Abstract
The autolysin AcmA of Lactococcus lactis was shown to be degraded by the extracellular lactococcal proteinase PrtP. Autolysis, as evidenced by reduction in optical density of a stationary-phase culture and concomitant release of intracellular proteins, was greatly reduced when L. lactis MG1363 cells expressed the cell wall-anchored lactococcal proteinase PrtP of the PI-type caseinolytic specificity (PI). On the other hand, lactococcal strains that did not produce the proteinase showed a high level of autolysis, which was also observed when the cells produced the secreted form of PI or a cell wall-anchored proteinase with PIII-type specificity. Autolysis was also increased when MG1363 expressed the cell wall-anchored hybrid PI/PIII-type proteinase PIac. Zymographic analysis of AcmA activity during stationary phase showed that AcmA was quickly degraded by PI and much more slowly by PrtP proteinases with PIII-type and intermediate specificities. Autolysis of L. lactis by AcmA was influenced by the specificity, amount, and location of the lactococcal proteinase. No autolysis was observed when the various proteinases were expressed in an L. lactis acmA deletion mutant, indicating that PrtP itself did not cause lysis of cells. The chain length of a strain was significantly shortened when the strain expressed a cell wall-anchored active proteinase.
Lactococcus lactis, like many other lactic acid bacteria, is a multiple-amino-acid auxotroph (6). For growth in milk, in which the concentrations of free amino acids and peptides are very low, these bacteria depend on an active proteolytic system which allows the degradation and release of amino acids from milk proteins (αS1-, β-, and κ-caseins). The initial step in degradation is carried out by the extracellular cell wall-bound proteinase PrtP. The extreme C terminus of this protein contains the typical LPXTG motif of gram-positive cell surface proteins. After cleavage between the threonine and glycine residues, such proteins are covalently bound to the cross bridge in the peptidoglycan (23). The genes encoding PrtP have been cloned from several L. lactis strains and sequenced. Although the deduced amino acid sequences are over 98% identical, the proteolytic specificity of the proteinases toward milk caseins can be quite different. The lactococcal proteinases were initially divided into two major classes on the basis of the caseinolytic specificity: PIII-type proteinases (PIII) degrade αS1-, β-, and κ-caseins, while the PI-type proteinases (PI) mainly degrade β-casein but with a specificity different from that of the PIII enzymes (30). Regions in the proteins determining cleavage specificity were identified by reciprocal exchange of fragments of the genes for PI of L. lactis Wg2 and PIII of strain SK11, and enzymes with new proteolytic properties were obtained (31). The proteinases of 16 different L. lactis strains have recently been classified into seven groups by examining their degradation of fragment f1-23 of αS1-casein (9).
L. lactis expresses one major autolysin, AcmA, which is responsible for cell separation and autolysis during the stationary phase of growth (4, 5). In a previous study on AcmA, we observed that the C-terminal repeats of AcmA are removed by an unknown proteolytic activity (3), without affecting the enzyme activity.
Several observations indicate that processes in which autolysins take part are influenced by proteolysis. Competence, cell separation, and motility in Bacillus subtilis all involve cell wall hydrolase activities and were shown to be sensitive to extracellular proteinases (1, 35). A multiple proteinase-deficient strain of B. subtilis had a higher rate of turnover of peptidoglycan than proteinase-proficient strains (12). Growth of the proteinase-deficient strain in the presence of subtilisin resulted in reduced peptidoglycan turnover and filament formation. Proteinase-hyperproducing strains showed an even more diminished turnover and formed filaments. Addition of a proteinase inhibitor to cultures of these strains resulted in an increase in peptidoglycan turnover. Coxon et al. (7) showed that protease deficiency of B. subtilis was associated with an increased tendency of cells to lyse as they approached stationary phase. Salt-induced lysis of exponential phase cells of Staphylococcus aureus was inhibited by a brief pretreatment with subtilisin. Analysis of cell wall-lytic enzymes by renaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) revealed that all lytic bands had disappeared after incubation with the proteinase (33). Loss of cell wall-lytic activity was also observed upon addition of protease to exponentially growing cells of Staphylococcus haemolyticus, which resulted in the formation of multicells (34). Lysis of Streptococcus pneumoniae induced by the addition of d-cycloserine or benzylpenicillin is inhibited by addition of trypsin to the growth medium (20). On the other hand, proteolytic activities have also been shown to be enhancers of cell wall-lytic activities. A muramidase of Enterococcus hirae is extracellular proteolytically activated from a 130-kDa latent form to a 87-kDa active form (13). The autolysin ATL of S. aureus is expressed as a bifunctional protein with amidase and glucosaminidase domains which is processed by an extracellular protease to generate two peptidoglycan hydrolases (17).
Haandrikman et al. (11) reported that the major secreted protein Usp45 of L. lactis MG1363 was absent in a culture of cells expressing the PI proteinase of strain Wg2, suggesting that this enzyme not only hydrolyzes caseins but is also able to degrade secreted proteins. In this paper, we show that PrtP degrades AcmA and that the rate and degree of autolysis of L. lactis are dependent on the specificity, location, and amount of PrtP produced.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. L. lactis was grown in M17 broth (Difco Laboratories, Detroit, Mich.) or whey-based medium (8) at 30°C as standing cultures. Twofold-diluted M17 (0.5× M17) agar plates contained 1.5% agar and 0.95% β-glycerolphosphate (Sigma Chemical Co., St. Louis, Mo.). In all cases, 0.5% glucose and 5 μg of erythromycin (Boehringer GmbH, Mannheim, Germany) per ml were added.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype(s) or genotype(s)a | Reference |
|---|---|---|
| Strains | ||
| L. lactis subsp. cremoris | ||
| MG1363 | Plasmid-free strain | 10 |
| MG1363acmAΔ1 | Derivative of MG1363 carrying a 701-bp SacI/SpeI deletion in acmA | 5 |
| Plasmids | ||
| pGK13 | Emr Cmr, pWV01-based lactococcal cloning vector | 14 |
| pGKV500 | Emr, pWV01 derivative specifying sPI of L. lactis Wg2 | 16 |
| pGKV552 | Emr, specifying aPI of L. lactis Wg2 | 11 |
| pGKV1552 | Emr, pGKV552 specifying aPI* | 11 |
| pGKV552abc | Emr, specifying aPIabc | 31 |
| pGKV552ac | Emr, specifying aPIac | 31 |
Cmr, chloramphenicol resistance; Emr, erythromycin resistance.
General DNA techniques and transformation.
Plasmid DNA was isolated by using a QIAGEN plasmid DNA isolation kit and protocol (QIAGEN GmbH, Hilden, Germany) or by the alkaline lysis method as described by Sambrook et al. (25), with modifications suggested by Seegers et al. (27). Electrotransformation of L. lactis was performed with a Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.) as described by Leenhouts and Venema (19). All chemicals were of analytical grade and were from Merck (Darmstadt, Germany) or BDH (Poole, United Kingdom).
SDS-PAGE and detection of AcmA.
Five milliliters of whey cultures and 2 ml of M17 cultures were subjected to centrifugation; 5 ml of the whey supernatants and 0.5 ml of the M17 supernatants were dialyzed against several changes of demineralized water, lyophilized, and dissolved in 1 and 0.25 ml, respectively, of denaturation buffer (2). The cell pellets were washed with 1 ml of corresponding fresh medium and resuspended in 1 ml of denaturation buffer. Cell extracts were prepared as described by van de Guchte et al. (29).
AcmA activity was detected by a zymogram staining technique using SDS–12.5% polyacrylamide gels containing 0.2% autoclaved, lyophilized Micrococcus lysodeikticus ATCC 4698 cells (Sigma) as described before (5).
SDS-PAGE (12.5% polyacrylamide gel) was carried out according to Laemmli (18) with the Protean II Minigel system (Bio-Rad). The Bio-Rad prestained high-range molecular weight marker and the Amersham high-range SDS-PAGE Rainbow colored protein molecular weight marker (Amersham Life Science Inc., Buckinghamshire, United Kingdom) were used as protein size references. SDS-polyacrylamide gels were stained with Coomassie brilliant blue (Bio-Rad).
Optical density measurements and enzyme assays.
Overnight cultures of L. lactis in M17 were diluted 100-fold in prewarmed M17, and the optical densities at 600 nm were followed over time in a Novaspec II spectrophotometer (Pharmacia Biotech AB, Uppsala, Sweden).
The presence of intracellular X-prolyl dipeptidyl aminopeptidase (PepX) in culture supernatants was measured by using the chromogenic substrate Ala-Pro-p-nitroanilide (Bachem Feinchemikalien AG, Bubendorf, Switzerland) as described before (3).
Proteinase activity was determined by using the chromogenic peptide MeO-Suc-Arg-Pro-Tyr-pNA (Chromogenix AB, Mölndal, Sweden) as described by Mierau et al. (22).
RESULTS
AcmA is degraded by PrtP.
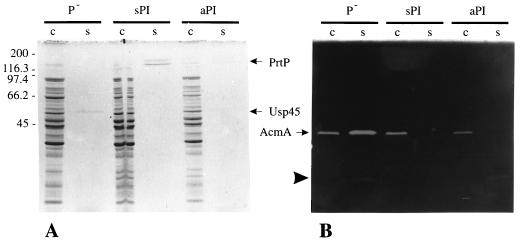
To examine the possible degradation of AcmA by the lactococcal proteinase PrtP, the PrtP-negative strain L. lactis MG1363(pGK13) [MG1363 (P−)] and two proteinase-producing strains were grown overnight in whey-based medium. L. lactis MG1363(pGKV552) expresses the PI-type PrtP (16). This full-length protein (cell wall-anchored PI [aPI]) is attached to the cell by a C-terminal cell wall anchor sequence (11). The proteinase produced by MG1363(pGKV500) lacks this anchor and PI; consequently, it is secreted into the growth medium (and hence referred to as sPI [secreted PI]). As expected, proteinase activity was absent in the cell extract and supernatant of MG1363 (P−), while all sPI activity was present in the supernatant. Most of the proteinase activity of MG1363 expressing aPI [MG1363 (aPI)] fractionated with the cells (results not shown). These activity measurements were corroborated by SDS-PAGE (12% polyacrylamide gel). Analysis of cell extract and supernatant samples of the three strains showed that PrtP is present in high amounts in the supernatant of MG1363 (sPI), while a lower amount is seen in that of MG1363 (aPI). In the latter strain, most of the proteinase is detected in the cell extract (Fig. 1A). The major secreted lactococcal protein Usp45 (28) is clearly present in the supernatant of MG1363 (P−) but absent in these fractions of the proteinase producers. When samples were run on an SDS-polyacrylamide gel containing M. lysodeikticus autoclaved cells, AcmA activity was detected in the cell extracts of all cultures (Fig. 1B). A band of cell wall hydrolytic activity corresponding to mature AcmA (40 kDa) is present in the supernatant of MG1363 (P−) but not in the supernatants of the sPI- and aPI-producing cultures. These results show that AcmA is subject to proteolytic degradation by PrtP.
FIG. 1.
(A) Protein profiles of cell (c) and supernatant (s) fractions of whey-grown L. lactis MG1363 containing pGK13 (P−), pGKV500 (sPI), or pGKV552 (aPI) in an SDS–12.5% polyacrylamide gel stained with Coomassie brilliant blue. PrtP, lactococcal proteinase; Usp45, secreted protein of unknown function (28). (B) Analysis of AcmA activity in the samples used for panel A by renaturing SDS-PAGE (12.5% polyacrylamide gel) in the presence of 0.15% M. lysodeikticus autoclaved cells. AcmA, N-acetylmuramidase of L. lactis; arrowhead, major degradation product of AcmA (5). Molecular masses (in kilodaltons) of standard proteins for both gels are shown on the left. Five microliters of each sample was loaded onto the gels.
Autolysis of various proteinase-producing strains of L. lactis.
To investigate the influence of AcmA degradation on autolysis of L. lactis, the optical densities at 600 nm and release of the intracellular proteins were followed over time. The strains used in this experiment were MG1363 (AcmA+) and MG1363acmAΔ1 (AcmA−) harboring either pGK13 (negative control), pGKV500 (producing sPI), pGKV552 (specifying aPI), and pGKV1552 (encoding an inactive [Asp30→Asn30] form of PI, aPI*). Plasmids pGKV552ac and pGKV552abc specify hybrid proteinases (aPIac and aPIabc, respectively) in which regions a and c and regions a, b, and c in PI have been replaced by the corresponding domains of the PIII-type proteinase of L. lactis SK11 (31). The hybrid proteinase specified by pGKV552abc has PIII-type specificity, whereas the hybrid proteinase specified by pGKV552ac has a specificity intermediate between that of PI and PIII (31). As MG1363 (P−) grows to a lower optical density in whey than a Prt+ strain, M17 medium was used to obtain identical culture densities for all strains.
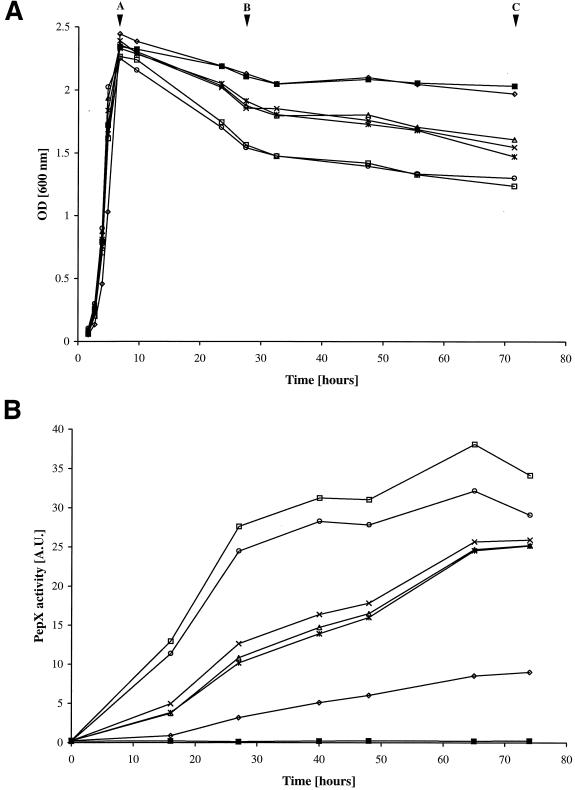
Overnight cultures of the strains were diluted 100-fold in fresh M17 medium, and the optical densities at 600 nm and release of the intracellular PepX of various strains were followed over time (Fig. 2). All strains grew equally well except for L. lactis MG1363 (aPI), which reached approximately the same maximum optical density but grew somewhat more slowly. Nearly no reduction in optical density during the stationary phase of growth was observed for any of the MG1363acmAΔ1 mutants, irrespective of which proteinase was produced, and for MG1363 (aPI). Reduction in optical density was highest for MG1363 expressing the hybrid proteinase aPIac or the inactive proteinase aPI* (Fig. 2A). An intermediate reduction in optical density was observed with the cultures of MG1363 producing no proteinase, sPI, or aPIabc.
FIG. 2.
(A) Growth and lysis by optical density (OD) measurement at 600 nm of cultures of L. lactis MG1363 containing pGK13 (P−; ×), pGKV500 (sPI; ∗), pGKV1552 (aPI*; ○), pGKV552 (aPI; ◊), pGKV552abc (aPIabc; ▵), or pGKV552ac (aPIac; □). L. lactis MG1363acmAΔ1 containing either of these plasmids exhibited very similar growth curves of which only one, MG1363acmAΔ1(pGK13), is shown (■). The strains were examined during 3 days of incubation at 30°C. Lettered arrowheads indicate the time points at which samples were taken from the cultures for analysis of AcmA activity (Fig. 3) and protein release (Fig. 2B and data not shown). (B) Analysis of PepX activity (in arbitrary units [A.U.]) in the culture supernatants of the strains presented in panel A. Symbols are as defined for panel A.
Release of PepX activity was in full agreement with the above results (Fig. 2B): the higher the reduction in optical density measured at 600 nm, the more PepX activity was detectable in the culture supernatant.
Release of intracellular proteins was also examined by SDS-PAGE (12.5% polyacrylamide gel) analysis of supernatant samples of the various MG1363 cultures taken at the end of the exponential growth phase and after 1 and 3 days of incubation at 30°C (Fig. 2A). Coomassie brilliant blue staining (results not shown) of supernatant fractions of all strains except MG1363 (aPI) revealed a protein banding pattern typical for that of intracellular proteins of L. lactis (4). Hardly any protein was visible in the supernatants of MG1363 (aPI). The largest amount of protein was detected in the supernatant of the 3-day samples of MG1363 (aPI*) and MG1363 (aPIac). No band was detected at the position of lactococcal PrtP, whereas in all samples a band corresponding to Usp45 was present, indicating that, as expected, only little PrtP is formed in M17 medium (21). This amount of activity was still detectable by an enzymatic assay using the chromogenic peptide substrate MeO-Suc-Arg-Pro-Tyr-pNA (not shown).
Apparently, when the proteinase remains attached to the cell wall, autolysis is strongly influenced by the specificity of the proteinase: a PI-type proteinase results in strongly reduced autolysis, whereas autolysis is maximal when cells produce a proteinase with a specificity intermediate between those of PI and PIII.
Proteinase specificity is an important parameter in AcmA degradation.
To examine whether proteinase specificity is important for AcmA degradation, we analyzed the pattern of active degradation products of AcmA. For this purpose, samples taken from all MG1363 cultures at the end of the exponential growth phase and after 1 and 3 days of incubation at 30°C (Fig. 2A) were examined by zymographic analysis. Figure 3 shows that the banding pattern of extracellular AcmA activity of MG1363 (P−) and of cells producing aPI* are very similar. Analysis of the supernatants of the other cultures shows that AcmA is degraded to various degrees during the 3-day incubation. In the supernatant of MG1363 (aPI), degradation of AcmA is already clearly observed at the end of the exponential growth phase. Degradation proceeds much more slowly in supernatant fractions of MG1363 producing aPIabc or aPIac.
FIG. 3.
Zymographic analysis of AcmA activity. Each lane contains 10 μl of the supernatant of an MG1363 culture in M17 producing the indicated proteinase sampled at the time points A, B, and C shown in Fig. 2A. Molecular masses (in kilodaltons) of standard proteins are shown on the left.
Similar amounts of both the unprocessed (46.1-kDa) and mature (40.3-kDa) forms of AcmA are present in the cell extracts of the three samples of all strains except MG1363 (aPI). In the latter strain, the amounts of both forms of AcmA were significantly lower at days 1 and 3. A band of activity of one of the major degradation products of AcmA was clearly present in the cell extracts of all three samples of MG1363 (aPI) and in the cell extract of the strain secreting sPI sampled at day 3 (not shown). These results indicate that degradation of AcmA is influenced by the specificity of the proteinase PrtP.
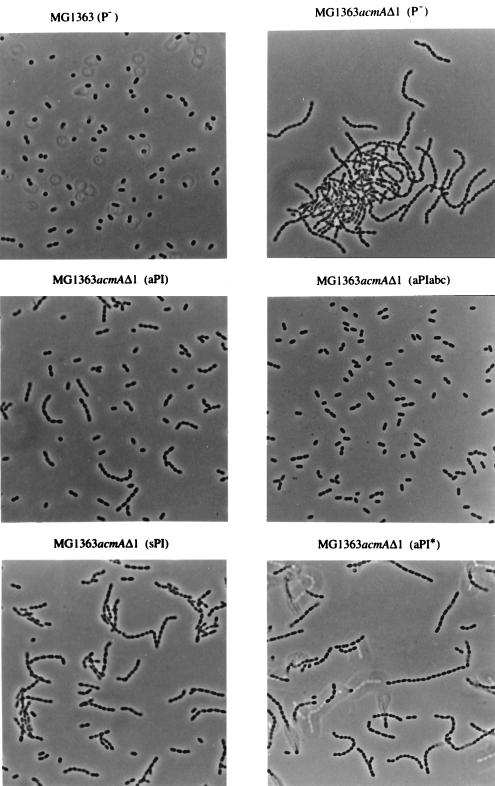
The proteinase PrtP is involved in cell separation.
In the course of the experiments described above, we observed a difference in sedimentation among MG1363acmAΔ1 strains producing the various proteinases. In contrast to MG1363 (AcmA+), the mutant MG1363acmAΔ1 grows in extremely long chains and sediments during overnight growth (5). Sedimentation of cells was also observed in overnight cultures of MG1363acmAΔ1 producing aPI* or sPI. This phenomenon was not seen with MG1363acmAΔ1 expressing an active cell wall-anchored proteinase (aPI, aPIabc, or aPIac). No sedimentation occurred in cultures of L. lactis MG1363 expressing either of these proteinases. Light microscopic analysis showed that all MG1363 strains had the same average chain length [Fig. 4; only shown for MG1363 (P−)]. As shown in Fig. 4, the length of the chains of MG1363acmAΔ1 is hardly influenced by the presence of an inactive cell-anchored proteinase [compare strains MG1363acmAΔ1 (P−) and MG1363acmAΔ1 (aPI*)]. In the presence of the unanchored, secreted form (sPI), the average chain length became only slightly shorter. However, the chains were found to be significantly shorter when cells produced an active proteinase that remained bound to the cell wall. The shortest chains (containing mainly two cells) were detected when MG1363acmAΔ1 expressed a hybrid anchored proteinase (aPIabc [Fig. 4] or aPIac [not shown]). In fact, the average chain length in these cultures was very similar to that of MG1363 (Prt− AcmA+). These results were highly reproducible, and the same effects were observed when the various strains were grown in whey-based medium.
FIG. 4.
Light microscopic analysis of the chain lengths in overnight M17 cultures of the indicated strains. Magnification, ×1,000.
DISCUSSION
The results presented in this work show that the autolysin AcmA of L. lactis is subject to degradation by the lactococcal proteinase (caseinase) PrtP. AcmA activity, which is normally present in the supernatant of a culture of L. lactis MG1363 grown on whey-based medium, was completely absent in the supernatant of a strain secreting sPI. One specific degraded form of AcmA, barely visible in Fig. 1B but clearly present after prolonged renaturation, was present in the supernatants of MG1363 (P−) and MG1363 producing aPI. This truncated form of AcmA is formed through cleavage of the C-terminal cell wall binding domain by an as yet unidentified proteolytic activity in L. lactis. The protein is still active but, depending on the exact position of the cleavage site, may have reduced cell binding capacity (3). Reduced binding may explain the presence of this protein in the supernatant of MG1363 (aPI), as there it would escape degradation by aPI. The extent of degradation of AcmA by sPI or aPI is much less when the strains were grown in M17 (compare Fig. 1 and 3). This is due to lower expression of PrtP in this medium compared to whey permeate: the PrtP protein was detected in the supernatant fraction of the whey cultures by SDS-PAGE but was not visible in this fraction of M17 cultures (reference 31 and data not shown). Also, Usp45 was still present in the M17 culture media but had been degraded in the proteinase-producing whey cultures.
The inactive mutant aPI* does not degrade AcmA, either when AcmA is present in the cell walls (not shown) or when it is secreted in the culture medium (Fig. 3). Zymographic analysis of AcmA activity, followed over time, in the supernatants of M17 cultures of MG1363 expressing different types of proteinases showed that AcmA degradation was highest when the cells expressed a PI-type proteinase. AcmA hydrolysis was significantly lowered when a hybrid proteinase was expressed (aPIabc or aPIac) with a caseinolytic specificity different from that of PI. The mature form of AcmA, which is normally located in the cell wall of L. lactis (5), was present in undiminished amounts in all samples in the cell walls of strains producing no PrtP, sPI, aPI*, or the two hybrid proteinases (results not shown). As AcmA is a good substrate for sPI and is fully degraded by the proteinase once the protein is released in the medium (Fig. 3), we assume that sPI has no access to the cell wall-located autolysin. When MG1363 expressed aPI, the amount of AcmA activity on the cells gradually decreased in time (not shown). While mature AcmA was still present on the cells at day 3, it was not detectable in the supernatant of these cells. It may be that AcmA, due to conformational constraints, is inaccessible for PrtP when the autolysin is bound to its substrate. After hydrolysis of peptidoglycan, AcmA would release and be subject to degradation by PrtP. Such a phenomenon has been described for partially purified glucosaminidase of B. subtilis. This enzyme preparation was unstable due to the action of contaminating proteinases. Addition of purified cell walls of B. subtilis resulted in partial resistance to proteolytic attack (24). Trypsin and α-chymotrypsin degradation of the pneumococcal LytA amidase and CPL1 lysozyme could be slowed down by the addition of choline, which induces a conformational change in these choline-dependent enzymes. Accordingly, the choline-independent CPL7 lysozyme was not protected by addition of this agent (26).
Due to AcmA degradation, autolysis was nearly absent in cells expressing a PI-type anchored proteinase. Although the reduction of the optical density at 600 nm did not reveal lysis, PepX and protein release in time from MG1363 (aPI) were evident. Detachment of the proteinase from the cell wall by removal of the cell wall anchor sequence (as in sPI) resulted in an extent of autolysis similar to that for the PrtP-negative strain MG1363. As AcmA is actively degraded in the supernatant of cells producing sPI, we presume that it is the cell wall-located autolysin that is involved in cell lysis. As detailed above, this fraction of AcmA is not affected by sPI. We expected autolysis of the aPI*-producing strain to be the same as in a PrtP-negative strain. The amounts of AcmA in the supernatant and on cells of MG1363 (aPI*) were the same as those for MG1363 (P−). Nevertheless, a higher level of cell lysis was observed with MG1363 (aPI*). This observation suggests that anchoring to the cell walls of PrtP destabilizes the cells in an as yet unknown way. The anchored hybrid proteinase aPIac also caused increased lysis of producing cells, while aPIabc had no effect on autolysis compared with MG1363 (P−). At this moment it is not clear what causes the difference in autolytic behavior of the two strains. The AcmA degradation patterns of cell extracts (not shown) and supernatants are very similar. The enzymes aPIabc and aPIac differ in only 9 of 1,902 amino acid residues. These residues are located in a domain in PrtP, which has previously been found to be of minor importance with respect to specificity of casein degradation (31) but may influence cell wall stability once the proteinase is anchored to the cells. When aPIac was expressed in an acmA deletion mutant, this strain did not autolyse; thus, the proteinase is not itself involved in degradation of the cell wall to an extent that cell lysis would occur.
The average chain length of MG1363acmAΔ1 expressing either of the hybrid proteinases aPIac and aPIabc was shorter than that of the same strain expressing aPI and nearly identical to that of MG1363. It is possible that cell wall proteins are degraded by the proteinases, thus possibly destabilizing the cell wall. This supposition seems to be strengthened by the observation that expression of sPI by MG1363acmAΔ1 does not result in such a drastic shortening of the average chain length.
The results presented in this report show that PrtP degrades AcmA and that the rate and degree of autolysis of L. lactis are dependent on the specificity, location, and amount of PrtP produced.
ACKNOWLEDGMENTS
We thank Henk Mulder for preparing the photographs and Anne de Jong for advice on and support of the computer work.
This study was supported by Unilever Research Laboratorium, Vlaardingen, The Netherlands. Jan Kok was the recipient of a fellowship from the Royal Netherlands Academy of Arts and Sciences.
REFERENCES
- 1.Ayusawa D, Yoneda Y, Yamane K, Maruo B. Pleiotropic phenomena in autolytic enzyme(s) content, flagellation, and simultaneous hyperproduction of extracellular α-amylase and protease in a Bacillus subtilis mutant. J Bacteriol. 1975;124:459–469. doi: 10.1128/jb.124.1.459-469.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Béliveau C, Potvin C, Trudel J, Asselin A, Bellemare G. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J Bacteriol. 1991;173:5619–5623. doi: 10.1128/jb.173.18.5619-5623.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buist G. AcmA of Lactococcus lactis, a cell-binding muramidase. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1997. [Google Scholar]
- 4.Buist G, Karsens H, Nauta A, van Sinderen D, Venema G, Kok J. Autolysis of Lactococcus lactis caused by induced overproduction of its major autolysin, AcmA. Appl Environ Microbiol. 1997;63:2722–2728. doi: 10.1128/aem.63.7.2722-2728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buist G, Kok J, Leenhouts K J, Dabrowska M, Venema G, Haandrikman A J. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopin A. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:21–37. doi: 10.1111/j.1574-6976.1993.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 7.Coxon R D, Harwood C R, Archibald A R. Protein export during growth of Bacillus subtilis: the effect of extracellular protease deficiency. Lett Appl Microbiol. 1991;12:91–94. [Google Scholar]
- 8.de Vos W M, Vos P, de Haard H, Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989;85:169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- 9.Exterkate F A, Alting A C, Bruinenberg P G. Diversity of cell envelope proteinase specificity among strains of Lactococcus lactis and its relationship to charge characteristics of the substrate-binding region. Appl Environ Microbiol. 1993;59:3640–3647. doi: 10.1128/aem.59.11.3640-3647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haandrikman A J, Meesters R, Laan H, Konings W N, Kok J, Venema G. Processing of the lactococcal extracellular serine proteinase. Appl Environ Microbiol. 1991;57:1899–1904. doi: 10.1128/aem.57.7.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jolliffe L K, Doyle R J, Streips U N. Extracellular proteases modify cell wall turnover in Bacillus subtilis. J Bacteriol. 1980;141:1199–1208. doi: 10.1128/jb.141.3.1199-1208.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kariyama R, Shockman G D. Extracellular and cellular distribution of muramidase-2 and muramidase-1 of Enterococcus hirae ATCC 9790. J Bacteriol. 1992;174:3236–3241. doi: 10.1128/jb.174.10.3236-3241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kok J. Special-purpose vectors for lactococci. In: Dunny G, Cleary P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. Washington, D.C: American Society for Microbiology; 1992. pp. 97–102. [Google Scholar]
- 15.Kok J, de Vos W M. The proteolytic system of lactic acid bacteria. In: Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, England: Blackie and Professional; 1994. pp. 169–210. [Google Scholar]
- 16.Kok J, van Dijl J M, van der Vossen J M B M, Venema G. Cloning and expression of a Streptococcus cremoris proteinase in Bacillus subtilis and Streptococcus lactis. Appl Environ Microbiol. 1985;50:94–101. doi: 10.1128/aem.50.1.94-101.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komatsuzawa H, Sugai M, Nakashima S, Yamada S, Matsumoto A, Oshida T, Suginaka H. Subcellular localization of the major autolysin, ATL and its processed proteins in Staphylococcus aureus. Microbiol Immunol. 1997;41:469–479. doi: 10.1111/j.1348-0421.1997.tb01880.x. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Leenhouts K J, Venema G. Lactococcal plasmid vectors. In: Hardy K G, editor. Plasmids: a practical approach. Oxford, England: Oxford University Press; 1993. pp. 65–94. [Google Scholar]
- 20.López R, Ronda-Lain C, Tapia A, Wacks S B, Tomasz A. Suppression of the lytic and bactericidal effects of cell wall inhibitory antibiotics. Antimicrob Agents Chemother. 1976;10:697–706. doi: 10.1128/aac.10.4.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meijer W, Marugg J D, Hugenholtz J. Regulation of proteolytic enzyme activity in Lactococcus lactis. Appl Environ Microbiol. 1996;62:156–161. doi: 10.1128/aem.62.1.156-161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mierau I, Kunji E R S, Leenhouts K J, Hellendoorn M A, Haandrikman A J, Poolman B, Konings W N, Venema G, Kok J. Multiple-peptidase mutants of Lactococcus lactis are severely impaired in their ability to grow in milk. J Bacteriol. 1996;178:2794–2803. doi: 10.1128/jb.178.10.2794-2803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navarre W W, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 24.Rashid M H, Mori M, Sekiguchi J. Glucosaminidase of Bacillus subtilis: cloning, regulation, primary structure and biochemical characterization. Microbiology. 1995;141:2391–2404. doi: 10.1099/13500872-141-10-2391. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sanz J M, Díaz E, García J L. Studies on the structure and function of the N-terminal domain of the pneumococcal murein hydrolases. Mol Microbiol. 1992;6:921–931. doi: 10.1111/j.1365-2958.1992.tb01542.x. [DOI] [PubMed] [Google Scholar]
- 27.Seegers J F M L, Bron S, Franke C M, Venema G, Kiewiet R. The majority of lactococcal plasmids carry a highly related replicon. Microbiology. 1994;140:1291–1300. doi: 10.1099/00221287-140-6-1291. [DOI] [PubMed] [Google Scholar]
- 28.van Asseldonk M, Rutten G, Oteman M, Seizen R J, de Vos W M, Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis ssp. lactis MG1363. Gene. 1990;95:155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 29.van de Guchte M, Kodde J, van der Vossen J M B M, Kok J, Venema G. Heterologous gene expression in Lactococcus lactis subsp. lactis: synthesis, secretion, and processing of the Bacillus subtilis neutral protease. Appl Environ Microbiol. 1990;56:2606–2611. doi: 10.1128/aem.56.9.2606-2611.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visser S, Exterkate F A, Slangen C J, de Veer G J C M. Comparative study of action of cell wall proteinases from various strains of Streptococcus cremoris on bovine αS1-, β-, and κ-casein. Appl Environ Microbiol. 1986;52:1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vos P, Boerrigter I J, Buist G, Haandrikman A J, Nijhuis M, de Reuver M B, Siezen R J, Venema G, de Vos W M, Kok J. Engineering of the Lactococcus lactis serine proteinase by construction of hybrid enzymes. Protein Eng. 1991;4:479–484. doi: 10.1093/protein/4.4.479. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson M G, Guinee T P, O’Callaghan D M, Fox P F. Autolysis and proteolysis in different strains of starter bacteria during Cheddar cheese ripening. J Dairy Res. 1994;61:249–262. [Google Scholar]
- 33.Yabu K, Kaneda S. Salt-induced cell lysis of Staphylococcus aureus. Curr Microbiol. 1995;30:299–303. doi: 10.1007/BF00295505. [DOI] [PubMed] [Google Scholar]
- 34.Yabu K, Nishiyama Y, Ochiai T. Protease-induced multicell formation in Staphylococcus haemolyticus. Microbiol Immunol. 1997;41:799–803. doi: 10.1111/j.1348-0421.1997.tb01930.x. [DOI] [PubMed] [Google Scholar]
- 35.Yoneda Y, Maruo B. Mutation of Bacillus subtilis causing hyperproduction of α-amylase and protease and its synergistic effect. J Bacteriol. 1975;124:48–54. doi: 10.1128/jb.124.1.48-54.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]