Abstract
The lysogenic bacteriophage MAV1, which is associated with the arthritogenicity of Mycoplasma arthritidis, was characterized. Several strains of M. arthritidis were examined for their ability to support growth of MAV1. A PFU assay was developed, and the sensitivity of phage to various chemical treatments was assayed. The most notable result was the resistance of MAV1 to proteinase K. The MAV1 genome is a double-stranded, linear DNA molecule of about 16 kb. The site of MAV1 DNA integration in the host chromosome was investigated. The ends of MAV1 DNA were cloned from three independent lysogens shown to have MAV1 DNA inserted at different sites in the host. The nucleotide sequences of the ends of the MAV1 genome and of the MAV1 DNA-chromosomal DNA junctions from each of three lysogens were determined. Sequences flanking the integrated prophage and the ends of native MAV1 DNA were determined, allowing the identification of the phage DNA (attP) and bacterial DNA (attB) recombination sites. Analysis of the left MAV1 DNA-chromosomal DNA junction sites showed a single-base heterogeneity located within MAV1 DNA sequences immediately adjacent to the attB sequence. A model for MAV1 integration-excision is proposed.
Mycoplasmas are members of the class Mollicutes and have the distinction of being the smallest organisms capable of self-replication. They are parasites and pathogens of plants, insects, and animals, including humans. Due to close parasite-host relationships, mycoplasmas have streamlined genomes with limited biosynthetic machinery and are dependent on their hosts for many of their nutritional requirements. The mycoplasma genome has been altered further during evolution by the conversion of the universal stop codon UGA into a codon for tryptophan (16).
While mycoplasmas are highly evolved parasites, they are not immune to being parasitized themselves. Extrachromosomal elements have been found within Mycoplasma species and several other genera within the class Mollicutes. The number of plasmids and viruses characterized for the mycoplasmas, however, is extremely small. Mycoplasma plasmids are very rare, having been found only within one species, Mycoplasma mycoides subsp. mycoides (2, 5). These plasmids are cryptic, circular, double-stranded DNA molecules of about 2 kb. Mycoplasma viruses have been identified for four species, Mycoplasma arthritidis (phage MAV1) (14), Mycoplasma bovirhinis (Br1) (7), Mycoplasma hyorhinis (Hr1) (8), and Mycoplasma pulmonis (P1) (6). Morphologically, they are nonenveloped viruses with either long (Br1) or short tails (Hr1 and P1). Except for phage P1, the nucleic acids of these phages have not been characterized, which has left them unclassified (9). P1 has a linear double-stranded DNA genome of 11.3 kb with protein covalently attached to the 5′ ends (17). Virulent and temperate viruses have been characterized within the Acholeplasma and Spiroplasma genera of the class Mollicutes. The virulent life cycle of these phages is usually not lytic since progeny phage particles bud through the cell membrane in a nonlytic manner similar to Ff phages and animal viruses.
Previously, we have shown that the M. arthritidis bacteriophage, MAV1, is required for the arthritis caused by this mycoplasma (14). Because very little is known about phages from the genus Mycoplasma and the significant role of MAV1 in the pathogenesis of M. arthritidis, we have further characterized this bacteriophage.
MATERIALS AND METHODS
Culture medium and bacterial strains.
M. arthritidis strains used for these studies have been described previously (14). Cultures were grown in EB medium consisting of 2% PPLO broth without crystal violet (Difco Laboratories, Detroit, Mich.) supplemented with 7.5% heat-inactivated horse serum (Gibco, Gaithersburg, Md.), 5% freshly prepared yeast extract, 1% PPLO serum fraction (Difco), 1% arginine-HCl (Sigma Chemical Co., St. Louis, Mo.), 0.5% IsoVitaleX (Fisher, Pittsburgh, Pa.), 0.02% DNA (degraded free acid type IV; Sigma), and 100 μg of ampicillin per ml. Solid medium, designated EA, consisted of EB medium supplemented with 1.4% agar. Cultures were grown at 37°C until late log phase.
Bacteriophage preparation.
Stocks of MAV1 were prepared by mixing 100 μl of MAV1 (106 PFU/ml) with 500 μl of log-phase M. arthritidis PG61 and were incubated at 37°C for 45 min. The infected-cell culture was added to 3 ml of precooled top agar, poured onto prewarmed EA plates, and incubated at 37°C until confluent lysis of the cell lawn was achieved. The top agar was scraped off, 2 ml of EB medium was added, 100 μl of chloroform was added, and the mixture was vortexed and incubated at 4°C for 4 h. The phage mixture was centrifuged at 8,000 × g for 10 min at 4°C, and the supernatant (representing the phage stock) was collected, filtered through a 0.2-μm-pore-size Acrodisc syringe filter (Gelman Sciences, Ann Arbor, Mich.), and stored at 4°C.
Host range and PFU assay.
The host range of MAV1 was screened by a megaplaque assay (12). Briefly, 3 ml of EB medium was inoculated with 30 μl of host strain and incubated at 37°C until mid-log to late log phase. Host cells (50 to 200 μl) were added to 1.5 ml of top agar (0.91% NaCl, 0.606% Tris-HCl [pH 7.0], 0.7% Select agar [Gibco]), which was cooled to 42°C, vortexed, and poured onto a prewarmed EA plate. Immediately, 10 μl of MAV1 phage stock (109 PFU/ml) was spotted onto the center of the plate before the top agar hardened. The plates were incubated at 37°C for 24 to 48 h and examined for the presence of a large zone of clearing (megaplaque) in the center of the plate, which was indicative of bacteriophage infection. A PFU assay was used to quantitate MAV1 titers. Samples to be screened were serially diluted 10-fold in 100 μl of EB medium. For each dilution, 10 μl was added to 50 to 200 μl of host cells and the mixture was incubated at 37°C for 30 min. The mixture was added to 1.5 ml of top agar, vortexed, and poured onto prewarmed EA plates. The plates were incubated at 37°C overnight and examined for plaques.
Isolation of phage nucleic acid.
MAV1 DNA was isolated by mixing 1.0 ml of phage stock (≥109 PFU/ml) with 333 μl of PEG solution (30% polyethylene glycol, 1.6 M NaCl) and incubated at room temperature for 1 h. The sample was centrifuged at 16,000 × g for 10 min, and the supernatant was discarded. The phage pellet was suspended in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Triton X-100 and MgCl2 were added to final concentrations of 1% and 10 mM, respectively. DNase I was added to a final concentration of 100 μg/ml, and the sample was incubated at 37°C for 1 h. The lysate was extracted twice with buffered phenol (pH 8.0), once with phenol-chloroform (1:1), and once with chloroform. Phage DNA was ethanol precipitated, suspended in 50 μl of TE buffer, and stored at −20°C. All DNA manipulations, restriction endonuclease digestion, and agarose gel electrophoresis were performed by using standard procedures (1).
Chemical and enzymatic treatments.
Phage MAV1 was subjected to treatment with the following substances at 37°C for 30 min: chloroform (50%), sodium dodecyl sulfate (1%), Triton X-100 (1%), and DNase I (100 μg/ml; Sigma) supplemented with 10 mM MgCl2, RNase A (100 μg/ml; Sigma), trypsin (100 μg/ml; Sigma), and proteinase K (100 μg/ml; Sigma). The titer of treated phage was compared with that of MAV1 that had been incubated in EB medium only.
Sequencing and PCR amplification.
The nucleotide sequence was determined by using double-stranded DNA or PCR products purified with a Wizard PCR preparation kit (Promega, Madison, Wis.) as a template and automated fluorescent dye terminator methods at the Sequencing Core Facility, University of Alabama at Birmingham. Primers used in sequencing and PCR were purchased from Genosys Biotechnologies, Inc. (The Woodlands, Tex.). PCR was performed in 50-μl reaction volumes with 200 ng of M. arthritidis genomic DNA, 25 pmol of each primer, 200 mM (each) deoxynucleoside triphosphates, 1.5 mM MgCl2, 1× Taq polymerase buffer, and 2.5 U of Taq polymerase (Promega). Amplification conditions consisted of a 1-min denaturation at 94°C, a 1-min annealing at 55°C, and a 1-min extension at 72°C for 30 cycles, followed by a 5-min extension at 72°C.
Nucleotide sequence accession numbers.
The nucleotide sequences of the left MAV1 DNA-chromosomal DNA junctions designated attL, the right MAV1 DNA-chromosomal DNA junctions designated attR, the attB site of MAV1 lysogens, and the attP site of MAV1 have been deposited in GenBank under accession nos. AF058272, AF058273, AF058274, AF058275, AF058276, AF058277, AF058278, AF058279, AF058280, and AF058281.
RESULTS AND DISCUSSION
Host range of MAV1.
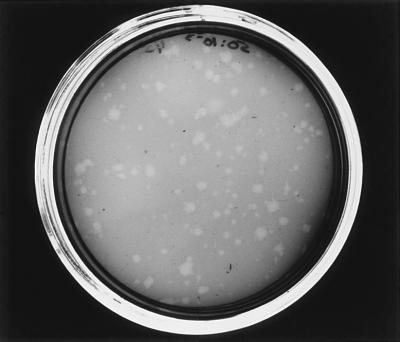
Several strains of M. arthritidis were examined for their ability to support growth of MAV1. When tested with a megaplaque assay, MAV1 produced a large zone of clearing on strains 1581, PG61, KD19611, and KDTru. Plaques were not produced by MAV1 on strains H6061, 13988, 19611, Tru, 14152*, 23192, H39, and 07, indicating that these strains are not capable of supporting phage growth. Strain PG61 was chosen for further studies and the development of a PFU assay. The optimal preincubation time for attachment of phage to host cells was found by mixing phage with PG61 cells, incubating at 37°C for various lengths of time, and assaying for PFU. MAV1 titers reached a plateau when the phage was incubated with host cells for 30 to 45 min before assaying for PFU. Lysogenic bacteriophages produce turbid plaques, which would be expected for MAV1 because MAV1 DNA has been shown to integrate into the host chromosome. Plaques produced by MAV1 appeared clear to the naked eye (Fig. 1), but individual colonies could be seen within the zone of clearing when examined with an inverted microscope. Strains 14124, 158p10, and 158p10p9 were shown previously to contain one or more copies of MAV1 DNA in their chromosomes (14). Plaques were not seen on any of these strains when the megaplaque assay was used. Similarly, plaques were not observed on lysogens previously constructed by infecting strain 158 with MAV1, showing that strains infected with MAV1 are immune to superinfection. Infection of PG61 with naked MAV1 DNA also did not produce plaques.
FIG. 1.
MAV1 plaques on a lawn of M. arthritidis PG61. The plate was stained with Dienes (Difco) for photographic purposes.
The inability to infect nonlysogenic strains may be due to the absence of a specific receptor required for phage attachment or the presence of a restriction-modification system that degrades MAV1 DNA upon injection into the host cell. The presence of a restriction-modification system (MarI) that is an isoschizomer of AluI has been described for M. arthritidis (13). However, some strains with this restriction system (1581 and PG61) are capable of supporting MAV1 growth while others lacking it (H606 and 13988) are not. If a restriction-modification system is responsible for the inability of MAV1 to infect some strains, it would have to be due to an additional uncharacterized system present in some strains but not all.
Productive infection versus lysogeny.
Although MAV1 was readily propagated on agar, no conditions that could reproducibly cause MAV1-infected cells to undergo productive infection in broth were found. Therefore, it was not possible to perform some basic experiments such as one-step growth curve assays. Broth infections resulted in essentially all CFU becoming lysogens with little, if any, detectable release of progeny phage. Attempts to induce lysogens by using mitomycin C were unsuccessful (6a).
Chemical and enzymatic treatment of MAV1.
The sensitivity of MAV1 to a variety of chemical and enzymatic treatments was examined. MAV1 infectivity was unaffected by treatment with DNase I, RNase A, chloroform, Triton X-100, trypsin, or proteinase K. The resistance of bacteriophages to proteinase K has been described in only two instances in the literature. One of these is the report on phage SpV4 from Spiroplasma citri, which is also a member of the class Mollicutes (11). Lack of the proper controls and possibly an insufficient amount of proteinase K bring into question whether SpV4 is truly resistant. The other case comes from the characterization of polysheaths from Alcaligenes eutrophus H16 (15). Because these are not functional phage particles, it cannot be ascertained whether this is an anomalous association with mutant phage forms or true resistance. We repeated the proteinase K treatment of MAV1 by using a second phage for an internal control. M. pulmonis virus P1 was chosen because its host will not grow in medium used to propagate M. arthritidis. This allowed a mixture of MAV1, P1, and proteinase K to be incorporated into a single reaction mixture. Because both phages can be propagated only on their respective hosts, their titers could be accurately determined without interference from each other. Incubation with proteinase K reduced the titer of P1 virus by greater than 99% while the titer of MAV1 was unchanged.
MAV1 infectivity was resistant to chloroform and nonionic detergents, suggesting that the phage is bound by protein and lacks lipid. Resistance to nuclease treatment and sensitivity to SDS confirmed that infectious particles are not naked nucleic acid and most likely contain a protein coat. Many phages are resistant to specific proteases such as trypsin and chymotrypsin, but proteinase K is the most general of the proteases and resistance is unlikely to be a result of a complete lack in MAV1 structural proteins of the amino acids recognized by this enzyme. A more plausible explanation is that MAV1 structural proteins have a tertiary structure which does not expose any of these amino acids to the surface. Proteins resistant to proteinase K have been described for prions and in mycoplasmas (3).
Restriction mapping.
A restriction map of MAV1 DNA was constructed by digesting purified phage DNA with various restriction endonucleases and selecting for further analysis those enzymes that gave a small number of restriction fragments. From the size of the restriction fragments, the MAV1 genome was estimated to be about 16 kb. Mapping data also indicated that the phage DNA was linear rather than circular. This was confirmed experimentally by digestion with λ exonuclease, which completely degraded MAV1 DNA (14a). The MAV1 restriction map is shown in Fig. 2. Interestingly, the MAV1 genome does not contain any sites for the Sau3AI and MboI enzymes, which recognize the sequence GATC. The inability of these enzymes to cleave MAV1 DNA is not due to the host cells having GATC-specific modifications or GATC-specific DNA methyltransferase activity, because M. arthritidis genomic DNA was readily digested with either enzyme. Computer analysis of approximately 2 kb of the MAV1 DNA sequence presented below confirms that no sites for either enzyme are present.
FIG. 2.
Restriction endonuclease map of native MAV1 DNA. The partial open reading frame designated int represents the putative MAV1 integrase gene. The arrows indicate the binding sites for primers used to PCR amplify the MAV1 attP site.
Sequence determination of MAV1 termini and integration sites.
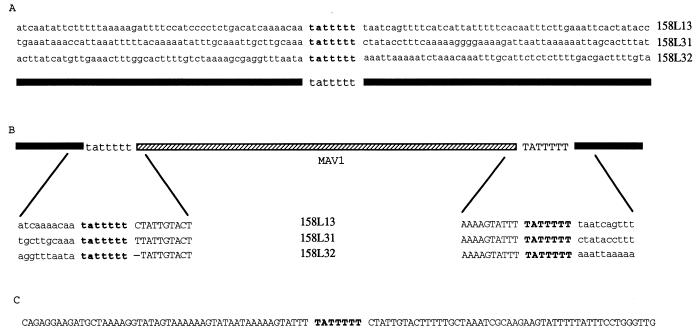
The sites of MAV1 DNA integration in the host chromosome were investigated. The ends of MAV1 DNA were cloned from three independent lysogens previously constructed and were shown to have single copies of MAV1 DNA inserted at different sites in the host chromosome (14). The strategy employed relied on the fact that the genome of MAV1 completely lacks sites for the restriction endonuclease Sau3AI. Genomic DNA from each lysogen was digested with Sau3AI. A large ∼16-kb MAV1 DNA fragment containing the entire MAV1 genome plus flanking chromosomal DNA was produced in addition to many small DNA fragments, all less than 3 kb. The 16-kb fragment from each lysogen was gel purified and subsequently digested with EcoRI or SpeI. The terminal Sau3AI-EcoRI or Sau3AI-SpeI fragments were then cloned into BamHI-EcoRI- or BamHI-SpeI-digested pBluescript SK+, respectively. For lysogens 158L13 and 158L31, Sau3AI-EcoRI fragments of 1.4 and 1.5 kb (158L13 left and right ends, respectively) and 2.4 and 2.2 kb (158L31 left and right ends, respectively) were cloned and completely sequenced. Comparison of the corresponding ends from the two lysogens showed a region of extensive nucleotide identity and a region of complete diversity (Fig. 3). Regions of nucleotide identity represent the ends of MAV1 DNA (Fig. 3) (uppercase letters), while the nonhomologous regions represent chromosomal DNA sequences flanking the integrated MAV1 genome (Fig. 3) (lowercase letters). To confirm this, we cloned the ends of MAV1 from a third lysogen (158L32) as Sau3AI-SpeI fragments of 10.3 and 5.6 kb. All of the flanking chromosomal DNA and ∼300 to 400 bp of MAV1 DNA were sequenced for each clone. A comparison of the left MAV1 DNA-chromosomal DNA junction and right MAV1 DNA-chromosomal DNA junction sites from all three lysogens (Fig. 3) delineated the ends of the MAV1 genome. Additionally, PCR primers that would target chromosomal DNA flanking MAV1 DNA from each lysogen were designed to amplify the integration sites from the original, uninfected host strain, 1581. The nucleotide sequence of the PCR products revealed the sequence (attB) into which MAV1 DNA had inserted to generate each lysogen and confirmed that the MAV1 DNA-chromosomal DNA junctions from each lysogen were correctly identified (Fig. 3).
FIG. 3.
(A) Alignment of sequences from integration sites in three different lysogens prior to insertion of MAV1 DNA. The conserved sequence (tattttt) which was designated attB is shown in boldface type. (B) Alignment of the left and right MAV1 DNA-chromosomal DNA junction sequences from the three lysogens shown in panel A. Sequences originating from the bacterial chromosome are shown in lowercase letters or are represented by a solid black line. Sequences originating from the phage genome are shown in uppercase letters or are represented by a hatched line. The tattttt sequence at the left end of MAV1 was arbitrarily designated as being of chromosomal origin, and the right-end TATTTTT sequence was designated as originating from MAV1. The 7-bp region representing the attB or attP sequence is in boldface type. The hyphen in the 158L32 sequence represents the deletion of a single T nucleotide resulting from possible imprecision in the cleavage mechanism as described in the text. (C) Sequence from circular intermediates isolated by PCR amplification of DNA from host cells infected with MAV1. The attP sequence is shown in boldface type.
The 7-bp sequence (TATTTTT), shown in boldface type in Fig. 3, is present in all MAV1 DNA-chromosomal DNA junctions and attB sites. To determine whether the TATTTTT sequence is present at the site of recombination (attP) in MAV1 DNA that leads to lysogeny, we designed a pair of PCR primers that bound near the ends of MAV1 DNA but were oriented in the outward direction (Fig. 2) such that a PCR product would be obtained only if MAV1 DNA were to circularize (or form linear head-to-tail dimers). A PCR product of the expected size was produced by using purified MAV1 DNA as the template. The sequence of the product coincided precisely with the predicted circularization of MAV1 DNA at the termini as determined from the MAV1 DNA-chromosomal DNA junction sequences. The junction of the circularized MAV1 DNA termini in the PCR product contained a single copy of the TATTTTT sequence representing the attP sequence (Fig. 3C). Integration most likely occurs by site-specific recombination between the viral attachment site (attP) and the attB site on the host chromosome.
MAV1 excision-integration model.
The nucleotide sequences of the right end of phage DNA inserted in all three lysogens are identical up to the MAV1 DNA sequence terminating in TATTTTT. Analysis of the left end of MAV1 DNA in the lysogens showed a single base heterogeneity located immediately adjacent to the tattttt sequence. Heterogeneity in the left end of MAV1 DNA may arise from the mechanism used for integration and excision. A partial open reading frame encoding 146 amino acids was identified on the right end of MAV1 DNA (int in Fig. 2). The results of BLAST analysis of the putative Int protein revealed a low level of amino acid sequence similarity to a variety of bacterial transposases and bacteriophage integrases. Further analysis of the highest scoring matches showed that the best match (26% amino acid sequence identity) was to the integrase of Tn916, which was calculated by using the Genetics Computer Group (University of Wisconsin, Madison) program GAP. Also, the Int protein contains the amino acid triad (H-R-Y) which is conserved in all phage integrases (10).
Heterogeneity in the left end of MAV1 DNA suggests that excision may occur by staggered cleavages as indicated by the arrows in Fig. 4, leaving two 6-bp single-stranded overhangs. The staggered ends could then be ligated and a circular intermediate could be formed. As described above, PCR products consistent with the formation of circular MAV1 DNA molecules have been obtained by using purified MAV1 DNA as the template. Identical PCR products were also obtained by using phage DNA isolated from host cells at various time points (5, 10, and 15 min) in the adsorption phase of a MAV1 infection. A single T nucleotide was deleted from the left end of MAV1 DNA in lysogen 158L32 (Fig. 3B). The missing base suggests a lack of cleavage precision due to the slippage of protein factors that bind to the stretch of five T nucleotides during recombination. Our model for MAV1 DNA excision-integration is reminiscent of the mechanism for transposition of conjugative transposon Tn916 (4). Tn916 has the sequence TATTTTT on its right end and is thought to be excised by a staggered cleavage reaction, usually resulting in 6-bp overhangs but sometimes resulting in excisants (class II) with a single base pair deletion in the circular intermediate that is subsequently formed. These features suggest that there may be a distant phylogenetic relationship between conjugative transposons and phages such as MAV1.
FIG. 4.
Proposed mechanism for MAV1 cleavage during excision from the host chromosome. Sequences corresponding to MAV1 are in uppercase letters, while those originating from the bacterial chromosome are in lowercase letters. The 7-bp sequence representing attB or attP is in boldface type. The positions of cleavage sites producing 5′ protruding ends are indicated by arrows above and below the sequence.
ACKNOWLEDGMENTS
This work was supported by PHS grant AR44252 to K.D. and training grant award AI07041 from the National Institutes of Health.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Bergemann A D, Finch L R. Isolation and restriction endonuclease analysis of a mycoplasma plasmid. Plasmid. 1988;19:68–70. doi: 10.1016/0147-619x(88)90064-9. [DOI] [PubMed] [Google Scholar]
- 3.Butler G H, Kotani H, Kong L, Frick M, Evancho S, Stanbridge E J, McGarrity G J. Identification and characterization of proteinase K-resistant proteins in members of the class Mollicutes. Infect Immun. 1991;59:1037–1042. doi: 10.1128/iai.59.3.1037-1042.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caparon M G, Scott J R. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell. 1989;59:1027–1034. doi: 10.1016/0092-8674(89)90759-9. [DOI] [PubMed] [Google Scholar]
- 5.Dybvig K, Khaled M. Isolation of a second cryptic plasmid from Mycoplasm mycoides subsp. mycoides. Plasmid. 1990;24:153–155. doi: 10.1016/0147-619x(90)90018-8. [DOI] [PubMed] [Google Scholar]
- 6.Dybvig K, Liss A, Alderete J, Cole R M, Cassell G H. Isolation of a virus from Mycoplasma pulmonis Isr. J Med Sci. 1987;23:418–422. [PubMed] [Google Scholar]
- 6a.Dybvig, K. Unpublished results.
- 7.Gourlay R N, Wyld S G, Garwes D J. Some properties of mycoplasma virus Br1. Arch Virol. 1983;75:1–15. doi: 10.1007/BF01314123. [DOI] [PubMed] [Google Scholar]
- 8.Gourlay R N, Wyld S G, Poulton M E. Some characteristics of mycoplasma virus Hr1, isolated from and infecting Mycoplasma hyorhinis. Arch Virol. 1983;77:81–85. doi: 10.1007/BF01314867. [DOI] [PubMed] [Google Scholar]
- 9.Maniloff J. Mycoplasma viruses. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 41–59. [Google Scholar]
- 10.Nunes-Düby S E, Kwon H J, Tirumalai R S, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renaudin J, Pascarel M C, Garnier M, Carle P, Bove J M. Characterization of spiroplasma virus group 4 (SV4) Isr J Med Sci. 1984;20:797–799. [PubMed] [Google Scholar]
- 12.Voelker L L, Dybvig K. Demonstration of extrachromosomal elements. In: Miles R, Nicholas R, editors. Methods in molecular biology: mycoplasma protocols. Totowa, N.J: Humana Press, Inc.; 1998. pp. 239–246. [DOI] [PubMed] [Google Scholar]
- 13.Voelker L L, Dybvig K. Gene transfer in Mycoplasma arthritidis: transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J Bacteriol. 1996;178:6078–6081. doi: 10.1128/jb.178.20.6078-6081.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voelker L L, Weaver K E, Ehle L J, Washburn L R. Association of lysogenic bacteriophage MAV1 with virulence of Mycoplasma arthritidis. Infect Immun. 1995;63:4016–4023. doi: 10.1128/iai.63.10.4016-4023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Voelker, L. L. Unpublished results.
- 15.Walther-Mauruschat A, Mayer F. Isolation and characterization of polysheaths, phage tail-like defective bacteriophages of Alcaligenes eutrophus H16. J Gen Virol. 1978;41:239–254. doi: 10.1099/0022-1317-41-2-239. [DOI] [PubMed] [Google Scholar]
- 16.Yamao F, Muto A, Kawauichi Y, Iwami M, Iwagami S, Azumi Y, Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci USA. 1985;82:2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou N X, Park K S, Dybvig K. Mycoplasma virus P1 has a linear, double-stranded DNA genome with inverted terminal repeats. Plasmid. 1995;33:41–49. doi: 10.1006/plas.1995.1005. [DOI] [PubMed] [Google Scholar]