformation protein expression was induced with 0.05 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and either incubated at 37 °C for 2 hours or at room temperature for 6 hours, with shaking. The cells grown on Terrific Broth (Thermo Scientific Cat# BP9728–2) were harvested by centrifugation at 4500× g at 4 °C for 20 min, and pellets were stored at −80 °C until use. For protein purification, protein pellets were resuspended in 30 ml Lysis Buffer containing 1X PBS, 0.5 mg/ml lysozyme, 1 mM PMSF, DNase, and EDTA-free protease cocktail inhibitor (Roche Cat# 11836170001) for 15 min on ice, briefly sonicated (3 sets with 33 strikes and 30-second breaks on ice between sets), and removed the insoluble material by centrifugation at 15,000 g at 4 °C for 30 min. The clarified lysate was incubated with 500 ml of glutathione-Sepharose 4B (Sigma Cat# 17-0756-01), preequilibrated with 1X PBS containing 0.1% Tween 20 and 5% glycerol (binding buffer), on a tumbler at 4 °C overnight. The GST-bound proteins were washed four times with 30 ml binding buffer and maintained at 4 °C for pull-down assays, in vitro phosphorylation, and in vitro dephosphorylation experiments. GST-bound proteins were occasionally eluted by TEV cleavage. In brief, 15 ug of the fusion protein was mixed with 5 ul of TEV protease reaction buffer (10X) and 1ul of TEV protease (New England Biolabs, Cat#P8112), followed by overnight incubation at 4 °C. DOI: dx.doi.org/10.17504/protocols.io.4r3l22y14l1y/v1.
In vitro dephosphorylation
100 μg of α-syn GST fusion protein on beads were resuspended in alkaline phosphatase (ALP) buffer supplemented with 250 units of ALP (Thermo Scientific Cat# EF0651). The mixes were incubated for 60 min or 180 min at 37 °C with orbital shaking (Eppendorf ThermoMixer F1.5). Samples were then analyzed by NuPAGE and immunoblotted or alternatively used in pull-down experiments with brain lysates. DOI: dx.doi.org/10.17504/protocols.io.kqdg3xb1pg25/v1.
In vitro phosphorylation
5 μg of α-syn GST fusion protein on beads were resuspended in a buffer containing 20 mM HEPES, 1 mM ATP, 10 mM MgCl2, 2 mM DTT, pH 7.4. The reaction was initiated by adding 1 ug of human recombinant PLK2 (Abcam, Cat#ab102108) and incubating at 30 °C with orbital shaking overnight (Eppendorf ThermoMixer F1.5). Samples were then analyzed by NuPAGE and immunoblotted or alternatively analyzed by LC-MS/MS. DOI: dx.doi.org/10.17504/protocols.io.ewov1q71ogr2/v1.
Pull-down Assays
To pull down proteins from brain lysates and LP2(P4) fractions, 1–2 mg of the sample was incubated with 25–50 μg of glutathione beads containing GST fusion proteins for 12–16 hrs. The mixtures were washed three times with 1X PBS with 0.15% Triton X-100, and then resuspended in 20 μl of 1X NuPAGE LDS sample buffer for NuPAGE and immunoblotted analysis. Alternatively, the samples were resuspended in 50μl of PBS for proteomics studies and stored at −20 °C before quality control and proteomics analysis (see below). DOI: dx.doi.org/10.17504/protocols.io.x54v9pw5pg3e/v1.
Proteomics
On-bead/in-solution digestion and phosphopeptide enrichment
Protein digestion of GST samples was performed following published protocols94. Digestion buffer (50 mM NH4HCO3) was added to the sample, followed by treatment with 1 mM dithiothreitol (DTT, Thermo Scientific Cat#R0861) at room temperature for 30 min. Subsequently, 5 mM iodoacetamide (IAA, Sigma Cat#I1149) was added to the mixture and incubated at room temperature for 30 min in the dark. Proteins were digested with 2 μg of lysyl endopeptidase (Wako, Cat# 125–05061) at room temperature (overnight). Further digestion was performed with 2 μg trypsin (Promega, Cat#VA9000) at room temperature (overnight). In contrast, for phosphopeptide enrichment, proteins were digested overnight at room temperature with 1 μg of Asp-N (Promega, Cat# VA1160) for samples in solution or 5 μg of Asp-N for the samples on-beads. The clean peptides were concentrated using a high-select Fe-NTA phosphopeptide enrichment kit (Thermo Scientific Cat#A32992) All resulting peptides were desalted and dried using an HLB column (Waters, Cat#186002034) and vacuum. DOI: dx.doi.org/10.17504/protocols.io.rm7vzx195gx1/v1.
Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS)
LC-MS/MS was used to determine the repertoire of proteins associating with α-syn in mouse brains, and to determine the phosphorylation status of GST-tagged α-syn proteins. Data acquisition by LC-MS/MS was adapted from a published procedure95. Derived peptides were resuspended in a loading buffer (0.1% trifluoroacetic acid, TFA) and separated on a Water’s Charged Surface Hybrid (CSH) column (150 μm internal diameter × 15 cm; particle size: 1.7 μm). The samples were run on an EVOSEP liquid chromatography system using the 15 samples per day preset gradient (88 min) and were monitored on a Q-Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific). The mass spectrometer cycle was programmed to collect one full MS scan followed by 20 data-dependent MS/MS scans. The MS scans (400–1600 m/z range, 3 × 106 AGC target, 100 ms maximum ion time) were collected at a resolution of 70,000 at m/z 200 in profile mode. The HCD MS/MS spectra (1.6 m/z isolation width, 28% collision energy, 1 × 105 AGC target, 100 ms maximum ion time) were acquired at a resolution of 17,500 at m/z 200. Dynamic exclusion was set to exclude previously sequenced precursor ions for 30 seconds. Precursor ions with +1, and +7, +8, or higher charge states were excluded from sequencing. DOI: dx.doi.org/10.17504/protocols.io.14egn3nrml5d/v1.
Proteome analysis
Mass spectrometry data was analyzed according to a published protocol96. Spectra were searched using Proteome Discoverer (RRID:SCR_014477) (https://www.thermofisher.com/order/catalog/product/IQLAAEGABSFAKJMAUH) software version 2.1 against 2020 mouse UniProtKB/Swiss-Prot (RRID:SCR_021164)(https://www.expasy.org/resources/uniprotkb-swiss-prot) (17,042 target sequences) along with the human α-synuclein protein sequence. Searching parameters included full tryptic or Asp-N restriction, precursor mass tolerance (± 20 ppm), and fragment mass tolerance (± 0.05 Da). Serine, threonine, and tyrosine phosphorylation (+79.9663 Da), methionine oxidation (+15.99492 Da), asparagine and glutamine deamidation (+0.98402 Da), and protein N-terminal acetylation (+42.03670 Da) were variable modifications (up to 3 allowed per peptide); cysteine was assigned a fixed carbamidomethyl modification (+57.021465 Da). Percolator was used to filter the peptide spectrum matches to a false discovery rate of 1%. Gene ontology was analyzed using The Database for Annotation, Visualization and Integrated Discovery DAVID (RRID:SCR_001881)(https://david.ncifcrf.gov/tools.jsp-DAVID97. Biological processes involving synaptic function were selected for grouping analysis. Functional grouping was based on Fisher’s exact test (p<0.05). Protein-protein interaction networks were then identified using the STRING (RRID:SCR_005223) (https://string-db.org/) version 11.598, and the protein class was determined using PANTHER (RRID:SCR_004869)(http://www.pantherdb.org/)99. DOI: dx.doi.org/10.17504/protocols.io.5jyl8pw86g2w/v1.
Multi-electrode array (MEA)
MEAs were used to determine global neuronal activity in primary mouse neurons. One day before plating, 6-well MEA plates (Axion Biosystems, M384-tMEA-6W) were coated with poly-D-lysine (Millipore Sigma P6407–5MG) in borate buffer, then washed six times with sterile water and aspirated prior use. 150,000 cells in a 15 ml drop were plated on a 6-well MEA plate. Cultures were maintained for 17–21 days at 37 °C in 5% CO2. On the day of recording, MEA plates were equilibrated for 10 min on the pre-warmer reader at 37 °C in 5% CO2. 2 mins of firing activity was recorded at a 12.5 kHz sampling rate and filtered with a bandpass filter from 200 Hz to 2.5 kHz. Each MEA well contained 64 electrodes that detected changes in extracellular field potentials reflecting the spiking activity of neurons. Spike detection was performed with the adaptive threshold crossing algorithm with a threshold of 6× standard deviation using the AxIS software version 2.4 (RRID:SCR_016308) https://www.axionbiosystems.com/products/axis-software. All spike trains were exported from AxIS software (DOI:dx.doi.org/10.17504/protocols.io.j8nlkonj6v5r/v1). DOI: dx.doi.org/10.17504/protocols.io.j8nlkonj6v5r/v1.
ColabFold Protein Structure Prediction and Membrane Interaction Modeling
Structure-prediction and modeling was used to evaluate putative effects of α-syn Ser129P on the C-terminus of α-syn. WT and phospho-mutant α-syn structures were predicted using ColabFold (https://colabfold.mmseqs.com/), which uses MMseqs2 (RRID:SCR_022962)(https://github.com/soedinglab/MMseqs2) for homology detection and multiple sequence alignment, and AlphaFold2 for simulated protein folding66. The full-length human α-synuclein isoform 1 sequence (Uniprot, P37840–1) was input into ColabFold with 3 recycles, and the model with the highest overall IDDT100 was chosen as representative. For modeling of the phospho-mimetic mutant of α-syn, Ser129 was replaced with an aspartate residue before input. To model the interaction between α-syn and SV-membranes, an all-atom simulation was set up, which included the previously selected ColabFold protein models and a phospholipid bilayer made up of 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG)71 spread in the x-y plane. The protein models were introduced as close to the membrane as possible, while ensuring no disruption of the bilayer, and then the simulation box was solvated with TIP3P water molecules, and charge neutralized by the required number of K+ and Cl− ions. Simulations were conducted using GROMACS v.2021.3 [https://www.gromacs.org/ (RRID:SCR_014565)] [doi: 10.5281/zenodo.5053201] with a CHARMM36 force field [(RRID:SCR_014892) https://www.charmm.org/archive/charmm/resources/charmm-force-fields/]101 at a temperature of 310.15K. Particle size of the simulations was 618946 and 324399 for wild-type and S129D α-synuclein, respectively. After the simulation setup, energy minimization was performed using the steepest descent minimization algorithm, and the LINCS algorithm [(RRID:SCR_016486) http://www.lincsproject.org/] was used to achieve the constraints for hydrogen bonds102. Equilibration of the system was performed with two steps of NVT with Berendsen temperature coupling used for temperature correction with a time step of 1 femtosecond, followed by four steps of NPT with Berendsen temperature and pressure coupling with a time-step of 1 femtosecond for the first NPT step and 2 femtoseconds for the following three. All equilibration steps were run for 125 picoseconds each. Finally, the molecular dynamics production run was performed, without any constraints, for approximately 200 ns using Nosé-Hoover temperature coupling103 and Parrinello-Rahman pressure coupling104, with a time-step of 2 femtoseconds. In the final model, the helicity of each residue is represented as a fraction of time spent either as a component of an α-helical segment of the protein, a 1, or otherwise, a 0. The depth profile of each protein model is represented as the position of each residue with respect to the phosphate head group and is shown as the mean and standard deviation of the last 20 nanoseconds of the production run. DOI: dx.doi.org/10.17504/protocols.io.rm7vzxod2gx1/v1.
Multiple Sequence Alignment
Multiple sequence alignment was used to determine the conservation of α-syn phosphorylation-sites across species. Protein sequences were retrieved from Uniprot or NCBI via Jalview’s sequence fetcher Jalview (RRID:SCR_006459)(https://www.jalview.org/help/html/features/seqfetch.html)105, and aligned with the Clustal Omega Webserver [(RRID:SCR_001591) http://www.ebi.ac.uk/Tools/msa/clustalo/] with default parameters106. For the α-synuclein alignment shown in Supp. Fig. 3F, human (uniprot OP37840), chimpanzee (uniprot P61145), baboons (uniprot A0A2I3N0Z9), marmoset (uniprot F7GY62), rat (uniprot P37377), mouse (uniprot O55042), rabbit (uniprot G1U0V2), cow (uniprot Q3T0G8), bat (uniprot A0A6J2L5S3), shrew (uniprot J7H0X3), armadillo (NCBI XP_004480597), elephant (uniprot G3T7Z3), tenrec (NCBI XP_004703317.1), chicken (uniprot Q9I9H1), zebra finch (uniprot Q4JHT6), frog (uniprot Q7SZ02), and fish (uniprot A0A090D865) sequences were used. DOI: dx.doi.org/10.17504/protocols.io.eq2lyj14wlx9/v1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Datasets were tested for normality using the Shapiro-Wilk test with α set at 0.05. All data is reported as mean±SEM and a p-value <0.05 was considered significant. Statistical analyses were performed using GraphPad Prism software [(RRID:SCR_002798) http://www.graphpad.com. Group mean differences between two groups were evaluated for all immunofluorescence, immunohistochemistry, western blots, and electron-microscopy data using an unpaired, two-tailed Student’s t-test. Group mean-differences between multiple groups in vGLUT1:pHluorin assays were assessed using one-way ANOVA with Kruskal-Wallis post-hoc test. All experiments were performed multiple times, and details of the data values (n – exact value and what it represents), sample sizes, and statistical measurements are provided in the figure legends. The complete set of tabular data was uploaded to Zenodo: https://zenodo.org/records/8059967.
Supplementary Material
Legend for Supplementary Table 2: List of proteins binding to α-syn S129D>S129A, related to Figure 6.
Legend for Supplementary Table 3: List of proteins binding to α-syn WT>S129A, related to Figure 6.
Legend for Supplementary Video 1: Live imaging of vGlut:pHl in cultured hippocampal neurons (related to Fig. 2B). Movie represents 180 seconds of total time, and each frame was taken at 3-second intervals. Scale bar = 5 mm.
Legend for Supplementary Video 2: Representative movies of simulations, related to Figure 8. Movies spanning 30–120 ns of the molecular simulations, capturing major transitions that occur during α-syn C-terminus folding – unphosphorylated state (left) and constitutively Ser-129 phosphorylated state (S129D, right). Note that initially, both scenarios start with an open structure. As the simulations progress, phosphorylated α-syn undergoes conformational changes due to the crowding of five positively-charged lysine residues (60K, 80K, 96K, 97K, and 102K) around the negatively-charged phosphorylated S129 residue. In contrast, the unphosphorylated molecule largely remains in an extended conformation over time. We speculate that the stability conferred by the phosphorylation-induced conformation changes in the C-terminus (96–140 residues) facilitates the interaction of VAMP2 and synapsin, which bind to this domain (see Supp. Fig. 5 and results).
Highlights:
Native α-syn Serine-129 phosphorylation is seen in discrete brain regions
Preventing α-syn Serine-129 phosphorylation blocks activity-dependent α-syn functions
Neuronal activity augments α-syn Serine-129 phosphorylation in cells and in vivo
α-syn Serine129 phosphorylation triggers protein-protein interactions at synapses
Acknowledgements:
We thank Dr. Patrick Kearney for the analysis and interpretation of immunostains. This work was funded by grants to Subhojit Roy from the NINDS (R01NS111978); the Farmer Family Foundation; and in part by Aligning Science Across Parkinson’s [ASAP-020495] through the Michael J. Fox Foundation for Parkinson’s Research (MJFF); as well as a NINDS P30NS047101 grant to the UCSD microscopy core. For the purpose of open access, the author has applied a CC BY public copyright license to all Author Accepted Manuscripts arising from this submission. Leonardo Parra-Rivas was supported by a postdoctoral fellowship from the American Parkinson’s Disease Association (APDA) and the Parkinson’s Foundation Launch award PF-LAUNCH-1046253. Proteomic studies were supported in part by the Emory Integrated Proteomics Core (EIPC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the Georgia Clinical & Translational Science Alliance of the National Institutes of Health under Award Number UL1TR002378. The authors thank the University of California, San Diego - Cellular and Molecular Medicine Electron Microscopy Core (UCSD-CMM-EM Core, RRID: SCR_022039) for equipment access and technical assistance (supported by the NIH Award number S10OD023527).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors have no competing interests.
REFERENCES
- 1.Anderson JP, Walker DE, Goldstein JM, de Laat R, Banducci K, Caccavello RJ, Barbour R, Huang J, Kling K, Lee M, et al. (2006). Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J Biol Chem 281, 29739–29752. M600933200 [pii] 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- 2.Fujiwara H, Hasegawa M, Dohmae N, Kawashima A, Masliah E, Goldberg MS, Shen J, Takio K, and Iwatsubo T (2002). alpha-Synuclein is phosphorylated in synucleinopathy lesions. Nat.Cell Biol 4, 160–164. [DOI] [PubMed] [Google Scholar]
- 3.Okochi M, Walter J, Koyama A, Nakajo S, Baba M, Iwatsubo T, Meijer L, Kahle PJ, and Haass C (2000). Constitutive phosphorylation of the Parkinson’s disease associated alpha-synuclein. J Biol Chem 275, 390–397. 10.1074/jbc.275.1.390. [DOI] [PubMed] [Google Scholar]
- 4.Beach TG, White CL, Hamilton RL, Duda JE, Iwatsubo T, Dickson DW, Leverenz JB, Roncaroli F, Buttini M, Hladik CL, et al. (2008). Evaluation of alpha-synuclein immunohistochemical methods used by invited experts. Acta Neuropathol 116, 277–288. 10.1007/s00401-008-0409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inglis KJ, Chereau D, Brigham EF, Chiou SS, Schobel S, Frigon NL, Yu M, Caccavello RJ, Nelson S, Motter R, et al. (2009). Polo-like kinase 2 (PLK2) phosphorylates alpha-synuclein at serine 129 in central nervous system. J Biol Chem 284, 2598–2602. 10.1074/jbc.C800206200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vancraenenbroeck R, Lobbestael E, Maeyer MD, Baekelandt V, and Taymans JM (2011). Kinases as targets for Parkinson’s disease: from genetics to therapy. CNS Neurol Disord Drug Targets 10, 724–740. 10.2174/187152711797247858. [DOI] [PubMed] [Google Scholar]
- 7.Oueslati A, Schneider BL, Aebischer P, and Lashuel HA (2013). Polo-like kinase 2 regulates selective autophagic alpha-synuclein clearance and suppresses its toxicity in vivo. Proc Natl Acad Sci U S A 110, E3945–3954. 10.1073/pnas.1309991110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azeredo da Silveira S, Schneider BL, Cifuentes-Diaz C, Sage D, Abbas-Terki T, Iwatsubo T, Unser M, and Aebischer P (2009). Phosphorylation does not prompt, nor prevent, the formation of alpha-synuclein toxic species in a rat model of Parkinson’s disease. Hum Mol Genet 18, 872–887. ddn417 [pii] 10.1093/hmg/ddn417. [DOI] [PubMed] [Google Scholar]
- 9.McFarland NR, Fan Z, Xu K, Schwarzschild MA, Feany MB, Hyman BT, and McLean PJ (2009). Alpha-synuclein S129 phosphorylation mutants do not alter nigrostriatal toxicity in a rat model of Parkinson disease. J Neuropathol Exp Neurol 68, 515–524. 10.1097/NEN.0b013e3181a24b5300005072-200905000-00007 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escobar VD, Kuo YM, Orrison BM, Giasson BI, and Nussbaum RL (2014). Transgenic mice expressing S129 phosphorylation mutations in alpha-synuclein. Neurosci Lett 563, 96–100. 10.1016/j.neulet.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck K, Landeck N, Ulusoy A, Majbour NK, El-Agnaf OM, and Kirik D (2015). Ser129 phosphorylation of endogenous alpha-synuclein induced by overexpression of polo-like kinases 2 and 3 in nigral dopamine neurons is not detrimental to their survival and function. Neurobiol Dis 78, 100–114. 10.1016/j.nbd.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Nestler EJ, Greengard P. (1999) Protein Phosphorylation is of Fundamental Importance in Biological Regulation. Lippincott-Raven. In: Siegel GJ, Agranoff BW, Albers RW, et al. , editors. Basic Neurochemistry: Molecular, Cellular and Medical Aspects. 6th edition. [Google Scholar]
- 13.Ramalingam N, Jin SX, Moors TE, Fonseca-Ornelas L, Shimanaka K, Lei S, Cam HP, Watson AH, Brontesi L, Ding L, et al. (2023). Dynamic physiological alpha-synuclein S129 phosphorylation is driven by neuronal activity. Npj Parkinsons Dis 9, 4. 10.1038/s41531-023-00444-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maroteaux L, Campanelli JT, and Scheller RH (1988). Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J.Neurosci 8, 2804–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinho R, Paiva I, Jercic KG, Fonseca-Ornelas L, Gerhardt E, Fahlbusch C, Garcia-Esparcia P, Kerimoglu C, Pavlou MAS, Villar-Pique A, et al. (2019). Nuclear localization and phosphorylation modulate pathological effects of alpha-synuclein. Hum Mol Genet 28, 31–50. 10.1093/hmg/ddy326. [DOI] [PubMed] [Google Scholar]
- 16.Killinger BA, Mercado G, Choi S, Tittle T, Chu Y, Brundin P, and Kordower JH (2023). Distribution of phosphorylated alpha-synuclein in non-diseased brain implicates olfactory bulb mitral cells in synucleinopathy pathogenesis. Npj Parkinsons Dis 9, 43. 10.1038/s41531-023-00491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koss DJ, Erskine D, Porter A, Palmoski P, Menon H, Todd OGJ, Leite M, Attems J, and Outeiro TF (2022). Nuclear alpha-synuclein is present in the human brain and is modified in dementia with Lewy bodies. Acta Neuropathol Commun 10, 98. 10.1186/s40478-022-01403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Zhang Y, Hu W, Xie S, Gong CX, Iqbal K, and Liu F (2015). Rapid alteration of protein phosphorylation during postmortem: implication in the study of protein phosphorylation. Sci Rep 5, 15709. 10.1038/srep15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott DA, Tabarean I, Tang Y, Cartier A, Masliah E, and Roy S (2010). A pathologic cascade leading to synaptic dysfunction in alpha-synuclein-induced neurodegeneration. J Neurosci 30, 8083–8095. 30/24/8083 [pii] 10.1523/JNEUROSCI.1091-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen KE, Schmitz Y, Troyer MD, Mosharov E, Dietrich P, Quazi AZ, Savalle M, Nemani V, Chaudhry FA, Edwards RH, et al. (2006). Alpha-synuclein overexpression in PC12 and chromaffin cells impairs catecholamine release by interfering with a late step in exocytosis. J Neurosci 26, 11915–11922. 26/46/11915 [pii] 10.1523/JNEUROSCI.3821-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemani VM, Lu W, Berge V, Nakamura K, Onoa B, Lee MK, Chaudhry FA, Nicoll RA, and Edwards RH (2010). Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron 65, 66–79. S0896–6273(09)01041–1 [pii] 10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burre J, Sharma M, and Sudhof TC (2018). Cell Biology and Pathophysiology of alpha-Synuclein. Cold Spring Harb Perspect Med 8. 10.1101/cshperspect.a024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gitler D, Xu Y, Kao HT, Lin D, Lim S, Feng J, Greengard P, and Augustine GJ (2004). Molecular determinants of synapsin targeting to presynaptic terminals. J.Neurosci 24, 3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weston LJ, Stackhouse TL, Spinelli KJ, Boutros SW, Rose EP, Osterberg VR, Luk KC, Raber J, Weissman TA, and Unni VK (2021). Genetic deletion of Polo-like kinase 2 reduces alpha-synuclein serine-129 phosphorylation in presynaptic terminals but not Lewy bodies. J Biol Chem 296, 100273. 10.1016/j.jbc.2021.100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghanem SS, Majbour NK, Vaikath NN, Ardah MT, Erskine D, Jensen NM, Fayyad M, Sudhakaran IP, Vasili E, Melachroinou K, et al. (2022). alpha-Synuclein phosphorylation at serine 129 occurs after initial protein deposition and inhibits seeded fibril formation and toxicity. Proc Natl Acad Sci U S A 119, e2109617119. 10.1073/pnas.2109617119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, and Feany MB (2005). Alpha-synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat.Neurosci 8, 657–663. [DOI] [PubMed] [Google Scholar]
- 27.Febbraro F, Sahin G, Farran A, Soares S, Jensen PH, Kirik D, and Romero-Ramos M (2013). Ser129D mutant alpha-synuclein induces earlier motor dysfunction while S129A results in distinctive pathology in a rat model of Parkinson’s disease. Neurobiol Dis 56, 47–58. 10.1016/j.nbd.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Perrin RJ, Woods WS, Clayton DF, and George JM (2000). Interaction of human alpha-Synuclein and Parkinson’s disease variants with phospholipids. Structural analysis using site-directed mutagenesis. J.Biol.Chem 275, 34393–34398. [DOI] [PubMed] [Google Scholar]
- 29.Fortin DL, Troyer MD, Nakamura K, Kubo S, Anthony MD, and Edwards RH (2004). Lipid rafts mediate the synaptic localization of alpha-synuclein. J.Neurosci 24, 6715–6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burre J, Sharma M, and Sudhof TC Systematic mutagenesis of alpha-synuclein reveals distinct sequence requirements for physiological and pathological activities. (2012). J Neurosci 32, 15227–15242. 32/43/15227 [pii] 10.1523/JNEUROSCI.3545-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Das U, Scott DA, Tang Y, McLean PJ, and Roy S (2014). alpha-synuclein multimers cluster synaptic vesicles and attenuate recycling. Curr Biol 24, 2319–2326. 10.1016/j.cub.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fenno LE, Ramakrishnan C, Kim YS, Evans KE, Lo M, Vesuna S, Inoue M, Cheung KYM, Yuen E, Pichamoorthy N, et al. (2020). Comprehensive Dual- and Triple-Feature Intersectional Single-Vector Delivery of Diverse Functional Payloads to Cells of Behaving Mammals. Neuron 107, 836–853 e811. 10.1016/j.neuron.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burrone J, Li Z, and Murthy VN (2006). Studying vesicle cycling in presynaptic terminals using the genetically encoded probe synaptopHluorin. Nat Protoc 1, 2970–2978. nprot.2006.449 [pii] 10.1038/nprot.2006.449. [DOI] [PubMed] [Google Scholar]
- 34.Royle SJ, Granseth B, Odermatt B, Derevier A, and Lagnado L (2008). Imaging phluorin-based probes at hippocampal synapses. Methods Mol Biol 457, 293–303. 10.1007/978-1-59745-261-8_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balaji J, and Ryan TA (2007). Single-vesicle imaging reveals that synaptic vesicle exocytosis and endocytosis are coupled by a single stochastic mode. Proc Natl Acad Sci U S A 104, 20576–20581. 0707574105 [pii] 10.1073/pnas.0707574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun J, Wang L, Bao H, Premi S, Das U, Chapman ER, and Roy S (2019). Functional cooperation of alpha-synuclein and VAMP2 in synaptic vesicle recycling. Proc Natl Acad Sci U S A 116, 11113–11115. 10.1073/pnas.1903049116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atias M, Tevet Y, Sun J, Stavsky A, Tal S, Kahn J, Roy S, and Gitler D (2019). Synapsins regulate alpha-synuclein functions. Proc Natl Acad Sci U S A 116, 11116–11118. 10.1073/pnas.1903054116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohansal-Nodehi M, Chua JJ, Urlaub H, Jahn R, and Czernik D (2016). Analysis of protein phosphorylation in nerve terminal reveals extensive changes in active zone proteins upon exocytosis. Elife 5. 10.7554/eLife.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silbern I, Pan KT, Fiosins M, Bonn S, Rizzoli SO, Fornasiero EF, Urlaub H, and Jahn R (2021). Protein Phosphorylation in Depolarized Synaptosomes: Dissecting Primary Effects of Calcium from Synaptic Vesicle Cycling. Mol Cell Proteomics 20, 100061. 10.1016/j.mcpro.2021.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thesleff S (1980). Aminopyridines and synaptic transmission. Neuroscience 5, 1413–1419. 10.1016/0306-4522(80)90002-0. [DOI] [PubMed] [Google Scholar]
- 41.Mbefo MK, Paleologou KE, Boucharaba A, Oueslati A, Schell H, Fournier M, Olschewski D, Yin G, Zweckstetter M, Masliah E, et al. (2010). Phosphorylation of synucleins by members of the Polo-like kinase family. J Biol Chem 285, 2807–2822. 10.1074/jbc.M109.081950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fedor FZ, Paraczky C, Ravasz L, Toth K, Borhegyi Z, Somogyvari Z, Juhasz G, and Fekete Z (2020). Electrophysiological and behavioral properties of 4-aminopyridine-induced epileptic activity in mice. Biol Futur 71, 427–434. 10.1007/s42977-020-00047-z. [DOI] [PubMed] [Google Scholar]
- 43.Ferguson SM, and De Camilli P (2012). Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol 13, 75–88. 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagy G, Matti U, Nehring RB, Binz T, Rettig J, Neher E, and Sorensen JB (2002). Protein kinase C-dependent phosphorylation of synaptosome-associated protein of 25 kDa at Ser187 potentiates vesicle recruitment. J Neurosci 22, 9278–9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian JH, Das S, and Sheng ZH (2003). Ca2+-dependent phosphorylation of syntaxin-1A by the death-associated protein (DAP) kinase regulates its interaction with Munc18. J Biol Chem 278, 26265–26274. 10.1074/jbc.M300492200. [DOI] [PubMed] [Google Scholar]
- 46.Rickman C, and Duncan RR (2010). Munc18/Syntaxin interaction kinetics control secretory vesicle dynamics. J Biol Chem 285, 3965–3972. 10.1074/jbc.M109.040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burre J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, and Sudhof TC (2010). Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 329, 1663–1667. science.1195227 [pii] 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Almandoz-Gil L, Persson E, Lindstrom V, Ingelsson M, Erlandsson A, and Bergstrom J (2018). In Situ Proximity Ligation Assay Reveals Co-Localization of Alpha-Synuclein and SNARE Proteins in Murine Primary Neurons. Front Neurol 9, 180. 10.3389/fneur.2018.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung CY, Khurana V, Yi S, Sahni N, Loh KH, Auluck PK, Baru V, Udeshi ND, Freyzon Y, Carr SA, et al. (2017). In Situ Peroxidase Labeling and Mass-Spectrometry Connects Alpha-Synuclein Directly to Endocytic Trafficking and mRNA Metabolism in Neurons. Cell Syst 4, 242–250 e244. 10.1016/j.cels.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffin TA, Schnier PD, Cleveland EM, Newberry RW, Becker J, and Carlson GA (2023). Fibril treatment changes protein interactions of tau and alpha-synuclein in human neurons. J Biol Chem 299, 102888. 10.1016/j.jbc.2023.102888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Betzer C, Movius AJ, Shi M, Gai WP, Zhang J, and Jensen PH (2015). Identification of synaptosomal proteins binding to monomeric and oligomeric alpha-synuclein. PLoS One 10, e0116473. 10.1371/journal.pone.0116473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McFarland MA, Ellis CE, Markey SP, and Nussbaum RL (2008). Proteomics analysis identifies phosphorylation-dependent alpha-synuclein protein interactions. Mol Cell Proteomics 7, 2123–2137. M800116–MCP200 [pii] 10.1074/mcp.M800116-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maor G, Dubreuil RR, and Feany MB (2023). alpha-synuclein promotes neuronal dysfunction and death by disrupting the binding of ankyrin to ss-spectrin. J Neurosci. 10.1523/JNEUROSCI.1922-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lautenschlager J, Stephens AD, Fusco G, Strohl F, Curry N, Zacharopoulou M, Michel CH, Laine R, Nespovitaya N, Fantham M, et al. (2018). C-terminal calcium binding of alpha-synuclein modulates synaptic vesicle interaction. Nat Commun 9, 712. 10.1038/s41467-018-03111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diao J, Burre J, Vivona S, Cipriano DJ, Sharma M, Kyoung M, Sudhof TC, and Brunger AT (2013). Native alpha-synuclein induces clustering of synaptic-vesicle mimics via binding to phospholipids and synaptobrevin-2/VAMP2. Elife 2, e00592. 10.7554/eLife.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fusco G, Pape T, Stephens AD, Mahou P, Costa AR, Kaminski CF, Kaminski Schierle GS, Vendruscolo M, Veglia G, Dobson CM, and De Simone A (2016). Structural basis of synaptic vesicle assembly promoted by alpha-synuclein. Nat Commun 7, 12563. 10.1038/ncomms12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janezic S, Threlfell S, Dodson PD, Dowie MJ, Taylor TN, Potgieter D, Parkkinen L, Senior SL, Anwar S, Ryan B, et al. (2013). Deficits in dopaminergic transmission precede neuron loss and dysfunction in a new Parkinson model. Proc Natl Acad Sci U S A 110, E4016–4025. 10.1073/pnas.1309143110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ciruelas K, Marcotulli D, and Bajjalieh SM (2019). Synaptic vesicle protein 2: A multi-faceted regulator of secretion. Semin Cell Dev Biol 95, 130–141. 10.1016/j.semcdb.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kahle PJ, Neumann M, Ozmen L, Muller V, Jacobsen H, Schindzielorz A, Okochi M, Leimer U, van der PH, Probst A, et al. (2000). Subcellular localization of wild-type and Parkinson’s disease-associated mutant alpha-synuclein in human and transgenic mouse brain. J.Neurosci 20, 6365–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, et al. (2006). Molecular anatomy of a trafficking organelle. Cell 127, 831–846. S0092–8674(06)01400–0 [pii] 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 61.Wilhelm BG, Mandad S, Truckenbrodt S, Krohnert K, Schafer C, Rammner B, Koo SJ, Classen GA, Krauss M, Haucke V, et al. (2014). Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028. 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- 62.Deak F, Schoch S, Liu X, Sudhof TC, and Kavalali ET (2004). Synaptobrevin is essential for fast synaptic-vesicle endocytosis. Nat Cell Biol 6, 1102–1108. ncb1185 [pii] 10.1038/ncb1185. [DOI] [PubMed] [Google Scholar]
- 63.Crawford DC, and Kavalali ET (2015). Molecular underpinnings of synaptic vesicle pool heterogeneity. Traffic 16, 338–364. 10.1111/tra.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guzikowski NJ, and Kavalali ET (2021). Nano-Organization at the Synapse: Segregation of Distinct Forms of Neurotransmission. Front Synaptic Neurosci 13, 796498. 10.3389/fnsyn.2021.796498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson LN, and Lewis RJ (2001). Structural basis for control by phosphorylation. Chem Rev 101, 2209–2242. 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 66.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, and Steinegger M (2022). ColabFold: making protein folding accessible to all. Nat Methods 19, 679–682. 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okochi M, Walter J, Koyama A, Nakajo S, Baba M, Iwatsubo T, Meijer L, Kahle PJ, and Haass C (2000). Constitutive phosphorylation of the Parkinson’s disease associated alpha-synuclein. J.Biol.Chem 275, 390–397. [DOI] [PubMed] [Google Scholar]
- 69.Bortolus M, Tombolato F, Tessari I, Bisaglia M, Mammi S, Bubacco L, Ferrarini A, and Maniero AL (2008). Broken helix in vesicle and micelle-bound alpha-synuclein: insights from site-directed spin labeling-EPR experiments and MD simulations. J Am Chem Soc 130, 6690–6691. 10.1021/ja8010429. [DOI] [PubMed] [Google Scholar]
- 70.Drescher M, Veldhuis G, van Rooijen BD, Milikisyants S, Subramaniam V, and Huber M (2008). Antiparallel arrangement of the helices of vesicle-bound alpha-synuclein. J Am Chem Soc 130, 7796–7797. 10.1021/ja801594s. [DOI] [PubMed] [Google Scholar]
- 71.Jao CC, Hegde BG, Chen J, Haworth IS, and Langen R (2008). Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc Natl Acad Sci U S A 105, 19666–19671. 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fusco G, De Simone A, Arosio P, Vendruscolo M, Veglia G, and Dobson CM (2016). Structural Ensembles of Membrane-bound alpha-Synuclein Reveal the Molecular Determinants of Synaptic Vesicle Affinity. Sci Rep 6, 27125. 10.1038/srep27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, Caldwell KA, Caldwell GA, Rochet JC, McCaffery JM, et al. (2008). The Parkinson’s disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc.Natl.Acad.Sci.U.S.A 105, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soper JH, Roy S, Stieber A, Lee E, Wilson RB, Trojanowski JQ, Burd CG, and Lee VM (2008). {alpha}-Synuclein-induced Aggregation of Cytoplasmic Vesicles in Saccharomyces cerevisiae. Mol Biol Cell 19, 1093–1103. E07-08-0827 [pii] 10.1091/mbc.E07-08-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tenreiro S, Reimao-Pinto MM, Antas P, Rino J, Wawrzycka D, Macedo D, Rosado-Ramos R, Amen T, Waiss M, Magalhaes F, et al. (2014). Phosphorylation modulates clearance of alpha-synuclein inclusions in a yeast model of Parkinson’s disease. PLoS Genet 10, e1004302. 10.1371/journal.pgen.1004302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abeliovich A, Schmitz Y, Farinas I, Choi-Lundberg D, Ho WH, Castillo PE, Shinsky N, Verdugo JM, Armanini M, Ryan A, et al. (2000). Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron 25, 239–252. [DOI] [PubMed] [Google Scholar]
- 77.Yavich L, Tanila H, Vepsalainen S, and Jakala P (2004). Role of alpha-synuclein in presynaptic dopamine recruitment. J.Neurosci 24, 11165–11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yavich L, Jakala P, and Tanila H (2006). Abnormal compartmentalization of norepinephrine in mouse dentate gyrus in alpha-synuclein knockout and A30P transgenic mice. J Neurochem 99, 724–732. JNC4098 [pii] 10.1111/j.1471-4159.2006.04098.x. [DOI] [PubMed] [Google Scholar]
- 79.Anwar S, Peters O, Millership S, Ninkina N, Doig N, Connor-Robson N, Threlfell S, Kooner G, Deacon RM, Bannerman DM, et al. (2011). Functional alterations to the nigrostriatal system in mice lacking all three members of the synuclein family. J Neurosci 31, 7264–7274. 31/20/7264 [pii] 10.1523/JNEUROSCI.6194-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Senior SL, Ninkina N, Deacon R, Bannerman D, Buchman VL, Cragg SJ, and Wade-Martins R (2008). Increased striatal dopamine release and hyperdopaminergic-like behaviour in mice lacking both alpha-synuclein and gamma-synuclein. Eur J Neurosci 27, 947–957. EJN6055 [pii] 10.1111/j.1460-9568.2008.06055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Greten-Harrison B, Polydoro M, Morimoto-Tomita M, Diao L, Williams AM, Nie EH, Makani S, Tian N, Castillo PE, Buchman VL, and Chandra SS (2010). alphabetagamma-Synuclein triple knockout mice reveal age-dependent neuronal dysfunction. Proc Natl Acad Sci U S A 107, 19573–19578. 1005005107 [pii] 10.1073/pnas.1005005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nussinov R, Tsai CJ, Xin F, and Radivojac P (2012). Allosteric post-translational modification codes. Trends Biochem Sci 37, 447–455. 10.1016/j.tibs.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 83.Laskowski RA, Gerick F, and Thornton JM (2009). The structural basis of allosteric regulation in proteins. FEBS Lett 583, 1692–1698. 10.1016/j.febslet.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 84.Nishi H, Shaytan A, and Panchenko AR (2014). Physicochemical mechanisms of protein regulation by phosphorylation. Front Genet 5, 270. 10.3389/fgene.2014.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fouke KE, Wegman ME, Weber SA, Brady EB, Roman-Vendrell C, and Morgan JR (2021). Synuclein Regulates Synaptic Vesicle Clustering and Docking at a Vertebrate Synapse. Front Cell Dev Biol 9, 774650. 10.3389/fcell.2021.774650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roy S, Yang G, Tang Y, and Scott DA (2011). A simple photoactivation and image analysis module for visualizing and analyzing axonal transport with high temporal resolution. Nat Protoc 7, 62–68. nprot.2011.428 [pii] 10.1038/nprot.2011.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scott D, and Roy S (2012). alpha-Synuclein inhibits intersynaptic vesicle mobility and maintains recycling-pool homeostasis. J Neurosci 32, 10129–10135. 32/30/10129 [pii] 10.1523/JNEUROSCI.0535-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, and Zhang F (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ogawa Y, and Rasband MN (2021). Endogenously expressed Ranbp2 is not at the axon initial segment. J Cell Sci 134. 10.1242/jcs.256180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao YD, Hisey E, Bradshaw TWA, Erata E, Brown WE, Courtland JL, Uezu A, Xiang Y, Diao YR, and Soderling SH (2019). Plug-and-Play Protein Modification Using Homology-Independent Universal Genome Engineering. Neuron 103, 583–+. 10.1016/j.neuron.2019.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bingham D, Jakobs CE, Wernert F, Boroni-Rueda F, Jullien N, Schentarra EM, Friedl K, Da Costa Moura J, van Bommel DM, Caillol G, et al. (2023). Presynapses contain distinct actin nanostructures. J Cell Biol 222. 10.1083/jcb.202208110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, et al. (2016). In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149. 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suzuki K, Yamamoto M, Hernandez-Benitez R, Li Z, Wei C, Soligalla RD, Aizawa E, Hatanaka F, Kurita M, Reddy P, et al. (2019). Precise in vivo genome editing via single homology arm donor mediated intron-targeting gene integration for genetic disease correction. Cell Res 29, 804–819. 10.1038/s41422-019-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soucek S, Zeng Y, Bellur DL, Bergkessel M, Morris KJ, Deng Q, Duong D, Seyfried NT, Guthrie C, Staley JP, et al. (2016). The Evolutionarily-conserved Polyadenosine RNA Binding Protein, Nab2, Cooperates with Splicing Machinery to Regulate the Fate of pre-mRNA. Mol Cell Biol 36, 2697–2714. 10.1128/MCB.00402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seyfried NT, Dammer EB, Swarup V, Nandakumar D, Duong DM, Yin L, Deng Q, Nguyen T, Hales CM, Wingo T, et al. (2017). A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer’s Disease. Cell Syst 4, 60–72 e64. 10.1016/j.cels.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wingo TS, Duong DM, Zhou M, Dammer EB, Wu H, Cutler DJ, Lah JJ, Levey AI, and Seyfried NT (2017). Integrating Next-Generation Genomic Sequencing and Mass Spectrometry To Estimate Allele-Specific Protein Abundance in Human Brain. J Proteome Res 16, 3336–3347. 10.1021/acs.jproteome.7b00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang da W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 98.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47, D607–D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mi H, Poudel S, Muruganujan A, Casagrande JT, and Thomas PD (2016). PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res 44, D336–342. 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mariani V, Biasini M, Barbato A, and Schwede T (2013). lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 29, 2722–2728. 10.1093/bioinformatics/btt473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang J, and MacKerell AD Jr. (2013). CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem 34, 2135–2145. 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hess B, Bekker H, Berendsen HJC, and Fraaije JGEM (1997). LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem 18, 1463–1472. . [DOI] [Google Scholar]
- 103.Evans DJ, and Holian BL (1985). The Nose–Hoover thermostat. Journal of Chemical Physics 83, 4069–4074. [Google Scholar]
- 104.Parrinello M, and Rahman A (1981). Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics 52, 7182–7190. 10.1063/1.328693. [DOI] [Google Scholar]
- 105.Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Higgins DG, and Sharp PM (1988). CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73, 237–244. 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legend for Supplementary Table 2: List of proteins binding to α-syn S129D>S129A, related to Figure 6.
Legend for Supplementary Table 3: List of proteins binding to α-syn WT>S129A, related to Figure 6.
Legend for Supplementary Video 1: Live imaging of vGlut:pHl in cultured hippocampal neurons (related to Fig. 2B). Movie represents 180 seconds of total time, and each frame was taken at 3-second intervals. Scale bar = 5 mm.
Legend for Supplementary Video 2: Representative movies of simulations, related to Figure 8. Movies spanning 30–120 ns of the molecular simulations, capturing major transitions that occur during α-syn C-terminus folding – unphosphorylated state (left) and constitutively Ser-129 phosphorylated state (S129D, right). Note that initially, both scenarios start with an open structure. As the simulations progress, phosphorylated α-syn undergoes conformational changes due to the crowding of five positively-charged lysine residues (60K, 80K, 96K, 97K, and 102K) around the negatively-charged phosphorylated S129 residue. In contrast, the unphosphorylated molecule largely remains in an extended conformation over time. We speculate that the stability conferred by the phosphorylation-induced conformation changes in the C-terminus (96–140 residues) facilitates the interaction of VAMP2 and synapsin, which bind to this domain (see Supp. Fig. 5 and results).
Data Availability Statement
All data to reproduce figure panels has been deposited and archived by Zenodo, DOI is listed in the key resource table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Ser129P α-syn | Abcam | Cat#ab51253,RRID:AB_869973 |
| NeuN | Abcam | Cat#ab236870,RRID:AB_2927651 |
| VAMP2 | Synaptic Systems | Cat#104211BT,RRID:AB_2619758 |
| α-syn | BD Biosciences | Cat# 610787,RRID:AB_398108 |
| β-syn | Abcam | Cat#ab76111,RRID:AB_1309981 |
| Tubulin | Abcam | Cat#ab7291,RRID:AB_2241126 |
| PLK2 | Thermo Fisher Scientific | Cat# PA5–14094,RRID:AB_2167742 |
| Synaptophysin-1 | Synaptic Systems | Cat#101011C2,RRID:AB_10890165 |
| SV2A | Abcam | Cat#ab32942,RRID:AB_778192 |
| Synapsin-1 | Abcam | Cat#ab254349,RRID:AB_2920663 |
| Synapsin-1 | Synaptic Systems | Cat#106104,RRID:AB_2721082 |
| c-myc | Sigma-Aldrich | Cat#M4439,RRID:AB_439694 |
| GFP | Abcam | Cat#ab290,RRID:AB_303395 |
| Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A32723,RRID:AB_2633275 |
| Alexa Fluor 594 | Thermo Fisher Scientific | Cat#A32740,RRID:AB_2762824 |
| Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A32733,RRID:AB_2633282 |
| α-syn(Syn 204) | Santa Cruz Biotechnology | Cat#sc-32280 |
| VAMP-2 | Abcam | Cat#ab214590 |
| DuoLink PLA technology probe | Sigma-Aldrich | Cat#DUO92002 |
| DuoLink PLA technology probe | Sigma-Aldrich | Cat#DUO92004 |
| DuoLink PLA technology probe | Sigma-Aldrich | Cat#DUO82049 |
| DuoLink PLA technology probe | Sigma-Aldrich | Cat#DUO92008 |
| DuoLink PLA technology probe | Sigma-Aldrich | Cat#DUO92014 |
| α-syn | Synaptic Systems | Cat#128211 |
| Ser129P α-syn | Cell Signaling | Cat #23706 |
| Goat anti-Rabbit HRP | Abcam | Cat#ab205718,RRID:AB_2819160 |
| Goat anti-mouse HRP | Abcam | Cat#ab205719,RRID:AB_2755049 |
| Bacterial and Virus Strains | ||
| Escherichia coli BL21(DE3) | New England Biolabs | Cat#C2530H |
| One Shot™ Stbl3™ Chemically Competent E. coli | Invitrogen | Cat#C737303 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) | TOCRIS bioscience | Cat#0190 |
| DuoLink mounting media | Sigma-Aldrich | Cat#DUO82040–5 |
| Recombinant human α-syn phospho-Ser129 protein | Proteos | Cat#RP-004 |
| Recombinant human α-syn | RnD systems | Cat#SP-485–500 |
| GST-α-syn | NovoPro | Cat#505565 |
| pGEX-KG myc | Addgene | Cat#209891 |
| Human recombinant PLK2 | Abcam | Cat#ab102108 |
| Lipofectamine 2000 | Invitrogen | Cat#11668019 |
| ProFection | Promega | Cat#E1200 |
| LentiX Concentrator | Takara | Cat#631232 |
| Isoflurane | VetOne | Cat#502017 |
| 4% Paraformaldehyde (PFA) | Thermo Scientific | Cat#J19943-K2 |
| Discovery Wash buffer | Ventana | Cat#07311079001 |
| CC1 buffer | Ventana | Cat#06414575001 |
| inhibitor CM buffer | Ventana | Cat#760–159 |
| HRP polymer | Ventana | Cat#05266548001 |
| Hematoxylin II | Ventana | Cat#05277965001 |
| Bluing reagent | Ventana | Cat#05266769001 |
| Lambda Protein Phosphatase | New England Biolab | Cat#P0753 |
| NotI restriction enzyme | New England Biolabs | Cat#R3189L |
| In-Fusion Snap Assembly Master Mix | Takara | Cat#638947 |
| DNA Ligation Kit Mighty Mix | Takara | Cat#6023 |
| PEI Max | Polysciences | Cat#24765 |
| Lucigen QuickExtract DNA extraction solution | Biosearch Technologies | Cat#QE09050 |
| Neuronal protein extraction reagent (N-PER) | Thermo Scientific | Cat#87792 |
| Protease/phosphatase inhibitors | Cell Signaling | Cat#5872 |
| Protein G-agarose beads | Thermo Scientific | Cat#20397 |
| NuPAGE LDS sample buffer | Thermo Scientific | Cat#NP007 |
| NuPAGE 4 to 12% Bis-Tris polyacrylamide gels | Thermo Scientific | Cat#NP0335BOX |
| 0.2 μM PVDF membrane | Thermo Scientific | Cat#LC2002 |
| Terrific Broth | Thermo Scientific | Cat# BP9728–2 |
| EDTA-free protease cocktail inhibitor | Roche | Cat#11836170001 |
| Glutathione-Sepharose 4B | Sigma | Cat#17-0756-01 |
| TEV protease | New England Biolabs | Cat#P8112 |
| Alkaline Phosphatase (ALP) | Thermo Scientific | Cat# EF0651 |
| Dithiothreitol (DTT) | Thermo Scientific | Cat#R0861 |
| Iodoacetamide (IAA) | Sigma | Cat#I1149 |
| Lysyl endopeptidase | Wako | Cat#125–05061 |
| Trypsin | Promega | Cat#VA9000 |
| Asp-N | Promega | Cat#VA1160 |
| HLB column | Waters | Cat#186002034 |
| Poly-D-lysine | Sigma | Cat#P6407–5MG |
| Critical Commercial Assays | ||
| Ventana Discovery ChromoMap DAB kit | Ventana | Cat#05266645001 |
| AAVpro Purification Kit (All Serotypes) | Takara | Cat#6666 |
| P3 Primary Cell 4D Nucleofector X Kit S | Lonza | Cat#V4XP-3032 |
| Monarch DNA gel extraction Kit | New England Biolabs | Cat#T1020S |
| High-select Fe-NTA Phosphopeptide enrichment kit | Thermo Scientific | Cat#A32992 |
| Deposited Data | ||
| ColabFold Protein model | This paper | Zenodo: https://zenodo.org/records/8059967 |
| Depth profile molecular simulation | This paper | Zenodo: https://zenodo.org/records/8059967 |
| Tabular data | This paper | Zenodo: https://zenodo.org/records/8059967 |
| Supplemental movies | This paper | Zenodo: https://zenodo.org/records/8059967 |
| Experimental Models: Cell Lines | ||
| HEK293T | Cellosaurus | RRID:CVCL_0063 |
| Neuro2A | Cellosaurus | Cat#TKG0509, RRID:CVCL_0470 |
| Experimental Models: Organisms/Strains | ||
| CD-1 IGS mouse (ICR) | Charles River Laboratories | Outbred Cat# 0220CD-1 |
| C57BL/6J | Jackson Labs | RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| α-syn forward primer (PCR#1): TGTGCTTTCTCTTCCCT CTCTG | IDT | N/A |
| α-syn forward primer (PCR#2): ATAACACTTCGTGCAG CACC | IDT | N/A |
| Reverse oScarlet primer (PCR#1 and PCR#2): ACAGGATGTCCCAGGA GAAG. | IDT | N/A |
| Recombinant DNA | ||
| pCCL-mScarlet | This paper | Addgene plasmid# 209889 |
| pCCL-Green Lantern | This paper | Addgene plasmid# 209890 |
| pGEX-KG myc | This paper | Addgene plasmid# 209891 |
| psPAX2 | Addgene | RRID:Addgene_12260 |
| pMD2.G | Addgene | RRID:Addgene_12259 |
| LentiCRISPR v2 | Addgene | Addgene plasmid#52961 |
| PX552 | Addgene | Addgene plasmid# 60958 |
| pAAV-SpCas9 | Addgene | Addgene plasmid# 60957 |
| pPHP.S | Addgene | Addgene plasmid #103005 |
| Software and Algorithms | ||
| GraphPad Prism Software | http://www.graphpad.com/ | RRID:SCR_002798 |
| MetaMorph Microscopy Automation and Image Analysis Software | Molecular Devices https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysissoftware/metamorph-microscopy#gref | RRID:SCR_002368 |
| MATLAB | MATLAB http://www.mathworks.com/products/matlab/ | RRID:SCR_001622 |
| CRISPick | Broad Institute https://portals.broadinstitute.org/gppx/crispick/public | N/A |
| Image Lab software version 6.1 | BioRad | RRID:SCR_014210 |
| STRING database version 11.5 | STRING | RRID:SCR_005223 |
| PANTHER database | PANTHER | RRID:SCR_004869 |
| AxIS software version 2.4 | Axion Biosystems | RRID:SCR_016308 |
| ColabFold | https://colabfold.mmseqs.com/ | N/A |
| GROMACS v.2021.3 | https://www.gromacs.org/ [doi: 10.5281/zenodo.5053201] | RRID:SCR_014565 |
| CHARMM36 force field | https://www.charmm.org/archive/charmm/resources/charmm-force-fields/ | RRID:SCR_014892 |
| LINCS algorithm | http://www.lincsproject.org/ | RRID:SCR_016486 |
| Jalview sequence fetcher | Jalview | RRID:SCR_006459 |
| Clustal Omega Webserv er | http://www.ebi.ac.uk/Tools/msa/clustalo/ | RRID:SCR_001591 |


