Abstract
Congenital myasthenic syndromes (CMS) are a rare group of inherited disorders caused by gene defects associated with the neuromuscular junction and potentially treatable with commonly available medications such as acetylcholinesterase inhibitors and β2 adrenergic receptor agonists. In this study, we identified and genetically characterized the largest cohort of CMS patients from India to date.
Genetic testing of clinically suspected patients evaluated in a South Indian hospital during the period 2014–19 was carried out by standard diagnostic gene panel testing or using a two-step method that included hotspot screening followed by whole-exome sequencing. In total, 156 genetically diagnosed patients (141 families) were characterized and the mutational spectrum and genotype-phenotype correlation described. Overall, 87 males and 69 females were evaluated, with the age of onset ranging from congenital to fourth decade (mean 6.6 ± 9.8 years). The mean age at diagnosis was 19 ± 12.8 (1–56 years), with a mean diagnostic delay of 12.5 ± 9.9 (0–49 years). Disease-causing variants in 17 CMS-associated genes were identified in 132 families (93.6%), while in nine families (6.4%), variants in genes not associated with CMS were found. Overall, postsynaptic defects were most common (62.4%), followed by glycosylation defects (21.3%), synaptic basal lamina genes (4.3%) and presynaptic defects (2.8%). Other genes found to cause neuromuscular junction defects (DES, TEFM) in our cohort accounted for 2.8%.
Among the individual CMS genes, the most commonly affected gene was CHRNE (39.4%), followed by DOK7 (14.4%), DPAGT1 (9.8%), GFPT1 (7.6%), MUSK (6.1%), GMPPB (5.3%) and COLQ (4.5%). We identified 22 recurrent variants in this study, out of which eight were found to be geographically specific to the Indian subcontinent. Apart from the known common CHRNE variants p.E443Kfs*64 (11.4%) and DOK7 p.A378Sfs*30 (9.3%), we identified seven novel recurrent variants specific to this cohort, including DPAGT1 p.T380I and DES c.1023+5G>A, for which founder haplotypes are suspected.
This study highlights the geographic differences in the frequencies of various causative CMS genes and underlines the increasing significance of glycosylation genes (DPAGT1, GFPT1 and GMPPB) as a cause of neuromuscular junction defects. Myopathy and muscular dystrophy genes such as GMPPB and DES, presenting as gradually progressive limb girdle CMS, expand the phenotypic spectrum. The novel genes MACF1 and TEFM identified in this cohort add to the expanding list of genes with new mechanisms causing neuromuscular junction defects.
Keywords: congenital myasthenic syndromes, neuromuscular junction, genetics, NGS, recurrent mutations
Polavarapu and Sunitha et al. describe the genetic characteristics of 156 Indian patients with suspected congenital myasthenic syndromes (CMS) and explore genotype-phenotype correlations. They show that 22 disease-causing variants account for about two-thirds of patients, and identify several novel genes associated with CMS.
Introduction
Congenital myasthenic syndromes (CMS) are a heterogeneous group of rare inherited neuromuscular junction (NMJ) disorders caused by defects in the proteins that play a pivotal role in neuromuscular transmission.1,2 Genetically, CMS are an evolving spectrum of disorders, and the advent of next-generation sequencing (NGS) technologies has resulted in a rapid increase in the identification of causative genes over the past decade.3 CMS is generally considered a very rare disorder, but published prevalence estimates of 9.2 cases per million children in the UK and 1.8 per million population in Spain4,5 are likely to be significant underestimates, due both to incomplete case ascertainment and the fact that diagnostic yield varies depending on the genetic testing methodology used, especially considering the recent adoption of NGS technologies like gene panel, exome and genome testing.3,6
Individual CMS subtypes are primarily classified as presynaptic, synaptic and postsynaptic according to the localization of the defective genes involved in the structure and function of the NMJ complex, while a fourth group of glycosylation genes has been found to cause CMS due to impaired glycosylation of various NMJ components.7 In the 2022 version of the gene table of neuromuscular disorders, 34 genes are reported to cause CMS, in addition to several myopathy genes, including RYR1, TPM3, DNM1 and DES, for which features of a NMJ defect have been described in addition to primary myopathy.8-12
To date, only three genetically confirmed CMS cohorts of over 100 patients have been published. These international cohorts were established over a number of years by groups from the Mayo Clinic (USA), the UK and Europe.1,4,13 Multiple smaller cohorts have also been reported from countries including Brazil, Turkey, Israel, Spain and China.5,14-17 Except for the Chinese cohort, in which GFPT1 defects constituted the most common genotype, CHRNE defects are by far the most common genetic cause of CMS identified worldwide. Postsynaptic defects in general are more common than presynaptic, synaptic and glycosylation defects. However, the frequencies of other gene defects do vary substantially between studies from different regions, indicating likely geographical variability in CMS genotypes.
The mutational spectrum and prevalence of CMS subtypes in India has not been studied in detail to date, with only a few reports describing genetically confirmed cases.18-21 A targeted NGS study from southern India, which analysed five CMS genes (CHRNE, RAPSN, COLQ, DOK7 and GFPT1), identified the genetic cause in 18 CMS patients in whom CHRNE defects were the most common cause.22 Another recent report from a South Indian hospital identified seven genetically confirmed cases in whom COLQ defects were predominant.23
In the present study, we describe a large cohort of 156 genetically confirmed CMS patients from 141 unique families who were seen in the neuromuscular speciality clinic of a quaternary referral centre in South India. We aimed to determine the genetic prevalence of CMS subtypes and characterize the recurrent or ‘regional hotspot’ variants among Indian patients in an effort to develop a cost-effective diagnostic pipeline. This is the first large genetically confirmed CMS cohort in the NGS era, only the fourth study worldwide to comprise over 100 patients and the largest from India to date. Our study confirms several previously reported epidemiological findings and introduces several novel findings that appear to be unique to India.
Materials and methods
Patients
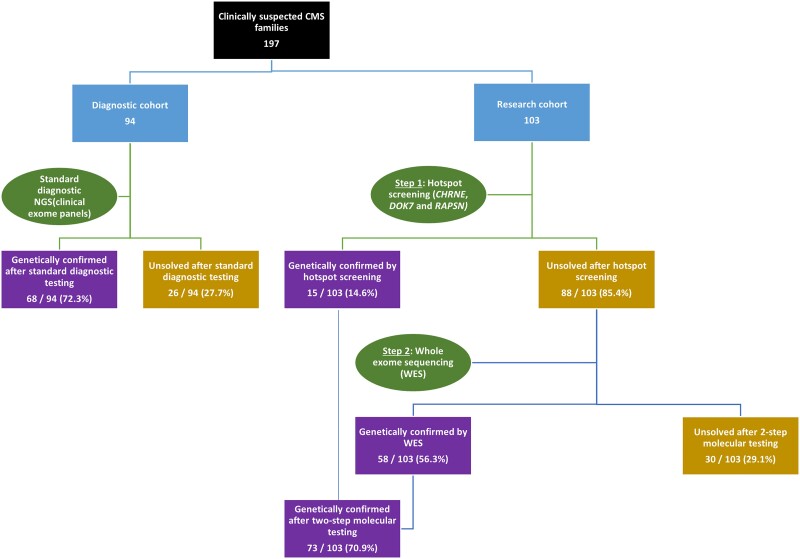
A total of 213 patients from 197 families with a clinical suspicion of CMS were divided into two independent sub-cohorts based on the genetic testing strategies adopted (Fig. 1). The first sub-cohort (diagnostic) comprised 97 suspected CMS patients (94 families) who underwent commercial genetic testing followed by review of the final genetic report as part of the standard diagnostic pathway. The second sub-cohort (research) comprised 116 patients (103 families) who underwent a two-step genetic testing process of hotspot screening followed by whole-exome sequencing (WES) and analysis through an international research collaboration. Of these, 156 patients (141 families) received a confirmed genetic diagnosis, while 57 (56 families) remained genetically undiagnosed and are not reported here in the further analysis. A detailed genotype-phenotype analysis was thus performed on 156 CMS patients (141 families) in whom disease-causing variants were identified. All patients were recruited, clinically evaluated by the lead author and neuromuscular specialist (A.N.) and investigated at the National Institute of Mental Health and Neuroscience (NIMHANS), Bangalore, India, during the period 2014–19. For the second sub-cohort, blood samples from 116 patients (103 families) were collected and DNA was extracted and subjected to quality control. Written consent was obtained from all individuals for the study, and DNA samples were submitted to the Newcastle MRC Centre Biobank for Neuromuscular Diseases, for which ethical approval was granted by the NRES Committee North East—Newcastle & North Tyneside 1 (reference 08/H0906/28). Detailed clinical data, including age of onset, progression, family history, symptoms and electrophysiology and histopathology results, if any, were collected, and all phenotypic data were uploaded onto the PhenoTips online software platform hosted by the RD-Connect Genome-Phenome Analysis Platform (GPAP), which makes use of Human Phenotype Ontology (HPO) terms to enable standardization and comparability across patients.
Figure 1.
Molecular diagnostic flow chart for suspected congenital myasthenic syndrome patients. CMS = congenital myasthenic syndrome; NGS = next generation sequencing.
Inclusion criteria
For a clinical suspicion of CMS, the following inclusion criteria were applied to individuals with isolated, or a combination of: (i) fatigable ocular (ptosis, ophthalmoparesis), bulbar, limb or respiratory muscle weakness with or without positive family history; (ii) infants/children with apnoeic spells; (iii) persistent myopathic/dystrophic features with fatigable weakness; (iv) electrophysiological evidence of normal nerve conduction studies and decrement/increment response on repetitive nerve stimulation (RNS) (slow/sub-tetanic/tetanic stimulation); or (v) improvement with acetylcholinesterase inhibitor drugs, salbutamol and/or fluoxetine.
Exclusion criteria
Sporadic patients with suspected autoimmune myasthenia gravis (positive anti-AChR or anti-MuSK antibodies with highly variable seasonal fluctuations, significant asymmetry in ocular findings, history of myasthenic crisis and significant previous response to steroids, immunomodulation or thymectomy were excluded.
Electrophysiology
All cooperative patients underwent standard motor and sensory nerve conduction studies of the median and ulnar nerves in the upper limbs and common peroneal and sural nerves in the lower limbs. Presence of double compound muscle action potential (CMAP) was carefully observed. RNS to detect NMJ transmission defect, pre- or postsynaptic, was performed at the routine 3 Hz stimulation rate and responses recorded from the orbicularis oculi, nasalis, trapezius, deltoid, quadriceps, tibialis anterior, abductor digiti minimi and anconeus muscles. For limb-girdle CMS (LG-CMS) patients, if slow-rate RNS did not reveal a decremental response, sub-tetanic stimulation at 10 Hz was performed in the proximal limb muscles. Where a presynaptic defect was suspected, tetanic stimulation was performed.
Genetic testing methodologies
Diagnostic sub-cohort
For the first sub-cohort of 97 CMS patients (94 families), molecular testing was performed as part of standard clinical diagnostics, with review of a final genetic report by the treating neurologist. The majority of these patients were sequenced in a commercial genetic testing laboratory using custom library capture kits for clinical exome sequencing as described previously24 (Supplementary material).
Research sub-cohort
Screening for hotspot variants
The sub-cohort of 116 patients (103 families) who were submitted to the Newcastle MRC Centre for a collaborative genetic study underwent a two-step process (Fig. 1). During Step 1, all patients were screened for the most common CMS causative variants: CHRNE c.1327delG; p.E443Kfs*6425 and c.130dupG; p.E44Gfs*3,26DOK7 c.1124_1127dupTGCC; p.A378Sfs*3026 and RAPSN c.264C>A; p.N88K.27CHRNE and DOK7 amplicons were Sanger sequenced and RAPSN amplicons were subjected to enzymatic restriction with BsrI and visualized by agarose gel electrophoresis. All negative patients were subjected to Step 2: WES.
Whole-exome sequencing and data analysis
WES in patients and family members was performed using the genomics platform at the Broad Institute of MIT and Harvard, Cambridge, USA. Library preparation and exome sequencing was performed as previously described.28 Exome data were then processed at the Centro Nacional de Análisis Genómico (CNAG), Barcelona, Spain, and variant prioritization was carried out on the RD-Connect GPAP (https://platform.rd-connect.eu).29 Likely pathogenic variants were identified by applying standard filtering criteria, including a moderate to high variant effect predictor (VEP) score (i.e. nonsense, splice site, frame-shift, in-frame and non-synonymous variants), minor allele frequency (MAF) <1% and CADD (Combined Annotation Dependent Depletion) score >20. WES data were sequentially analysed first for known CMS genes (n = 34), followed by screening of the 611 genes in the 2022 neuromuscular gene table (https://www.musclegenetable.fr/) and other known disease-causing genes from the Online Mendelian Inheritance in Man (OMIM) database.8,30,31 Patients in whom no defects in known genes were found were further screened for putative pathogenic variants in novel candidate genes. Shortlisted variants were interrogated for their predicted in silico deleteriousness and previous known association with human disease. Novel variants that had not previously been reported in the literature were assessed by in silico methods, including mutation taster, UMD-predictor, PolyPhen-2, SIFT, PROVEAN, CADD and/or human splicing finder, and were classified individually according to American College of Medical Genetics (ACMG) standards and guidelines for interpretation of variants.32,33 Segregation of the identified variants was carried out where possible and genetic diagnosis was confirmed with the referring clinician.
The identified causative genes in this cohort were classified as either ‘CMS’ or ‘non-CMS’ genes based on strong evidence of association with a CMS phenotype as reported in the OMIM database, neuromuscular gene table and/or literature. As the cohort includes multiple affected members in some of the families, the percentages from here on were calculated among ‘patients’ for clinical findings and among ‘families’ for genetic data.
Results
Demographic details
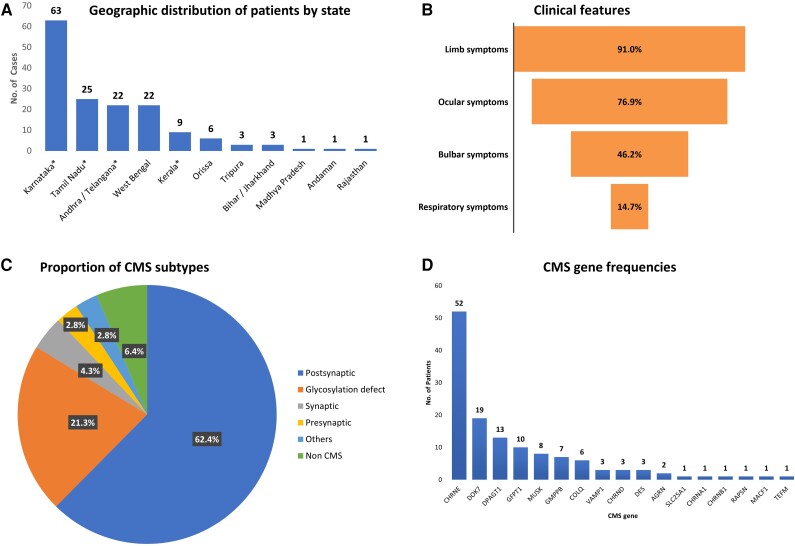
To understand the phenotypic and mutational pattern of CMS in India, we studied the 156 CMS patients (from 141 families) in whom disease-causing variants had been identified. Most patients were from the three South India states of Karnataka (40.4%), Tamil Ndu (16%) and Andhra/Telangana (14.1%), which correlated with the frequency of referral of these populations to the quaternary referral hospital located in Bengaluru (Bangalore), Karnataka. Apart from these southern states, the hospital also receives sizeable referrals from the eastern state of West Bengal, which contributed 14.1% of the cases to the cohort (Fig. 2A). Seventy (49.6%) of the families were known to be consanguineous, all but two of whom came from South India. A positive family history of other affected individuals with a similar phenotype was present in 39/141 families (27.6%), while the remaining 102 were sporadic cases. Recessive genotypes were identified in 131 families (92.9%).
Figure 2.
Demographic, clinical features, congenital myasthenic syndrome subtypes and gene frequencies. (A) Geographic distribution by their various Indian states of origin of congenital myasthenic syndrome (CMS) patients reported in this study. (B) Clinical features of CMS patients: percentage of four common symptom groups (limb, ocular, bulbar and respiratory). (C) Pie chart showing frequencies of various CMS subtypes based on location of defect at neuromuscular junction and/or underlying pathomechanism. (D) Frequencies of individual CMS genes identified in the current cohort.
Clinical details
The sex ratio was 87 (male): 69 (female). The age of onset ranged from as early as at birth to as late as the fourth decade, with a mean age of 6.6 ± 9.8 (0–40 years). Onset was predominantly congenital or infantile (34.6%) followed by early childhood <5 years (29.5%). Juvenile or adult onset >12 years was reported in 33 patients (21.2%). The mean age at diagnosis was 19 ± 12.8 (1–56 years). There was a mean delay of 12.5 ± 9.9 (0–49 years) between onset and diagnostic confirmation of CMS. All patients presented with fatigable weakness of either ocular, cranial or limb muscles. Limb symptoms (predominantly lower limb fatigable weakness) were the most common, occurring in 142 patients (91%), followed by ocular symptoms (ptosis/ophthalmoparesis) in 120 (76.9%). Bulbar involvement was reported in 72 patients (46.1%), while 23 patients (14.7%) had respiratory symptoms (Fig. 2B). Patients with acetylcholine receptor subunit defects and DPAGT1 glycosylation defects presented with predominantly early onset of symptoms, while those with DOK7, GFPT1 and GMPPB glycosylation defects had a later onset. Similarly, ocular symptoms were more common at onset in patients with receptor defects, while DOK7 and all glycosylation defects had a predominantly LG-CMS phenotype (Fig. 3D and Table 1).
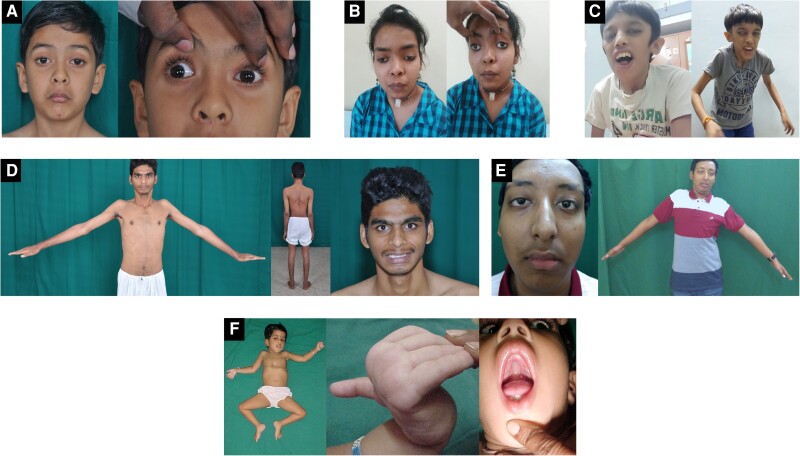
Figure 3.
Clinical images of genetically confirmed congenital myasthenic syndrome patients. (A) An 8-year-old male with homozygous CHRNE c.1052C>T; p.P351L showing ptosis, ophthalmoparesis and facial weakness. (B) A 14-year-old female with homozygous COLQ c.1319G>A; p.C440Y presented with history of recurrent apneic spells, ptosis, limb weakness and respiratory distress from birth. She was ventilator-dependent until 13 years of age and showed significant improvement with salbutamol. (C) A 12-year-old male with compound heterozygous RAPSN variants c.-210A>G (5′UTR)/c.1144T>C; p.C382R with dysmorphic facial features, ptosis, open mouth and severe proximal contractures. (D) A 23-year-old male with homozygous GFPT1 c.1103C>T; p.T368I presented with limb girdle weakness and muscle wasting with no ocular or bulbar involvement mimicking the limb girdle muscular dystrophy phenotype. (E) A 14-year-old male with compound heterozygous AGRN c.5302G>A; p.A1768T/c.6057_6060delCGTGinsT p.V2022del presented with easy fatigability from childhood and profound proximal muscle weakness. (F) A 2-year-old female with homozygous VAMP1 c.97C>T (p.R33*) showing severe hypotonia, pectus excavatum, myopathic facies, hyperextensible fingers and high arched palate (3F published previously in Polavarapu et al.24).
Table 1.
Phenotype details of individual congenital myasthenic syndrome genes
| Gene | No. of patients (families) | No. of consang. cases | M:F | Mean age at onset (years) ± SD | Mean age at diagnosis (years) ± SD | Mean diagnostic delay (years) ± SD | Symptoms | CK | Treatment response | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Initial symptom | Ocular | Bulbar | Respiratory | Limb | PYR | SAL | FLU | ||||||||
| CHRNE | 59 (52) | 30 | 31:28 | 1.5 ± 5.1 (0–35) | 15.1 ± 11.5 (1–55) | 13.4 ± 11.3 (0.5–55) | Ptosis | 59 (100%) | 39 (66.1%) | 9 (15.2%) | 49 (83%) | N | ↑ | ↑ | – |
| DOK7 | 20 (19) | 6 | 14:6 | 11.8 ± 12.2 (0–34) | 25.2 ± 11.9 (5–48) | 13.3 ± 7.8 (2–32) | LGW | 15 (75%) | 6 (30%) | 2 (10%) | 19 (95%) | N/↑ | ↓ | ↑ | ↑ |
| GMPPB | 12 (7) | 5 | 6:6 | 25.4 ± 9 (10–40) | 41.7 ± 9.4 (27–56) | 14 ± 9.7 (1.6–34) | LGW | 3 (25%) | 7 (58.3%) | – | 12 (100%) | ↑↑ | ↑ | ↑ | – |
| DPAGT1 | 13 (13) | 3 | 8:5 | 2.5 ± 3.6 (0–13) | 11.6 ± 7.1 (4–26) | 8.6 ± 7 (2–26) | LGW | 7 (53.8%) | 5 (38.5%) | 3 (23.1%) | 13 (100%) | N/↑ | ↑ | ↑ | – |
| GFPT1 | 10 (10) | 5 | 6:4 | 11.1 ± 6.2 (2–25) | 15.6 ± 5.7 (9–28) | 4.4 ± 4.4 (1.0–16) | LGW | 4 (40%) | – | – | 10 (100%) | N/↑ | ↑ | ↑ | – |
| MUSK | 8 (8) | 4 | 4:4 | 7.4 ± 5.9 (0–18) | 18 ± 6.8 (6–24) | 10.6 ± 7.2 (1–21) | LGW | 6 (75%) | 2 (25%) | – | 8 (100%) | N | ↑ | ↑ | – |
| COLQ | 6 (6) | 2 | 4:2 | 0.8 ± 0.9 (0–2) | 20.8 ± 15.8 (7–50) | 19.4 ± 15.5 (5.6–48) | Ptosis/LGW | 3 (50%) | 1 (16.7%) | 4 (66.7%) | 6 (100%) | N | NR | ↑ | ↑ |
| CHRND | 3 (3) | 1 | 3:0 | 7.7 ± 11.6 (0–21.0) | 31.0 ± 23.6 (5–51) | 23.3 ± 22.9 (5–49) | Ptosis/BW | 3 (100%) | 3 (100%) | 2 (66.7%) | 2 (66.7%) | N | ↑ | ↑ | ↑ |
| VAMP1 | 3 (3) | 3 | 0:3 | 0 | 2.5 ± 2.1 (0.75–5) | 2.5 ± 2.1 (0.75–5) | CH/BW | 2 (66.7%) | 3 (100%) | 2 (66.7%) | 3 (100%) | N | ↑ | ↑ | – |
| DES | 3 (3) | 3 | 2:1 | 2.0 ± 2.0 (0–4) | 17 ± 4.6 (13–22) | 15 ± 5.6 (9–20) | LGW | 3 (100%) | 3 (100%) | – | 3 (100%) | ↑↑ | ↑ | ↑ | – |
| AGRN | 2 (2) | 1 | 1:1 | 13 ± 8.5 (7–19) | 17 ± 5.6 (13–21) | 3.7 ± 3.2 (1.4–6) | LGW | 1 (50%) | – | – | 2 (100%) | N | ↓ | ↑ | – |
| SLC25A1 | 2 (1) | 1 | 1:1 | 1 | 25.5 ± 0.7 (25–26) | 24.5 ± 0.7 (24–25) | LGW | 2 (100%) | 1 (50%) | – | 2 (100%) | N | NR | NR | – |
| TEFM | 2 (1) | no | 2:0 | 5 | 18.5 ± 3.5 (16–21) | 13.5 ± 3.5 (11–16) | LGW/Ptosis | 2 (100%) | – | – | 2 (100%) | N | NR | ↑ | – |
| RAPSN | 1 (1) | no | 1:0 | 0 | 12 | 12 | Ptosis/Contractures | 1 (100%) | 1 (100%) | 1 (100%) | 1 (100%) | N | ↑ | ↑ | – |
| CHRNA1 | 1 (1) | no | 0:1 | 22 | 29 | 7 | Dropped head | 1 (100%) | – | 1 (100%) | 1 (100%) | N | ↓ | ↑ | – |
| CHRNB1 | 1 (1) | yes | 0:1 | 3 | 3.5 | 0.5 | Ptosis | 1 (100%) | 1 (100%) | – | 1 (100%) | N | NR | ↑ | – |
| MACF1 | 1 (1) | no | 0:1 | 29 | 35 | 6 | LGW | – | – | – | 1 (100%) | ↑ | ↑ | ↑ | – |
BW = bulbar weakness; CH = congenital hypotonia; CK = creatine kinase; consang. = consanguineous; F = female; FLU = fluoxetine; LGW = limb girdle weakness; M = male; N = normal; NR = no response; PYR = pyridostigmine; SAL = salbutamol; SD = standard deviation; ↑ = mildly elevated (CK) or improved (treatment response); ↑↑ = high; ↓ = worsened (treatment response).
Electrophysiology data comprising nerve conduction and RNS studies were available for 138/156 (88.5%) of patients. A positive decrement response of ≥10% was observed in 124 patients (79.5%). Eight patients did not cooperate for RNS, and records were not available for another 10 individuals. Of 14 individuals in whom RNS did not show a positive decrement, five were found to have defects in non-CMS genes.
Treatment response with either pyridostigmine (acetylcholinesterase inhibitor) and/or salbutamol (β2 adrenergic receptor agonist) was assessed by oral or telephonic follow-up after a treatment period of at least 3 months.
Genetic results
Diagnostic sub-cohort
Among the 97 patients (94 families) screened through the standard diagnostic pathway, a confirmed genetic diagnosis was obtained in 71 (68 families or 72.3%) (Fig. 1). Patients from 26 families (27.7%) remained unsolved, and further genetic testing was not performed as part of this study.
Research sub-cohort
Hotspot screening results
As described above, 116 suspected CMS patients (103 families) were subjected to screening of the four most common CMS-causing variants in CHRNE, DOK7 and RAPSN. In 16 patients (13 families), the two hotspot CHRNE frameshift variants (c.1327delG and c.130dupG) were identified in either the homozygous or compound heterozygous state, thus confirming their diagnosis. In another seven unrelated patients, the DOK7 hotspot variant c.1124_1127dupTGCC was identified; two of these patients carried the variant in homozygosity and were considered to have a confirmed diagnosis. The remaining five were heterozygous, and their diagnosis remained inconclusive. None of the patients carried the RAPSN hotspot variant. Thus, hotspot screening provided a genetic diagnosis in 18 patients from 15 families (14.6%).
Whole-exome screening results
After hotspot screening, 98 patients (88 families) who remained undiagnosed, including five patients with a single heterozygous DOK7 hotspot variant, were screened by WES. WES identified disease-causing variants in an additional 69 patients (58 families). Overall, the two-step molecular screening contributed to diagnosis in 73/103 of the suspected CMS families analysed: a diagnostic yield of 70.9%.
Mutational spectrum
Of the 156 patients (141 families) who received a confirmed diagnosis, 147 patients (132 families or 93.6%) were found to have causative variants in CMS genes, while nine families (6.4%) had causative variants in non-CMS genes. Overall, 101 unique variants were identified in our cohort (Tables 2 and 3). While 57 have previously been reported in other patients, 54 are novel variants identified as disease-causing in this cohort following in silico predictions and phenotype correlation along with segregation wherever familial samples were available. The most common group of CMS genes affected were postsynaptic (88/141 or 62.4%) followed by glycosylation defects (30/141 or 21.3%) (Fig. 2C). Among the CMS genes, the most commonly affected was CHRNE (52 or 39.4%), followed by DOK7 (19 or 14.4%), DPAGT1 (13 or 9.8%), GFPT1 (10 or 7.6%), MUSK (8 or 6.1%), GMPPB (7 or 5.3%), COLQ (6 or 4.5%), VAMP1 (3 or 2.3%), CHRND (3 or 2.3%), DES (3 or 2.3%), AGRN (2 or 1.8%) and one each (0.8%) of SLC25A1, CHRNA1, CHRNB1, RAPSN, MACF1 and TEFM (Fig. 2D). Affected individuals from nine families (6.4%) carried likely disease-causing variants in non-CMS genes. Six variants were found in genes associated with myopathy or muscular dystrophy: TTN (n = 2), DYSF, TRAPPC11, ADSSL1 and CAPN3. In two patients, reported pathogenic variants were identified in mitochondrial genes (MT-TL1 and NF1). The MT-TL1 variant was heteroplasmic in the patient, while the NF1 variant was de novo. One patient had a homozygous splice site variant in a known mental retardation gene, ANK3. The majority of the patients (all but 12/156) had a recessive genotype with either homozygous or likely compound heterozygous variants (Tables 2 and 3 and Supplementary material). Two sporadic patients (one CHRNE and one CHRNA1) had reported heterozygous slow-channel variants in acetylcholine receptor subunit genes. Segregation analysis was performed in 59 families where additional family samples were available. Of 38 patients with suspected compound heterozygous variants, segregation was confirmed in 27 patients where family samples were available. Further segregation was not performed in 11 patients with likely compound heterozygous variants and two patients with reported dominant slow channel variants due to lack of parental samples. In another seven families (eight patients), a single heterozygous variant was identified in DOK7 (2), GFPT1 (2), CHRNE (1), CHRND (1) and CHRNB1 (1) genes, respectively, with a good phenotypic fit and likely recessive pattern of inheritance, but a second heteroallelic variant remained elusive. However, these patients were considered as likely solved by respective genes and were included in the variant analysis. A total of 22 recurrent variants were observed in eight genes (CHRNE, DPAGT1, COLQ, GMPPB, GFPT1, MUSK, DES and DOK7). Of these, 15 were previously reported as disease-causing in CMS patients, while seven novel recurrent variants were identified in this cohort (Table 4).
Table 2.
Variants identified in congenital myasthenic syndrome patients diagnosed with postsynaptic defects
| Gene | Variant 1 (cDNA) | Variant 1 (p.) | Variant 2 (cDNA) | Variant 2 (p.) | Zygosity | Patients |
|---|---|---|---|---|---|---|
| Acetylcholine receptor defects | ||||||
| CHRNE | c.1327delG | p.E443Kfs*64 | c.1327delG | p.E443Kfs*64 | Hom | P1–17a |
| CHRNE | c.130dupG | p.E44Gfs*3 | c.130dupG | p.E44Gfs*3 | Hom | P18 |
| CHRNE | c.130dupG | p.E44Gfs*3 | c.1327delG | p.E443Kfs*64 | C.Het | P19–20 |
| CHRNE | c.1204C>T | p.Q402* | c.1204C>T | p.Q402* | Hom | P21–23 |
| CHRNE | c.991C>T | p.R331W | c.991C>T | p.R331W | Hom | P24 |
| CHRNE | c.1371delC | p.C458Afs*49 | c.1371delC | p.C458Afs*49 | Hom | P25–28 |
| CHRNE | c.1052C>T | p.P351L | c.1052C>T | p.P351L | Hom | P29–30 |
| CHRNE | c.250C>T | p.R84* | c.501-10_504dup | p.Q169Gfs*19 | C.Het | P31 |
| CHRNE | c.799C>T | p.Q267* | c.799C>T | p.Q267* | Hom | P32 |
| CHRNE | c.860_861insT | p.L287Ffs*110 | c.860_861insT | p.L287Ffs*110 | Hom | P33–34 |
| CHRNE | c.689T>A | p.V230D | c.729C>G | p.Y243* | C.Het | P35–36 |
| CHRNE | c.235-1G>A | – | c.1216_1219+19del | – | C.Het | P37 |
| CHRNE | c.467C>T | p.P156L | c.467C>T | p.P156L | Hom | P38–39 |
| CHRNE | c.1219+1G>T | – | c.1219+1G>T | – | Hom | P40 |
| CHRNE | c.1216_1219+19del | – | c.1216_1219+19del | – | Hom | P41–42 |
| CHRNE | c.501-10_504dup | p.Q169Gfs*19 | c.235-1G>C | – | Likely C.Het | P43b |
| CHRNE | c.713G>A | p.R238Q | c.713G>A | p.R238Q | Hom | P44 |
| CHRNE | c.729C>G | p.Y243* | c.729C>G | p.Y243* | Hom | P45 |
| CHRNE | c.712C>T | p.R238W | c.712C>T | p.R238W | Hom | P46 |
| CHRNE | c.183_187dup | p.L63Pfs*3 | c.183_187dup | p.L63Pfs*3 | Hom | P47 |
| CHRNE | c.684_687del | p.D229Sfs*70 | c.684_687del | p.D229Sfs*70 | Hom | P48 |
| CHRNE | c.686_687dup | p.V230Tfs*71 | c.686_687dup | p.V230Tfs*71 | Hom | P49 |
| CHRNE | c.905C>T | p.P302L | c.905C>T | p.P302L | Hom | P50 |
| CHRNE | c.501-10_504dup | p.Q169Gfs*19 | c.501-10_504dup | p.Q169Gfs*19 | Hom | P51 |
| CHRNE | c.293T>C | p.L98P | c.293T>C | p.L98P | Hom | P52 |
| CHRNE | c.501-10_504dup | p.Q169Gfs*19 | c.293T>C | p.L98P | Likely C.Het | P53b |
| CHRNE | c.209_210del | p.L70Hfs*3 | c.209_210del | p.L70Hfs*3 | Hom | P54 |
| CHRNE | c.1002_1008dup | p.A337Hfs*62 | c.1216_1219+19del | – | Likely C.Het | P55b |
| CHRNE | c.799C>T | p.Q267* | c.500+40G>C | – | Likely C.Het | P56b |
| CHRNE | c.854T>C | p.V285A | – | – | Het (D) | P57b |
| CHRNE | c.326A>T | p.E109V | – | – | Het (R) | P58–59 |
| CHRND | c.1334T>C | p.I445T | c.1334T>C | p.I445T | Hom | P60 |
| CHRND | c.1204G>A | p.E402K | c.1204G>A | p.E402K | Hom | P61 |
| CHRND | c.1390C>T | p.R464* | – | – | Het (R) | P62 |
| CHRNA1 | c.517G>A | p.G173S | – | – | Het (D) | P63b |
| CHRNB1 | c.432_437dup | p.G145_I146insMG | – | – | Het (R) | P64 |
| Development and maintenance gene defects | ||||||
| DOK7 | c.1124_1127dup | p.A378Sfs*30 | c.1124_1127dup | p.A378Sfs*30 | Hom | P65–71 |
| DOK7 | c.1124_1127dup | p.A378Sfs*30 | c.1263dupC | p.S422Lfs*97 | Likely C.Het | P72b |
| DOK7 | c.1124_1127dup | p.A378Sfs*30 | c.957delC | p.K320Sfs*136 | Likely C.Het | P73b |
| DOK7 | c.1124_1127dup | p.A378Sfs*30 | c.437C>T | p.P146L | C.Het | P74–76 |
| DOK7 | c.1124_1127dup | p.A378Sfs*30 | c.773-1G>C | - | C.Het | P77 |
| DOK7 | c.1197_1212del | p.S400Mfs*51 | c.1197_1212del | p.S400Mfs*51 | Hom | P78 |
| DOK7 | c.437C>T | p.P146L | c.1112C>A | p.S371* | Likely C.Het | P79b |
| DOK7 | c.331+1G>T | c.1324_1357del | p.C442Afs*3 | Likely C.Het | P80b | |
| DOK7 | c.1322_1347dup | p.R450Gfs*15 | c.472C>T | p.R158W | C.Het | P81 |
| DOK7 | c.652G>A | p.D218N | c.652G>A | p.D218N | Hom | P82 |
| DOK7 | c.1124_1127dup | p.A378Sfs*30 | – | – | Het (R) | P83 |
| DOK7 | c.1083_1086del | p.V362Gfs*93 | – | – | Het (R) | P84 |
| MUSK | c.496C>T | p.R166* | c.1634T>C | p.L545P | Hom | P85 |
| MUSK | c.1742T>A | p.I581N | c.1742T>A | p.I581N | Likely C.Het | P86,P87–90 |
| MUSK | c.1925T>A | p.L642* | c.2114T>A | p.I705N | Likely C.Het | P91b |
| MUSK | c.1904_1912dup | p.N635_N637dup | c.1904_1912dup | p.N635_N637dup | C.Het | P92 |
| AGRN | c.3413T>C | p.L1138P | c.3413T>C | p.L1138P | C.Het | P93 |
| AGRN | c.5302G>A | p.A1768T | c.6057_6060delinsT | p.V2022del | Hom | P94 |
| RAPSN | c.-210A>G | – | c.1144T>C | p.C382R | Likely C.Het | P95 |
| MACF1 | c.4687G>A | p.V1563M | c.4687G>A | p.V1563M | Likely C.Het | P96 |
C.Het = compound heterozygous; Hom = homozygous; Het (R) = heterozygous in recessive; Het (D) = heterozygous in dominant. Bold text represents single heterozygous variants in possible recessive phenotypes.
aP17 had homozygous pathogenic mutations in both CHRNE and DYSF.
bSegregation not confirmed.
Table 3.
Variants identified in congenital myasthenic syndrome patients diagnosed with glycosylation, synaptic, presynaptic and non-congenital myasthenic syndrome gene defects
| Gene | Variant 1 (cDNA) | Variant 1 (p.) | Variant 2 (cDNA) | Variant 2 (p.) | Zygosity | Patients |
|---|---|---|---|---|---|---|
| Glycosylation defect | ||||||
| DPAGT1 | c.299G>A | p.G100D | c.299G>A | p.G100D | Hom | P97 |
| DPAGT1 | c.185G>C | p.S62T | c.85A>T | p.I29F | Likely C.Het | P98a |
| DPAGT1 | c.652C>T | p.R218W | c.652C>T | p.R218W | Hom | P99 |
| DPAGT1 | c.1139C>T | p.T380I | c.324G>A | p.M108I | C.Het | P100 |
| DPAGT1 | c.1139C>T | p.T380I | c.85A>T | p.I29F | C.Het | P101–107 |
| DPAGT1 | c.853A>C | p.N285H | c.1139C>T | p.T380I | C.Het | P108 |
| DPAGT1 | c.769T>C | p.F257L | c.769T>C | p.F257L | Hom | P109 |
| GMPPB | c.1000G>A | p.D334N | c.1000G>A | p.D334N | Hom | P110–121 |
| GFPT1 | c.1103C>T | p.T368I | c.1103C>T | p.T368I | Hom | P122 |
| GFPT1 | c.589C>A | p.Q197K | c.589C>A | p.Q197K | Hom | P123 |
| GFPT1 | c.158A>G | p.N53S | c.158A>G | p.N53S | Hom | P124–125 |
| GFPT1 | c.479A>G | p.Y160C | – | – | Het (R) | P126 |
| GFPT1 | c.44C>T | p.T15M | c.44C>T | p.T15M | Hom | P127 |
| GFPT1 | c.586G>A | p.G196R | c.586G>A | p.G196R | Hom | P128 |
| GFPT1 | c.1276A>T | p.N426Y | c.1276A>T | p.N426Y | Hom | P129 |
| GFPT1 | c.881T>C | p.L294P | c.881T>C | p.L294P | Hom | P130 |
| GFPT1 | c.1280C>G | p.T427R | – | – | Het (R) | P131 |
| Synaptic defect | ||||||
| COLQ | c.1061G>A | p.W354* | c.1061G>A | p.W354* | Hom | P132 |
| COLQ | c.1319G>A | p.C440Y | c.1319G>A | p.C440Y | Hom | P133–134 |
| COLQ | c.1228C>T | p.R410W | c.1204C>T | p.R402C | C.Het | P135 |
| COLQ | c.1228C>T | p.R410W | c.870A>T | p.R290S | C.Het | P136 |
| COLQ | c.1228C>T | p.R410W | c.1228C>T | p.R410W | Hom | P137 |
| Presynaptic defect | ||||||
| VAMP1 | c.66delT | p.G23Afs*6 | c.66delT | p.G23Afs*6 | Hom | P138 |
| VAMP1 | c.202C>T | p.R68* | c.202C>T | p.R68* | Hom | P139 |
| VAMP1 | c.97C>T | p.R33* | c.97C>T | p.R33* | Hom | P140 |
| SLC25A1 | c.740G>A | p.R247Q | c.740G>A | p.R247Q | Hom | P141–142 |
| DES | c.1023+5G>A | – | c.1023+5G>A | – | Hom | P143–145 |
| TEFM | c.469C>G | p.P157A | c.469C>G | p.P157A | Hom | P146–P147 |
| Non-CMS genes | ||||||
| TTN | c.86065G>T | p.E28689* | c.30683-2A>T | Likely C.het | P148a | |
| TTN | c.41595C>A | p.C13865* | c.15544_15546del | p.G5182del | C.het | P149 |
| ANK3 | c.114+1G>T | c.114+1G>T | Hom | P150 | ||
| DYSF | c.3113G>A | p.R1038Q | c.3113G>A | p.R1038Q | Hom | P151 |
| DYSF | c.1116C>A | p.S372R | c.1116C>A | p.S372R | Hom | P17b |
| TRAPPC11 | c.2938G>A | p.G980R | c.2938G>A | p.G980R | Hom | P152 |
| ADSSL1 | c.794G>A | p.G265E | c.794G>A | p.G265E | Hom | P153 |
| CAPN3 | c.2288A>G | p.Y763C | c.2288A>G | p.Y763C | Hom | P154 |
| MT-TL1 | m.3243A>Gc | – | – | – | Heteroplasmy | P155 |
| NF1 | c.3721C>T | p.R1241* | – | – | Het (D) | P156 |
C.Het = compound heterozygous; Hom = homozygous; Het (R) = heterozygous in recessive; Het (D) = heterozygous in dominant. Bold text represents single heterozygous variants in possible recessive phenotypes.
aSegregation not confirmed.
bP17 had homozygous pathogenic mutations in both CHRNE and DYSF.
cHeteroplasmic MT-TL1 variant.
Table 4.
Recurrent congenital myasthenic syndrome mutations
| Gene | Variant | Number of patients (families) | Reported as disease-causing/novel | GnomAD MAF (no. of mutant alleles) | SA MAF (no. of mutant alleles) | IndiGenomes (no. of mutant alleles) | GenomeAsia 1000k (no. of mutant alleles) |
|---|---|---|---|---|---|---|---|
| CHRNE | c.1327delG; p.E443Kfs*64 | 19 (16) | Abicht et al.25 | 0.0001276 (24) | 0.0008729 (22) | 0.0010 (2) | 0 |
| CHRNE | c.1052C>T; p.P351L | 2 (2) | Croxen et al.34 | 0 | 0 | 0 | 0 |
| CHRNE | c.860_861insT; p.L287Ffs*110 | 2 (2) | Novel | 0 | 0 | 0 | 0 |
| CHRNE | c.293T>C; p.L98P | 2 (2) | Croxen et al.,35 Selvam et al.22 | 0.00001591 (4) | 0.0001307 (4) | 0 | 0 |
| CHRNE | c.1204C>T; p.Q402*) | 3 (2) | Kraner et al.36 | 0.000008568 (2, European) |
0 | 0 | 0 |
| CHRNE | c.729C>G; p.Y243*) | 3 (2) | Novel | 0 | 0 | 0 | 0 |
| CHRNE | c.130dupG; p.E44Gfs*3 | 3 (3) | Ohno et al.26 | 0.0001034 (26) | 0.00003266 (1) | 0 | 0 |
| CHRNE | c.1371delC; p.C458Afs*49 | 4 (4) | Ganapathy et al.37 | 0 | 0 | 0 | 0 |
| CHRNE | c.1216_1219+19del | 4 (4) | Selvam et al.22 | 0.000009466 (2, south and east Asian) | 0.00003567 (1) | 0 | 0 |
| CHRNE | c.501-10_504dup; p.Q169Gfs*19 | 4 (4) | Novel (aClinVar:465862) | 0.00002844 (8) | 0.0001307 (4) | 0 | 0 |
| CHRNE | c.235-1G>A (3′ splice variant) | 2 (2) | Novel | 0 | 0 | 0 | 0 |
| CHRNE | c.799C>T; p.Q267*) | 2 (2) | Ganapathy et al.37 | 0 | 0 | 0 | 0 |
| COLQ | c.1319G>A; p.C440Y | 2 (2) | Novel (aClinVar: 1679828) | 0 | 0 | 0 | 0 |
| COLQ | c.1228C>T; p.R410W | 3 (3) | Wargon et al.,38 Guo et al.,39 Selvam et al.22 | 0.00002886 (8) | 0.00006739 (2) | 0 | 0 |
| DOK7 | c.1124_1127dupTGCC; p.A378Sfs*30) | 14 (13) | Beeson et al.40 | 0.0007024 (158) | 0.0003376 (9) | 0 | 0 |
| DOK7 | c.437C>T; p.P146L | 4 (3) | Ben Ammar et al.41 | 0.00002393 (6) | 0 | 0 | 0.000575 (2) |
| DPAGT1 | c.85A>T; p.I29F | 8 (8) | Iqbal et al.42 | 0.0001034 (26) | 0.0008492 (26) | 0 | 0.000287 (1) |
| DPAGT1 | c.1139C>T; p.T380I | 9 (9) | Novel (aClinVar: 1373663) | 0.00003535 (10) | 0.0002940 (9) | 0 | 0 |
| GFPT1 | c.158A>G; p.N53S | 2 (2) | Selvam et al.22 | 0 | 0 | 0 | 0 |
| GMPPB | c.1000G>A; p.D334N | 12 (7) | Carss et al.43 | 0.00007568 (19) | 0.0006206 (19) | 0.0005 (1) | 0 |
| MUSK | c.1742T>A; p.I581N | 5 (5) | Owen et al.44 | 0 | 0 | 0 | 0 |
| DES | c.1023+5G>A | 3 (3) | Novel | 0.000004018 (1) | 0.00003287 (1) | 0 | 0 |
MAF = minor allele frequency.
avariants of uncertain significance/conflicting interpretations in ClinVar with no associated literature available.
Discussion
CMS is a clinically identifiable phenotype with characteristic fatigable weakness and variable oculo-bulbar, limb muscles and respiratory involvement. Many patients respond well to acetylcholinesterase inhibitors (pyridostigmine or neostigmine). However, patients with defects in certain genes, including slow-channel CMS (SCCMS), as well as DOK7 and COLQ-related CMS, can deteriorate when treated with acetylcholinesterase inhibitors but often improve with β-sympathomimetics such as ephedrine and salbutamol (albuterol) or fluoxetine. Hence, it is imperative to establish the underlying genetic defect in any suspected CMS patient to effectively assign the appropriate therapy and thereby improve the prognosis. The outcome in these patients varies and depends on how early they are identified and appropriately treated.45
This study is the first comprehensive report of the mutational spectrum and phenotypic pattern of CMS in India. Here we attempt to highlight the salient features and novel findings in this large CMS cohort comprising 141 unique families in which disease-causing variants were identified.
Congenital myasthenic syndrome genes
CMS genes are broadly classified as postsynaptic, presynaptic, synaptic basal lamina and glycosylation gene defects.46 Postsynaptic defects, including those in acetylcholine receptor subunit genes and genes associated with NMJ development and maintenance, form the largest group in our cohort (88/141; 62.4%), of which acetylcholine receptor defects are the most common, as found in previous larger cohorts worldwide1,4,13 (Table 5). Patients with receptor subunit defects had predominantly early onset (congenital or early childhood) and presented with ocular features of fatigable ptosis and ophthalmoparesis with variable limb and bulbar involvement (Fig. 3A). While receptor deficiency or fast-channel defects seem to be the most common of the subunit defects, both displaying a predominantly recessive inheritance pattern, only two patients (Patients P57 and P63) in this cohort were confirmed to have SCCMS with previously reported slow channel heterozygous variants in CHRNE (p.V285A) and CHRNA1 (p.G173S), respectively.47,48 Among the endplate maintenance and development defects, those in RAPSN and DOK7 are usually described as the most common, with RAPSN being the second most frequent CMS gene after CHRNE in the majority of previously published studies.1,5,13,14 The high frequency of RAPSN CMS can be attributed to the common founder mutation p.N88K in European patients.27 In our cohort, DOK7 was the second most common gene (13.5%) after CHRNE (36.8%). Unlike other studies, we identified only one confirmed RAPSN case, and the p.N88K variant is conspicuously absent in our cohort. This may be explained by the fact that the majority of previous large cohort studies have been from Europe and the Americas, as p.N88K is largely lacking from the literature and from population databases from the wider Asian region. Only three Indian patients have previously been reported with p.N88K in the literature.22,49,50 A study reporting 18 genetically confirmed CMS patients from southern India found one patient harbouring p.N88K in the compound heterozygous state, while another study from western India did not report any RAPSN cases.22,51 Further cohorts from China (35 patients) and Turkey (43 patients) also reported no RAPSN cases, suggesting a lower prevalence of RAPSN defects in Asia more broadly.15,17 Interestingly, the Chinese cohort reported DOK7 as one of the least common genes (3.4%), and defects in DOK7 were not identified in Turkish patients,15,17 which contrasts with its prevalence in our Indian cohort. While our RAPSN patient, Patient P95, presented with a severe congenital-onset phenotype with oculobulbar and respiratory involvement, patients with DOK7, MUSK and AGRN variants presented predominantly with a limb-onset fatigable weakness (LG-CMS) and relatively late onset compared to receptor defects (Fig. 3C and E).
Table 5.
Congenital myasthenic syndrome gene frequencies in current and selected previously published cohorts
| Abicht et al.13 (predominantly European) | Engel et al.1 (Worldwide) | Parr et al.4 (UK) | Estephan et al.14 (Brazil) | Yis et al.15 (Turkey) | Aharoni et al.16 (Israel) | Natera-de Benito et al.5 (Spain) | Zhao et al.17 (China) | Current study (India) | |
|---|---|---|---|---|---|---|---|---|---|
| Total no. of CMS families with molecular diagnosis | 299 | 356 | 123 | 52 | 43 | 35 | 47 | 29 | 132 |
| Acetylcholine receptor subunit genes | |||||||||
| CHRNE | 148 (49.5%) | 180 (50.5%) | 46 (37.4%) | 25 (48.1%) | 14 (32.5%) | 7 (20%) | 10 (21.3%) | 5 (17.2%) | 52 (39.4%) |
| CHRNA1 | 5 (1.7%) | 5 (4.1%) | 1 (1.9%) | 1 (2.1%) | 1 (3.4%) | 1 (0.8%) | |||
| CHRNB1 | 3 (1%) | 1 (2.1%) | 1 (0.8%) | ||||||
| CHRND | 3 (1%) | 2 (1.6%) | 1 (1.9%) | 1 (2.3%) | 1 (3%) | 3 (2.3%) | |||
| CHRNG | 4 (8.5%) | ||||||||
| Neuromuscular junction development and maintenance genes | |||||||||
| DOK7 | 31 (10.4%) | 35 (9.8%) | 22 (17.9%) | 3 (5.8%) | 1 (3%) | 5 (10.6%) | 1 (3.4%) | 19 (14.4%) | |
| MUSK | 1 (0.3%) | 1 (0.3%) | 2 (4.6%) | 8 (6.1%) | |||||
| AGRN | 1 (2.3%) | 5 (17.2%) | 2 (1.8%) | ||||||
| RAPSN | 43 (14.4%) | 51 (14.3%) | 34 (27.6%) | 6 (11.5%) | 13 (37%) | 9 (19.1%) | 1 (0.8%) | ||
| MACF1 | 1 (0.8%) | ||||||||
| Synaptic basal lamina genes | |||||||||
| COLQ | 38 (12.7%) | 45 (12.6%) | 12 (9.7%) | 2 (3.8%) | 14 (32.5%) | 11 (31%) | 9 (19.1%) | 4 (13.8%) | 6 (4.5%) |
| COL13A1 | 3 (5.8%) | ||||||||
| Glycosylation genes | |||||||||
| DPAGT1 | 2 (0.6%) | 13 (9.8%) | |||||||
| GFPT1 | 12 (2%) | 11 (3.1%) | 2 (3.8%) | 1 (2.3%) | 1 (3%) | 5 (10.6%) | 8 (27.6%) | 10 (7.6%) | |
| GMPPB | 1 (1.9%) | 1 (2.1%) | 2 (6.9%) | 7 (5.3%) | |||||
| COG7 | 1 (3.4%) | ||||||||
| Presynaptic genes | |||||||||
| CHAT | 15 (5%) | 18 (5.1%) | 2 (1.6%) | 1 (1.9%) | 8 (18.6%) | 1 (3%) | 2 (4.2%) | 1 (3.4%) | |
| MYO9A | 2 (4.6%) | ||||||||
| VAMP1 | 3 (2.3%) | ||||||||
| SLC25A1 | 1 (3.4%) | 1 (0.8%) | |||||||
| Other CMS genes | |||||||||
| SCN4A | 1 (0.3%) | 2 (3.8%) | |||||||
| PLEC | 2 (0.6%) | ||||||||
| PREPL | 1 (0.3%) | ||||||||
| DES | 3 (2.3%) | ||||||||
| TEFM | 1 (0.8%) | ||||||||
CMS = congenital myasthenic syndrome.
Patient P96 is one of the two CMS patients worldwide found to harbour variants in a novel CMS gene, MACF1, which is a scaffolding protein at the NMJ shown to play a crucial role in synaptic differentiation and transmission in a mouse model.52 While our Patient P96 presented with a mild adult-onset LG-CMS phenotype and harboured a homozygous missense variant p.V1563M in the plakin domain, the other known patient to date, who is of Serbian origin and harbours compound heterozygous variants p.T1799A and p.L2208F in the plakin domain and spectrin repeats, respectively, had a more severe early-onset phenotype.52 Although de novo heterozygous MACF1 variants, especially in the distal GAR domain, have previously been reported in patients with lissencephaly and complex brain malformations, the recessive CMS patients and family members did not have any brain abnormalities, suggesting a domain-specific phenotype expression.53 However, to our knowledge, no further patients with recessive variants in MACF1 have yet been reported, which restricts our ability to establish the exact phenotypic association and convincingly validate MACF1 as a CMS gene.
Glycosylation defects (GFPT1, DPAGT1 and GMPPB) represented the second largest category in our cohort and were causative in 30 families (21.3%). In previous larger studies, predominantly from Europe and the Americas, the frequency of glycosylation defects was comparatively lower, accounting for less than 10% of genetically diagnosed CMS.1,5,13-16 However, in the small cohort of 29 Chinese CMS families mentioned previously, Zhao et al.17 reported GFPT1 and GMPPB variants in eight and two families, respectively, accounting for 34.5% of the diagnosed CMS cases. This suggests the possibility of geographically and ethnically higher prevalence of glycosylation defects among Asian populations compared to those in European regions. Among the glycosylation genes, DPAGT1 was most common, with 13 unrelated patients having biallelic variants. This contrasts with previous cohorts in which GFPT1 has been the predominant glycosylation CMS defect.1,5,13,17 While a LG-CMS pattern is characteristic of all glycosylation CMS patients, DPAGT1 patients presented with a more severe and early onset phenotype with CNS abnormalities including seizures and intellectual disability reported in 7 of 13 patients (53.8%). GFPT1-CMS presented as a relatively milder LG-CMS phenotype with predominantly early-to-late childhood onset and no CNS abnormalities (Fig. 3D). All patients with GMPPB-CMS carried the identical Indian founder mutation p.D334N in homozygosity and displayed variable onset and a slowly progressive mixed phenotype of limb girdle muscular dystrophy (LGMD) with CMS as described in our previous publication.21 While our centre has previously diagnosed a number of patients with other recessive GMPPB variants who presented with a pure muscular dystrophy phenotype, the p.D334N variant is the only one in our cohort associated with CMS features and seems to be the only CMS-associated GMPPB mutation reported in Indian patients to date.
Defects in the synaptic basal lamina gene COLQ are also among the more commonly reported defects in previous cohorts with frequencies ranging from 6–14%, while in Israel and Turkey the reported frequency has been >30%.1,5,13,15,16 In our cohort, COLQ variants were identified in only six families (4.3%). Interestingly, the Brazilian cohort had only two COLQ cases out of 54 (3.8%), similar to our study, while the Chinese study reported COLQ in 13.8% of cases, similar to other larger studies from Europe.14,17 Early onset with a variably progressive course was characteristic of our COLQ patients, and four of six patients had respiratory defects (Fig. 3B).
While presynaptic gene defects are the least common cause of CMS worldwide, recessive CHAT variants form the majority of presynaptic cases reported to date, with a frequency of 2–5% reported in the European and Mayo Clinic cohorts.1,5,13 However, CHAT variants were not identified in any of our patients and CHAT-CMS is apparently extremely rare in the Indian subcontinent, with a complete lack of reported cases to our knowledge. Two recently identified rare CMS genes, VAMP1 and SLC25A1, were the only presynaptic genes in this cohort and are reported for the first time from India. We previously reported three unrelated children with severe congenital onset and characteristic severe hypotonia, feeding difficulties and respiratory distress identified to have homozygous loss-of-function variants in the VAMP1 gene, a component of the SNARE complex, essential for vesicular docking and fusion at NMJ (Fig. 3F).24 Two siblings with SLC25A1 were among the six patients reported in 2020 with a characteristic mild phenotype of fatigable weakness with variable involvement of ocular, proximal limb and bulbar muscles and mild intellectual disability in all cases. The p.R247Q variant in these siblings was reported as a recurrent CMS variant with no founder effect identified in four families from unrelated ethnicities including Europe and Asia.54 A further detailed genotype-phenotype description of individual CMS genes identified in this cohort can be found in the Supplementary material.
Taken together, these results suggest that, while CHRNE defects appear to be the most common cause of CMS worldwide, the frequencies of other gene defects vary markedly between different populations. These differences—and the increased prevalence of certain recessive CMS subtypes—can in part be attributed to the high frequency of consanguineous marriages in our source population, estimated in the 2010–14 census55 to be 23% in South India, from where most of our patients originate. A related factor is the prevalence of recurrent mutations with possible founder effects as discussed below. Variable severity, with phenotypes ranging from early death to late-onset mild presentations, as well as clinical presentations overlapping with other neuromuscular disorders, especially in patients with LG-CMS, may also lead to a certain level of ascertainment bias in CMS cohorts recruited exclusively from specialist neuromuscular clinics as in this cohort. These factors underscore the need for larger global studies to fully understand the ethnic and geographical distribution of CMS genetic subtypes.
‘Other’ congenital myasthenic syndrome genes
Among the CMS genes, we also included patients with recessive variants in DES and TEFM genes, which could not be classified into any of the four CMS subgroups. However, we consider DES and TEFM to be CMS genes based on the available functional evidence of NMJ defects and association with specific CMS phenotypes.12,56 In three unrelated patients from South India, Patients P143, P144 and P145, we identified a rare novel intronic homozygous variant in DES, c.1023+5G>A, which likely affects normal splicing of exon 5. While all three patients presented with a fairly uniform phenotype of childhood onset, gradually progressive limb-girdle weakness, variable oculo-bulbar involvement and elevated serum creatine kinase, two of the patients showed significant decrement on RNS, confirming the presence of a NMJ defect and resembling a LG-CMS phenotype. Muscle biopsy in one of the patients showed myopathic features and partial loss of desmin on immunostaining. While the parents were unaffected in all three cases, segregation and functional validation of the novel intronic variant is planned to investigate the possibility of a novel founder haplotype associated with a milder LG-CMS phenotype. CMS features have previously been reported in two recessive desmin null patients who had a severe combined phenotype of CMS, myopathy and cardiomyopathy.12 A recessive DES knock-in mouse model has been shown to have NMJ endplate pathology.12 While further functional characterization is necessary to understand the underlying NMJ pathophysiology, our patients further confirm the presence of a mixed myopathy-CMS phenotype specific to recessive DES variants, and we suggest that all recessive desminopathy patients should be evaluated for NMJ defects.
Searching for novel candidate genes in unsolved CMS patients led to the identification of a new CMS associated gene, TEFM, which encodes the mitochondrial transcription elongation factor.57 In two siblings (Patients P146 and P147), a homozygous missense variant p.P157A was identified in TEFM. Both siblings had childhood onset fatigable limb weakness with variably progressive ocular involvement (more severe in the younger sibling), accompanied by CNS involvement in the form of epilepsy in both siblings and ataxia with behavioural abnormalities in the elder sibling. Serum lactate elevation, positive decrement on RNS and biopsy showing mitochondrial respiratory chain complex I and IV deficiencies confirmed the mitochondrial disease together with the NMJ defect in the siblings. Additional patients with recessive TEFM defects were subsequently identified in other cohorts worldwide with variable neurological phenotypes and severity, suggesting a wide clinical spectrum. A knockdown zebrafish model showed abnormalities in mitochondrial function and the NMJ, further confirming the genotype-phenotype association and establishing TEFM as a novel mitochondrial disease gene with or without a CMS phenotype.56
‘Non-congenital myasthenic syndrome’ genes
We identified causative variants in non-CMS genes in nine clinically suspected CMS patients (6.4%). Of these nine unrelated patients, six were found to have recessive variants in known myopathy/muscular dystrophy genes including TTN, DYSF, CAPN3, TRAPPC11 and ADSSL1. All these patients presented with a predominantly limb-girdle phenotype with fatigability. Elevated serum creatine kinase and dystrophic features correlated with DYSF, TRAPPC11 and CAPN3 variants in Patients P151, P152 and P154, respectively. Patients P148 and P149 likely had compound heterozygous variants in TTN. The phenotype in both patients did not resemble characteristic recessive TTN myopathy or muscular dystrophy; instead, both presented with a LG-CMS phenotype with ocular involvement. Patient P153, who presented with congenital onset bulbar symptoms that improved over time, but subsequently developed gradually progressive proximo-distal fatigable limb weakness with muscle wasting and ocular symptoms, was found to have a homozygous missense variant in ADSSL1, p.G265E, previously reported in a Japanese myopathy patient. While initially reported in distal myopathy patients, recessive ADSSL1 variants are now attributed to a broader pathophenotype of nemaline myopathy characterized by proximo-distal limb involvement with prominent fatigability which can be attributed to defective ATP synthesis.58
Reverse phenotyping and segregation confirmed the genetic diagnosis in two non-CMS patients (Patients P155 and P156) with a predominantly ocular phenotype, with heteroplasmic MT-TL1 substitution (m.3243A>G) and heterozygous NF1 nonsense variant (p.R1241*), respectively. While the presence of prominent clinical fatigability prompted a provisional CMS diagnosis in all non-CMS patients, three of these patients—those with CAPN3 (Patient P154), TTN (Patient P148) and ADSSL1 (Patient P153) variants—also had significant decrement on RNS. A double-trouble scenario with a second undiscovered variant causing the CMS phenotype cannot be ruled out in these cases. Interestingly, in one of our patients, Patient P17, a double-trouble scenario was in fact observed, with the patient having both CHRNE and DYSF recessive variants, which correlated with the mixed pattern of ocular and limb-girdle phenotype along with significant decrement, high creatine kinase and biopsy-confirmed dysferlin loss. One patient, Patient P145, who presented with predominantly oculobulbar involvement with delayed motor milestones at 1 year of age, had a homozygous splice site variant (c.114+1G>T) in the ANK3 gene. Recessive loss-of-function variants in the ANK3 gene have previously been reported in patients with intellectual disability (OMIM:615493). A detailed neuropsychological assessment and RNS study was not completed for this patient due to missed follow-up. Although no association with either muscle or NMJ disease in humans has been reported to date, Drosophila Ank2, the closest homologue to ankyrin-G-producing human ANK3, is reported to be crucial for NMJ stabilization, and loss of presynaptic Ank2 can disrupt NMJ morphology.59 Based on the identification of similar cases with ANK3 variants, it may be necessary to further explore the role of ankyrin-G in NMJ dysfunction. Overall, although our cohort does not present sufficient grounds to reclassify these ‘non-CMS’ genes as new CMS genes, we consider that reporting such cases of clear NMJ involvement in genes where this has not previously been established is of significant value to understanding likely CMS mimics as well as identifying new pathways for NMJ dysfunction.
While we have made efforts to exclude possible autoimmune cases across the entire cohort, we acknowledge that autoimmune aetiology due to uncommon antibodies or seronegative myasthenia gravis, etc. cannot be ruled out conclusively in sporadic patients, especially in uncommon CMS and non-CMS genotypes.
Treatment response
In the present cohort, worsening or no response to pyridostigmine was seen predominantly in DOK7, MUSK and AGRN patients as well as those with SCCMS, but all showed improvement with salbutamol. Patients with glycosylation defects (GMPPB, GFPT1 and DPAGT1) responded well to combination therapy of pyridostigmine plus salbutamol. Among the new and rarer CMS genotypes in the cohort, while SLC25A1 patients did not show any response to either pyridostigmine or salbutamol, MACF1, VAMP1, TEFM and DES patients showed good initial response to pyridostigmine with added benefit from salbutamol (Table 1). Six patients with non-CMS variants also reported significant improvement with pyridostigmine and/or salbutamol, which further points towards a possible alternate NMJ pathomechanism in these patients. While treatment response was subjectively recorded based on patient feedback after at least 3 months of continuous intake of pyridostigmine and/or salbutamol, the lack of objective and long-term assessment of improvement is a limitation in this study.
Recurrent congenital myasthenic syndrome variants and diagnostic implication
We identified 22 recurrent disease-causing variants in our cohort (Table 4). Based on a comparison of MAFs from the gnomAD worldwide and South Asian population databases, eight recurrent variants appear to be predominantly specific to the South Asian region (Table 4). Unsurprisingly, the two most frequently recurring variants in our cohort were the common frameshift variants CHRNE p.E443Kfs*64 in 16 families (11.4%) and DOK7 p.A378Sfs*30 in 13 families (9.3%). CHRNE p.E443Kfs*64 (c.1327delG), with its high South Asian-specific gnomAD MAF of 0.087%, was originally discovered in the migrant Roma population in Europe13 and has already been established as a founder mutation predominantly affecting both the Roma population and the population from the Indian subcontinent. In contrast, the common DOK7 frameshift p.A378Sfs*30 (c.1124_1127dupTGCC) is more universally prevalent according to gnomAD, with a 0.07% worldwide MAF and 158 alleles in total, of which only nine came from South Asia. The CHRNE gene had nine recurrent variants in our cohort, of which we also identified three novel recurrent CHRNE variants including a frameshift p.L287Ffs*110, a nonsense p.Y243* and a 3′ splice site change c.235-1G>A. These were neither previously reported in CMS patients nor present in population or internal variant databases (Table 4). Our two patients (Patients P52 and P53) with the CHRNE variant p.L98P hail from the states of Jharkhand and West Bengal in the eastern part of India. Interestingly, the variant has previously been reported in patients from Bangladesh, which is adjacent to West Bengal and Jharkhand, suggesting a possible founder haplotype specific to the eastern region of the Indian subcontinent.22,35 This is also supported by gnomAD data in which only four control alleles were reported, all from Southeast Asia.
Apart from the expected common CHRNE and DOK7 variants, it is interesting to note that in our cohort, the other frequent variants were found in glycosylation genes, with three recurrent variants being found in DPAGT1 and GMPPB. Among these, a novel recurrent missense variant, p.T380I, located in DPAGT1 transmembrane helix 10, was found in nine unrelated patients exclusively in compound heterozygous state, seven of which harboured the previously reported helix 1 missense variant p.I29F on the second allele.42 We observed that the p.T380I/p.I29F genotype occurred only in patients from the eastern state of West Bengal, suggesting the possibility of founder haplotypes specific to Bengalis. The MAFs of c.1139C>T (p.T380I) and c.85A>T (p.I29F) in gnomAD also suggest a relative South Asian predominance at 0.03% and 0.08%, respectively, in the region (Table 4). Another recurrent recessive variant in GMPPB, p.D334N, is also exclusive to South Asia (South Asian MAF 0.06%) and was recently reported by us as a founder mutation causing a slowly progressive LGMD-CMS phenotype in 12 patients from seven consanguineous families of South Indian descent when present in homozygosity.21,60 Interestingly, the p.D334N variant has previously been reported in the compound heterozygous state in congenital muscular dystrophy patients from India and Pakistan.43,61 Overall, these recurrent variants accounted for 61% of the CMS families (86/141) in this cohort.
In this cohort, molecular testing of suspected CMS patients was performed either as part of standard diagnostic testing (predominantly NGS clinical exome panels in commercial labs) or a two-step research process with hotspot screening followed by WES. The overall diagnostic rate was 71.6%, as 156 patients (141 families) were genetically diagnosed out of 213 (197 families) suspected clinically as having CMS (Fig. 1). Based on the two-step molecular testing we employed in the ‘research’ sub-cohort (104 families), hotspot screening of four previously reported common variants in CHRNE, DOK7 and RAPSN yielded a variant detection rate of 14.4% (15 families) with an additional five LG-CMS cases in which a single heterozygous DOK7 variant was identified. The RAPSN hotspot variant p.N88K commonly reported in European patients was not identified in any of our patients. While p.N88K has been associated with an ancient Indo-European haplotype and may not be entirely absent in India, given individual previous reports of Indian RAPSN patients harbouring the mutation, the prevalence of genetically confirmed RAPSN patients in India as reported in the literature is low overall.22,50 This coincides with variant frequencies obtained from Asian cohorts in population databases in which the p.N88K variant is either absent or present at a much lower prevalence (Supplementary material). In particular, the gnomAD prevalence of p.N88K is 3.5-fold lower in the South Asian population (0.075%) compared with the European population (0.26%), which underscores the variability of genotypic patterns between different populations and suggests that the variations seen in this cohort cannot exclusively be ascribed to ascertainment bias, although that may still be a factor as noted above.
The mean delay from onset to molecular diagnosis was 12.5 ± 9.9 (0–49 years) for CMS patients in this cohort. While severe presynaptic VAMP1 cases had a shorter average diagnostic delay of 2.5 ± 2.1 years, patients affected by other common CMS subtypes had to wait a decade or more on average to receive diagnostic confirmation. The diagnostic delay was especially long (41.7 ± 9.4 years) for GMPPB patients in this cohort, predominantly due to the delayed clinical diagnosis of their complicated LGMD-CMS phenotype. By including the recurrent variants identified in this study, we anticipate that the diagnostic rate can be increased significantly through cost-effective mutational screening, which will in turn reduce the diagnostic delay by enabling screening even in smaller centres without access to expensive NGS facilities. Given the high frequency of consanguineous and intra-community marriages in various regions, such recurrent variants with population-specific founder haplotypes may also contribute to the different prevalence of recessive CMS genotypes in India as seen in genes like DPAGT1 and GMPPB in the present cohort. Based on these findings, we suggest that screening for common variants is likely to be most effective when tailored to population-specific founder effects.
Conclusion
This uniquely large CMS cohort from a single quaternary neurology centre in South India provides crucial insights into the mutational spectrum and prevalence of individual CMS subtypes in India with an in-depth genotype-phenotype correlation. The clear differences identified in the mutational spectrum and relative frequencies of defects in specific genes in this Indian cohort compared to previous CMS cohorts emphasises the need for additional global large-scale genetic studies to better capture these regional variations in terms of prevalence. As our cohort consists predominantly of south Indian patients, we are limited in our ability to assess true frequencies across the whole Indian subcontinent, and broader population studies would be of value even within this region. In our study, exome-wide analysis not only led to the identification of novel CMS-causing genes including TEFM and MACF1 but also further expanded global knowledge of rare genotype associations including DES, GMPPB, VAMP1 and SLC25A1 in CMS. The finding of gradually progressive and treatment-responsive LG-CMS phenotypes in the known myopathy and muscular dystrophy genes DES and GMPPB expand the phenotype spectrum and suggest a hidden NMJ dysfunction mechanism in previously known NMD genes. The higher frequency of glycosylation CMS patients in our cohort suggests not only that this CMS subgroup may be more prevalent in India but also that many such patients with this limb-girdle CMS phenotype might have remained undiagnosed or misdiagnosed earlier due to overlapping clinical features, and this may be true in other cohorts as well. While CMS is often a clinical diagnosis, muscle fatigability can often be confounding especially in other myopathy and LGMD cases necessitating faster molecular diagnosis. Identification and screening of CMS-associated variants in suspected LGMD/myopathy patients is important to enable prompt initiation of treatment, which can improve fatigable weakness and delay progression of disability. This study also identifies 22 unique recurrent disease-causing CMS variants which account for about two-thirds of patients. In highly consanguineous populations like India, the identification of recurrent and founder disease-causing variants may be a valuable and cost-effective strategy enabling early diagnostic screening.
Supplementary Material
Acknowledgements
Sequencing was provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG). Data were analysed using the RD-Connect Genome-Phenome Analysis platform.
Contributor Information
Kiran Polavarapu, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India; Children’s Hospital of Eastern Ontario Research Institute, Ottawa, ON K1H 8L1, Canada.
Balaraju Sunitha, John Walton Muscular Dystrophy Research Centre, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne NE1 3BZ, UK; Department of Clinical Neurosciences, University of Cambridge School of Clinical Medicine, Cambridge CB2 0SP, UK; Division of Cardiovascular Medicine, Radcliffe Department of Medicine, University of Oxford, Oxford OX3 9DU, UK.
Ana Töpf, John Walton Muscular Dystrophy Research Centre, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne NE1 3BZ, UK.
Veeramani Preethish-Kumar, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India; Department of Neurology, Neurofoundation, Salem, Tamil Nadu 636009, India.
Rachel Thompson, Children’s Hospital of Eastern Ontario Research Institute, Ottawa, ON K1H 8L1, Canada.
Seena Vengalil, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Saraswati Nashi, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Mainak Bardhan, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Sai Bhargava Sanka, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Akshata Huddar, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India; Department of Neurology, St Johns Medical College Hospital, Bangalore 560034, India.
Gopikrishnan Unnikrishnan, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India; Department of Neurology, Amruta Institute of Medical Sciences, Kochi 682041, India.
Gautham Arunachal, Department of Human Genetics, National Institute of Mental Health and Neurosciences, Bengaluru 560029, India.
Manu Santhappan Girija, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Anna Porter, John Walton Muscular Dystrophy Research Centre, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne NE1 3BZ, UK.
Yoshiteru Azuma, John Walton Muscular Dystrophy Research Centre, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne NE1 3BZ, UK.
Paulo José Lorenzoni, Neuromuscular Disorders Division, Service of Neurology, Department of Internal Medicine, Hospital de Clínicas, Universidade Federal do Paraná, Rua General Carneiro, Curitiba - PR 80060-900, Brazil.
Dipti Baskar, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Ram Murthy Anjanappa, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Madassu Keertipriya, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Hansashree Padmanabh, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Ganaraja Valakunja Harikrishna, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Steve Laurie, Centro Nacional de Análisis Genómico (CNAG-CRG), Center for Genomic Regulation, Barcelona Institute of Science and Technology (BIST), Barcelona, Catalonia 08028, Spain.
Leslie Matalonga, Centro Nacional de Análisis Genómico (CNAG-CRG), Center for Genomic Regulation, Barcelona Institute of Science and Technology (BIST), Barcelona, Catalonia 08028, Spain.
Rita Horvath, Department of Clinical Neurosciences, University of Cambridge School of Clinical Medicine, Cambridge CB2 0SP, UK.
Atchayaram Nalini, Department of Neurology, National Institute of Mental Health and Neurosciences, Bangalore 560029, India.
Hanns Lochmüller, Children’s Hospital of Eastern Ontario Research Institute, Ottawa, ON K1H 8L1, Canada; Centro Nacional de Análisis Genómico (CNAG-CRG), Center for Genomic Regulation, Barcelona Institute of Science and Technology (BIST), Barcelona, Catalonia 08028, Spain; Brain and Mind Research Institute, University of Ottawa, Ottawa, ON K1H 8M5, Canada; Division of Neurology, Department of Medicine, The Ottawa Hospital, Ottawa, ON K1H 8M5, Canada; Department of Neuropediatrics and Muscle Disorders, Medical Center–University of Freiburg, Faculty of Medicine, Freiburg 79110, Germany.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material. Genomic and phenotypic data for all research cohort cases analysed are available to registered users through the secure-access RD-Connect GPAP.
Funding
Broad Center for Mendelian Genomics was funded by the National Human Genome Research Institute, the National Eye Institute and the National Heart, Lung and Blood Institute grant UM1 HG008900 and in part by National Human Genome Research Institute grant R01 HG009141. The RD-Connect Genome-Phenome Analysis platform was developed under the RD-Connect project funded from 2007–2013 under the European Union’s Seventh Framework Programme (FP7) (grant agreement no. 305444) and with additional funding from the European Union’s Horizon 2020 European Joint Programme on Rare Diseases (grant agreement no. 825575), the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 779257: Solve-RD) and institutional funding from Instituto Nacional de Bioinformática and ELIXIR ES. H.L. receives support from the Canadian Institutes of Health Research (CIHR) for Foundation Grant FDN-167281 (Precision Health for Neuromuscular Diseases), Transnational Team Grant ERT-174211 (ProDGNE) and Network Grant OR2-189333 (NMD4C), the Canada Foundation for Innovation (CFI-JELF 38412), the Canada Research Chairs program (Canada Research Chair in Neuromuscular Genomics and Health, 950-232279), the European Union's Horizon Europe programme (grant agreement no. 101080249) and the Canada Research Coordinating Committee New Frontiers in Research Fund (NFRFG-2022-00033) for SIMPATHIC, and from the Government of Canada's Canada First Research Excellence Fund (CFREF) for the Brain-Heart Interconnectome (2022-00007). K.P. holds a Canadian Institutes of Health Research postdoctoral fellowship award under grant no. MFE-491707. R.T. holds a Canadian Institutes of Health Research postdoctoral fellowship award under grant no. MFE-171275. B.S. was a Newton International postdoctoral fellow; the work was funded by the Newton fund (UK–NIF003\1002). A.T. has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 779257 (Solve-RD).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Engel AG, Shen XM, Selcen D, Sine SM. Congenital myasthenic syndromes: Pathogenesis, diagnosis, and treatment. Lancet Neurol. 2015;14:420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McMacken G, Abicht A, Evangelista T, Spendiff S, Lochmüller H. The increasing genetic and phenotypical diversity of congenital myasthenic syndromes. Neuropediatrics. 2017;48:294–308. [DOI] [PubMed] [Google Scholar]
- 3. Ramdas S, Beeson D. Congenital myasthenic syndromes: Where do we go from here? Neuromuscul Disord. 2021;31:943–954. [DOI] [PubMed] [Google Scholar]
- 4. Parr JR, Andrew MJ, Finnis M, Beeson D, Vincent A, Jayawant S. How common is childhood myasthenia? The UK incidence and prevalence of autoimmune and congenital myasthenia. Arch Dis Child. 2014;99:539–542. [DOI] [PubMed] [Google Scholar]
- 5. Natera-de Benito D, Töpf A, Vilchez JJ, et al. . Molecular characterization of congenital myasthenic syndromes in Spain. Neuromuscul Disord. 2017;27:1087–1098. [DOI] [PubMed] [Google Scholar]
- 6. Schuler BA, Nelson ET, Koziura M, Cogan JD, Hamid R, Phillips JA. Lessons learned: Next-generation sequencing applied to undiagnosed genetic diseases. J Clin Invest. 2022;132:e154942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Finsterer J. Congenital myasthenic syndromes. Orphanet J Rare Dis. 2019;14:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen E, Bonne G, Rivier F, Hamroun D. The 2022 version of the gene table of neuromuscular disorders (nuclear genome). Neuromuscul Disord. 2021;31:1313–1357. [DOI] [PubMed] [Google Scholar]
- 9. Illingworth MA, Main M, Pitt M, et al. . RYR1-related congenital myopathy with fatigable weakness, responding to pyridostigimine. Neuromuscul Disord. 2014;24:707–712. [DOI] [PubMed] [Google Scholar]
- 10. Munot P, Lashley D, Jungbluth H, et al. . Congenital fibre type disproportion associated with mutations in the tropomyosin 3 (TPM3) gene mimicking congenital myasthenia. Neuromuscul Disord. 2010;20:796–800. [DOI] [PubMed] [Google Scholar]
- 11. Gibbs EM, Clarke NF, Rose K, et al. . Neuromuscular junction abnormalities in DNM2-related centronuclear myopathy. J Mol Med Berl Ger. 2013;91:727–737. [DOI] [PubMed] [Google Scholar]
- 12. Durmuş H, Ayhan Ö, Çırak S, et al. . Neuromuscular endplate pathology in recessive desminopathies: Lessons from man and mice. Neurology. 2016;87:799–805. [DOI] [PubMed] [Google Scholar]
- 13. Abicht A, Dusl M, Gallenmüller C, et al. . Congenital myasthenic syndromes: Achievements and limitations of phenotype-guided gene-after-gene sequencing in diagnostic practice: A study of 680 patients. Hum Mutat. 2012;33:1474–1484. [DOI] [PubMed] [Google Scholar]
- 14. Estephan EP, Zambon AA, Thompson R, et al. . Congenital myasthenic syndrome: Correlation between clinical features and molecular diagnosis. Eur J Neurol. 2022;29:833–842. [DOI] [PubMed] [Google Scholar]
- 15. Yiş U, Becker K, Kurul SH, et al. . Genetic landscape of congenital myasthenic syndromes from Turkey: Novel mutations and clinical insights. J Child Neurol. 2017;32:759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aharoni S, Sadeh M, Shapira Y, et al. . Congenital myasthenic syndrome in Israel: Genetic and clinical characterization. Neuromuscul Disord. 2017;27:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao Y, Li Y, Bian Y, et al. . Congenital myasthenic syndrome in China: Genetic and myopathological characterization. Ann Clin Transl Neurol. 2021;8:898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maramattom BV, Patil R, Thomas J. Familial dok7 congenital myasthenic syndrome responsive to salbutamol. Neurol India. 2014;62:313. [DOI] [PubMed] [Google Scholar]
- 19. Khadilkar S, Bhutada A, Nallamilli B, Hegde M. Limb girdle weakness responding to salbutamol: An Indian family with DOK7 mutation. Indian Pediatr. 2015;52:243–244. [DOI] [PubMed] [Google Scholar]
- 20. Padmanabha H, Saini AG, Sankhyan N, Singhi P. COLQ-Related Congenital myasthenic syndrome and response to salbutamol therapy. J Clin Neuromuscul Dis. 2017;18:162–163. [DOI] [PubMed] [Google Scholar]
- 21. Polavarapu K, Mathur A, Joshi A, et al. . A founder mutation in the GMPPB gene [c.1000G>A (p.Asp334Asn)] causes a mild form of limb-girdle muscular dystrophy/congenital myasthenic syndrome (LGMD/CMS) in South Indian patients. Neurogenetics. 2021;22:271–285. [DOI] [PubMed] [Google Scholar]
- 22. Selvam P, Arunachal G, Danda S, et al. . Congenital myasthenic syndrome: Spectrum of mutations in an Indian cohort. J Clin Neuromuscul Dis. 2018;20:14–27. [DOI] [PubMed] [Google Scholar]
- 23. Wadwekar V, Nair SS, Tandon V, Kuruvilla A, Nair M. Congenital myasthenic syndrome: Ten years clinical experience from a quaternary care south-Indian hospital. J Clin Neurosci. 2020;72:238–243. [DOI] [PubMed] [Google Scholar]
- 24. Polavarapu K, Vengalil S, Preethish-Kumar V, et al. . Recessive VAMP1 mutations associated with severe congenital myasthenic syndromes–A recognizable clinical phenotype. Eur J Paediatr Neurol. 2021;31:54–60. [DOI] [PubMed] [Google Scholar]
- 25. Abicht A, Stucka R, Karcagi V, et al. . A common mutation (epsilon1267delG) in congenital myasthenic patients of Gypsy ethnic origin. Neurology. 1999;53:1564–1569. [DOI] [PubMed] [Google Scholar]
- 26. Ohno K, Anlar B, Özdirim E, Brengman JM, DeBleecker JL, Engel AG. Myasthenic syndromes in Turkish kinships due to mutations in the acetylcholine receptor. Ann Neurol. 1998;44:234–241. [DOI] [PubMed] [Google Scholar]
- 27. Müller JS, Mildner G, Müller-Felber W, et al. . Rapsyn N88K is a frequent cause of congenital myasthenic syndromes in European patients. Neurology. 2003;60:1805–1810. [DOI] [PubMed] [Google Scholar]
- 28. Hiz Kurul S, Oktay Y, Töpf A, et al. . High diagnostic rate of trio exome sequencing in consanguineous families with neurogenetic diseases. Brain. 2022;145:1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laurie S, Piscia D, Matalonga L, et al. . The RD-connect genome-phenome analysis platform: Accelerating diagnosis, research, and gene discovery for rare diseases. Hum Mutat. 2022;43:717–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.Org: Online Mendelian inheritance in man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(Database issue):D789–D798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abicht A, Müller JS, Lochmüller H. Congenital myasthenic syndromes overview. In: Adam MP, Everman DB, Mirzaa GM, et al., eds. Genereviews®. University of Washington; 1993. [Google Scholar]
- 32. Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Richards S, Aziz N, Bale S, et al. . Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Croxen R, Young C, Slater C, et al. . End-plate γ- and Ɛ-subunit mRNA levels in AChR deficiency syndrome due to Ɛ-subunit null mutations. Brain. 2001;124:1362–1372. [DOI] [PubMed] [Google Scholar]
- 35. Croxen R, Hatton C, Shelley C, et al. . Recessive inheritance and variable penetrance of slow-channel congenital myasthenic syndromes. Neurology. 2002;59:162–168. [DOI] [PubMed] [Google Scholar]
- 36. Kraner S, Burgunder JM, Rösler KM, Steinlein OK, Sieb JP. Congenital myasthenic syndrome due to heteroallelic nonsense/missense mutations in the acetylcholine receptor epsilon subunit gene. Eur J Neurol. 2002;9:694–695. [DOI] [PubMed] [Google Scholar]
- 37. Ganapathy A, Mishra A, Soni MR, et al. . Multi-gene testing in neurological disorders showed an improved diagnostic yield: Data from over 1000 Indian patients. J Neurol. 2019;266:1919–1926. [DOI] [PubMed] [Google Scholar]
- 38. Wargon I, Richard P, Kuntzer T, et al. . Long-term follow-up of patients with congenital myasthenic syndrome caused by COLQ mutations. Neuromuscul Disord. 2012;22:318–324. [DOI] [PubMed] [Google Scholar]
- 39. Guo Y, Menezes MJ, Menezes MP, et al. . Delayed diagnosis of congenital myasthenia due to associated mitochondrial enzyme defect. Neuromuscul Disord. 2015;25:257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beeson D, Higuchi O, Palace J, et al. . Dok-7 mutations underlie a neuromuscular junction synaptopathy. Science. 2006;313:1975–1978. [DOI] [PubMed] [Google Scholar]
- 41. Ben Ammar A, Petit F, Alexandri N, et al. . Phenotype genotype analysis in 15 patients presenting a congenital myasthenic syndrome due to mutations in DOK7. J Neurol. 2010;257:754–766. [DOI] [PubMed] [Google Scholar]
- 42. Iqbal Z, Shahzad M, Vissers LELM, et al. . A compound heterozygous mutation in DPAGT1 results in a congenital disorder of glycosylation with a relatively mild phenotype. Eur J Hum Genet. 2013;21:844–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carss KJ, Stevens E, Foley AR, et al. . Mutations in GDP-mannose pyrophosphorylase B cause congenital and limb-girdle muscular dystrophies associated with hypoglycosylation of α-dystroglycan. Am J Hum Genet. 2013;93:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Owen D, Töpf A, Preethish-Kumar V, et al. . Recessive variants of MuSK are associated with late onset CMS and predominant limb girdle weakness. Am J Med Genet A. 2018;176:1594–1601. [DOI] [PubMed] [Google Scholar]
- 45. Lee M, Beeson D, Palace J. Therapeutic strategies for congenital myasthenic syndromes. Ann N Y Acad Sci. 2018;1412:129–136. [DOI] [PubMed] [Google Scholar]
- 46. Thompson R, Abicht A, Beeson D, et al. . A nomenclature and classification for the congenital myasthenic syndromes: Preparing for FAIR data in the genomic era. Orphanet J Rare Dis. 2018;13:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shen XM, Okuno T, Milone M, et al. . Mutations causing slow-channel myasthenia reveal that a valine ring in the channel pore of muscle AChR is optimized for stabilizing channel gating. Hum Mutat. 2016;37:1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sine SM, Ohno K, Bouzat C, et al. . Mutation of the acetylcholine receptor α subunit causes a slow-channel myasthenic syndrome by enhancing agonist binding affinity. Neuron. 1995;15:229–239. [DOI] [PubMed] [Google Scholar]
- 49. Burke G, Cossins J, Maxwell S, et al. . Rapsyn mutations in hereditary myasthenia: Distinct early- and late-onset phenotypes. Neurology. 2003;61:826–828. [DOI] [PubMed] [Google Scholar]
- 50. Muller J, Abicht A, Burke G, et al. . The congenital myasthenic syndrome mutation RAPSN N88K derives from an ancient indo-European founder. J Med Genet. 2004;41:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khadilkar SV, Oza H, Dhonde P, et al. . Congenital myasthenic syndromes: A study of 15 cases. Neurol Clin Neurosci. 2023;11:127–133. [Google Scholar]
- 52. Oury J, Liu Y, Töpf A, et al. . MACF1 Links Rapsyn to microtubule- and actin-binding proteins to maintain neuromuscular synapses. J Cell Biol. 2019;218:1686–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dobyns WB, Aldinger KA, Ishak GE, et al. . MACF1 Mutations encoding highly conserved zinc-binding residues of the GAR domain cause defects in neuronal migration and axon guidance. Am J Hum Genet. 2018;103:1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Balaraju S, Töpf A, McMacken G, et al. . Congenital myasthenic syndrome with mild intellectual disability caused by a recurrent SLC25A1 variant. Eur J Hum Genet. 2020;28:373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sahoo H, Debnath P, Mandal C, Nagarajan R, Appunni S. Changing trends of consanguineous marriages in South India. J Asian Afr Stud. 2022;57:209–225. [Google Scholar]
- 56. Van Haute L, O’Connor E, Díaz-Maldonado H, et al. . TEFM Variants impair mitochondrial transcription causing childhood-onset neurological disease. Nat Commun. 2023;14:1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Minczuk M, He J, Duch AM, et al. . TEFM (C17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39:4284–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saito Y, Nishikawa A, Iida A, et al. . ADSSL1 Myopathy is the most common nemaline myopathy in Japan with variable clinical features. Neurology. 2020;95:e1500–e1511. [DOI] [PubMed] [Google Scholar]
- 59. Pielage J, Cheng L, Fetter RD, Carlton PM, Sedat JW, Davis GW. A presynaptic giant ankyrin stabilizes the NMJ through regulation of presynaptic microtubules and transsynaptic cell adhesion. Neuron. 2008;58:195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Siddiqui S, Polavarapu K, Bardhan M, et al. . Distinct and recognisable muscle MRI pattern in a series of adults harbouring an identical GMPPB gene mutation. J Neuromuscul Dis. 2022;9:95–109. [DOI] [PubMed] [Google Scholar]
- 61. Belaya K, Rodríguez Cruz PM, Liu WW, et al. . Mutations in GMPPB cause congenital myasthenic syndrome and bridge myasthenic disorders with dystroglycanopathies. Brain. 2015;138:2493–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material. Genomic and phenotypic data for all research cohort cases analysed are available to registered users through the secure-access RD-Connect GPAP.